Abstract
Glycoproteins produced by non-engineered insects or insect cell lines characteristically bear truncated, paucimannose N-glycans in place of the complex N-glycans produced by mammalian cells. A key reason for this difference is the presence of a highly specific N-glycan processing β-N-acetylglucosaminidase in insect, but not in mammalian systems. Thus, reducing or abolishing this enzyme could enhance the ability of glycoengineered insects or insect cell lines to produce complex N-glycans. Of the three insect species routinely used for recombinant glycoprotein production, the processing β-N-acetylglucosaminidase gene has been isolated only from Spodoptera frugiperda. Thus, the purpose of this study was to isolate and characterize the genes encoding this important processing enzyme from the other two species, Bombyx mori and Trichoplusia ni. Bioinformatic analyses of putative processing β-N-acetylglucosaminidase genes isolated from these two species indicated that each encoded a product that was, indeed, more similar to processing β-N-acetylglucosaminidases than degradative or chitinolytic β-N-acetylglucosaminidases. In addition, over-expression of each of these genes induced an enzyme activity with the substrate specificity characteristic of processing, but not degradative or chitinolytic enzymes. Together, these results demonstrated that the processing β-N-acetylglucosaminidase genes had been successfully isolated from Trichoplusia ni and Bombyx mori. The identification of these genes has the potential to facilitate further glycoengineering of baculovirus-insect cell expression systems for the production of glycosylated proteins.
Keywords: glycoengineering, fused lobes, N-glycan processing β-N-acetylglucosaminidase, baculovirus expression system
Introduction
Glycoproteins produced by mammalian and insect cells are structurally identical if their N-glycans are of the minimally processed, oligomannose type.1 However, if those N-glycans are destined to undergo more extensive processing, the glycoproteins produced by these two cell types are structurally distinct. Specifically, a glycoprotein produced by mammalian cells can acquire complex, terminally sialylated N-glycans, while the same glycoprotein produced by insect cells typically acquires paucimannose N-glycans at the same sites.2 For example, native human serum transferrin has mainly biantennary, bisialylated N-glycans,3 but recombinant human serum transferrin produced by Trichoplusia ni cells has mainly paucimannose N-glycans.4 Similarly, natural human interferon-β and recombinant human interferon-β produced by mammalian cells have terminally sialylated biantennary and tri-antennary N-glycans,5,6 but recombinant murine interferon-β produced by baculovirus-infected Bombyx mori larvae has mostly paucimannose N-glycans.7 Finally, native human secreted placental alkaline phosphatase has biantennary, mainly sialylated N-glycans,8 whereas the same protein isolated from B. mori and T. ni cell lines and larvae has only paucimannose and oligomannose N-glycans.9,10
These differences in N-glycan structure are pharmacologically significant because the serum half-life of glycoproteins lacking terminal sialic acids is very short.11 Consequently, recombinant glycoproteins produced in insect cells are not suitable for therapeutic applications in which the product is required to remain in the circulation for an extended time. This problem was nicely illustrated in a study demonstrating that recombinant thyrotropin produced in insect cells had a higher level of activity in vitro, but a far lower level of activity in vivo than the same protein produced in CHO cells.12 The inability of insect cells to produce glycoproteins with terminally sialylated N-glycans is a major limitation that justifies the use of transgenic mammalian cells for the production of therapeutic glycoproteins. However, this limitation can be overcome by glycoengineering, which involves the incorporation of mammalian genes into the insect cell genome. Expression of these genes extends the endogenous N-glycan processing pathway and allows insect cells to produce glycoproteins with complex N-glycans.13–15 Thus, glycoengineering of insect cell systems is an approach that can be used to overcome their major limitation and exploit their advantages relative to transgenic mammalian cell systems for recombinant glycoprotein production, which include faster product development times and increased biosafety.
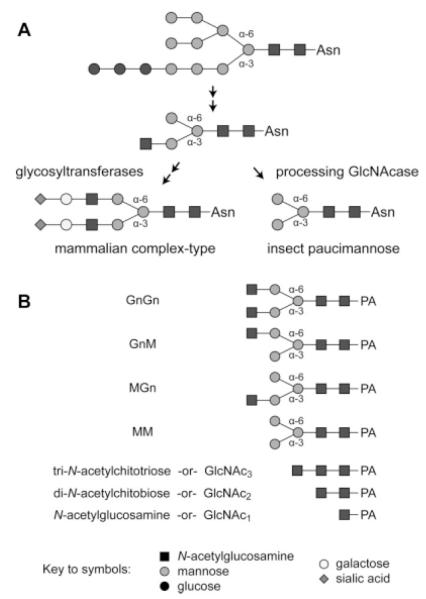
Despite the major structural differences between their fully processed N-glycan products, the N-glycan processing pathways of mammalian and insect cells are actually quite similar.14,16 The steps involving the assembly and transfer of the dolichol-linked glycan precursor appear to be the same in both cell types. The subsequent processing of the N-glycan, which involves the action of the ER glucosidases, the ER and Golgi class I α-mannosidases, N-acetylglucosaminyltransferase I, and Golgi mannosidase II, are also functionally identical in both cell types. These steps produce a common intermediate (Figure 1A), and it is only beyond this point that the mammalian and insect N-glycan processing pathways diverge. Mammalian cells elongate the common intermediate through the addition of N-acetylglucosamine, galactose, and sialic acid residues, thereby converting the intermediate to a complex N-glycan. However, the glycosyltransferase activities required for these additions are absent or negligible in the insect cell lines used as hosts for baculovirus expression vectors (BEVs). Moreover, insect cells express a processing β-N-acetylglucosaminidase that specifically removes the terminal N-acetylglucosamine residue from the common intermediate, thereby converting it to a paucimannose N-glycan (Figure 1A).17 Unlike typical β-N-acetylglucosaminidases, this enzyme is highly specific and can remove only the terminal N-acetylglucosamine linked to the (lower) α-3 mannose branch of an N-glycan. This substrate specificity distinguishes the N-glycan processing enzyme from related, broad-spectrum β-N-acetylglucosaminidases, which remove N-acetylglucosamine from both the α-6 and α-3 mannose branches of N-glycans.
