Summary
Lysosomal storage diseases (LSDs) are often caused by mutations compromising lysosomal enzyme folding in the endoplasmic reticulum (ER), leading to degradation and loss of function. Mass spectrometry analysis of Gaucher fibroblasts treated with mechanistically distinct molecules that increase LSD enzyme folding, trafficking and function resulted in the identification of 9 commonly down-regulated and 2 jointly up-regulated proteins, which we hypothesized would be critical proteostasis network components for ameliorating loss-of-function diseases. LIMP-2 and FK506 binding protein 10 (FKBP10) were validated as such herein. Increased FKBP10 levels accelerated mutant glucocerebrosidase (GC) degradation over folding and trafficking, whereas decreased ER FKBP10 concentration led to more LSD enzyme partitioning into the calnexin profolding pathway, enhancing folding and activity to levels thought to ameliorate LSDs. Thus, targeting FKBP10 appears to be a heretofore unrecognized therapeutic strategy to ameliorate LSDs.
Keywords: endoplasmic reticulum associated degradation, lysosomal storage diseases, protein homeostasis
Introduction
The secreted eukaryotic proteome is folded in the endoplasmic reticulum (ER) (Brodsky and Skach, 2011), wherein folded proteins such as N-glycosylated lysosomal enzymes engage their trafficking receptors, triggering vesicular packaging for transport to the Golgi and on to the lysosome (Brodsky and Skach, 2011; Edmunds, 2010; Futerman and van Meer, 2004; Hebert, et al., 2010; Mazzulli, et al., 2011; Reczek, et al., 2007; Sun, et al., 2010; Vembar and Brodsky, 2008; Vitner, et al., 2010). Excessive misfolding and degradation of a mutant lysosomal enzyme results in a deficiency of lysosomal enzyme activity leading to substrate accumulation in several cell types, which manifests in a lysosomal storage disease (LSD) (Futerman and van Meer, 2004; Sun, et al., 2010; Vitner, et al., 2010; Zhao and Grabowski, 2002). The delicate balance between LSD enzyme folding, trafficking and function vs. degradation (Miyata, et al., 2011) is achieved by the ER proteostasis network comprising >75 proteins (Adachi, et al., 2008; Lee, et al., 2003; Okada, et al., 2002; Yamamoto, et al., 2007).
Treating LSD patient-derived fibroblasts, e.g., Gaucher fibroblasts, with either of two mechanistically distinct proteostasis regulators (PRs), molecules that enhance proteostasis network capacity without eliciting cytotoxicity at the levels used herein, led to more efficient folding and trafficking of several mutant misfolding-prone lysosomal enzymes, increasing their lysosomal activity (Mu, et al., 2008; Mu, et al., 2008; Ong, et al., 2010). The first mechanistic class of PRs, which includes MG-132, activates the unfolded protein response (UPR) (Mu, et al., 2008) stress-responsive signaling pathway that transcriptionally remodels the ER by up-regulating ER chaperones (Chen, et al., 2011; Hartl, et al., 2011) and degradation components, while also attenuating translation and expanding the ER (Ron and Walter, 2007; Schroder and Kaufman, 2005). While MG-132 is a proteasome inhibitor and induces additional stress-responsive signaling, we established that UPR activation is sufficient to account for MG-132's PR function (Mu, et al., 2008). The second mechanistic category of PRs, exemplified by the drug diltiazem, post-translationally enhances ER folding capacity at the expense of ER-associated degradation (ERAD) (Brodsky and Skach, 2011; Hebert, et al., 2010) by increasing the ER Ca2+ concentration, likely enhancing the chaperoning activity of a subset of Ca2+-dependent chaperones including calnexin, which is critical for the folding of glucocerebrosidase (GC), the lysosomal enzyme associated with Gaucher's disease, and other N-glycosylated lysosomal enzymes (Mu, et al., 2008; Ong, et al., 2010). Proteomic analyses of Gaucher fibroblasts treated with these compounds identified FK506 binding protein 10 (FKBP10), which when depleted, decreased the degradation of mutant GC, increasing its folding, trafficking, and lysosomal function. Depletion of FKBP10 also increased the lysosomal activity of α-mannosidase, suggesting that targeting FKBP10 may be a general therapeutic strategy for ameliorating LSDs.
Results
Comparative proteomics identifies additional GC ER proteostasis network components
Herein, we employed mass spectrometry, namely Multidimensional Protein Identification Technology (MudPIT), to compare the proteomes of Gaucher's disease patient-derived fibroblasts harboring the L444P GC mutation treated with MG-132 (0.8 μM, 3d) or diltiazem (10 μM, 5d), the PRs described above (Mu, et al., 2008; Mu, et al., 2008; Ong, et al., 2010). Cellular proteins whose levels were either significantly up- or down-regulated by PR treatment were identified using the spectral counting method (Figure 1A and Table S1 available online) (Eng, et al., 1994; Liao, et al., 2007; Liu, et al., 2004; Rikova, et al., 2007; Washburn, et al., 2001). A protein of interest was defined as one that had to be detected in both pharmacologically-treated and vehicle-treated fibroblast lysates (at least two peptides per protein). The concentration of the majority of the fibroblast proteome was unaltered by MG-132 or diltiazem treatment (Figure S1A), suggesting that most proteins were folding efficiently (Powers, et al., 2009) and thus their concentration was not significantly altered by proteostasis network modulation.
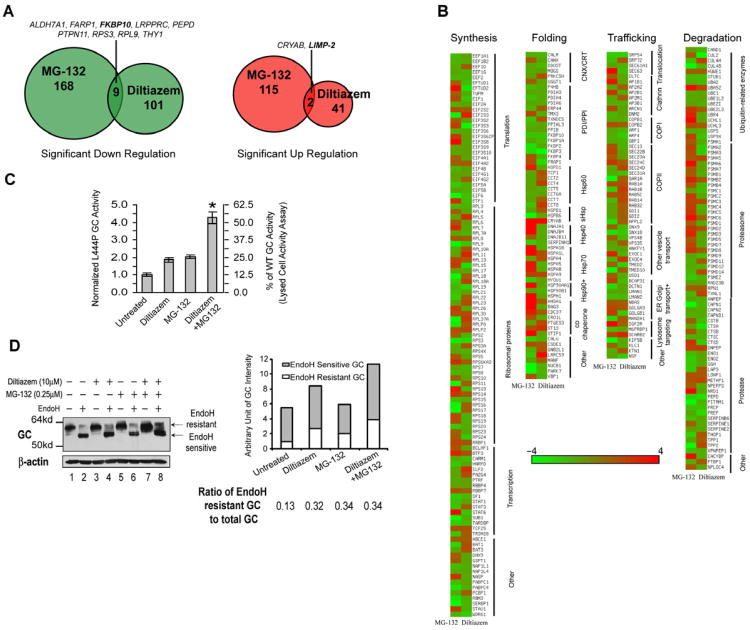
Figure 1.
Comparative proteomics identified additional GC proteostasis network components. (A) A Venn diagram showing the proteins that were significantly upregulated or downregulated by MG-132 or diltiazem in L444P GC fibroblasts. (B) Proteins that appeared in both MG-132- or diltiazem-treated fibroblasts are sorted into four categories, namely synthesis, folding, trafficking and degradation. (C and D) Diltiazem and MG-132 are mechanistically distinct and combining the two PRs synergistically increases GC activity (C) and the mature GC glycoform (D). Quantification of endo H resistant and sensitive GC bands is shown on the right of (D).The experiments were done three times. The data in (C) are reported as mean ± SEM (n=8) and any statistical significance was calculated using a two-tailed Student's t-Test. * p < 0.01. See also Figure S1.
Notably, the majority of proteins modulated by diltiazem treatment were not altered by MG-132 application and vice versa, confirming their largely distinct PR functions (Figures 1A and 1B). Oppositely up- or down-regulated proteostasis network components included those involved in (i) protein synthesis; (ii) folding (Bukau, et al., 2006; Deuerling and Bukau, 2004; Tang, et al., 2008); (iii) trafficking (Gurkan, et al., 2006); and (iv) degradation (Vembar and Brodsky, 2008) (Table S2). That MG-132 and diltiazem function as mechanistically distinct PRs was further supported by the 4.3-fold increase in L444P GC intact cell activity achieved by their co-application to fibroblasts, a slight synergistic effect (Figure 1C, * p<0.01). The endoglycosidase H (endo H) resistant GC band, representing the population of GC that had properly folded, exited the ER and trafficked at least through the Golgi compartment, was significantly increased upon diltiazem and MG-132 co-application (Figure 1D) and the GC transcript level was increased by 2.5 fold (Figure S1B), presumably owing to MG-132 treatment as diltiazem did not alter the GC mRNA level.
