Abstract
Triple-negative breast cancer (TNBC) studies have shown that neoadjuvant chemotherapy before surgery was effective in the minority of women, whereas the majority who had residual tumor had a relatively poor outcome. To identify the mechanism by which residual cancer cells survive chemotherapy, we initially performed gene expression profiling using the CRL2335 TNBC cell line derived from a squamous breast carcinoma before and after treatment with cisplatin plus TRAIL. We found a significant increase in the expression of FZD8, one of Wnt receptors, and its downstream targets LEF1 and TCF in residual CRL2335 tumor cells after treatment with cisplatin plus TRAIL. Increased FZD8 levels were further confirmed in other TNBC breast carcinoma cell lines. Inhibition of FZD8 by siRNA in CRL2335 cells in the presence of cisplatin plus TRAIL reduced β-catenin and survivin levels, and increased apoptosis compared to scrambled siRNA-treated cells. In vivo data demonstrate that cisplatin plus TRAIL treatment significantly reduces tumor volume in nod/SCID mice. However, we found that cisplatin plus TRAIL treatment predominantly eliminated non-tumor initiating cells (non-TICs), as demonstrated by whole-body fluorescent imaging of mice injected with mammosphere-forming CRL2335 cells stably transfected with DsRed. This led to TIC enrichment in residual tumors, as confirmed by immunostaining for TIC markers. Moreover, an increase in FZD8 expression was observed in residual tumors treated with cisplatin and TRAIL. Taken together, our findings suggest that FZD8-mediated Wnt-signaling may play a major role in mediating resistance to chemotherapy, making it a potential target to enhance chemotherapeutic efficacy in patients with TNBC.
Keywords: FZD8, Tumor initiating cells, Triple-negative breast cancer, Cisplatin, TRAIL
Introduction
In breast cancer patients, the survival rates have improved steadily over the past two decades. However, triple-negative breast cancer (TNBC) exhibits aggressive characteristics associated with shorter disease-free survival (1). TNBC tumors are characterized by the absence of expression of estrogen receptor (ER) and progesterone receptor (PR), as well as HER-2 amplification. Therefore, TNBC patients do not benefit from commonly used antiestrogen- and herceptin-based therapies (2, 3). Although chemotherapy is currently the mainstay of systemic treatment for breast cancer, patients with TNBC disease have a worse outcome after chemotherapy than patients with other subtypes of breast cancer (4, 5). Previous studies suggested that neoadjuvant treatment involving the administration of chemotherapy before surgery was effective in a minority of women with TNBC, who show a complete pathological response and an excellent outcome; however, the outcome for the majority of them, who still have a residual disease after treatment, is relatively poor (6).
Several alternative hypotheses have been proposed to explain the treatment failure and recurrence. It has been suggested that tumor initiating cells (TICs), also commonly called cancer stem-like cells (CSCs), are resistant to chemotherapy and radiotherapy, and surviving TICs can reinitiate tumor growth after treatment (7–10). TICs can be enriched by FACS using specific cell-surface markers, by either CD44+/CD24−/low staining and/or aldehyde dehydrogenase 1 (ALDH1) activity (8, 11, 12). In addition, it was shown that human breast cancer cells that are CD44+/CD24−/low or ALDH1-positive could efficiently regenerate tumors containing an array of cell types similar to those found in the original tumor (8, 10, 13).
Defining the signaling pathways in residual tumors that survived chemotherapy could help to design new treatments for patients with TNBC. Here, we evaluated CRL-2335 TNBC cells before and after treatment with cisplatin plus tumor-necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL), with the assumption that changes seen in gene expression in residual tumors that survived chemotherapy could provide new therapeutic targets for those breast cancer patients who do not benefit from antiestrogen- and herceptin-based therapies.
Materials and methods
Cell culture
The human breast triple-negative breast cell line CRL2335 (derived from squamous carcinoma of the breast), CRL-2336, MDA-MB-468, and MDA-231 was obtained from the American Type Culture Collection (ATCC). Cells were maintained in RPMI-1640 Medium (GIBCO/Life Technologies) supplemented with fetal bovine serum (GIBCO/Life Technologies) to a final concentration of 10%. The cells were immediately expanded and frozen after being obtained from ATCC, and restarted every 3 to 4 months from a frozen vial of the same batch of cells and no additional authentication was done on these cells. All cell lines were free of mycoplasma infection tested by PCR.
RT2 Profiler PCR array system
The expression of 84 genes related to human stem cell signaling pathway was analyzed using RT2 Profiler PCR array technology (SABiosciences) using CRL2335 TNBC cells treated with 10 µg/ml cisplatin (Sigma) and 10 ng/ml TRAIL (R&D systems) for 24 hours compared to vehicle-treated CRL2335 cells. Total RNA was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. The RT2 Profiler PCR array kit was used to convert mRNA into cDNA, and then mixed with LightCycler® 480 SYBR Green I Master mix (Stratagene) and dispensed to each well of the PCR array plate containing pre-filled gene-specific primer sets, and PCR was performed, according to the manufacturer’s instructions. An MS Excel sheet from the manufacturer's Web site was used to include the PCR data, which were analyzed using the PCR Array Data Analysis Web Portal, and then normalized by correcting the Ct (threshold cycle) values obtained with respect to average Ct values of housekeeping genes present on the array to obtain fold change in expression for each gene. A four-fold change in gene expression between vehicle- and cisplatin-TRAIL-treated cells was used as a threshold.