Figure 1. A comparison of insect and mammalian protein N-glycan processing pathways and the structures of the substrates used for β-N-acetylglucosaminidase assays.
(A) The initial steps of N-glycan processing, which are mediated by glucosidase I and II, ER mannosidase I, β-N-acetylglucosaminyltransferase I and Golgi mannosidase II, are common to both cell types and result in the production of a common intermediate with a N-acetylglucosamine residue on the lower, α-3 mannose branch. Mammalian cells express glycosyltransferases, which can add more branches and extend this intermediate to yield terminally sialylated complex N-glycans. Conversely, insect cells express a processing β-N-acetylglucosaminidase that specifically removes the N-acetylglucosamine residue from the α-3 mannose branch to generate the paucimannose structures commonly found on recombinant glycoproteins produced in the BEVS. (B) This panel shows the structures of the glycans used as substrates for the β-N-acetylglucosaminidase assays.
Considering the function of this enzyme, inhibition of the processing β-N-acetylglucosaminidase would be expected to enhance the efficiency of complex N-glycan production by glycoengineered insect cells. Such inhibition could be achieved by adding the small molecule inhibitor 2-acetamido-1,2-dideoxynojirimycin (2-ADN) to the culture medium.18,19 However, this approach is not economically feasible at production scales due to the high cost of 2-ADN. Other potential ways to inhibit the processing β-N-acetylglucosaminidase include deleting the gene encoding this enzyme or repressing its expression using RNAi. Both of these approaches require the isolation and unambiguous identification of the target gene encoding the processing β-N-acetylglucosaminidase in the insect cell of choice.
The first processing β-N-acetylglucosaminidase gene to be isolated and functionally characterized was the fused lobes (fdl) gene of Drosophila melanogaster, which is not a host for BEVs.20 Subsequently, an fdl gene ortholog was isolated and characterized from Sf9 cells, which are widely used as hosts for BEVs.21 Additional genes encoding β-N-acetylglucosaminidases have been molecularly cloned from the two other organisms that are also widely used as BEV hosts, Bombyx mori and Trichoplusia ni (highlighted in Figures 4–5). However, characterization of the gene products showed that none had the specificity of the glycan processing enzymes. Instead, each was a chitinase or a broad-spectrum hexosaminidase with activity against a wide variety of sub-strates. Bioinformatic analyses also showed that these genes were only distantly related to the fdl genes encoding the bona-fide processing β-N-acetylglucosaminidases in Drosophila and Sf9 cells. Thus, despite their biotechnological significance as hosts for baculovirus-mediated recombinant glycoprotein production, the processing β-N-acetylglucosaminidase genes have not been isolated from either B. mori or T. ni. This prompted us to isolate the fdl genes of these two organisms and functionally confirm the gene products to set the stage for future subtractive glycoengineering of their N-glycan processing pathways.
Figure 4. Multiple sequence alignment of insect β-N-acetylglucosaminidases.
This Figure shows an alignment of the predicted amino acid sequences of BmFDL, TnFDL, SfFDL, DmFDL, and other cloned β-N-acetylglucosaminidase (GNase) sequences. The shading is proportional to the degree of amino acid conservation. The boxed region indicates the family 20 glycosyl hydrolase catalytic motif HXGGDEVXXXCW. The solid triangles indicate the aspartate residue likely to stabilize the transition state and the glutamate residue likely to serve as a proton donor during hydrolysis.26 The arrows above the sequences indicate the peptides targeted by degenerate primers used in the degenerate and semi-degenerate PCR reactions to isolate the Tn-fdl gene (see Supporting Information Table 1 for the nucleotide sequences). The amino acid sequences used in this alignment include: Tn FDL (FJ695479): Trichoplusia ni processing GNase (this study), Bm FDL (FJ695481): Bombyx mori processing GNase (this study), Sf FDL (ACA30398): Spodoptera frugiperda processing GNase,21 Dm FDL (Q8WSF3): Drosophila melanogaster processing GNase,20 Bm GNAse (BAF52531): Bombyx mori BmGlcNAcase1, a non-specific GNase,27 Tn GNase (AAL82580): an uncharacterized, putative Trichoplusia ni GNase. Amino acid sequences were aligned using ClustalX 2.0.10 with the default settings and the image was generated using DSGene 1.5.
Figure 5. Phylogenetic tree of lepidopteran and other insect β-N-acetylglucosaminidases.
