Abstract
NON-EXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1) is the central regulator of the pathogen defense reaction systemic acquired resistance (SAR). NPR1 acts by sensing the SAR signal molecule salicylic acid (SA) to induce expression of PATHOGENESIS-RELATED (PR) genes. Mechanistically, NPR1 is the core of a transcription complex interacting with TGA transcription factors and NIM1-INTERACTING (NIMIN) proteins. Arabidopsis NIMIN1 has been shown to suppress NPR1 activity in transgenic plants. The Arabidopsis NIMIN family comprises four structurally related, yet distinct members. Here, we show that NIMIN1, NIMIN2, and NIMIN3 are expressed differentially, and that the encoded proteins affect expression of the SAR marker PR-1 differentially. NIMIN3 is expressed constitutively at a low level, but NIMIN2 and NIMIN1 are both responsive to SA. While NIMIN2 is an immediate early SA-induced and NPR1-independent gene, NIMIN1 is activated after NIMIN2, but clearly before PR-1. Notably, NIMIN1, like PR-1, depends on NPR1. In a transient assay system, NIMIN3 suppresses SA-induced PR-1 expression, albeit to a lesser extent than NIMIN1, whereas NIMIN2 does not negatively affect PR-1 gene activation. Furthermore, although binding to the same domain in the C-terminus, NIMIN1 and NIMIN2 interact differentially with NPR1, thus providing a molecular basis for their opposing effects on NPR1. Together, our data suggest that the Arabidopsis NIMIN proteins are regulators of the SAR response. We propose that NIMINs act in a strictly consecutive and SA-regulated manner on the SA sensor protein NPR1, enabling NPR1 to monitor progressing threat by pathogens and to promote appropriate defense gene activation at distinct stages of SAR. In this scenario, the defense gene PR-1 is repressed at the onset of SAR by SA-induced, yet instable NIMIN1.
Keywords: NIM1-INTERACTING (NIMIN) proteins, NON-EXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1), PATHOGENESIS-RELATED GENE1 (PR-1), plant defense gene activation, protein–protein interaction, salicylic acid (SA), systemic acquired resistance (SAR)
INTRODUCTION
Plants have evolved different layers of defense to recognize and combat invading microbes (Jones and Dangl, 2006). The immune response systemic acquired resistance (SAR) is launched after primary infection and activation of effector-triggered immunity (ETI) accompanied by formation of necrosis at the sites of pathogen invasion. SAR becomes effective in non-infected plant tissue far away from the pathogen penetration sites (Ross, 1961). The response fends off secondary infections by diverse types of biotrophic pathogens and is long-lasting. The local signal to induce SAR in non-infected leaves is salicylic acid (SA;Vernooij et al., 1994). Levels of free and conjugated SA rise not only in infected necrotic tissue, but also systemically in non-infected leaves (Malamy et al., 1990;Métraux et al., 1990). This increase in SA concentration is paralleled by local and systemic induction of various PATHOGENESIS-RELATED (PR) genes (van Loon and van Kammen, 1970;Ward et al., 1991;van Loon et al., 2006). Some PR genes, e.g., PR-1, can be induced solely by exogenous application of SA or its functional analogs 2,6-dichloroisonicotinic acid (INA) and benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester (BTH;White, 1979;Vernooij et al., 1995;Friedrich et al., 1996;Lawton et al., 1996). Furthermore, it has been shown that SA-treated tobacco and Arabidopsis plants expressing PR-1 genes display SAR (White, 1979;Uknes et al., 1992, 1993). Thus, accumulation of PR-1 transcripts and PR-1 proteins either in non-infected parts of plants exhibiting necrosis or in response to exogenous application of SA serves as marker for the SAR resistance reaction.