Real-time RT-PCR was done to confirm the results for certain genes that revealed significant changes in expression in the RT2 Profiler PCR array, and to analyze additional genes that code for Wnt receptors and their ligand. Briefly, one microgram of each total RNA sample was reverse transcribed using iScript cDNA Synthesis Kit (BioRad). Brilliant SYBR Green QPCR master mix (Stratagene) was mixed with 1 μl of cDNA and 150 nM of the following primers:
| FZD1: | Forward: 5'-GCCCATGAGCCCGGACTTCAC-3' |
| Reverse: 5'-TCAGACTGTAGTCTCCCCTTG-3' | |
| FZD2 | Forward: 5'-TACCCAGAGCGGCCTATCATTTTT-3' |
| Reverse: 5'- ACGAAGCCGGCCAGGAGGAAGGAC-3' | |
| FZD3: | Forward: 5'-AAGGCTTCCACAGTGACACAAGG-3' |
| Reverse: 5'-AGAGGAGAGAAACCCCAACTACCAC-3' | |
| FZD7: | Forward: 5'-ACAGACTTAGCCACAGCAGCAAGG-3' |
| Reverse: 5'-TTTCCAAATCACCCCTCGCC-3' | |
| FZD8: | Forward: 5'-TGGAGTGGGGTTACCTGTTG-3' |
| Reverse: 5'-AGCGGCTTCTTGTAGTCCTC-3' | |
| Rspo1: | Forward: 5'-CAGCCATAACTTCTGCACCA-3' |
| Reverse: 5'-AGAGCTGCTGCTTCTTGGAG-3' | |
| Rspo2: | Forward: 5'-GTACCAAGTGCAAAGTAGGC-3' |
| Reverse: 5'-AACTTCACATCCTTCCACAC-3'. |
The real-time PCR thermal profile was: 1 cycle of 10 min at 95 °C and then 40 cycles of 30 s at 95 °C, 1 min at 55 °C and 1 min at 72 °C followed by 1 cycle of 1 min at 95 °C, and finally 41 cycles of increasing the temperature 1°C every 30 s starting at 55 °C. Ct values were obtained using the built-in Eppendorf program “realplex” of the Eppendorf Realplex2 Mastercycler machine, and normalized with respect to average Ct values of GAPDH housekeeping gene.
RNA interference assay
CRL2335 cells were plated in 6-well tissue culture plates at a density of 3×105/well in complete growth culture medium, and 24h later transfected with 100 pmol of siRNA directed against FZD8 or random siRNA with scrambled sequence (Novus Biologicals) using Lipofectamine transfection reagent (Invitrogen) according to the manufacturer’s instructions. Forty eight hours after transfection, cells were treated with cisplatin (10 µg/ml) and TRAIL (10 ng/ml) for additional 24h.
Western blot analysis
CRL2335 cells transiently transfected with FZD8-siRNA or random siRNA and treated with or not with cisplatin and TRAIL as explained above were grown near confluence. Cells were lysed and Western blotting was performed as described previously (14) using a standard protocol. In brief, cell extracts were obtained by lysing the cells in RIPA buffer (20 mM Hepes, 100 mM NaCl, 0.1% SDS, 1% Nonidet P-40, 1% deoxycholate, 1 mM Na3VO4, 1 mM EGTA, 50 mM NaF, 10% glycerol, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 1x protease inhibitor mixture) (all reagents from Sigma). Samples containing 100 µg of total protein were electrophoresed on 8 % or 15% SDS-polyacrylamide gels and transferred to PVDF membrane by electroblotting. Membranes were probed with specific antibodies against FZD8 (Aviva System Biology), β-catenin (Santa Cruz Biotechnology), survivin (Cell Signaling Technology), or ALDH (BD Transduction Laboratories) followed by HRP-conjugated mouse or rabbit secondary antibodies (Amersham) accordingly. The specific bands were developed on autoradiography films by treatment of the membranes with enhanced chemiluminescent substrate (Pierce). Tissue homogenates from tumor xenografts obtained as described below were also used for Western blot analysis of ALDH. As a loading control, β-actin expression levels were determined with an anti-actin antibody (BD Biosciences). The intensities of specific bands in immunoblots were measured using NIH free software ImageJ and normalized to the corresponding actin levels. Values for treated groups were presented as percentage compared to untreated group.
Apoptosis assay
Apoptosis was assessed using the Cell Death Detection ELISAplus kit (Roche Applied Science) according to the manufacturer’s instructions. This kit quantitatively detects cytosolic histone-associated DNA fragments. In brief, cells were treated with cisplatin and TRAIL for 16 h after FZD8-siRNA or random siRNA treatment. Apoptosis was quantified by ELISA and normalized to values measured in untreated cells. Data are mean + SE of triplicate determination.
Isolation of ALDH1+ cells by fluorescence-activated cell sorting
ALDH activity of CRL2335 cells was measured using the ALDEFLOUR assay kit (STEMCELL Technologies Inc.), according to the manufacturer’s instructions. Briefly, cells were incubated in an ALDEFLUOR assay buffer containing ALDH substrate (1 µmol/L per 1x106 cells). In each experiment, a sample of cells was stained under identical conditions with 50 mmol/L of diethylaminobenzaldehyde, a specific ALDH inhibitor, as a negative control.
For fluorescent-activated cell sorting, CRL2335 cells were labeled with the ALDEFLUOR kit and sorted at the Imaging and Cytometry resource’s core at The Karmanos Cancer Institute, Wayne State University. The sorting gates were established using propidium iodide-stained cells for viability.
Mouse Experiments
Eight weeks old female NOD scid mice were purchased from Taconic Farms, and maintained under aseptic conditions in accordance with Animal care and use guidelines of Wayne State University. ADEFLUOR-negative CRL2335 cells (non-TIC) sorted as explained above, were mixed at the ratio found in the original cell population (~75% ADEFLUOR-negative non-TICs and ~ 25% ADEFLUOR-positive TICs) with ALDH1+ CRL2335 cells stably transfected with pCMV-DsRed-express vector (PDsRed2-N1, Clontech) using the Lipofectamine 2000 reagent (Invitrogen), as per manufacturer’s recommendations. A total of 5×105 cells of the mixture of ALDH1− and DsRed-expressing ALDH1+ were suspended in 50 µl of cold liquid growth factor- reduced matrigel (BD Biosciences) and injected subcutaneously into the inguinal region of mammary fat pad [presence of ALDH1 is an accepted marker for TICs (14–16)]. Once the tumors reached a mean size of ~ 50 mm3, a group of four mice began to be treated once a week with cisplatin (4 mg/kg) and TRAIL (15 mg/kg) intraperitoneally for three weeks. A control group consisting of the same number of mice was treated with vehicle. The tumors were measured twice a week with calipers, and tumor volumes in mm3 were calculated by the formula (width2×length/2. The mice were fluoroimaged once every 10 days with Kodak IS4000MM Live Animal Multimodal Imager under anesthesia to monitor growth of tumor areas formed by DsRed-expressing CRL2335 cells. The quantification of DsRed expression in the tumors was an average of integrated intensity that was calculated by using built in software Metamorph (version 7.6.4.0.) associated with the Kodak image station. Mice were euthanized 30 days after tumor injection, and their tumor xenografts harvested. Half of each tumor was fixed in 4% paraformaldehyde and then paraffin-embedded for immunohistochemical analysis, while the other half was snap frozen in liquid nitrogen and stored at −80°C for further homogenate preparation. Briefly, tumor lysates were obtained by adding the 5 mg of tissue into RIPA buffer and homogenizing using an electric homogenizer on ice. The lysate was centrifuged and the supernatant was used to determine the protein concentration, and used for Western blotting as explained before.
