Abstract
Background
Tumor-derived cytokines and their receptors usually take important roles in the disease progression and prognosis of cancer patients. In this survey, we aimed to detect the expression levels of MIF and CXCR4 in different cell populations of tumor microenvironments and their association with survivals of patients with esophageal squamous cell carcinoma (ESCC).
Methods
MIF and CXCR4 levels were measured by immunochemistry in tumor specimens from 136 resected ESCC. Correlation analyses and independent prognostic outcomes were determined using Pearson’s chi-square test and Cox regression analysis.
Results
The expression of CXCR4 in tumor cells was positively associated with tumor status (P = 0.045) and clinical stage (P = 0.044); whereas the expression of CXCR4 in tumor-infiltrating lymphocytes (TILs) and the expression of MIF in tumor cells and in TILs were not associated with clinical parameters of ESCC patients. High MIF expression in tumor cells or in TILs or high CXCR4 expression in tumor cells was significantly related to poor survival of ESCC patients (P < 0.05). Multivariate analysis showed that the expression of MIF or CXCR4 in tumor cells and the expression of MIF in TILs were adverse independent factors for disease-free survival (DFS) and overall survival (OS) in the whole cohort of patients (P < 0.05). Furthermore, the expression of MIF and CXCR4 in tumor cells were independent factors for reduced DFS and OS in metastatic/recurrent ESCC patients (P < 0.05). Interestingly, the expressions of MIF and CXCR4 in tumor cells and in TILs were significantly positively correlated (P < 0.05), and the combined MIF and CXCR4 expression in tumor cells was an independent adverse predictive factor for DFS and OS (P < 0.05).
Conclusion
The expressions of MIF and CXCR4 proteins in tumor cells and TILs have different clinically predictive values in ESCC.
Keywords: Esophageal squamous cell carcinoma, Tumor microenvironment, MIF, CXCR4, Prognosis
Background
Esophageal squamous cell carcinoma (ESCC) is one of the major histopathological subtypes of esophageal cancer. ESCC is the fourth most prevalent malignancy in China and a leading cause of cancer-related death, and its overall five-year survival rate is less than 30% [1]. It has been reported that the molecular markers related to tumor cell growth and metastasis, the function of the tumor infiltrating-lymphocytes (TILs), and the interaction between tumor cells and infiltrated immune cells in tumor microenvironments have been evaluated for their contribution to the prognoses of ESCC patients in recent studies, except the traditional prognostic factors determined at diagnosis, such as TNM stage and cell differentiation [2-7]. However, reliable markers for disease development and prognosis are still lacking in ESCC. To date, it has been revealed that the expression levels of some over-expressed genes within tumor microenvironments are related to the prognosis of ESCC, such as interleukin 17 (IL-17), SKP2, Foxp3 and Tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL); however the results are still conflicting [8-13].
The macrophage migration inhibitory factor (MIF) is a 115-amino acid secreted cytokine that is involved in a number of pathological conditions, including autoimmunity, obesity and cancer [14]. The primary MIF receptor is CD74, and CD74 can bind to CD44 to form a receptor complex and mediate the transduction of MIF signaling [15]; However, CD74 can also form complexes with the C-X-C chemokine receptor type 2 (CXCR2) and type 4 (CXCR4) to transmit MIF signals to integrins in inflammatory cells [16,17]. Recent studies have demonstrated that MIF and CXCR4 were overexpressed in a number of cancers, including gastric cancer, breast cancer, prostate cancer, colon cancer and nasopharyngeal carcinoma [18-26]. However, the expression pattern of MIF and CXCR4 proteins in tumor microenvironments and their impact on the survival of cancer patients are still unclear.
Therefore, we evaluated the expression of MIF and its receptor CXCR4 protein in tumor cells and TILs of tumor microenvironment in 136 resected ESCC specimens using immunohistochmeistry staining. The correlations between the expression levels of MIF and CXCR4 in different cell subsets in tumor microenvironment and prognostic factors were assessed to determine the clinical relevance and predictive value of the MIF and CXCR4 expression in different cell subsets of tumor microenvironments of ESCC.
Methods
Patient selection
One hundred and thirty-six ESCC patients who underwent surgery at Sun Yat-Sen University Cancer Center in Guangzhou City of China from November of 2000 to December of 2002 were involved in this retrospective study. None of the patients had received anticancer treatment prior to surgery, and all of the patients had histologically confirmed primary ESCC. The patients had a median age of 62 years (range, 35 to 90 years); 111 (81.6%) were males and 25 (18.4%) were females. There were 74 (54.4%) cases of Stage I and II tumors and 62 (45.6%) cases of Stage III and IV tumors based on the International Union against Cancer 2002 TNM staging system and WHO classification criteria [27]. Of the 136 patients, 103 (75.7%) had died. The patients’ clinical parameters are detailed in Additional file 1: Table S1. The tumor specimens were obtained as paraffin blocks from the Pathology Department of our cancer center and clinical data were obtained from hospital records after surgery. The follow-up data from the 136 patients with ESCC in this study were available and complete. The OS was defined as the time interval from the date of surgery to the date of cancer-related death or the end of follow-up (December 2011), and the DFS was defined as the time interval from the date of surgery to the date of tumor recurrence or tumor metastasis. This study was approved by the Research Ethics Committee of the Sun Yat-Sen University Cancer Center.
Reagents and antibodies
The following primary antibodies were used in this study: mouse anti-human MIF (ab55445; Abcam, USA), mouse anti-human CXCR4 (Clone 44716; R&D Systems, Minneapolis, MN), and horseradish peroxidase-labeled goat antibody against a mouse/rabbit IgG antibody (Envision; Dako, Glostrup, Denmark).