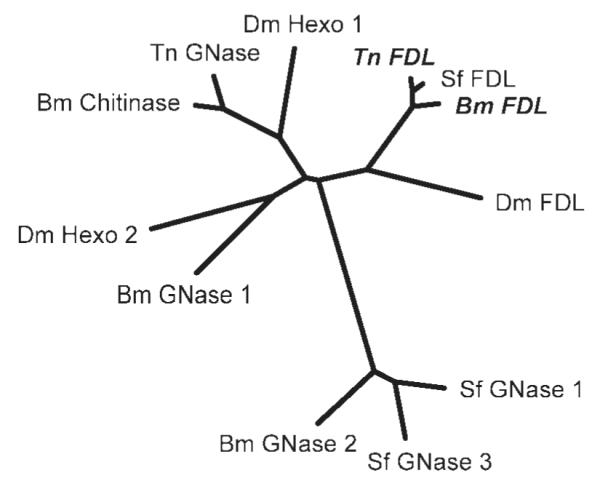
This unrooted tree demonstrates the phylogenetic relationships between cloned insect GNases. The distances between sequences are inversely proportional to their similarity. To produce this tree, a multiple sequence alignment was performed by ClustalX 2.0.10 using the default settings, which was then used to generate a distance matrix using Protdist (PHYLIP version 3.68) with the Jones-Taylor-Thornton model. An unrooted tree was then drawn by the PHYLIP drawtree postscript generator from the distance matrix using the neighbor-joining method (Neighbor, PHYLIP version 3.68). The GNase proteins and their Genbank accession numbers used in this tree are: Tn FDL (FJ695479): Trichoplusia ni processing GNase (this study), Bm FDL (FJ695481): Bombyx mori processing GNase (this study), Sf FDL (ACA30398): Spodoptera frugiperda processing GNase,21 Dm FDL (Q8WSF3): Drosophila melanogaster processing GNase,20 Bm GNase1 (BAF52531) and 2 (BAF52532): Bombyx mori non-specific GNases,27 Dm Hexo1 and 2: Drosophila melanogaster chitinolytic GNases,20 Bm Chitinase (AAC60521): Bombyx mori chitinolytic GNase,28 Tn GNase (AAL82580): uncharacterized, putative Trichoplusia ni GNase, Sf GNase 1 (ABB76924) and 3 (ABB76926): Spodoptera frugiperda non-specific GNases.22
Materials and Methods
Cell culture and viruses
AcBm-FDL and AcTn-FDL were produced by the Baculo-Direct™ method as indicated in the Supporting Online Information and subjected to one round of plaque purification. AcSf-FDL,21 AcSfGlcNAcase-3,22 and the E2 strain of the parental Autographa californica nucleopolyhedrovirus (AcMNPV) were isolated in previous studies. Each of these viruses was amplified and titered as described earlier.23 Sf9 cells were maintained at 28°C in TNMFH medium supplemented with 10% FCS (complete TNMFH) as described earlier.23
Expression of the native, full-length fdl gene products in insect cells
Sf9 cells were seeded into 100 mL of complete TNMFH medium in 250 mL DeLong flasks (Corning Glass Works, Corning, NY) at a density of 0.5 × 106 cells/mL and allowed to grow overnight to a density of about 1.0 × 106 cells/mL at 28°C and 125 rpm in a model 4580 rotary platform shaker-incubator (Forma Scientific, Marietta, OH). The cells were then infected by adding the volume of titered baculovirus needed for a multiplicity of infection of about two plaque-forming units/cell. After 1 h, the cells were pelleted by centrifugation at 200g for 5 min, resuspended in 100 mL of fresh complete TNMFH (without virus), and incubated for another 48 h under the same conditions.
Preparation of solubilized microsomal fractions
Baculovirus-infected cells were pelleted by centrifugation at 200g for 5 min, resuspended in 100 mL of ice-cold phosphate-buffered saline, and repelleted. The cell pellet was then resuspended in 10 mL of lysis buffer (5 mM imidazole, pH 7.0; 250 mM sucrose, 0.5 mM mercaptoethanol) supplemented with one Complete Mini™ protease inhibitor cocktail tablet (Roche, Indianapolis, IN). The cells were subsequently homogenized on ice in a Dounce homogenizer with pestle A, after which nuclei and remaining intact cells were pelleted by centrifugation at 4°C at 2,000g for 10 min. The crude microsomal preparation was then layered onto a sucrose cushion (5 mM imidazole, pH 7.0; 1.3 M sucrose, 0.1 mM EDTA), covered with sucrose overlay (125 mM sucrose, 5 mM imidazole, pH 7.0, 0.1 mM EDTA), and then centrifuged in a Beckman SW28 rotor at 100,000gav for 1 h at 4°C. Subsequently, the microsomal band was harvested, diluted with sucrose overlay, and recentrifuged in a SW28 rotor at 110,000gav for 30 min at 4°C. The microsomal pellet was resuspended in 200 μL of storage buffer (100 mM citrate-phosphate, pH 6.0; 0.02% NaN3, 125 mM sucrose), divided into two aliquots, and stored at −80°C for up to 2 weeks. The thawed microsomes were pelleted by centrifugation for 10 min at 13,000g in a microcentrifuge, resuspended in β-N-acetylglucosaminidase assay buffer (100 mM citrate-phosphate, pH 6.0; 0.5% v/v Triton-X-100) and solubilized by vortexing for 10 s and incubating at room temperature for 5 min. Insoluble debris was subsequently pelleted by centrifugation in a microcentrifuge at 13,000g for 15 min at 4°C and the supernatant was then used for β-N-acetylglucosaminidase assays as described below.
β-N-acetylglucosaminidase assays
Total protein concentrations in the solubilized microsomal preparations were determined using the Pierce® BCA Protein Assay Kit (Thermo Scientific, Waltham, MA) with chicken ovalbumin as the standard. Twenty microgram of total protein were used in enzyme assays with a final volume of 100 μL of β-N-acetylglucosaminidase assay buffer containing 50 pmoles of PA-tagged glycan substrate (GnGn, MGn, GnM or GlcNAc3, Figure 1B). The N-glycan substrates were a generous gift from Drs. Friedrich Altmann and Iain Wilson of the Glycobiology group in the Department of Chemistry, University of Natural Resources and Applied Life Sciences, Vienna. The substrates were prepared from biantennary, PA-derivatized glycans of bovine fibrin by sequential mild acid de-sialylation, enzymatic de-galactosylation using A. oryzae galactosidase, and partial removal of terminal N-acetylglucosamine using jack bean hexosaminidase, followed by HPLC purification of the reaction products. GlcNAc3 was prepared by PA-derivatization of chitotriose (Northstar BioProducts, East Falmouth, MA) followed by HPLC purification. Each reaction was incubated at 37°C for 16 h and then stored at −20°C. The frozen reactions were quick-thawed and centrifuged for 10 min at 13,000g to remove insoluble debris. Subsequently, the clarified reaction products were analyzed by reverse-phase HPLC using a Dionex Acclaim® 120 column with fluorescent detection (EX: 320 nm, EM: 400 nm) and a constant flow rate of 1 mL/min. Samples were loaded for 5 min with 88% buffer A (0.1 M ammonium acetate, pH 4.0) and 12% buffer B (30% v/v MeOH) and then eluted using a linear gradient that increased the amount of buffer B to 20% over 10 min followed by a linear gradient that increased the amount of buffer B to 100% over the next 15 min.