Immunohistochemical Analysis
Adjacent 5-µm paraffin sections were used for expression analysis of human CD44, CD24, ALDH1, Survivin, and FZD8. Briefly, tissue sections were deparaffinized, rehydrated, subjected to antigen retrieval using Ag Citrus Plus Retrieval Solution (BioGenex) and microwave treatment. Primary antibody detection was with Vectastain Elite ABC Systems (Vector Laboratories) accordingly chosen based on the species of origin for each antibody, or M.O.M. Immunodetection kit peroxidase (Vector Laboratories) if mouse IgG. Color was developed with diaminobenzidine peroxidase substrate SIGMAFAST™ DAB(Sigma), and sections were counterstained with hematoxylin (Sigma). Primary antibodies and dilutions used were: rabbit anti-CD44 (Abcam) at 1:50, 60 min at room temperature (RT), rabbit anti-CD24 (Abcam) at 1:100, overnight at 4°C, mouse anti-ALDH1 (BD Biosciences) at 1:500, 60 min at RT, goat anti-FZD8 (Aviva System Biology) at 3µg/ml, overnight at 4°C, and rabbit anti-survivin (R&D system) at 10µg/ml, overnight at 4°C. TICs were identified in xenograft tumors by CD44+ and CD24low/− staining and/or ALDH+ staining.
Statistical Analysis
All data are expressed as mean ± SEM, and statistically analyzed using unpaired Student’s t-test. Differences were considered statistically significant when P < 0.05.
Results
Gene expression profiles of residual cells after cisplatin plus TRAIL treatment
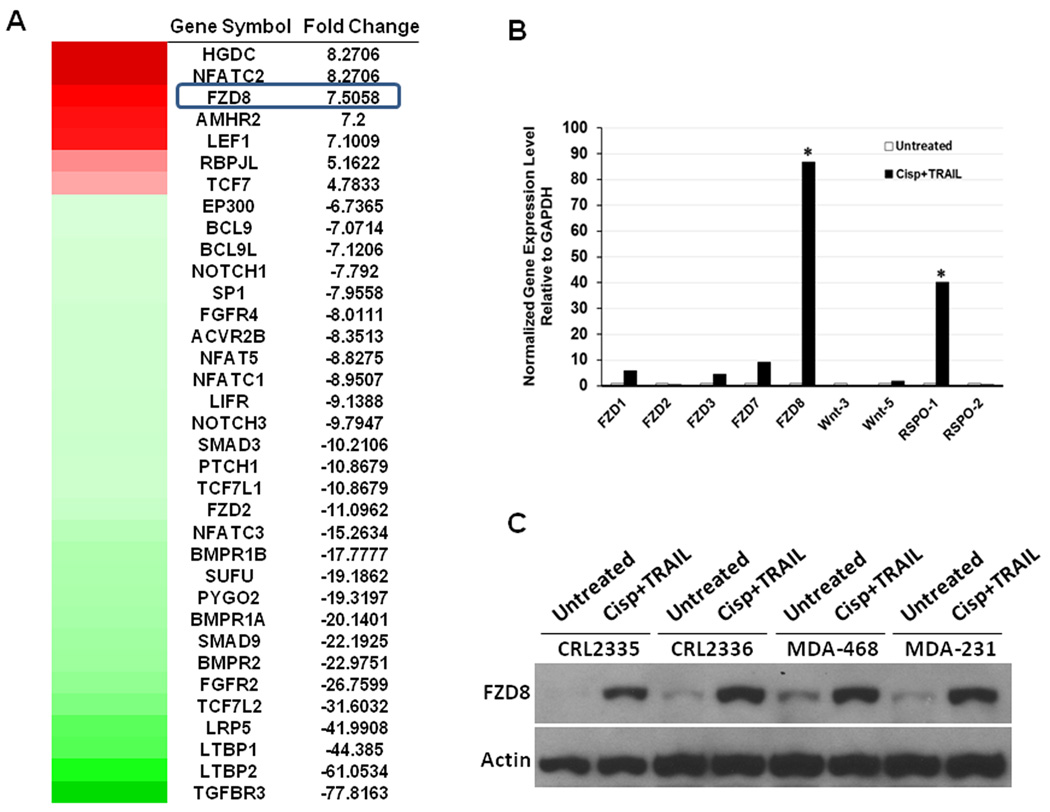
In order to identify the major molecular pathways that are involved in drug resistance to cisplatin plus TRAIL treatment, gene expression profiling of CRL2335 cells before and after cisplatin plus TRAIL treatment was done using a stem cell signaling PCR array. We found that the expressions of FZD8, one of the Wnt receptors, and its downstream targets LEF1 and TCF7 were increased in CRL2335 cells treated by cisplatin plus TRAIL compared with untreated controls (Figure 1A heatmap). Data from PCR array also showed that the treatment significantly inhibited other signaling pathways such as FGF, Hedgehog, Notch, and TGF-β (heatmap). Using the same samples analyzed with the stem cell signaling PCR array, we validated the results obtained for some of the genes that revealed significant changes in the expression and analyzed additional genes that code for Wnt receptors and their ligand using real-time RT-PCR. As shown in Figure 1B, treatment did not change significantly the gene expression levels of most of the Wnt-receptors and their ligands, except for FZD8 and R-spondin-1 (RSPO-1), which is one of its ligands. It was previously shown that RSPOs bind to FZD8 and low-density lipoprotein-related receptor (LRP)-5 and/or -6 to initiate downstream signaling that involves β-catenin, survivin, and c-myc, among others (17, 18). Consistent with these observations our data shows an increased expression of c-myc protein levels in cisplatin plus TRAIL treated TNBC cell lines compared to untreated controls (Supplementary Figure 1). Increase in FZD8 protein expression levels was further confirmed by Western blot analysis in TNBC CRL2335, CRL2336, MDA-468 and MDA-231 cells after treatment with cisplatin plus TRAIL (Figure 1C). Taken together, these results suggest that the cells resistant to the combined treatment with cisplatin and TRAIL showed increased expression of FZD8 and its ligand Rspo-1.
