Abstract
The proteasome is a proteolytic machinery that executes the degradation of polyubiquitinated proteins to maintain cellular homeostasis. Proteasome inhibition is a unique and effective way to kill cancer cells because they are sensitive to proteotoxic stress. Indeed, the proteasome inhibitor bortezomib is now indispensable for the treatment of multiple myeloma and other intractable malignancies, but is associated with patient inconvenience due to intravenous injection and emerging drug resistance. To resolve these problems, we attempted to develop orally bioavailable proteasome inhibitors with distinct mechanisms of action and identified homopiperazine derivatives (HPDs) as promising candidates. Biochemical and crystallographic studies revealed that some HPDs inhibit all three catalytic subunits (ß 1, ß 2 and ß 5) of the proteasome by direct binding, whereas bortezomib and other proteasome inhibitors mainly act on the ß5 subunit. Proteasome-inhibitory HPDs exhibited cytotoxic effects on cell lines from various hematological malignancies including myeloma. Furthermore, K-7174, one of the HPDs, was able to inhibit the growth of bortezomib-resistant myeloma cells carrying a ß5-subunit mutation. Finally, K-7174 had additive effects with bortezomib on proteasome inhibition and apoptosis induction in myeloma cells. Taken together, HPDs could be a new class of proteasome inhibitors, which compensate for the weak points of conventional ones and overcome the resistance to bortezomib.
Introduction
The paradigm of cancer treatment has been dramatically changed by the introduction of small molecular compounds that target the “Achilles' heel” of cancer cells [1]. The proteasome is a proteolytic machinery that executes the degradation of polyubiquitinated proteins to maintain cellular homeostasis [2]. Cancer cells are very sensitive to proteotoxic stress because of intracellular protein overload due to rapid cell cycling and apoptosis inhibition. This feature makes proteasome inhibition a unique and effective way to kill cancer cells that can tolerate conventional therapies [3].
Bortezomib is the first proteasome inhibitor (PI) approved for clinical application, which preferentially targets ß1 and ß5 subunits of the proteasome [3], [4]. This drug is particularly effective for multiple myeloma (MM), because it accelerates the unfolded protein response (UPR) via down-regulation of histone deacetylases (HDACs) [5], [6] and targets cell adhesion-mediated drug resistance via down-regulation of very late antigen-4 [7], [8]. Accordingly, bortezomib is now indispensable for the treatment of MM in combination with other anti-cancer drugs including alkylating agents, corticosteroids and HDAC inhibitors [9]–[11].
Although bortezomib therapy is a major advance in clinical oncology, there are at least three major problems to be resolved as soon as possible. First, bortezomib has several possible off-target toxicities [12], [13]. Second, the development of intrinsic and acquired resistance to bortezomib is an emerging problem [14]–[19]. Third, bortezomib should be administered intravenously on biweekly schedules with treatment periods extending for 6 months or more. The development of orally bioavailable PIs with distinct mode of action is a possible way to circumvent these issues.
Homopiperazine-derived compounds have been developed as orally active agents because of their superb bioavailability. Among them, dilazep, an inhibitor of nucleoside transporters, has been clinically used for the treatment of cardiac dysfunction via post-oral administration [20]. Some homopiperazine derivatives (HPDs), such as K-7174 and K-11706, were shown in pre-clinical studies to inhibit cell adhesion [21] and to rescue anemia of chronic disorders via the activation of erythropoietin production in vitro and in vivo [22], [23]. In addition, K-7174 was reported to exert anti-inflammatory action via induction of the UPR [24]. These observations prompted us to consider that HPDs could be orally active PIs; however, this possibility has not been tested so far. In this study, we demonstrated that HPDs, including K-7174, have the ability to inhibit proteasome activity via different mechanisms of action from bortezomib and other conventional PIs.
Materials and Methods
Cells and Cell Culture
We used cell lines from acute lymphoblastic leukemia (KOPM30 and Jurkat), mantle cell lymphoma (HBL-2, Granta519 and NCEB-1), Burkitt lymphoma (Daudi and Namalwa), multiple myeloma (KMS12-BM, RPMI8226, U266, KMM1 and Delta47) [25], acute myeloid leukemia (HL60, KG1a and U937) and chronic myeloid leukemia (K562) in this study. These cell lines were purchased from the Health Science Research Resources Bank (Osaka, Japan) except for KOPM30, which was provided by Dr. Takeshi Inukai (University of Yamanashi, Yamanashi, Japan) [26], and Granta519 and NCEB-1, which were provided by Professor Martin J. S. Dyer (Leicester University, Leicester, UK) [27], and maintained in RPMI1640 medium (Sigma Co., St. Louis, MO) supplemented with 10% heat-inactivated fetal calf serum (Sigma) and antibiotics.
Drugs
HPDs and bortezomib were provided by Kowa (Tokyo, Japan) and Millennium Pharmaceuticals (Cambridge, MA), respectively. The compounds were dissolved in dimethyl sulfoxide (DMSO), diluted with 0.9% NaCl at appropriate concentrations and stored at −80°C until use. The chemical structures of K-7174 (N,N’-bis-(E)-(5-(3,4,5-trimethoxy-phenyl)-4-pentenyl) homopiperazine) and other HPDs are shown in Table 1.
Table 1. Chemical structures of homopiperazine derivatives used in this study.
| Compound | Chemical formula | Number of carbon atom chains |
| K-7174 | N,N’-bis-(E)-[5-(3,4,5-trimethoxy-phenyl)-4-pentenyl] homopiperazine | C = 5 |
| K-7259 | N,N’-bis-[4-(3,4,5-trimethoxy-phenyl)-butyl] homopiperazine | C = 4 |
| K-7220 | N,N’-bis-[3-(3,4,5-trimethoxy-phenyl)-propyl] homopiperazine | C = 3 |
| K-10310 | N,N’-bis-(E)-[6-(3,4,5-trimethoxy-phenyl)-4- oxo-5-hexenyl] homopiperazine | C = 6 |
| K-10252 | N,N’-bis-(E)-[7-(3,4,5-trimethy-phenyl)-5-oxo-6- heptenyl] homopiperazine | C = 7 |
| K-10228 | N,N’-bis-(E)-[5-(3,4,5-trimethyl-phenyl)-4- pentenyl] homopiperazine | C = 5 |
| K-10256 | N,N’-bis-(E)-[5-(2,3,4,5-tetramethoxy-phenyl)-4- pentenyl] homopiperazine | C = 5 |
| K-10487 | N,N’-bis-(E)-[4-(3,4,5-trimethoxy-naphthyl)-butyl] homopiperazine | C = 5 |
| K-10552 | N,N’-bis-(E)-[5-(3,4,5-trimethoxy-phenyl)-4- pentenyl]-6-methoxy-homopiperazine | C = 5 |
Cell Proliferation Assay
Cell proliferation was measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) reduction assay using a Cell Counting Kit (Wako Biochemicals, Osaka, Japan). Absorbance at 450 nm was analyzed with a microplate reader and expressed as a percentage of the value of corresponding untreated cells.