However, there were a few proteins that were either commonly up-regulated (2 proteins) or down-regulated (9 proteins) upon treatment with MG-132 or diltiazem (Figure 1A). Of these 11 proteins, we followed up on FKBP10, a jointly down-regulated candidate not previously associated with LSD enzyme proteostasis (Figure 1A), and LIMP-2 (also called SCARB2), a previously identified GC proteostasis network component (Reczek, et al., 2007) that was commonly up-regulated.
Validating LIMP-2 as an important GC proteostasis network component
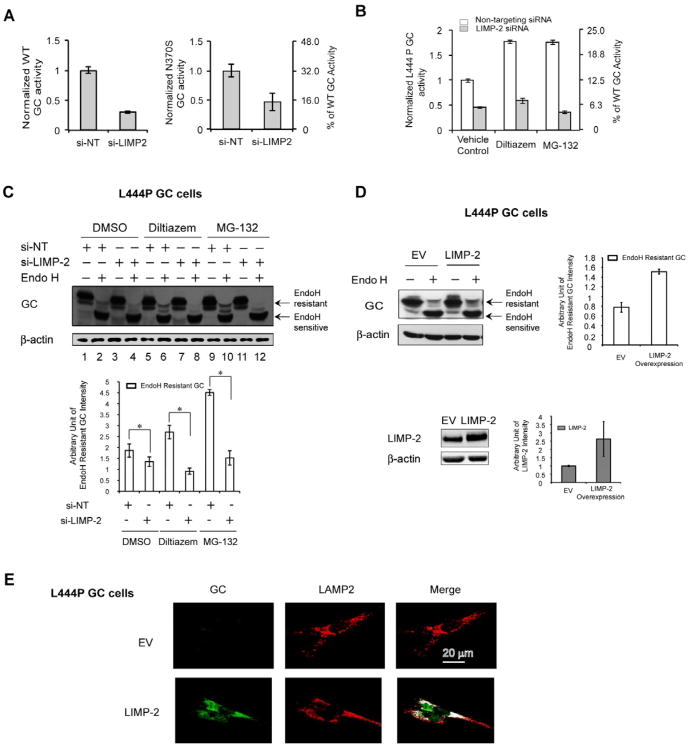
Consistent with the previously reported role of LIMP-2 as a GC trafficking receptor (Reczek, et al., 2007), LIMP-2 siRNA administration (> 90% knockdown, Figure S2) decreased GC activity by 3.3-fold in WT GC fibroblasts (Figure 2A, left), by 2.2-fold in N370S GC fibroblasts (Figure 2A, right), and by 2.2-fold in L444P GC fibroblasts (Figure 2B, vehicle control) relative to the non-targeting (NT) siRNA control. Moreover, co-application of LIMP-2 siRNA with either diltiazem or MG-132 in L444P GC fibroblasts substantially reduced the increase in L444P GC activity that was afforded by PR treatment (Figure 2B). Silencing LIMP-2 also decreased the endo H resistant GC band intensity (Figure 2C, * p<0.05).
Figure 2.
LIMP-2 is an important GC proteostasis trafficking network component. (A) The knockdown of LIMP-2 significantly decreases the WT and N370S GC activity in the fibroblasts when compared to the NT siRNA control. (B and C) Silencing LIMP-2 diminishes the increase in L444P GC activity (B) and the endo H resistant post-ER GC glycoform in western blot analysis (C) that is afforded by either MG-132 or diltiazem alone. Quantification of endo H resistant GC bands is shown at the bottom of (C).(D and E) The modest transient overexpression of LIMP-2 in L444P GC fibroblasts increases the endo H resistant post-ER GC glycoform in western blot analysis (D) and the lysosomal trafficking of L444P GC in indirect immunofluorescence study (E). Quantification of endo H resistant GC bands and LIMP-2 bands is shown on the right of (D).The experiments were done three times. The data in (A)-(D) are reported as mean ± SEM (n=8 for A and B) and any statistical significance was calculated using a two-tailed Student's t-Test. * p < 0.05. See also Figure S2.
In contrast, transient overexpression of LIMP-2 in L444P GC fibroblasts increased the endo H resistant post-ER GC glycoform (Figure 2D) and its lysosomal trafficking as detected by indirect immunofluorescence (Figure 2E), indicating enhanced ER exit and lysosomal trafficking. These data confirm the prior report of the importance of LIMP-2 in GC trafficking (Reczek, et al., 2007), and indicate that LIMP-2 is likely a limiting proteostasis network component for ER-to-Golgi-to-lysosome GC trafficking.
A decreased ER FKBP10 concentration enhanced lysosomal enzyme proteostasis
We next turned our attention to the ER FKBP10 protein, harboring four peptidylprolyl cis-trans isomerase (PPIase) domains and two C-terminal Ca2+-binding EF hand domains (Figure 3A). There are at least five FKBPs in the ER, and some of them (e.g., FKBP10, FKBP2 (or FKBP13) and FKBP7 (or FKBP23)) have been proposed to act as molecular chaperones or cochaperones (Feng, et al., 2011; Ishikawa, et al., 2008). FKBP10 (and possibly the highly homologous protein FKBP9) has been shown to inhibit the aggregation of denatured citrate synthase and rhodanese, suggesting that it may be a holdase chaperone (Ishikawa, et al., 2008). Perhaps the most convincing evidence that FKBP10 may be a chaperone is that mutations in FKBP10 cause collagen misfolding diseases (Alanay, et al., 2010; Kelley, et al., 2011; Shaheen, et al., 2011; Venturi, et al., 2012) and delay type I collagen fibril formation in vitro. The best understood FKBP protein, not in the FKBP9/10 family, is trigger factor, a ribosome-associated cytosolic chaperone that is clearly a holdase and a PPIase (Deuerling and Bukau, 2004; Stoller, et al., 1995). Given the redundancy in ER PPIases, one would expect that not all of them function solely as PPIases (Jakob, et al., 2009; Kang, et al., 2008). We set out to test the hypothesis that FKBP10 could influence the ER LSD enzyme degradation vs. folding decision, not a previously proposed role for FKBP10, but one that is consistent with the notion that PPIases can bind to proteins undergoing folding (Ishikawa, et al., 2008).
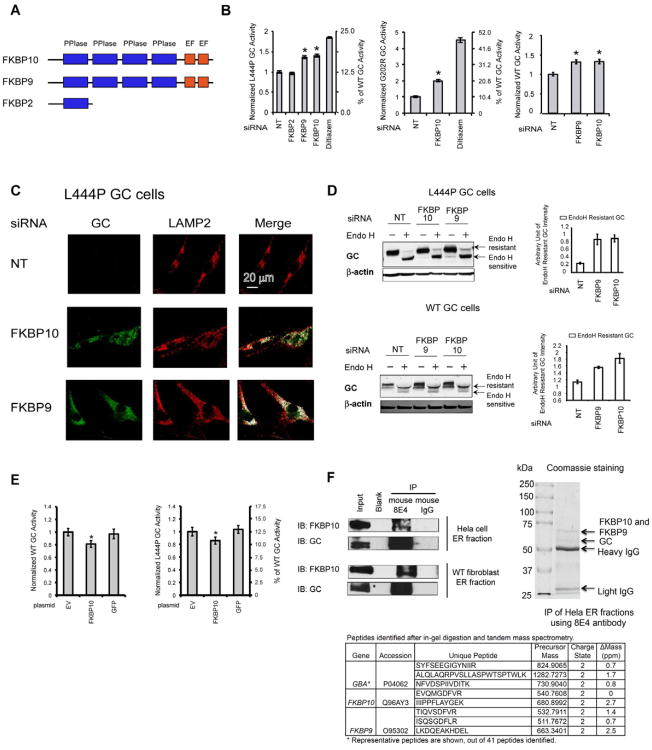
Figure 3.