Results
Isolation of the B. mori and T. ni N-glycan processing β-N-acetylglucosaminidase genes
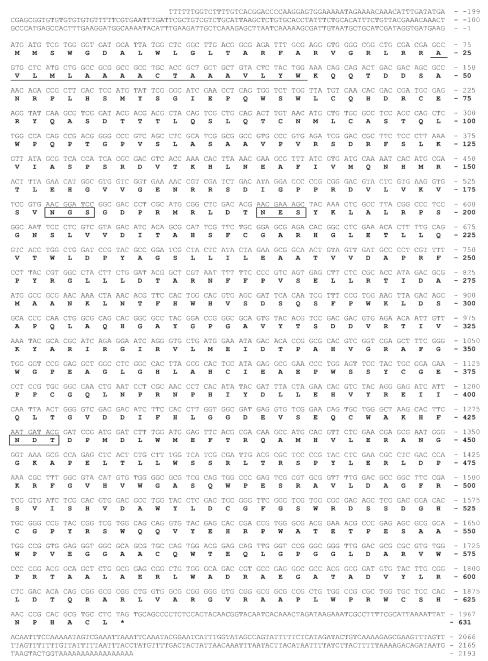
Our efforts to isolate genes encoding the processing β-N-acetylglucosaminidase from B. mori and T. ni were informed by the availability of partial B. mori genomic sequences as well as the previous isolation of an fdl cognate from S. frugiperda.21 tBLASTn searches of B. mori genome sequences (p50/Daizo strain) yielded two putative coding exons, including a translational initiation and termination site, which could be joined by splicing in silico. Amplification of the full-length Bm-fdl open reading frame with primers designed using the predicted coding sequence appeared to validate the prediction, as essentially the same sequence was recovered from cDNA. However, we actually isolated two distinct alleles with nucleotide substitutions in the intron and in both exons. The conceptual translation of one of these two alleles yielded a product that was identical to the conceptual translation product of the putative fdl gene identified in the p50/Daizo genomic sequences. The other allele had three nucleotide changes that resulted in the L138I, G404E, and H481Q amino acid substitutions. Because of the agreement between the conceptual translations of the p50/daizo genomic data-base sequence and the first of the two experimentally determined alleles, we used this allele for further experiments designed to determine the specificity of the Bm-fdl gene product. Finally, we verified and extended the nucleotide sequence of the putative Bm-fdl gene using 5′- and 3′-RACE. The resulting nucleotide sequence of the Bm-fdl gene and the amino acid sequence of its conceptual translation product are shown in Figure 2. Consistent with the expectation that a processing β-N-acetylglucosaminidase would be a membrane-bound, secretory pathway protein, the Bm-fdl gene product appears to be a type II membrane protein with a signal peptide, as predicted by signalP 3.0,24 that also functions as an amino-terminal transmembrane domain, as predicted by TMHMM 2.0.25
Figure 2. Nucleotide and predicted amino acid sequences of an fdl gene from B. mori.
The nucleotide sequence of one allele of the Bm-fdl gene is shown together with its derived amino acid sequence. We chose to present this allele because its derived amino acid sequence matched that of the putative p50/daizo gene, unlike the other allele isolated in this study. The predicted transmembrane domain (underlined) and consensus N-glycosylation sites (boxed) are indicated. The GenBank accession number for this Bm-fdl allele is FJ695481 and the accession number for the other allele isolated in this study is FJ695480.
In contrast to the approach used to isolate the Bm-fdl gene, a different approach was used to isolate the T. ni fdl gene because no T. ni genomic sequences were available. Initially, degenerate PCR was used to obtain a partial sequence. Subsequently, we attempted to use 3′- and 5′-RACE with gene-specific primers to obtain the full-length T. ni gene sequence, but this effort failed (data not shown). Thus, semi-degenerate PCR with a combination of gene-specific and degenerate primers was used to extend the partial putative Tn-fdl sequence obtained by degenerate PCR. The results of the semi-degenerate PCR experiments extended the sequence of the Tn-fdl gene and allowed us to design new gene-specific primers located closer to the ends of the coding sequence. Subsequently, these gene-specific primers were used in 5′- and 3′-RACE reactions to determine the full-length Tn-fdl open reading frame and 3′-UTR, which are shown together with the conceptual translation product in Figure 3. Bioinformatic analyses indicated that the predicted Tn-fdl gene product, like the predicted Bm-fdl gene product, is likely to be a type II membrane-bound, secretory pathway glycoprotein.
Figure 3. Nucleotide and predicted amino acid sequences of an fdl gene from T. ni.
The nucleotide sequence of the Tn-fdl gene is shown together with its derived amino acid sequence. The predicted transmembrane domain (underlined) as well as consensus N-glycosylation sites (boxed) are indicated. Surprisingly, we did not find a 5′-UTR, as the sequence of the 5′-RACE product immediately following the GeneRacer™ 5′ nested primer started with the shown initiation codon followed by the contiguous open reading frame. The GenBank accession number for the Tn-fdl nucleotide sequence is FJ695479.
