Abstract
In a neural integrator, the variability and topographical organization of neuronal firing rate persistence can provide information about the circuit’s functional architecture. Here we use optical recording to measure the time constant of decay of persistent firing (“persistence time”) across a population of neurons comprising the larval zebrafish oculomotor velocity-to-position neural integrator. We find extensive persistence time variation (10-fold; coefficients of variation 0.58–1.20) across cells within individual larvae. We also find that the similarity in firing between two neurons decreased as the distance between them increased and that a gradient in persistence time was mapped along the rostrocaudal and dorsoventral axes. This topography is consistent with the emergence of persistence time heterogeneity from a circuit architecture in which nearby neurons are more strongly interconnected than distant ones. Collectively, our results can be accounted for by integrator circuit models characterized by multiple dimensions of slow firing rate dynamics.
Introduction
Neural integrators are brain circuits specialized for performing the mathematical operation of integration on a time-varying signal1. This temporal integration is fundamental to a range of motor, memory, and decision-making tasks2–4. In the velocity-to-position neural integrator for horizontal eye movements (hVPNI) in the vertebrate oculomotor system, neurons temporally integrate brief eye velocity-encoding burst signals that drive saccades. This integration generates steps in the firing rate of hVPNI neurons that encode an eye position signal necessary for stabilizing gaze5,6. The steps in firing rate of an hVPNI neuron can persist following saccadic input for tens of seconds, ensuring that eye position changes only slowly (decay time constants are typically 10–100 s) during fixations between saccades. The cellular and circuit mechanisms that explain temporal integration and this persistent neural activity remain poorly understood7. Understanding these mechanisms in the hVPNI may provide a general framework for understanding integration in other neural systems.
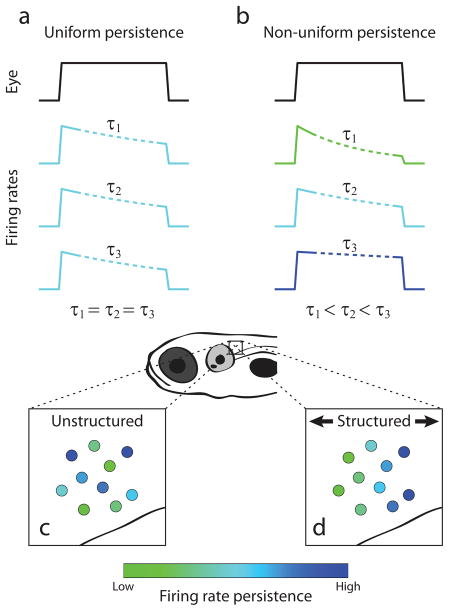
The firing rates of any ipsilateral pair of hVPNI neurons are approximately linearly related to each other over eye positions at which both are active8–10. This observation of the mutual linearity of firing rates lead to the idea that the circuit dynamics could be considered as that of a one-dimensional line attractor11. In addition to capturing this linearity, the most commonly considered line attractor-based model of the hVPNI has two other defining characteristics, both of which are aspects of the firing rate persistence in component neurons following a transient stimulus. First, consistent with experimental observations, the firing rates of neurons in the circuit decay slowly toward a baseline level, persisting far longer than typical membrane and synaptic time constants (1–100 ms). Second, this decay is dominated by a single time constant so that the rate of decay is uniform across neurons (Fig. 1a).
Figure 1.
Potential dynamical and spatial structure amongst hVPNI neurons. (a),(b) Eye position-dependent firing rates given uniform (a) or non-uniform (b) firing rate persistence across neurons. Colored traces show hypothetical firing rates relative to a hypothetical eye position measurement (black). Dotted segments correspond to firing rate during fixation from which firing rate persistence is measured. (c),(d) Spatial distribution of neurons in terms of firing rate persistence given unstructured (c) or structured (d) arrangements. Cells are pseudocolored according to their firing rate persistence.
Although line attractor dynamics have provided a general framework for conceptualizing parametric memory in neural systems12,13 and are approximated by several proposed models of the hVPNI14–16, other neuronal circuit architectures have also been proposed. Importantly, these architectures make different predictions of firing rate persistence across the neuronal population. In particular, recurrent network models with circuit architectures or intrinsic neuronal biophysics giving rise to not one but multiple long time scales of firing rate decay have been suggested to be more consistent than line attractor-based models with firing during vestibular stimulation17 and after detuning18,19 and partial inactivation20 of the hVPNI, and may be more robust to perturbation11,21. A network with such an architecture could display heterogeneous firing rate persistence across cells (Fig. 1b). Similarly, recently described integrating circuits that differ from line attractor models by emphasizing feedforward connectivity22 could also display heterogeneous firing rate persistence across cells, with cells further down the feedforward chain showing progressively longer persistence. Despite its capacity to help distinguish circuit model architectures in terms of the firing dynamics they generate, a systematic experimental study of the similarity of firing rate persistence across hVPNI neurons has not been reported. This is primarily because simultaneous measurement from many neurons has been difficult in adult preparations studied previously with electrophysiological methods. Recently, we have developed an optical recording-based method for identifying and measuring calcium changes in populations of hVPNI neurons in larval zebrafish23; these calcium changes can be used to estimate firing rate dynamics. This created an opportunity to investigate the similarity of persistence times within these populations, which is a principal focus of the present work.
Imaging methods enable not only simultaneous measurement from many neurons suitable for examining the diversity of dynamics, but also precise anatomical localization of the recorded cells. Electrophysiological approaches to studying the hVPNI have been limited in resolving the relative locations of neurons from which action potentials were recorded. This has prevented assessment of topography in firing rate dynamics amongst hVPNI neurons on short spatial scales (≤ ~100 μm), yet such structure could also indicate functional organization of the circuit these neurons comprise. For example, in a neuronal population showing diverse firing rate persistence, cells could be randomly arranged with respect to this persistence (Fig. 1c), or they could be ordered along one or more spatial dimensions (Fig. 1d). This sort of order might reflect the time course of neuronal maturation during development24 or differential connectivity between neurons25, as have been observed in other neuronal circuits. Because in general there is a relationship between connectivity and circuit dynamics22, characterization of topography could facilitate the understanding of circuit mechanisms underlying neural integration. Observation of structure on these scales in terms of firing patterns could also help elucidate genetic determination or organization of the hVPNI, since genetically defined cell types can be ordered on short spatial scales26,27.
Here we report a quantitative analysis of the variation in firing rate persistence, and the anatomical organization of this variation, among hVPNI neurons optically recorded with cellular resolution in awake larval zebrafish during spontaneous eye movements23. Firing rate persistence is quantified for these neurons as the persistence time, the time constant of the decay in firing during fixations. Persistence times were measured from cellular fluorescence time series deconvolved with calcium impulse response functions estimated for individual cells from data. These times varied over ten-fold across cells within individual larvae. A corresponding spatial organization on a short length scale (< 100 μm) was also observed in which persistence times were graded along the rostrocaudal and dorsoventral axes. This spatial organization in functional activity is consistent with a dependence of coupling strength on proximity, resulting in a circuit architecture ordered in terms of firing rate persistence.
Results
Identification and variability of eye movement-related hindbrain neurons
We used a recently described23 optical recording-based method to identify and characterize putative hVPNI neurons in awake larval zebrafish based on the temporal correlation of calcium concentration changes with simultaneously measured eye position. Hindbrain neurons were bolus-loaded with Oregon Green BAPTA-1 AM (OGB-1) and a custom-built two-photon microscope was used to collect image time series from sagittal windows 20 to 70 μm lateral of the midline that spanned from 100–130 μm to 200–230 μm caudal of the Mauthner cell soma (rhombomere 7/8; Supplementary Fig. 1) during spontaneous eye movements (Fig. 2a). These imaging windows spanned the lower ~2/3 of the dorsoventral extent of the hindbrain. In larval zebrafish, spontaneous eye movements (both in the light and in the dark) frequently consist of a stereotypical back and forth scanning pattern of sequential saccades and fixations. Eye positions are often similar across fixations following saccades in a given direction, producing eye position distributions that are somewhat bimodal. However, gaze stability is demonstrated at multiple eye positions (Fig. 2b, “Left Eye Position”), indicating that eye position in larval zebrafish is governed by an hVPNI capable of achieving at least an approximate continuum of stable eye positions, rather than just two.
Figure 2.
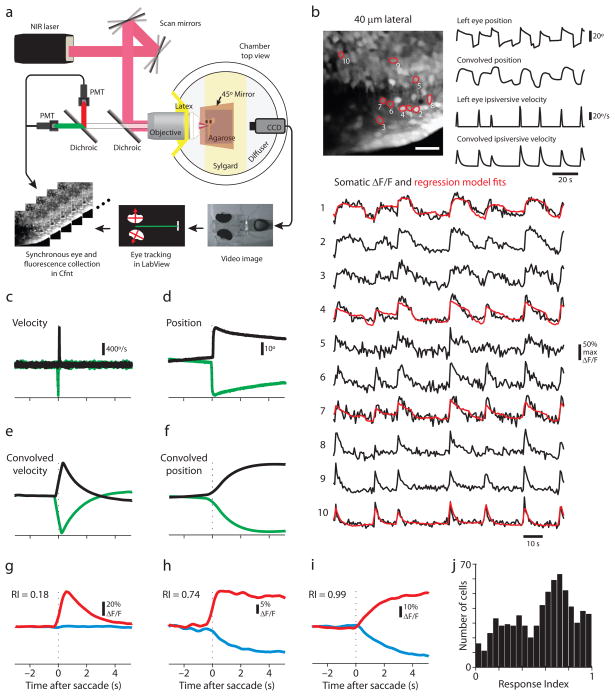
Saccade-related calcium fluctuations in optically-identified hindbrain somata. (a) Schematic of the custom-built microscope allowing simultaneous and synchronous eye tracking and two-photon laser-scanning imaging. (b) Calcium fluctuations in somata imaged 40 μm lateral of the midline. Gray scale image is the time projection of a sagittal image time series (scale bar = 20 μm). Ten of the identified somata are outlined in red, labeled with their rank in terms of the correlation between their ROI-averaged fluorescence (ΔF/F) time series and CIRF-convolved ipsilateral eye position. 100 seconds of their fluorescence time series are plotted in black with a regression model fit overlaid in red for several examples. Corresponding measured and CIRF-convolved eye variables are also shown. (c,d) Representative STA ipsilateral eye velocity (c) and position (d) measurements for ipsiversive (black) and contraversive (green) saccades. In (c)-(i), the mean of the first 1.5 s of each trace is subtracted off, and the dotted line indicates saccade time. (e,f) CIRF-convolved STA eye velocity (e) and position (f) calculated from (c) and (d), respectively. (g-i) Ipsiversive (red) and contraversive (blue) STA fluorescence responses for cells (not among those depicted in (b)) with a range of response indices (RI). (j) Histogram of response indices. Data collected in both the dark (n = 306 cells) and the light (n = 385 cells) were pooled.
For each ipsiversive saccade and subsequent fixation, ipsilateral eye position-encoding neurons in the caudal hindbrain regions we imaged, like integrator neurons in other species, show a burst of action potentials followed by persistent action potential firing at an elevated rate23. Typically, bursts scale with saccade amplitude (“velocity sensitivity”) while the persistent firing rate scales with eye position (“position sensitivity”). We have shown that this firing rate behavior produces somatic OGB-1 fluorescence time series that can be understood as a sum of ipsilateral eye position and thresholded eye velocity (“ipsiversive velocity”) convolved with a temporal filter that captures the mapping between action potential firing and intracellular calcium concentration changes, the calcium impulse response function (CIRF). We use CIRFs with the form of an instantaneous increase in calcium followed by an exponential decay. A regression-based approach in which CIRF-convolved eye position and ipsiversive velocity variables served as regressors in a linear model fit to calcium-sensitive fluorescence image series can be used to identify in an unbiased way putative hVPNI neurons23. Selection is based on significance measurements of the correlation of each pixel’s fluorescence time series with these regressors.