Immunohistochemistry and assessment
The paraffin-embedded tissues were sectioned continuously into 4-μm-thick sections. The tissue sections were dewaxed in xylene, rehydrated and rinsed in graded ethanol solutions. The antigens were retrieved by heating the tissue sections at 100°C for 30 min in citrate (10 mmol/L, pH 6.0) or EDTA (1 mmol/L, pH 9.0) solution when necessary. The sections were then immersed in a 0.3% hydrogen peroxide solution for 30 min to block endogenous peroxidase activity, rinsed in phosphate-buffered saline (PBS) for 5 min, and incubated with the primary antibodies, including MIF, CXCR4 at 4°C overnight. A negative control was performed by replacing the primary antibody with a normal murine IgG antibody. The sections were then incubated with a horseradish peroxidase-labeled goat antibody against a mouse/rabbit secondary antibody at room temperature for 30 min. Finally, the signal was developed for visualization with 3, 3′-diaminobenzidine tetrahydrochloride (DAB), and all of the slides were counterstained with hematoxylin.
Two independent observers blinded to the clinicopathological information scored the MIF and CXCR4 expression levels in tumor cells by assessing (a) the proportion of positively stained cells :(0, <5%; 1, 6 to 25%; 2, 26 to 50%; 3, 51 to 75%; 4, >75%) and (b) the signal intensity: (0, no signal; 1, weak; 2, moderate; 3, strong). The score was the product of a × b. The levels of MIF and CXCR4 expression in lymphocytes were obtained by counting the positively and negatively stained lymphocytes in five to ten separate 400× high-power microscopic fields and calculating the mean percentage of positively stained lymphocytes among the total lymphocytes per field.
Statistical analysis
All analyses were conducted with SPSS 16.0 (SPSS Inc., Chicago, IL, USA). The patients were divided into two subgroups (a high-level group and a low-level group) based on the median values of various immunohistochemical variables in our data. Pearson’s chi-square test and Fisher’s chi-square test were used to analyze the correlation between immunohistochemical variants in different cell subsets and the patients’ clinicopathological parameters. The MIF and CXCR4 expression levels were examined in tumor cells and in TILs in relation to the patients’ clinical prognosis using the Kaplan-Meier method and the log-rank survival analysis. Prognostic factors were assessed by univariate and multivariate analyses using the Cox proportional hazards model. The relationships among the expression levels of MIF and CXCR4 were assessed using Pearson’s correlation coefficient and linear regression analyses. A two-tailed P-value <0.05 was considered statistically significant in this study.
Results
Expression patterns of MIF and CXCR4 in ESCC and their correlations with clinicopathological and immunohistochemical variables
In the present study, the protein expression levels of MIF and CXCR4 were examined in tumor specimens from 136 patients with ESCC. MIF was expressed in the cytoplasm of tumor cells and TILs, and CXCR4 was expressed in the nucleus or cytoplasm and cell membrane of tumor cells and the cell membrane and cytoplasm of TILs (Figure 1). Based on the criteria described in the Methods section, high expression levels of MIF and CXCR4 in tumor cells were noted in samples from 73 (53.7%) and 47 (34.6%) of the 136 patients, respectively. The mean percentage and the range of the percentage of patients with TILs positive for MIF or CXCR4 expression per high-power light microscopic field were 33% (range, 0 to 92%) and 20% (range, 0 to 78%), respectively, among the 136 patients assessed (Additional file 2: Table S2).
Figure 1.
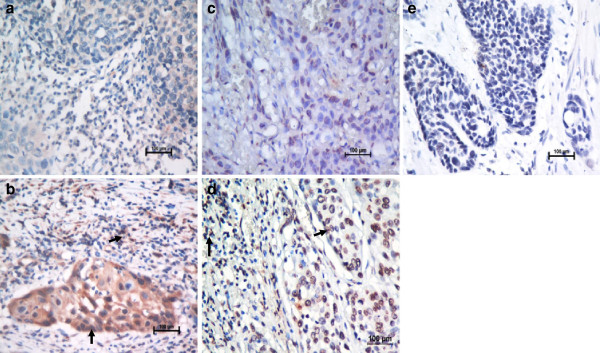
Immunohistochemical staining for MIF and CXCR4 in human esophageal carcinoma. Our data showed low expression levels of MIF (a) and CXCR4 (c) (X 400) and high expression levels of MIF (b) and CXCR4 (d), compared with the negative control (e) (X 400), in tumor tissues from patients with ESCC. The arrows point to the positive staining of tumor cells or TILs.
The associations between clinicopathological features and immunohistochemical variables in different cell subsets of the tumor microenvironment in samples from 136 ESCC patients are summarized in Table 1. In the present study, the patients were divided into two groups (a high-level group and a low-level group) based on the medians of immunohistochemical variable values in diverse cell subsets. High expression levels of MIF in tumor cells were not correlated with clinicopathological variables, whereas high expression levels of CXCR4 in tumor cells were positively closely correlated with T status (P = 0.045) and clinical stage (P = 0.044). Furthermore, the expression levels of MIF and CXCR4 in TILs were not related to any of the clinicopathological parameters, including age, gender, WHO grade, T status, N status and clinical stage.
Table 1.
