Abstract
Spider silk has been evolutionarily optimized for contextual mechanical performance over the last 400 million years. Despite precisely balanced mechanical properties, which have yet to be reproduced, the underlying molecular architecture of major ampullate spider silk can be simplified being viewed as a versatile block copolymer. Four primary amino acid motifs: polyalanine, (GA)n, GPGXX, and GGX (X = G,A,S,Q,L,Y) will be considered in this study. Although synthetic mimetics of many of these amino acid motifs have been produced in several biological systems, the source of spider silk’s mechanical integrity remains elusive. Mechanical robustness may be a product not only of the amino acid structure but also of the tertiary structure of the silk. Historically, solid state Nuclear Magnetic Resonance (ssNMR) has been used to reveal the crystalline structure of the polyalanine motif; however, limitations in amino acid labeling techniques have obscured the structures of the GGX and GPGXX motifs thought to be responsible for the structural mobility of spider silk. We describe the use of metabolic pathways to label tyrosine for the first time as well as to improve the labeling efficiency of proline. These improved labeling techniques will allow the previously unknown tertiary structures of major ampullate silk to be probed.
1. Introduction
Silk production is fundamental to spiders and is hypothesized to have evolved at least 400 million years ago (Gatesy et al. 2001). Although not the only organisms that produce silk, spiders are exceptional in their ability to produce silk throughout their lifetime. Spider silk has been specialized to perform diverse ecological functions from egg case protection, to lining their living areas, to a structural fiber for prey capture, allowing them to develop a broad ecological niche. Based on morphological, ecological, and genetic differences, these pervasive arachnids can be subdivided into orb-weavers such as the Argiope and Nephila species and cobweb weavers such as the Latrodectus species (Hayashi et al. 1999). Orb-weaving species have evolved to produce up to six solid silk fibers and an aqueous silk glue (Peters 1955). Historically, major ampullate silk has been the most extensively studied of all the fibers because of its dichotomous mechanical properties that combine strength and elasticity. Major ampullate silk is a blend of two proteins, major ampullate spidroin protein 2 (MaSp2) and major ampullate spidroin protein 1 (MaSp1) (Hinman et al. 1992). Although partial genetic sequences of the major ampullate silk proteins for many spider species were revealed over a decade ago, very little is known about the overall spider genome and metabolic pathways.
Although in an evolutionary context metabolic pathways may be impacted by natural ecological variations in a spider’s diet, the genetically encoded metabolic pathways remain unchanged by amino acid enrichment or deprivation (Zax et al. 2004). Regardless since silk is critical for many ecological functions and its production is not confined to specific maturation states, much of the spider’s metabolic balance is expended on the production of silk. Therefore, isotopically labeled amino acids and glucose provide a glimpse into some of the major amino acid metabolic pathways in three distinct spider families, Argiope, Nephila, and Latrodectus. Solid state nuclear magnetic resonance (ssNMR) can be used, in the context of known major and minor ampullate amino acid sequences, to assess the fidelity of amino acid labeling as an indication of metabolic processing. Importantly, major ampullate fibers can be harvested from the spider without endangering the spiders’ life (Work et al. 1982). Thus, it is possible to metabolically label (Michal et al. 1998; Holland et al. 2008b) silk fibers from an individual spider through a time course (up to several months) allowing a unique look into the amino acid processing and metabolic pathways employed during spider silk production. Using a novel labeling technique, developed in the course of this study, amino acid components of major ampullate silk fibers were targeted and isotopically enriched and ssNMR was used as a tool to reveal unique metabolic pathways.
SsNMR pulse sequences have been used in past studies to reveal secondary, tertiary and quaternary protein structure; however, samples relying on natural isotopic abundance are useful for only the very basic experiments. A mere increase of 9% of the total 13C in a sample allows an extended repertoire of pulse sequences to be feasible. As the amino acid label increases the signal to noise ratio also increases while the number of scans decreases (decreasing the overall time required for the experiment) according to the square of the signal to noise. Thus, to enable the use of more complex ssNMR pulse sequences and provide additional structural information isotopically enriched samples are needed. Although earlier techniques to enrich the sequence with isotopically labeled alanine and glycine enabled seminal ssNMR to probe the backbone (Asakura et al. 1994; van Beek et al. 2002) geometry of the silk sequence as well as the LGXQ region (Michal et al. 1998), low concentration aqueous isotopic labeling solutions, label infidelity, and poor labeling efficiency as evidenced by mass spectrometry investigations, have prevented a systemic study of all repetitive elements. Instead of trying to overcome biology, amino acid metabolic pathways can be utilized to precisely incorporate an amino acid label on key silk amino acids such as tyrosine and proline. For the first time, these samples offer a glance into previously obscured amino acid sequence secondary structures (Holland et al. 2008b) and also allow the study of amino acid metabolic pathways of spiders.