We employed this method to identify neurons, focusing all subsequent analysis on somatic regions of interest (ROIs) whose fluorescence time series had a Pearson correlation of > 0.5 with either CIRF-convolved eye position or ipsiversive velocity (n = 691 “identified” cells, combined dark and light conditions). Examples of identified somata are outlined in the time projection of an image time series in Fig. 2b. Fluorescence time series for these cells show clear correspondence with the CIRF-convolved eye position and ipsiversive velocity time series. We observed a great deal of diversity across cells in the response structure during fixations. Some cells showed relatively stable fluorescence levels during fixations (e.g. Fig. 2b–1 and 2b–2), while others showed decaying fluorescence (e.g. Fig. 2b–4 and 2b–5).
Figure 4.
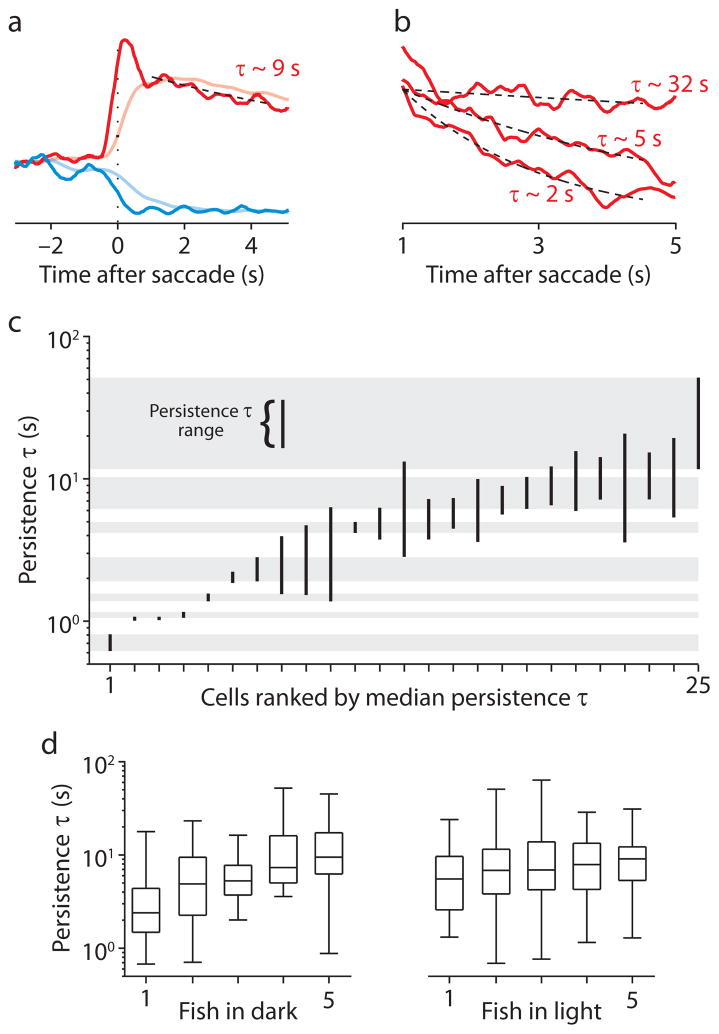
The distribution of persistence time ranges within individual larvae. (a) Representative ipsiversive (red) and contraversive (blue) STA fluorescence responses, before (translucent) and after (opaque) deconvolution with a CIRF. The mean of the first 1.5 s of each trace is subtracted off. The black dashed line is the fit of an exponential function (τ= 9.26 s) to the deconvolved ipsiversive STA response from 1 to 4.5 s after saccade time. (b) Ipsiversive STA fluorescence responses from three cells in one fish from 1 to 5 s after saccade time. Black dashed lines show exponential fits with time constants indicated. Traces are normalized to the value of the fit 1 s after saccade time. (c) Persistence time ranges for 25 cells imaged in one fish in the dark. The vertical extent of each bar spans the persistence time range for each cell. Gray bars indicate non-overlapping persistence time ranges. Cells are ordered according to their rank in terms of the median of their persistence time distribution. (d) Box plots representing the distribution of median persistence times for fish imaged in the dark (left panel) or the light (right panel). Horizontal lines indicate the minimum, 25th percentile, median, 75th percentile, and maximum values of each distribution.
Figure 5.
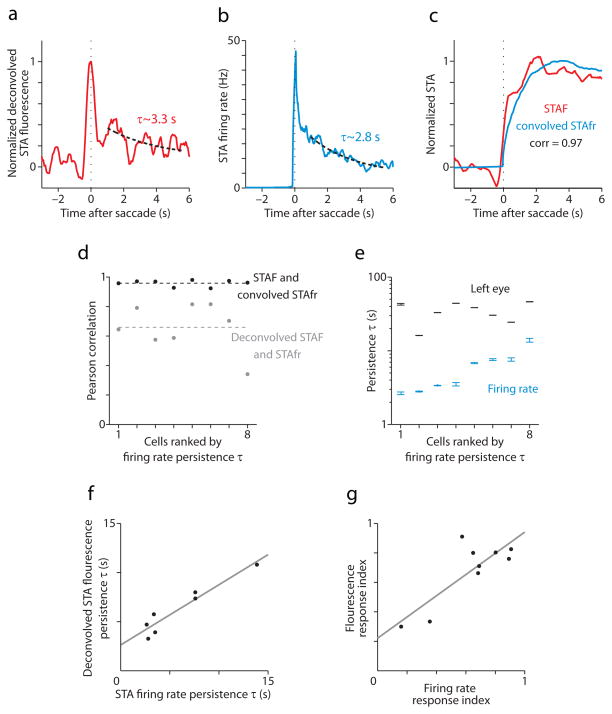
Agreement between electrical and optical recording-based parameterizations of saccade-related activity (a) CIRF-deconvolved ipsiversive STA fluorescence response for an identified position-sensitive neuron prior to electrical recording. The black dotted lines in (a) and (b) are fits of exponential functions to STAs from 1 to 4.5 s after saccade time. (b) Ipsiversive STA firing rate from targeted electrical recording of the cell from which (a) was calculated. (c) Ipsiversive STA fluorescence response (red) and CIRF-convolved STA firing rate (blue) for the same neuron. The convolved trace was normalized to have a maximum of 1. The fluorescence trace had the mean of the first 1.5 s subtracted off, then it was scaled by a constant to minimize its mean squared difference from the convolved trace (this did not affect correlation measurements). (d) Pearson correlation for ipsiversive STA fluorescence and CIRF-convolved STA firing rate (black circles) and for CIRF-deconvolved STA fluorescence and STA firing rate (gray circles) for 8 electrically-recorded position cells. Dashed lines represent the mean correlation for each comparison. (e) 95% confidence intervals for the persistence time estimates (blue) of 8 electrically-recorded position cells, along with 95% confidence intervals for the persistence time of left eye position (black) measured simultaneously. (f) Scatterplot of persistence times estimated from deconvolved STA fluorescence and STA firing rate for cells for which both estimates were > 0 (7/8 position cells). In (f) and (g), the gray line is a least-squares best fit to the data. (g) Scatterplot of response indices calculated from STA fluorescence and STA firing rate (9 total cells).
Halorhodopsin-mediated hVPNI localization
Before further analyzing the variability in identified eye movement related neurons, it is important to demonstrate that these neurons are part of the hVPNI. We have previously shown that laser ablation within the hindbrain region imaged here produces deficits in the ability of zebrafish larvae to hold their eyes at eccentric positions23 without eliminating saccades. This supports the notion that neurons within this region belong to the hVPNI. In adult preparations, transient pharmacological inactivation has been used to identify the hVPNI20,28. However, such experiments are difficult to perform in a larval nervous system. Given the importance of demonstrating this for the present work, we decided to provide additional evidence using a method for transient optical silencing of neuronal firing analogous to pharmacological inactivation.
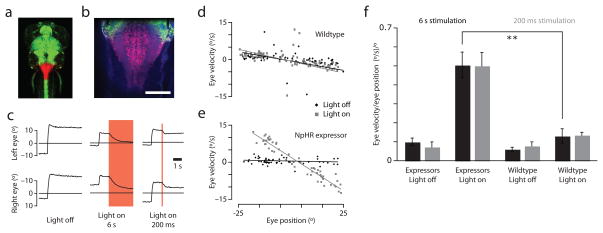
We used previously described transgenic zebrafish expressing halorhodopsin (NpHR) in most neurons29 to perturb firing in the hindbrain region imaged above. A similar method was used previously to localize saccade-generating neurons in rhombomere 530. 633 nm illumination from a low NA optical fiber was used to focally stimulate NpHR in an ~200 micron diameter column29 covering the caudal hindbrain (primarily rhombomere 7/8) while eye position was measured (Fig. 3a,b). Both long (6 s) and short (200 ms) activations markedly reduced eye position stability, causing the eyes to drift rapidly towards the center of gaze (Fig. 3c); centripetal drift was observed regardless of which side of this center the eyes were on prior to activation (Fig. 3e). The changes in induced eye position were persistent, as the eyes remained at their new positions following the termination of illumination until the subsequent saccade (Fig. 3c, right panels). Eye stability was quantified as the slope of linear fits to plots of eye velocity versus eye position (PV plots; Fig. 3d,e). During illumination, eye positions in NpHR expressing animals were significantly less stable than those in wildtype larvae (Fig. 3f; p = 0.0007 for short and p = 0.0015 for long activations), as is observed with pharmacological inactivation of the hVPNI in adults20,28.
Figure 3.
NpHR-mediated silencing of the caudal hindbrain reduces eye position stability. (a) Dorsal view of a 6 dpf larva transgenic for Et(E1b:Gal4-VP16)s1101t, Tg(UAS:NpHR-mCherry)s1989t, Tg(UAS:Kaede)s1999t. The photoconverted region (red) in the caudal hindbrain indicates the volume illuminated in experiments analyzed in (c-f). (b) Close-up of the photo-activated region with autofluoresence in blue. Scale bar = 100 μm. (c) Eye traces of the animal in (a,b). Left: A spontaneous saccade without illumination. Middle: NpHR is activated in the caudal hindbrain one second after a spontaneous saccade (red shade). The eyes drift back toward the center of gaze. Right: Short NpHR activation (red shade) induces transient eye drift. (d,e) Position-Velocity plots for 200 ms illumination experiments with least-squares best fit lines of slope k. (d) No significant eye velocity increase is observed in a wildtype animal during illumination (Light On: k = −0.16, Light Off: k = −0.16). (e) Illumination of the hindbrain in the NpHR-expressing animal shown in (a) induces large increases in eye velocity across eye positions (Light On: k = −0.54, Light Off: k = −0.02). (f) Quantification of k across animals (n = 4–5 each). Error bars = sem.
Based on measurements of the effect of localized optical fiber-based NpHR activation on firing in hindbrain neurons in the strains used for these experiments29, we expect such activation to markedly reduce the average firing rate across these neurons. The demonstration that NpHR activation in neurons of the caudal hindbrain diminishes gaze stability and permanently alters eye position, when combined with our previous results obtained with laser ablation, demonstrates that the neurons we imaged include the hVPNI.