Figure 1.
FZD8 levels are increased in Cisplatin plus TRAIL-treated CRL2335 cells. (A) Gene expression profiles were performed to identify the major pathways involved in drug resistance using stem cell signaling PCR array. Note upregulation of Wnt receptor FZD8, and downstream targets LEF1 and TCF7 in cells treated with cisplatin plus TRAIL in heatmap. (B) Real-time RT-PCR analysis show no significant change in gene expression levels for most Wnt-receptors and ligands analyzed, with exception of FZD8 and RSPO-1 (* P< 0.05). (C) FZD8 increase in residual cells after cisplatin plus TRAIL treatment was further confirmed at the protein level by Western blot analysis.
FZD8 inhibition reduces β-catenin and survivin levels leading to increased apoptosis
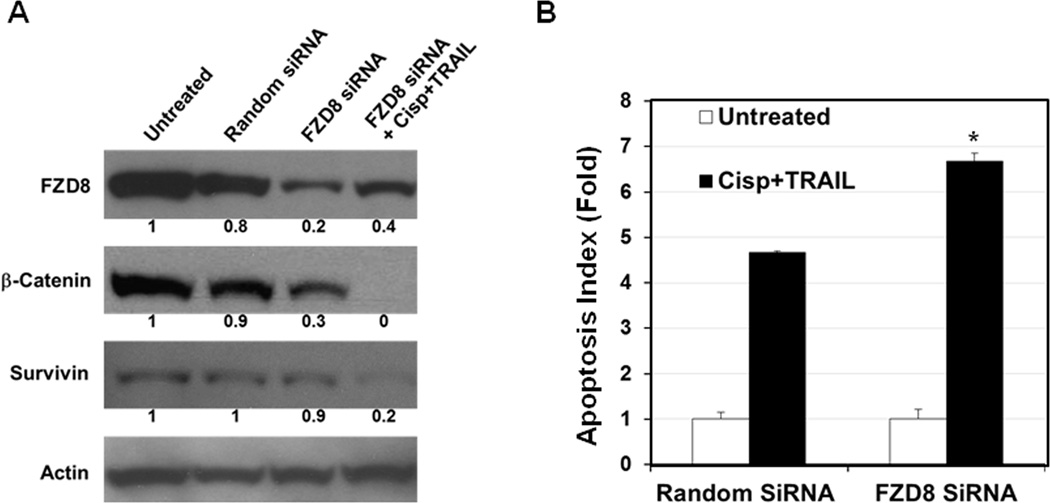
The expression of FZD8 protein in CRL2335 cells was knocked down using FZD8-siRNA to determine whether inhibition of this protein could inhibit Wnt-signaling and increases sensitivity to cisplatin plus TRAIL-induced apoptosis. The results presented in Figure 2A suggest that the inhibition of FZD8 by siRNA inhibited β-catenin and survivin protein levels compared to random siRNA-treated cells. However, maximum inhibition was observed in cells treated with FZD8-siRNA and cisplatin plus TRAIL. The results presented in Figure 2B suggest that FZD8-siRNA in combination with cisplatin plus TRAIL treatment significantly induced apoptosis, as compared to cells treated with random siRNA and cisplatin plus TRAIL. These results suggest that inhibition of FZD8 sensitizes the cells to cisplatin plus TRAIL-induced apoptosis.
Figure 2.
FZD8 inhibition increases apoptosis in the presence of cisplatin plus TRAIL. (A) Western blot analysis showing that FZD8 inhibition by siRNA reduces β-catenin and survivin protein levels in CRL2335 cells as compared to random siRNA-treated cells. Maximum inhibition was seen in cells transfected with FZD8 siRNA and treated with cisplatin plus TRAIL. (B) FZD8 siRNA in combination with cisplatin plus TRAIL treatment significantly increases apoptosis compared to random siRNA and cisplatin plus TRAIL treated cells. Results are expressed as mean ± SE (*P<0.05).
Cisplatin plus TRAIL treatment inhibits tumor growth as a consequence of non-TICs’ cell death
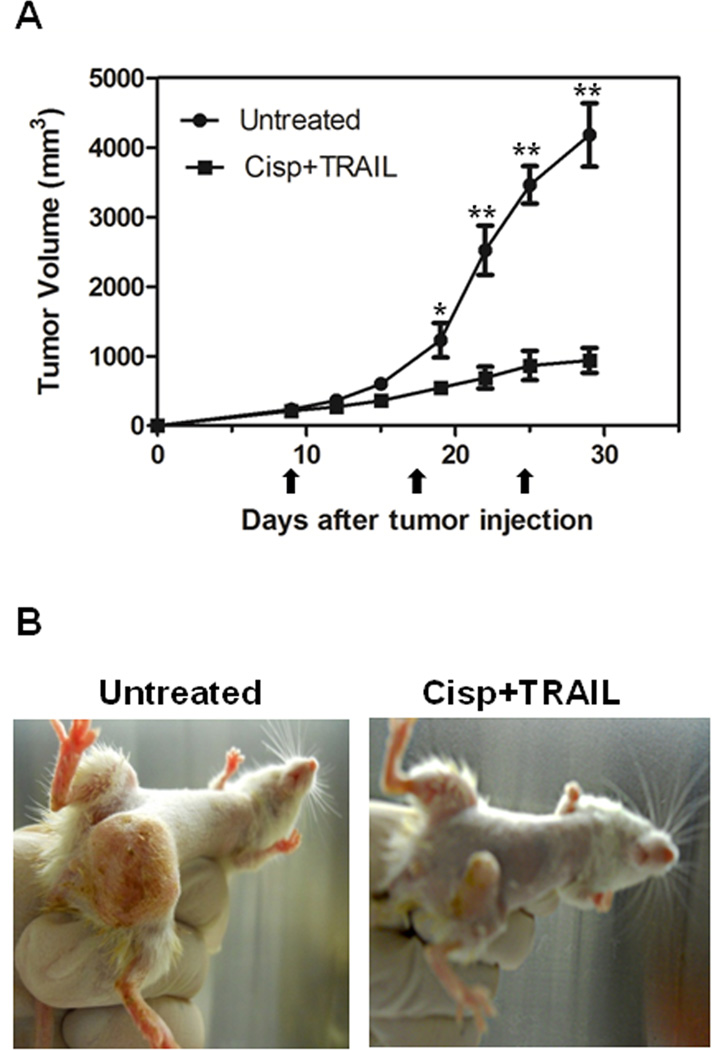