Bioinformatic analyses
A multiple sequence alignment that includes the newly identified fdl gene products, the previously isolated and experimentally characterized Sf-fdl and Dm-fdl gene products, and the conceptual translation products of two other β-N-acetylglucosaminidase genes from T. ni and B. mori, is shown in Figure 4. The conceptual translation products of both of the newly identified genes are most similar to the Sf-FDL and Dm-FDL proteins. In contrast, the conceptual products of the other T. ni and B. mori β-N-acetylglucosaminidase genes are much less similar to the Sf-FDL and Dm-FDL proteins. Interestingly, the predicted Tn-FDL protein has a large glycine-rich sequence in the carboxy-terminal region that has no similarity to any of the other insect β-N-acetylglucosaminidase sequences and, therefore, appears to be the result of a random DNA insertion. The inferred phylogenetic relationships between the newly identified gene products and other insect β-N-acetylglucosaminidases are shown in Figure 5. This phylogenetic tree clearly illustrates the close relationship between the putative Bm-fdl and Tn-fdl β-N-acetylglucosaminidase gene products and the Sf-FDL and Dm-FDL proteins. In contrast, the other B. mori and T. ni β-N-acetylglucosaminidase gene products cluster with proteins that have been characterized as degradative and/or chitinolytic enzymes. The much closer phylogenetic relationship of the newly isolated Tn-fdl and Bm-fdl gene products to the Sf-fdl and Dm-fdl gene products, when compared with degradative and chitinolytic enzymes, supports the conclusion that these genes encode N-glycan processing β-N-acetylglucosaminidases.
β-N-acetylglucosaminidase assays
To test the hypothesis that these newly isolated genes encode N-glycan processing β-N-acetylglucosaminidases, we directly examined the substrate specificity of the over-expressed Bm-fdl and Tn-fdl gene products. If these gene products are involved in N-glycan processing, they should specifically remove N-acetylglucosamine from the α-3, but not the α-6 mannose branch of N-glycans. Thus, we assayed the Bm-fdl and Tn-fdl gene products using substrates (GnGn, GnM, and MGn; Figure 1B) that allowed us to discriminate between specific and non-specific cleavage of terminal N-acetylglucosamine residues on N-glycans. We also assessed the chitinolytic activities of these gene products using chitotriose (GlcNAc3; Figure 1B), a small chitin-like compound.
The full-length Bm-fdl and Tn-fdl gene products were over-expressed in Sf9 cells using baculovirus expression vectors. Microsomal membranes were then isolated and assayed for activity using various glycan substrates, as described in Materials and Methods. Microsomes from cells over-expressing the Sf-FDL protein served as a specific, processing enzyme control and microsomes from insect cells expressing SfGlcNAcase-3 served as a non-specific, degradative enzyme control. Microsomes from cells infected with wild-type AcMNPV served as a control to determine background β-N-acetylglucosaminidase activity levels.
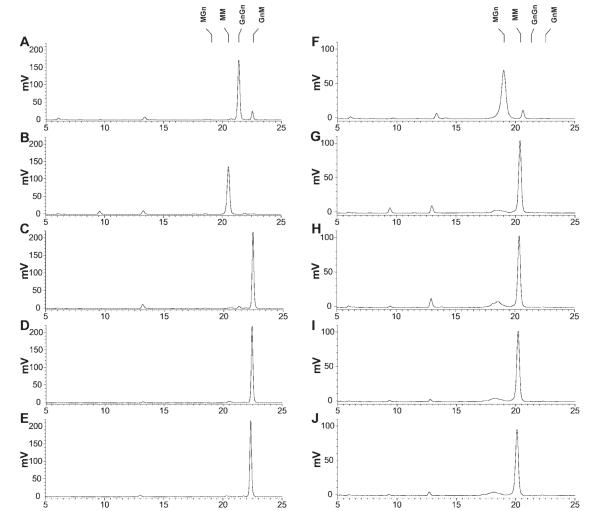
Baculovirus-infected Sf9 cells have low levels of specific, processing β-N-acetylglucosaminidase activity, which is derived from the endogenous N-glycan processing machinery.17,21,22 Accordingly, microsomes from Sf9 cells infected with wild-type baculovirus converted about 10% of MGn to MM and about 11% of GnGn to GnM (Figure 6). On the other hand, microsomes from cells infected with AcSf-FDL, AcTn-FDL, and AcBm-FDL quantitatively converted GnGn to GnM and MGn to MM, reflecting an increase of at least 10-fold in the levels of specific, processing β-N-acetylglucosaminidase activity. The specificity of these enzymes for terminal N-acetylglucosamine on the α-3 mannose branch was evidenced by the complete absence of any MM or MGn reaction products in the GnGn digests. In contrast to these highly specific enzymes, microsomes from cells infected with AcSfGlcNAcase-3 quantitatively degraded both GnGn and MGn to MM, reflecting the non-specific nature of this enzyme, as described earlier.21,22
Figure 6. Activity of recombinant insect β-N-acetylglucosaminidases on glycan substrates with an N-acetylglucosamine residue on the α1,3 mannose branch.