Distributed firing rate persistence times
To quantify the dynamics of fluorescence changes, ipsiversive and contraversive saccade-triggered average (STA) fluorescence responses were first computed to represent these changes with an improved signal-to-noise ratio. Only fluorescence measurements surrounding saccades for which the previous saccade was in the opposite direction were included in these averages. This ensured that STAs captured fluorescence changes averaged over repeats of highly similar behaviors. When compared to STA convolved eye velocity (Fig. 2e) and eye position (Fig. 2f), these averages reveal a range of relative ipsiversive velocity and position dependences, from the highly ipsiversive velocity-correlated (e.g. Fig. 2g) to the highly position correlated (e.g. Fig. 2i). Most responses resided between these two extremes (e.g. Fig. 2h).
To quantify this variation in relative dependences, a response index was computed for all identified cells (see Methods) that is zero for purely velocity-correlated time series and one for those purely position-correlated (Fig. 2j). Index values varied widely within individual fish; the width of the middle 95% of their distribution within fish ranged from 0.59 to 0.93 in the dark (n = 6 larvae with ≥ 18 identified cells) and 0.83 to 0.97 in the light (n = 6 larvae with ≥ 18 identified cells). The wide range of sensitivities among cells demonstrated by this distribution is similar to that observed in sequential single unit recordings of putative hVPNI neurons in cats31 and monkeys9. Our simultaneous measurements of activity in many cells confirm that such heterogeneity is characteristic of population activity in the hVPNI.
Because the fixations typical in our larval preparation are long (generally > 5 s), we were able to use OGB-1 STA fluorescence responses to examine the uniformity of firing rate persistence across many identified hVPNI neurons in individual larvae. Our strategy was to first compute STA firing rate estimates from STA fluorescence responses by deconvolving fluorescence responses with the CIRF determined individually for each cell (Supplementary Fig. 2). The CIRF decay time constant τ was calculated for each neuron from an analysis of the fluorescence decay following contraversive saccades23 (see Methods). This analysis was limited to cells whose fluorescence had a Pearson correlation of > 0.5 with CIRF-convolved eye position (n = 455 “position” cells) to include only cells having robust persistent activity, and further to cells with CIRF τ estimates of reasonable quality (R2 > 0.5, n = 416).
The estimated firing rate for most cells appeared to decay over several seconds during fixation toward a baseline level associated with the center of gaze. However, across cells within individual fish, STA firing rate estimates displayed extensive variation in the rate of this decay, and thus in the degree of firing rate persistence. To quantify this persistence, time constants from exponential fits to firing rate estimates were used to measure persistence time (Fig. 4a,b; Supplementary Fig. 3; see Methods). Because of some ambiguity in CIRF τ and the baseline level to which firing rate estimates decay, each estimate is consistent with a distribution of persistence times. We refer here to the extent of the middle 99% of this distribution as the persistence time range.
Because the canonical line attractor model predicts that persistence times among hVPNI neurons would be similar (Fig. 1a), we sought explicit evidence for non-overlapping persistence time ranges in individual larvae. Several non-overlapping persistence time ranges were observed in each larvae both in the dark (n = 5 larvae with ≥ 16 position cells) and the light (n = 5 larvae with ≥19 position cells), consistent with the presence of multiple distinct persistence times across cells within individual fish. Fig. 4c shows the persistence time ranges calculated for 25 cells recorded in one larvae in the dark. The gray bars span the ranges for seven cells for which the ranges do not overlap. For each larva, persistence time distributions were statistically unlikely to be drawn from a single underlying distribution (Kruskal-Wallis test, p < 10−10 for each larvae, n = 5 in the dark and n = 5 in the light). The median values from persistence time distributions varied by approximately an order of magnitude or more within individual larvae, both in the dark and in the light (Fig. 4c,d; Supplementary Fig. 4a). The width of the middle 95% of the distribution of these medians within larvae ranged from 13.41 to 48.00 s in the dark (n = 5) and 21.63 to 62.76 s in the light (n = 5). The shape and width of these distributions of medians was not sensitive to the choice of thresholds for data inclusion (Supplementary Table 1). The coefficient of variation for these distributions of medians ranged from 0.58 to 0.94 in the dark (n = 5) and 0.70 to 1.20 in the light (n = 5).
The presence of multiple distinct persistence times within individual larvae is further illustrated by average STA firing estimates for cells grouped according to median persistence time (Supplementary Fig. 5). Persistence time heterogeneity is still observed if firing rate estimates are calculated for all cells with a single CIRF instead of CIRFs for which τ was determined cell-specifically (Supplementary Fig. 5g–i).
We validated the finding of persistence time heterogeneity using a second approach in which a function modeling the convolution of underlying firing with a CIRF was fit directly to STA fluorescence responses (see Methods). This approach yielded similar heterogeneity in persistence time estimates (Supplementary Fig. 4b). Although single exponential decays do not capture all of the structure in ipsiversive STA firing rate estimates (Supplementary Fig. 6a), they adequately capture the magnitude of firing rate decay during fixation, which is sufficient to reveal heterogeneity in persistence time across cells. Similar heterogeneity is also observed via a different metric of persistence time, the fractional change of STA firing rate estimates during fixation (Supplementary Fig. 6b). The presence of multiple distinct persistence times is inconsistent with neural integrator models characterized by uniform firing rate persistence across cells, like the canonical line attractor-based circuit model.
Validation of fluorescence-based firing rate estimates
To confirm that persistence time estimates from fluorescence reflect the actual dynamics of action potential firing, these estimates were compared with persistence times determined from electrical recordings from the same cell. Following image time series acquisition, single-unit loose-patch recordings were made from cells identified using our online regression-based strategy23. Persistence times were calculated for recorded neurons both from deconvolved STA fluorescence (Fig. 5a) and STA firing rates (Fig. 5b). The correlation between deconvolved STA fluorescence responses and STA firing rates was strong (Fig. 5a,b, and d). Similarly, CIRF-convolved STA firing rate was highly correlated with STA fluorescence (Fig. 5c,d). The somewhat lower correlation for deconvolved STA fluorescence and STA firing rate is likely due to the amplification of noise in fluorescence measurements upon deconvolution.
Persistence times calculated from STA firing rates ranged from 2.65 to 13.87 s (n = 8; Fig. 5e). The correlation between these values and persistence times calculated in a similar fashion for the simultaneously measured ipsilateral (left) eye position was 0.30, suggesting that only a small amount (~10%) of the variance in persistence times can be explained by variation in the stability of the eyes themselves. This fact, coupled with the greater than 5-fold difference between the shortest and longest persistence times is itself strong evidence of heterogeneity in firing rate persistence across hVPNI cells. Moreover, persistence times estimated from fluorescence and firing rate for the same neurons show strong agreement (Fig. 5f; slope = 0.62; R2 = 0.91, rms error = 0.88 s). These results confirm that the distribution of persistence times measured from STA fluorescence responses reflects a similar distribution of persistence times for the underlying firing rates. An analogous validation was performed for response indices. Correlations between firing rate time series and time series of both eye position and ipsiversive velocity were used to calculate a firing rate-based response index for recorded neurons. These values agree well with response indices calculated from fluorescence for the same neurons (Fig. 5g; slope = 0.77; R2 = 0.69, rms error = 0.13). This suggests that fluorescence-based response indices report relative burst and position sensitivities in firing rates with an accuracy sufficient to conclude that the distribution of response indices we observed corresponds to a distribution of relative sensitivities in firing rates.
Topography of pairwise correlation, persistence time, and response index
In addition to providing measurements of activity from many cells simultaneously, optical recording identifies the precise anatomical location of each recorded cell. We used this information to examine whether the variation in saccade-related firing presented above corresponded to a topographic organization among neurons. Topography was observed in terms of three quantities: (1) correlation between fluorescence time series, (2) firing rate persistence time, and (3) response index.
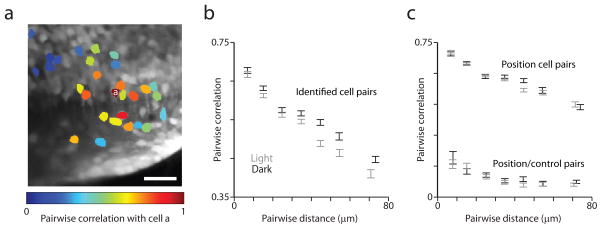
The Pearson correlation for fluorescence time series was calculated for all pairs of somata of identified cells within individual image time series. When the correlation with a particular cell was plotted using color-coded somata for all other identified cells--this included position cells and those that may be exclusively ipsiversive velocity-sensitive--nearby cells appeared to be more highly correlated (Fig. 6a). When the data from all image time series were combined, pairwise correlation was significantly negatively correlated with the distance separating the cells (“pairwise distance”) in three dimensions both in the dark and light (Fig. 6b; in the dark: Spearman rank correlation (src) = −0.36, p < 10−9; in the light: src = −0.33, p < 10−9). The same held true for pairs of position cells (Fig. 6c; in the dark: src = −0.36, p < 10−9; in the light: src = −0.34, p < 10−9). Significant dependence of pairwise correlation among position cells was seen for pairwise distance along both the rostrocaudal (in the dark: src = −0.29, p < 10−9; in the light: src = −0.24, p < 10−9) and dorsoventral (in the dark: src = −0.23, p < 10−9; in the light: src = −0.27, p < 10−9) axes.
Figure 6.
Activity correlations between cells depend on their pairwise distance. (a) Time projection of an image time series with 29 identified neurons color-coded according to the Pearson correlation of their fluorescence time series with that of the cell labeled “a”. (b) Pairwise correlation of fluorescence time series for pairs of identified cells versus pairwise distance for data collected in the light (gray) and in the dark (black). For plots (b) and (c), pairs were grouped according to pairwise distance, data are plotted along the x-axis according to the mean pairwise distance for each group, and error bars = mean ± sem. (c) Pairwise correlation of fluorescence time series for position cell pairs and position/control cell pairs versus pairwise distance for data collected in the light (gray) and in the dark (black).
In order to rule out the possibility that these correlations stemmed from specimen motion-induced correlations or fluorescence from processes overlapping cellular ROIs out of the image plane, we examined the proximity dependence of the correlation between position cells and a control population of non-position-dependent cells (absolute value of Pearson correlation with CIRF-convolved eye position < 0.15). Pairs of position and control cells showed pairwise correlation that was weakly distance dependent at short distances but was not significantly dependent on distance beyond 35 μm (Fig. 6c). For position/control cell pairs ≥ 35 μm apart, the src of pairwise correlation and pairwise distance was −0.02 (p = 0.60) in the dark and −0.02 (p = 0.50) in the light. However, for position cell pairs ≥ 35 μm apart, the src of pairwise correlation and pairwise distance was −0.29 (p < 10−9) in the dark and −0.16 (p < 10−9) in the light. These results suggest that the proximity dependence of correlation between identified position neurons is unlikely to be explained by motion-induced fluorescence correlations or overlapping processes.