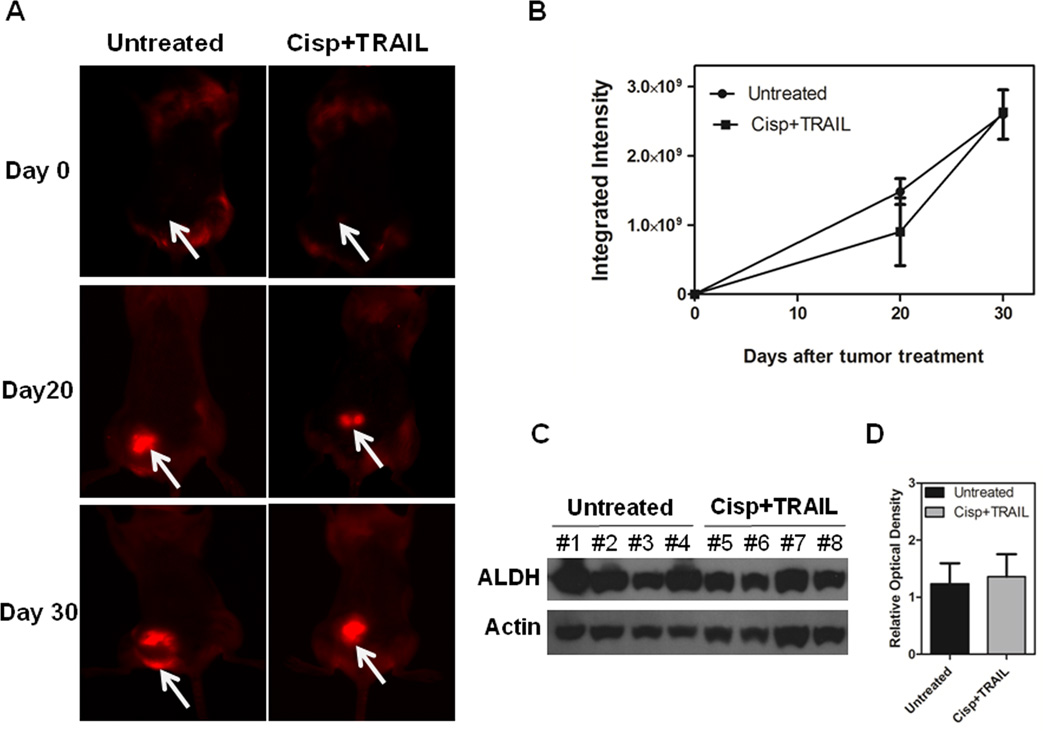
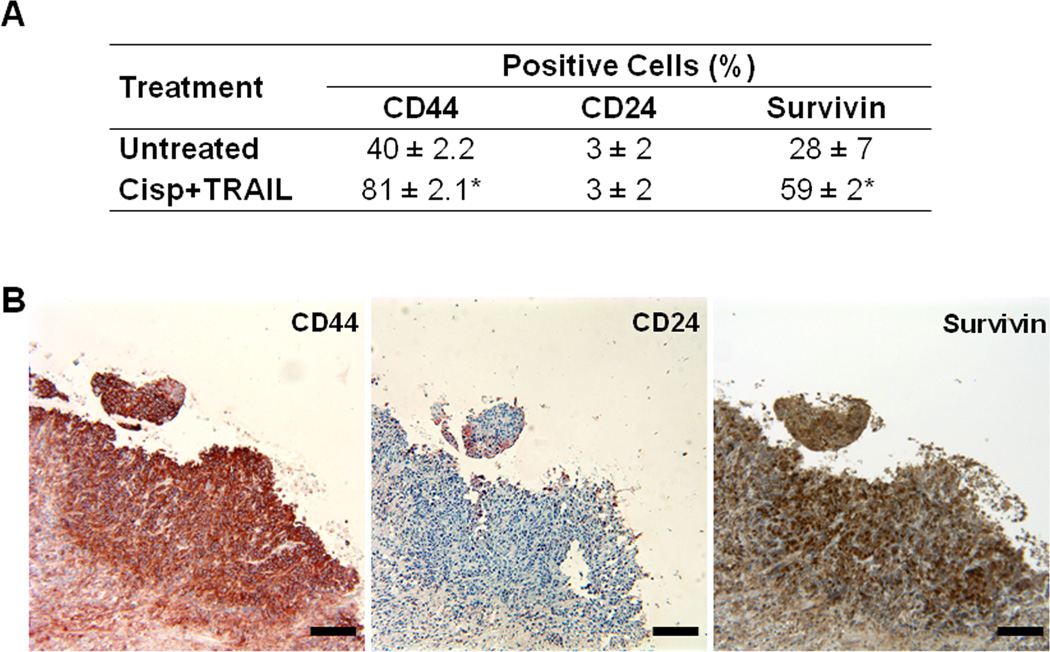
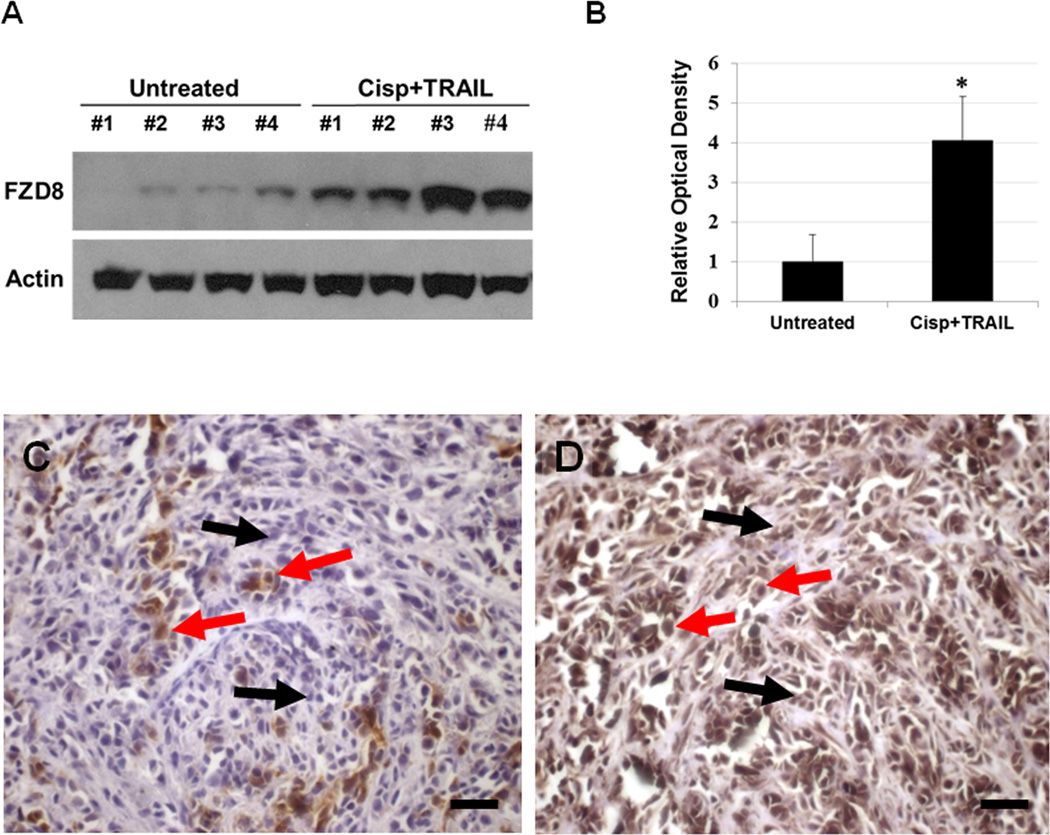
To determine the relevance of our in vitro data in a more clinically-related setting, we treated CRL2335 xenograft tumors formed by co-injection of ALDH1− and DsRed-transfected ALDH1+ CRL2335 cells with cisplatin (4 mg/kg) and TRAIL (15 mg/kg) once a week for three weeks. We confirmed the inhibitory effect of the combined treatment on the growth of the tumors, as shown in Figures 3A and 3B. However, this highly effective therapeutic effect, as documented by a significant reduction in tumor growth, was not reflected in the reduction of the DsRed-labeled tumor area detected by whole-body fluorescent imaging. In fact, the red fluorescent areas in tumors from mice treated with cisplatin and TRAIL did not show a significant reduction with respect to fast growing tumors in control mice (Figs. 4A and B). Homogenates obtained from the tumors showed no significant change in ALDH1 protein levels, one of the markers used for TICs, in cisplatin plus TRAIL-treated tumors compared to untreated tumors (Figs. 4C and D). Since our previous studies have shown that cisplatin plus TRAIL treatment is very effective in eliminating TICs in vitro (19), we were interested in determining how this treatment could affect TICs in vivo. Therefore, we analyzed the expression of CD44 and CD24 in the tumors using immunohistochemistry, considering that the CD44+/CD24−/low phenotype has been associated with tumor-initiating properties. We observed a higher percentage of CD44+/CD24−/low cells in tumors harvested from cisplatin plus TRAIL-treated animals than untreated animals (Figs. 5A and B. Moreover, the percentage of cells that immunostained positive for the anti-apoptotic protein and downstream target for Wnt/β-catenin signaling survivin in cisplatin plus TRAIL-treated tumors was significantly higher than in control tumors (Figs. 5A and B). Homogenates obtained from tumors were used to analyze the expression of FZD8 using Western blot. As depicted in Figures 6A and B, FZD8 protein levels were significantly increased in tumors treated with cisplatin plus TRAIL with respect to untreated tumors. In order to determine if FZD8 expression is localized to TICs, non-TICs, or both, we analyzed