The central regulator of SAR is NON-EXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1). The gene was identified from Arabidopsis mutants compromised in chemical induction of PR genes and in resistance to fungal infection (Cao et al., 1997;Ryals et al., 1997;Shah et al., 1997). Overexpression experiments strongly suggest that NPR1 is active only after SA induction (Cao et al., 1998;Friedrich et al., 2001). The Arabidopsis NPR1 family encompasses six members, NPR1 to NPR6, and recent evidence indicates that SA signals directly through some members. However, the mechanism of how SA acts on NPR1 family proteins is controversial. First, it has been demonstrated that NPR1 from Arabidopsis (At) and two NPR1 family members from tobacco (Nt) alter some of their biochemical capabilities in response to the SA signal molecule in a heterologous yeast system in absence of any other plant protein (Maier et al., 2011). For example, Nt NPR1 gains transcription activity, when SA is added to yeast growth medium. The data indicate that NPR1 family proteins are able to sense SA, and that they undergo an alteration upon perception of SA. Consequently, Arabidopsis NPR1 family members have been found to bind SA in vitro (Fu et al., 2012;Wu et al., 2012), albeit with very different affinities. While NPR4 is a high affinity receptor and NPR3 is a lower affinity receptor, SA appears to bind only very weakly to NPR1. It has been proposed that PR-1 gene activation in the course of SAR is regulated through availability of NPR1, which, in turn, is controlled by cytoplasmic oligomer–nuclear monomer shuttling and by differential interaction of NPR1 with SA-perceiving NPR4 and NPR3 in the nucleus (Mou et al., 2003;Fu et al., 2012). In two other models, SA perception during SAR has, however, been attributed to the NPR1 protein, itself.Wu et al. (2012) have suggested that NPR1 binds SA via the transition metal copper in a complex with two cysteine residues, Cys-521 and Cys-529, and that, upon SA binding, a C-terminal transactivation domain is released from the N-terminal autoinhibitory BTB/POZ (broad complex, tramtrack, and bric à brac/pox virus and zinc finger) domain. Curiously, only Arabidopsis NPR1 contains two closely spaced cysteine residues in its C-terminus. In a third model, based on biochemical evidence obtained in the heterologous yeast system, two distinct domains in the C-terminus of NPR1 proteins have been implicated in sensing the SA signal (Maier et al., 2011). These domains are highly conserved in NPR1 proteins from diverse species and they are also conserved in the NPR1 paralogs NPR2, NPR3, and NPR4 from Arabidopsis and in tobacco NPR3 (also known as NIM1-LIKE1). One domain comprises the penta-amino acid motif LENRV (amino acids 429–433). The LENRV motif imposes SA sensitivity on NPR1 proteins from Arabidopsis and tobacco in yeast. The signature is altered in the non-functional nim1-4 mutant (R432K;Ryals et al., 1997). The latter model is corroborated by genetic evidence provided through an en masse in planta screen for Arabidopsis insensitive to the functional SA analog BTH (Canet et al., 2010). In this screen, dozens of npr1 alleles were identified, and the mutants have been found to be clustered in the same two regions identified independently by biochemical dissection of NPR1 family proteins in yeast. The nim1-4 mutant was isolated three times. On the contrary, Cys-521 and Cys-529 were not uncovered genetically.
The SA sensor protein NPR1 interacts with two groups of proteins. TGA transcription factors connect NPR1 with SA-responsive as-1-like cis-acting elements present in the promoters of PR-1 genes from tobacco and Arabidopsis (Lebel et al., 1998;Strompen et al., 1998;Zhang et al., 1999;Després et al., 2000;Zhou et al., 2000). This finding is consistent with several reports showing that NPR1 proteins from Arabidopsis, tobacco, and rice promote transcription activation in diverse systems (Rochon et al., 2006;Maier et al., 2011;Chern et al., 2012). The data imply that NPR1 is the core of a transcription complex on PR gene promoters. In addition to TGA factors, NPR1 interacts with the group of small NIM1-INTERACTING (NIMIN) proteins (Weigel et al., 2001). Like NPR1, NIMIN genes are dispersed in the whole plant kingdom (Chern et al., 2005;Zwicker et al., 2007). NIMIN proteins harbor nuclear localization signals, and thus target NPR1 in the nucleus (Weigel et al., 2001;Chern et al., 2005;Zwicker et al., 2007). However, their functional significance was not evident, when NIMINs were first identified.