Immunoblotting
Immunoblotting was carried out according to the standard method using the following antibodies: anti-ubiquitin, anti-ubiquityl histone H2A (Lys119), anti-K48-linked polyubiquitin (Cell Signaling Technology, Beverly, MA), anti-proteasome ß5 subunit (PSMB5) (Enzo Life Sciences, Farmingdale, NY), and anti-GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA) [28].
20 S Proteasome Activity Assay
Proteasome activity assays were performed using 20 S proteasome assay kits (Enzo Life Sciences and Cayman Chemical, Ann Arbor, MI). Proteasomal chymotrypsin-like, trypsin-like and caspase-like activities were determined using fluorogenic substrates suc-LLVY-amc, boc-LRR-amc, and z-LLE-amc (Enzo Life Sciences), respectively, with purified human erythrocyte-derived 20 S proteasome (Enzo Life Sciences) or MM cell lysates. Reactions were initiated by enzyme or lysate addition, and monitored for amc product formation at 30°C on a spectrofluorometer (SpectraMax Gemini EM; Molecular Devices, Sunnyvale, CA) using excitation of 360 nm and emission of 480 nm [29], [30].
X-ray Crystallographic Analysis of the K-7174/proteasome Complex
Single crystals of 20 S proteasome from Saccharomyces cerevisiae (Enzo Life Sciences) in complex with K-7174 were grown using the sitting drop vapor diffusion method at 20°C by mixing 8 µl of protein and 8 µl of reservoir solution. The protein concentration used for crystallization was 10 mg/ml in 10 mM Tris-HCl (pH 7.5) and 1 mM EDTA. The reservoir solution contained 4.5% (v/v) 2-methyl-2,4-pentanediol (MPD), 36 mM magnesium acetate, 90 mM morpholino-ethane-sulphonic acid (MES, pH 7.2), 10% (v/v) DMSO, and 12.5 mM K-7174. Crystals were soaked in cryoprotectant buffer containing 30% (v/v) MPD and flash frozen in liquid nitrogen. X-ray data were collected at beamline BL44XU of Spring-8 (Hyogo, Japan) equipped with an MAR CCD detector 225 mm at 100 K under a nitrogen gas stream. The wavelength of the incident X-ray was 1.0 Å. Diffraction data sets were processed with HKL2000, and scaled with SCALEPACK [31]. The crystals belonged to the space group P2 1 with unit cell parameters of a = 134.26, b = 301.36, c = 143.96 and ß = 112.9°. The structures were solved by molecular replacement using MOLREP [32] with the previously reported structure of 20 S proteasome (Protein Data Bank code 1RYP) as a starting model. The best solution using data from 40 to 3.5 Å resolution range yielded a correlation coefficient of 0.7 and an R-factor of 0.33 for structure, after rigid body refinement. At this stage, crystallographic refinement was pursued in PHENIX [33]. After an initial round of simulated annealing refinement, several macrocycles that included bulk solvent correction, anisotropic scaling of the data, individual coordinate refinement with minimization, and individual isotropic ADP (atomic displacement parameters) refinement were carried out with maximum likelihood as a target. In the course of the refinement, K-7174 compound and water molecules were added to the models by manual inspection of their positions in both 2Fo-Fc and Fo-c maps, and individual ADP refinement was carried out in the final stages. Map fitting and other manipulations with molecular models were performed using the graphic software COOT [34]. The stereochemistry of the final models was assessed using MolProbity [35]. Data collection and refinement statistics are summarized in Table 2. Atomic coordinates and structure factors of the complex have been deposited in the Protein Data Bank under accession code 4EU2.
Table 2. Statistics of crystallographic analysis.
| Space group/unit cell(Å) | P21/a = 134.26, b = 301.36, c = 143.96, ß = 112.9° |
| Resolution range (Å) | 50.0–2.5 |
| Reflections (Measured/Unique) | 313,772/962,020 |
| Completeness (Overall/Outer Shell, %) a | 88.5/72.3 |
| R-merge (Overall/Outer Shell, %)a, b | 10.1/28.0 |
| Redundancy (Overall) | 3.1 |
| Mean <Ió/(I)> (Overall) | 12.2 |
| Overall B-factor from Wilson plot (Å2) | 22 |
| Refinement statistics | |
| R-work c/R-free (%)c | 20.2/25.5 |
| R.m.s.d. bond lengths/bond angles (Å) | 0.008/1.178 |
| Average B-factor (protein/water/compound, Å2) | 39/31/82 |
| Ramachandran plot | |
| Favored/allowed/outlier (%) | 95.86/3.82/0.32 |
Completeness and R-merge, are given for overall data and for the highest resolution shell.
The highest resolution shells for the dataset were 2.54–2.50 Å.
Rmerge = ∑ | Ii – <I> |/∑|Ii|; where Ii is intensity of an observation and <I> is the mean value of that reflection and the summations are over all equivalents.
R-work = ∑h || Fo(h) | – | Fc(h) ||/∑hFo(h); where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. The R-free was calculated with 5% of the data excluded from the refinement.
Establishment of Bortezomib-resistant MM Cell Lines
To confer bortezomib resistance to MM cells, we transduced with mutated PSMB5 cDNA as described previously [17]. A mutation was inserted at nucleotide position 322 (G/A) by PCR-based site directed mutagenesis using wild-type PSMB5 cDNA (obtained from OpenBiosystems, Thermo Fisher Scientific, Huntsville, AL) as a template. Mutated PSMB5 cDNA was inserted into a lentiviral vector CSII-CMV-MCS-IRES-VENUS (kindly provided by Dr. Hiroyuki Miyoshi, RIKEN BioResorce Center, Ibaraki, Japan) [36]. Wild-type PSMB5 cDNA was also inserted into the same vector and used as a control. These vectors were co-transfected into 293FT cells with packaging plasmids (Invitrogen, Carlsbad, CA) to produce infective lentiviruses in culture supernatants as previously described [37]. RPMI8226 cells were infected with each viral supernatant for 24 h. We collected VENUS-positive cells using a FACSaria flow cytometer (Becton Dickinson, Bedford, MA) and seeded them at one cell/well in a 96-well plate to obtain single cell clones.
