Abstract
A previous genome-wide linkage study of alcohol dependence (AD) in multiplex families found a suggestive linkage result for a region on Chromosome 1 near microsatellite markers D1S196 and D1S2878. The ASTN1 gene is in this region, a gene previously reported to be associated with substance abuse, bipolar disorder and schizophrenia. Using the same family data consisting of 330 individuals with phenotypic data and DNA, finer mapping of a 26 cM region centered on D1S196 was undertaken using SNPs with minor allele frequency (MAF) ≥ 0.15 and pair-wise linkage disequilibrium (LD) of r2 <0.8 using the HapMap CEU population. Significant FBAT P-values for SNPs within the ASTN1 gene were observed for four SNPs (rs465066, rs228008, rs6668092, and rs172917), the most significant, rs228008, within intron 8 had a P-value of 0.001. Using MQLS, which allows for inclusion of all families, we find three of these SNPs with MQLS P-values <0.003. In addition, two additional neighboring SNPs (rs10798496 and rs6667588) showed significance at P = 0.002 and 0.03, respectively. Haplotype analysis was performed using the haplotype-based test function of FBAT for a block that included rs228008, rs6668092, and rs172917. This analysis found one block (GCG) over-transmitted and another (ATA) under-transmitted to affected offspring. Linkage analysis identified a region consistent with the association results. Family-based association analysis shows the ASTN1 gene significantly associated with alcohol dependence. The potential importance of the ASTN1 gene for AD risk may be related its role in glial-guided neuronal migration.
Keywords: ASTN1, alcohol dependence, multiplex families
INTRODUCTION
Excessive use of alcohol is the third leading cause of preventable death [Mokdad et al., 2004] in the US. The economic and social costs have been estimated to be $184 billion due to alcohol-related accidents, lost productivity, incarceration and other alcohol related morbidity [Harwood, 2000]. In spite of the fact that use of alcohol is quite common, a smaller proportion of the population drink in sufficient quantity and with associated health, family, and work-related problems to be considered alcohol dependent (AD). Data from the National Comorbidity Survey, a survey of respondents ages 15–54 found 20.1% of men and 8.2% of women meeting criteria for alcohol dependence (AD) [Kessler et al., 1997]. There is now evidence that those individuals with the greatest propensity for AD may carry an increased genetic risk for developing alcohol dependence.
Although there is considerable heritability for alcohol dependence (0.49–0.64) in males [Caldwell and Gottesman, 1991; Heath et al., 1997] and females (0.56–0.59) [Kendler et al., 1992; Prescott et al., 1999], few genes have been identified that reliably confer susceptibility. However, studies employing well-designed sampling strategies that over sample families with a high density of cases have revealed important clues for gene finding as seen in the Collaborative Study on the Genetics of Alcoholism (COGA) studies [Reich et al., 1998; Edenberg et al., 2004]. Genome-wide association (GWAS) studies have also revealed potentially important loci but require large samples to detect loci having genome-wide significance. A meta-analysis of two GWAS studies of alcohol dependence totaling 4,979 cases and controls has identified three loci with statistical significance of α′<5 × 10−7 [Wang et al., 2011].
In a genome wide scan of multiplex families ascertained through a pair of affected probands [Hill et al., 2004], we found evidence for linkage in multiple chromosomal regions. The present report is based on efforts to follow up on linkage findings for a region on Chromosome 1q23.3–1q25.1 that included a maximal LOD score of 3.46 (P = 0.002) at marker D1S196 and at an adjacent marker D1S2878 with a LOD value of 3.45 (P = 0.002).
MATERIALS AND METHODS
Study Sample
All members of the multiplex families who participated in the study gave their written consent to do so after the nature and purpose of the study was fully explained to them. (Consent forms were approved by the University of Pittsburgh Institutional Review Board.)
Multiplex Families
Multiplex families were selected on the basis of the presence of a pair of alcohol dependent brothers or sisters. The probands were selected from among individuals in treatment for alcohol dependence in the Pittsburgh area. Probands were eligible if they met DSM-III criteria for AD and had a same sex sibling who similarly met criteria for AD. All proband pairs and their cooperative relatives (siblings and parents) were personally interviewed using a structured psychiatric interview (Diagnostic Interview Schedule [DIS]). The DIS provides good reliability and validity [Helzer et al., 1985] for alcohol dependence and alcohol abuse by DSM-III and IIIR criteria [American Psychiatric Association, 1980, 1987] and alcoholism by Feighner Criteria [Feighner et al., 1972], an early diagnostic set of criteria used in the Collaborative Studies on the Genetics of Alcoholism (COGA) family [Reich et al., 1998].
Families were excluded if the probands or any first-degree relative were considered to be primary for drug dependence (preceded alcohol dependence onset by at least 1 year), or the proband or first-degree relative met criteria for schizophrenia, or a recurrent major depressive disorder. Probands and relatives with mental retardation or physical illness precluding participation were excluded. Complete details regarding participant selection may be seen in Hill et al. [2004]. The majority of probands (80%) had three or more siblings who contributed DNA, consented to a clinical interview, and provided family history. These large sibships resulted in a total of 418 sib pairs of all types (201 affected–affected, 172 unaffected–affected, and 45 unaffected–unaffected. One or both parents have been genotyped in 86% of the families. An average of 5.1 individuals per family were genotyped.
SNP Selection
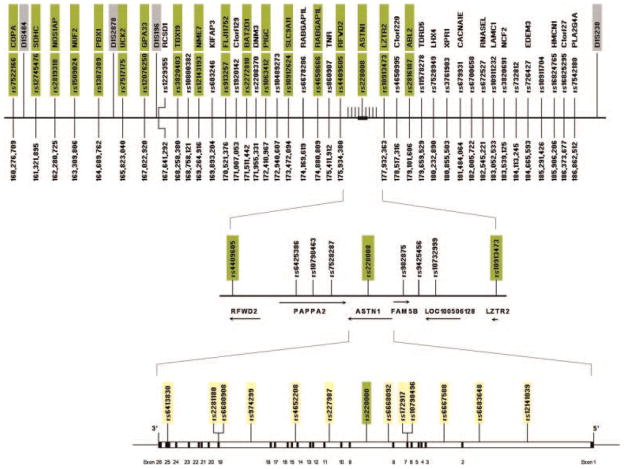
Previously, we carried out a genome-wide linkage analysis finding potentially important linkage results for multiple regions including Chromosome 1 [Hill et al., 2004]. Our study included genotyping in a 26.6 cM region on Chromosome 1 that centered on the microsatellite marker D1S196. Using a binary alcohol dependence phenotype and including relevant covariates (age, gender, and Constraint) a LOD score of 3.46 was obtained. To study this region further, SNPs were chosen with minor allele frequency (MAF) ≥ 0.15 and pair-wise linkage disequilibrium (LD) of r2 <0.8 using the HapMap CEU population at approximately 1 cM intervals in this region. The gentoyping and analysis was completed in three stages. First, we focused on a 19 cM region (Fig. 1) extending from rs7522166 to rs2816187. This region, bounded by these SNPs was chosen because rs7522166 is 7 cM proximal to D1S196 and rs2816187 is 13 cM distal to D1S196. We genotyped 18 SNPs at approximately 1 cM intervals in this region. Preliminary analysis revealed that the most significant SNP was rs228008 located in ASTN1 gene. In step 2, 31 SNPs were chosen to cover a region from rs1229355 near D1S196 distally to rs7542180, covering a 19 cM region at approximately 500 kb, and including three SNPs proximal and three distal to the ASTN1 gene at 125 kb intervals. Twelve additional SNPs were then chosen within the ASTN1 gene at an average distance of 28.9 kb.