FKBP10 is a novel GC proteostasis network component. (A) Schematic representation of the domain structures of FKBP10, FKBP9 and FKBP2. (B) Depleting the levels of FKBP10 (and FKBP9) significantly increases L444P, G202R and WT GC activity in fibroblasts. (C) Depleting the levels of FKBP10 and FKBP9 increases GC lysosomal trafficking in L444P GC fibroblasts. (D) Depleting the levels of FKBP10 and FKBP9 increases the endo H resistant GC glycoform in L444P and WT GC fibroblasts. Quantification of endo H resistant GC bands is shown on the right of (D). (E) The FKBP10 overexpression significantly decreases WT and L444P GC activity in fibroblasts. (F) The GC-FKBP10 endogenous interaction is confirmed using ER fractions from Hela cells or WT fibroblasts. Both Western blot analysis and tandem MS analysis identified FKBP10 after immunoprecipitation of Hela ER fractions using 8E4 GC antibody. The experiments were done three times. The data in (B),(D) and (E) are reported as mean ± SEM (n=8 for B and E) and any statistical significance was calculated using a two-tailed Student's t-Test. * p < 0.01. See also Figure S3.
FKBP10 levels were reduced 2-fold in either diltiazem– or MG-132–treated L444P GC fibroblasts (Table S1 and Figure S3A). The concentration of other FKBP family members was also suppressed by the application of these PRs (Table S2 and Figure 1B). FKBP10 mRNA levels in L444P GC fibroblasts decreased when treated with diltiazem (reduced 1.4-fold) or MG-132 (reduced 3-fold) or both (reduced 3-fold) vs. vehicle (Figure S3B), indicating that FKBP10 is primarily transcriptionally down-regulated. It is also established that FKBP10 is directed towards ERAD in fibroblasts in response to ER stress, known to result from MG-132 treatment (Murphy, et al., 2011).
SiRNA silencing of FKBP10 in L444P GC fibroblasts (>70% knockdown, Figure S3C) resulted in a 1.4-fold increase in GC activity (∼18 % of WT GC activity) vs. the NT siRNA control (Figure 3B, left, * p<0.01). While this increase may seem modest, it is thought to be sufficient to ameliorate Gaucher's disease (Schueler, et al., 2004). FKBP9 was not detected by MudPIT, but the similarity of its domain architecture with FKBP10 (Figure 3A) and its ER localization led us to also explore its potential role in lysosomal enzyme proteostasis. siRNA silencing of FKBP9 (>70% knockdown, Figure S3C) increased cellular L444P GC activity 1.4-fold (Figure 3B, left, * p<0.01). In contrast, silencing FKBP2, harboring only one PPIase domain (Figure 3A), did not alter L444P GC activity (Figure 3B, left). Silencing FKBP10 (>60% knockdown, Figure S3D) increased the activity of the Gaucher's G202R mutant 2.0-fold (Figure 3B, middle, * p<0.01). Silencing FKBP10 or FKBP9 (>60% knockdown, Figure S3E) also increased WT GC activity 1.3-fold (Figure 3B, right, * p<0.01). Indirect immunofluorescence microscopy studies revealed that knockdown of FKBP10 or FKBP9 dramatically increased the ability to detect cellular L444P GC, normally almost completely degraded by ERAD (Figure 3C; cf. green fluorescence in left panels), and importantly partially restored GC lysosomal colocalization, artificially colored white (Figure 3C; right panels). Moreover, knockdown of FKBP9 or FKBP10 increased the endo H resistant post-ER GC glycoform concentrations in L444P and WT GC fibroblasts (Figure 3D). Conversely, overexpressing FLAG-tagged FKBP10 (Figure S3F) decreased both WT and L444P GC activity by ∼ 20 % in patient-derived fibroblasts (Figure 3E, * p<0.01).
We measured the mRNA levels of GC and several established UPR markers (spliced XBP1, CHOP and BiP), as well as the protein levels of several established UPR markers (p-eIF2α, BiP and HRD1) and verified that FKBP10 depletion did not alter GC mRNA abundance or induce the UPR (Figures S3G and S3H). Nor did knockdown of FKBP10 (or FKBP9 and FKBP10) significantly alter L444P GC fibroblast viability (resazurin reduction assay; Figure S3I). Treating the cells (post-FKBP knockdown) with tunicamycin or thapsigargin to induce ER proteome misfolding/aggregation did not significantly exacerbate proteotoxicity. However, there was a consistent trend towards decreased cell viability upon UPR activation when both FKBP9 and FKBP10 were down regulated (Figure S3I, cf. left and right panels).
An endogenous GC-FKBP10 interaction in the ER
We detected a GC-FKBP10 endogenous interaction in immunoprecipitates of GC analyzed by western blot from the ER fractions of HeLa cells and WT GC fibroblasts (Figure 3F). Coomassie blue staining showed two strong bands between 50 kDa and 75 kDa in the GC immunoprecipitate of Hela ER fractions (Figure 3F). In-gel digestion and tandem MS analysis revealed that the top band contained both FKBP10 and FKBP9, whereas the bottom band contained GC. The unique peptides identified are listed in Figure 3F. Collectively, these results demonstrated that decreasing FKBP9 or FKBP10 levels in the ER enhanced mutant GC proteostasis in patient-derived fibroblasts through a direct interaction and without any involvement of an UPR.
FKBP10 binds to misfolding-prone GC directing it to ERAD
For mechanistic investigations, we used an overexpression system, focusing on FKBP10, since FKBP10 and FKBP9 appear to be structurally and functionally equivalent. Co-overexpression of FKBP10 together with GC decreased the steady state levels of VSVG-WT and -L444P GC (Figure 4A, input), not due to UPR induction (Figure S4A). Immunoprecipitating overexpressed WT or L444P VSVG-tagged GC from HeLa lysate showed that they bound similarly to glycosylated overexpressed FLAG-tagged FKBP10 (Figure 4A). Inhibiting proteasomal degradation using lactacystin led to more endogenous WT GC binding to overexpressed FKBP10 (Figure 4B), consistent with the hypothesis that FKBP10 is partitioning GC to ERAD. p53 was used as a control since it is degraded by the proteasome (Maki, et al., 1996).
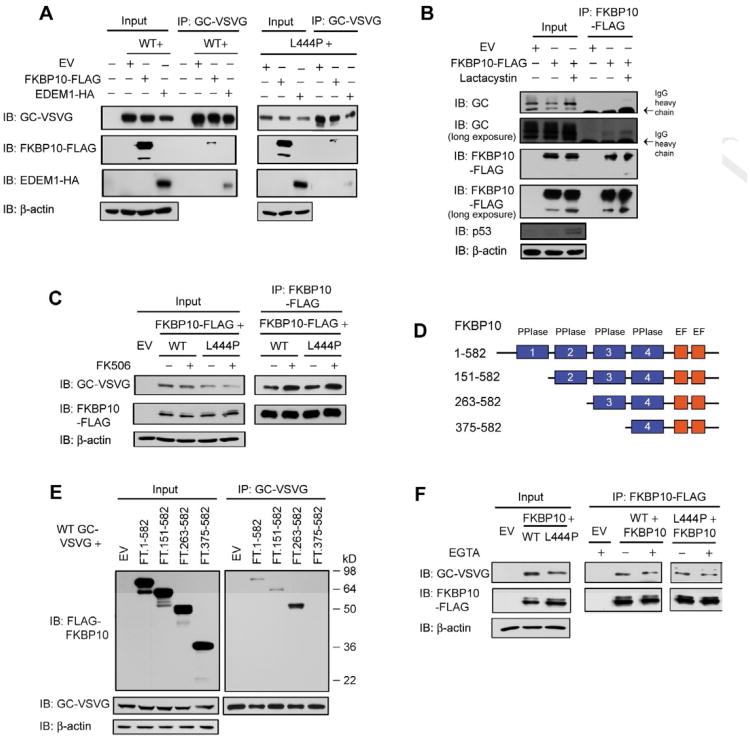
Figure 4.