Approximately 50 pmoles of the 2-aminopyridine derivatized N-glycan substrates GnGn (left column) and MGn (right column) were incubated for 16 h with solubilized microsomal fractions containing 20 μg of total protein from Sf9 cells infected with wild-type AcMNPV (panels A and F), AcSfGlcNAcase-3 (panels B and G), AcSf-FDL (panels C and H), AcBm-FDL (panels D and I), or AcTn-FDL (panels E and J). The reaction products were then analyzed by reverse phase HPLC with fluorescent detection, as described in Materials and Methods. The markers indicate the elution times of the relevant glycan standards. This reverse phase HPLC method resolves glycans according to the degree of their interaction with the hydrophobic stationary phase. Hence, this method can be used to separate very similar glycans, even if they differ only in the linkages connecting individual sugars. The relative elution positions of the glycans have previously been validated in several studies.17,20,21
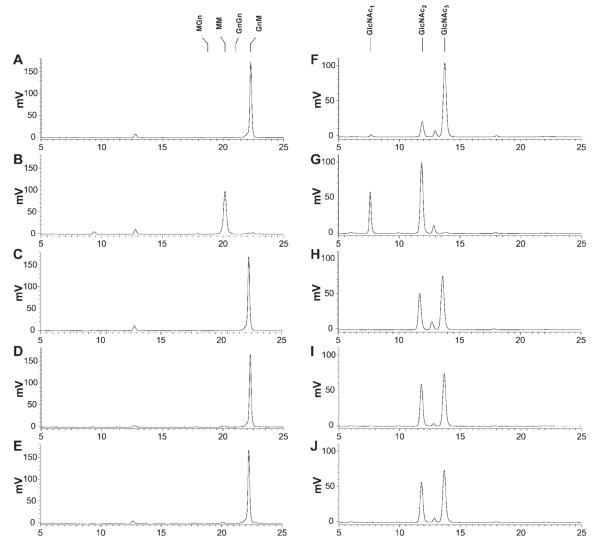
Microsomes from cells infected with wild-type baculovirus had no detectable activity against GnM (Figure 7). Similarly, microsomes from cells infected with AcSf-FDL, AcTn-FDL, and AcBm-FDL converted only a tiny fraction of GnM to MM (1.0, 2.3, and 1.7%, respectively). This was in stark contrast to the control reaction with microsomes from cells infected with AcSfGlcNAcase-3, which, as expected due to its nonspecificity,21,22 quantitatively converted GnM to MM.
Figure 7. Activity of recombinant insect β-N-acetylglucosaminidases on glycan substrates without an N-acetylglucosamine residue on an α1,3 mannose branch.
Approximately 50 pmoles of the 2-aminopyridine derivatized glycan substrates GnM (left column) and tri-N-acetylchitotriose (right column) were incubated for 16 h with solubilized microsomal fractions containing 20 μg of total protein from Sf9 cells infected with wild-type AcMNPV (panels A and F), AcSfGlcNAcase-3 (panels B and G), AcSf-FDL (panels C and H), AcBm-FDL (panels D and I), or AcTn-FDL (panels E and J). The reaction products were then analyzed by reverse phase HPLC with fluorescent detection, as described under Materials and Methods. The markers indicate the elution times of the relevant glycan standards.
Microsomes from Sf9 cells infected with a wild-type baculovirus have some chitinolytic activity, which presumably reflects expression of the baculovirus-encoded chitinase and/or an endogenous low-level cellular chitinase activity.29 Reactions containing microsomes from wild-type baculovirus-infected cells converted 15% of GlcNAc3 to GlcNAc2, but did not cleave the produced GlcNAc2 further to produce any detectable GlcNAc1. Microsomes from cells infected with AcSf-FDL, AcTn-FDL, and AcBm-FDL had higher levels of chitinolytic activity, converting 38%, 42%, and 41% of GlcNAc3 to GlcNAc2, respectively. However, none of these reactions showed any evidence of further degradation of GlcNAc2 to GlcNAc1. On the other hand, microsomes from AcSfGlcNAcase-3 infected cells not only quantitatively degraded GlcNAc3 to GlcNAc2, but also converted about 30% of the resulting GlcNAc2 to GlcNAc1, reflecting the non-specificity of this enzyme.
Discussion
T. ni, B. mori, and S. frugiperda are the most widely used hosts for baculovirus expression vectors, as larvae and cell lines derived from these species have the capacity to produce high levels of recombinant proteins. Accordingly, the N-glycan processing potential of these hosts has been thoroughly investigated in previous studies. The results of these studies have shown that recombinant glycoproteins produced by S. frugiperda, T. ni, and B. mori acquire mainly paucimannose N-glycans in place of the complex N-glycans found on mammalian products.9,10 To address this limitation, mammalian glycosyltransferase genes have been incorporated into the genomes of S. frugiperda and T. ni cell lines to extend their protein N-glycan processing pathways13–15 and B. mori cells could be glycoengineered in the same fashion. Furthermore, the N-glycan processing capabilities of these insect cell lines could be enhanced by reducing the endogenous processing β-N-acetylglucosaminidase activity, as the N-glycan trimming function of this enzyme competes with the N-glycan elongation function of the glycosyltransferases. In support of this notion, it has been shown that a non-glycoengineered insect cell line derived from Estigmene acrea, which has a naturally low level of processing β-N-acetylglucosaminidase activity, can produce N-glycans bearing a terminal N-acetylglucosamine residue.30 An essential prerequisite for efforts to similarly reduce the processing β-N-acetylglucosaminidase activities in S. frugiperda, T. ni, and B. mori through glycoengineering was to isolate the fdl genes of these three widely used hosts for BEVs.