FIG. 1.
The figure depicts a 26.6 cM region of Chromosome 1q 25.2 that includes the human ASTN1 gene and single nucleotide polymorphism (SNP) sites included in the present analyses. Distances are based on physical maps from Ensembl 63.
DNA Isolation and Genotyping
Genomic DNA was extracted from whole blood with a second aliquot prepared for EBV transformation and cryopreservation. PCR conditions were as described in Hill et al. [2004]. Genotyping was completed on a Biotage PSQ 96MA Pyrosequencer (Biotage AB, Uppsala, Sweden). Each polymorphism was analyzed by PCR amplification incorporating a biotinylated primer. Thermal cycling included 45 cycles at an annealing temperature of 60°C. The Biotage workstation was used to isolate the biotinylated single strand from the double strand PCR products. The isolated product was then sequenced using the complementary sequencing primer.
Quality Control
SNP genotyping quality control involved ongoing monitoring of SNP signals provided by Qiagen software. Output is provided using three categories for each SNP: pass, fail and check. Data analysis was performed for only those signals meeting the “pass” criterion. Signals that failed or were returned as needing further checking were rerun. If after three attempts the SNP did not meet the “pass” criterion, it was eliminated from the analysis and another SNP chosen as a replacement.
Statistical Methods
Mendelian inconsistency
The PedCheck program [O’Connell and Weeks, 1998] was used to evaluate individual SNPs for Mendelian inconsistencies based on the pedigree structures. As a result of the evaluation, 43 marker genotypes from among 19,470 were coded as missing to resolve the reported inconsistencies.
Hardy–Weinberg equilibrium (HWE)
Estimates of population allele frequencies were calculated using MENDEL version 11 [Lange et al., 2001]. Files required by the MENDEL program were generated via the program Mega2 [Mukhopadhyay et al., 2005]. Marker allele frequencies were tested for departures from Hardy–Weinberg equilibrium using the allele frequency option in MENDEL. None of the 59 SNPs analyzed were found to have P-values below the Bonferroni adjusted threshold (<0.00085) that would indicate significant HWE departures.
Genetic maps
Our Genetic Map Interpolator (GMI) software [Mukhopadhyay et al., 2010] was used to retrieve current physical map positions from Ensembl (Ensembl 63); these physical positions were then used to linearly interpolate genetic map positions based on the Rutgers Combined Linkage-Physical Map [Kong et al., 2004; Matise et al., 2007].
Family based association test (FBAT)
Transmission rates of marker alleles were examined using the family-based association test program, FBAT [Laird et al., 2000; Rabinowitz and Laird, 2000], assuming an additive genetic model with robust variance estimation (-e option) to account for the relatedness. This family-based method is a generalization of the transmission disequilibrium test (TDT) [Spielman et al., 1993] which provides a valid test of association even if admixture present. FBAT converts each pedigree into nuclear families, which are then treated as independent families for the test statistic calculation. Informative families consisting of parent-child trios are utilized in the FBAT analysis.
More powerful quasi-likelihood score (MQLS)
We also computed the “more powerful quasi-likelihood score” (MQLS) test [Thornton and McPeek, 2007], which is designed to test for case–control association on data sets containing related individuals. This method uses kinship coefficients to account for relatedness in the sample, using these coefficients to assign weights to individuals within pedigrees. The MQLS test, which uses the intact pedigree structures was performed assuming 10% population prevalence for alcohol dependence. Results did not differ in a meaningful way when we varied the assumed prevalence from 5% to 20%.
LODPAL linkage analysis
Nonparametric linkage analysis of 106 affected sibpairs was performed using each subject’s gender and age, and their scores on the Constraint scale of the Multidimensional Personality Questionnaire [Tellegen, 1982; Tellegen and Waller, 1982] as covariates and implemented using the LODPAL program [S.A.G.E 6.1.0, 2010]. The Constraint scale measures behavioral constraint, a construct that overlaps with risk-taking behavior, a tendency that has been linked to adverse health consequences including alcohol and drug dependence [DiClemente et al., 1995]. Using data from twins reared apart and together, Tellegen et al. [1988] reported that among the personality traits tested Constraint was among those with the greatest genetic variance (0.58). Adding covariates to the linkage analysis can increase the power to capture linkage by accounting for potential disease heterogeneity. Use of the personality trait Constraint appears to reduce heterogeneity among substance users. McGue et al. [1999] reported that within a sample of alcohol dependent individuals a subset with drug use disorder showed elevated Constraint scale scores, concluding that behavioral disinhibition among alcohol dependent individuals may be attributable to those who abuse drugs other than alcohol. LODPAL uses a general conditional logistic model to test for identity by descent (IBD) allele sharing [Olson, 1999]. The covariates chosen were those utilized in a previous genome-wide linkage analysis [Hill et al., 2004]. Estimated multipoint marker IBD allele sharing for the affected sibpairs was obtained from GENIBD (S.A.G.E. 6.1.0) and utilized as input for LODPAL analysis.
