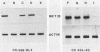
Abstract
In Saccharomyces cerevisiae, the MET25 gene encodes O-acetylhomoserine sulfhydrylase. Synthesis of this enzyme is repressed by the presence of S-adenosylmethionine (AdoMet) in the growth medium. We identified cis elements required for MET25 expression by analyzing small deletions in the MET25 promoter region. The results revealed a regulatory region, acting as an upstream activation site, that activated transcription of MET25 in the absence of methionine or AdoMet. We found that, for the most part, repression of MET25 expression was due to a lack of activation at this site, reinforced by an independent repression mechanism. The activation region contained a repeated dyad sequence that is also found in the promoter regions of other unlinked but coordinately regulated genes (MET3, MET2, and SAM2). We show that the presence of the two dyads is necessary for maximal gene expression. Moreover, we demonstrate that in addition to this transcriptional regulation, a posttranscriptional regulation, probably targeted at the 5' region of mRNA, is involved in MET25 expression.
Full text
PDF


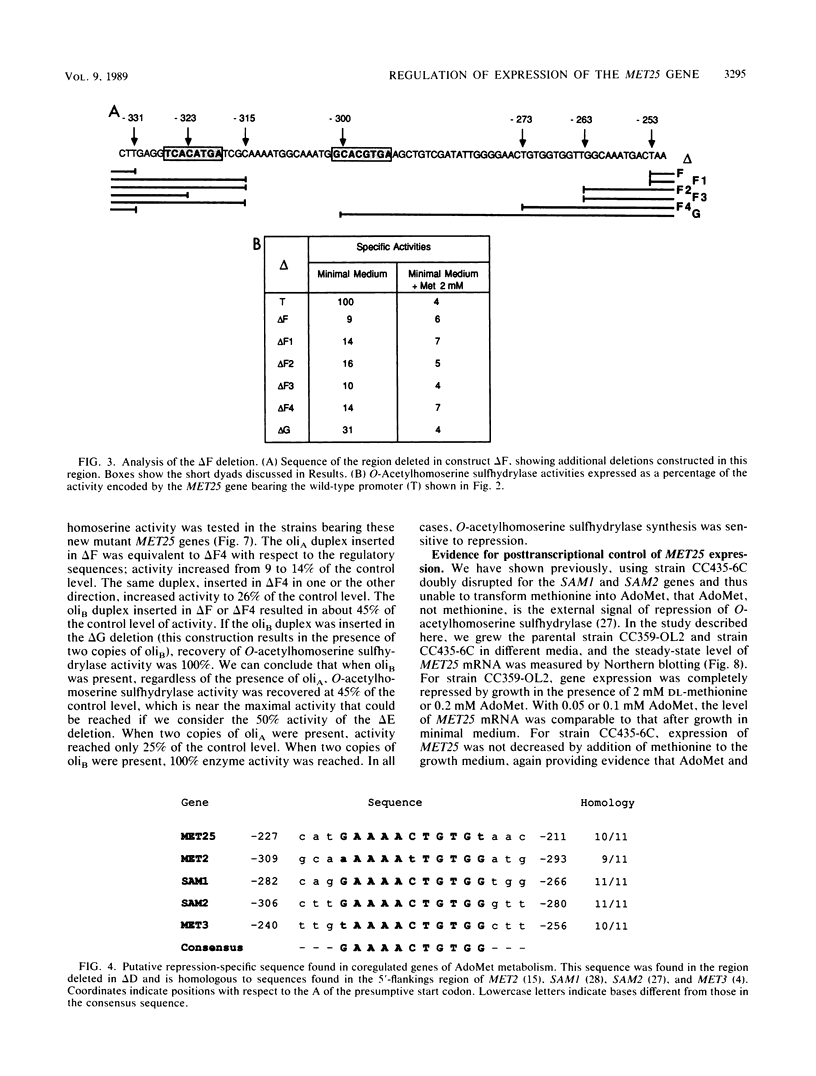



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brent R. Repression of transcription in yeast. Cell. 1985 Aug;42(1):3–4. doi: 10.1016/s0092-8674(85)80091-x. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Martinez-Arias A., Shapira S. K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Cherest H., Kerjan P., Surdin-Kerjan Y. The Saccharomyces cerevisiae MET3 gene: nucleotide sequence and relationship of the 5' non-coding region to that of MET25. Mol Gen Genet. 1987 Dec;210(2):307–313. doi: 10.1007/BF00325699. [DOI] [PubMed] [Google Scholar]
- Cherest H., Surdin-Kerjan Y., Antoniewski J., Robichon-Szulmajster H. S-adenosyl methionine-mediated repression of methionine biosynthetic enzymes in Saccharomyces cerevisiae. J Bacteriol. 1973 Jun;114(3):928–933. doi: 10.1128/jb.114.3.928-933.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Regulatory proteins in yeast. Annu Rev Genet. 1987;21:425–452. doi: 10.1146/annurev.ge.21.120187.002233. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerjan P., Cherest H., Surdin-Kerjan Y. Nucleotide sequence of the Saccharomyces cerevisiae MET25 gene. Nucleic Acids Res. 1986 Oct 24;14(20):7861–7871. doi: 10.1093/nar/14.20.7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kredich N. M., Tomkins G. M. The enzymic synthesis of L-cysteine in Escherichia coli and Salmonella typhimurium. J Biol Chem. 1966 Nov 10;241(21):4955–4965. [PubMed] [Google Scholar]