2. Materials and Experimental Details
2.1 Silking
Argiope argentata, Nephila clavipes, and Latrodectus hesperus major ampullate silk was collected by forcibly silking two adult female spiders of each species at 2 cm/s (Work et al. 1982) every other day for each labeling scheme. The spiders were anesthetized using CO2 and silk collection did not occur until spiders were able to drink 20 μL of an aqueous solution to ensure that they were revived. The silking process was monitored under a dissection microscope to ensure that only major ampullate silk was collected. Each silking session was one hour in length and constituted a single “silking” in Figures 1-3 and Table 1.
Figure 1.
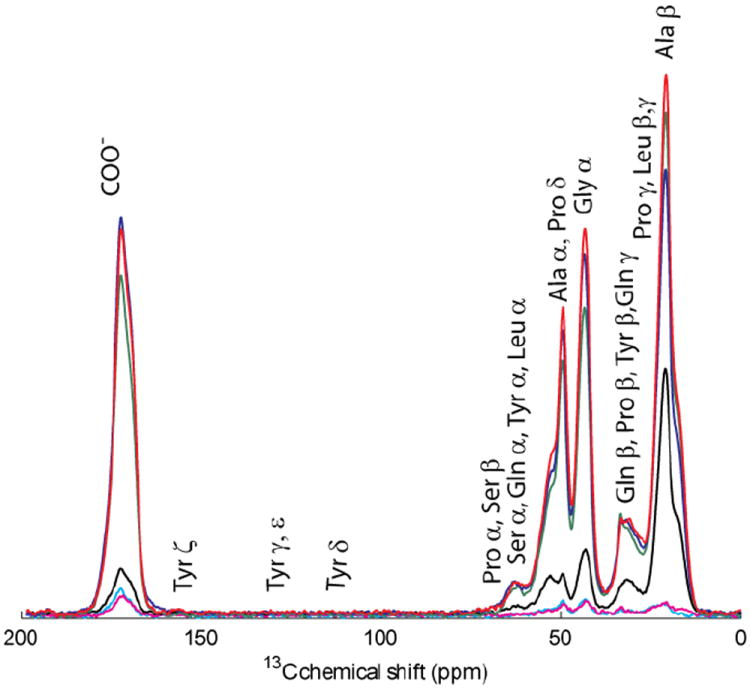
CP/MAS of the 3-13C alanine labeled silks at different silking increments, Natural, Silkings 1-2, Silkings 3-5, Silkings 6-9, Silkings 11-12, and Silkings 13-16. Silking refers to each individual one hour session during which silk was forcibly collected from the spider.
Figure 3.

CP/MAS of natural and U-15N-13C proline labeled (in black) major ampullate silk from Nephila clavipes (bottom), Latrodectus hesperus (middle), and Argiope argentata (top).
Table 1.