Haplotype analysis
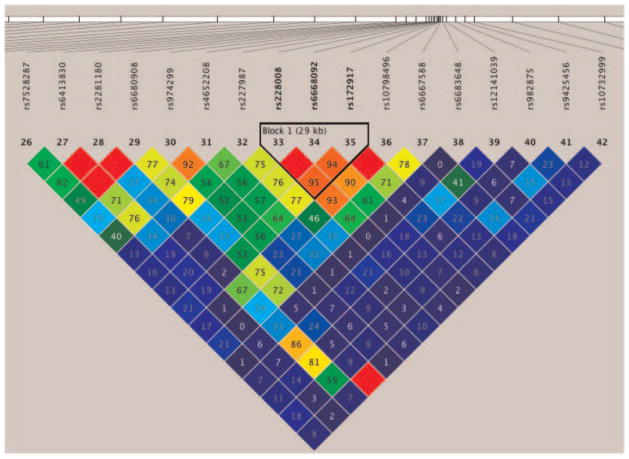
Linkage disequilibrium (LD) analysis was performed using the HAPLOVIEW program version 4.2 [Barrett et al., 2005]. The LD block structure was defined by calculating D′ values pairwise between SNPs. SNP haplotype blocks were created using the HAPLOVIEW default block determination method [Gabriel et al., 2002]. Only one haplotype block was identified. This haplotype block contained three SNPs (rs228008, rs6668092, and rs172917), spanning 0.03 cM within ASTN1. Pairwise linkage disequilibrium between the SNPs within ASTN1 and the LD block are shown in Figure 2.
FIG. 2.
Linkage disequilibrium analysis was performed using HAPLOVIEW (version 4.2). The block structure was defined by calculating D′ values pairwise between SNPs. One block was identified containing three SNPs within the ASTN1 gene.
RESULTS
Stage 1 Association Results
The initial 19 cM region (Fig. 1) extending from rs7522166 to rs2816187 included 18 SNPs at approximately 1 cM intervals. FBAT results revealed significant results for rs465066 P = 0.013 and for rs228008 P = 0.012.
Stage 2 Association Results
An additional 31 SNPs were chosen to cover a 19 cM region from rs1229355 near D1S196 distally to rs7542180 at approximately 500 kb intervals, and including three SNPs proximal and three distal to the ASTN1 gene at 125 kb intervals. FBAT results for the 49 SNPs revealed significant results for only two SNPs, rs465066 and rs228008. The only SNP showing significance in an MQLS analysis was rs228008.
Stage 3 Association Results
Ten additional SNPs were included at this stage to allow for finer mapping of the region immediately proximal and distal to rs228008 at 25 kb intervals to include rs6413830 proximally and rs1241039 distally. Analysis of the 59 SNPs revealed that the most strongly associated SNP for alcohol dependence affected status was rs228008 (MQLS P = 0.0009, FBAT P = 0.012) located within an intron of ASTN1. Four additional SNPs spanning a 0.03 cM region within the same intron were identified by MQLS as statistically significant (Table I). Results of the FBAT and MQLS analyses are summarized in Table I. LocusZoom was used to generate the association plot (http://csg.sph.umich.edu/locuszoom/; Fig. 3).
TABLE I.
Results of FBAT, MQLS, and LODPAL Analyses
| Marker | cM | LOD | Informative families—FBAT | FBAT P-value | MQLS P-value | Major/minor Allelea | Nucleotide | Allele freq.
|
Gene | |
|---|---|---|---|---|---|---|---|---|---|---|
| Affected | Unaffected | |||||||||
| rs7522166 | 164.75 | 0.089 | 23 | 0.629 | 0.233 | 2 | C | 0.478 | 0.523 | COPA |
| rs12745476 | 167.01 | 0.024 | 18 | 0.709 | 0.529 | 1 | C | 0.774 | 0.785 | SDHC |
| rs2819318 | 168.15 | 0.033 | 21 | 0.779 | 0.554 | 1 | C | 0.552 | 0.560 | NOS1AP |
| rs1509024 | 170.13 | 0.435 | 27 | 0.148 | 0.250 | 1 | G | 0.724 | 0.705 | NUF2 |
| rs1387389 | 173.39 | 1.771 | 25 | 0.455 | 0.344 | 1 | C | 0.687 | 0.723 | PBX1 |
| rs7517175 | 174.66 | 2.676 | 27 | 0.366 | 0.129 | 1 | T | 0.537 | 0.504 | UCK2 |
| rs12076250 | 176.07 | 3.250 | 24 | 0.135 | 0.152 | 1 | C | 0.600 | 0.547 | GPA33 |
| rs1229355 | 177.46 | 3.193 | 26 | 0.915 | 0.979 | 1 | T | 0.542 | 0.563 | RCSD1 |
| rs3820403 | 178.71 | 3.471 | 20 | 0.220 | 0.834 | 1 | A | 0.629 | 0.593 | TBX19 |
| rs10800382 | 179.46 | 3.459 | 30 | 0.238 | 0.376 | 1 | A | 0.582 | 0.516 | |
| rs12143193 | 180.03 | 3.262 | 19 | 0.205 | 0.367 | 1 | T | 0.684 | 0.642 | NME7 |
| rs603246 | 180.76 | 2.876 | 26 | 0.609 | 0.400 | 1 | G | 0.612 | 0.591 | KIFAP3 |
| rs913257 | 181.51 | 2.926 | 25 | 0.851 | 0.477 | 1 | A | 0.498 | 0.523 | FLJ11752 |
| rs1920142 | 182.01 | 2.660 | 25 | 0.359 | 0.409 | 2 | C | 0.343 | 0.356 | C1orf129 |
| rs2272810 | 182.46 | 2.557 | 26 | 0.864 | 0.458 | 2 | A | 0.490 | 0.485 | BAT2D1 |
| rs2208370 | 182.79 | 2.572 | 17 | 0.726 | 0.842 | 1 | G | 0.612 | 0.623 | DNM3 |
| rs1063412 | 183.12 | 2.650 | 29 | 0.970 | 0.302 | 1 | T | 0.525 | 0.609 | PIGC |
| rs10489273 | 183.53 | 2.916 | 24 | 0.676 | 0.584 | 1 | G | 0.570 | 0.557 | |
| rs10912624 | 183.86 | 2.817 | 20 | 0.501 | 0.599 | 1 | T | 0.780 | 0.797 | SLC9A11 |