FKBP10 partitions GC to the ERAD pathway, independent of its PPIase or Ca2+ binding activity. (A) Overexpressing FKBP10 or EDEM1 decreases the steady state levels of VSVG-tagged GC in HeLa cells. Both FKBP10 and EDEM1 bind to GC. (B) Inhibiting proteasomal degradation using lactacystin leads to more endogenous GC binding to overexpressed FKBP10. (C) FK506 treatment modestly decreases GC levels and enhances the GC–FKBP10 interaction in HeLa cells. (D) Schematic representation of the various FKBP10 truncations used in the immunoprecipitation experiment in (E). (E) A minimum of two PPIase domains is required for FKBP10 to associate with WT GC. (F) Chelating Ca2+ ions in the lysate using EGTA does not affect the GC–FKBP10 interaction. The experiments were done three times. See also Figure S4.
We hypothesized that FKBP10 enhances mutant GC degradation in fibroblasts and HeLa cells by binding to more misfolding-prone mutant GC and directing more of it to ERAD by a pathway that is distinct from the calnexin-EDEM1 ERAD pathway. Overexpressed EDEM1 (an acceptor of misfolded glycoproteins released from calnexin) (Molinari, et al., 2003; Oda, et al., 2003) bound to WT and L444P GC and decreased the GC steady state levels (Figure 4A, input). However, reducing the level of EDEM1 (Figures S4B and S4C) or ER α-mannosidase I (required for ERAD of terminally misfolded glycoproteins) (Figure S4D), or inhibiting the latter (Figure S4E) did not enhance L444P GC proteostasis in fibroblasts. Inhibiting ER α-mannosidase I noticeably increased the endo H sensitive GC glycoform, indicating ER accumulation of L444P GC without productive ER folding and export (Figures S4D and S4E).
FKBP10's function in GC proteostasis does not require its PPIase activity or Ca2+ binding
We investigated whether the PPIase activities of human FKBP10 affected its ability to influence GC proteostasis. Treatment with FK506, a general (not FKBP10-selective) PPIase inhibitor, for 3 d (0.1-1 nM) resulted in a modest, but significant decrease in L444P GC activity (Figure S4F) and a decrease in the total GC band as well as the endo H sensitive GC band, without significantly altering FKBP10 levels (Figure S4G). This contrasts with the increase in GC proteostasis observed upon FKBP10 silencing. FK506 treatment (0.1 nM) enhanced the GC-FKBP10 interaction, as more GC was detected in the FKBP10 immunoprecipitates by western blot analysis (Figure 4C, IP panels). Successive deletions of the PPIase domains from the N-terminus of FKBP10 (Figure 4D) did not disrupt the interaction between FLAG-tagged FKBP10 and VSVG-tagged WT GC (Figure 4E), with the exception of the 375-582 construct wherein all but one of the PPIase domains were deleted, suggesting that a minimum of two PPIase domains are required for binding to GC.
We also scrutinized the role of Ca2+ in the GC-FKBP10 interaction because mouse FKBP9 has been shown to bind Ca2+ in vitro (Shadidy, et al., 1999). Chelating Ca2+ ions from the lysate prior to immunoprecipitating FKBP10 revealed that the GC-FKBP10 interaction appeared to be Ca2+ independent (Figure 4F). Our data suggest that FKBP10 directs misfolding-prone GC into ERAD by associating with it in a manner that does not appear to require FKBP10's PPIase activity or Ca2+ binding.
FKBP10 overexpression accelerates the ERAD of mutant glucocerebrosidase
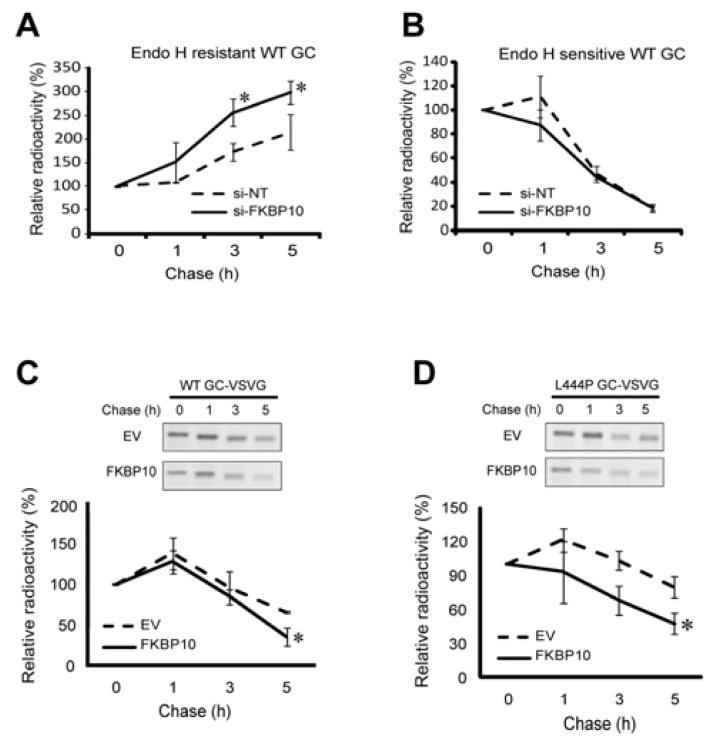
To examine whether FKBP10 influenced the degradation vs. folding and trafficking decision, we employed pulse-chase experiments, taking advantage of the fact that radiolabeled, endo H treated, WT GC will afford both endo H resistant (reflecting the rate of GC folding and trafficking; Figure 5A) and endo H sensitive GC (reflecting the rate of GC degradation; Figure 5B) bands (Jonsson, et al., 1987; Schmitz, et al., 2005). Upon FKBP10 silencing of WT GC fibroblasts, the rate of appearance of radiolabeled endo H resistant WT GC was significantly faster in comparison to the NT siRNA control (5h chase, Figure 5A), possibly due to increased association of WT GC with calnexin (see below). In contrast, there was no significant difference in the rate of disappearance of radiolabeled endo H sensitive WT GC between the FKBP10 siRNA and NT siRNA treated samples over the same chase period (Figure 5B). When VSVG-WT or -L444P GC and FKBP10-FLAG were co-overexpressed in HeLa cells, the rates of WT and L444P GC degradation were significantly faster with FKBP10 overexpression than with the empty vector control after a 5 h chase period (* p<0.05) (Figures 5C and 5D). A GC immunoisolation revealed that FKBP10 bound to newly synthesized WT and L444P GC at the outset of the chase period (0 h), and binding continued over a 5 h chase period, consistent with the notion that FKBP10 bound to and enhanced ERAD of GC (Figures S5A and S5B).
Figure 5.
The level of FKBP10 influences the degradation rate of both WT and L444P GC. (A) FKBP10 knockdown increases the rate of WT GC folding and trafficking (as reflected by the endo H resistant GC band) in fibroblasts when compared to the NT siRNA control. (B) Quantification of the endo H sensitive GC band intensity upon FKBP10 knockdown. (C and D) Overexpression of FKBP10 significantly increases the rate of WT (C) and L444P GC (D) degradation in HeLa cells compared to the empty vector control. Quantification of the bands was performed using ImageJ (NIH software). The experiments were done three times. The data are reported as mean ± SD (n>5) and any statistical significance was calculated using a two-tailed Student's t-Test. * p < 0.05. See also Figure S5.