| Silkings | ppm | 1-2 | 3-5 | 6-9 | 11-12 | 13-16 |
|---|---|---|---|---|---|---|
| Ala Cβ | 20.9 | 1.4 | 34.8 | 50.6 | 49.4 | 63.9 |
| Gln Cγ | 33.3 | 1.6 | 6.6 | 16.5 | 16.8 | 18.3 |
| Gly Cα | 43.5 | 1.1 | 6.0 | 20.8 | 26.8 | 30.8 |
| Ala Cα | 49.6 | 0.9 | 4.0 | 18.1 | 22.2 | 25.6 |
| Gln & Ser Cα | 52.8 | 0.6 | 8.6 | 21.6 | 26.5 | 30.4 |
| Ser Cβ | 61.8 | 0.6 | 9.3 | 25.3 | 27.7 | 29.2 |
| COO- | 172.3 | 1.1 | 3.1 | 16.9 | 22.0 | 23.1 |
| Amount of Silk (mg) | 5.8 | 4.6 | 6.5 | 4.9 | 4.6 | |
| Amount of 3-13C alanine in 16.5% w/v aqueous solution (μL/spider) | ~160 | ~240 | ~240 | ~160 | ~320 |
2.2 Labeling
Two Nephila clavipes were used for each labeling strategy; a total of 24 spiders were used for these experiments. The spiders were fed one cricket per silking in addition to a solution of one of a variety of isotopically labeled amino acids or sugar: 3-13C1 alanine, U-13C3-15N alanine, 2-13C1-15N glycine, U-13C5-15N proline, U-13C6-15N leucine, U-13C9-15N phenylalanine, U-13C6 glucose (Cambridge Isotope Laboratories, Andover, MA, USA) 3, 5-13C2 tyrosine or 4-13C1 tyrosine (Los Alamos National Laboratory, Los Alamos NM, USA). Two Argiope argentata and two Latrodectus hesperus were fed saturated aqueous U-13C5-15N proline (Cambridge Isotope Laboratories) solutions. All aqueous solutions were made using distilled H2O. Spiders were only fed an aqueous solution of labeled amino acids or sugar (up to 80 μL) while actively being silked; however, they were misted daily with unlabelled room temperature water. The amount collected during each session varied from 0.5 mg to 1 mg, depending on: 1) the diameter of the silk, 2) if silk was obtained from both major ampullate spigots, and 3) the duration of the silking (up to one hour).
2.3 NMR
Solid-state NMR spectra were collected on a Varian VNMRS (Palo Alto, CA, USA) 400 MHz wide-bore spectrometer equipped with a 3.2 mm triple resonance MAS probe operating in double resonance mode (1H/13C). The major ampullate silk samples were packed in zirconia rotors. Notably, the thermal properties of spider silk indicate that the fibers will not be impacted by heating effects from MAS and/or 1H decoupling (Kaplan 1998). 1H→13C (Cross Polarization/Magic Angle Spinning) CP/MAS spectra were collected at 10 kHz MAS with the CP condition matched to the -1 spinning sideband from the Hartmann-Hahn profile. 13C CP/MAS NMR spectra of 13C-labeled samples were collected using a 4 μs 1H pulse, a 1 ms CP pulse at 62.5 kHz rf field strength, 100 kHz two pulse phase modulated (TPPM) (Bennet et al. 1995) 1H decoupling during acquisition, 1024 data points, 128 scans, a 50 kHz sweep width, and a 4 s recycle delay. Processing parameters for 13C CP/MAS spectra were zero-filling to 4096 points and 50 Hz of exponential line broadening. Chemical shifts were attained utilizing an external adamantane standard setting the downfield peak at 38.56 ppm.
3. Theory
The structure of spider silk has long remained obscured by the same repetitive amino acid sequences that lend it to genetic manipulation for commercial and medical applications. Incorporating a labeled amino acid with engineering-like precision can be accomplished by taking advantage of the spider’s metabolic pathways and administering saturated amino acid solutions. For the first time the tyrosine component of the GGX motif, where X=tyrosine, can now be labeled and probed with NMR by taking advantage of phenylalanine metabolism as can the proline component of the GPGXX motif.
4. Results and Discussion
Saturated isotopically labeled amino acid solutions provided enrichment in silk isotope characterization. Previously lower amounts of isotope enriched amino acids were used to look at the backbone structures of silk using ssNMR (Michal et al. 1998; van Beek et al. 2002). Traditionally, amino acid analysis (AAA) has been used to identify changes in amino acid composition from different spiders of the same species (Andersen 1970) and different regions of the same major ampullate fiber (data not shown). Unfortunately, a combination of natural variations in amino acid percentages, a relatively large coefficient of variation for AAA (Sarwar et al. 1985; Woolfitt et al. 2009), and an inability to precisely differentiate a labeled carbon (i.e. Ala Cα versus Ala Cβ) all limit the utility of this technique. Therefore, in addition to previous literature studies that indicated no change in amino acid composition with amino acid enrichment or deprivation (Zax et al. 2004), it is improbable that AAA would reveal any changes between either MaSp1 or MaSp2 resulting from the isotope enriched diet or due to additional amino acid(s) or sugar. Nevertheless, AAA performed on other isotopically labeled major ampullate silk not included in this study did not reveal any differences, thus confirming the inadequacy of this technique (data not shown). In addition to limited utility AAA, as well as liquid state NMR or ESI-MS methods, the samples must undergo hydrolysis preventing further studies on the fiber. Additionally, previous mass spectroscopy studies (van Beek et al. 2002), which are particularly susceptible to label infidelity, using isotopically labeled amino acids suffer from the inability to identify the precise nature of the incorporation (13Cα vs 13Cβ). Hence, solid state NMR was chosen as it conserved the isotopically labeled major ampullate silk samples and provided the necessary level of structural detail to probe spider metabolic pathways. Although 1H→13C CP/MAS NMR is notorious for not being quantitative (Dudley et al. 1982), it provided a relative comparison of the incorporation based on increasing peak amplitude (Nakao et al. 1995). Importantly, any differential mobility in the sample can cause great error in quantification. Nevertheless, it was clear that the percent label incorporation correlated with an increased isotopic label administered, and that the label was metabolized or scrambled into other amino acids. Thus, percent incorporation reported (Table 1) is relative and does not represent an absolute quantity but does reveal important metabolic pathways. The novelty of the techniques presented here lies in the ability to incorporate a label into previously obscured tyrosine and proline containing major ampullate silk motifs.