Isobologram Analysis of Drug Interaction
The cytotoxic interaction of bortezomib and K-7174 was evaluated at the point of IC80 by the isobologram of Steel and Peckham. The IC80 was defined as the concentration of drugs that produced 80% inhibition of cell growth. The theoretical basis of the isobologram method has been described in detail previously [38]. Briefly, the envelope of additivity is constructed from dose–response curves of the combined drugs. The combination is regarded as additive when the data points of the drug combination fall within the envelope of additivity. The drug combination is regarded as supra-additive (synergism) and antagonistic when the data points fall to the left and the right of the envelope, respectively.
Results
Homopiperazine Derivatives Inhibit Proteasome Activities
The ability of K-7174 to elicit the UPR prompted us to speculate that this drug could inhibit proteasome activity [5], [6], [24]. To test this idea, we incubated purified human erythrocyte-derived 20 S proteasome with K-7174 and eight HPDs, which are structurally similar to K-7174 (Table 1), and measured chymotrypsin-like (ß5), caspase-like (ß1), and trypsin-like (ß2) activities of the proteasome using specific fluorogenic peptides. As shown in Fig. 1A, six out of nine HPDs significantly inhibited proteasome activities of the purified 20 S proteasome. We performed the same experiments with cell-based assays using RPMI8226 cells to confirm the proteasome-inhibitory effect of the six HPDs (Fig. 1B). Interestingly, HPDs inhibited all three catalytic subunits with similar kinetics, suggesting a different mechanism of action from bortezomib.
Figure 1. Inhibition of 20 S proteasome activity by homopiperazine derivatives.
A. Purified erythrocyte-derived 20 S proteasome was incubated in the absence (control) or presence of the indicated HPDS at 5 µM. Chymotrypsin-like, caspase-like and trypsin-like activities were determined by measuring fluorescence generated from the cleavage of specific substrates. Results are represented as relative fluorescence units (RFU) with control set at 100%. The means ± S.D. (bars) of three independent experiments are shown. P-values were calculated by one-way ANOVA with the Student-Newman-Keuls multiple comparisons test. Asterisks indicate p<0.05 against corresponding controls. B. RPMI8226 cells were treated with or without 5 µM HPDs, and analyzed for proteolytic activities as described above. C. RPMI8226 cells were cultured in the absence (control) or presence of 10 µM HPDs for 24 hours, and subjected to immunoblotting for ubiquitinated proteins and GAPDH (internal control).
Among the HPDs examined, K-7174, K-10256, K-10487 and K-10552 showed relatively strong effects on proteasome activities. They commonly possess methoxy-phenyl groups and pentenyl arms (with five carbon atom chains; C = 5) (Table 1). In contrast, proteasome-inhibitory activity was weak or none in K-7259 and K-7220, which have shorter arms (C = 4 and 3, respectively), and K-10310 and K-10252, which have longer arms (C = 6 and 7, respectively), although all of them possess trimethoxy-phenyl groups (Table 1). The inhibitory effect was also undetected in K-10228, which has trimethyl-phenyl groups instead of trimethoxy-phenyl groups and pentenyl arms (C = 5) (Fig. 1A and B). Consistent with these data, immunoblot analysis revealed a marked accumulation of ubiquitinated proteins in RPMI8226 cells treated with K-7174, K-10256, K-10487 and K-10552 but not K-7259, K-7220, K-10252 and K-10228 (Fig. 1C). These results suggest that the trimethoxy-phenyl group at the tip of the pentenyl arm (C = 5) is a critical structure of homopiperazine-derived proteasome inhibitors. Based on this finding, K-7174 was selected as the most promising candidate for pharmaceutical development as a PI and was further characterized in this study.
K-7174 Inhibits Proteasome Activities in a Distinct Manner from Bortezomib
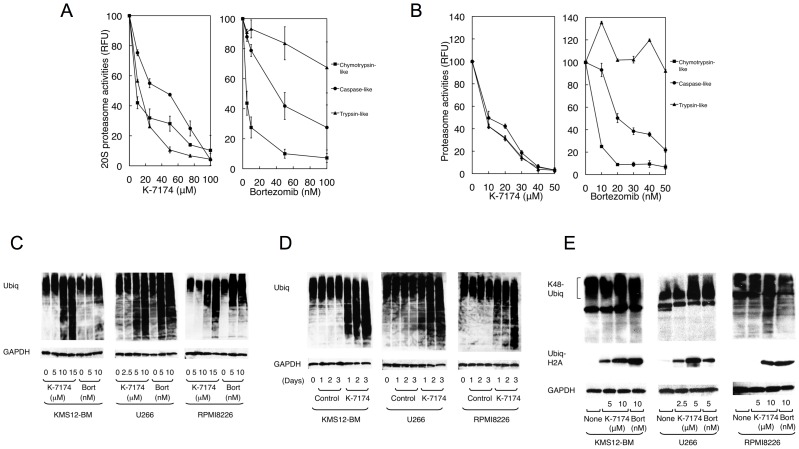
First, we compared the proteasome-inhibitory activity of K-7174 with that of bortezomib. As shown in Fig. 2A, K-7174 similarly inhibited all three subunits in a dose-dependent fashion, whereas bortezomib did not affect trypsin-like activity but efficiently inhibited chymotrypsin- and caspase-like activities. This pattern was readily reproducible in cell-based assays using RPMI8226 (Fig. 2B) and other myeloma cell lines (data not shown).
Figure 2. Inhibition of 20S proteasome activity by K-7174.
A. We treated purified erythrocyte-derived 20 S proteasome with either K-7174 or bortezomib at the indicated doses and determined chymotrypsin-like, caspase-like and trypsin-like activities as described in the legend of Fig. 1. B. RPMI8226 cells were treated with either K-7174 or bortezomib at the indicated doses, and analyzed for proteolytic activities. C. MM cell lines (KMS12-BM, U266, and RPMI8226) were cultured with K-7174 or bortezomib (Bort) at the indicated doses for 48 hours. Whole cell lysates were subjected to immunoblotting for ubiquitinated proteins and GAPDH (internal control). D. MM cell lines were cultured with either K-7174 (5 µM for U266 and 10 µM for KMS12-BM and RPMI8226) or the vehicle alone (Control) for up to 3 days. Whole cell lysates were prepared at given time points and subjected to immunoblotting as described above. E. MM cell lines were cultured in the absence (None) or presence of K-7174 or bortezomib (Bort) at the indicated doses for 48 hours, and subjected to immunoblotting for lysine48-linked polyubiquitinated proteins (K48-Ubiq), ubiquityl histone H2A (Ubiq-H2A), and GAPDH (internal control).