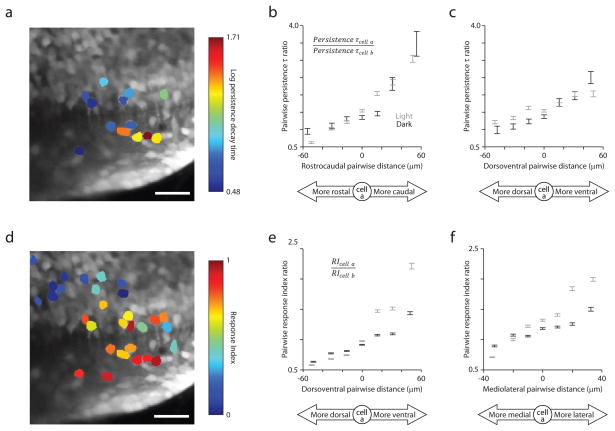
Topography was also seen among cells in terms of persistence time. Strikingly, we observed gradients of persistence time along spatial dimensions. For pairs of position cells, we quantified the similarity between persistence times using their ratio. To introduce directionality, the ratio’s denominator was the value for the cell relative to which pairwise distance was measured. We found a clear dependence of this ratio on pairwise distance along individual spatial dimensions (Fig. 7a–c; Supplementary Fig. 7a). The src of this persistence time ratio with rostrocaudal, dorsoventral, and mediolateral distance were 0.22, 0.28, and 0.06, respectively in the dark; and 0.35, 0.11, and −0.21, respectively in the light. The magnitude of these correlation values depends only weakly on the choice of thresholds for data inclusion (Supplementary Table 2). The dependence is most prominent across both dark and light conditions along the rostrocaudal axis (Fig. 7a,b; Supplementary Figs. 8,9). In the dark, higher persistence time cells tend to be located more caudally and ventrally than low persistence time cells.
Figure 7.
Persistence time and response index similarity depend on pairwise distance along spatial dimensions. (a) Time projection of an image time series with 13 position neurons color- coded according to the log10 of their persistence time. (b),(c) Persistence time ratio versus pairwise distance along the rostrocaudal (b) and dorsoventral (c) axes for pairs of position neurons. Data are included from cells for which persistence time estimates were > 0. For plots (b),(c),(e), and (f), pairs identified in the light (gray) or dark (black) were grouped according to pairwise distance along particular spatial dimensions, data are plotted along the x-axis according to the mean pairwise distance for each group, and error bars = mean ± sem. (d) Time projection of the same image time series with 29 identified neurons colored-coded according to their response index. (e),(f) Response index ratio versus pairwise distance along the dorsoventral (e) and mediolateral (f) axes for pairs of identified neurons.
Response index similarity had a somewhat different spatial dependence across all identified cells (Fig. 7d–f; Supplementary Fig. 7b). The response index ratio varied most prominently over the dorsoventral and mediolateral axes. The src of response index ratio with rostrocaudal, dorsoventral, and mediolateral distance were 0.05, 0.30, and 0.19, respectively in the dark; and −0.10, 0.45, and 0.32, respectively in the light. Collectively, these dependencies between saccade-related firing and proximity mark the first observation of spatial structure on the length scale of our measurements (< 100 μm) among neurons implicated in neural integration.
Discussion
We report two major results on functional organization of the hVPNI. First, our optical recordings reveal a heterogeneity in firing rate persistence. Second, we observed persistence times graded along the rostrocaudal and dorsoventral axes, implying that cells with similar persistence times tended to be closer together in space. This topography represents the first elucidation of functional organization within a neuronal circuit subserving temporal integration or parametric short-term memory in general on a sub-100 μm length scale.
Advances in methodology are also demonstrated by our study. The heterogeneity we observed in the relative position and ipsiversive velocity sensitivity is similar to findings from electrode studies in adult vertebrates, supporting larval zebrafish as a model for investigating the mechanisms and development of neural integration. In addition, the correspondence demonstrated between electrode recordings and CIRF-deconvolved optical recordings establishes that the latter can be a reliable reporter of collective neuronal dynamics.
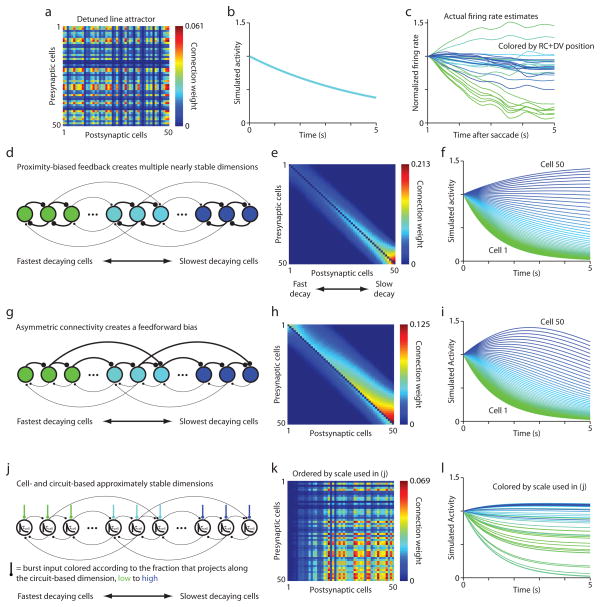
To explain the apparent continuum of stable eye positions observed during fixations in adult vertebrates, hVPNI firing rate dynamics have been modeled as approximating a line attractor11,14. In a state space where each axis represents one neuron’s firing rate, a line attractor is a continuous line of stable points, each representing the set of firing rates corresponding to one horizontal eye position (Fig. 1 in reference 12). This line represents the continuum of eye positions possible during fixations. Both eye position and hVPNI neuronal firing rates are observed to slowly drift centripetally during fixations, suggesting that positions along the line are only semi-stable, with the circuit’s representation in state space drifting slowly along the line toward a single fixed point corresponding to the center for gaze. The firing of all hVPNI neurons would then decay at a uniform rate (Fig. 8a,b), implying a one-dimensional circuit dynamics. However, we observed hVPNI neuron firing decaying on multiple time scales (Fig. 8c), inconsistent with the line attractor model as originally formulated11 and directly demonstrating that the dynamics are multidimensional.
Figure 8.
Mechanistic implications of heterogeneity in dynamics. (a),(b) Connectivity matrix (a) and numerical simulation results (b) for a circuit constructed to generate a line attractor, then subjected to uniform detuning of connection weights to achieve a persistence time of ~5 seconds (approximately the median for the cells from which we estimated this quantity). The traces in (b), which all overlap, show the time courses for 50 neurons from simulations. (c) Firing rate estimates for 29 neurons imaged in one larvae in the dark. Saccade-triggered fluorescence was smoothed by projection onto principal components capturing > 99% of the variance in the data before deconvolution. Traces are colored according to the sum of the rostrocaudal and dorsoventral location of each neuron, revealing again the correspondence between persistence time and location along these dimensions. (d) Recurrent feedback circuit in which neurons nearby in space with more similar persistence times are more strongly coupled. This generates multiple approximately stable dimensions in firing rate state space. The colors of cells in (d) and (g) correspond to the colors of simulation results in (f) and (i), respectively. (e),(f) Connectivity matrix (e) and numerical simulation results (f) for a network constructed with the features of (d). Cells are ordered in (e) and (h) from 1 to 50 in order of increasing persistence. (g) Asymmetric recurrent connectivity with a feedforward bias. Strong local coupling creates an ordering in which the activity in cells is more persistent further down the feedforward chain partially due to progressive input filtration. (h),(i) Connectivity matrix (h) and numerical simulation results (i) for a network constructed with the features of (g). (j) Cell-intrinsic processes generate one time scale of persistence decay (τcell) and tuned recurrent feedback generates another. By tuning the inputs to cells to differentially express these two decay times, a continuum of apparent persistence times can be achieved. (k),(l) Connectivity matrix (k) and numerical simulation results (l) for a network constructed with the features of (j). Cells are ordered in (k) from 1 to 50 and colored in (l) according to the fraction of their burst input that projects along the circuit-based dimension, from lowest to highest.
Previous studies of the adult hVPNI have not systematically examined the uniformity of firing rate drift dynamics across neurons during fixations. Our discovery of persistence heterogeneity was facilitated by optical recording, which provided simultaneous activity measurements from groups of hVPNI neurons. Heterogeneity could also be masked in adults by the fact that persistence times increase while the duration of individual fixations decreases, making it more challenging to observe and then quantify differences across neurons. However, a wide range of persistence times (~4 s to −21 s) from cells measured across many adult goldfish has been previously reported18. These measurements were not simultaneous and bounding these estimates or quantifying their relationship to variation in integrator performance across fish would be required for a direct comparison with our results. The authors further found extensive evidence of persistence heterogeneity in sequential electrode recordings in goldfish whose hVPNI were detuned via erroneous visual feedback18. The larval zebrafish we examined here show fixation stability like that of a somewhat detuned adult, raising the possibility that persistence heterogeneity becomes less prominent in a properly tuned adult hVPNI. This would imply that the persistence time distribution may narrow during development.
Exploring this idea quantitatively, we re-analyzed previously published simultaneous recordings from pairs of adult goldfish hVPNI neurons during spontaneous eye movements32 to estimate the percent difference in persistence time between each pair. The median percent difference was 34% (n = 6 ipsilateral pairs). We then calculated the percent difference between persistence times for pairs of simultaneously imaged zebrafish neurons and found a median percent difference of 69% (n = 1576 ipsilateral pairs). The two data sets were statistically unlikely to be drawn from a common distribution (Wilcoxon test, p = 0.048). Our results suggest both that some persistence heterogeneity exists in adult goldfish and that larval heterogeneity may be comparatively more exaggerated. However, a more systematic characterization of adult persistence times present would be necessary to directly compare larval and adult persistence time distributions.
Persistence time heterogeneity is consistent with other findings in adults beyond those mentioned above, further supporting its existence in the adult hVPNI. First, during sinusoidal vestibular stimulation, the dependence on stimulus frequency of putative hVPNI neurons’ firing phase relative to eye position9,33 is captured by models in which cells show non-uniform firing rate decay 17,34. Second, hysteresis in the dependence between firing rates of hVPNI neurons during fixation is inconsistent with canonical line attractor dynamics35 yet would be expected if multiple time scales of decay were present to varying degrees in these firing rates. Third, the results of experiments on monocular optokinetic stimulation point to a lack of uniformity in firing dynamics across hVPNI neurons36. Finally, evidence for a broad distribution of relaxation times in the eye plant37 suggests that diverse persistence times in hVPNI neurons could contribute to gaze stabilization. Optimal control of such a plant would necessitate neuronal drive to eye muscles with a similar distribution of relaxation times to stabilize gaze; this could be provided by appropriately patterned innervation of motor neurons from hVPNI neurons with different persistence times.
The topography of hVPNI neuronal persistence we observed on sub-100 μm length scales has not been previously reported; however, the poor spatial resolution of electrode recording would preclude characterization of topography on this scale. The response index gradients we found are analogous to the rostrocaudal gradients in relative position and velocity sensitivity seen among hVPNI neurons in cats 31, although similar organization was not observed among goldfish hVPNI neurons10. Looking on a shorter length scale, we did not see a prominent rostrocaudal gradient in relative sensitivity, yet saw a strong dependence of persistence time along this dimension.
The topography we observed may reflect developmental patterning within the nascent hVPNI circuit. A recent study found that older cells reside more ventrally in the caudal hindbrain38 during early larval stages, suggesting that relative position sensitivity and persistence time may increase with neuronal age during these stages. This could reflect synaptic input tuning that improves firing rate persistence as neurons mature. This study also found that neuropil clusters according to neuronal age in the hindbrain38. This clustering may increase the connectivity likelihood for similarly-aged neurons, thereby increasing this likelihood for neurons with similar persistence. Such clustering could contribute to a proximity bias in connectivity likelihood that may explain the spatial gradients we observed. Spatial gradients in the anatomical position of spinal motor neurons and premotor interneurons subserving fast and slow swimming have also been reported in the larval zebrafish24,39. Our observations add to the evidence that spatial gradients in the dynamical properties of neurons within circuits may be a general property of anatomical organization in the developing vertebrate central nervous system. Though continued migration may alter the spatial arrangement of cells, the synaptic connections forged during development can contribute to the functional organization of adult circuits.