4.1 Alanine
The abundance of alanine in major ampullate silk in the species investigated here varies between 18 and 30% (Andersen 1970; Casem et al. 1999). Alanine is found in three structural motifs and is the main component of the rigid β-sheet structure of silk (Asakura et al. 1994; Holland et al. 2008a). Initially, a 3-13C1-alanine label was incorporated via feeding a 0.1% w/v aqueous solution of labeled alanine; unfortunately, this method resulted in <3% incorporation. Complex pulse sequences used to determine structures are unrealistic at this percentage of 13C. This labeling percent is dismal compared to labels achieved in previous experiments performed by van Beek et al. (2002) and Michal et al (1998), where they obtained 11.8±6 - 27.2±17% (van Beek et al. 2002) and 11±4 - 14±7% (Michal et al. 1998) label incorporation on alanine. Thus, incorporation of an alanine label was then attempted two additional ways: (1) using a uniform label of 13C and (2) using labeled methyl carbon, where both labels were fed at saturated 16.5% w/v aqueous solutions. Although these labeling strategies led to high label incorporation, both labeling strategies demonstrated a lack of label fidelity or indiscriminant amino acid labeling indicating that alanine is metabolized in the production of other amino acids, i.e. the alanine labeled carbon is scrambled into other amino acids. This is seen in Figure 1 where silkings 1-16 and a natural silk are compared through CP/MAS spectra overlaid on one another. Table 1 shows the relative percent of 13C from the incorporations of the saturated aqueous solution of the 3-13C1-alanine gathered from Figure 1. This label was traced through ~6 day iterations to follow the percentages of incorporation to other amino acids. Notably, the peak at ~23 ppm for Cγ, which has been attributed to leucine is absent in the isotopic alanine labeled sample (Holland et al. 2008a). This is not surprising in light of other known metabolic pathways where leucine and valine are synthesized from threonine. Additionally, samples from the first two silkings are analyzed agreeing with previous data that it takes at least three silkings to obtain an appreciable amount of label incorporation (Michal et al. 1998). The uniform 13C alanine surpassed the percentages from the 3-13C1 alanine and yielded >60% 13C on the α- and β-carbons of alanine, and ~45% label on glycine.
4.2 Glycine
Glycine is the most abundant amino acid in the major ampullate silks of all spiders, being found in every motif; it ranges from 42-49% (Andersen 1970; Casem et al. 1999). Spiders were fed the 13C-glycine label using a 10% w/v aqueous solution with the label on the α-carbon only (Figure 2a). The glycine α-carbon showed an increase ~38% to 13C. Not surprisingly, the α-carbon of glycine was also shown to be scrambled into the α- and β-carbons of alanine, as well as to the α- and β-carbons of serine. Although the COO- group was also labeled, scrambling could not be determined because the overlap of the other amino acids’ carbonyl groups.
Figure 2.

CP/MAS of natural and labeled (in black) Nephila clavipes major ampullate silk a) 15N-2-13C glycine, b) U-15N-13C leucine and c) U-15N-13C phenylalanine→tyrosine labeling schemes.
4.3 Leucine
Leucine makes up only 0.8 to 4.6% (Andersen 1970; Casem et al. 1999) of the amino acids in major ampullate silk. It is primarily found in the GGX motif (Xu et al. 1990; Hinman et al. 1992; Hayashi et al. 1999) and is four times more abundant in Nephila clavipes when compared to the other spiders in this study. The uniform 13C leucine solution was saturated at 2.2% w/v. The label was incorporated into the leucine at about ~20% label on all of its carbons (Figure 2b). The leucine also scrambled into alanine on the Cα and Cβ and glycine on the Cα with 15% label. The general overall carbonyl region had a ~15% label incorporation.