FKBP10 may collaborate with OS-9 to deliver GC for degradation
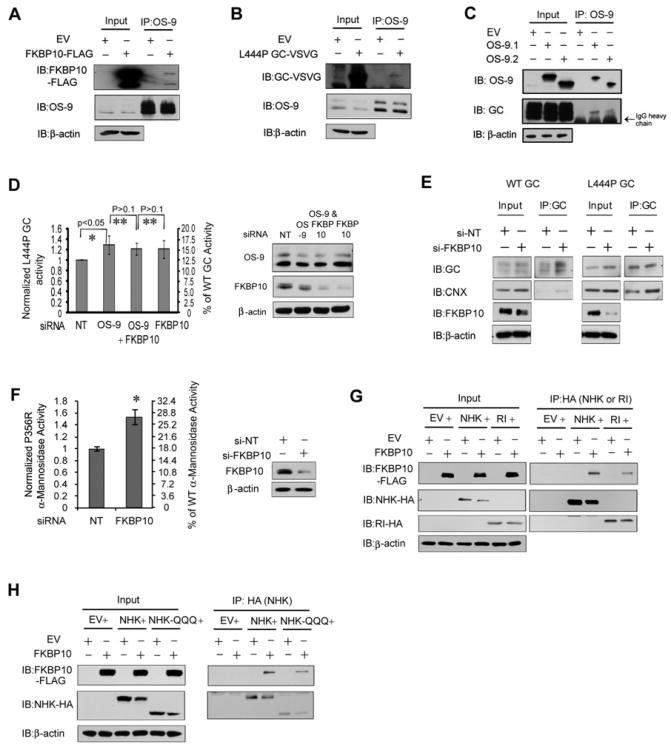
To further explore the role of FKBP10, we investigated the possible involvement of OS-9 in ERAD of GC. OS-9 is an ERAD lectin that interacts with the membrane-embedded ubiquitin ligase HRD1–SEL1L complex (Bernasconi, et al., 2010; Christianson, et al., 2008). Current models posit that OS-9 can recognize demannosylated oligosaccharides on glycoproteins and/or the misfolded non-glycosylated structures of proteins, resulting in ERAD of glycoproteins (Hebert, et al., 2010). Co-overexpressed FKBP10 and L444P GC co-immunoprecipitated in HeLa cell lysates with endogenous OS-9 employing an OS-9 antibody (Figures 6A and 6B, respectively). Moreover, overexpressed OS-9 interacted with endogenous WT GC (Figure 6C) and immunoprecipitating GC and OS-9 sequentially from the HeLa cell lysates enabled the detection of FKBP10 (Figure S6A). This suggested that ERAD of GC may involve the association of GC with both FKBP10 and OS-9, possibly as a ternary complex. An interaction between glycosylated FKBP10 and XTP3-B, which can substitute for OS-9 (Bernasconi, et al., 2010; Christianson, et al., 2008), was not detected. Notably, silencing OS-9 in GC fibroblasts increased L444P GC function 1.2 to 1.4-fold (Figure 6D, * p<0.05), supporting the notion that OS-9 may be involved in the FKBP10-mediated GC degradation. The lack of additivity in L444P GC activity when OS-9 and FKBP10 siRNA were co-applied (Figure 6D, ** p>0.1) suggests that FKBP10 and OS-9 function in the same pathway for GC ERAD, possibly through a direct interaction.
Figure 6.
The level of FKBP10 influences the proteostasis of other substrates besides GC. (A and B) Overexpressed FKBP10 (A) and overexpressed L444P GC (B) co-immunoprecipitate with endogenous OS-9. (C) Endogenous WT GC co-immunoprecipitates with overexpressed OS-9. (D) The knockdown of OS-9, FKBP10 or the combination of OS-9 and FKBP10 in fibroblasts results in a similar increase in L444P GC activity (left). OS-9 and FKBP10 are effectively depleted in the L444P GC fibroblasts (right). (E) Silencing FKBP10 promotes the interaction between GC and calnexin. (F) Depleting the FKBP10 level significantly increases P356R lysosomal α-mannosidase activity in patient-derived fibroblasts. FKBP10 is effectively depleted in the fibroblasts (right). (G) FKBP10 overexpression modestly decreases the steady state levels of NHK but not RI332 in HeLa cells. FKBP10 associates with both NHK and RI332. (H) FKBP10 overexpression modestly decreases the steady state levels of NHK-QQQ in HeLa cells. FKBP10 associates with NHK-QQQ. The experiments were done three times. The data in (D) and (F) are reported as mean ± SD (n=8) and any statistical significance was calculated using a two-tailed Student's t-Test. *p < 0.05, **p > 0.1. See also Figure S6.
FKBP10 competes with calnexin for GC binding early in the progression of GC through the ER proteostasis network
Calnexin binding to GC enables its productive folding and ER exit (Ong, et al., 2010). We hypothesized that FKBP10 and calnexin compete for binding to GC undergoing maturation in the ER, resulting in a bias towards degradation or folding, respectively (Figure 7). As such, more GC should be bound to calnexin when FKBP10 concentrations are reduced. Indeed, fibroblast lysates with silenced FKBP10 exhibited an increased amount of calnexin associated with the immunoprecipitated WT and L444P GC (Figure 6E).
Figure 7.
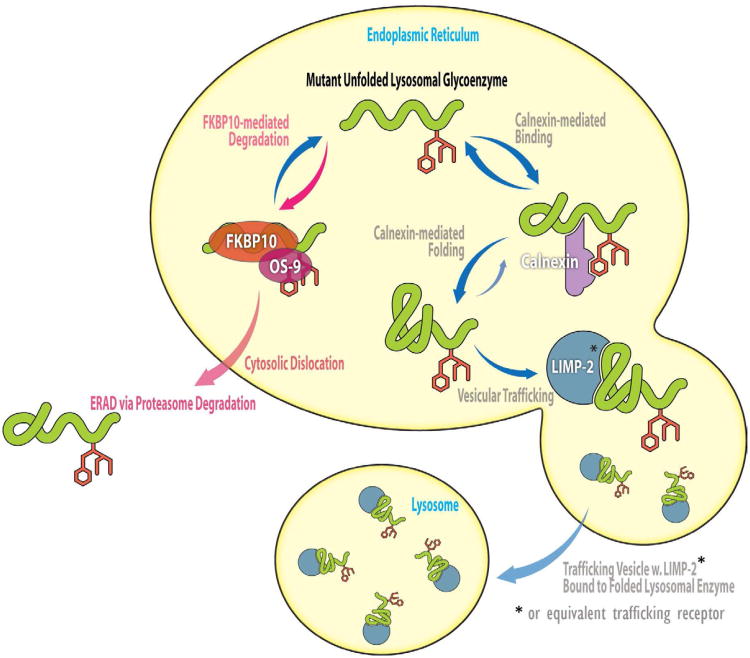
FKBP10 serves as an ERAD partitioning factor for mutant lysosomal enzymes. In the model, the newly synthesized unfolded mutant lysosomal glycoenzyme encounters either FKBP10 or calnexin in the ER, resulting in its degradation or folding, respectively. FKBP10 bound-lysosomal glycoenzyme appears to associate with OS-9 that delivers misfolded lysosomal enzyme to the ERAD machinery for cytosolic dislocation and proteasome-mediated degradation. In the competitive pathway, the calnexin bound-lysosomal enzymes are folded into their native state, which enables LIMP-2 (or an equivalent trafficking receptor) binding and lysosomal trafficking.
It is possible that FKBP10 could associate directly with calnexin to accelerate mutant GC degradation, blocking the profolding calnexin-GC interaction. Immunoprecipitating overexpressed FLAG-tagged FKBP10 from HeLa cell lysate revealed that FKBP10 associated with calnexin (Figure S6B). However, in the reciprocal calnexin immunoprecipitation, calnexin associated only with unglycosylated FKBP10 (Figure S6B), which is not the physiologically relevant form. Furthermore, treating HeLa cells with castanospermine to inhibit the interaction between calnexin and its glycosylated substrates did not increase FKBP10-mediated L444P GC degradation rate when compared to the empty vector control (Figure S6C, lanes 10 to 12 cf 7 to 9, and Figure S6D, right). Therefore, it appears that calnexin is not required for FKBP10's function to promote GC degradation in the ER.
FKBP10 influences the proteostasis of other misfolding-prone proteins
FKBP10 knockdown (>80%; Figure 6F, right) increased P356R α-mannosidase lysosomal activity 1.5-fold in α-mannosidosis patient-derived fibroblasts (Figure 6F, *p<0.05), demonstrating that the proteostasis of other LSD-associated enzymes can also be enhanced. Enhancing calnexin function by increasing ER Ca2+ levels also increased P356R α-mannosidase proteostasis (Mu, et al., 2008), consistent with our hypothesis that calnexin and FKBP10 compete for LSD-associated enzymes undergoing maturation (Figure 7).