In general, correlation-proximity relations can be ascribed to convergent afferent input. However, in the hVPNI, saccadic burst input is transient, sustained vestibular input is not eye position dependent40,41, and persistent firing encoding eye position must emerge locally via recurrent excitation and/or cell-intrinsic mechanisms20. Convergent afferent input from neurons external to the hVPNI does not seem a tenable explanation. However, if a new source of eye position feedback to the caudal hindbrain was discovered, convergent input would warrant reconsideration.
Persistence heterogeneity is consistent with hVPNI circuit models dynamically distinct from the canonical line attractor model. The classic line attractor exhibits one approximately stable dimension in firing rate state space along which firing rates change slowly, while persistence heterogeneity requires mechanisms generating slow firing rate dynamics along multiple dimensions. Interestingly, integrator models with slow dynamics along multiple dimensions will be more robust than line attractor-type models to local perturbations of the circuit such as the loss of a neuron11,21.
One possibility is that locally-biased synaptic feedback amongst hVPNI neurons generates a multidimensional attractor with several approximately stable dimensions (Fig. 8d). Simulations of such a model can produce persistence time distributions qualitatively similar to those experimentally observed (Fig. 8e,f). Here, the variation in persistence emerges from structured connectivity in which cells with more similar (disparate) persistence times tend to be more strongly (weakly) coupled. If the spatial gradients we observed emerge from a proximity bias in the likelihood any two neurons are connected, they would imply exactly this type of circuit structure.
A second possibility is that persistence heterogeneity arises by a progressive filtering of activity propagating down a feedforward cascade22. Such circuits need not be purely feedforward, and may involve a mixture of feedback and feedforward connections, or even functionally feedforward interactions. An example of a circuit utilizing feedforward dynamics is shown in Fig. 8g, where locally-biased synaptic connectivity is modified with a feedforward bias, generating firing rate dynamics that resemble those seen in our data (Fig. 8h,i). In general, the activity of all neurons in this circuit type eventually decreased during simulated fixations if the contribution of feedforward activity was substantial. Further work would be needed to address whether this circuit type is capable of generating persistence on longer time scales when the number of neurons is of the order considered here.
A third possibility is that two distinct time scales of firing rate decay are generated separately by recurrent feedback and a cell-intrinsic mechanism (Fig. 8j). Previous results following partial inactivation of the goldfish hVPNI are consistent with the presence of a cell-intrinsic mechanism generating 1–2 s firing rate persistence. Differential manifestation across cells of this persistence and a longer time scale persistence due to recurrent feedback could create varying persistence times. The topography we observed would then suggest that saccadic burst input to the hVPNI is spatially structured to vary the degree to which each time scale dictates a cell’s persistence. Simulations demonstrate that such a model can recapitulate spatially-graded persistence heterogeneity (Fig. 8k,l).
Recent theoretical results have demonstrated that randomly-connected recurrent networks with heterogeneous unit activity can execute functional transformations like temporal integration19,42. Investigating such networks may identify further circuit architectures that can generate the firing dynamics we observed. In general, while our measurements in larvae expose a multidimensional firing dynamics that can reconcile a number of observations from adults, the precise hVPNI circuit architecture instantiating these dynamics remains the subject of future work. Emerging techniques for comprehensive connectivity reconstruction could further these efforts43.
hVPNI neurons by definition must have eye position-dependent firing, and eye position, but not saccades themselves, must causally depend on that firing. Previous results demonstrate that laser ablation in the caudal hindbrain region studied here, which contains neurons with position-dependent firing, reduces gaze stability23. Here we additionally show that halorhodopsin-mediated perturbation of firing in an ~200 micron diameter focal area encompassing the same region induces rapid centripetal drift in eye position, causing position changes that persist following termination of halorhodopsin activation. These complementary results localize hVPNI functionality to neurons in this hindbrain region.
An important question is whether all neurons we have designated as position cells and estimated persistence times for belong to the hVPNI, since other distinct functional oculomotor populations display neuronal firing with related parameter sensitivity. We expect few if any of our position cells to be exclusively ipsiversive velocity-sensitive like saccadic burst neurons, since electrical recordings from cells meeting our criteria for classification as position cells revealed eye position-dependent firing in all cases23. We further expect the fraction of designated position cells participating instead in the velocity storage integrator to be very low. Neurons in this population show firing that depends only weakly on eye position during spontaneous eye movements in adult goldfish44 and its anatomical location in larval zebrafish45 overlaps minimally with the region we imaged.
Assuming that halorhodopsin activation directly hyperpolarizes hVPNI neurons rather than just their tonic inputs (as suggested by centripetally drifting exponential relaxation in eye position), the effects of halorhodopsin activation are inconsistent with some hypothesized cell-intrinsic neural integration mechanisms . Though circuit mechanisms of integration have been strongly implicated in the hVPNI20,46,47, a contribution from cell-intrinsic mechanisms cannot be excluded7. In contrast to proposed membrane voltage-independent mechanisms48–50, the persistent change of firing in hVPNI neurons following transient hyperpolarization we observed suggests that both cellular and circuit contributions to integration are membrane-voltage dependent. Further studies using optogenetic probes to perturb activity in hVPNI neurons could be fruitful in elucidating the cellular and circuit mechanisms underlying neural integration in the hVPNI.
Methods
All experiments were performed in compliance with protocols approved by the Princeton University Institutional Animal Care and Use Committee. mitfa−/− (nacre) mutant zebrafish larvae aged 6–12 days post-fertilization (dpf) ranging in length from 4.0 to 4.8 mm were used for all experiments. Methods for functional imaging, eye tracking, and targeted electrical recording are described elsewhere23.
Functional inactivation
Animals (5–6 dpf) were transgenic for a combination of transgenes: Et(E1b:Gal4-VP16)s1101t, Tg(UAS:NpHR-mCherry)s1989t, Tg(UAS:Kaede)s1999t. “Wildtype” refers to siblings of NpHR expressors that were potentially expressing Gal4 and Kaede. Animals were mounted in agarose in a 35 mm dish and the agarose was removed around the eyes. A 200 μm diameter optic fiber was placed above the caudal hindbrain to activate NpHR (633 nm, 50 mW/mm2). Animals were imaged at 20–30 frames/s under infrared illumination. Eye tracking was performed using a custom LabView program that detected saccades and triggered fiber illumination one s after half of saccades (both ipsiversive and contraversive) Only animals that showed a high frequency of saccades into both directions were used. Experiments were not performed in the dark, since scattered light from the fiber sometimes triggered saccades. Instead, a white backlight was used in addition to the infrared light in order to reduce the salience of the fiber light. 400 ms segments of eye traces were fit using linear regression in order to measure eye velocity for PV plots. Control measurements of eye stability in the absence of illumination found no significant difference between NpHR expressors and wildtypes.
Data analysis
All analysis was completed in Matlab v.7.8 or 7.10 (Mathworks). The identification of somata corresponding the eye position- and ipsiversive velocity encoding neurons from fluorescence image time series is described elsewhere23.ROI time series were calculated as the average of pixel time series for all included pixels. Eye position correlation, cp , and ipsiversive velocity correlation, cv , for each somata were defined as the Pearson correlation coefficient between the ROI time series and CIRF-convolved eye position and CIRF- convolved ipsiversive velocity, respectively. The significance threshold used in cell identification was set empirically so that identified somata had cp ranging below 0.5 for some somata. This ensured that all somata with cp > 0.5 were likely identified. The cellular response index, RI, was defined as
Response indices were calculated for somata for which either cp > 0.5 or cv > 0.5. Response indices were calculated from firing rate data using the same formula but with cp and cv defined using firing rate and unconvolved eye position and ipsiversive velocity measurements. Saccade identification and STA generation are described elsewhere23. We reiterate here that data inclusion criteria were employed that ensured included saccades had reasonably uniform sizes and preceding histories, as is indicated by measurements of the distribution of eye positions surrounding included saccades. For each STA response, we calculated the coefficient of variation (CV) for eye positions immediately preceding (mean from 400 to 300 ms before) and following (mean from 500 to 600 ms after) included saccades. For all ipsiversive STAs, the mean CV was 0.26 for pre-saccadic eye positions and 0.14 for post-saccadic eye positions. For all contraversive STAs, the mean CV was 0.18 for pre-saccadic eye positions and 0.16 for post-saccadic eye positions. Only 1.7% of included saccades did not cross the center of gaze. STA fluorescence responses and firing rates were convolved with a 200 ms box filter before subsequent analysis.
A deconvolution-based approach was used to calculate firing rate estimates for optically-recorded cells. To do so, CIRFs were first estimated for cells23 for which cp > 0.5 (n = 455 “position” cells; this eliminates cells that may not have strong persistent activity during fixations) Confidence intervals (Matlab function confint) ranging between 0 and 100% in width were calculated for the CIRF τ estimate from the fit in order to populate a distribution representing this estimate. This method may systematically underestimate the CIRF τ for cells whose fluorescence has not completely decayed prior to subsequent saccades. However, we expect this underestimation to have minimal impact on firing rate estimation since the median CIRF τ · 3 is less than the mean saccade frequency of ~10 seconds23 for 94% of position cells. Deconvolutions (Matlab function deconv) and convolutions (Matlab function conv) were performed with STAs padded on either end with 5 second long stretches of zeros using a 5 second long stretch of an exponential decay of a given CIRF τ.
Persistence time estimates were made for cells for which R2 > 0.5 for the fit to estimate CIRF τ (416/455 cells) according to the following four steps, which were repeated 1000 times for each cell (Supplementary Figure 3). First, ipsiversive and contraversive STA fluorescence responses were deconvolved with a CIRF defined by a τ chosen randomly from that cell’s CIRF τ estimate distribution, generating firing rate estimates. Second, these estimates were used to define the possible range of the baseline null value to which the firing rate decays after saccades in either direction. The lower bound for the null value was estimated as the mean of the ipsiversive firing rate estimate between 1 and 2 seconds before the saccade time. The upper bound was estimated as the average of the means of ipsiversive and contraversive STA firing rate estimates 4 to 4.5 seconds following the saccade time (Supplementary Figure 3). These bounds were chosen because they account for the possibility that neurons may be hyperpolarized below firing-threshold after contraversive saccades. Third, a null value was chosen randomly from a uniform distribution of values between the bounds calculated in the previous step. Lastly, Ae−t/τ was fit to the ipsiversive STA firing rate estimate from 1 to 4.5 seconds following saccade time after subtracting off the null value. The decay time τ from this fit is an estimate of the persistence time. Confidence intervals ranging between 0 and 100% in width were calculated for the persistence τ estimate in order to populate a distribution representing the estimate. These estimate distributions from all 1000 iterations were collected into one distribution that characterizes the persistence time for a given cell. We defined the middle 99% of this collective distribution as a cell’s persistence time range. Negative persistence time ranges, indicating a firing rate rising during fixation, were obtained for a fraction of optically-recorded cells (50/416). These cells and cells for which persistence time ranges straddled 0 (49/416) were excluded from further analysis. The persistence time distributions for individual larvae shown in Figure 4d were populated with the median of persistence time distributions for cells with positive persistence time ranges. Fractional change in fluorescence during fixation was calculated for these same cells. It was defined as the fractional change between the mean of the first 512 ms and the last 512 ms of the segment of the STA firing rate estimate used for persistence time estimation.