4.4 Proline
Proline varies in the species from 1-11% of the amino acids. Proline is found almost exclusively in the GPGXX (Hayashi et al. 1999) motif, which is thought to give silk its elasticity. Proline, uniformly labeled with 13C using a 16.5% w/v aqueous solution, labeled the proline to ~20% and was scrambled into glutamine obtaining ~17-20% incorporation (Figure 3) in both the Argiope and Nephila species. Importantly, the Latrodectus species cannot ingest the quantity of the aqueous solutions as the other two species so the labeling obtained was ~10% on the proline and 5-7% on the glutamine. Surprisingly, a proline diet had the unintentional side effect of killing the spider within two weeks of beginning the labeling. After 4-5 feedings, their exoskeletons lost their functional rigidity, becoming appreciably softened. In retrospect, this is not unique to the arachnid family. At levels of 10mM and above, free proline, which is associated with an increase in apoptosis, has recognized toxicity in plants (Mani et al. 2002; Nanjo et al. 2003; Deuschle et al. 2004). Notably, the cytotoxic concentration of free proline in Arabidopsis is over 80 fold less than the relative amount used to feed the spiders. Isotopic labeled proline was fed to the spiders for approximately five silkings, until the spider seemed ill, as evidenced by an appreciably softer exoskeleton as well as lethargic behavior. Subsequently, the labeling was ceased and the spider was fed only crickets for a week at which time the labeling was resumed for another five feedings. The same procedure was performed with both the Nephila and the Argiope, allowing an increase of the 13C in proline to ~32-37%. The differences in the ratio of the MaSp1 to MaSp2 proteins does not make a difference in the amount of proline label obtained or the amount of time it took the label to make the spiders ill. The peaks attributed by proline (Figure 3) clearly show the label incorporation. Silk obtained from Nephila clavipes and the Latrodectus hesperus spiders whose diet was enriched with isotopic proline showed distinct proline peaks where in conditions of natural isotopic proline abundance these peaks are hard to distinguish above the baseline. The Argiope argentata does show a peak at the Pro Cα in the natural abundance silk but the intensity of the peak is increased substantially with the label.
4.5 Tyrosine
Tyrosine is found in the GGX and the GPGXX motifs and makes up 3 to 7% of amino acids in major ampullate silk. Labeled tyrosine was fed using a saturated 0.046 % w/v aqueous solution. The label was incorporated at ~ 1-2% but no higher incorporation was obtained. Presumably, the low incorporation was due to the extremely low solubility of the tyrosine. Based on other known metabolic pathways, (Nelson et al. 2000) it was hypothesized that labeled phenylalanine would be converted to tyrosine; thus incorporating a more pervasive label while confirming and capitalizing on the phenylalanine metabolic processing (Figure 2c).
4.6 Phenylalanine
Phenylalanine varies from trace amounts to 1% of the silk composition. It is found mainly in the c-terminal non-repetitive region in MaSp1. Uniform 13C phenylalanine was used in a saturated 2.79% w/v aqueous solution. This represented a 60-fold increase over the tyrosine label. The label stayed in phenylalanine uniformly labeling it. As predicted, the label quickly converted to tyrosine also, where it was seen clearly within the 3rd and 4th silkings, eventually providing a 22% label on all of the carbons in tyrosine (Figure 2c). Importantly, it did not scramble into the other amino acids in any discernable amount within 10 silkings.
4.7 Glucose
In an attempt to uniformly 13C label major ampullate silk, U-13C-glucose was fed to the spider’s daily with a cricket to supplement. On average ~6% of the 13C label was incorporated into the silk; whereas, a higher percent of 13C label ~8.5% (Holland et al. 2008b) was identified in alanine and glycine. Again, if glucose was dosed at greater than 20% w/v the spiders died; however, they thrived when provided with a 10% w/v solution.
