Summary
Insect photoreceptor function is dependent on precise placement of the rhabdomeres, elaborated apical domains specialized for capturing light, within each facet of a compound eye [1]. In Diptera, an asymmetric arrangement of rhabdomeres, combined with a particular pattern of axonal connections, enhances light sensitivity through the principle of neural superposition [2–3]. To achieve the necessary retinal geometry, different photoreceptors (R cells) have distinct shapes. The Crumbs and Bazooka complexes play critical roles in directing rhabdomere development [4–9], but whether they might direct cell-type specific apical architectures is unknown. We demonstrate that while mutations in Bazooka complex members cause pleiotropic morphogenesis defects in all R cell sub-types, Crumbs (Crb) and Stardust (Sdt) function cell-autonomously to direct early stages in rhabdomere assembly in specific subsets of R cells. This requirement is reflected in the cell-type specific expression of Crb protein, and demonstrates that Sdt and Crb can act independently to similar effect. These two genes are also required for zonula adherens (ZA) assembly, but display an unusual pattern of cellular redundancy for this function, as each gene is required in only one of two adjoining cells. Thus our results provide a direct link between fate specification and morphogenetic patterning, and suggest a model for ZA assembly.
Results
R cell rhabdomeres in the outer photoreceptors, R1–R6, form a trapezoidal shape in each ommatidium, surrounding those of the inner R cells, R7 and R8 (Figure 1A; [1]). Each rhabdomere comprises a stack of apical microvilli that contains the signaling components necessary for phototransduction. Morphogenetic events that take place within the apical domain of each R cell during pupal development cause each rhabdomere to occupy a specific location relative to its neighbors [10]. During the third larval stage, R cell types are specified at invariant relative positions, and are morphologically indistinguishable (Figure 1A). Approximately 30h after puparium formation (APF), the apical domains begin to elaborate differently in each cell type, changes that can be detected through patterned increases in Chaoptin expression, a protein involved in rhabdomere assembly (Figure 1A; [11]). By 46h APF, the cell-type specific pattern of Chaoptin localization is clearly visible, defining actin-rich fields in which the microvilli that make up the adult rhabdomere will extend. Immediately basal to these domains are the developing ZA, discrete regions that contain β-Catenin/Armadillo and the classical cadherins N-cadherin and E-cadherin (Figure 1A; [4, 5, 12–13]). By examining these markers, R2, R3, R4, and R5 can be distinguished by their shapes; R1 and R6 define a mirror symmetric pair of cells, representing a fifth morphologically defined cell type.
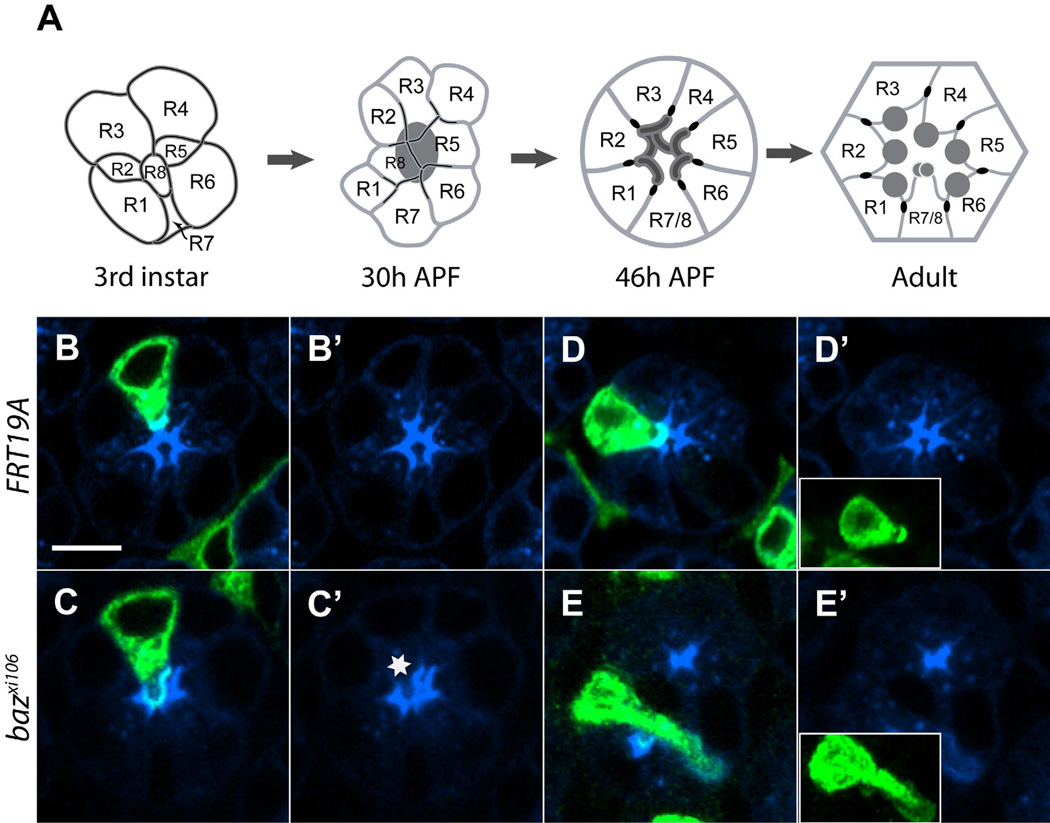
Figure 1. bazooka is a Par3 polarity complex gene required for photoreceptor development.