the expression of ALDH1 and FZD8 in tumors harvested from cisplatin plus TRAIL-treated and untreated control tumors. We observed that FZD8 was expressed both in TICs as well as non-TICs (Fig. 6C and D). Taken together, our results suggest that cisplatin plus TRAIL treatment significantly enhances cell death in non-TICs, while the effects on DsRed-expressing TICs are minimal.
Figure 3.
Efficacy of cisplatin plus TRAIL treatment on growth of tumor xenografts. (A) Significant inhibition of tumor xenografts formed by co-injection of non-TICs (ALDH1−) and DsRed-transfected TICs (ALDH1+) and treated with cisplatin plus TRAIL with respect to those in untreated control mice. Results are expressed as mean ± SE (*P<0.05; **P<0.005). Arrows indicate days of treatment. (B) Representative pictures of untreated and cisplatin and TRAIL-treated tumor-bearing mice 20 after treatment initiation showing differences in tumor size.
Figure 4.
Cisplatin plus TRAIL reduces in vivo tumor volume by increasing non-TICs death. (A) Representative pictures of whole-body fluorescent imaging showing that, despite the reduction in tumor volume reflected by dial caliper measurements (Fig. 3), DsRed-labeled tumor area is not reduced after treatment with cisplatin and TRAIL. (B) Quantification of DsRed expression in the tumors as integrated intensity in the untreated and treated groups. Results are expressed as mean ± SE. (C) Western blot showing ALDH1 protein levels in untreated and treated tumors on day 20 of treatment. (D) Densitometric analysis of ALDH1 bands obtained with Western blot showing relative levels in untreated and treated groups after normalization for equal protein loading using actin as loading control. Results are expressed as mean ± SE.
Figure 5.
Cisplatin plus TRAIL treatment increases TICs and survivin in vivo. (A) Percentage of CD44+/CD24−/low cells used as a marker for TICs in tumors harvested from cisplatin plus TRAIL-treated and untreated tumors. In addition to an increase in TICs, cisplatin plus TRAIL-treated tumors show an increase in the anti-apoptotic protein survivin compared to untreated tumors. Results are expressed as mean ± SE (*P<0.05). (B) Representative immunostaining for CD44, CD24, and survivin in tumor xenografts from cisplatin plus TRAIL-treated mice. Bars = 200 µm
Figure 6.