5. Conclusion
Based on genotypic variation in spider silk proteins, it was assumed that metabolic pathways may differ between species; however, this manuscript demonstrates the conservation of metabolic pathways. The spider’s metabolic pathways follow very closely the reported amino acid metabolic pathways of plants and microorganisms. Although tyrosine is not essential, its saturations level is so low that the de novo amino acid precursor phenylalanine was used. This holds true in other organisms like Bombyx mori. These pathways can thus be exploited to study the major ampullate silks of many species. This important revelation can now be used to precisely incorporate amino acid labels leading to a more in depth determination of major ampullate silk tertiary structure and a probative tool to investigate the impact of silk protein sequence variation. Due to the governing structure/function relationship of spider silk, mimicking natural tertiary structure with a synthetic analog is critical for recapitulating the unique mechanical properties of spider silk. This study allows a label to be placed on specific amino acids via the metabolic pathway of the isotopically labeled amino acid. Using this principle, tyrosine was labeled at an appreciable level for the first time and the GGX (X=tyrosine) motif can now be probed. In fact this labeling scheme was recently used to investigate the structure and dynamics of the tyrosine in Nephila clavipes major ampullate silk (Izdebski et al. 2010). Additionally, the GPGXX motif can be investigated more fully due to the higher 13C uniform incorporation into proline as evidenced by the recent structural investigation comparing the structure of the proline region in Argiope aurantia major ampullate silk with elastin (Jenkins et al. 2010). Using the improved labeling methods described here a more thorough understanding of the structure function relationships that govern spider silk can now be more directly probed. Importantly, despite evolutionary differences between Argiope argentata, which has a high ratio of MaSp2 to MaSp1 proteins (Brooks et al. 2005), and Nephila clavipes, and Latrodectus hesperus, which present a higher MaSp1 to MaSp2 ratio, there were no apparent differences in the metabolic pathways in any of the spiders (Figure 3).
Acknowledgments
This work was supported by grants from the US National Science Foundation (NSF-DMR 0805197 and NSF-DMR 0805197), the US National Institute of Health (NIH-EB000490) and the US Department of Defense (DOD-AFOSR FA9550-10-1-0275). We would like to thank Dr. Jeff Yarger, Dr. Brian Cherry and Dr. Janelle Jenkins for their help with NMR experiments and the Magnetic Resonance Research Center at Arizona State University for the use of the NMR facility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen SO. Amino acid composition of spider silks. Comp Biochem Physiol. 1970;35:705–711. [Google Scholar]
- Asakura T, Demura M, Hiraishi Y, Ogawa K, Uyama A. Determination of the structure of [1-13C]glycine-[15N]alanine double labeled Bombyx mori silk fibroin fibers using solid state 15N NMR. Chem Letts. 1994;23:2249–2252. [Google Scholar]
- Bennet AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- Brooks AE, Steinkraus HB, Nelson SR, Lewis RV. An investigation of the divergence of major ampullate silk fibers from Nephila clavipes and Argiope aurantia. Biomacromolecules. 2005;6:3095–3099. doi: 10.1021/bm050421e. [DOI] [PubMed] [Google Scholar]
- Casem ML, Turner D, Houchin K. Protein and amino acid composition of silks from the cob weaver, Latrodectus hesperus (black widow) Int J Biol Macromol. 1999;24:103–108. doi: 10.1016/s0141-8130(98)00078-6. [DOI] [PubMed] [Google Scholar]
- Deuschle K, Funck D, Forlani G, Stransky H, Biehl A, Leister D, van der Graaff E, Kunze R, Frommer WB. The role of Δ1-pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell. 2004;16:3413–3425. doi: 10.1105/tpc.104.023622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley RL, Fyfe CA. Evaluation of the quantitative reliability of the 13C CP/MAS technique for the analysis of coals and related materials. Fuel. 1982;61:651–657. [Google Scholar]
