Abstract
In Saccharomyces cerevisiae, methylation of the principal membrane sterol at C-24 produces the C-28 methyl group specific to ergosterol and represents one of the few structural differences between ergosterol and cholesterol. C-28 in S. cerevisiae has been suggested to be essential for the sparking function (W. J. Pinto and W. R. Nes, J. Biol. Chem. 258:4472-4476, 1983), a cell cycle event that may be required to enter G1 (C. Dahl, H.-P. Biemann, and J. Dahl, Proc. Natl. Acad. Sci. USA 84:4012-4016, 1987). The sterol biosynthetic pathway in S. cerevisiae was genetically altered to assess the functional role of the C-28 methyl group of ergosterol. ERG6, the putative structural gene for S-adenosylmethionine: delta 24-methyltransferase, which catalyzes C-24 methylation, was cloned, and haploid strains containing erg6 null alleles (erg6 delta 1 and erg6 delta ::LEU2) were generated. Although erg6 delta cells are unable to methylate ergosterol precursors at C-24, they exhibit normal vegatative growth, suggesting that C-28 sterols are not essential in S. cerevisiae. However, erg6 delta cells exhibit pleiotropic phenotypes that include defective conjugation, hypersensitivity to cycloheximide, resistance to nystatin, a severely diminished capacity for genetic transformation, and defective tryptophan uptake. These phenotypes reflect the role of ergosterol as a regulator of membrane permeability and fluidity. Genetic mapping experiments revealed that ERG6 is located on chromosome XIII, tightly linked to sec59.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aoyama Y., Yoshida Y. The 14alpha-demethylation of lanosterol by a reconstituted cytochrome P-450 system from yeast microsomes. Biochem Biophys Res Commun. 1978 Nov 14;85(1):28–34. doi: 10.1016/s0006-291x(78)80006-0. [DOI] [PubMed] [Google Scholar]
- Bard M. Biochemical and genetic aspects of nystatin resistance in saccharomyces cerevisiae. J Bacteriol. 1972 Sep;111(3):649–657. doi: 10.1128/jb.111.3.649-657.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard M., Lees N. D., Burrows L. S., Kleinhans F. W. Differences in crystal violet uptake and cation-induced death among yeast sterol mutants. J Bacteriol. 1978 Sep;135(3):1146–1148. doi: 10.1128/jb.135.3.1146-1148.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton D. H., Corrie J. E., Bard M., Woods R. A. Biosynthesis of terpenes and steroids. IX. The sterols of some mutant yeasts and their relationship to the biosynthesis of ergosterol. J Chem Soc Perkin 1. 1974;11(0):1326–1333. [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. An electrophoretic karyotype for yeast. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3756–3760. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl C., Biemann H. P., Dahl J. A protein kinase antigenically related to pp60v-src possibly involved in yeast cell cycle control: positive in vivo regulation by sterol. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4012–4016. doi: 10.1073/pnas.84.12.4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaber R. F., Mathison L., Edelman I., Culbertson M. R. Frameshift Suppression in SACCHAROMYCES CEREVISIAE VI. Complete Genetic Map of Twenty-Five Suppressor Genes. Genetics. 