Next, we monitored the accumulation of ubiquitinated proteins in K-7174-treated MM cells in comparison with bortezomib. As shown in Fig. 2C and D, K-7174 induced a marked accumulation of ubiquitinated proteins in all three cell lines in a dose- and time-dependent fashion, as did bortezomib. In addition, we detected the accumulation of lysine 48-linked polyubiquitinated proteins and ubiquityl histone H2A, both of which represent specific and critical modifications leading to proteasomal degradation, in both K-7174- and bortezomib-treated MM cells (Fig. 2E). These results indicate that K-7174 is a novel PI distinct from bortezomib in its chemical structure and effects on proteasome activity.
K-7174 Interacts with the Catalytic Domains of ß1, ß2 and ß5 Subunits of the Proteasome in a Distinct Manner from Bortezomib
To understand the mechanisms of proteasome inhibition by K-7174, we determined the X-ray crystal structure of the yeast 20 S proteasome in complex with K-7174 at 2.5 Å resolution (Table 2). Analysis of the structure revealed that three molecules of K-7174 bind to and block the active sites of all three catalytic ß-type subunits, ß1, ß2 and ß5, with a similar binding mode (Fig. 3A), consistent with the biochemical data. Figure 3B shows the conformation and binding mode of K-7174 near the ß5 active site for example. The electron density of K-7174 was well-defined except for one trimethoxy-phenyl group near the ß4 subunit, which was partially discorded (Fig. 3B). The overall binding is determined largely by hydrophobic interactions between K-7174 and Gly47, Met97, Asp118, Gly130 and Ser131 of the ß5 subunit as well as Arg22 and Gly23 of the ß4 subunit (Fig. 3B). Importantly, the oxygen atom of the methoxy group of K-7174 makes a hydrogen bond (with a distance of 2.9 Å) to the OH group of the N-terminal threonine residue (Thr1), which acts as a nucleophile in hydrolysis. We also observed that, despite some difference in binding interactions, the active sites of the ß1 and ß2 subunits are blocked by the trimethoxy-phenyl group of K-7174 in a similar fashion to that observed in the ß5 subunit (Fig. 3C). These findings are fully compatible with our conclusion from biochemical analyses, and confirmed that the trimethoxy-phenyl group, but not the trimethyl-phenyl group, interacts with the active sites of three catalytic subunits via hydrogen bonding and the pentenyl arm (C - 5) fits the hydrophobic grooves between ß subunits via hydrophobic interaction. It is highly likely that K-10256, K-10487 and K-10552 interact with ß-subunits in a similar manner. Taken together, these results provide the molecular basis of HPDs’ ability to inhibit all three catalytic subunits at the same time.
Figure 3. Crystallographic structure of the K-7174/proteasome complex.
A. An overall ribbon diagram showing the folds of ß1 to ß7 subunits in the proteasome. The ß4 and ß5 subunits are colored yellow and blue, respectively. The K-7174 molecules bound to each ß subunit are shown as a space-filling representation colored green. Red spots indicate oxygen atoms. B. The final averaged electron density map (2Fo-Fc) covering K-7174 is shown (contoured at 1∂) (upper panel). A schematic diagram showing the interactions between K-7174 and the ß4 and ß5 subunits. The residues of ß4 and ß5 subunits associated with K-7174 are colored yellow and blue, respectively. A hydrogen bond is shown as a green dotted line (lower panel). C. A schematic diagram showing the interactions between K-7174 and the ß1 and ß2 subunits. The residues of ß1 and ß2 subunits associated with K-7174 are shown. A hydrogen bond is shown as a green dotted line.
Cytotoxic Effects of Homopiperazine Derivatives on Hematological Malignancies in vitro
Given the proteasome-inhibitory action of HPDs, we investigated whether these agents exert cytotoxic activity against myeloma and other hematological malignancies. To this end, we cultured various cell lines with K-7174 and K-10487, and determined IC50 values using MTT assays. As shown in Fig. 4A, both agents showed remarkable cytotoxicity at ∼10 µM in most cell lines. The cytotoxic doses were virtually identical to the proteasome-inhibitory concentrations, suggesting that K-7174 and K-10487 exert cytotoxic effects mainly via proteasome inhibition. Consistent with this notion, the cytotoxic activity of both K-7174 and bortezomib was mediated via the caspase-8-dependent pathway (manuscript in preparation). However, the sensitivity pattern was obviously different between HPDs and bortezomib, implying a difference in their mechanisms of action.
Figure 4. Comparison of K-7174 and bortezomib in cytotoxic activity and proteasome binding.
A. Cell proliferation was measured by MTT assays after culturing with serially diluted K-7174, K-10487 and bortezomib for 72 hours. Absorbance at 450 nm was analyzed with a microplate reader, and expressed as a percentage of the value of corresponding untreated cells. The IC50 value was defined as the concentration of each drug that produces 50% inhibition of cell growth. The means ± S.D. (bars) of three independent experiments are shown. Asterisks indicate “not determined”. B. Overall crystallographic structures showing the folds of ß1 to ß7 subunits in the proteasome bound with K-7174 (left panel) and bortezomib (right panel). Mutation sites observed in bortezomib-resistant cells are circled. C. Structure of the proteasome in complex with bortezomib (PDB cord 2F16) overlapped that with K-7174 described here. Only the protein atoms of the bortezomib-bound form are shown. Bortezomib-resistant mutant residues (Ala49, Thr21, Cys52 and Met45) are colored red and shown as a space-filling diagram.