| rs6678286 | 184.23 | 3.048 | 19 | 0.248 | 0.227 | 1 | A | 0.721 | 0.764 | RABGAP1L |
| rs4650666 | 184.62 | 2.589 | 21 | 0.014 | 0.802 | 1 | A | 0.610 | 0.617 | RABGAP1L |
| rs860907 | 185.00 | 2.390 | 26 | 0.992 | 0.199 | 1 | G | 0.622 | 0.650 | TNR |
| rs4409605 | 185.58 | 2.212 | 29 | 0.382 | 0.836 | 1 | A | 0.580 | 0.590 | RFWD2 |
| rs6425386 | 186.10 | 2.039 | 22 | 0.480 | 0.316 | 2 | T | 0.403 | 0.377 | PAPPA2 |
| rs10798463 | 186.22 | 2.032 | 27 | 0.559 | 0.620 | 1 | A | 0.704 | 0.728 | PAPPA2 |
| rs7528287 | 186.33 | 2.034 | 30 | 0.587 | 0.323 | 1 | G | 0.620 | 0.663 | PAPPA2 |
| rs6413830 | 186.44 | 2.035 | 22 | 0.828 | 0.365 | 1 | G | 0.597 | 0.547 | ASTN1 |
| rs2281180 | 186.47 | 2.035 | 17 | 0.677 | 0.852 | 1 | C | 0.830 | 0.862 | ASTN1 |
| rs6680908 | 186.47 | 2.035 | 7 | NA | 0.227 | 1 | C | 0.910 | 0.937 | ASTN1 |
| rs974299 | 186.49 | 2.035 | 28 | 0.248 | 0.105 | 1 | C | 0.525 | 0.469 | ASTN1 |
| rs4652208 | 186.52 | 2.035 | 27 | 0.426 | 0.321 | 2 | C | 0.498 | 0.457 | ASTN1 |
| rs227987 | 186.53 | 2.035 | 27 | 0.185 | 0.074 | 1 | T | 0.530 | 0.472 | ASTN1 |
| rs228008 | 186.56 | 1.943 | 26 | 0.012 | 0.001 | 1 | G | 0.652 | 0.547 | ASTN1 |
| rs6668092 | 186.57 | 1.874 | 25 | 0.021 | 0.002 | 1 | C | 0.637 | 0.532 | ASTN1 |
| rs172917 | 186.58 | 1.826 | 29 | 0.027 | 0.003 | 1 | G | 0.652 | 0.547 | ASTN1 |
| rs10798496 | 186.58 | 1.826 | 28 | 0.128 | 0.002 | 1 | C | 0.500 | 0.421 | ASTN1 |
| rs6667588 | 186.60 | 1.544 | 33 | 0.134 | 0.030 | 1 | G | 0.545 | 0.508 | ASTN1 |
| rs6683648 | 186.63 | 1.450 | 23 | 0.778 | 0.592 | 1 | A | 0.512 | 0.535 | ASTN1 |
| rs12141039 | 186.66 | 1.343 | 30 | 0.147 | 0.312 | 1 | T | 0.635 | 0.661 | ASTN1 |
| rs982875 | 186.76 | 1.458 | 27 | 0.059 | 0.429 | 1 | C | 0.528 | 0.528 | FAM5B |
| rs9425456 | 186.85 | 2.453 | 26 | 0.243 | 0.388 | 1 | C | 0.503 | 0.548 | |
| rs10732999 | 186.94 | 1.953 | 21 | 0.007 | 0.184 | 1 | G | 0.719 | 0.693 | |
| rs10913473 | 187.31 | 0.662 | 22 | 0.194 | 0.087 | 1 | T | 0.547 | 0.563 | LZTR2 |
| rs4650995 | 187.66 | 0.560 | 23 | 0.986 | 0.459 | 1 | T | 0.555 | 0.560 | C1orf220 |
| rs2816187 | 187.97 | 0.469 | 22 | 0.408 | 0.962 | 1 | C | 0.614 | 0.646 | ABL2 |
| rs11578278 | 188.18 | 0.480 | 28 | 0.984 | 0.632 | 1 | C | 0.639 | 0.673 | TDRD5 |
| rs7528949 | 188.47 | 0.458 | 24 | 0.611 | 0.635 | 1 | G | 0.557 | 0.540 | LHX4 |
| rs3761903 | 189.08 | 0.611 | 30 | 0.967 | 0.699 | 1 | C | 0.527 | 0.548 | XPR1 |
| rs679931 | 189.91 | 0.612 | 20 | 0.684 | 0.711 | 1 | A | 0.653 | 0.650 | CACNA1E |
| rs6700658 | 190.66 | 0.465 | 19 | 0.791 | 0.367 | 1 | C | 0.557 | 0.595 | |
| rs672527 | 191.50 | 0.286 | 21 | 0.227 | 0.910 | 1 | C | 0.702 | 0.701 | RNASEL |
| rs10911232 | 192.21 | 0.248 | 26 | 0.332 | 0.098 | 1 | C | 0.557 | 0.599 | LAMC1 |
| rs3820691 | 192.70 | 0.207 | 22 | 0.787 | 0.410 | 1 | T | 0.525 | 0.524 | NCF1 |
| rs732812 | 193.15 | 0.206 | 29 | 0.557 | 0.796 | 1 | T | 0.532 | 0.532 | |
| rs726427 | 193.58 | 0.227 | 22 | 0.994 | 0.453 | 1 | C | 0.542 | 0.598 | EDEM3 |
| rs10911704 | 194.04 | 0.225 | 26 | 0.322 | 0.386 | 1 | T | 0.575 | 0.598 | |
| rs16824765 | 194.39 | 0.219 | 19 | 0.171 | 0.433 | 1 | C | 0.731 | 0.701 | HMCN1 |
| rs16825295 | 194.61 | 0.194 | 23 | 0.895 | 0.867 | 1 | C | 0.592 | 0.602 | C1ord27 |
| rs7542180 | 194.94 | 0.154 | 17 | 0.880 | 0.866 | 2 | G | 0.500 | 0.480 | PLA2G4A |
1, major allele and 2, minor allele (designation based on NCBI allele frequencies European Caucasians).
FIG. 3.
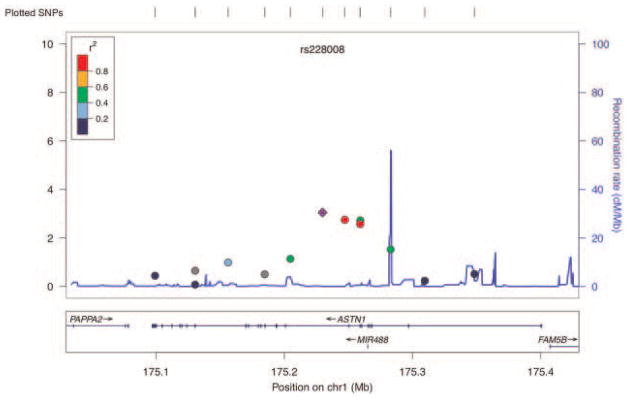
Association plot (−log 10 of the p values from MQLS) for SNPs within 300 kb of rs228008, the SNP with the maximum association observed.
Stage 3 Haplotype Results
A within-family association analysis between alcohol dependence and the revealed haplotype was tested using haplotype FBAT [Laird et al., 2000] assuming an additive genetic model and using a robust estimate of variance (Fig. 2). The GCG haplotype block within ASTN1 which included rs228008, rs6668092, and rs172917, with a frequency of 0.56, was found to be over-transmitted (P = 0.041) to affected offspring while the ATA haplotype block with a frequency of 0.40 was under-transmitted to affected offspring (P = 0.014).