Arabidopsis contains four NIMIN genes, NIMIN1, NIMIN1b, NIMIN2, and NIMIN3 (Weigel et al., 2001). Of these, NIMIN1 and NIMIN2 have been studied in some detail. Both genes are strongly up-regulated by SA. In contrast, the two genes are not induced significantly in pathogen-infected necrotic tissue displaying ETI (Glocova et al., 2005). Hence, NIMIN1 and NIMIN2 seem to be specifically linked to the SA-dependent SAR response, rather than to ETI. Similarly, tobacco NIMIN2-type mRNAs accumulate in response to the SA signal molecule (Horvath et al., 1998;Zwicker et al., 2007). Although clearly structurally related, the Arabidopsis NIMIN proteins are distinct from each other. For example, they interact differentially with NPR1 (Weigel et al., 2001). NIMIN3 interacts with the At NPR1 N-terminal half, whereas NIMIN1, NIMIN1b, and NIMIN2 possess similar motifs by which they bind to the At NPR1 C-terminal third. In the C-terminus of Nt NPR1, the binding region of SA-induced NIMIN2-type proteins has been mapped from amino acids 494 to 510 (Maier et al., 2011). Notably, several npr1 mutant alleles have been uncovered in the corresponding region of At NPR1, all of which affect responsiveness to BTH in planta (Canet et al., 2010). Furthermore, occurrence of the interaction domain for inducible NIMIN2-type proteins and the LENRV domain is coincident in NPR1 proteins and its paralogs from many species. Thus, these two domains appear to be intimately connected with the SA response.
The functional significance of NIMIN proteins for NPR1 activity has been addressed in overexpression experiments. Both Arabidopsis NIMIN1 and NEGATIVE REGULATOR OF DISEASE RESISTANCE (NRR), a NIMIN homolog from rice, are able to suppress induction of PR genes and to cause enhanced susceptibility to bacterial pathogens in transgenic plants (Chern et al., 2005, 2008;Weigel et al., 2005). From these data, it has been concluded that NIMIN proteins are repressors of NPR1. However, in tobacco, constitutive overexpression of Nt NIMIN2a produced only a delay in PR-1 protein accumulation, and it has been suggested that NIMIN proteins, although negatively affecting NPR1 activity, are, at bottom, positive regulators of NPR1-mediated PR gene induction (Zwicker et al., 2007). Apart from NIMIN1, the biological significance of other Arabidopsis NIMIN family members has not yet been addressed. Here, we provide evidence that the Arabidopsis NIMIN proteins affect NPR1 differentially at distinct stages of SAR, thus enabling the plant to strictly control defense gene activation in tissue distant from sites of pathogen entry undergoing ETI.