Comparison of the Binding Modes of K-7174 and Bortezomib
To gain mechanistic insights, we compared the binding mode of K-7174 with that of bortezomib using X-ray crystallographic data. As shown in Fig. 4B, three molecules of K-7174 bind to the active pockets of the ß1, ß2 and ß5 subunits along hydrophobic grooves in the direction of the ß7, ß1 and ß4 subunits, respectively. In contrast, bortezomib is attached to the ß5 subunit by a hydrogen-bond network composed of Ala49, Thr21 and Gly44 [39] (Fig. 4B and C). Mutations of amino acids within or near the bortezomib-binding pocket in the ß5 subunit, such as Ala49, Thr21, Met45 and Cys52, were reported to cause bortezomib resistance by reducing the affinity to the drug (14–19). Among them, Ala49 makes a direct hydrogen bond to bortezomib, explaining why this position is most frequently mutated in bortezomib-resistant cells. Furthermore, a Cys52Phe or Met45Val substitution results in a steric clash between the side chains of these two residues (Fig. 4C), leading to repulsion of bortezomib from the binding pocket [14], [16], [17]. In contrast, these mutations should not affect the affinity of K-7174 to ß5 subunit, because the binding site of K-7174 is spatially distinct from the bortezomib-binding pocket (Fig. 4B and C).
K-7174 Overcomes Bortezomib Resistance caused by a ß5-subunit Mutation
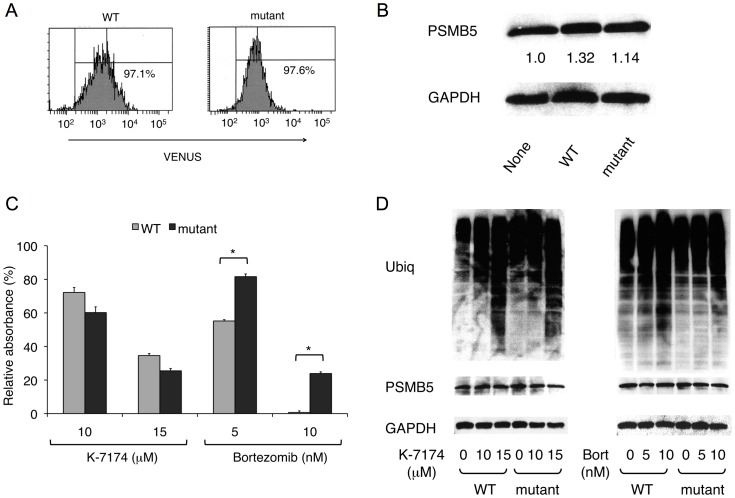
Because K-7174 appears to inhibit proteasome activity with a distinct mode from bortezomib, it is anticipated that K-7174 is effective for bortezomib-resistant cells. Previous studies revealed that a mutation of the PSMB5 gene at nucleotide position 322 (G322A), which corresponds to the substitution of Ala49 to Thr (Ala49Thr), induced conformational changes in the bortezomib-binding pocket of ß5 subunit and was responsible for acquired bortezomib resistance in T-cell acute lymphoblastic leukemia and myeloid leukemia cells [14]–[16]. Recently, Ri et al. [17] reported the establishment of bortezomib-resistant MM cell lines by transduction with G322A-mutated PSMB5 cDNA. Taking the same approach, we established three mutant sublines (mutant-5B, 5F and 9B) from RPMI8226 cells lentivirally transduced with mutated PSMB5 along with a marker gene VENUS. As controls, three wild-type sublines (WT-4G, 8F and 9G) were concomitantly established by mock transfection. We selected mutant-5F and WT-9G as representative sublines after determining the sensitivity to bortezomib (data not shown). The expression levels of VENUS and PSMB5 were virtually identical in these sublines (Fig. 5, A and B). As expected, sensitivity to bortezomib was significantly lower in the mutant subline than in the WT subline in MTT assays (Fig. 5C). In striking contrast, K-7174 induced cytotoxicity equally in WT and mutant sublines (Fig. 5C). In correlation with the results of MTT assays, K-7174 inhibited chymotrypsin-like (ß5) activity similarly in both sublines, whereas bortezomib could only partially inhibit the activity in the mutant subline (data not shown). These results were fully reproducible in other WT and mutant sublines (data not shown). To confirm the effects on proteasome activities, we determined the accumulation of ubiquitinated proteins in these sublines. As shown in Fig. 5D, ubiquitinated proteins were accumulated to a lesser extent in mutant cells than WT cells when they were treated with bortezomib (right panel). In contrast, K-7174 similarly induced intracellular protein ubiquitination in WT and mutant sublines (Fig. 5D, left panel). These results suggest that K-7174 can overcome bortezomib resistance.
Figure 5. Cytotoxic effects of K-7174 on bortezomib-resistant MM cells.
We established wild-type (WT) and mutant (mutant) sublines from RPMI8226 by transducing with wild-type and mutated PSMB5 cDNA, respectively, and analyzed the expression of VENUS by flow cytometry (A) and proteasome ß5 subunit by immunoblotting (B). The signal intensities of ß5 subunit (PSMB5) were quantified, normalized to those of the corresponding GAPDH, and shown as relative values in the panel B. C. Cell proliferation was measured by MTT assays after culturing each subline with either K-7174 or bortezomib at the indicated doses for 72 hours. Results are represented as relative absorbance with untreated control set at 100%. The means ± S.D. (bars) of three independent experiments are shown. P-values were calculated by one-way ANOVA with the Student-Newman-Keuls multiple comparisons test. Asterisks indicate p<0.01 against the WT subline. D. Each subline was cultured with either K-7174 or bortezomib (Bort) at the indicated doses for 48 hours. Whole cell lysates were subjected to immunoblotting for cellular protein ubiquitination, proteasome ß5 subunit (PSMB5) and GAPDH (internal control).
K-7174 and Bortezomib Exert Additive Cytotoxicity Against MM Cells
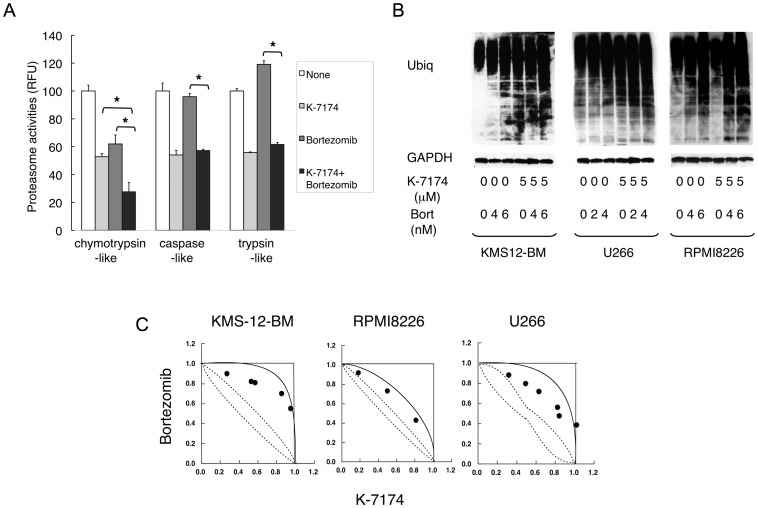
As K-7174 inhibits proteasome activity in a distinct manner from bortezomib, their combination would be additive and useful for dose reduction of bortezomib. Indeed, K-7174 additively enhanced the proteasomal ß5-inhibitory effect of bortezomib (Fig. 6A) as well as bortezomib-induced accumulation of ubiquitinated proteins (Fig. 6B). Furthermore, isobologram analysis of drug interaction revealed that K-7174 and bortezomib showed additive cytotoxicity in MM cell lines (Fig. 6C).