(A) Schematic illustrations of various stages of R cell morphogenesis, including late 3rd instar larval, 30h after pupal formation (APF; equivalent to 30% pupal development when grown at 25°C), 46h APF (equivalent to 46% pup al development), and adult retina. Developing apical domain, dark gray; ZA, black. (B-E) 44h retinas, stained with anti-Chaoptin (mAB24B10, blue). Labeled cells (UASmCD8GFP, green) were made homozygous mutant for FRT19A (B, B’, D, D’), bazxi106 (C, C’, E, E’). (B, C). R3 cells labeled. (D, E) R2 cells labeled. Inset panels in (B’-E’) are maximum intensity projections of the corresponding clones. Scale bar, 5um.
Apical – basal polarity proteins direct photoreceptor morphogenesis [4–9]. The Crumbs complex comprises three interacting genes, crumbs (crb), stardust (sdt), and Drosophila Pals associated tight junction protein (dPatj). All of these proteins are expressed in the apical domain, immediately apical to the ZA, and mutants in these genes have defects in rhabdomere and ZA formation [4–7, 9, 12]. The Par complex comprises bazooka/par3 (baz), par6, and aPKC, and often includes the small GTPase cdc42 (reviewed in [14]). In R cells, both complexes are localized to particular apical domains at different developmental stages, mutations in these genes cause defects in rhabdomere morphogenesis, and critical regulatory interactions occur between the two complexes [4–9, 15, 16].
Previous studies of apical-basal polarity protein function in R cells examined clones in which many cells were simultaneously made mutant, and concluded that these genes function similarly in all photoreceptors. However, whether they might function differently in specific subsets of R1–R6 cells was not directly tested. We reasoned that since individual R cells have cell-type specific apical domain morphologies, genes involved in specifying apical structures might act differently in distinct R cell subsets. We therefore extended previous studies to the level of individual photoreceptor sub-types using single cell mosaic clones.
Bazooka, Par6 and cdc42 are required in all R cell sub-types
We used heat shock-induced expression of the FLP recombinase, combined with the MARCM method [17, 18] to generate single homozygous cells positively labeled with mCD8GFP. Mosaic cells homozygous for control chromosomes were invariably normal, displaying fields of microvilli of normal extent and cell bodies of normal morphology (Figure 1B, 1B’, 1D, 1D’; Figure S1). In contrast, R cells homozygous mutant for baz, par6, or cdc42 displayed a range of defects, with some cells displaying misplaced fields of microvilli (Figure 1C and 1C’; Figure S1), while in more severe cases, the shape and orientation of the cell was defective (Figure 1E, 1E’; Figure S1). Quantitatively, in clones homozygous for the strong reduction of function allele bazxi106, 11% of R cells displayed defects affecting only the apical region, while an additional 7% had broad morphological defects; 82% of R cells displayed no defects (Figure S1; n=216). In clones homozygous for the null allele par6Δ226, 27% of mutant R cells displayed defects specific to the apical region, while 24% of cells displayed broader defects in morphogenesis; 49% of R cells were morphologically normal (Figure S1; n=154). Finally, in clones homozygous for cdc424, 35% and 16% of cells had defects specific to the apical region, or more generally within the cell body, respectively, and 49% of R cells were normal (Figure S1; n=109). All R cell sub-types were affected in all of these mutant backgrounds (data not shown), demonstrating that these three genes act cell-autonomously in all R cells to control morphogenesis.
Crumbs and Stardust act cell-type specifically
We next examined single R cells that were made homozygous for null alleles of crb or sdt. Compared to control clones (Figure 2A-2G, 2A’-2G’, Figure S2), we observed defects in apical domain structure in R2 and R4 cells homozygous for the null allele crb11A22, while all other R cells were almost invariably normal (Figure 2H-2N). In affected R2 and R4 cells, developing fields of microvilli were missing, or fragmented, as demonstrated by the loss or mis-localization of chaoptin staining. Quantitatively, these phenotypes were of moderate expressivity in R2, with 24% of clones displaying phenotypes (n=33; Figure S2), while highly expressive phenotypes were detected in R4 (with 73% of cells displaying defects, n=40; Figure S2). We obtained similar results when we stained for F-actin (Figure S2).