RESULTS
NIMIN3 IS NOT RESPONSIVE TO PLANT DEFENSE SIGNALS
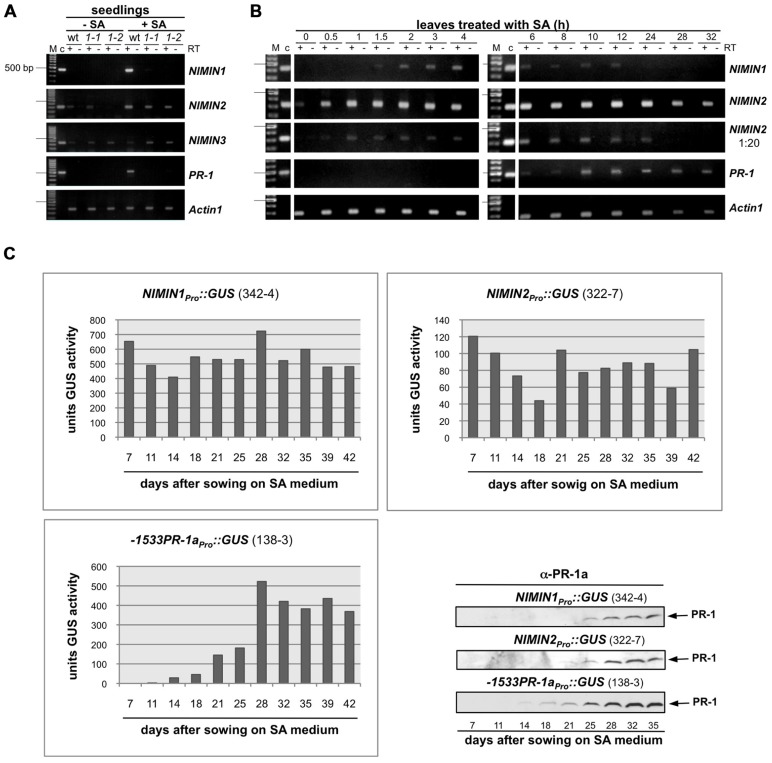
Previously, we have shown that NIMIN1 and NIMIN2 are strongly induced by treatment of Arabidopsis plants with SA or Bion®, a commercial plant growth regulator containing the functional SA analog BTH, and that this induction is due to transcriptional gene activation (Weigel et al., 2001, 2005;Glocova et al., 2005). To further elucidate the functional relevance of NIMIN genes, we have now analyzed expression of NIMIN3 in response to diverse signal molecules involved in plant defense reactions. Initially, transcript accumulation was monitored using reverse transcriptase-polymerase chain reaction (RT-PCR) analyses. The primers used and the sizes of fragments generated by PCR from plasmids carrying cDNAs for NIMIN3 and various control genes are listed in Table 1. NIMIN3 transcript levels were compared to expression of the NIMIN1, NIMIN2, and PR-1 genes. Unlike NIMIN1, NIMIN2, and PR-1, expression of NIMIN3 was neither induced by SA nor BTH (Figure 1A). Moreover, jasmonate (JA), another plant defense signal, had no effect on either of the NIMIN genes (data not shown). However, we were able to detect NIMIN3 transcripts in several independent RNA preparations irrespective of whether they had been isolated from control or chemically induced plant tissue (Figures 1A and 2A), suggesting that NIMIN3 may be expressed constitutively at a low level. To address this question, we isolated 1.4 kb of the NIMIN3 5′-upstream region and fused it to the β -glucuronidase (GUS) reporter gene. The chimeric gene was transferred to the tobacco genome, and GUS enzyme activity was determined in seven independent primary transformants, all containing intact copies of the reporter gene construct (data not shown). As compared to transgenic tobacco plants carrying analogous NIMIN1Pro::GUS or NIMIN2Pro::GUS constructs (0.8 and 0 GUS units on an average, respectively;Glocova et al., 2005), untreated plants containing NIMIN3Pro::GUS exhibited constitutive GUS enzyme activity (14.7 GUS units on an average; Figure 1B). Reporter gene expression from the NIMIN3Pro::GUS construct in the tobacco genome was not enhanced significantly by treatment of plants with SA (0.3 and 1 mM; 17.6 GUS units on an average), BTH (0.34 mM), methyl JA (MeJA; 0.1 mM), or H2O2 (0.1 and 1 mM; data not shown). Likewise, gene expression from the NIMIN3 promoter was not elevated by elicitation of HR or by exogenous application of the phytohormones 2,4-dichlorophenoxy acetic acid (2,4-D), gibberellic acid (GA), indole-3-acetic acid (IAA), or 1-naphthalene acetic acid (NAA; 0.01 and 0.1 mM each; data not shown). As determined by histochemical staining, NIMIN3-mediated reporter enzyme activity is mainly localized in leaf tissue (Figure 1B). Of note, NIMIN3 gene expression is independent from an intact NPR1 gene (Figure 2A).
Table 1.