Since deconvolution of fluorescence can result in an amplification of noise that is unrelated to the firing rate, we developed an alternative method for persistence time estimation that circumvented this undesirable effect. We calculated persistence time distributions for cells as described above a second time, but in the last step of each iteration fit the function below to the ipsiversive STA fluorescence. STA fluorescence responses were fit with a model of fluorescence that would result from a firing profile consisting of an initial burst followed by an exponential decay during fixation. The fluorescence, f(t), resulting from a decaying firing rate, r(t) , will be given by the convolution of the firing rate with the CIRF so that
where T is the time constant of the CIRF. Since the fluorescence at time t = 0 also depends on the firing rate in the past, an extra term must be included which accounts for the fluorescence resulting from past activity, so that the fluorescence during a fixation is given by
The parameters a and τ can be estimated by fitting the STA fluorescence with this function at some time after the end of the burst. Typical bursts lasted for 100–500 ms; therefore fits began 1 second after the saccade. The parameter c , representing the null value, was held fixed at the randomly selected level during each fit.
To determine whether cellular persistence time distributions differed statistically within individual larvae, we tested the null hypothesis that the distributions for a given larvae had equivalent medians using the nonparametric Kruskal-Wallis test (Matlab function kruskalwallis). Because persistence time distributions are not populated by independent measurements, the test was not performed directly on these distributions. Instead, we first determined approximately how many independent measurements were present in the segment of the ipsiversive STA firing rate estimate used for persistence time estimates. This was done by iteratively downsampling the residuals of an exponential fit to this segment until serial correlation between adjacent measurements was negligible. This method involved the following steps:
For all cells in the data set, fit the segment of the ipsiversive STA firing rate estimate used for persistence time estimates with Ae−t/τ, generating a residual time series.
-
For all cells in the data set, similarly fit and generate residuals from 500 random permutations of this segment.
For each cell:
Calculate the serial correlation for the residual time series from step 1. Serial correlation was measured for a sequence of residuals x = x1, x2, ..., xn by calculating the Pearson correlation between x = x1, x2, ..., xn–1 and x2, x3,..., xn.
Calculate the serial correlation similarly for 1000 random permutations of this residual time series.
Calculate the fraction of the 1000 correlation measurements from step 4 larger than the correlation measurement from step 3.
Perform the calculations from steps 3 to 5 again for each of the residual time series generated in step 2. The resulting 500 values define a cell-specific empirical distribution for the fractions calculated in step 5 under a null hypothesis of no serial correlation.
-
Calculate a p value for the fraction calculated in step 5 assuming a two-tailed test against the null distribution generated in step 6. This is:
The use of a two-tailed test here makes the test sensitive to both correlation and anti- correlation.
Repeat steps 3 to 7 for all cells in the data set. Calculate the fraction of p values for all cells from step 7 that fall below 0.3.
Downsample the residuals from steps 1 and 2 to half the previous sampling frequency (Matlab function downsample). Repeat steps 3 to 8.
In the absence of serial correlation (including anti-correlation), the fraction calculated in step 8 should equal ~0.3. We observed that as the time series were progressively downsampled, this fraction approached and then plateaued at ~0.3. Serial correlation was deemed negligible when this fraction first fell below 0.3. At this point, 15 samples remained. Thus the Kruskal-Wallis test was performed using 15 random samples from each cell’s persistence time distribution.
Somal position was defined using the ROI centroid. Cell positions were measured in distances caudal of the Mauthner cell soma, dorsal of the MLF, and lateral of the midline. For the plots shown in Figures 6b and c, cell pairs were grouped based on pairwise distance into bins divided at 10, 20, 30, 40, 50, and 60 μm. For the plots in Figures 7b, c, and e and Supplementary Figure 7b, bins were divided at −40, −24, −8, 8, 24, and 40 μm. For the plots in Figure 7f and Supplementary Figure 7a, divisions were made at −30, −18, −6, 6, 18, and 30 μm. Directional distances were calculated by subtracting the position of one cell of a pair, cell b, from the other, cell a, where cell a was the cell whose value was used in the numerator when calculating response index or persistence time ratios. Group means were plotted as error bars representing the mean ± sem versus the mean pairwise distance for each cohort. Outlying ratios were eliminated by including only values between the 2.5th and 97.5th percentile ratio values. Although data was binned for the purpose of plotting, correlation coefficients quoted for data presented in Figures 6 and 7 and Supplementary Figure 7 were calculated from individual data points. p values were calculated from two-tailed Students’ t-tests.
Previously described32 simultaneous firing rate measurements from pairs of adult goldfish hVPNI neurons were re-analyzed to extract persistence time estimates. Segments of firing rate time series were used in calculating STA firing rates if they corresponded to a fixation lasting > 4.5 seconds following the time of the saccade and if the average eye position from 0.5 to 1 second following the saccade fell in the top quartile of the distribution of such values for fixations following both ipsiversive and contraversive saccades. This second criteria focused the measurement on data collected during fixations more equivalent to those in larval zebrafish that largely saccade back and forth between positions near the extremes of their eye position range. The firing rate corresponding to the center of gaze was estimated as the y-intercept of a linear function best-fit in the least-squares sense to the relation between firing rate and eye position. This null value was subtracted off the STA firing rate, and the function Ae−t/τ was fit to the result. τ from this fit was used as the persistence time estimate. Percent differences for pairs of persistence times were computed as their absolute difference divided by their mean. One cell pair out of seven was excluded from percent difference calculation because one cell of the pair yielded a negative persistence time. Only zebrafish neurons having exclusively positive persistence time ranges were used to estimate pairwise percent differences.
Simulations
Network simulations used the linear system
where ri represents the firing rate of neuron i and wij represents the weight of the connection from neuron j to neuron i . All network simulations were generated using the 4th order Runge-Kutta method with 0.002 s time steps. τr was set to 1 s, the same value assumed based on experimental results for adult goldfish hVPNI neurons in reference 22, and N = 50. All rates were set to 1 at the first time point.
The weight matrix for the uniformly detuned line attractor network was generated as the outer product of two random vectors scaled uniformly so that the persistence time of simulated units was ~5 s, approximately the median value for optically recorded cells for which we calculated persistence times. The weight matrix for the feedback network with multiple stable dimensions was constructed by setting the weights between neurons equal to max{0,11− |i − j|} for i ≠ j (this effects a proximity bias to the connectivity weights) and 0 for i=j (no autapses). Entries were then divided by the sum of all entries in their respective columns and scaled by 0.185+0.985 (i/N). The constants in this expression were chosen empirically to create a distribution of persistence times qualitatively similar to those experimentally observed. The weight matrix for the asymmetric network with a feedforward bias was constructed by setting the weights between neurons equal to max{0, 20 − (j − i)} for i < j, max{0, 10 − (i − j)} for i > j, and 0 for i = j. Entries were then divided by the sum of all entries in their respective columns and scaled by 0.21+1.11(i/N).
Supplementary Material
Supplementary Figure 1 Dorsal-view schematic of the caudal hindbrain region from which image time series were collected. The cyan region in the upper left inset spans the schematized area. Sagittal imaging windows (gray) were spaced in 10 μm increments ranging from 20 to 70 μm lateral of the midline. Their rostrocaudal position relative to reticulospinal neurons (red) are calculated based on calibration measurements reported in Miri et al., 2011. In general, image windows ran from 100–130 μm to 200–230 μm caudal of the Mauthner cell soma. Rhombomeres 4 though 8 (r4 – r8) are labeled. The rostral image window boundary was measured relative to the ear peak, the dorsal peak of the otic vesicle in the sagittal plane in which the posterior cerebral vein bends caudally around the ear. The ear peak is easily identified under two-photon excitation in dye-loaded larvae. This allowed rostrocaudal distances to be measured relative to the average position of the ipsilateral Mauthner cell soma, as previous work has mapped reticulospinal neuron positions relative to the ear peak23. The midline from which mediolateral positions were measured was defined as the midpoint between the medial longitudinal fasciculi (MLFs) running in either hemisphere. Dorsoventral distances were measured from the ipsilateral MLF.
Supplementary Figure 2 Examples of STA firing rate estimates from deconvolution of fluorescence with CIRFs. (a)–(c) Ipsiversive (red) and contraversive (blue) STA fluorescence responses shown in Figure 2(g)–(i) deconvolved with CIRFs to provide an estimate of STA firing rate. Response indices (RI) calculated from fluorescence time series are indicated. Traces are aligned to their mean values over the first 1.5 seconds of each average.
Supplementary Figure 3 Schematic of the method used to calculate the persistence time distribution for individual cells. Four steps are repeated 1000 times: 1. STA fluorescence responses were deconvolved with a CIRF defined by a τ chosen randomly from the previously calculated distribution of CIRF τ, generating STA firing rate estimates. 2. The range of possible null values to which firing rate estimates decay is calculated from these estimates. 3. A null value is chosen randomly from this range. 4. Using this value, a persistence time is estimated from fitting the ipsiversive STA firing rate with an exponential function. This fit defines a distribution for the persistence time which is added to the overall persistence time distribution. See Methods for more details regarding this method.
Supplementary Figure 4 Further persistence time ranges. (a) Persistence time ranges for fits to firing rate estimates from 55 cells recorded in the light in the larva from which data is shown in Figure 4c. The vertical extent of each bar spans the middle 99% of the calculated persistence time distribution for each cell. In both (a) and (b), cells are ordered according to their rank in terms of the median of this distribution and gray bars span non-overlapping persistence time ranges. (b) Persistence time ranges for fits directly to STA fluorescence for the same cells depicted in Figure 4c, except one for which the fits yielded an extremely large persistence time range (≫ 1000 s).
Supplementary Figure 5 Group-averaged traces for an individual larvae, illustrating persistence time heterogeneity. Data were collected in the dark. Dotted lines indicate saccade time. (a)–(c) Group-averaged ipsiversive STA fluorescence for the lowest, middle, and highest 20% of cells in terms of median persistence time. (d)–(f) Group-averaged ipsiversive STA firing rate estimates for the lowest, middle, and highest 20% of cells in terms of median persistence time. Cell-specific CIRFs were used for deconvolution. (g)–(i) Group-averaged ipsiversive STA firing rate estimates for the lowest, middle, and highest 20% of cells in terms of median persistence time. A common CIRF having decay time equal to the median for the distribution of CIRF decay times across all cells was used for deconvolution.
Supplementary Figure 6 Accounting for the quality of exponential fits to firing rate estimates. (a) Histogram of R2 values for exponential fits used for persistence time estimation when these estimates where > 0. (b) Box plots representing the distribution of fractional change in STA firing rate estimates during fixation segments fit to calculate persistence times for larvae imaged in the dark (left panel) or the light (right panel). Horizontal lines indicate the minimum, 25th percentile, median, 75th percentile, and maximum values of each distribution.
Supplementary Figure 7 Weaker dependence of persistence time and response index similarity on certain spatial dimensions. Pairs identified in the light (gray) or dark (black) were grouped according to pairwise distance, data are plotted along the x-axis according to the mean distance for each group, and error bars = mean ± sem. (a) Persistence time ratio versus pairwise distance along the mediolateral axis. Data are included from cells for which persistence time estimates were > 0. (b) Response index ratio versus pairwise distance along the rostrocaudal axis for pairs of identified neurons.