Figure 6. K-7174 and bortezomib exert additive cytotoxicity against MM cell.
A. We treated RPMI8226 cells with K-7174 (10 µM), bortezomib (5 nM) or both agents, and determined chymotrypsin-like, caspase-like and trypsin-like activities. Results are represented as relative fluorescence units (RFU) with vehicle controls set at 100%. The means ± S.D. (bars) of three independent experiments are shown. Asterisks indicate p<0.01 by paired Student’s t-test. B. We cultured KMS12-BM, U266 and RPMI8226 cells in the absence or presence of K-7174, bortezomib or both agents for 48 hours at the indicated doses. Whole cell lysates were subjected to immunoblotting for ubiquitinated proteins and GAPDH (internal control). Data are representative of multiple independent experiments. C. Isobolograms of simultaneous exposure of three MM cell lines to K-7174 and bortezomib. The concentrations that produced 80% growth inhibition are expressed as 1.0 on the ordinate and abscissa of isobolograms. The envelope of additivity, surrounded by solid and broken lines, was constructed from dose–response curves of bortezomib and K-7174. The combination is regarded as additive, because all data points fall within the envelope of additivity. The isobolograms shown are representative of at least three independent experiments. Each point represents the mean of at least three independent experiments; standard deviations were less than 25% and were omitted.
Discussion
In the present study, we show that HPDs constitute a novel class of PIs with a unique mode of proteasome binding. Although many kinds of small molecular PIs with various chemical structures have been developed [40], [41], this is the first demonstration of the proteasome-inhibitory activity of HPDs. In addition, most of the previous PIs mainly acted on one or two catalytic subunits and their mechanisms of action are not fully understood [40], [41]. In contrast, we have demonstrated that HPDs act on all three catalytic subunits of the proteasome by direct binding to the active pockets of the ß1, ß2 and ß5 subunits with a similar binding mode and kinetics. These results indicate the unique features of homopiperazine-derived PIs in chemical structures and effects on the proteasome. Moreover, we have identified the critical chemical structure of homopiperazine-derived PIs; therefore, these observations may contribute to the development of novel PIs with higher activity and specificity.
The high concentrations to trigger cytotoxicity might be the obstacle for clinical application of K-7174. Crystal structure analyses revealed that K-7174 interacts with ß subunits largely via hydrophobic interaction, whereas bortezomib binds to the ß5 subunit via a hydrogen-bond network, explaining why higher concentrations are required for HPDs compared with bortezomib. Therefore, the development of novel HPDs with higher activity and specificity is essential for clinical translation. Our finding on the chemical structure of homopiperazine-derived PIs may be of great help in this regard.
Despite the great success of bortezomib in the treatment of refractory malignancies such as MM and mantle cell lymphoma [3], [4], we still intend to develop orally bioavailable PIs with distinct mechanisms of action from bortezomib. Several novel PIs, such as carfilzomib [42], [43], NPI-0052 [44], CEP-18770 [45], MLN9708 [46], and ONX-0912 [47], are now undergoing clinical trials and show considerable benefits for refractory/relapsed cases as well as untreated MM patients. Among them, carfilzomib and its derivative ONX-0912 are peptide derivatives and have greater selectivity for the ß5 subunit than bortezomib. Although NPI-0052 is a non-peptide PI targeting all three proteasome subunits, its effect was strong for chymotrypsin-like (ß5), moderate for trypsin-like (ß2), and weak for caspase-like (ß1) activities [44]. In addition, NPI-0052 is intravenously administered in clinical studies [48], although it is expected to have oral bioactivity [44]. MLN9708 is orally available and its efficacy has been demonstrated in phase I clinical trials with oral administration [49]; however, this drug is speculated to be ineffective for MM carrying ß5-subunit mutations because of its boronate-based structure similar to bortezomib. Recently, in contrast to our speculation, Chauhan et al. [50] reported the effectiveness of MLN9708 to overcome bortezomib resistance. As several mechanisms have been proposed for bortezomib resistance in addition to ß5 subunit mutations [51], MLN9708 may be effective for such cases.
HPDs are expected to compensate for the weak points of bortezomib as well as the second generation PIs described above, because HPDs are non-peptide agents that inhibit all three catalytic subunits of the proteasome with equal kinetics and could be orally bioactive. Moreover, crystal structure analyses indicate that the binding mode is completely different from that of bortezomib [39] and NPI-0052 [52]. This ensures the activity of this agent against bortezomib-resistant cells, which was experimentally proven in this study, and probably against cells developing the resistance to NPI-0052. Moreover, we have found that oral administration of K-7174 is indeed effective and is not associated with obvious toxicities, including leukocytopenia, in a murine xenograft model (manuscript in preparation). These features provide a rationale for the clinical translation of HPDs as novel PIs with effectiveness for the treatment of bortezomib-resistant patients, a low probability of acquired drug resistance, and flexibility in dosing schedules.
Acknowledgments
We are grateful to Dr. Hiroaki Kimura (Jichi Medical University) for helpful discussions and technical advice. We are indebted to Ms. Akiko Yonekura for excellent technical assistance.