Primers and control plasmids used in RT-PCR analyses.
| Gene | Control plasmid | Primer | Sequence | Fragment size (bp) |
|---|---|---|---|---|
| NIMIN1 | pGBT9/NIMIN1 | N1fwd N1bck |
5′-CGGGATCCATATGTATCCTAAACAATTTAG 5′-AACCCGGGCTACTACAATGCAAGATTAAGATC |
449 |
| NIMIN2 | pGBT9/NIMIN2 | N2fwd N2bck |
5′-ACGCGTAGAAGAAGATAACGG 5′-CTAACGCTGTCTGGTTCCGGT |
330 |
| NIMIN3 | pGBT9/NIMIN3 | N3fwd N3bck |
5′-GGGGATCCATATGGACAGAGACAGAAAGAG 5′-TTCCCGGGCTACAGAGAAAGATTCAAGTC |
357 |
| PR-1 | pUC19/AtPR-1 | PR1fwd PR1bck |
5′-GGGGATCCATATGAATTTTACTGGC 5′-CTGAGCTCTTAGTATGGCTTCTCG |
504 |
| Actin1 | – | Act1 Act2 |
5′-CGATGAAGCTCAATCCAAACGA 5′-CAGAGTCGAGCACAATACCG |
302 |
FIGURE 1.
ArabidopsisNIMIN3 is expressed constitutively. (A) RT-PCR analyses of NIMIN3 expression in Arabidopsis whole seedlings and leaf tissue. Expression of NIMIN3 is compared to expression of NIMIN1, NIMIN2, and PR-1. RNA samples were isolated from 2-week-old whole seedlings grown either on MS medium or MS medium with addition of 0.3 mM SA and from leaves of 4-week-old plants 24 h after spraying with water or a suspension of Bion® containing 0.34 mM BTH. RT-PCR analyses were performed on DNase I-treated total RNA preparations in presence or absence of reverse transcriptase (RT) with primer combinations listed in Table 1. In lanes c, PCR products from 1 ng of plasmid DNAs carrying the respective cDNAs were loaded. The amplification of Actin1 mRNA serves as an internal standard for different RNA samples used in the amplification reactions. (B) Expression of a NIMIN3Pro::GUS reporter gene in transgenic tobacco seedlings. Expression from the NIMIN3 promoter is compared to reporter gene expression from the NIMIN1, NIMIN2, and Nt PR-1a promoters. Tobacco seedlings (T1 generation) transformed with the indicated reporter genes were grown on MS medium with kanamycin or on selective medium supplemented with 0.3 mM SA. Two independent lines for each construct or, as in case of the Nt PR-1a promoter, two different constructs were analyzed. Seedlings were stained for GUS reporter enzyme activity when 4-weeks-old.
FIGURE 2.
Salicylic acid-induced ArabidopsisNIMIN1 and NIMIN2 are expressed differentially from each other and from PR-1. RNA samples were isolated from Arabidopsis seedlings or Arabidopsis leaves and analyzed as described in Figure 1A. Expression of NIMIN1 and NIMIN2 is compared to expression of NIMIN3 and PR-1. (A) RT-PCR analyses of RNAs from wild-type (Col-0) and npr1-1 and npr1-2 mutant seedlings. 1-1, npr1-1; 1-2, npr1-2. (B) RT-PCR analyses of RNAs from leaf tissue at different times after spraying plants with 1 mM SA. (C) Time course of SA-induced GUS reporter enzyme activities and PR-1 protein accumulation in tobacco seedlings transformed with NIMIN1Pro::GUS or NIMIN2Pro::GUS. Expression from the two NIMIN promoters is compared to reporter gene expression from the Nt -1533PR-1a promoter. For immunodetection of endogenous PR-1 proteins, equal amounts of protein were loaded in each lane of the SDS gels. Seedlings (T1 generation) were grown on selective medium with 0.3 mM SA. Similar results were obtained with independent lines of NIMIN1Pro::GUS, NIMIN2Pro::GUS and -1533PR-1aPro::GUS.