Stage 3 Linkage results
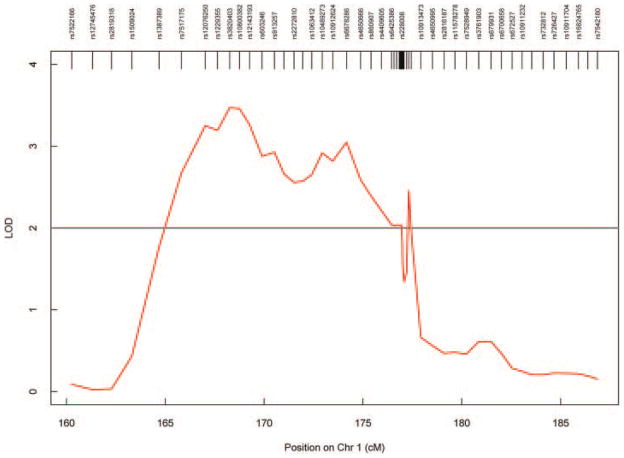
With 59 SNPs available for linkage analysis, we performed a LODPAL analysis using the same covariates used in the initial linkage analysis of this region that had also utilized LODPAL analyses [Hill et al., 2004]. Results of the LODPAL linkage analysis revealed LOD values greater than 1.95 in a 12 cM region extending from rs7517175 to rs1073299 that included the ASTN1 gene (Fig. 4).
FIG. 4.
Nonparametric linkage results using LODPAL with covariates (age, sex, and constraint).
DISCUSSION
Linkage analysis (LODPAL) and within family association (FBAT and MQLS) analyses were performed for 59 SNPs in this region of Chromosome 1. FBAT analysis requires heterozygosity in parents for families to be informative and included in the analysis. Accordingly, only a subset of families could be included in our FBAT analyses potentially reducing the power to detect within-family variation. Therefore, MQLS was included in our data analytic plan to allow for use of all of our data.
The first goal of our Chromosome 1 search was determine if genes might be uncovered in a 26 cM region that included D1S196 and D1S2878, microsatellite markers that had provided the strongest evidence for linkage on Chromosome 1. Linkage analysis of the 59 SNPs in this region found LOD scores between 1.83 and 3.47 in a region extending from rs7517175 to rs10798496, an approximately 11 cM region. Following the first stage analysis of Chromosome 1, our next goal was determine if association would be seen within this region using our family data. Our results point to SNPs within the ASTN1 gene being significantly associated with alcohol dependence.
Based on both linkage and association analyses, our results suggest that variation in the ASTN1 gene is associated with risk for alcohol dependence within multiplex for alcohol dependence families. In a previous study investigating 306 genes involved in neurotransmission and development, Gratacos and colleagues found rs2281180 within exon 19 associated with substance abuse. The present results confirm the Gratacos et al. [2008] case/control findings by identifying within family variation in alcohol dependence to be related to ASTN1 variation though an association with exonic rs2281180 was not seen.
Because our results point to the importance of the astrotactin neuronal protein (ASTN1) gene in the development of alcohol dependence, it may be useful to speculate on the origin of this relationship. ASTN1 has been extensively documented to be a receptor for glial-guided neuronal migration [Edmondson et al., 1988; Fishell and Hatten, 1991; Zheng et al., 1996; Adams et al., 2002]. ASTN1 along with a recently identified member of the astrotactin gene family ASTN2 [Wilson et al., 2010] has been shown to directly alter neuronal migration along glial fibers in the developing cerebellum. The early development of the mammalian brain is crucially dependent on the migration of neuronal precursors from germinal zones into the formation of neuronal laminae where synaptic connections are formed [Rakic, 1978; Hatten, 1999]. Molecular control of neuronal migration associated with the ASTN1 neuronal protein appears to hold promise for understanding a variety of human brain disorders. ASTN1 has been implicated in autism [Glessner et al., 2009], schizophrenia [Vrijenhoek et al., 2008; Kahler et al., 2008], bipolar disorder [Gratacos et al., 2008] and attention deficit hyperactivity disorder (ADHD) [Lesch et al., 2008]. One previous report has found a significant association between ASTN1 and substance use disorder contrasting 165 cases and 937 controls [Gratacos et al., 2008].
The potential importance of variation in a neuronal migration protein on alcohol dependence is apparent when one considers the multitude of studies now suggesting that volumetric differences in key brain areas are associated with risk for psychiatric disorders. An emerging literature has identified familial loading for alcohol dependence as a factor influencing brain structure and function [Hill et al., 2001, 2007, 2009, 2010a; Benegal et al., 2007; Herting et al., 2011]. Structural variation has been identified for the amygdala, orbitofrontal cortex and other components of the limbic network involved in emotion regulation as well as for the cerebellum [see Tessner and Hill, 2010 for review]. These anatomical alterations may provide the neurological substrate for excessive use of alcohol and development of alcohol dependence (AD) as a result of altered personality variation and cognitive functioning [see Hill, 2010b for review]. Therefore, it appears plausible that variation in ASTN1 gene may be related to brain morphological changes that could influence risk for AD. Genetic variation in other genes responsible for neuronal growth and differentiation has been found for the orbitofrontal cortex and cerebellum and risk for alcohol dependence [Hill et al., 2009, 2010a].
The present results should be interpreted in the context of some limitations, however. First, the linkage peak originally reported [Hill et al., 2004] for this region of Chromosome 1 is relatively large though large peaks are typical for complex traits. Because our peak was large, it can be presumed that it contains many genes. For example, one potentially important gene, KIAA0040, that was not included in our planned analysis, has recently been reported with genome-wide significance for alcohol dependence [Zuo et al., 2011]. This gene is within our originally identified linkage peak [Hill et al., 2004].
Although the originally identified linkage peak was confirmed in the present analysis, the LOD score obtained was modest and required inclusion of covariates to reveal the obtained results. However, selection of covariates for the current linkage analysis which included personality variation was based on phenotypic variation in AD that had proved important in our earlier analysis which had suggested that this region might harbor genes for alcohol dependence susceptibility [Hill et al., 2004]. Personality variation in traits such as Constraint, a determinant of behavioral tendencies to engage in risk-taking behavior [see Hill, 2010b for review], may have provided useful information for reducing heterogeneity among those with addictive behavior. Previous studies suggest that Constraint may be an endophenotype for addictive behavior [Hill et al., 1990; McGue et al., 1999].