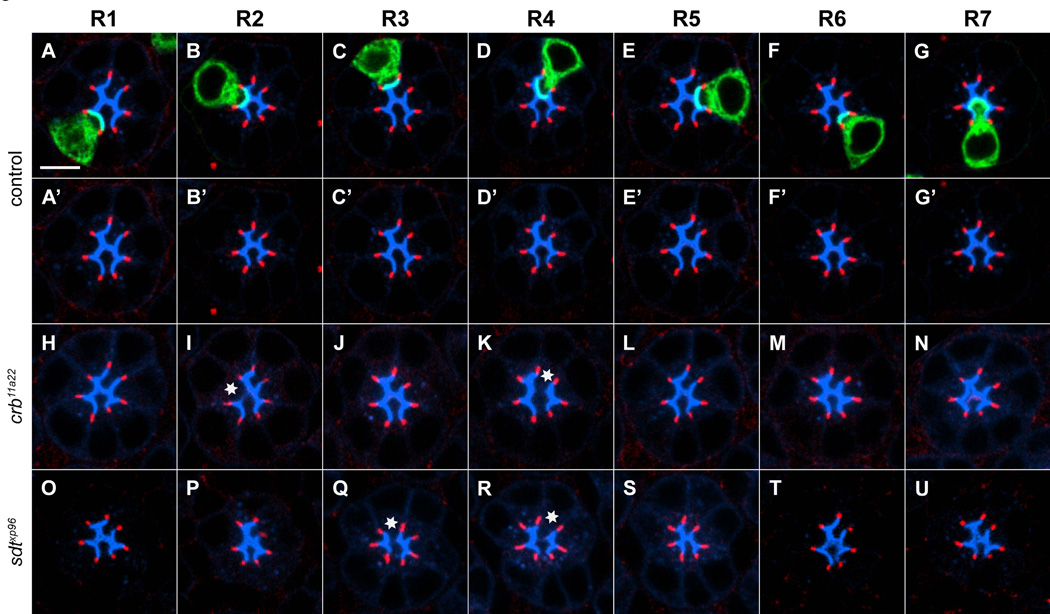
Figure 2. Subtype specific requirements of crumbs and stardust in rhabdomere development.
46h APF pupae. R1 cells (A, A’ H, O), R2 cells (B, B’, I, P), R3 cells (C, C’, J, Q), R4 cells (D, D’, K, R), R5 cells (E, E’, L, S), R6 cells (F, F’, M, T), and R7 cells (G, G’, N, U) labeled with (mCD8GFP, green), homozygous for a control chromosome (A-G), crb11a22 (H-N), and sdtxp96 (O-U). Apical regions are stained with anti-Chaoptin (mAB24B10, blue). ZA stained with anti-Armadillo, red. Asterisk, defective apical region. Scale bar, 5um.
To exclude the possibility that these phenotypes were the result of a mutation affecting another gene, rather than crb itself, we expressed a crb rescuing transgene [21] specifically in homozygous mutant crb clones. We observed full rescue of apical domain phenotypes in both R2 and R4 (Figure S2 and data not shown). To test whether these crb mutant phenotypes might reflect transient effects on R cell development, we examined a later stage of development, 55h APF. Consistent with the notion that these phenotypes are not simply developmental delays, we observed qualitatively and quantitatively indistinguishable phenotypes in R2 (57%, n=7) and R4 (70%, n=20) while other R cell types remained unaffected (Figure S2). Thus Crb function is required for normal morphogenesis of the apical domains of R2 and R4, but is largely dispensable for the early development of apical domains in other R cell subtypes.
We next performed the same analysis for a null allele of sdt, sdtXP96. Intriguingly, we detected qualitatively similar phenotypes to those associated with crb mutant clones, but in an overlapping but distinct subset of R cell types (Figure 2O-2U). In particular, R3 and R4 cells that were homozygous mutant for sdt frequently displayed abnormal Chaoptin staining as defects were detected in 61% of R3 cells (n=38; Figure S2) and 39% of R4 cells (n=44; Figure S2). Other R cell sub-types were only rarely affected. Thus, Crb and Sdt have distinct roles in R cell development from those associated with Baz, Par6, and Cdc42, and are prominently required in only specific R cell types.
Cell subtype specific expression of Crumbs and Stardust in R cells
We next considered whether Crb and Sdt might be expressed cell-type specifically at this developmental stage. As previously described, Crb was strongly expressed in a sub-apical region, as well as at slightly lower levels in the apical-most region (Figure S3 [4, 5, 12,19]). A Crb GFP knock-in line showed expression in an indistinguishable pattern (Figure S3 [20]). Sdt was also expressed in this subapical region, as well as in a more diffuse pattern within the apical domain (Figure S3; [6]). Colocalization of Crb and Sdt staining suggested that their expression only partially overlaps (Figure S3; [19]). Finally, given that Crb and Sdt protein is largely localized to the borders between adjacent cells, and given the resolution limits of optical microscopy, direct visualization cannot definitively determine whether each R cell expresses each protein.