Association of the expression of MIF, CXCR4 and clinicopathologic parameters in 136 patients with ESCC
|
Clinicopathologic parameter |
Case |
Expression in tumor cells |
Expression in TILs |
||||||
|---|---|---|---|---|---|---|---|---|---|
| High level expression of MIF (%) | Pa | High level expression of CXCR4 (%) | Pa | High level expression of MIF (%) | Pa | High level expression of CXCR4 (%) | Pa | ||
|
Age |
|
|
|
|
|
|
|
|
|
|
≤62 (y) |
71 |
35 (49.3%) |
0.284 |
26 (36.6%) |
0.597 |
31 (43.7%) |
0.122 |
38 (53.5%) |
0.391 |
|
>62 (y) |
65 |
38 (58.5%) |
|
21 (32.3%) |
|
37 (56.9%) |
|
30 (46.2%) |
|
|
Gender |
|
|
|
|
|
|
|
|
|
|
Female |
25 |
11 (44.0%) |
0.283 |
9 (36.0%) |
0.867 |
14 (56.0%) |
0.507 |
17 (68.0%) |
0.075b |
|
Male |
111 |
62 (55.9%) |
|
38 (34.2%) |
|
54 (48.6%) |
|
51 (45.9%) |
|
|
WHO grade |
|
|
|
|
|
|
|
|
|
|
G1 |
40 |
27 (67.5%) |
0.087 |
13 (32.5%) |
0.669 |
22 (55.0%) |
0.579 |
19 (47.5%) |
0.448 |
|
G2 |
59 |
30 (50.8%) |
|
19 (32.2%) |
|
30 (50.8%) |
|
33 (55.9%) |
|
|
G3 |
37 |
16 (43.2%) |
|
15 (40.5%) |
|
16 (43.2%) |
|
16 (43.2%) |
|
|
T status |
|
|
|
|
|
|
|
|
|
|
T1-2 |
44 |
22 (50.0%) |
0.552 |
10 (22.7%) |
0.045* |
19 (43.2%) |
0.271 |
24 (54.5%) |
0.463 |
|
T3-4 |
92 |
51 (55.4%) |
|
37 (40.2%) |
|
49 (53.3%) |
|
44 (47.8%) |
|
|
N status |
|
|
|
|
|
|
|
|
|
|
N0 |
69 |
36 (52.2%) |
0.721 |
20 (29.0%) |
0.165 |
32 (46.4%) |
0.391 |
37 (53.6%) |
0.391 |
|
N1 |
67 |
37 (55.2%) |
|
27 (40.3%) |
|
36 (53.7%) |
|
31 (46.3%) |
|
|
Clinical stage |
|
|
|
|
|
|
|
|
|
|
I-II |
74 |
41 (55.4%) |
0.659 |
20 (27.0%) |
0.044* |
37 (50.0%) |
1.00 |
41 (55.4%) |
0.168 |
| III-IV | 62 | 32 (51.6%) | 27 (43.5%) | 31 (50.0%) | 27 (43.5%) | ||||
Note: *P < 0.05, a, Pearson’s X2 test. b, Fisher’s X2 test.
Immunohistochemical variables in diverse cell subsets and patient survival
Among the 136 patients with ESCC, the median survival time was 25 months (range: 0 to 33 months). The cumulative five-year OS rate and DFS rate of the patients in this study were 29 and 31%, respectively (data not shown). The statistical analysis showed a significant negative correlation between DFS, OS and the expression levels of MIF in tumor cells and TILs and CXCR4 in tumor cells (P < 0.05, Figure 2).
Figure 2.
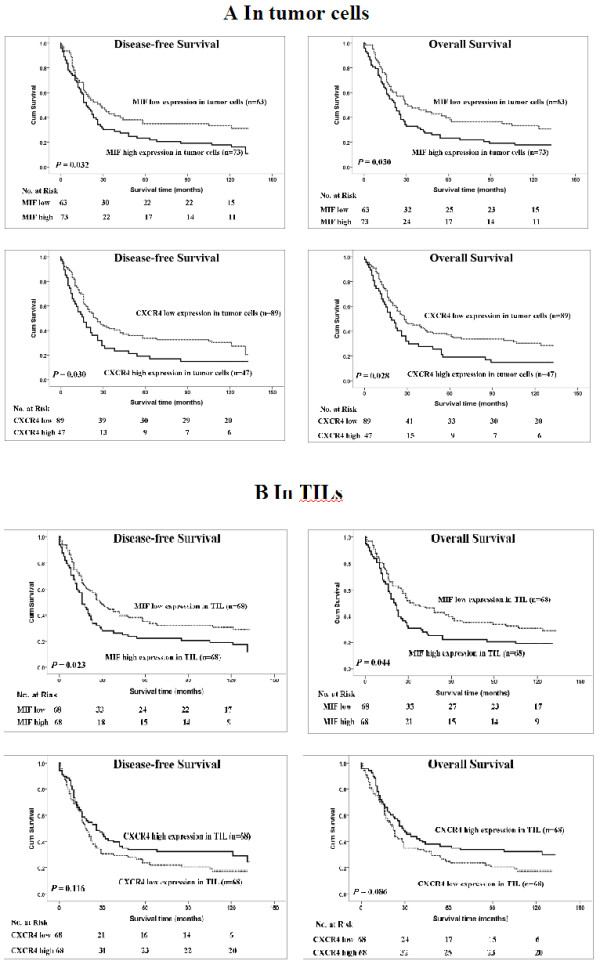
Kaplan-Meier survival analysis in patients with ESCC. (A) Disease-free survival and overall survival curves for patients according to the low and high expression levels of immunohistochemical variables in tumor cells. (B) Disease-free survival and overall survival curves for patients according to the low and high expression levels of immunohistochemical variables in TILs.
The univariate analysis demonstrated that high expression levels of MIF (P = 0.032 and P = 0.030) or CXCR4 (P = 0.030 and P = 0.028) in tumor cells were noticeably correlated with reduced DFS and OS and that high expression level MIF (P = 0.023 and P = 0.044) in TILs were also significantly associated with decreased DFS and OS; however, the high expression of CXCR4 was weakly correlated with improved DFS and OS (P > 0.05) (Table 2). As expected, and as shown in Table 2, clinicopathological parameters such as gender, WHO grade, nodal status and TNM stage are also of prognostic value. Furthermore, we determined that, with the exception of classical prognostic factors such as gender and WHO grade, the expression of MIF or CXCR4 in tumor cells and the MIF expression in TILs were independent predictors of DFS and OS according to the multivariate Cox model analysis (Table 3).
Table 2.
Univariate analysis of DFS and OS in 136 patients with ESCC
|
Variables |
DFS (n=136) |
OS (n=136) |
||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
|
Age, years (≤62/>62) |
0.682 (0.462-1.005) |
0.053 |
0.714 (0.483-1.054) |
0.090 |
|
Gender (male/female) |
0.452 (0.256-0.798) |
0.006* |
0.417 (0.232-0.747) |
0.003* |
|
WHO Grade (1/2/3) |
1.362 (1.053-1.763) |
0.019* |
1.324 (1.023-1.715) |
0.033* |
|
Tumor (T) status (1-2/3-4) |
1.569 (1.019-2.416) |
0.041* |
1.508 (0.976-2.332) |
0.064 |
|
Nodal (N) status (0/1) |
2.095 (1.415-3.101) |
<0.001* |
1.998 (1.346-2.965) |
0.001* |
|
TNM stage (I-II/III-IV) |
1.346 (1.110-1.633) |
0.003* |
1.318 (1.085-1.601) |
0.005* |
|
MIF in tumor cells (low/high) |
1.518 (1.028-2.242) |
0.036* |
1.532 (1.034-2.269) |
0.033* |
|
CXCR4 in tumor cells (low/high) |
1.537 (1.035-2.283) |
0.033* |
1.550 (1.041-2.307) |
0.031* |
|
MIF in lymphocytes (low/high) |
1.548 (1.053-2.275) |
0.026* |
1.481 (1.003-2.185) |
0.048* |
| CXCR4 in lymphocytes (low/high) | 0.738 (0.501-1.085) | 0.122 | 0.715 (0.485-1.055) | 0.091 |
Note: * P < 0.05.