SALICYLIC ACID-MEDIATED INDUCTION OF NIMIN1 AND NIMIN2 PROCEEDS THROUGH SEPARATE PATHWAYS
RNA analyses as depicted in Figure 1A had shown that NIMIN1 was expressed only after induction, just as PR-1, while NIMIN2 expression was occasionally observed prior to chemical treatment of plants. This finding was unexpected since the NIMIN2 promoter exhibits clear chemical induction in transgenic tobacco plants (0 GUS units and 265.0 GUS units on an average for water and SA treatment, respectively, n = 10; Figure 1B;Glocova et al., 2005). It therefore seemed of interest to analyze regulation of the NIMIN1 and NIMIN2 genes in closer detail.
Initially, we used two npr1 mutants, npr1-1 and npr1-2, which are not able to support PR-1 gene induction (Cao et al., 1994;Glazebrook et al., 1996). Surprisingly, NIMIN1, like PR-1, was inactive in absence of a functional NPR1 gene (Figure 2A). Yet, NIMIN2 expression was clearly detectable in both npr1 mutants, although, in some experiments, NIMIN2 transcript levels appeared to accumulate to lower overall levels in npr1 than in wild-type plants (Figure 2A and data not shown). Our data are in conflict with another report.Blanco et al. (2009) have described that expression of both NIMIN1 and NIMIN2 is abolished in the npr1-1 mutant. To support our results, we verified the identity of the NIMIN2 RT-PCR products by digestion with restriction enzymes (data not shown). Hence, NIMIN2 expression, unlike NIMIN1 and PR-1 expression, may be either independent or only partly dependent on NPR1. Furthermore, the kinetics of gene induction turned out to be different between NIMIN1 and NIMIN2. Both genes are expressed transiently after SA application (Figure 2B). Yet, NIMIN2 gene expression started immediately (0.5 h) after SA treatment, reached its maximum early (after 1 h) and was maintained at a high level for 24 h (Figure 2B). Thus, NIMIN2 seems to be an immediate early SA responsive gene, as suggested previously for the tobacco NIMIN2a gene (Horvath et al., 1998). NIMIN1 transcripts, on the other side, became most abundant only around 2 h after SA application (Figure 2B). This is clearly later than the onset of NIMIN2 expression, yet earlier than the onset of PR-1 induction. Notably, NIMIN1 expression appeared even more transient than NIMIN2 expression and was already shut down when PR-1 transcripts began to accumulate. The time course of NIMIN1 gene induction shown here is in accordance with previous results obtained by northern blotting (Weigel et al., 2005). Together, our data strongly suggest that SA-mediated induction of the NIMIN1 and NIMIN2 genes proceeds through separate pathways.
The kinetics of gene induction were also monitored in tobacco seedlings containing NIMINPro::GUS reporter gene constructs. Transgenic seeds were germinated on SA-containing medium. The germination of seeds occurred simultaneously for all lines analyzed, and the development of seedlings progressed similarly. GUS enzyme activities were first determined 7 days after sowing when small seedlings had emerged. With both NIMIN2Pro::GUS and NIMIN1Pro::GUS, we did not observe a clear induction profile (Figure 2C). GUS enzyme activity was already switched on to high levels early after germination. In contrast, PR-1a promoter activation and accumulation of the endogenous PR-1 proteins occurred with significant delay (Figure 2C). Thus, the kinetics of reporter gene activation from the NIMIN1 and NIMIN2 promoters in SA-treated tobacco seem to parallel the transcript accumulation patterns observed in Arabidopsis, i.e., NIMIN genes are induced by SA prior to PR-1 genes. The data indicate that the molecular cues for early induction during the SAR response are contained within the 1 kb 5′-flanking regions of NIMIN1 and NIMIN2, and that these cues are recognized in the heterologous species tobacco. Reporter gene expression from both the NIMIN1 and NIMIN2 promoters occurred in leaf and root tissue (Figure 1B). Likewise, green fluorescent protein (GFP) expression from the 0.8 kb NIMIN1 promoter has been observed in roots, petioles, and leaves in transgenic Arabidopsis plants (Fonseca et al., 2010). This expression pattern distinguishes the SA-inducible NIMIN1 and NIMIN2 promoters from the NIMIN3 promoter and the tobacco PR-1a promoter which are predominantly active in leaf tissue (Figure 1B).