To define Crb and Sdt expression in each R cell subtype, we therefore made single-cell mutants and examined Crb and Sdt expression at 46h APF. For clarity, only clones in which apical domain morphogenesis was normal were examined. Remarkably, when R2, R4, R5, and R7 cells were individually homozygous mutant for crb11A22, we observed loss of Crb expression in the apical domain relative to control clones, while no change was detected when R1, R3, or R6 were mutant (Figure 3A1-3B7). Thus, at this stage, Crb is strongly expressed only in R2, R4, R5 and R7 (but may also be expressed at levels we cannot detect in other R cells), results that are in line with previous observations [21].
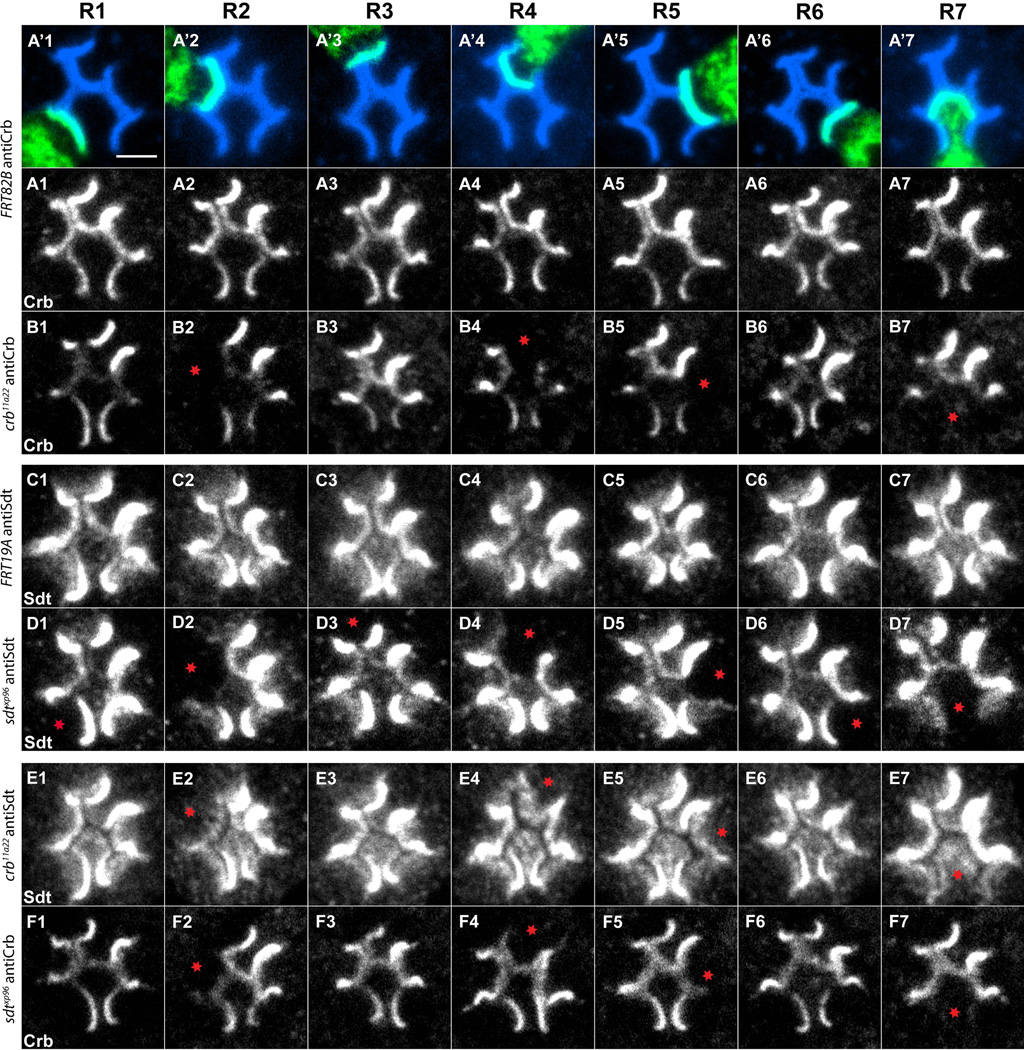
Figure 3. Mapping the cell-type specific expression patterns of crb and sdt.
46h APF pupae. (A’1-A’7). Control clones labeled with mCD8GFP (green), and anti-Chaoptin (mAb24B10, blue). (A1-A7) Control clones labeled with anti-Crb (white). (B1-B7) Crb mutant clones stained with anti-Crb (white). Control (C1-C7) and sdtxp96 (D1-D7) R cells, stained with anti-Sdt (white). (E1-E7) crb11a22 R cells, stained with anti-Sdt. (F1-F7) sdtxp96 R cells, stained with anti-Crb. Red asterisks label clones in which staining is lost. Scale bar, 2um.