In a previous study, we isolated the S. frugiperda fdl gene and experimentally confirmed its biochemical function.21 In this study, we fulfilled the prerequisite noted above by isolating the T. ni and B. mori fdl genes and experimentally confirming the biochemical function of the gene products. The nucleotide sequences of these lepidopteran insect fdl genes can now be used for focused glycoengineering efforts to reduce or eliminate the processing β-N-acetylglucosaminidase activity from all three host systems. For example, these sequences can be used to introduce specific double-stranded RNAs targeting the fdl genes into all three insect cell species to trigger the degradation of endogenous fdl mRNAs via the RNA interference machinery. A simple approach would be to transfect these cells with the relevant double-stranded RNAs. In a recent study, however, the use of this approach had very little impact on N-glycan processing in the Drosophila S2 cell system.19 Serious limitations of the transfection approach include the transient nature of the reduction in cellular mRNA levels and the severe economic constraints associated with transfecting cells at production scales. These limitations could be circumvented by generating transgenic insect cell lines designed to constitutively express double-stranded RNAs31 specific for the endogenous fdl gene. Indeed, in a preliminary study, we used this approach to reduce the specific N-glycan processing β-N-acetylglucosaminidase activity in Sf9 cells by about 60%.21
In addition to the cell lines derived from these species, T. ni and B. mori larvae are also used as hosts for BEVs. Transgenic variants of these insects can also be used as bioreactors to produce recombinant proteins.32 For example, B. mori larvae have recently shown great promise for tissue-specific recombinant protein production and secretion from their silk glands.33 Transgenic silk worms with a glycoengineered protein N-glycan processing pathway would be an invaluable resource for the production of recombinant therapeutic glycoproteins with complex N-glycans. Based upon the substantial increase in N-glycans terminating in N-acetylglucosamine in an fdl knockout strain of Drosophila,20 a targeted deletion of the Bm-fdl gene might be expected to permit the silkworm to produce recombinant glycoproteins with hybrid N-glycans. Targeted recombination to delete entire genes has recently become routine in insects, including the silkworm B. mori.34 Thus, through the identification of the fdl genes in T. ni and B. mori, this study has set the stage for future subtractive glycoengineering designed to create improved insect hosts for the production of recombinant glycoproteins with mammalian-like N-glycans.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants RO1 GM49734 and RO1 GM80672.
Footnotes
Additional Supporting Information may be found in the online version of this article.
Literature Cited
- 1.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 2.Marchal I, Jarvis DL, Cacan R, Verbert A. Glycoproteins from insect cells: sialylated or not? Biol Chem. 2001;382:151–159. doi: 10.1515/BC.2001.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu D, van Halbeek H. N-glycosylation site mapping of human serotransferrin by serial lectin affinity chromatography, fast atom bombardment-mass spectrometry, and 1H nuclear magnetic resonance spectroscopy. Anal Biochem. 1992;206:53–63. doi: 10.1016/s0003-2697(05)80010-7. [DOI] [PubMed] [Google Scholar]
- 4.Ailor E, Takahashi N, Tsukamoto Y, Masuda K, Rahman BA, Jarvis DL, Lee YC, Betenbaugh MJ. N-glycan patterns of human transferrin produced in Trichoplusia ni insect cells: effects of mammalian galactosyltransferase. Glycobiology. 2000;10:837–847. doi: 10.1093/glycob/10.8.837. [DOI] [PubMed] [Google Scholar]
- 5.Conradt HS, Egge H, Peter-Katalinic J, Reiser W, Siklosi T, Schaper K. Structure of the carbohydrate moiety of human interferon-β secreted by a recombinant Chinese hamster ovary cell line. J Biol Chem. 1987;262:14600–14605. [PubMed] [Google Scholar]
- 6.Kagawa Y, Takasaki S, Utsumi J, Hosoi K, Shimizu H, Kochibe N, Kobata A. Comparative study of the asparagine-linked sugar chains of natural human interferon-β1 and recombinant human interferon-β1 produced by three different mammalian cells. J Biol Chem. 1988;263:17508–17515. [PubMed] [Google Scholar]
- 7.Misaki R, Nagaya H, Fujiyama K, Yanagihara I, Honda T, Seki T. N-linked glycan structures of mouse interferon-β produced by Bombyx mori larvae. Biochem Biophys Res Commun. 2003;311:979–986. doi: 10.1016/j.bbrc.2003.10.094. [DOI] [PubMed] [Google Scholar]
- 8.Endo T, Ohbayashi H, Hayashi Y, Ikehara Y, Kochibe N, Kobata A. Structural study on the carbohydrate moiety of human placental alkaline phosphatase. J Biochem. 1988;103:182–187. doi: 10.1093/oxfordjournals.jbchem.a122228. [DOI] [PubMed] [Google Scholar]
- 9.Kulakosky PC, Hughes PR, Wood HA. N-Linked glycosylation of a baculovirus-expressed recombinant glycoprotein in insect larvae and tissue culture cells. Glycobiology. 1998;8:741–745. doi: 10.1093/glycob/8.7.741. [DOI] [PubMed] [Google Scholar]
- 10.Kulakosky PC, Shuler ML, Wood HA. N-glycosylation of a baculovirus-expressed recombinant glycoprotein in three insect cell lines. In Vitro Cell Dev Biol Anim. 1998;34:101–108. doi: 10.1007/s11626-998-0091-0. [DOI] [PubMed] [Google Scholar]
- 11.Morell AG, Gregoriadis G, Scheinberg IH, Hickman J, Ashwell G. The role of sialic acid in determining the survival of glycoproteins in the circulation. J Biol Chem. 1971;246:1461–1467. [PubMed] [Google Scholar]