Cisplatin plus TRAIL treatment increases FZD8 expression in vivo. (A) Western blot analysis of tumor homogenates show significant increase in FZD8 protein levels in tumors from cisplatin plus TRAIL-treated mice compared to tumors from untreated mice. (B) ) Densitometric analysis of FZD8 bands obtained with Western blot showing relative levels in untreated and treated groups after normalization for equal protein loading using actin as loading control. Results are expressed as mean ± SE (*P<0.05). (C) Immunodetection of ALDH1 and (D) FZD8 in a representative tumor specimen harvested from a mouse treated with cisplatin and TRAIL. Red arrows show positive immunostaining for both ALDH and FZD8, while black arrows point cells which are positive for FZD8 but negative for ALDH1. Bars = 50 µm.
Discussion
TNBC is the most aggressive form of breast cancer and chemotherapy is currently the mainstay of systemic treatment for breast cancer, patients with TNBC disease have a worse outcome after chemotherapy than patients with other subtypes of breast cancer (4, 5). In spite of exciting new developments in treatment options including PARP inhibitors, cisplatin, angiogenesis inhibitor bevacizumab, and paclitaxel, many affected women still experience a relapse, and metastatic breast cancer remains a largely incurable disease.
We have previously shown that cisplatin plus TRAIL treatment was most effective in eliminating both TICs and non-TIC cells compared to PARP inhibitors, cisplatin, paclitaxel and docetaxel in vitro (19, 20). In order to identify the major pathways involved in drug resistance to cisplatin plus TRAIL treatment, here we analyzed gene expression profiles using CRL2335 TNBC cells before and after treatment with cisplatin plus TRAIL. The results from the present investigation suggest that cisplatin plus TRAIL treatment leads to a significant increase in gene expression of FZD8, and its downstream targets LEF1 and TCF7. In, addition, we found that RSPO-1 gene expression is also increased in cisplatin plus TRAIL-treated CRL2335 cells, suggesting the possibility that FZD8-mediated Wnt-signaling may play a major role in drug resistance in this system. It was previously shown that RSPOs can physically interact with the extracellular domains of LRP6/LRP5, and FZD8 and activate Wnt-reporter genes (17). Consistent with these observations, our data showed that inhibition of FZD8 by siRNA significantly reduces β-catenin, TCF/LEF and survivin protein expressions in the presence of cisplatin plus TRAIL. We have previously shown that suppression of survivin significantly increases TRAIL-induced cell death in CRL2335 TNBC cells (19). Different groups, in addition to our group, reported that increased survivin expression enhances tumor resistance to various apoptotic stimuli, primarily through caspase-dependent mechanism and its inhibition increase’s apoptosis (19, 21).
Our study using a new model that allows tracking of dsRed-expressing cells with TIC characteristics in vivo showed that treatment with cisplatin plus TRAIL reduces xenograft tumor volume by predominantly eliminating non-TICs, while having a minimal effect on DsRed-expressing cells. Consistent with these findings, immunostaining studies showed an increase in cells with CD44+/CD24−/low phenotype, which is typically associated with tumor-initiating properties, in residual tumor xenografts after treatment with cisplatin plus TRAIL. The commonly accepted criteria for clinical efficacy in phase II trials are tumor shrinkage, as defined by Response Evaluation Criteria in Solid Tumors (RECIST) (22), being the premise that tumor regression equates with the clinical benefits. However, our observations suggest that tumor shrinkage may not always correlate with clinical benefits because TICs enriched in residual tumors after treatment may reconstitute the primary tumor leading to tumor recurrence, drug resistance, and metastasis. Consistent with our observation, some studies have suggested that there is only a modest overall survival advantage for metastatic breast cancer patients even when tumor regression occurs (23–25).
Previous studies have demonstrated the involvement of several FZD receptors such as FZD2, FZD4, FZD7 and FZD10 in different types of cancers (26–30). It was recently shown that FZD-8 play a major role in human lung cancer, and its inhibition by FZD8-shRNA was shown to sensitize lung cancer cells to taxotere chemotherapy (31). Recently, it was reported that Wnt- signaling can be inhibited using the extracellular cysteine-rich domain of FZD8 fused to human Fc domain. This soluble receptor fused protein was shown to inhibit tumor growth in xenograft models (32). Our in vitro and in vivo results indicate that FZD8-mediated Wnt-signaling is up-regulated in residual tumors after cisplatin and TRAIL treatment in TNBC, leading to an increase in β-catenin and survivin levels, and reduced apoptosis.
In conclusion, our results suggest that the concept of equating tumor shrinkage with clinical benefits should be revisited, considering that a possible enrichment of cells with tumor-initiating-like characteristics may occur after treatment of TNBCs, leading to more malignant chemoresistant tumors. Based on our findings, the use of novel therapies targeting FZD8-mediated Wnt-signaling in combination with chemotherapeutic agents could be beneficial to improve the treatment outcome of patients diagnosed with TNBC.
Supplementary Material
Acknowledgments
Grant Support. This work was supported in part by Department of Pathology, WSU and the National Institute of Health Grant RO1 CA 118089 (to K.B. Reddy). The microscopy, imaging and cytometry resources core is supported, in part, by NIH center grant P30CA22453 (to G. Bepler) to The Karmanos Cancer Institute, WSU and Perinatology research branch of the National Institutes of Child health and development, Wayne State University.
Abbreviations
- ALDH
aldehyde dehydrogenase
- DsRed
Discosoma sp. red fluorescent protein
- FZD8
Frizzled receptor-8
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- LRP
low-density lipoprotein-related receptor, RSPO, R-spondin
- siRNA
small interfering RNA
- TCF/LEF
T-cell factor/lymphoid enhancer factor
- TIC
tumor initiating cells
- TNBC
triple-negative breast cancer
- TNF
tumor necrosis factor
- TRAIL
TNF-related apoptosis-inducing ligand
Footnotes
Conflicts of interests: The authors declare that they have no potential conflicts of interest.