We next mapped the expression pattern of Sdt. As was seen with crb mutants, when R2, R4, R5, and R7 were made homozygous mutant for sdtXP96, we observed a loss of bright, localized Stardust expression in the apical region, relative to control clones (Figure 3C1-3D7). However, we also observed changes in Stardust expression when R1, R3, and R6 cells were made homozygous mutant, in that these sdt mutant cells lost a relatively diffuse, weak staining in sub-apical regions. Thus, at this stage, Crb and Sdt display distinct expression patterns. In particular, Crb expression is detectable in only a subset of R cells, while Sdt is expressed in all R cells, but displays distinct sub-cellular localization in R2, R4, R5 and R7 cells, relative to R1, R3 and R6 (Figure S3).
Crb and Sdt maintain each other’s expression as homozygous crb mutant retinas display reduced Sdt staining, and vice versa [6–7]. We examined this interdependence at the single-cell level. Consistent with earlier experiments, Crb expression was dependent on Sdt function in all cell subtypes that express Crumbs, namely R2, R4, R5, and R7 (Figure 3F1-3F7). Sdt localization, however, displayed a more complicated dependence on Crb activity. While the vast majority of Sdt expression was lost in R2, R4, R5, and R7 cells homozygous for crb11A22, some staining remained (Figure 3E2, 3E4, 3E5, 3E7). This residual Sdt staining was diffuse, and reminiscent of that seen in wild-type R1, R3, and R6 cells (compare Figure 3D2, 3D4, 3D5, 3D7 with 3E2, 3E4, 3E5, 3E7). These results are consistent with the existence of two populations of Sdt protein within R cells, as previously described [22], with only the more highly expressed apical pool dependent on Crb function. Finally, consistent with the undetectable levels of Crb expression in R1, R3 and R6, Sdt staining was unaffected by the removal of Crb from these cells. Thus Crumbs and Stardust are partially, but not completely, required for maintaining each other’s expression.
Crumbs and Stardust are required in neighboring cells to regulate ZA marker localization
Previous work demonstrated that Crb and Sdt are required for the formation of ZA in epithelial cells [4–6,12]. The ZA forms as a narrow band of contact between neighboring R cell bodies, lying just below the apical surface, and extending along the proximal distal length of the cell. Previous work demonstrated that this region was defined by the localization of the proteins E-cadherin and b-catenin (Armadillo), as well as Bazooka [4, 5, 6, 23–25]. We therefore examined the cell-type specific requirements for Crb and Sdt activity in ZA remodeling, by staining somatic mosaic clones with antibodies directed against Armadillo and Bazooka (Figure 4).
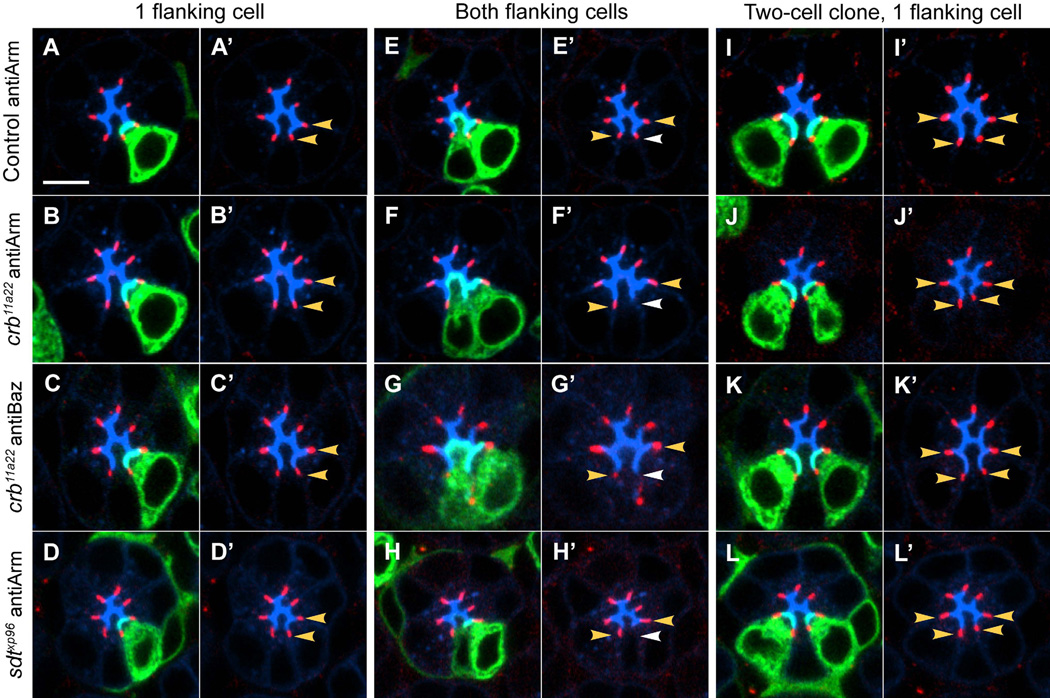
Figure 4. Crb and Sdt are required in neighboring R cells for ZA formation.