Second, the families upon which the present report is based were ascertained through affected sib pairs, possibly rendering the results atypical for the general population of alcohol dependent families. Multiplex families appear to differ from alcohol dependent families in the general population by having greater transmission of alcohol dependence across generations. Follow up of offspring from these multiplex families indicates an exceptionally high rate of AD and associated substance use by young adulthood [Hill et al., 2008]. Although these families may not be representative of AD families in the general population, the study of multiplex families appears to provide an efficient means for identifying genes because of the greater likelihood that salient genes may be segregating within these families [Hill, 2010b]. Third, alcohol dependence was defined by DSM-III criteria a diagnostic system that requires the presence of tolerance and physical dependence. Use of the DSM IV diagnostic scheme may have provided differing results. Because subjects were selected at a time when DSM-III was the current diagnostic scheme, it was not possible to include the newer DSM systems.
Finally, the ascertainment of multiplex families along with assessment of multiple family members presents logistical challenges that limit the number of sibling pairs that can realistically be included. Our sample size was modest with 418 sibling pairs. Over the past decade there has been a shift toward large-scale genome-wide association studies (GWAS) instead of family-based methods where sample sizes are typically more modest. This trend was predicated on the notion that association studies are sometimes more powerful than linkage studies [Risch and Merikangas, 1996]. However, some have questioned whether GWAS methods that are designed to detect common rather than rare variants will explain a substantial portion of heritability in psychiatric disorders [Maher, 2008]. Others have argued that the GWAS approach may find common variants that provide statistically significant results, but only modest population attributable risk, comparing less favorably with focused investigations of families where genes can be identified with high predictive value [Mitchell and Porteous, 2009].
No one statistical genetic method can be expected to completely characterize the genetic underpinnings of complex phenotypes such as alcohol dependence. As Suarez et al. [2007] illustrated in simulations carried out using linkage, case–control association and family-based tests, each approach has limitations that are best addressed by using multiple methods. The potential value of family-based methods for detection of multiple rare variants within one locus or several more weakly associated loci within the same chromosomal region has been demonstrated for other complex psychiatric disorders using family data [Ng et al., 2009]. Recently, Ott et al. [2011] proposed that evaluating linkage and association simultaneously while taking combinations of data from pedigrees with different relationship structures (extended pedigrees and sibships) and case–control samples may provide maximal power to identify new genetic variants for trait loci beyond those that can be identified in genome-wide association case–control designs. Although we did not simultaneously evaluate linkage and association, we did use both approaches with the same data set to uncover evidence for both. Also, because the power to detect association increases with available data from related individuals [Sahana et al., 2010], it appears that family-based data collection will remain an important source for detecting both linkage and association. Availability of parental genotypes is especially important for accurate linkage analyses [Suarez et al., 2007] though such data are often unavailable where parents of the proband generation are >50 years of age. Although the parents of our probands were older, our analyses were performed with the benefit of DNA for 86% of the parents. All of the parents including those without DNA had phenotypic information available through direct interview, or if deceased through multiple family history reports. However, parental DNA was not available for 9 of the 65 families.
Although the present study has limitations, the approach taken was hypothesis-driven and included genotyping successively smaller chromosomal regions in order to confirm or refute the direction of previous analyses. Each step of our investigation pointed to the ASTN1 gene as being associated with alcohol dependence within these multiplex families. Haplotype analysis utilizing SNP variation within the ASTN1 gene confirmed the influence of variation in this gene on risk for alcohol dependence. Finally, linkage analysis using the same methodology utilized in the initial genome-wide linkage report [Hill et al., 2004] (LODPAL with covariates) finds a region with suggestive LOD scores that overlaps the region in which family-based association results showed maximal significance.
Footnotes
The authors have no conflicts of interest regarding this manuscript.
References
- Adams NC, Tomoda T, Cooper M, Dietz G, Hatten ME. Mice that lack astrotactin have slowed neuronal migration. Development. 2002;129:965–972. doi: 10.1242/dev.129.4.965. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders (DSM-III) Washington, DC: American Psychiatric Association; 1980. [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders-revised (DSM-III-R) Washington, DC: American Psychiatric Association; 1987. [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Benegal V, Antony G, Venkatasubramanian G, Jayakumar PN. Grey matter abnormalities and externalizing symptoms in subjects at high risk for alcohol dependence. Addict Biol. 2007;12:122–132. doi: 10.1111/j.1369-1600.2006.00043.x. [DOI] [PubMed] [Google Scholar]
- Caldwell CB, Gottesman II. Sex differences in the risk for alcoholism: A twin study. Behav Genet. 1991;21:563(abstract). [Google Scholar]
- DiClemente RJ, Hansen WB, Ponton LE. Handbook of adolescent health risk behavior. New York: Plenum; 1995. [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li T-K, Nurnberger JI, Jr, O’Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson JC, Liem RK, Kuster JE, Hatten ME. Astrotactin: A novel neuronal cell surface antigen that mediates neuron–astroglial interactions in cerebellar microcultures. J Cell Biol. 1988;106:505–517. doi: 10.1083/jcb.106.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feighner JP, Robins E, Guze SB, Woodruff RA, Jr, Winokur G, Munoz R. Diagnostic criteria for use in psychiatric research. Arch Gen Psychiatry. 1972;26:57–63. doi: 10.1001/archpsyc.1972.01750190059011. [DOI] [PubMed] [Google Scholar]