NIMIN1 AND NIMIN3 SUPPRESS SALICYLIC ACID-INDUCED EXPRESSION FROM THE TOBACCO PR-1a PROMOTER
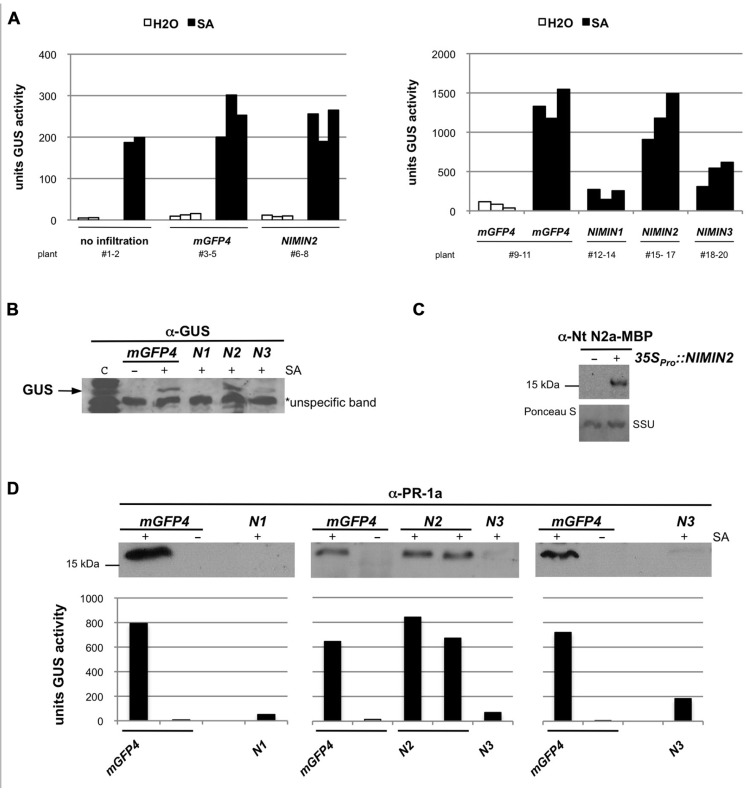
To unravel the functional significance of NIMIN gene expression at different times during the SAR response, we have developed an in planta assay for NIMIN activity. The gene coding for GUS under control of the tobacco PR-1a promoter (-1533PR-1aPro::GUS;Grüner and Pfitzner, 1994) was stably integrated in the genome of Nicotiana benthamiana. Several primary transformants were obtained all of which exhibited very strong and stringent induction of the reporter gene upon SA treatment of leaf tissue (data not shown). One typical line (3 GUS units uninduced and 1100 GUS units after SA treatment) was propagated, and T2 plants were used for infiltration experiments with an Agrobacterium strain carrying the gene for GFP (mGFP4) driven by the Cauliflower mosaic virus (CaMV) 35S RNA promoter (35SPro::mGFP4). Infiltration of 35SPro::mGFP4 Agrobacteria yielded GUS enzyme activities only slightly above the background levels of non-infiltrated control leaves, showing that agroinfiltration alone is not sufficient for efficient activation of the PR-1aPro::GUS reporter gene (Figures 3A and 4A).
FIGURE 3.