46h APF pupae. (A-D, A’-D’) ZA with one flanking cell homozygous. (E-H, E’-H’) ZA with both flanking cells homozygous. (I-L, I’-L’) ZA with one flanking cell homozygous, in a two-cell clone. (A, A’ E, E’, I, I’) Control cells labeled with anti-Armadillo, red. (B, B’, F, F’, J, J’) crb11a22 cells labeled with anti-Armadillo, red. (C, C’, G, G’, K, K’) crb11a22 cells labeled with anti-Bazooka, red. (D, D’, H, H’, L, L’) sdtxp96 cells labeled with anti-Armadillo, red. Apical regions (mAb24B10), blue. Clones (mCD8GFP), green. Yellow arrowhead demarks a ZA with one mutant cell neighbor. White arrowheads denote ZA with two mutant neighbors. Scale bar, 5um.
In control clones, expression of Armadillo was normal, regardless of whether one or both of the cells flanking the ZA were homozygous (Figure 4A, 4E; Figure S4). Similarly, ZA where only one flanking neighbor was homozygous for crb11A22 never displayed defects in Armadillo localization (1.5%, n=458; Figure 4B, 4B’; Figure S4). Similar results were observed with antibodies directed against Bazooka (3.6%, n=194; Figure 4C, 4C’; Figure S4). However, we observed that ZA that were flanked by two cells homozygous for crb11A22 frequently displayed loss or disruption in Armadillo and Bazooka staining (41%, n=56 for Armadillo, 50% n=12/24 for Bazooka; Figure 4F, 4F’, 4G, 4G’; Figure S4). To rule out differences in protein perdurance as a possible cause of these observations, we restricted our analysis to two-cell clones only, which should have identical levels of Crb protein; an example of a ZA in a two-cell clone in which each ZA has no more than one mutant neighbor is shown in Figure 4I, 4J, 4K. This analysis yielded the same result, where ZA with one flanking cell homozygous for crb11A22 were normal, (1.6%, n=125; Figure 4I; Figure S4) while those where both neighboring cells were homozygous for crb11A22 were often defective (43%, n=30 for Armadillo and 60%, n=15, for Bazooka; Figure S4). We obtained similar results with Sdt, as localization of Armadillo was normal when one adjoining neighbor was mutant for sdt (1.3%, n=228; Figure 4D, 4D’, 4L, 4L’; Figure S4) but defective when both neighbors were mutant (52%, n=29; Figure 4H; Figure S4). Finally, there was no apparent cell-type specificity for this function, as the ZA phenotypes observed in sdt and crb clones were indistinguishable, regardless of which pair of neighboring R cells were mutant. As we described cell-type specific expression of Crb at this stage of pupal development (Figure 3; Figure S3), the function of Crb in ZA assembly likely reflects the fact that Crb is expressed in all R cell types at earlier developmental stages (when ZA assembly begins [4, 5]). Thus, Crb and Sdt function is required for ZA remodeling in all R cells, but only in one of two cells neighboring the structure.
Discussion
Our results reveal that Crb and Sdt act cell-type specifically to direct the localization of regulators of photoreceptor apical domain morphogenesis (Figure S4). Interestingly, Crb and Sdt are cell-autonomously required in different cells. In particular, R3 cells are primarily dependent on Sdt, while R4 cells are equally dependent on both Crb and Sdt and R2 cells have a weak requirement for Crb alone. While technical limitations prevent us from generating single cell clones homozygous for both genes, we hypothesize that in other R cell sub-types, Crb and Sdt function redundantly. However, these cells may use still other molecules to guide localization of regulators of apical morphogenesis. In addition, our data demonstrate that Crb and Sdt are expressed at high levels during mid-pupal development only in cells that have relatively long stalk regions (namely, R2, R4, R5, and R7), whereas expression is either undetectable (for Crb), or low (for Sdt), at this stage in cells that have short stalks, namely R1, R3, and R6 (following observations described in [21]). While mutations in Crb are associated with highly expressive phenotypes in R4 (affecting Chp, actin and Sdt localization), as well as weakly expressive phenotypes in R2, the cell-autonomous effect of Crb mutations on the other R cell subtypes was more modest. In addition, while mutations in Sdt cause expressive phenotypes in R4, where Sdt is highly expressed, they also cause highly expressive phenotypes in R3, where Sdt is expressed at only low levels. Thus, the Sdt and Crb mutant phenotypes are neither correlated with the level of protein expression, nor with the known morphological differences between R cell types, namely the relative apportionment of the apical domain into rhabdomeric regions and stalk regions. Thus, the genetic programs that determine the localization of critical regulators of apical domain morphogenesis in each R cell subtype are different.