Table 3.
Multivariate Cox analyses for DFS and OS of 136 patients with ESCC
|
Variables |
DFS (n=136) |
OS (n=136) |
||
|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | |
|
MIF in Tumor cells (low/high) |
1.689 (1.132-2.521) |
0.010* |
1.619 (1.084-2.418) |
0.018* |
|
CXCR4 in Tumor cells (low/high) |
1.708 (1.126-2.591) |
0.012* |
1.612 (1.072-2.425) |
0.022* |
|
MIF in Lymphocytes (low/high) |
1.473 (0.999-2.172) |
0.050* |
1.523 (1.027-2.259) |
0.037* |
| Combination of MIF and CXCR4 in tumor cells (low/mid/high) | 1.338 (1.064-1.683) | 0.013* | 1.263 (1.009-1.583) | 0.042* |
Note: The Cox proportional hazards regression model contained the significantly different factors in univariate analysis, including gender, WHO grade, T status, N status and TNM stage. HR, hazard ratio; CI, confidence interval; * means P < 0.05.
The expression levels of MIF and CXCR4 in diverse cell subsets and the survival of patients with metastatic/recurrent ESCC
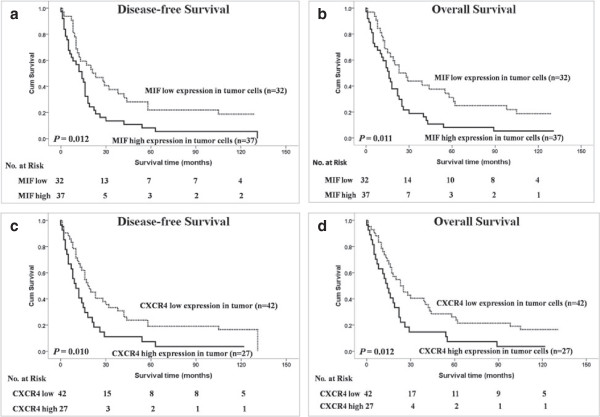
Among the 136 patients with ESCC, there were 67 (49.3%) patients with locoregional ESCC and 69 cases (50.7%) with metastatic/recurrent ESCC. The univariate analysis demonstrated that the high expression levels of MIF (P = 0.012 and P = 0.011) or CXCR4 (P = 0.010 and P = 0.012) in tumor cells were significantly correlated with poor DFS and OS in patients with metastatic/recurrent ESCC (Figure 3) but no association with locoregional ESCC (Data not shown).
Figure 3.
Kaplan-Meier survival analysis in patients with metastatic/recurrent ESCC. Disease-free survival and overall survival curves for metastatic/recurrent ESCC patients with low and high expression levels of MIF in tumor cells (a and b). Disease-free survival and overall survival curves for metastatic/recurrent ESCC patients with low and high expression levels of CXCR4 in tumor cells (c and d).
The correlation of combined expression levels of MIF and CXCR4 in diverse cell populations and survivals of patients
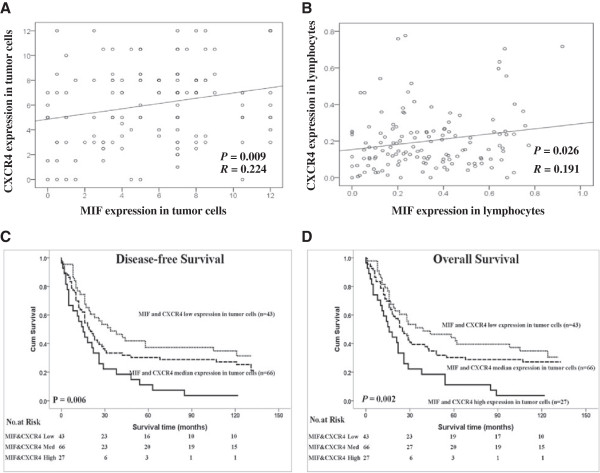
In the current study, Pearson’s correlation coefficient and a linear regression analysis were applied to evaluate the correlations between the expression levels of MIF and CXCR4 in tumor cells and TILs. The MIF expression levels in tumor cells and in TILs were significantly positively correlated with the CXCR4 expression levels in tumor cells and in TILs (P = 0.009, R = 0.224 and P = 0.026, R = 0.191, respectively), as shown in Figure 4A and 4B. Furthermore, the patients with the double high expression levels of MIF and CXCR4 in tumor cells had the worst DFS and OS compared to the patients with single high expression level of MIF or CXCR4 in tumor cells or double low expression level of MIF and CXCR4 in tumor cells (P = 0.002 and P = 0.006, respectively, Figure 4C and 4D). Furthermore, the combined expression of MIF and CXCR4 in tumor cells was an independent predictive factor for DFS and OS according to the multivariate Cox model analysis (Table 2).
Figure 4.
Correlation analysis between the MIF and CXCR4 expressions in different cell populations and survival curves for ESCC patients according to their expression levels of MIF and CXCR4 in tumor cells. (A) The expression levels of MIF and CXCR4 in tumor cells were significantly positively correlated (P = 0.009, R = 0.224). (B) The expression levels of MIF and CXCR4 in TILs were significantly positively correlated (P = 0.026, R = 0.191). (C and D) Disease-free survival and overall survival curves for patients according to the combined low expression level, single high and combined high expression level of MIF and CXCR4 in tumor cells and TILs.