Funding Statement
This work was supported in part by the High-Tech Research Center Project for Private Universities: Matching Fund Subsidy from MEXT, a Grant-in-Aid for Scientific Research from JSPS (to Y.F. and J.K.), Adaptable and Seamless Technology transfer Program (A-STEP) from JST, Japan Leukemia Research Fund, Takeda Science Foundation, Kano Foundation, and Mitsui Life Social Welfare Foundation (to J.K.). Y.F. is the winner of the Award in Aki’s Memory from the International Myeloma Foundation Japan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sawyers C (2004) Targeted cancer therapy. Nature 432: 294–297. [DOI] [PubMed] [Google Scholar]
- 2. Weissman AM, Shabek N, Ciechanover A (2011) The predator becomes the prey: regulating the ubiquitin system by ubiquitination and degradation. Nat Rev Mol Cell Biol 12: 605–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Frankland-Searby S, Bhaumik SR (2012) The 26S proteasome complex: An attractive target for cancer therapy. Biochem Biophys Acta 1825: 64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Richardson PG, Mitsiades C, Schlossman R, Ghobrial I, Hideshima T, et al. (2008) Bortezomib in the front-line treatment of multiple myeloma. Expert Rev Anticancer Ther 8: 1053–1072. [DOI] [PubMed] [Google Scholar]
- 5. Kikuchi J, Wada T, Shimizu R, Izumi T, Akutsu M, et al. (2010) Histone deacetylases are critical targets of bortezomib-induced cytotoxicity in multiple myeloma. Blood 116: 406–417. [DOI] [PubMed] [Google Scholar]
- 7. Yanamandra N, Colaco NM, Parquet NA, Buzzeo RW, Boulware D, et al. (2006) Tipifarnib and bortezomib are synergistic and overcome cell adhesion-mediated drug resistance in multiple myeloma and acute myeloid leukemia. Clin Cancer Res 12: 591–599. [DOI] [PubMed] [Google Scholar]
- 8. Noborio-Hatano K, Kikuchi J, Takatoku M, Shimizu R, Wada T, et al. (2009) Bortezomib overcomes cell-adhesion-mediated drug resistance through downregulation of VLA-4 expression in multiple myeloma. Oncogene 28: 231–242. [DOI] [PubMed] [Google Scholar]
- 9. San Miguel JF, Schlag R, Khuageva NK, Dimopoulos MA, Shpilberg O, et al. (2008) Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. New Engl J Med 359: 906–917. [DOI] [PubMed] [Google Scholar]
- 10. Palumbo A, Bringhen S, Rossi D, Cavalli M, Larocca A, et al. (2010) Bortezomib-melphalan-prednisone-thalidomide followed by maintenance with bortezomib-thalidomide compared with bortezomib-melphalan-prednisone for initial treatment of multiple myeloma: A randomized controlled trial. J Clin Oncol 28: 5101–5109. [DOI] [PubMed] [Google Scholar]
- 11. Harrison SJ, Quach H, Link E, Seymour JF, Ritchie DS, et al. (2011) A high rate of durable responses with romidepsin, bortezomib, and dexamethasone in relapsed or refractory multiple myeloma. Blood 118: 6274–6283. [DOI] [PubMed] [Google Scholar]
- 12. Lonial S, Waller EK, Richardson PG, Jagannath S, Orlowski RZ, et al. (2005) Risk factors and kinetics of thrombocytopenia associated with bortezomib for relapsed, refractory multiple myeloma. Blood 106: 3777–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Richardson PG, Briemberg H, Jagannath S, Wen PY, Barlogie B, et al. (2006) Frequency, characteristics, and reversibility of peripheral neuropathy during treatment of advanced multiple myeloma with bortezomib. J Clin Oncol 24: 3113–3120. [DOI] [PubMed] [Google Scholar]
- 14. Lu S, Yang J, Song X, Gong S, Zhou H, et al. (2008) Point mutation of the proteasome 5 subunit gene is an important mechanism of bortezomib resistance in bortezomib-selected variants of Jurkat T cell lymphoblastic lymphoma/leukemia line. J Pharmacol Exp Ther 326: 423–431. [DOI] [PubMed] [Google Scholar]
- 15. Oerlemans R, Franke NE, Assaraf YG, Assaraf YG, Cloos J, et al. (2008) Molecular basis of bortezomib resistance: proteasome subunit ß5 (PSMB5) gene mutation and over-expression of PSMB5 protein. Blood 112: 2489–2499. [DOI] [PubMed] [Google Scholar]
- 16. Lu S, Yang J, Chen Z, Gong S, Zhou H, et al. (2009) Different mutants of PSMB5 confer varying bortezomib resistance in T lymphoblastic lymphoma/leukemia cells derived from the Jurkat cell line. Exp Hematol 37: 831–837. [DOI] [PubMed] [Google Scholar]
- 17. Ri M, Iida S, Nakashima T, Miyazaki T, Mori F, et al. (2010) Bortezomib-resistant myeloma cell lines: a role for mutated PSMB5 in preventing the accumulation of unfolded proteins and fatal ER stress. Leukemia 24: 1506–1512. [DOI] [PubMed] [Google Scholar]
- 18. Suzuki E, Demo S, Deu E, Keats J, Arastu-Kapur S, et al. (2011) Molecular mechanisms of bortezomib resistant adenocarcinoma cells. PLoS One 6: e27996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Franke NE, Niewerth D, Assaraf YG, van Meerloo J, Vojtekova K, et al. (2012) Impaired bortezomib binding to mutant ß5 subunit of the proteasome is the underlying basis for bortezomib resistance in leukemia cells. Leukemia 26: 757–768. [DOI] [PubMed] [Google Scholar]
- 20. Kitakaze M, Minamino T, Node K, Takashima S, Funaya H, et al. (1999) Adenosine and cardioprotection in the diseased heart. Jpn Circ J 63: 231–243. [DOI] [PubMed] [Google Scholar]
- 21. Umetani M, Nakao H, Doi T, Iwasaki A, Ohtaka M, et al. (2000) A novel cell adhesion inhibitor, K-7174, reduces the endothelial VCAM-1 induction by inflammatory cytokines, acting through the regulation of GATA. Biochem Biophys Res Commun 272: 370–374. [DOI] [PubMed] [Google Scholar]
- 22. Imagawa S, Nakano Y, Obara N, Suzuki N, Doi T, et al. (2003) A GATA-specific inhibitor (K-7174) rescues anemia induced by IL-1ß, TNF-α, or L-NMMA. FASEB J 17: 1742–1744. [DOI] [PubMed] [Google Scholar]
- 23. Nakano Y, Imagawa S, Matsumoto K, Stockmann C, Obara N, et al. (2004) Oral administration of K-11706 inhibits GATA binding activity, enhances hypoxia-inducible factor 1 binding activity, and restores indicators in an in vivo mouse model of anemia of chronic disease. Blood 104: 4300–4307. [DOI] [PubMed] [Google Scholar]