Our data also shed new light on the role that Crb and Sdt play in ZA formation, through our demonstration that Crb and Sdt are required in only one of two adjacent cells to ensure appropriate localization of regulators of the ZA between them (Figure S4). This type of non-autonomy is unusual, in that the function of each gene is apparently “redundant” between cells. This pattern of requirement demands communication between neighboring cells to determine whether one or both cells lack Crb or Sdt function. As Sdt is a cytosolic protein lacking an extracellular domain, the signal passing between Sdt mutant cells must be mediated by another molecule. Moreover, this signal cannot be conveyed by homophilic interactions mediated by Crb, as loss of Crb function in either cell would then be indistinguishable from the loss of Crb in both cells. We therefore propose that Crb and Sdt define the ZA localization of a third molecule, which mediates homophillic interactions between neighboring photoreceptors that are required for ZA formation or stabilization. In this view, the ZA is stabilized cell autonomously and independently in each cell through the joint action of Crb and Sdt, and is also stabilized non-cell autonomously by homophillic interactions across the ZA, mediated by a third molecule (E-cadherin/Adhesion X). In the absence of Crb or Sdt in both cells, the non-cell autonomous interaction is not sufficient to maintain a ZA, but if Crb or Sdt function remains in one cell (stabilizing the interaction on one side), the non-cell autonomous homophillic signal can then recruit and maintain the interaction across the ZA to the neighboring cell. While E-cadherin is the most promising candidate for the signal that stabilizes the ZA, given its known roles [24–26], we have been unable to cell-specifically manipulate E-cadherin levels in R cells without inducing pleiotropic phenotypes and cannot exclude the possible involvement of other molecules (J.J.H. and T.R.C., unpublished observations). Nonetheless, these studies provide evidence for an unusual form of genetic redundancy underpinning the formation of the ZA.
Supplementary Material
Highlights.
Two sets of apical-basal polarity proteins have distinct functions in photoreceptors.
crumbs and stardust direct apical domain formation in specific photoreceptor types.
Different genetic programs create distinct photoreceptor morphologies.
Genetic mosaic analysis defines a new model for zonula adherens remodeling.
Acknowledgments
We wish to thank Chris Doe for Par6 and Baz antibodies as well as the parΔ226 allele, Elisabeth Knust for the UAS-CRB transgene, bazxi106 allele, and Stardust and Crumbs antibodies, Uli Tepass for the crb11A22 allele and Crumbs antibody, as well as Yang Hong for the sdtXP96 allele and the Crb-GFP knock-in lines. We also thank U. Tepass for extensive guidance, and Marion Silies, Tina Schwabe, Jessica Tsai and Jennifer Esch for helpful comments on the manuscript. This work was supported by R01 EY015231 (T.R.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- 2.Kirschfeld K. Die Projektion der optischen Umwelt auf das Raster der Rhabdomere im Komplexauge von Musca. Exp. Brain Res. 1967;3:248–270. doi: 10.1007/BF00235588. [DOI] [PubMed] [Google Scholar]
- 3.Braitenberg V. Patterns of projection in the visual system of the flyIRetinalamina projections. Exp Brain Res. 1967;3:81–98. doi: 10.1007/BF00235589. [DOI] [PubMed] [Google Scholar]
- 4.Pellikka M, Tanentzapf G, Pinto M, Smith C, McGlade CJ, Ready DF, Tepass U. Crumbs, the Drosophila homologue of human CRB1/RP12, is essential for photoreceptor morphogenesis. Nature. 2002;416:143–149. doi: 10.1038/nature721. [DOI] [PubMed] [Google Scholar]
- 5.Izaddoost S, Nam SC, Bhat MA, Bellen HJ, Choi KW. Drosophila Crumbs is a positional cue in photoreceptor adherens junctions and rhabdomeres. Nature. 2002;416:178–183. doi: 10.1038/nature720. [DOI] [PubMed] [Google Scholar]
- 6.Hong Y, Ackerman L, Jan LY, Jan YN. Distinct roles of Bazooka and Stardust in the specification of Drosophila photoreceptor membrane architecture. Proc. Natl. Acad. Sci. U S A. 2003;100:12712–12717. doi: 10.1073/pnas.2135347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nam SC, Choi KW. Interaction of Par-6 and Crumbs complexes is essential for photoreceptor morphogenesis in Drosophila. Development. 2003;130:4363–4372. doi: 10.1242/dev.00648. [DOI] [PubMed] [Google Scholar]
- 8.Richard M, Grawe F, Knust E. DPATJ plays a role in retinal morphogenesis and protects against light-dependent degeneration of photoreceptor cells in the Drosophila eye. Dev. Dyn. 2006;235:895–907. doi: 10.1002/dvdy.20595. [DOI] [PubMed] [Google Scholar]
- 9.Nam SC, Choi KW. Domain-specific early and late function of Dpatj in Drosophila photoreceptor cells. Dev. Dyn. 2006;235:1501–1507. doi: 10.1002/dvdy.20726. [DOI] [PubMed] [Google Scholar]