- 12.Grossmann M, Wong R, Teh NG, Tropea JE, East-Palmer J, Weintraub BD, Szkudlinski MW. Expression of biologically active human thyrotropin (hTSH) in a baculovirus system: effect of insect cell glycosylation on hTSH activity in vitro and in vivo. Endocrinology. 1997;138:92–100. doi: 10.1210/endo.138.1.4897. [DOI] [PubMed] [Google Scholar]
- 13.Harrison RL, Jarvis DL. Protein N-glycosylation in the baculovirus-insect cell expression system and engineering of insect cells to produce “mammalianized” recombinant glycoproteins. Adv Virus Res. 2006;68:159–191. doi: 10.1016/S0065-3527(06)68005-6. [DOI] [PubMed] [Google Scholar]
- 14.Geisler C, Jarvis DL. Insect cell glycosylation patterns in the context of biopharmaceuticals. In: Walsh G, editor. Post-Translational Modifications in the Context of Biopharmaceuticals. 1st ed Wiley-VCH; Weinheim: 2009. pp. 165–191. [Google Scholar]
- 15.Shi X, Jarvis DL. Protein N-glycosylation in the baculovirus-insect cell system. Curr Drug Targets. 2007;8:1116–1125. doi: 10.2174/138945007782151360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomiya N, Narang S, Lee YC, Betenbaugh MJ. Comparing N-glycan processing in mammalian cell lines to native and engineered lepidopteran insect cell lines. Glycoconj J. 2004;21:343–360. doi: 10.1023/B:GLYC.0000046275.28315.87. [DOI] [PubMed] [Google Scholar]
- 17.Altmann F, Schwihla H, Staudacher E, Glössl J, März L. Insect cells contain an unusual, membrane-bound β-N-acetylglucosaminidase probably involved in the processing of protein N-glycans. J Biol Chem. 1995;270:17344–17349. doi: 10.1074/jbc.270.29.17344. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe S, Kokuho T, Takahashi H, Takahashi M, Kubota T, Inumaru S. Sialylation of N-glycans on the recombinant proteins expressed by a baculovirus-insect cell system under β-N-acetylglucosaminidase inhibition. J Biol Chem. 2002;277:5090–5093. doi: 10.1074/jbc.M110548200. [DOI] [PubMed] [Google Scholar]
- 19.Kim YK, Kim KR, Kang DG, Jang SY, Kim YH, Cha HJ. Suppression of β-N-acetylglucosaminidase in the N-glycosylation pathway for complex glycoprotein formation in Drosophila S2 cells. Glycobiology. 2009;19:301–308. doi: 10.1093/glycob/cwn138. [DOI] [PubMed] [Google Scholar]
- 20.Léonard R, Rendic D, Rabouille C, Wilson IB, Preat T, Altmann F. The Drosophila fused lobes gene encodes an N-acetylglucosaminidase involved in N-glycan processing. J Biol Chem. 2006;281:4867–4875. doi: 10.1074/jbc.M511023200. [DOI] [PubMed] [Google Scholar]
- 21.Geisler C, Aumiller JJ, Jarvis DL. A fused lobes gene encodes the processing β-N-acetylglucosaminidase in Sf9 cells. J Biol Chem. 2008;283:11330–11339. doi: 10.1074/jbc.M710279200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aumiller JJ, Hollister JR, Jarvis DL. Molecular cloning and functional characterization of β-N-acetylglucosaminidase genes from Sf9 cells. Protein Expr Purif. 2006;47:571–590. doi: 10.1016/j.pep.2005.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Summers MD, Smith GE. A Manual of Methods for Baculovirus Vectors and Insect Cell Culture Procedures. Texas Agricultural Experiment Station; College Station, TX: 1987. [Google Scholar]
- 24.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3. 0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 25.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 26.Prag G, Papanikolau Y, Tavlas G, Vorgias CE, Petratos K, Oppenheim AB. Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540. J Mol Biol. 2000;300:611–617. doi: 10.1006/jmbi.2000.3906. [DOI] [PubMed] [Google Scholar]
- 27.Okada T, Ishiyama S, Sezutsu H, Usami A, Tamura T, Mita K, Fujiyama K, Seki T. Molecular cloning and expression of two novel β-N-acetylglucosaminidases from silkworm Bombyx mori. Biosci Biotechnol Biochem. 2007;71:1626–1635. doi: 10.1271/bbb.60705. [DOI] [PubMed] [Google Scholar]
- 28.Nagamatsu Y, Yanagisawa I, Kimoto M, Okamoto E, Koga D. Purification of a chitooligosaccharidolytic β-N-acetylglucosaminidase from Bombyx mori larvae during metamorphosis and the nucleotide sequence of its cDNA. Biosci Biotechnol Biochem. 1995;59:219–225. doi: 10.1271/bbb.59.219. [DOI] [PubMed] [Google Scholar]
- 29.Hawtin RE, Arnold K, Ayres MD, Zanotto PM, Howard SC, Gooday GW, Chappell LH, Kitts PA, King LA, Possee RD. Identification and preliminary characterization of a chitinase gene in the Autographa californica nuclear polyhedrosis virus genome. Virology. 1995;212:673–685. doi: 10.1006/viro.1995.1525. [DOI] [PubMed] [Google Scholar]
- 30.Wagner R, Geyer H, Geyer R, Klenk HD. N-acetyl-β-glucosaminidase accounts for differences in glycosylation of influenza virus hemagglutinin expressed in insect cells from a baculovirus vector. J Virol. 1996;70:4103–4109. doi: 10.1128/jvi.70.6.4103-4109.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 32.Tamura T, Thibert C, Royer C, Kanda T, Abraham E, Kamba M, Komoto N, Thomas JL, Mauchamp B, Chavancy G, Shirk P, Fraser M, Prudhomme JC, Couble P. Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nat Biotechnol. 2000;18:81–84. doi: 10.1038/71978. [DOI] [PubMed] [Google Scholar]
- 33.Tomita M, Munetsuna H, Sato T, Adachi T, Hino R, Hayashi M, Shimizu K, Nakamura N, Tamura T, Yoshizato K. Transgenic silkworms produce recombinant human type III procollagen in cocoons. Nat Biotechnol. 2003;21:52–56. doi: 10.1038/nbt771. [DOI] [PubMed] [Google Scholar]
- 34.Yamao M, Katayama N, Nakazawa H, Yamakawa M, Hayashi Y, Hara S, Kamei K, Mori H. Gene targeting in the silkworm by use of a baculovirus. Genes Dev. 1999;13:511–516. doi: 10.1101/gad.13.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.