- 24. Takano Y, Hiramatsu N, Okamura M, Hayakawa K, Shimada T, et al. (2007) Suppression of cytokine response by GATA inhibitor K-7174 via unfolded protein response. Biochem Biophys Res Commun 360: 470–475. [DOI] [PubMed] [Google Scholar]
- 25. Drexler HG, Matsuo Y, MacLeod RA (2003) Persistent use of false myeloma cell lines. Hum Cell 16: 101–105. [DOI] [PubMed] [Google Scholar]
- 26. Uno K, Inukai T, Kayagaki N, Goi K, Sato H, et al. (2003) TNF-related apoptosis-inducing ligand (TRAIL) frequently induces apoptosis in Philadelphia chromosome-positive leukemia cells. Blood 101: 3658–3667. [DOI] [PubMed] [Google Scholar]
- 27. de Leeuw RJ, Davies JJ, Rosenwald A, Bebb G, Gascoyne RD, et al. (2004) Comprehensive whole genome array CGH profiling of mantle cell lymphoma model genomes. Hum Mol Genet 13: 1827–1837. [DOI] [PubMed] [Google Scholar]
- 28. Mitsunaga K, Kikuchi J, Wada T, Furukawa Y (2012) Latexin Regulates the abundance of multiple cellular proteins in hematopoietic stem cells. J Cell Physiol 227: 1138–1147. [DOI] [PubMed] [Google Scholar]
- 29. Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, et al. (1999) Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci USA 96: 10403–10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith DM, Wang Z, Kazi A, Li LH, Chan,TH, et al (2002) Synthetic analogs of green tea polyphenols as proteasome inhibitors. Mol Med 8: 382–392. [PMC free article] [PubMed] [Google Scholar]
- 31. Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collection in oscillation mode. Method Enzymol 276: 307–326. [DOI] [PubMed] [Google Scholar]
- 32. Vagin A, Teplyakov A (2000) An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr 56: 1622–1624. [DOI] [PubMed] [Google Scholar]
- 33. Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, et al. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954. [DOI] [PubMed] [Google Scholar]
- 34. Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- 35. Davis IW, Murray LW, Richardson JS, Richardson DC (2004) MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32: W615–619 doi:10.1093/nar/gkh398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kikuchi J, Shimizu R, Wada T, Ando H, Nakamura M, et al. (2007) E2F-6 suppresses growth-associated apoptosis of human hematopoietic progenitor cells by counteracting proapoptotic activity of E2F-1. Stem Cells 25: 2439–2447. [DOI] [PubMed] [Google Scholar]
- 37. Wada T, Kikuchi J, Furukawa Y (2012) Histone deacetylase 1 enhances microRNA processing via deacetylation of DGCR8. EMBO Rep 13: 142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Furukawa Y, Vu HA, Akutsu M, Odgerel T, Izumi T, et al. (2007) Divergent cytotoxic effects of PKC412 in combination with conventional antileukemic agents in FLT3 mutation-positive versus-negative leukemia cell lines. Leukemia 21: 1005–1014. [DOI] [PubMed] [Google Scholar]
- 39. Groll M, Berkers CR, Ploegh HL, Ovaa H (2006) Crystal structure of the boronic acid-based proteasome inhibitor bortezomib in complex with the yeast 20 S proteasome. Structure 14: 451–456. [DOI] [PubMed] [Google Scholar]
- 40. De Bettignies G, Coux O (2010) Proteasome inhibitors: Dozens of molecules and still counting. Biochimie 92: 1530–1545. [DOI] [PubMed] [Google Scholar]
- 41. Ruschak AM, Slassi M, Kay LE, Schimmer AD (2011) Novel proteasome inhibitors to overcome bortezomib resistance. J Natl Cancer Inst 103: 1007–1017. [DOI] [PubMed] [Google Scholar]
- 42. Kuhn DJ, Chen Q, Voorhees PM, Strader JS, Shenk KD, et al. (2007) Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood 110: 3281–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. O'Connor OA, Stewart AK, Vallone M, Molineaux CJ, Kunkel LA, et al. (2009) A phase I dose escalation study of the safety and pharmacokinetics of the novel proteasome inhibitor carfilzomib (PR-171) in patients with hematological malignancies. Clin Cancer Res 15: 7085–7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chauhan D, Catley L, Li G, Podar K, Hideshima T, et al. (2005) A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from Bortezomib. Cancer Cell 8: 407–419. [DOI] [PubMed] [Google Scholar]
- 45. Piva R, Ruggeri B, Williams M, Costa G, Tamagno I, et al. (2008) CEP-18770: A novel, orally active proteasome inhibitor with a tumor-selective pharmacologic profile competitive with bortezomib. Blood 111: 2765–2775. [DOI] [PubMed] [Google Scholar]
- 46. Kupperman E, Lee EC, Cao Y, Bannerman B, Fitzgerald M, et al. (2010) Evaluation of the proteasome inhibitor MLN9708 in preclinical models of human cancer. Cancer Res 70: 1970–1980. [DOI] [PubMed] [Google Scholar]
- 47. Chauhan D, Singh AV, Aujay M, Kirk CJ, Bandi M, et al. (2010) A novel orally active proteasome inhibitor ONX 0912 triggers in vitro and in vivo cytotoxicity in multiple myeloma. Blood 116: 4906–4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Millward M, Price T, Townsend A, Sweeney C, Spencer A, et al. (2012) Phase 1 clinical trial of the novel proteasome inhibitor marizomib with the histone deacetylase inhibitor vorinostat in patients with melanoma, pancreatic and lung cancer based on in vitro assessments of the combination. Invest New Drugs 30: 2303–2317. [DOI] [PubMed] [Google Scholar]
- 49. Moreau P, Richardson PG, Cavo M, Orlowski RZ, San Miguel JF, et al. (2012) Proteasome inhibitors in multiple myeloma: 10 years later. Blood 120: 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chauhan D, Tian Z, Zhou B, Kuhn D, Orlowski R, et al. (2011) In vitro and in vivo selective antitumor activity of a novel orally bioavailable proteasome inhibitor MLN9708 against multiple myeloma cells. Clin Cancer Res 17: 5311–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mujtaba T, Dou QP (2011) Advances in the understanding of mechanisms and therapeutic use of bortezomib. Discov Med 12: 471–480. [PMC free article] [PubMed] [Google Scholar]
- 52. Groll M, Huber R, Potts BC (2006) Crystal structures of salinosporamide A (NPI-0052) and B (NPI-0047) in complex with the 20 S proteasome reveal important consequences of ß-lactone ring opening and a mechanism for irreversible binding. J Am Chem Soc 128: 5136–5141. [DOI] [PubMed] [Google Scholar]