Discussion
A serial of inflammatory cytokine and its receptor genes overexpress in different cell subsets of tumor microenvironments, including tumor cells and immune cells, to control the “cross talk” between tumor cells and immune cells and impact on the disease progression and clinical outcome of cancer patients [28]. MIF, a cytokine overexpressed in tumor microenvironments plays a critical role in several inflammatory conditions, as well xas in oncogenic transformation and tumor progression [29-32]. CXCR4 is the receptor of stromal cell-derived factor-1 (CXCL12/SDF-1α) and can also bind to MIF, and takes an important role in tumor progression and anti-tumor immunity. However, the association between the expression levels of MIF and CXCR4 in diverse cell populations of the tumor microenvironment and the survival of cancer patients remains ambiguous. In this context, we examined the expression pattern of MIF and CXCR4 in different cell populations in tumor tissues from 136 patients with ESCC, to determine the predictive value of the MIF and CXCR4 expressions in different cell populations within tumor microenvironment of ESCC.
High MIF levels were found in the tumors and sera of patients with different types of cancer, and MIF production has been consistently associated with the aggressiveness and metastatic potential of human tumors [33-36]. Our results suggest that MIF could be expressed in the cytoplasm of tumor cells and TILs (Figure 1). In the present study, our results demonstrated for the first time that high MIF expression in tumor cells and TILs is significantly and independently associated with poor DFS and OS in patients with ESCC (Figure 2A), as well as that high MIF expression in tumor cells is an adverse independent factor for DFS and OS in patients with metastatic/recurrent ESCC (Figure 3). Many studies have demonstrated that the biological function of MIF in tumor cells is to promote the growth of tumor cells; however, the expression of MIF in tumor tissues and patients’ clinical outcomes differed for different types of cancers [35,37-41]; our previous study showed that the increased expression of MIF in TILs within tumor microenvironments was correlated with improved outcomes for patients with nasopharyngeal carcinoma (NPC) [25]. Recent studies have indicated that MIF can induce the generation and homing of Th17 cells to the tumor microenvironments [25,42]; however, the function and clinical relevance of Th17 cells in tumor microenvironments were conflicting in different cancers [43,44]. Therefore, we think that although the expression of MIF in tumor cells is to promote the tumor cell growth as an ‘oncogenic gene’, the MIF expression in immune cells is associate with intratumoral immune response; this may explain the different impact of MIF expressions within tumor microenvironments on the survival of patients in different cancers.
CXCR4 promotes tumor progression at different levels of malignancy, including tumor growth, angiogenesis, metastatic dissemination, and homing in CXCL12-enriched cellular niches in metastatic tissues [45-47]. CXCR4 expression is a prognostic marker in various types of cancer, including acute myelogenous leukemia, breast and colon carcinomas [48,49]. Our data revealed that CXCR4 could be expressed in the nucleus, cytoplasm and cell membrane of tumor cells and TILs (Figure 1). In the current study, CXCR4 expression levels in tumor cells were positively associated with primary tumor invasion and clinical stage progression. Our results were consistent with other researchers’ findings regarding the biological functions of CXCR4 in malignant cells; namely, CXCR4 promoted malignant cell proliferation, anti-apoptosis and metastasis [45]. However, the expression of CXCR4 in tumor cells was significantly associated with poor DFS and OS in patients with ESCC or metastatic/recurrent ESCC, whereas CXCR4 expression in TILs was associated with a slightly improved DFS and OS (P = 0.122 and P = 0.091, respectively) in the ESCC patients in our study (Figure 2 and Figure 3). Our results imply that CXCR4 has different biological functions in tumor cells and lymphocytes and that high CXCR4 expression in lymphocytes can induce the homing of immune cells to tumor microenvironments to improve the number of TILs and the anti-tumor immunity of TILs in ESCC. Therefore, the combination of CXCR4 expression in both tumor cells and TILs was not associated with the survival of ESCC patients in this study (data not shown), and other studies on CXCR4 expression in tumor tissues and the clinical outcomes of ESCC patients also have reported conflicting results [50,51].
Importantly, MIF and CXCR4 expression levels in tumor cells and in TILs were positively associated (Figure 4). Our results suggest that the expression levels of MIF and CXCR4 were altered in the same way in different cell populations in the tumor microenvironments of ESCC and that the CXCR4 protein was a receptor response to MIF signaling in both immune cells and tumor cells. Interestingly, our results showed for the first time that the combined expression of MIF and CXCR4 in tumor cells was also an independent prognostic marker for ESCC patients and was strongly associated with reduced survival (Figure 4 and Table 3).
Conclusions
The expression of tumor-derived cytokine MIF and its receptor CXCR4 were significantly associated with poor survivals of patients with ESCC; and the MIF and CXCR4 expression levels in tumor cells were independent predictive factors of survivals in patients with ESCC, as were the MIF expression level in TILs. Furthermore, the expression levels of MIF and CXCR4 in tumor cells were independent predictive factors for survivals in patients with metastatic/recurrent ESCC. Interestingly, the MIF and CXCR4 expression levels in tumor cells and in TILs were positively correlated, and the combined expression of MIF and CXCR4 in tumor cells was an adverse independent factor for survivals of ESCC patients. Therefore, the protein levels of MIF and CXCR4 in diverse cell populations within the tumor microenvironment have different clinically prognostic values in ESCC. Further studies are required to confirm our results in a large number of patients with ESCC.
Abbreviations
ESCC: Esophageal squamous cell carcinoma; TILs: Tumor-infiltrating lymphocytes; DFS: Disease-free survival; OS: Overall survival; TNM: Tumor node-metastasis; WHO: World Health Organization; T stage: Tumor status; N status: Lymph node metastasis.
Competing interests
The authors have declared that they have no competing interest. The sources that funded this study played no role in the study design, data collection, data analysis, decision to publish, or preparation of the manuscript.
Authors’ contributions
Conceived and designed the experiments: JL, YXZ. Performed the experiments: LZ, XFT, SPC, JH. Analyzed the data: JL, SBY, GM. Contributed reagents/materials/analysis tools: WLL and DX. Wrote the manuscript: JL, SBY. All authors read and approved the final manuscript.
Supplementary Material
Clinical characteristics of 136 patients with ESCC.
Descriptive statistics of immunohistochemical variables.
Contributor Information
Lin Zhang, Email: zhanglin@sysucc.org.cn.
Shu-Biao Ye, Email: Yeshb@sysucc.org.cn.