Supplementary Figure 8 Group-averaged traces for an individual larvae, illustrating the relationship between persistence time and rostrocaudal location. Data collected in the dark. Dotted lines indicate saccade time. (a)-(c) Group-averaged ipsiversive STA fluorescence for the lowest, middle, and highest 20% of cells in terms of rostrocaudal location. (d)-(f) Group-averaged ipsiversive STA firing rate estimates for the lowest, middle, and highest 20% of cells in terms of rostrocaudal location. Cell-specific CIRFs were used for deconvolution. (g)-(i) Group-averaged ipsiversive STA firing rate estimates for the lowest, middle, and highest 20% of cells in terms of rostrocaudal location. A common CIRF having decay time equal to the median for the distribution of CIRF decay times across all cells was used for deconvolution.
Supplementary Figure 9 A rostrocaudal gradient of firing persistence. (a)-(e) Time projection of image time series with cells color-coded according to the log10 of their persistence time for data collected 20 μm (a), 30 μm (b), 40 μm (c), 50 μm (d), and 60 μm from the midline in an individual fish. (f) Scatterplot of persistence times versus position along the rostrocaudal axis for the 54 cells colored in (a)-(e). The gray line is a least-squares linear fit to the data.
Supplementary Table 1. Mean 95% widths of the persistence time distributions for individual larvae using different thresholds for data inclusion. The values populating these distributions are the medians of persistence time distributions calculated for individual cells. 95% width is the distance between the 2.5th and the 97.5th quantiles of the distribution. Widths were calculated for larvae in which ≥ 16 cells (dark) or ≥ 19 cells (light) fell above two thresholds: one on a cell’s Pearson correlation with CIRF-convolved eye position, and one on the R2 value for its exponential CIRF estimate fit. Widths were normalized by the width of the distribution obtained using the thresholds used in the manuscript for selecting cells for persistence time estimation (both 0.5). Normalized widths were then averaged across larvae to compute the values in the table. For all cases in which the correlation threshold was ≤ 0.5, n = 5 larvae. When the correlation threshold was 0.6, n ranged from 2 to 4. Little sensitivity to threshold variation is observed.
Supplementary Table 2. Spearman correlation of persistence time ratios and pairwise distance using different thresholds for data inclusion. Correlations were calculated as described in the manuscript using persistence time ratios calculated for all pairs of cells falling above two thresholds: one on a cell’s Pearson correlation with CIRF-convolved eye position, and one on the R2 value for its exponential CIRF estimate fit. The values shown in red are those reported in the manuscript when both thresholds equaled 0.5. Spearman correlation scores are similar across threshold pairs, changing slowing and smoothly as thresholds are changed. Moreover, all p values for the scores listed in the table are < 10−4, suggesting that the significance of these correlations does not depend on threshold choice.
Acknowledgments
This work was supported by an NSF predoctoral fellowship (A.M.), NIH Training grant EY007138-16 (K.D.), a Burroughs Wellcome Career Award at the Scientific Interface, a Searle Scholar award, and the Frueauff Foundation (E.A.), the Human Frontier Science Program, and US National Institutes of Health grants R01 MH060651 (D.W.T.) and R01 NS053358 (H.B.). We thank D. Dombeck for technical advice, F. Collman for motion-correction software, and M. Goldman, A. Kinkhabwala, and P. Bradley for helpful discussions.
Footnotes
Author Contributions
A.M. collected functional imaging and electrical recording data under the supervision of D.W.T. A.M., E.A., and D.W.T. developed the preparation, experimental procedures, and instrumentation for the imaging and electrophysiological studies. A.M. and K.D. analyzed this data with guidance from E.A. and D.W.T. A.B.A. and H.B. designed the NpHR study; A.B.A. collected and analyzed the data in H.B.’s laboratory. A.M., K.D., E.A., and D.W.T. wrote the paper.
References
- 1.Robinson DA. Integrating with neurons. Ann Rev Neurosci. 1989;12:33–45. doi: 10.1146/annurev.ne.12.030189.000341. [DOI] [PubMed] [Google Scholar]
- 2.Oestreich J, Dembrow NC, George AA, Zakon HH. A “sample-and-hold” pulse-counting integrator as a mechanism for graded memory underlying sensorimotor adaptation. Neuron. 2006;49:577–588. doi: 10.1016/j.neuron.2006.01.027. [DOI] [PubMed] [Google Scholar]
- 3.Huk AC, Shadlen MN. Neural activity in macaque parietal cortex reflects temporal integration of visual motion signals during perceptual decision making. J Neurosci. 2005;25:10420–10436. doi: 10.1523/JNEUROSCI.4684-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taube JS, Bassett JP. Persistent neural activity in head direction cells. Cereb Cortex. 2003;13:1162–1172. doi: 10.1093/cercor/bhg102. [DOI] [PubMed] [Google Scholar]
- 5.Cohen B, Komatsuzaki A. Eye movements induced by stimulation of the pontine reticular formation: evidence for integration in oculomotor pathways. Exp Neurol. 1972;36:101–117. doi: 10.1016/0014-4886(72)90139-2. [DOI] [PubMed] [Google Scholar]
- 6.Skavenski AA, Robinson DA. Role of abducens neurons in vestibuloocular reflex. J Neurophysiol. 1973;36:724–738. doi: 10.1152/jn.1973.36.4.724. [DOI] [PubMed] [Google Scholar]
- 7.Major G, Tank D. Persistent neural activity: prevalence and mechanisms. Curr Opin Neurobiol. 2004;14 :675–684. doi: 10.1016/j.conb.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 8.Lopez-Barneo J, Darlot C, Berthoz A, Baker R. Neuronal activity in prepositus nucleus correlated with eye movement in the alert cat. J Neurophysiol. 1982;47:329–352. doi: 10.1152/jn.1982.47.2.329. [DOI] [PubMed] [Google Scholar]
- 9.McFarland JL, Fuchs AF. Discharge patterns in nucleus prepositus hypoglossi and adjacent medial vestibular nucleus during horizontal eye movement in behaving macaques. J Neurophysiol. 1992;68:319–332. doi: 10.1152/jn.1992.68.1.319. [DOI] [PubMed] [Google Scholar]
- 10.Aksay E, Baker R, Seung HS, Tank DW. Anatomy and discharge properties of pre-motor neurons in the goldfish medulla that have eye-position signals during fixations. J Neurophysiol. 2000;84 :1035–1049. doi: 10.1152/jn.2000.84.2.1035. [DOI] [PubMed] [Google Scholar]
- 11.Seung HS. How the brain keeps the eyes still. Proc Natl Acad Sci USA. 1996;93:13339–13344. doi: 10.1073/pnas.93.23.13339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Machens CK, Romo R, Brody CD. Flexible control of mutual inhibition: a neural model of two-interval discrimination. Science. 2005;307:1121–1124. doi: 10.1126/science.1104171. [DOI] [PubMed] [Google Scholar]
- 13.Singh R, Eliasmith C. Higher-dimensional neurons explain the tuning and dynamics of working memory cells. J Neurosci. 2006;26:3667–3678. doi: 10.1523/JNEUROSCI.4864-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seung HS, Lee DD, Reis BY, Tank DW. Stability of the memory of eye position in a recurrent network of conductance-based model neurons. Neuron. 2000;26:259–271. doi: 10.1016/s0896-6273(00)81155-1. [DOI] [PubMed] [Google Scholar]
- 15.Koulakov AA, Raghavachari S, Kepecs A, Lisman JE. Model for a robust neural integrator. Nat Neurosci. 2002;5:775–782. doi: 10.1038/nn893. [DOI] [PubMed] [Google Scholar]
- 16.Goldman MS, Levine JH, Major G, Tank DW, Seung HS. Robust persistent neural activity in a model integratorwith multiple hysteretic dendrites per neuron. Cereb Cortex. 2003;13:1185–1195. doi: 10.1093/cercor/bhg095. [DOI] [PubMed] [Google Scholar]
- 17.Anastasio TJ. The fractional-order dynamics of brainstem vestibulo-oculomotor neurons. Biol Cybern. 1994;72:69–79. doi: 10.1007/BF00206239. [DOI] [PubMed] [Google Scholar]
- 18.Major G, Baker R, Aksay E, Seung HS, Tank DW. Plasticity and tuning of the time course of analog persistent firing in a neural integrator. Proc Natl Acad Sci USA. 2004;101:7745–7750. doi: 10.1073/pnas.0401992101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maass W, Joshi P, Sontag ED. Computational aspects of feedback in neural circuits. PLoS Comput Biol. 2007;3:e165. doi: 10.1371/journal.pcbi.0020165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aksay E, et al. Functional dissection of circuitry in a neural integrator. Nat Neurosci. 2007;10:494–504. doi: 10.1038/nn1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cannon SC, Robinson DA, Shamma S. A proposed neural network for the integrator of the oculomotor system. Biol Cybern. 1983;49:127–136. doi: 10.1007/BF00320393. [DOI] [PubMed] [Google Scholar]
- 22.Goldman MS. Memory without feedback in a neural network. Neuron. 2009;61:621–634. doi: 10.1016/j.neuron.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miri A, Daie K, Burdine RD, Aksay E, Tank DW. Regression-based identification of behavior-encoding neurons during large-scale optical imaging of neural activity at cellular resolution. J Neurophysiol. 2011;105:964–980. doi: 10.1152/jn.00702.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLean DL, Fan J, Higashijima S, Hale ME, Fetcho JR. A topographic map of recruitment in spinal cord. Nature. 2007;446:71–75. doi: 10.1038/nature05588. [DOI] [PubMed] [Google Scholar]
- 25.Weiler N, Wood L, Yu J, Solla SA, Shepherd GM. Top-down laminar organization of the excitatory network in motor cortex. Nat Neurosci. 2008;11:360–366. doi: 10.1038/nn2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higashijima S, Mandel G, Fetcho JR. Distribution of prospective glutamatergic, glycinergic, and GABAergic neurons in embryonic and larval zebrafish. J Comp Neurol. 2004;480:1–18. doi: 10.1002/cne.20278. [DOI] [PubMed] [Google Scholar]
- 27.Dasen JS, Jessell TM. Hox networks and the origins of motor neuron diversity. Curr Top Dev Biol. 2009;88:169–200. doi: 10.1016/S0070-2153(09)88006-X. [DOI] [PubMed] [Google Scholar]
- 28.Pastor AM, de La Cruz RR, Baker R. Eye position and eye velocity integrators reside in separate brainstem nuclei. Proc Natl Acad Sci USA. 1994;91:807–811. doi: 10.1073/pnas.91.2.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arrenberg AB, Del Bene F, Baier H. Optical control of zebrafish behavior with halorhodopsin. Proc Natl Acad Sci U S A. 2009;106:17968–17973. doi: 10.1073/pnas.0906252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoonheim PJ, Arrenberg AB, Del Bene F, Baier H. Optogenetic localization and genetic perturbation of saccade-generating neurons in zebrafish. J Neurosci. 2010;30:7111–7120. doi: 10.1523/JNEUROSCI.5193-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Escudero M, de la Cruz RR, Delgado-Garcia JM. A physiological study of vestibular and prepositus hypoglossi neurones projecting to the abducens nucleus in the alert cat. J Physiol. 1992;458:539–560. doi: 10.1113/jphysiol.1992.sp019433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aksay E, Baker R, Seung HS, Tank DW. Correlated discharge among cell pairs within the oculomotor horizontal velocity-to-position integrator. J Neurosci. 2003;23:10852–10858. doi: 10.1523/JNEUROSCI.23-34-10852.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shinoda Y, Yoshida K. Dynamic characteristics of responses to horizontal head angular acceleration in vestibuloocular pathway in the cat. J Neurophysiol. 1974;37:653–673. doi: 10.1152/jn.1974.37.4.653. [DOI] [PubMed] [Google Scholar]
- 34.Anastasio TJ. Nonuniformity in the linear network model of the oculomotor integrator produces approximately fractional-order dynamics and more realistic neuron behavior. Biol Cybern. 1998;79:377–391. doi: 10.1007/s004220050487. [DOI] [PubMed] [Google Scholar]