- Kronstad J. W., Holly J. A., MacKay V. L. A yeast operator overlaps an upstream activation site. Cell. 1987 Jul 31;50(3):369–377. doi: 10.1016/0092-8674(87)90491-0. [DOI] [PubMed] [Google Scholar]
- Langin T., Faugeron G., Goyon C., Nicolas A., Rossignol J. L. The MET2 gene of Saccharomyces cerevisiae: molecular cloning and nucleotide sequence. Gene. 1986;49(3):283–293. doi: 10.1016/0378-1119(86)90364-1. [DOI] [PubMed] [Google Scholar]
- Machida M., Uemura H., Jigami Y., Tanaka H. The protein factor which binds to the upstream activating sequence of Saccharomyces cerevisiae ENO1 gene. Nucleic Acids Res. 1988 Feb 25;16(4):1407–1422. doi: 10.1093/nar/16.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangsoda S., Cherest H., Surdin-Kerjan Y. The expression of the MET25 gene of Saccharomyces cerevisiae is regulated transcriptionally. Mol Gen Genet. 1985;200(3):407–414. doi: 10.1007/BF00425724. [DOI] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surdin-Kerjan Y., de Robichon-Szulmajster H. Existence of two levels of repression in the biosynthesis of methionine in Saccharomyces cerevisiae: effect of lomofungin on enzyme synthesis. J Bacteriol. 1975 May;122(2):367–374. doi: 10.1128/jb.122.2.367-374.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor C. W., Tabor H., Hafner E. W. Escherichia coli mutants completely deficient in adenosylmethionine decarboxylase and in spermidine biosynthesis. J Biol Chem. 1978 May 25;253(10):3671–3676. [PubMed] [Google Scholar]
- Thomas D., Rothstein R., Rosenberg N., Surdin-Kerjan Y. SAM2 encodes the second methionine S-adenosyl transferase in Saccharomyces cerevisiae: physiology and regulation of both enzymes. Mol Cell Biol. 1988 Dec;8(12):5132–5139. doi: 10.1128/mcb.8.12.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., Surdin-Kerjan Y. SAM1, the structural gene for one of the S-adenosylmethionine synthetases in Saccharomyces cerevisiae. Sequence and expression. J Biol Chem. 1987 Dec 5;262(34):16704–16709. [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebers J. L., Garner H. R. Acyl derivatives of homoserine as substrates for homocysteine synthesis in Neurospora crassa, yeast, and Escherichia coli. J Biol Chem. 1967 Dec 10;242(23):5644–5649. [PubMed] [Google Scholar]