1983 Mar;103(3):389–407. doi: 10.1093/genetics/103.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaber R. F., Styles C. A., Fink G. R. TRK1 encodes a plasma membrane protein required for high-affinity potassium transport in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jul;8(7):2848–2859. doi: 10.1128/mcb.8.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollub E. G., Liu K. P., Dayan J., Adlersberg M., Sprinson D. B. Yeast mutants deficient in heme biosynthesis and a heme mutant additionally blocked in cyclization of 2,3-oxidosqualene. J Biol Chem. 1977 May 10;252(9):2846–2854. [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees N. D., Bard M., Kemple M. D., Haak R. A., Kleinhans F. W. ESR determination of membrane order parameter in yeast sterol mutants. Biochim Biophys Acta. 1979 Jun 2;553(3):469–475. doi: 10.1016/0005-2736(79)90302-x. [DOI] [PubMed] [Google Scholar]
- McCammon M. T., Hartmann M. A., Bottema C. D., Parks L. W. Sterol methylation in Saccharomyces cerevisiae. J Bacteriol. 1984 Feb;157(2):475–483. doi: 10.1128/jb.157.2.475-483.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molzahn S. W., Woods R. A. Polyene resistance and the isolation of sterol mutants in Saccharomyces cerevisiae. J Gen Microbiol. 1972 Sep;72(2):339–348. doi: 10.1099/00221287-72-2-339. [DOI] [PubMed] [Google Scholar]
- Pierce A. M., Unrau A. M., Oehlschlager A. C., Woods R. A. Azasterol inhibitors in yeast. Inhibition of the delta 24-sterol methyltransferase and the 24-methylene sterol delta 24(28)-reductase in sterol mutants of Saccharomyces cerevisiae. Can J Biochem. 1979 Mar;57(3):201–208. doi: 10.1139/o79-025. [DOI] [PubMed] [Google Scholar]
- Pinto W. J., Lozano R., Sekula B. C., Nes W. R. Stereochemically distinct roles for sterol in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1983 Apr 15;112(1):47–54. doi: 10.1016/0006-291x(83)91795-3. [DOI] [PubMed] [Google Scholar]
- Pinto W. J., Nes W. R. Stereochemical specificity for sterols in Saccharomyces cerevisiae. J Biol Chem. 1983 Apr 10;258(7):4472–4476. [PubMed] [Google Scholar]
- Ramgopal M., Bloch K. Sterol synergism in yeast. Proc Natl Acad Sci U S A. 1983 Feb;80(3):712–715. doi: 10.1073/pnas.80.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R. J., Low C., Bottema C. D., Parks L. W. Multiple functions for sterols in Saccharomyces cerevisiae. Biochim Biophys Acta. 1985 Dec 4;837(3):336–343. doi: 10.1016/0005-2760(85)90057-8. [DOI] [PubMed] [Google Scholar]
- Rodriguez R. J., Parks L. W. Structural and physiological features of sterols necessary to satisfy bulk membrane and sparking requirements in yeast sterol auxotrophs. Arch Biochem Biophys. 1983 Sep;225(2):861–871. doi: 10.1016/0003-9861(83)90099-1. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
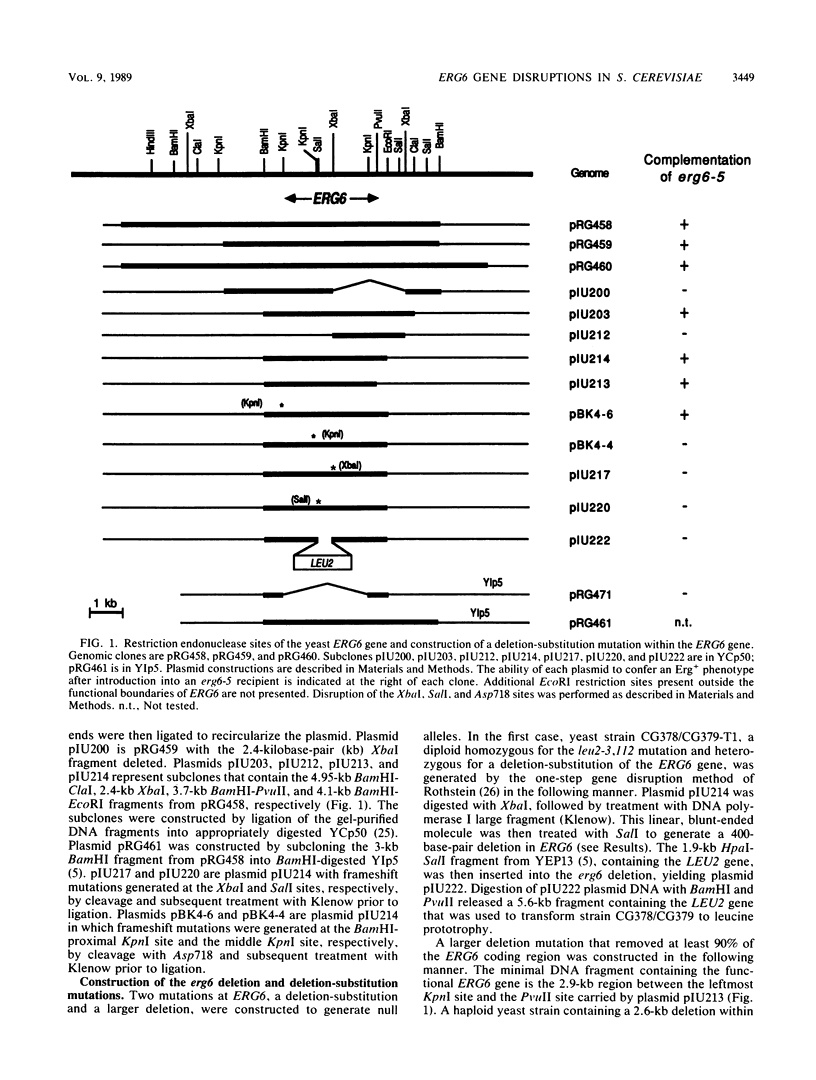
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Taylor F. R., Parks L. W. An assessment of the specificity of sterol uptake and esterification in Saccharomyces cerevisiae. J Biol Chem. 1981 Dec 25;256(24):13048–13054. [PubMed] [Google Scholar]
- Trueheart J., Boeke J. D., Fink G. R. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987 Jul;7(7):2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]