- 35.Aksay E, et al. History dependence of rate covariation between neurons during persistent activity in an oculomotor integrator. Cereb Cortex. 2003;13:1173–1184. doi: 10.1093/cercor/bhg099. [DOI] [PubMed] [Google Scholar]
- 36.Debowy O, Baker R. Encoding of eye position in the goldfish horizontal oculomotor neural integrator. J Neurophysiol. 2011;105:896–909. doi: 10.1152/jn.00313.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sklavos S, Porrill J, Kaneko CR, Dean P. Evidence for wide range of time scales in oculomotor plant dynamics: implications for models of eye-movement control. Vision Res. 2005;45:1525–1542. doi: 10.1016/j.visres.2005.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinkhabwala A, et al. A structural and functional ground plan for neurons in the hindbrain of zebrafish. Proc Natl Acad Sci U S A. 2011;108:1164–1169. doi: 10.1073/pnas.1012185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLean DL, Masino MA, Koh IY, Lindquist WB, Fetcho JR. Continuous shifts in the active set of spinal interneurons during changes in locomotor speed. Nat Neurosci. 2008;11:1419–1429. doi: 10.1038/nn.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldberg JM, Fernandez C. Physiology of peripheral neurons innervating semicircular canals of the squirrel monkey. I Resting discharge and response to constant angular acceleration. J Neurophysiol. 1971;34:635–660. doi: 10.1152/jn.1971.34.4.635. [DOI] [PubMed] [Google Scholar]
- 41.Miles FA, Braitman DJ. Long-term adaptive changes in primate vestibuloocular reflex. II Electrophysiological observations on semicircular canal primary afferents. J Neurophysiol. 1980;43:1426. doi: 10.1152/jn.1980.43.5.1426. [DOI] [PubMed] [Google Scholar]
- 42.Sussillo D, Abbott LF. Generating coherent patterns of activity from chaotic neural networks. Neuron. 2009;63:544–557. doi: 10.1016/j.neuron.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Helmstaedter M, Briggman KL, Denk W. 3D structural imaging of the brain with photons and electrons. Curr Opin Neurobiol. 2008;18:633–641. doi: 10.1016/j.conb.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 44.Beck JC, Rothnie P, Straka H, Wearne SL, Baker R. Precerebellar hindbrain neurons encoding eye velocity during vestibular and optokinetic behavior in the goldfish. J Neurophysiol. 2006;96:1370–1382. doi: 10.1152/jn.00335.2006. [DOI] [PubMed] [Google Scholar]
- 45.Ma LH, Punnamoottil B, Rinkwitz S, Baker R. Mosaic hoxb4a neuronalpleiotropism in zebrafish caudal hindbrain. PLoS One. 2009;4:e5944. doi: 10.1371/journal.pone.0005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Z. Princeton University. 2009. [Google Scholar]
- 47.Aksay E, Gamkrelidze G, Seung HS, Baker R, Tank DW. In vivo intracellular recording and perturbation of persistent activity in a neural integrator. Nat Neurosci. 2001;4:184–193. doi: 10.1038/84023. [DOI] [PubMed] [Google Scholar]
- 48.Teramae JN, Fukai T. A cellular mechanism for graded persistent activity in a model neuron and its implications in working memory. J Comput Neurosci. 2005;18:105–121. doi: 10.1007/s10827-005-5474-6. [DOI] [PubMed] [Google Scholar]
- 49.Fransen E, Tahvildari B, Egorov AV, Hasselmo ME, Alonso AA. Mechanism of graded persistent cellular activity of entorhinal cortex layer v neurons. Neuron. 2006;49:735–746. doi: 10.1016/j.neuron.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 50.Loewenstein Y, Sompolinsky H. Temporal integration by calcium dynamics in a model neuron. Nat Neurosci. 2003;6:961–967. doi: 10.1038/nn1109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Dorsal-view schematic of the caudal hindbrain region from which image time series were collected. The cyan region in the upper left inset spans the schematized area. Sagittal imaging windows (gray) were spaced in 10 μm increments ranging from 20 to 70 μm lateral of the midline. Their rostrocaudal position relative to reticulospinal neurons (red) are calculated based on calibration measurements reported in Miri et al., 2011. In general, image windows ran from 100–130 μm to 200–230 μm caudal of the Mauthner cell soma. Rhombomeres 4 though 8 (r4 – r8) are labeled. The rostral image window boundary was measured relative to the ear peak, the dorsal peak of the otic vesicle in the sagittal plane in which the posterior cerebral vein bends caudally around the ear. The ear peak is easily identified under two-photon excitation in dye-loaded larvae. This allowed rostrocaudal distances to be measured relative to the average position of the ipsilateral Mauthner cell soma, as previous work has mapped reticulospinal neuron positions relative to the ear peak23. The midline from which mediolateral positions were measured was defined as the midpoint between the medial longitudinal fasciculi (MLFs) running in either hemisphere. Dorsoventral distances were measured from the ipsilateral MLF.
Supplementary Figure 2 Examples of STA firing rate estimates from deconvolution of fluorescence with CIRFs. (a)–(c) Ipsiversive (red) and contraversive (blue) STA fluorescence responses shown in Figure 2(g)–(i) deconvolved with CIRFs to provide an estimate of STA firing rate. Response indices (RI) calculated from fluorescence time series are indicated. Traces are aligned to their mean values over the first 1.5 seconds of each average.
Supplementary Figure 3 Schematic of the method used to calculate the persistence time distribution for individual cells. Four steps are repeated 1000 times: 1. STA fluorescence responses were deconvolved with a CIRF defined by a τ chosen randomly from the previously calculated distribution of CIRF τ, generating STA firing rate estimates. 2. The range of possible null values to which firing rate estimates decay is calculated from these estimates. 3. A null value is chosen randomly from this range. 4. Using this value, a persistence time is estimated from fitting the ipsiversive STA firing rate with an exponential function. This fit defines a distribution for the persistence time which is added to the overall persistence time distribution. See Methods for more details regarding this method.
Supplementary Figure 4 Further persistence time ranges. (a) Persistence time ranges for fits to firing rate estimates from 55 cells recorded in the light in the larva from which data is shown in Figure 4c. The vertical extent of each bar spans the middle 99% of the calculated persistence time distribution for each cell. In both (a) and (b), cells are ordered according to their rank in terms of the median of this distribution and gray bars span non-overlapping persistence time ranges. (b) Persistence time ranges for fits directly to STA fluorescence for the same cells depicted in Figure 4c, except one for which the fits yielded an extremely large persistence time range (≫ 1000 s).
Supplementary Figure 5 Group-averaged traces for an individual larvae, illustrating persistence time heterogeneity. Data were collected in the dark. Dotted lines indicate saccade time. (a)–(c) Group-averaged ipsiversive STA fluorescence for the lowest, middle, and highest 20% of cells in terms of median persistence time. (d)–(f) Group-averaged ipsiversive STA firing rate estimates for the lowest, middle, and highest 20% of cells in terms of median persistence time. Cell-specific CIRFs were used for deconvolution. (g)–(i) Group-averaged ipsiversive STA firing rate estimates for the lowest, middle, and highest 20% of cells in terms of median persistence time. A common CIRF having decay time equal to the median for the distribution of CIRF decay times across all cells was used for deconvolution.
Supplementary Figure 6 Accounting for the quality of exponential fits to firing rate estimates. (a) Histogram of R2 values for exponential fits used for persistence time estimation when these estimates where > 0. (b) Box plots representing the distribution of fractional change in STA firing rate estimates during fixation segments fit to calculate persistence times for larvae imaged in the dark (left panel) or the light (right panel). Horizontal lines indicate the minimum, 25th percentile, median, 75th percentile, and maximum values of each distribution.
Supplementary Figure 7 Weaker dependence of persistence time and response index similarity on certain spatial dimensions. Pairs identified in the light (gray) or dark (black) were grouped according to pairwise distance, data are plotted along the x-axis according to the mean distance for each group, and error bars = mean ± sem. (a) Persistence time ratio versus pairwise distance along the mediolateral axis. Data are included from cells for which persistence time estimates were > 0. (b) Response index ratio versus pairwise distance along the rostrocaudal axis for pairs of identified neurons.
Supplementary Figure 8 Group-averaged traces for an individual larvae, illustrating the relationship between persistence time and rostrocaudal location. Data collected in the dark. Dotted lines indicate saccade time. (a)-(c) Group-averaged ipsiversive STA fluorescence for the lowest, middle, and highest 20% of cells in terms of rostrocaudal location. (d)-(f) Group-averaged ipsiversive STA firing rate estimates for the lowest, middle, and highest 20% of cells in terms of rostrocaudal location. Cell-specific CIRFs were used for deconvolution. (g)-(i) Group-averaged ipsiversive STA firing rate estimates for the lowest, middle, and highest 20% of cells in terms of rostrocaudal location. A common CIRF having decay time equal to the median for the distribution of CIRF decay times across all cells was used for deconvolution.
Supplementary Figure 9 A rostrocaudal gradient of firing persistence. (a)-(e) Time projection of image time series with cells color-coded according to the log10 of their persistence time for data collected 20 μm (a), 30 μm (b), 40 μm (c), 50 μm (d), and 60 μm from the midline in an individual fish. (f) Scatterplot of persistence times versus position along the rostrocaudal axis for the 54 cells colored in (a)-(e). The gray line is a least-squares linear fit to the data.
Supplementary Table 1. Mean 95% widths of the persistence time distributions for individual larvae using different thresholds for data inclusion. The values populating these distributions are the medians of persistence time distributions calculated for individual cells. 95% width is the distance between the 2.5th and the 97.5th quantiles of the distribution. Widths were calculated for larvae in which ≥ 16 cells (dark) or ≥ 19 cells (light) fell above two thresholds: one on a cell’s Pearson correlation with CIRF-convolved eye position, and one on the R2 value for its exponential CIRF estimate fit. Widths were normalized by the width of the distribution obtained using the thresholds used in the manuscript for selecting cells for persistence time estimation (both 0.5). Normalized widths were then averaged across larvae to compute the values in the table. For all cases in which the correlation threshold was ≤ 0.5, n = 5 larvae. When the correlation threshold was 0.6, n ranged from 2 to 4. Little sensitivity to threshold variation is observed.
Supplementary Table 2. Spearman correlation of persistence time ratios and pairwise distance using different thresholds for data inclusion. Correlations were calculated as described in the manuscript using persistence time ratios calculated for all pairs of cells falling above two thresholds: one on a cell’s Pearson correlation with CIRF-convolved eye position, and one on the R2 value for its exponential CIRF estimate fit. The values shown in red are those reported in the manuscript when both thresholds equaled 0.5. Spearman correlation scores are similar across threshold pairs, changing slowing and smoothly as thresholds are changed. Moreover, all p values for the scores listed in the table are < 10−4, suggesting that the significance of these correlations does not depend on threshold choice.