We also examined the effect of FKBP10 overexpression on the null Hong Kong variant of α1-antitrypsin (NHK) (Hirao, et al., 2006; Oda, et al., 2003) and RI332 (a mutant of ribophorin I) (de Virgilio, et al., 1998), established ERAD substrates. FKBP10 overexpression decreased the steady state levels of NHK, but not RI332 (Figure 6G, left panels), although FKBP10 co-immunoprecipitated from HeLa cell lysates with both these ERAD substrates (Figure 6G, right panels). Interestingly, OS-9 is required for the degradation of NHK (Christianson, et al., 2008) and given that FKBP10 and OS-9 interact (Figures 6B-6D), these data are consistent with FKBP10 enhancing degradation for a variety of misfolding-prone substrates. To assess whether the FKBP10-substrate interaction is N-glycan dependent, we co-expressed FKBP10 with non-glycosylated, mutant NHK-QQQ (Hirao, et al., 2006). FKBP10 co-immunoprecipitated with NHK-QQQ and its overexpression reduced the steady state levels of NHK-QQQ (Figure 6H), suggesting that FKBP10 does not distinguish its substrates by N-glycan recognition.
Discussion
Proteomic assessment of patient-derived LSD fibroblasts treated with mechanistically distinct PRs (MG-132 or diltiazem) led to the discovery of a new LSD enzyme proteostasis network component, FKBP10, which when silenced enhances GC proteostasis. FKBP9 appears to play a similar ERAD partitioning role, although we focused largely on FKBP10.
The data demonstrate that increased ER FKBP10 levels accelerated GC ERAD, apparently by collaborating with OS-9 (Figure 7). Since OS-9 has been shown to deliver ERAD substrates to the membrane-embedded HRD1-SEL1L ubiquitin ligase complex for ERAD (Bernasconi, et al., 2010; Christianson, et al., 2008), we hypothesize that OS-9 may then transfer the GC–FKBP10 (GC–FKPB9) complex to HRD1-SEL1L to mediate ERAD. Decreasing the concentration of FKBP10 appears to decrease its competitiveness for misfolded GC, allowing pro-folding components such as calnexin (levels increased by MG-132 or diltiazem treatment) to more effectively compete for unfolded glycosylated enzymes, resulting in their ER folding and exit (Figure 7). The modest increases in mutant lysosomal GC or P356R α-mannosidase activities observed with FKBP10 knockdown are thought to be sufficient to ameliorate pathology in patients (Schueler, et al., 2004).
It appears that there is an early delicate balance between calnexin-mediated glycoprotein folding vs. FKBP 9- or FKBP 10-mediated ERAD as lysosomal enzymes like GC begin their journey through the ER proteostasis network. Generally speaking, knocking down or inhibiting established ERAD proteostasis network components does not lead to more folding, trafficking and function of mutant LSD-associated enzymes because they are too far along the ERAD pathway. That perturbation of FKBP9 or FKBP10 can alter LSD mutant enzyme proteostasis suggests that FKBP9 or FKBP10 makes an early degradation vs. folding decision before mannose trimming, after which ERAD seems inevitable.
Significance
In summary, silencing FKBP9 or FKBP10 enhances the folding, trafficking, and lysosomal function of LSD-associated mutant enzymes to levels thought to be sufficient to ameliorate disease. This suggests that this approach is worth exploring as a LSD therapeutic strategy. Moreover, our data reveals that a subset of FKBPs can identify as yet unfolded proteins and direct them to ERAD, suggesting that some FKBPs act as ERAD partitioning factors. As such, FKBP silencing could be efficacious in other loss-of-function diseases.
Experimental Procedures
Reagents
Diltiazem hydrochloride was from Tocris Bioscience (Ellisville, MO). MG-132 and lactacystin was from Calbiochem (San Diego, CA). 4-methylumbelliferyl β-D-glucoside (MUG), 4-methylumbelliferyl α-D-mannopyranoside, FK506 monohydrate, castanospermine and EGTA were from Sigma (St. Louis, MO). Conduritol B epoxide (CBE) and kifunensine were from Toronto Research Chemicals (Downsview, ON, Canada). Cell culture media were purchased from Gibco (Grand Island, NY).
Cell cultures
Primary skin fibroblast cultures were established from Gaucher's patients homozygous for either the G202R GC (c.721G>A) mutation (kindly provided by Dr. K.P. Zimmer (Children's Hospital of the University of Munster, Munster)) or the N370S GC (c.1226A>G) mutation (kindly provided by Dr. E. Beutler). An apparently normal fibroblast cell line (GM00498), the Gaucher's disease fibroblast cell line homozygous for the L444P (c.1448T>C) mutation (GM08760) and a homozygous α-mannosidosis fibroblast cell line containing the P356R α-mannosidase mutation (GM04518) were obtained from Coriell Cell Repositories (Camden, NJ). Fibroblasts were grown in minimal essential medium with Earle's salts supplemented with 10% heat-inactivated fetal bovine serum and 1% glutamine Pen-Strep at 37°C in 5% CO2. Cell medium was replaced every 3 or 4 days. Monolayers were passaged upon reaching confluency with TrypLE Express.
Relative quantification of protein expression level changes by MudPIT
Proteins from L444P GC fibroblasts treated with diltiazem (10 μM) for 5 d, MG-132 (0.8 μM) for 3 d, or the respective vehicle controls were subjected to MudPIT mass spectrometry proteomics analysis (Link, et al., 1999; Washburn, et al., 2001). See Supplemental Experimental Procedures for detailed method. Estimation of protein abundance based on spectral count was used as the relative quantification method (Liu, et al., 2004) which has been widely applied (Cao, et al., 2008; Liao, et al., 2007; Rikova, et al., 2007). Spectral counts from the three replicates of each sample were merged to average the run-to-run variation and a normalization factor (F = Total number of spectra in control sample/Total number of spectra in treated sample) was applied. We used stringent, albeit arbitrary, criteria for the significantly altered protein expression level after drug treatment. If a protein is detected in both vehicle- and PR-treated samples, proteins with expression level changes were filtered according to the following criteria: (1) if the same protein was identified in both samples with spectra counts greater than 10, normalized spectra count ratios of 2 or above were considered as increased; likewise, 0.5 or less as decreased; (2) if the same protein was identified in both samples with a spectra count from either of them less than 10 but the difference between the two was greater than 10, normalized spectra count ratios of 2.5 or above were considered as increased; likewise, 0.4 or less as decreased. If a protein was identified in only one sample, a spectra count of greater than 20 was considered a significant change. Details about the identified proteins are given in the Supplemental Experimental Procedures and Table S1 (Significant up- or down-regulation) and Table S2 (Functional characterization).
Enzyme activity assays
The intact cell GC activity and lysed cell GC activity assays for WT, L444P, N370S and G202R GC fibroblasts using the MUG substrate were described previously (Mu, et al., 2008; Sawkar, et al., 2002). The activity of lysosomal α-mannosidase was determined as previously described with the minor modification of using 2 mM 4-methylumbelliferyl α-D-mannopyranoside as the substrate (Gotoda, et al., 1998). Each data point reported was evaluated at least in triplicate in each plate, and on three different days. See Supplemental Experimental Procedures for detailed method.
Western blot analysis
Cells were lysed with Roche complete lysis-M buffer containing complete protease inhibitor cocktail. Company specifications were followed for protein treatment with Endo H or PNGase F (New England Biolabs). Aliquots of cell lysates were separated in a 10% SDS-PAGE gel and western blot analysis was performed using appropriate antibodies. Details of antibodies used are given in the Supplemental Experimental Procedures.
Immunofluorescence
Immunofluorescence was performed as previously described (Mu, et al., 2008; Sawkar, et al., 2006). The experiments were done three times and similar results were obtained. See Supplemental Experimental Procedures for detailed method.
Plasmid and siRNA transfection
HeLa cells and patient-derived GC fibroblasts were seeded at approximately 2 × 105 cells per well in 6-well plates and allowed to reach ∼80% confluency before transient transfection using Fugene 6 transfection reagent (Roche) for plasmids or HiPerfect Transfection Reagent (Qiagen) for siRNA, according to the manufacturers' instructions. Details of the plasmids and siRNA duplexes used can be found in the Supplemental Experimental Procedures.
Quantitative RT-PCR
The relative expression levels of target genes were analyzed by quantitative RT-PCR using the forward and reverse primers for the genes analyzed as described previously (Mu, et al., 2008; Mu, et al., 2008; Ong, et al., 2010). See Supplemental Experimental Procedures for detailed method. Each data point was evaluated in triplicate, and measured three times.