Gang Ma, Email: magang@sysucc.org.cn.
Xiao-Feng Tang, Email: Tangxf@sysucc.org.cn.
Shi-Ping Chen, Email: chensp@sysucc.org.cn.
Jia He, Email: hejia@sysucc.org.cn.
Wan-Li Liu, Email: liuwl@sysucc.org.cn.
Dan Xie, Email: xiedan@sysucc.org.cn.
Yi-Xin Zeng, Email: zengyx@sysucc.org.cn.
Jiang Li, Email: lijiang2@mail.sysu.edu.cn.
Acknowledgements
This work was supported by the National Natural Science Foundation of China [grant number 81172164, JL], the Guangdong Province Natural Science Foundation [grant number 10151008901000156, JL], the Key Program of Guangzhou City Science Foundation [grant number 2011Y100036, JL], and the Education Administration Starting Foundation for scientists returning to China from overseas [grant number 43, JL].
References
- Yan W, Wistuba II, Emmert-Buck MR, Erickson HS. Squamous cell carcinoma - similarities and differences among anatomical sites. American journal of cancer research. 2011;1:275–300. [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Hu N, Han XY, Ding T, Giffen C, Goldstein AM, Taylor PR. Risk factors for esophageal and gastric cancers in Shanxi Province, China: a case–control study. Cancer Epidemiol. 2011;35:e91–e99. doi: 10.1016/j.canep.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghcheli K, Marjani HA, Nasrollahzadeh D, Islami F, Shakeri R, Sotoudeh M, Abedi-Ardekani B, Ghavamnasiri MR, Razaei E, Khalilipour E. Prognostic factors for esophageal squamous cell carcinoma–a population-based study in Golestan Province, Iran, a high incidence area. PLoS One. 2011;6:e22152. doi: 10.1371/journal.pone.0022152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Hu Z, He Z, Jia W, Wang F, Zhou Y, Liu Z, Zhan Q, Liu Y, Yu D. Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations. Nat Genet. 2011;43:679–684. doi: 10.1038/ng.849. [DOI] [PubMed] [Google Scholar]
- Zhang H, Chen Z, Cheng J, Zhu X, Guo W, Hu A, Du Y, Zhou Y, Wang Y. The high incidence of esophageal cancer in parts of China may result primarily from genetic rather than environmental factors. Diseases of the esophagus: official journal of the International Society for Diseases of the Esophagus / ISDE. 2010;23:392–397. doi: 10.1111/j.1442-2050.2009.01020.x. [DOI] [PubMed] [Google Scholar]
- Akita H, Doki Y, Miyata H, Hirao T, Yano M, Takachi K, Miyashiro I, Sasaki Y, Ishikawa O, Ohigashi H, Imaoka S. Clinical significance of the second cycle response to cisplatin-based chemotherapy as preoperative treatment for esophageal squamous cell carcinoma. J Surg Oncol. 2006;93:401–409. doi: 10.1002/jso.20501. [DOI] [PubMed] [Google Scholar]
- Cao W, Chen X, Dai H, Wang H, Shen B, Chu D, McAfee T, Zhang ZF. Mutational spectra of p53 in geographically localized esophageal squamous cell carcinoma groups in China. Cancer. 2004;101:834–844. doi: 10.1002/cncr.20437. [DOI] [PubMed] [Google Scholar]
- Lv L, Pan K, Li XD, She KL, Zhao JJ, Wang W, Chen JG, Chen YB, Yun JP, Xia JC. The accumulation and prognosis value of tumor infiltrating IL-17 producing cells in esophageal squamous cell carcinoma. PLoS One. 2011;6:e18219. doi: 10.1371/journal.pone.0018219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Fu ZX. Localization of IL-17+Foxp3+ T cells in esophageal cancer. Immunol Investig. 2011;40:400–412. doi: 10.3109/08820139.2011.555489. [DOI] [PubMed] [Google Scholar]
- Liang Y, Hou X, Cui Q, Kang TB, Fu JH, Zhang LJ, Luo RZ, He JH, Zeng YX, Yang HX. Skp2 expression unfavorably impacts survival in resectable esophageal squamous cell carcinoma. J Transl Med. 2012;10:73. doi: 10.1186/1479-5876-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XC, Tian LL, Tian J, Jiang XY. Overexpression of SKP2 promotes the radiation resistance of esophageal squamous cell carcinoma. Radiat Res. 2012;177:52–58. doi: 10.1667/RR2679.1. [DOI] [PubMed] [Google Scholar]
- Fukuchi M, Masuda N, Nakajima M, Fukai Y, Miyazaki T, Kato H, Kuwano H. Inverse correlation between expression levels of p27 and the ubiquitin ligase subunit Skp2 in early esophageal squamous cell carcinoma. Anticancer Res. 2004;24:777–783. [PubMed] [Google Scholar]
- Kondo K, Yamasaki S, Inoue N, Sugie T, Teratani N, Kan T, Shimada Y. Prospective antitumor effects of the combination of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and cisplatin against esophageal squamous cell carcinoma. Surg Today. 2006;36:966–974. doi: 10.1007/s00595-006-3295-5. [DOI] [PubMed] [Google Scholar]
- Morand EF. New therapeutic target in inflammatory disease: macrophage migration inhibitory factor. Intern Med J. 2005;35:419–426. doi: 10.1111/j.1445-5994.2005.00853.x. [DOI] [PubMed] [Google Scholar]
- Gore Y, Starlets D, Maharshak N, Becker-Herman S, Kaneyuki U, Leng L, Bucala R, Shachar I. Macrophage migration inhibitory factor induces B cell survival by activation of a CD74-CD44 receptor complex. J Biol Chem. 2008;283:2784–2792. doi: 10.1074/jbc.M703265200. [DOI] [PubMed] [Google Scholar]
- Schwartz V, Lue H, Kraemer S, Korbiel J, Krohn R, Ohl K, Bucala R, Weber C, Bernhagen J. A functional heteromeric MIF receptor formed by CD74 and CXCR4. FEBS Lett. 2009;583:2749–2757. doi: 10.1016/j.febslet.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhagen J, Krohn R, Lue H, Gregory JL, Zernecke A, Koenen RR, Dewor M, Georgiev I, Schober A, Leng L. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. 2007;13:587–596. doi: 10.1038/nm1567. [DOI] [PubMed] [Google Scholar]