References
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.Cleator S, Heller W, Coombes RC. Triple-negative breast cancer: therapeutic options. Lancet Oncol. 2007;8:235–244. doi: 10.1016/S1470-2045(07)70074-8. [DOI] [PubMed] [Google Scholar]
- 3.Reddy KB. Triple-negative breast cancers: an updated review on treatment options. Curr Oncol. 2011;18:e173–e179. doi: 10.3747/co.v18i4.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 5.Tan DS, Marchio C, Jones RL, Savage K, Smith IE, Dowsett M, et al. Triple negative breast cancer: molecular profiling and prognostic impact in adjuvant anthracycline-treated patients. Breast Cancer Res Treat. 2008;111:27–44. doi: 10.1007/s10549-007-9756-8. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Pinilla SM, Sarrio D, Honrado E, Hardisson D, Calero F, Benitez J, et al. Prognostic significance of basal-like phenotype and fascin expression in node-negative invasive breast carcinomas. Clin Cancer Res. 2006;12:1533–1539. doi: 10.1158/1078-0432.CCR-05-2281. [DOI] [PubMed] [Google Scholar]
- 7.Al-Hajj M, Becker MW, Wicha M, Weissman I, Clarke MF. Therapeutic implications of cancer stem cells. Curr Opin Genet Dev. 2004;14:43–47. doi: 10.1016/j.gde.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 11.Ginestier C, Liu S, Diebel ME, Korkaya H, Luo M, Brown M, et al. CXCR1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. J Clin Invest. 2010;120:485–497. doi: 10.1172/JCI39397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K. Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res. 2009;69:7507–7511. doi: 10.1158/0008-5472.CAN-09-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Hajj M, Clarke MF. Self-renewal and solid tumor stem cells. Oncogene. 2004;23:7274–7282. doi: 10.1038/sj.onc.1207947. [DOI] [PubMed] [Google Scholar]
- 14.Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA, et al. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J Cell Mol Med. 2009;13:2236–2252. doi: 10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nam JS, Turcotte TJ, Smith PF, Choi S, Yoon JK. Mouse cristin/R-spondin family proteins are novel ligands for the Frizzled 8 and LRP6 receptors and activate beta-catenin-dependent gene expression. J Biol Chem. 2006;281:13247–13257. doi: 10.1074/jbc.M508324200. [DOI] [PubMed] [Google Scholar]
- 18.Wei Q, Yokota C, Semenov MV, Doble B, Woodgett J, He X. R-spondin1 is a high affinity ligand for LRP6 and induces LRP6 phosphorylation and beta-catenin signaling. J Biol Chem. 2007;282:15903–15911. doi: 10.1074/jbc.M701927200. [DOI] [PubMed] [Google Scholar]
- 19.Xu L, Yin S, Banerjee S, Sarkar F, Reddy KB. Enhanced anticancer effect of the combination of cisplatin and TRAIL in triple-negative breast tumor cells. Mol Cancer Ther. 2011;10:550–557. doi: 10.1158/1535-7163.MCT-10-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin S, Xu L, Bandyopadhyay S, Sethi S, Reddy KB. Cisplatin and TRAIL enhance breast cancer stem cell death. Int J Oncol. 2011;39:891–898. doi: 10.3892/ijo.2011.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 22.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 23.Abratt RP, Brune D, Dimopoulos MA, Kliment J, Breza J, Selvaggi FP, et al. Randomised phase III study of intravenous vinorelbine plus hormone therapy versus hormone therapy alone in hormone-refractory prostate cancer. Ann Oncol. 2004;15:1613–1621. doi: 10.1093/annonc/mdh429. [DOI] [PubMed] [Google Scholar]
- 24.Huff CA, Matsui W, Smith BD, Jones RJ. The paradox of response and survival in cancer therapeutics. Blood. 2006;107:431–434. doi: 10.1182/blood-2005-06-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huff CA, Matsui WH, Douglas Smith B, Jones RJ. Strategies to eliminate cancer stem cells: clinical implications. Eur J Cancer. 2006;42:1293–1297. doi: 10.1016/j.ejca.2006.01.045. [DOI] [PubMed] [Google Scholar]
- 26.Fukukawa C, Nagayama S, Tsunoda T, Toguchida J, Nakamura Y, Katagiri T. Activation of the non-canonical Dvl-Rac1-JNK pathway by Frizzled homologue 10 in human synovial sarcoma. Oncogene. 2009;28:1110–1120. doi: 10.1038/onc.2008.467. [DOI] [PubMed] [Google Scholar]
- 27.Jin X, Jeon HY, Joo KM, Kim JK, Jin J, Kim SH, et al. Frizzled 4 regulates stemness and invasiveness of migrating glioma cells established by serial intracranial transplantation. Cancer Res. 2011;71:3066–3075. doi: 10.1158/0008-5472.CAN-10-1495. [DOI] [PubMed] [Google Scholar]
- 28.Ueno K, Hazama S, Mitomori S, Nishioka M, Suehiro Y, Hirata H, et al. Down-regulation of frizzled-7 expression decreases survival, invasion and metastatic capabilities of colon cancer cells. Br J Cancer. 2009;101:1374–1381. doi: 10.1038/sj.bjc.6605307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L, Wu X, Wang Y, Zhang K, Wu J, Yuan YC, et al. FZD7 has a critical role in cell proliferation in triple negative breast cancer. Oncogene. 2011;30:4437–4446. doi: 10.1038/onc.2011.145. [DOI] [PubMed] [Google Scholar]
- 30.Zeng G, Germinaro M, Micsenyi A, Monga NK, Bell A, Sood A, et al. Aberrant Wnt/beta-catenin signaling in pancreatic adenocarcinoma. Neoplasia. 2006;8:279–289. doi: 10.1593/neo.05607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang HQ, Xu ML, Ma J, Zhang Y, Xie CH. Frizzled-8 as a putative therapeutic target in human lung cancer. Biochem Biophys Res Commun. 2012;417:62–66. doi: 10.1016/j.bbrc.2011.11.055. [DOI] [PubMed] [Google Scholar]
- 32.DeAlmeida VI, Miao L, Ernst JA, Koeppen H, Polakis P, Rubinfeld B. The soluble wnt receptor Frizzled8CRD-hFc inhibits the growth of teratocarcinomas in vivo. Cancer Res. 2007;67:5371–5379. doi: 10.1158/0008-5472.CAN-07-0266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.