Pulse-chase and immunoprecipitation
WT GC fibroblasts treated with FKBP10 siRNA for 4 d were starved for 45 min, metabolically labeled with 60 μCi of 35S EasyTag™ (Perkin-Elmer) for 1 h as previously described (Jonsson, et al., 1987; Schmitz, et al., 2005) and chased for the indicated periods. HeLa cells 24 h post-transfection were starved for 45 min before being metabolically labeled for 15 min. See Supplemental Experimental Procedures for detailed method.
Enrichment of the ER fraction
Cells were collected, washed in PBS, and crosslinked using 1mM DSP (Pierce) according to the manufacturer's instructions. After undergoing three freeze-thaw cycles, cells were lysed in lysis buffer (50 mM Tris, 150 mM NaCl, pH 7.5) containing protease inhibitor cocktail (Roche). For enrichment of the ER fraction, lysates were depleted of nuclei via centrifugation at 800 g for 10 min. The supernatant was then centrifuged at 10,000 g for 10 min to pellet the mitochondria, and the supernatant was then ultracentrifuged at 100,000 g for 65 min to pellet the ER. The ER pellet was resuspended in lysis buffer plus 1% Triton X-100 and Roche protease inhibitor cocktail. The ER protein concentration was quantified using a microBCA assay (Pierce).
The effect of calcium on the FKBP10-GC protein interaction
Lysates were incubated with 2 mM EGTA for 15 min at 37 °C prior to the addition of the mouse monoclonal anti-FLAG antibody for immunoprecipitation as described previously (Le, et al., 1994; Ong, et al., 2010).
The effect of FK506 on the FKBP10-GC interaction
HeLa cells 24 h post-transfection were treated with FK506 (0.1 nM) for 24 h. The mouse monoclonal anti-FLAG antibody was added to the lysates prior to immunoprecipitation.
The effect of proteasomal degradation on the FKBP10-GC interaction
HeLa cells 24 h post-transfection were treated with lactacystin (5 μM) for 24 h prior to lysis. The lysates were subjected to immunoprecipitation with the mouse monoclonal anti-FLAG antibody.
Sequential immunoprecipitation of GC and OS-9
GC proteins were immunoprecipitated from HeLa cells 48 h post-transfection using the anti-VSVG antibody and the bound proteins were eluted using VSVG peptides (Roche) at 37 °C for 30 min. The eluted proteins were subjected to a second round of immunoprecipitation using the anti-OS-9 antibody and the resulting bound proteins were eluted by boiling in SDS loading buffer in the presence of DTT.
Statistical analysis
All data are presented as mean ± SEM or mean ± SD as stated and any statistical significance was calculated using two-tailed Student's t-Test.
Supplementary Material
Highlights.
Whole-cell proteomics identifies FKBP10 as key to lysosomal enzyme proteostasis
FKBP10 knockdown increases mutant lysosomal enzyme folding, trafficking and function
FKBP10 overexpression accelerates ERAD of mutant lysosomal enzymes
FKBP10 appears to act as an “ER degradation vs. folding partitioning factor”
Acknowledgments
This work was supported by the NIH (DK075295), the Skaggs Institute for Chemical Biology and the Lita Annenberg Hazen Foundation. Y.L. Tan is supported by a predoctoral fellowship from the Agency for Science, Technology and Research (A*STAR) Singapore. We thank the following for their generosity in providing us with plasmids: N. Hosokawa (Kyoto University) for pCMV-SPORT2-EDEM1-HA and R. Kopito (Stanford University) for pcDNA3.1-RI-322-HA, pcDNA3.1-α1-antitrypsin NHK-HA, pcDNA3.1-OS-9.1 and pcDNA3.1-OS-9.2. We thank R Kopito for the rabbit polyclonal anti-OS-9, J. Aerts (Academic Medical Center, Amsterdam) for the mouse monoclonal anti-GC 8E4, W.E. Balch (The Scripps Research Institute) for mouse monoclonal anti-VSVG, M. Fukuda (Burnham Institute) for the rabbit anti-LAMP2, and C. Fearns for critical feedback on the manuscript.
Footnotes
Supplemental Information: Supplemental Information contains Supplemental Experimental Procedures, six figures, and three Tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- Alanay Y, Avaygan H, Camacho N, Utine GE, Boduroglu K, Aktas D, Alikasifoglu M, Tuncbilek E, Orhan D, Bakar FT, et al. Mutations in the Gene Encoding the RER Protein FKBP65 Cause Autosomal-Recessive Osteogenesis Imperfecta. Am J Hum Genet. 2010;86:551–559. doi: 10.1016/j.ajhg.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi R, Galli C, Calanca V, Nakajima T, Molinari M. Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-L-S substrates. J Cell Biol. 2010;188:223–235. doi: 10.1083/jcb.200910042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Skach WR. Protein folding and quality control in the endoplasmic reticulum: Recent lessons from yeast and mammalian cell systems. Curr Opin Cell Biol. 2011;23:464–475. doi: 10.1016/j.ceb.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Cao R, He QY, Zhou J, He QZ, Liu Z, Wang XC, Chen P, Xie JY, Liang SP. High-throughput analysis of rat liver plasma membrane proteome by a nonelectrophoretic in-gel tryptic digestion coupled with mass spectrometry identification. J Proteome Res. 2008;7:535–545. doi: 10.1021/pr070411f. [DOI] [PubMed] [Google Scholar]
- Chen B, Retzlaff M, Roos T, Frydman J. Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol. 2011;3:a004374. doi: 10.1101/cshperspect.a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha 1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Virgilio M, Weninger H, Ivessa NE. Ubiquitination is required for the retro-translocation of a short-lived luminal endoplasmic reticulum glycoprotein to the cytosol for degradation by the proteasome. J Biol Chem. 1998;273:9734–9743. doi: 10.1074/jbc.273.16.9734. [DOI] [PubMed] [Google Scholar]
- Deuerling E, Bukau B. Chaperone-assisted folding of newly synthesized proteins in the cytosol. Crit Rev Biochem Molec Biol. 2004;39:261–277. doi: 10.1080/10409230490892496. [DOI] [PubMed] [Google Scholar]
- Edmunds T. In: Gaucher disease In Protein Misfolding Diseases: Current and Emerging Principles and Therapies. Ramirez-Alvarado M, Kelly JW, Dobson CM, editors. Hoboken, New Jersey: John Wiley & Sons, Inc.; 2010. pp. 469–485. [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Feng M, Gu C, Ma S, Wang Y, Liu H, Han R, Gao J, Long Y, Mi H. Mouse FKBP23 mediates conformer-specific functions of BiP by catalyzing Pro117 cis/trans isomerization. Biochem Biophys Res Commun. 2011;408:537–540. doi: 10.1016/j.bbrc.2011.04.050. [DOI] [PubMed] [Google Scholar]
- Futerman AH, van Meer G. The cell biology of lysosomal storage disorders. Nat Rev Mol Cell Biol. 2004;5:554–565. doi: 10.1038/nrm1423. [DOI] [PubMed] [Google Scholar]
- Gotoda Y, Wakamatsu N, Kawai H, Nishida Y, Matsumoto T. Missense and nonsense mutations in the lysosomal alpha-mannosidase gene (MANB) in severe and mild forms of alpha-mannosidosis. Am J Hum Genet. 1998;63:1015–1024. doi: 10.1086/302048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurkan C, Stagg SM, LaPointe P, Balch WE. The COPII cage: unifying principles of vesicle coat assembly. Nat Rev Mol Cell Biol. 2006;7:727–738. doi: 10.1038/nrm2025. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Bernasconi R, Molinari M. ERAD substrates: Which way out? Semin Cell Dev Biol. 2010;21:526–532. doi: 10.1016/j.semcdb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Hirao K, Natsuka Y, Tamura T, Wada I, Morito D, Natsuka S, Romero P, Sleno B, Tremblay LO, Herscovics A, et al. EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming. J Biol Chem. 2006;281:9650–9658. doi: 10.1074/jbc.M512191200. [DOI] [PubMed] [Google Scholar]
- Ishikawa Y, Vranka J, Wirz J, Nagata K, Bachinger HP. The Rough Endoplasmic Reticulum-resident FK506-binding Protein FKBP65 Is a Molecular Chaperone That Interacts with Collagens. J Biol Chem. 2008;283:31584–31590. doi: 10.1074/jbc.M802535200. [DOI] [PubMed] [Google Scholar]