- Dumitru CA, Gholaman H, Trellakis S, Bruderek K, Dominas N, Gu X, Bankfalvi A, Whiteside TL, Lang S, Brandau S. Tumor-derived macrophage migration inhibitory factor modulates the biology of head and neck cancer cells via neutrophil activation. Int J Cancer. 2011;129:859–869. doi: 10.1002/ijc.25991. [DOI] [PubMed] [Google Scholar]
- Larsen M, Tazzyman S, Lund EL, Junker N, Lewis CE, Kristjansen PE, Murdoch C. Hypoxia-induced secretion of macrophage migration-inhibitory factor from MCF-7 breast cancer cells is regulated in a hypoxia-inducible factor-independent manner. Cancer Lett. 2008;265:239–249. doi: 10.1016/j.canlet.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Rendon BE, Roger T, Teneng I, Zhao M, Al-Abed Y, Calandra T, Mitchell RA. Regulation of human lung adenocarcinoma cell migration and invasion by macrophage migration inhibitory factor. J Biol Chem. 2007;282:29910–29918. doi: 10.1074/jbc.M704898200. [DOI] [PubMed] [Google Scholar]
- Wilson JM, Coletta PL, Cuthbert RJ, Scott N, MacLennan K, Hawcroft G, Leng L, Lubetsky JB, Jin KK, Lolis E. Macrophage migration inhibitory factor promotes intestinal tumorigenesis. Gastroenterology. 2005;129:1485–1503. doi: 10.1053/j.gastro.2005.07.061. [DOI] [PubMed] [Google Scholar]
- Chen G, Chen SM, Wang X, Ding XF, Ding J, Meng LH. Inhibition of chemokine (CXC motif) ligand 12/chemokine (CXC motif) receptor 4 axis (CXCL12/CXCR4)-mediated cell migration by targeting mammalian target of rapamycin (mTOR) pathway in human gastric carcinoma cells. J Biol Chem. 2012;287:12132–12141. doi: 10.1074/jbc.M111.302299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigo A, Gottardi M, Zamo A, Mauri P, Bonifacio M, Krampera M, Damiani E, Pizzolo G, Vinante F. Macrophages may promote cancer growth via a GM-CSF/HB-EGF paracrine loop that is enhanced by CXCL12. Mol Cancer. 2010;9:273. doi: 10.1186/1476-4598-9-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MC, Luker KE, Garbow JR, Prior JL, Jackson E, Piwnica-Worms D, Luker GD. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res. 2004;64:8604–8612. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- Li J, Mo HY, Xiong G, Zhang L, He J, Huang ZF, Liu ZW, Chen QY, Du ZM, Zheng LM. Tumor microenvironment MIF directs the accumulation of IL-17-producing tumor-infiltrating lymphocytes and predicts favorable survival in nasopharyngeal carcinoma patients. J Biol Chem. 2012;28:35484–35495. doi: 10.1074/jbc.M112.367532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B, Zhong BL, Li Z, Tian XY, Li Y, Li B. Macrophage migration inhibitory factor contributes angiogenesis by up-regulating IL-8 and correlates with poor prognosis of patients with primary nasopharyngeal carcinoma. J Surg Oncol. 2010;102:844–851. doi: 10.1002/jso.21728. [DOI] [PubMed] [Google Scholar]
- O’Sullivan B, Shah J. New TNM staging criteria for head and neck tumors. Semin Surg Oncol. 2003;21:30–42. doi: 10.1002/ssu.10019. [DOI] [PubMed] [Google Scholar]
- Domschke C, Schuetz F, Ge Y, Seibel T, Falk C, Brors B, Vlodavsky I, Sommerfeldt N, Sinn HP, Kuhnle MC. Intratumoral cytokines and tumor cell biology determine spontaneous breast cancer-specific immune responses and their correlation to prognosis. Cancer Res. 2009;69:8420–8428. doi: 10.1158/0008-5472.CAN-09-1627. [DOI] [PubMed] [Google Scholar]
- Liu A, Fang H, Dirsch O, Jin H, Dahmen U. Early release of macrophage migration inhibitory factor after liver ischemia and reperfusion injury in rats. Cytokine. 2012;57:150–157. doi: 10.1016/j.cyto.2011.11.009. [DOI] [PubMed] [Google Scholar]
- Seike T, Fujita K, Yamakawa Y, Kido MA, Takiguchi S, Teramoto N, Iguchi H, Noda M. Interaction between lung cancer cells and astrocytes via specific inflammatory cytokines in the microenvironment of brain metastasis. Clin Exp Metastasis. 2011;28:13–25. doi: 10.1007/s10585-010-9354-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach JP, Deuster O, Balzer-Geldsetzer M, Meyer B, Dodel R, Bacher M. The role of macrophage inhibitory factor in tumorigenesis and central nervous system tumors. Cancer. 2009;115:2031–2040. doi: 10.1002/cncr.24245. [DOI] [PubMed] [Google Scholar]
- Bach JP, Rinn B, Meyer B, Dodel R, Bacher M. Role of MIF in inflammation and tumorigenesis. Oncology. 2008;75:127–133. doi: 10.1159/000155223. [DOI] [PubMed] [Google Scholar]
- Funamizu N, Hu C, Lacy C, Schetter A, Zhang G, He P, Gaedcke J, Ghadimi BM, Ried T, Yffantis HG. Macrophage Migration Inhibitory Factor (MIF) induces epithelial to mesenchymal transition, enhances tumor aggressiveness and predicts clinical outcome in resected pancreatic ductal adenocarcinoma. Int J Cancer. 2012;132:785–794. doi: 10.1002/ijc.27736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant P, Sainsbury A, Reed KR, Pollard SG, Scott N, Clarke AR, Coletta PL, Hull MA. Rectal epithelial cell mitosis and expression of macrophage migration inhibitory factor are increased 3 years after Roux-en-Y gastric bypass (RYGB) for morbid obesity: implications for long-term neoplastic risk following RYGB. Gut. 2011;60:893–901. doi: 10.1136/gut.2010.230755. [DOI] [PubMed] [Google Scholar]