- Gatesy J, Hayashi C, Motriuk D, Woods J, Lewis R. Extreme diversity, conservation, and convergence of spider silk fibroin sequences. Science. 2001;291:2603–2605. doi: 10.1126/science.1057561. [DOI] [PubMed] [Google Scholar]
- Hayashi CY, Shipley NH, Lewis RV. Hypotheses that correlate the sequence, structure, and mechanical properties of spider silk proteins. Int J Biol Macromol. 1999;24:271–275. doi: 10.1016/s0141-8130(98)00089-0. [DOI] [PubMed] [Google Scholar]
- Hinman M, Lewis RV. Isolation of a clone encoding a second dragline silk fibroin. Nephila clavipes dragline silk is a two-protein fiber. J of Biol Chem. 1992;267:19320–19324. [PubMed] [Google Scholar]
- Holland GP, Creager MS, Jenkins JE, Lewis RV, Yarger JL. Determining secondary structure in spider dragline silk by carbon-carbon correlation solid-state NMR spectroscopy. J Am Chem Soc. 2008a;130:9871–9877. doi: 10.1021/ja8021208. [DOI] [PubMed] [Google Scholar]
- Holland GP, Jenkins JE, Creager MS, Lewis RV, Yarger JL. Solid-state NMR investigation of major and minor ampullate spider silk in the native and hydrated states. Biomacromolecules. 2008b doi: 10.1021/bm700950u. [DOI] [PubMed] [Google Scholar]
- Izdebski T, Akhenblit P, Jenkins JE, Yarger JL, Holland GP. Structure and dynamics of aromatic residues in spider silk: 2D carbon correlation NMR of dragline fibers. Biomacromolecules. 2010;11:168–174. doi: 10.1021/bm901039e. [DOI] [PubMed] [Google Scholar]
- Jenkins JE, Creager MS, Butler EB, Lewis RV, Yarger JL, Holland GP. Solid-state NMR evidence for elastin-like beta-turn structure in spider dragline silk. Chem Commun (Camb) 2010;46:6714–6716. doi: 10.1039/c0cc00829j. [DOI] [PubMed] [Google Scholar]
- Kaplan DL. Fibrous proteins--silk as a model system. Polymer Degrad Stability. 1998;59:25–32. [Google Scholar]
- Mani S, Van de Cotte B, Van Montagu M, Verbruggen N. Altered levels of proline dehydrogenase cause hypersensitivity to proline and its analogs in Arabidopsis. Plant Physiol. 2002;128:73–83. [PMC free article] [PubMed] [Google Scholar]
- Michal CA, Jelinski LW. Rotational-echo double-resonance in complex biopolymers: a study of Nephila clavipes dragline silk. J Biomol NMR. 1998;12:231–241. doi: 10.1023/a:1008286004222. [DOI] [PubMed] [Google Scholar]
- Nakao Y, Yeung BKS, Yoshida WY, Scheuer PJ, Kelly-Borges M. Kapakahine B, a cyclic hexapeptide with an α-carboline ring system from the marine sponge Cribrochalina olemda. J Am Chem Soc. 1995;117:8271–8272. [Google Scholar]
- Nanjo T, Fujita M, Seki M, Kato T, Tabata S, Shinozaki K. Toxicity of free proline revealed in an arabidopsis T-DNA-tagged mutant deficient in proline dehydrogenase. Plant Cell Physiol. 2003;44:541–548. doi: 10.1093/pcp/pcg066. [DOI] [PubMed] [Google Scholar]
- Nelson DL, Cox MM. Lehninger Principles of Biochemistry. Worth Publishers; New York: 2000. [Google Scholar]
- Peters HM. Ueber den Spinnapparat von Nephila madagascariensis. Z Naturforsch. 1955;10:95. [Google Scholar]
- Sarwar G, Blair R, Friedman M, Gumbmann MR, Hackler LR, Pellett PL, Smith TK. Comparison of interlaboratory variation in amino acid analysis and rat growth assays for evaluating protein quality. J Assoc Anal Chem. 1985;68:52–56. [PubMed] [Google Scholar]
- van Beek JD, Hess S, Vollrath F, Meier BH. The molecular structure of spider dragline silk: folding and orientation of the protein backbone. Proc Natl Acad Sci USA. 2002;99:10266–10271. doi: 10.1073/pnas.152162299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfitt AR, Solano MI, Williams TL, Pirkle JL, Barr JR. Amino acid analysis of peptides using isobaric-tagged isotope dilution LC-MS/MS. Anal Chem. 2009;81:3979–3985. doi: 10.1021/ac900367q. [DOI] [PubMed] [Google Scholar]
- Work RW, Emerson PD. An apparatus and technique for the forcible silking of spiders. J Arachnol. 1982;10:1–10. [Google Scholar]
- Xu M, Lewis R. Structure of a Protein Superfiber: Spider Dragline Silk. Proc Natl Acad Sci USA. 1990;87:7120–7124. doi: 10.1073/pnas.87.18.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zax DB, Armanios DE, Horak S, Malowniak C, Yang Z. Variation of mechanical properties with amino acid content in the silk of Nephila clavipes. Biomacromolecules. 2004;5:732–738. doi: 10.1021/bm034309x. [DOI] [PubMed] [Google Scholar]