- Jakob RP, Zoldak G, Aumuller T, Schmid FX. Chaperone domains convert prolyl isomerases into generic catalysts of protein folding. Proc Natl Acad Sci U S A. 2009;106:20282–20287. doi: 10.1073/pnas.0909544106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson LM, Murray GJ, Sorrell SH, Strijland A, Aerts JF, Ginns EI, Barranger JA, Tager JM, Schram AW. Biosynthesis and maturation of glucocerebrosidase in Gaucher fibroblasts. Eur J Biochem. 1987;164:171–179. doi: 10.1111/j.1432-1033.1987.tb11008.x. [DOI] [PubMed] [Google Scholar]
- Kang CB, Ye H, Dhe-Paganon S, Yoon HS. FKBP Family Proteins: Immunophilins with Versatile Biological Functions. Neurosignals. 2008;16:318–325. doi: 10.1159/000123041. [DOI] [PubMed] [Google Scholar]
- Kelley BP, Malfait F, Bonafe L, Baldridge D, Homan E, Symoens S, Willaert A, Elcioglu N, Van ML, Verellen-Dumoulin C, et al. Mutations in FKBP10 cause recessive osteogenesis imperfecta and Bruck syndrome. J Bone Miner Res. 2011;26:666–672. doi: 10.1002/jbmr.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le AQ, Steiner JL, Ferrell GA, Shaker JC, Sifers RN. Association between calnexin and a secretion-incompetent variant of human alpha(1)-antitrypsin. J Biol Chem. 1994;269:7514–7519. [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L, Pilotte J, Xu T, Wong CCL, Edelman GM, Vanderklish P, Yates JR. BDNF induces widespread changes in synaptic protein content and up-regulates components of the translation machinery: An analysis using high-throughput proteomics. J Proteome Res. 2007;6:1059–1071. doi: 10.1021/pr060358f. [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Liu HB, Sadygov RG, Yates JR. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Maki CG, Huibregtse JM, Howley PM. In vivo ubiquitination and proteasome-mediated degradation of p53(1) Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- Mazzulli JR, Xu YH, Sun Y, Knight AL, McLean PJ, Caldwell GA, Sidransky E, Grabowski GA, Krainc D. Gaucher Disease Glucocerebrosidase and α-Synuclein Form a Bidirectional Pathogenic Loop in Synucleinopathies. Cell. 2011;146:37–52. doi: 10.1016/j.cell.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata Y, Koren J, Kiray J, Dickey CA, Gestwicki JE. Molecular chaperones and regulation of tau quality control: strategies for drug discovery in tauopathies. Future Med Chem. 2011;3:1523–1537. doi: 10.4155/fmc.11.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari M, Calanca V, Galli C, Lucca P, Paganetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299:1397–1400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- Mu TW, Fowler DM, Kelly JW. Partial restoration of mutant enzyme homeostasis in three distinct lysosomal storage disease cell lines by altering calcium homeostasis. Plos Biology. 2008;6:e26. doi: 10.1371/journal.pbio.0060026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu TW, Ong DST, Wang YJ, Balch WE, Yates JR, Segatori L, Kelly JW. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 2008;134:769–781. doi: 10.1016/j.cell.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy LA, Ramirez EA, Trinh VT, Herman AM, Anderson VC, Brewster JL. Endoplasmic reticulum stress or mutation of an EF-hand Ca2+-binding domain directs the FKBP65 rotamase to an ERAD-based proteolysis. Cell Stress Chaperones. 2011;16:607–619. doi: 10.1007/s12192-011-0270-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y, Hosokawa N, Wada I, Nagata K. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science. 2003;299:1394–1397. doi: 10.1126/science.1079181. [DOI] [PubMed] [Google Scholar]
- Okada T, Yoshida H, Akazawa R, Negishi M, Mori K. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochemical J. 2002;366:585–594. doi: 10.1042/BJ20020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong DST, Mu TW, Palmer AE, Kelly JW. Endoplasmic reticulum Ca2+ increases enhance mutant glucocerebrosidase proteostasis. Nat Chem Biol. 2010;6:424–432. doi: 10.1038/nchembio.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and Chemical Approaches to Diseases of Proteostasis Deficiency. Annu Rev Biochem. 2009;78:23.21–33. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- Reczek D, Schwake M, Schroder J, Hughes H, Blanz J, Jin XY, Brondyk W, Van Patten S, Edmunds T, Saftig P. LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of beta-Glucocerebrosidase. Cell. 2007;131:770–783. doi: 10.1016/j.cell.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Sawkar AR, Cheng WC, Beutler E, Wong CH, Balch WE, Kelly JW. Chemical chaperones increase the cellular activity of N370S beta-glucosidase: A therapeutic strategy for Gaucher disease. Proc Natl Acad Sci USA. 2002;99:15428–15433. doi: 10.1073/pnas.192582899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawkar AR, D'Haeze W, Kelly JW. Therapeutic strategies to ameliorate lysosomal storage disorders - a focus on Gaucher disease. Cell Mol Life Sci. 2006;63:1179–1192. doi: 10.1007/s00018-005-5437-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz M, Alfalah M, Aerts JM, Naim HY, Zimmer KP. Impaired trafficking of mutants of lysosomal glucocerebrosidase in Gaucher's disease. Int J Biochem Cell Biol. 2005;37:2310–2320. doi: 10.1016/j.biocel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Schueler UH, Kolter T, Kaneski CR, Zirzow GC, Sandhoff K, Brady RO. Correlation between enzyme activity and substrate storage in a cell culture model system for Gaucher disease. J Inherited Metab Dis. 2004;27:649–658. doi: 10.1023/b:boli.0000042959.44318.7c. [DOI] [PubMed] [Google Scholar]
- Shadidy M, Caubit X, Olsen R, Seternes OM, Moens U, Krauss S. Biochemical analysis of mouse FKBP60, a novel member of the FKPB family. Biochim Biophys Acta-Gene Structure and Expression. 1999;1446:295–307. doi: 10.1016/s0167-4781(99)00080-9. [DOI] [PubMed] [Google Scholar]
- Shaheen R, Al-Owain M, Faqeih E, Al-Hashmi N, Awaji A, Al-Zayed Z, Alkuraya FS. Mutations in FKBP10 cause both Bruck syndrome and isolated osteogenesis imperfecta in humans. Am J Med Genet, Part A. 2011;155:1448–1452. doi: 10.1002/ajmg.a.34025. [DOI] [PubMed] [Google Scholar]
- Stoller G, Ruecknagel KP, Nierhaus KH, Schmid FX, Fischer G, Rahfeld JU. A ribosome-associated peptidyl-prolyl cis/trans isomerase identified as the trigger factor. EMBO J. 1995;14:4939–4948. doi: 10.1002/j.1460-2075.1995.tb00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Liou B, Ran HM, Skelton MR, Williams MT, Vorhees CV, Kitatani K, Hannun YA, Witte DP, Xu YH, et al. Neuronopathic Gaucher disease in the mouse: viable combined selective saposin C deficiency and mutant glucocerebrosidase (V394L) mice with glucosylsphingosine and glucosylceramide accumulation and progressive neurological deficits. Hum Molec Genet. 2010;19:1088–1097. doi: 10.1093/hmg/ddp580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Chang HC, Chakraborty K, Hartl FU, Hayer-Hartl M. Essential role of the chaperonin folding compartment in vivo. EMBO J. 2008;27:1458–1468. doi: 10.1038/emboj.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–U930. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi G, Monti E, Dalle CL, Corradi M, Gandini A, Valenti MT, Boner A, Antoniazzi F. A novel splicing mutation in FKBP10 causing osteogenesis imperfecta with a possible mineralization defect. Bone. 2012;50:343–349. doi: 10.1016/j.bone.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Vitner EB, Platt FM, Futerman AH. Common and uncommon pathogenic cascades in lysosomal storage diseases. J Biol Chem. 2010;285:20423–20427. doi: 10.1074/jbc.R110.134452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Zhao H, Grabowski GA. Gaucher disease: Perspectives on a prototype lysosomal disease. Cell Mol Life Sci. 2002;59:694–707. doi: 10.1007/s00018-002-8458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.