- Bandres E, Bitarte N, Arias F, Agorreta J, Fortes P, Agirre X, Zarate R, Diaz-Gonzalez JA, Ramirez N, Sola JJ. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res. 2009;15:2281–2290. doi: 10.1158/1078-0432.CCR-08-1818. [DOI] [PubMed] [Google Scholar]
- Howard BA, Zheng Z, Campa MJ, Wang MZ, Sharma A, Haura E, Herndon JE 2nd, Fitzgerald MC, Bepler G, Patz EF Jr. Translating biomarkers into clinical practice: prognostic implications of cyclophilin A and macrophage migratory inhibitory factor identified from protein expression profiles in non-small cell lung cancer. Lung Cancer. 2004;46:313–323. doi: 10.1016/j.lungcan.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Mittelbronn M, Platten M, Zeiner P, Dombrowski Y, Frank B, Zachskorn C, Harter PN, Weller M, Wischhusen J. Macrophage migration inhibitory factor (MIF) expression in human malignant gliomas contributes to immune escape and tumour progression. Acta Neuropathol. 2011;122:353–365. doi: 10.1007/s00401-011-0858-3. [DOI] [PubMed] [Google Scholar]
- Yao K, Shida S, Selvakumaran M, Zimmerman R, Simon E, Schick J, Haas NB, Balke M, Ross H, Johnson SW, O’Dwyer PJ. Macrophage migration inhibitory factor is a determinant of hypoxia-induced apoptosis in colon cancer cell lines. Clin Cancer Res. 2005;11:7264–7272. doi: 10.1158/1078-0432.CCR-05-0135. [DOI] [PubMed] [Google Scholar]
- Legendre H, Decaestecker C, Nagy N, Hendlisz A, Schuring MP, Salmon I, Gabius HJ, Pector JC, Kiss R. Prognostic values of galectin-3 and the macrophage migration inhibitory factor (MIF) in human colorectal cancers. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2003;16:491–504. doi: 10.1097/01.MP.0000068235.45178.C1. [DOI] [PubMed] [Google Scholar]
- Tomiyasu M, Yoshino I, Suemitsu R, Okamoto T, Sugimachi K. Quantification of macrophage migration inhibitory factor mRNA expression in non-small cell lung cancer tissues and its clinical significance. Clin Cancer Res. 2002;8:3755–3760. [PubMed] [Google Scholar]
- Shkolnik T, Livni E, Reshef R, Lachter J, Eidelman S. The macrophage migration inhibition (MIF) assay as a marker of colorectal cancer. Studies in patients with colorectal cancer, noncolonic neoplasms, and conditions predisposing to colorectal cancer. Dis Colon Rectum. 1987;30:101–105. doi: 10.1007/BF02554942. [DOI] [PubMed] [Google Scholar]
- Stojanovic I, Cvjeticanin T, Lazaroski S, Stosic-Grujicic S, Miljkovic D. Macrophage migration inhibitory factor stimulates interleukin-17 expression and production in lymph node cells. Immunology. 2009;126:74–83. doi: 10.1111/j.1365-2567.2008.02879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I, Banerjee M, Cheng P, Vatan L, Szeliga W, Wei S, Huang E, Finlayson E, Simeone D, Welling TH. Phenotype, distribution, generation, and functional and clinical relevance of Th17 cells in the human tumor environments. Blood. 2009;114:1141–1149. doi: 10.1182/blood-2009-03-208249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreni M, Mantovani A, Allavena P. Tumor-associated Macrophages (TAM) and inflammation in colorectal cancer. Cancer microenvironment: official journal of the International Cancer Microenvironment Society. 2011;4:141–154. doi: 10.1007/s12307-010-0052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron N, Deuster O, Noelker C, Stuer C, Strik H, Schaller C, Dodel R, Meyer B, Bacher M. Role of macrophage migration inhibitory factor in primary glioblastoma multiforme cells. J Neurosci Res. 2011;89:711–717. doi: 10.1002/jnr.22595. [DOI] [PubMed] [Google Scholar]
- Tarnowski M, Grymula K, Liu R, Tarnowska J, Drukala J, Ratajczak J, Mitchell RA, Ratajczak MZ, Kucia M. Macrophage migration inhibitory factor is secreted by rhabdomyosarcoma cells, modulates tumor metastasis by binding to CXCR4 and CXCR7 receptors and inhibits recruitment of cancer-associated fibroblasts. Mol Cancer Res. 2010;8:1328–1343. doi: 10.1158/1541-7786.MCR-10-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulbe H, Thompson R, Wilson JL, Robinson S, Hagemann T, Fatah R, Gould D, Ayhan A, Balkwill F. The inflammatory cytokine tumor necrosis factor-alpha generates an autocrine tumor-promoting network in epithelial ovarian cancer cells. Cancer Res. 2007;67:585–592. doi: 10.1158/0008-5472.CAN-06-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoo AC, Lubbert M, Wierda WG, Burger JA. CXCR4 is a prognostic marker in acute myelogenous leukemia. Blood. 2007;109:786–791. doi: 10.1182/blood-2006-05-024844. [DOI] [PubMed] [Google Scholar]
- Burger JA, Kipps TJ. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood. 2006;107:1761–1767. doi: 10.1182/blood-2005-08-3182. [DOI] [PubMed] [Google Scholar]
- Gockel I, Schimanski CC, Heinrich C, Wehler T, Frerichs K, Drescher D, von Langsdorff C, Domeyer M, Biesterfeld S, Galle PR. Expression of chemokine receptor CXCR4 in esophageal squamous cell and adenocarcinoma. BMC Cancer. 2006;6:290. doi: 10.1186/1471-2407-6-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaifi JT, Yekebas EF, Schurr P, Obonyo D, Wachowiak R, Busch P, Heinecke A, Pantel K, Izbicki JR. Tumor-cell homing to lymph nodes and bone marrow and CXCR4 expression in esophageal cancer. J Natl Cancer Inst. 2005;97:1840–1847. doi: 10.1093/jnci/dji431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical characteristics of 136 patients with ESCC.
Descriptive statistics of immunohistochemical variables.