- Fishell G, Hatten ME. Astrotactin provides a receptor system for CNS neuronal migration. Development. 1991;113:755–765. doi: 10.1242/dev.113.3.755. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, Defelice M, Lochner A, Faggart M. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, Imielinski M, Frackelton EC, Reichert J, Crawford EL, Munson J, Sleiman PMA, Chiavacci R, Annaiah K, Thomas K, Hou C, Glaberson W, Flory J, Otieno F, Garris M, Soorya L, Klei L, Piven J, Meyer KJ, Evdokia Anagnostou E, Sakurai T, Game RM, Rudd DS, Zurawiecki D, McDougle CJ, Davis LK, Miller J, Posey DJ, Michaels S, Kolevzon A, Silverman JM, Bernier R, Levy SE, Schultz RT, Dawson G, Owley T, McMahon WM, Wassink TH, Sweeney JA, Nurnberger JI, Jr, Coon H, Sutcliffe JS, Minshew NJ, Grant SFA, Bucan M, Cook EH, Jr, Buxbaum JD, Devlin B, Schellenberg GD, Hakonarson H. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratacos M, Costas J, deCid R, Bayes M, Gonzalez JR, Baca-Garcia E, deDiego Y, Frernandez-Aranda F, Fernandez-Piqueras J, Guitart M, Martin-Santos R, Martorell L, Menchon JM, Roca M, Saiz-Ruiz J, Sanjuan J, Torrens M, Urretavizcaya M, Valero J, Vilella E, Estivill X, Carracedo A the Psychiatric Genetics Network Group. Identification of new putative suceptibility genes for several psychiatric disorders by asssociation analysis of regulatory and non-synonmous SNPs of 306 genes involved in neurotranmission and neurodevelopment. Am J Med Genet Part B. 2008;150B:808–816. doi: 10.1002/ajmg.b.30902. [DOI] [PubMed] [Google Scholar]
- Harwood H. Updating estimates of the economic costs of alcohol abuse in the United States: Estimates, update methods, and data. Report prepared by The Lewin Group for the National Institute on Alcohol Abuse and Alcoholism. Based on estimates, analyses, and data reported. In: Harwood H, Fountain D, Livermore G, editors. The economic costs of alcohol and drug abuse in the United States 1992. Report prepared for the National Institute on Drug Abuse and the National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health, Department of Health and Human Services; Rockville, MD: National Institutes of Health; 2000. NIH Publication No. 98-4327. [Google Scholar]
- Hatten ME. Central nervous system migration. Annu Rev Neurosci. 1999;22:511–539. doi: 10.1146/annurev.neuro.22.1.511. [DOI] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PAF, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: Consistency of findings in women and men. Psychol Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Helzer JE, Robins LN, McEvoy LT, Spitznagel EL, Stoltzman RK, Farmer A, Brockington IF. A comparison of clinical and diagnostic interview schedule diagnoses. Physician reexamination of lay-interviewed cases in the general population. Arch Gen Psychiatry. 1985;42(7):657–666. doi: 10.1001/archpsyc.1985.01790300019003. [DOI] [PubMed] [Google Scholar]
- Herting MM, Fair D, Nagel BJ. Altered fronto-cerebellar connectivity in alcohol-naïve youth with a family history of alcoholism. Neuroimage. 2011;54:2582–2589. doi: 10.1016/j.neuroimage.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SY. Neural plasticity, human genetics, and risk for alcohol dependence. Functional plasticity and genetic variation. In: Reilly MT, Lovinger DM, editors. International review of neurobiology. Vol. 91. San Diego, CA: Academic Press; 2010b. pp. 53–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SY, Zubin J, Steinhauer SR. Personality resemblance in relatives of male alcoholics: A comparison with families of male control cases. Biol Psychiatry. 1990;27:1305–1322. doi: 10.1016/0006-3223(90)90501-r. [DOI] [PubMed] [Google Scholar]
- Hill SY, DeBellis MD, Keshavan MS, Lowers L, Shen S, Hall J, Pitts T. Right amygdala volume in adolescent and young adult offspring from families at high risk for developing alcoholism. Biol Psychiatry. 2001;49:894–905. doi: 10.1016/s0006-3223(01)01088-5. [DOI] [PubMed] [Google Scholar]
- Hill SY, Shen S, Zezza N, Hoffman EK, Perlin M, Allan W. A genome-wide search for alcoholism susceptibility genes. Am J Med Genet Part B. 2004;128B:102–113. doi: 10.1002/ajmg.b.30013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SY, Muddasani S, Prasad K, Steinhauer S, Scanlon J, McDermott M, Keshavan M. Cerebellar volume in offspring from multiplex alcohol dependence families. Biol Psychiatry. 2007;61:41–47. doi: 10.1016/j.biopsych.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SY, Shen S, Locke-Wellman J, Lowers L. Psychopathology in childhood and adolescence: A prospective study of offspring from multiplex alcoholism families. Psychiatry Res. 2008;160:155–166. doi: 10.1016/j.psychres.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SY, Wang S, Kostelnik B, Carter H, Holmes B, McDermott M, Zezza N, Stiffler S, Keshavan MS. Disruption of orbitofrontal cortex laterality in offspring from multiplex alcohol dependence families. Biol Psychiatry. 2009;65:129–136. doi: 10.1016/j.biopsych.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SY, Wang S, Carter H, Tessner K, Holmes B, McDermott M, Zezza N, Stiffler S. Cerebellum development in offspring from multiplex alcohol dependence families: Allelic variation in GABRA2 and BDNF. Psychiatry Res Neuroimaging. 2011;194:304–313. doi: 10.1016/j.pscychresns.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahler AK, Djurovic S, Kulle B, Jonsson EG, Agartz I, Hall H, Opjordsmoen S, Jacobsen KD, Hansen T, Melle I, Werge T, Steen VM, Andreassen OA. Association analysis of schizophrenia on 18 genes involved in neuroal migration: MDGA1 as a new susceptibility gene. Am J Med Genet Part B. 2008;147B:1089–1100. doi: 10.1002/ajmg.b.30726. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Heath AC, Neale MC, Kessler RC, Eaves LJ. A population-based twin study of alcoholism in women. JAMA. 1992;268:1877–1882. [PubMed] [Google Scholar]
- Kessler RC, Crum RM, Warner LA, Nelson CB, Schulenberg J, Anthony J. Lifetime co-occurrence of DSM-III-R alcohol abuse and dependence with other psychiatric disorder in the national comorbidity survey. Arch Gen Psychiatry. 1997;54:3123–3321. doi: 10.1001/archpsyc.1997.01830160031005. [DOI] [PubMed] [Google Scholar]
- Kong X, Murphy K, Raj T, He C, White PS, Matise TC. A combined linkage-physical map of the human genome. Am J Hum Genet. 2004;75:1143–1148. doi: 10.1086/426405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19:S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Lange K, Cantor R, Horvath S, Perola M, Sabatti C, Sinsheimer J, Sobel E. MENDEL version 4.0: A complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am J Hum Genet. 2001;69(Suppl):504. [Google Scholar]