- 10.Wolff T, Ready DF. Pattern formation in the Drosophila retina. In: Bate M, Arias AM, editors. The development of Drosophila melanogaster. Volume II. Cold Spring Harbor Laboratory Press; 1993. pp. 1277–1325. [Google Scholar]
- 11.Van Vactor D, Jr, Krantz DE, Reinke R, Zipursky SL. Analysis of mutants in chaoptin, a photoreceptor cell-specific glycoprotein in Drosophila, reveals its role in cellular morphogenesis. Cell. 1988;52:281–290. doi: 10.1016/0092-8674(88)90517-x. [DOI] [PubMed] [Google Scholar]
- 12.Bulgakova NA, Kempkens O, Knust E. Multiple domains of Stardust differentially mediate localisation of the Crumbs-Stardust complex during photoreceptor development in Drosophila. J Cell Sci. 2008;121:2018–2026. doi: 10.1242/jcs.031088. [DOI] [PubMed] [Google Scholar]
- 13.Tepass U, Harris KP. Adherens junctions in Drosophila retinal morphogenesis. Trends Cell Biol. 2007;17:26–35. doi: 10.1016/j.tcb.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walther RF, Pichaud F. Crumbs/DaPKC-dependent apical exclusion of Bazooka promotes photoreceptor polarity remodeling. Curr. Biol. 2010;20:1065–1074. doi: 10.1016/j.cub.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 16.Muschalik N, Knust E. Increased levels of the cytoplasmic domain of Crumbs repolarise developing Drosophila photoreceptors. J. Cell Sci. 2011;124:3715–3725. doi: 10.1242/jcs.091223. [DOI] [PubMed] [Google Scholar]
- 17.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 18.Prakash S, Caldwell JC, Eberl DF, Clandinin TR. Drosophila N-cadherin mediates an attractive interaction between photoreceptor axons and their targets. Nat Neurosci. 2005;8:443–450. doi: 10.1038/nn1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wodarz A, Hinz U, Engelbert M, Knust E. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell. 1995;82:67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]
- 20.Huang J, Zhou W, Dong W, Watson AM, Hong Y. Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc Natl Acad Sci U S A. 2009;106:8284–8289. doi: 10.1073/pnas.0900641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karagiosis SA, Ready DF. Moesin contributes an essential structural role in Drosophila photoreceptor morphogenesis. Development. 2004;131:725–732. doi: 10.1242/dev.00976. [DOI] [PubMed] [Google Scholar]
- 22.Bulgakova NA, Rensch M, Knust E. Antagonistic functions of two stardust isoforms in Drosophila photoreceptor cells. Mol. Biol. Cell. 2010;21:3915–3925. doi: 10.1091/mbc.E09-10-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richard M, Muschalik N, Grawe F, Ozüyaman S, Knust E. A role for the extracellular domain of Crumbs in morphogenesis of Drosophila photoreceptor cells. Eur J Cell Biol. 2009;88:765–777. doi: 10.1016/j.ejcb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 24.Peifer M. The product of the Drosophila segment polarity gene armadillo is part of a multi-protein complex resembling the vertebrate adherens junction. J Cell Sci. 1993;105:993–1000. doi: 10.1242/jcs.105.4.993. [DOI] [PubMed] [Google Scholar]
- 25.Oda H, Uemura T, Harada Y, Iwai Y, Takeichi M. A Drosophila homolog of cadherin associated with armadillo and essential for embryonic cell-cell adhesion. Dev Biol. 1994;165:716–726. doi: 10.1006/dbio.1994.1287. [DOI] [PubMed] [Google Scholar]
- 26.Harris TJ, Peifer M. Adherens junction-dependent and –independent steps in the establishment of epithelial cell polarity in Drosophila. J Cell Biol. 2004;167:135–147. doi: 10.1083/jcb.200406024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oda H, Uemura T, Takeichi M. Phenotypic analysis of null mutants for DEcadherin and Armadillo in Drosophila ovaries reveals distinct aspects of their functions in cell adhesion and cytoskeletal organization. Genes Cells. 1997;2:29–40. doi: 10.1046/j.1365-2443.1997.d01-284.x. [DOI] [PubMed] [Google Scholar]
- 28.Jurgens G, Wieschaus E, Nusslein-Volhart C, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster II. Zygotic loci on the X chromosome and the third chromosome. Roux Arch. Dev. Biol. 1984;193:283–295. doi: 10.1007/BF00848157. [DOI] [PubMed] [Google Scholar]
- 29.Wieschaus E, Nusslein-Volhart C, Jurgens G. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster III. Zygotic loci on the X chromosome and the fourth chromosome. Roux Arch. Dev. Biol. 1984;193:296–307. doi: 10.1007/BF00848158. [DOI] [PubMed] [Google Scholar]
- 30.Petronczki M, Knoblich JA. DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat Cell Biol. 2001;3:43–49. doi: 10.1038/35050550. [DOI] [PubMed] [Google Scholar]
- 31.Fehon RG, Oren T, Lajeunesse DR, Melby TE, McCartney BM. Isolation of mutations in the Drosophila homologues of the human Neurofibromatosis 2 and yeast CDC42 genes using a simple and efficient reverse-genetic method. Genetics. 1997;146:245–252. doi: 10.1093/genetics/146.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wodarz A, Ramrath A, Grimm A, Knust E. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J. Cell Biol. 2000;150:1361–1374. doi: 10.1083/jcb.150.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.