- Lesch KP, Timmesfeld N, Renner TJ, Halperin R, Roser C, Nguyen TT, Craig DW, Romanos J, Heine M, Meyer J, Freitag C, Warnke A, Romanos M, Schafer H, Walitza S, Reif A, Stephan DA, Jacob C. Molecular genetics of adult ADHD: Converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm. 2008;115:1573–1585. doi: 10.1007/s00702-008-0119-3. [DOI] [PubMed] [Google Scholar]
- Maher B. Personal genomes: The case of missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- Matise TC, Chen F, Chen W, De La Vega FM, Hansen M, He C, Hyland FC, Kennedy GC, Kong X, Murray SS, Ziegle JS, Stewart WC, Buyske S. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17:1783–1786. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Slutske W, Iacono WG. Personality and substance use disorders. J Consult Clin Psychol. 1999;67:394–404. doi: 10.1037//0022-006x.67.3.394. [DOI] [PubMed] [Google Scholar]
- Mitchell KJ, Porteous DJ. GWAS for psychiatric disease: Is the framework built on a solid foundation. Mol Psychiatry. 2009;14:740–745. doi: 10.1038/mp.2009.17. [DOI] [PubMed] [Google Scholar]
- Mokdad AH, Marks JS, Stroup DF, Gerberding JL. Actual causes of death in the United States, 2000. JAMA. 2004;291:1238–1245. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill WP, Weeks DE. Mega2: Data-handling for facilitating genetic linkage and association analyses. Bioinformatics. 2005;21:2556–2557. doi: 10.1093/bioinformatics/bti364. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay N, Tang X, Weeks DE. Genetic map interpolator. Paper presented at the annual meeting of the American Society of Human Genetics; Washington, D.C. 2010. [Google Scholar]
- Ng MYM, Levinson DF, Faraone SV, Suarez BK, DeLisi LE, Arinami T, Riley B, Paunio T, Pulver AE, Irmansyah, Holmans PA, Escamilla M, Wildenauer DB, Williams NM, Laurent C, Mowry BJ, Brzustowicz LM, Maziade M, Sklar P, Garver DL, Abecasis GR, Lerer B, Fallin MD, Gurling HMD, Gejman PV, Lindholm E, Moises HW, Byerley W, Wijsman EM, Forabosco P, Tsuang MT, Hwu H-G, Okazaki Y, Kendler KS, Wormley B, Fanous A, Walsh D, O’Neill FA, Peltonen L, Nestadt G, Lasseter VK, Liang KY, Papadimitriou GM, Dikeos DG, Schwab SG, Owen MJ, O’Donovan MC, Norton N, Hare E, Raventos H, Nicolini H, Albus M, Maier W, Nimgaonkar VL, Terenius L, Mallet J, Jay M, Godard S, Nertney D, Alexander M, Crowe RR, Silverman JM, Bassett AS, Roy M-A, Mérette C, Pato CN, Pato MT, Louw Roos J, Kohn Y, Amann-Zalcenstein D, Kalsi G, McQuillin A, Curtis D, Brynjolfson J, Sigmundsson T, Petursson H, Sanders AR, Duan J, Jazin E, Myles-Worsley M, Karayiorgou M, Lewis CM. Meta-analysis of 32 genome-wide linkage studies of schizophrenia. Mol Psychiatry. 2009;14:774–785. doi: 10.1038/mp.2008.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: A program for identifying genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JM. A general conditional-logistic model for affected-relative-pair linkage studies. Am J Hum Genet. 1999;65:1760–1769. doi: 10.1086/302662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J, Kamatani Y, Lathrop M. Family-based designs for genome-wide association studies. Nat Rev: Genet. 2011;12:465–474. doi: 10.1038/nrg2989. [DOI] [PubMed] [Google Scholar]
- Prescott CA, Aggen SH, Kendler KS. Sex differences in the sources of genetic liability to alcohol abuse and dependence in a population based sample of US twins. Alcohol Clin Exp Res. 1999;23:1136–1144. doi: 10.1111/j.1530-0277.1999.tb04270.x. [DOI] [PubMed] [Google Scholar]
- Rabinowitz D, Laird N. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Human Heredity. 2000;504:227–233. doi: 10.1159/000022918. [DOI] [PubMed] [Google Scholar]
- Rakic P. Neuronal migration and contact guidance in the primate telencephalon. Postgrad Med J. 1978;54(Suppl 1):25–40. [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li T-K, Conneally PM, Nurnberger JI, Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- S.A.G.E 6.1.0. Statistical analysis for genetic epidemiology. 2010 http://darwin.cwru.edu/sage/
- Sahana G, Guldbrandtsen B, Janss C, Ott J. Comparison of association mapping methods in a complex pedigreed population. Genet Epidemiol. 2010;34:455–462. doi: 10.1002/gepi.20499. [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: The insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Suarez BK, Culverhouse R, Jin CH, Hinrichs A. Linkage, case–control association, and family-based association tests for complex disorders. BMS Proc. 2007;1(Suppl 1):S43. doi: 10.1186/1753-6561-1-s1-s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellegen A. Brief manual for the multidimensional personality questionnaire. Minneapolis: University of Minnesota. C; 1982. Unpublished manuscript. [Google Scholar]
- Tellegen A, Waller NG. Exploring personality through test construction: Development of the multidimensional personality questionnaire. Minneapolis: University of Minnesota Press; 1982. [Google Scholar]
- Tellegen A, Lykken DT, Bouchard TJ, Jr, Wilcox KJ, Segal NL, Rich S. Personality similarity in twins reared apart and together. J Pers Soc Psychol. 1988;54:1031–1039. doi: 10.1037//0022-3514.54.6.1031. [DOI] [PubMed] [Google Scholar]
- Tessner K, Hill SY. Neural circuitry associated with risk for alcohol dependence. Neuropsychol Rev. 2010;20:1–20. doi: 10.1007/s11065-009-9111-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton T, McPeek MS. Case–control association testing with related individuals: A more powerful quasi-likelihood score test. Am J Hum Genet. 2007;81:321–337. doi: 10.1086/519497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, Strengman E, Sabatti C, Geurts van Kessel A, Brunner HG, Ophoff RA, Veltman JA. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet. 2008;83:504–510. doi: 10.1016/j.ajhg.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K-S, Liu X, Zhang Q, Pan Y, Aragam N, Zeng M. A meta-analysis of two genome-wide association studies identifies 3 new loci for alcohol dependence. J Psychiatric Res. 2011;45:1419–1425. doi: 10.1016/j.jpsychires.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Wilson PM, Fryer RH, Fan Y, Hatten ME. ASTN2, A novel member of the astrotactin gene family regulates the trafficking of ASTN1 during glial-guided neuronal migration. J Neurosci. 2010;30:8529–8540. doi: 10.1523/JNEUROSCI.0032-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Heintz N, Hatten ME. CNS gene encoding astrotactin which supports neuronal migration along glial fibers. Science. 1996;272:417–419. doi: 10.1126/science.272.5260.417. [DOI] [PubMed] [Google Scholar]
- Zuo L, Gelernter J, Zhang CK, Zhao H, Lu L, Kranzler HR, Malison RT, Li C-SR, Wang F, Zhang X-Y, Deng H-W, Krystal JH, Zhang F, Luo X. Genome-Wide association study of alcohol dependence implicates KIAA0040 on Chromosome 1q. Neuropsychopharmacolgy. 2012;37:557–566. doi: 10.1038/npp.2011.229. [DOI] [PMC free article] [PubMed] [Google Scholar]