Arabidopsis NIMIN1 and NIMIN3 suppress salicylic acid-induced gene expression from the tobacco PR-1a promoter in N. benthamiana. (A) Effects of transient expression of 35SPro::NIMIN1 and 35SPro::NIMIN3 in an N. benthamiana reporter line with integrated -1533PR-1aPro::GUS. Three plants were infiltrated in parallel for each gene construct with Agrobacterium strains as indicated. For a better direct comparison, the two halves of the same leaf were infiltrated with Agrobacteria harboring 35SPro::NIMIN1 and 35SPro::NIMIN3, respectively. Leaf disks excised from infiltrated leaf areas were floated on water or on 1 mM SA before determination of GUS enzyme activity. The three bars for each construct and treatment represent GUS activities from the three agroinfiltration experiments performed in parallel. Representative results are shown. N1, NIMIN1; N3, NIMIN3. (B) Immunodetection of NIMIN3 in extracts from agroinfiltrated and SA-floated leaf tissue. NIMIN3 accumulation was detected with a specific antiserum in an extract shown in Figure 3A. An unspecific band marked on the X-ray serves as loading control. Exposure of the X-ray film was for 1 min. (C) Immunodetection of NIMIN1 after agroinfiltration. Results from two independent time course experiments are shown. Leaf tissue was extracted after infiltration as indicated. Extracts were analyzed for protein accumulation with a specific antibody. As loading control, the region of the nitrocellulose filters with the small subunit of RuBisCO (SSU) stained with Ponceau S is shown. Exposure of the X-ray films was over night. dpi, days post-infiltration. (D) Immunodetection of green fluorescent protein (GFP) after agroinfiltration. Leaf tissue was extracted after infiltration as indicated. Exposure of the X-ray film was for 1 min. (E) Immunodetection of NIMIN1- and NIMIN3-Gal4 DNA binding domain (GBD) fusion proteins in extracts from transformed yeast. The NIMIN1 and NIMIN3 fusions were detected with the specific antisera used in Figures 3B,C.
FIGURE 4.
Arabidopsis NIMIN2 does not affect salicylic acid-induced gene expression from the tobacco PR-1a promoter in N. benthamiana. Transient expression assays and immunodetection were performed as described in Figure 3. N1, NIMIN1; N2, NIMIN2; N3, NIMIN3. (A) Effects of transient expression of 35SPro::NIMIN2 in the N. benthamiana -1533PR-1aPro::GUS reporter line. The effects of NIMIN2 on the PR-1a::GUS reporter are compared to effects produced by NIMIN1 and NIMIN3. Representative results are shown. (B) Effects of transient expression of 35SPro::NIMIN1, 35SPro::NIMIN2, and 35SPro::NIMIN3 on accumulation of the GUS reporter protein in SA-treated leaf tissue. GUS accumulation was detected in extracts shown in Figure 4A. Lane c contains an extract from a tobacco plant stably transformed with 35SPro::GUS. An unspecific band marked on the X-ray serves as loading control. (C) Immunodetection of NIMIN2 in agroinfiltrated tissue. NIMIN2 accumulation was detected with a specific antiserum in an extract shown in Figure 4A. (D) Effects of transient expression of 35SPro::NIMIN1, 35SPro::NIMIN2, and 35SPro::NIMIN3 on accumulation of the endogenous PR-1 protein in SA-treated N. benthamiana leaf tissue. GUS reporter enzyme activities of extracts analyzed for PR-1 protein accumulation are given below the immunodetections.
Next, we tested the influence of different NIMIN proteins on PR-1a gene induction after agroinfiltration of N. benthamiana. It has been shown previously that overexpression of NIMIN1 suppresses SA-mediated PR gene induction and SAR in transgenic Arabidopsis plants (Weigel et al., 2005). However, the functional roles of NIMIN2 and NIMIN3 are not known. Initially, Agrobacteria adjusted to equal cell densities were infiltrated into leaves of individual N. benthamiana plants with the -1533PR-1aPro::GUS reporter. In each experiment, three plants were infiltrated in parallel with the same Agrobacterium strain. After 4–5 days, disks were cut from leaf areas close to the infiltration sites. At this time, strong fluorescence was typically observed in tissue infiltrated with 35SPro::mGFP4 Agrobacteria, demonstrating efficient expression of the GFP reporter. GUS activity assays revealed that none of the NIMIN proteins is able to activate the PR-1aPro::GUS reporter gene on its own (Figures 3A and 4A and data not shown). The excised leaf disks were then floated for 2 days on water or on a 1 mM SA solution. As controls, disks from non-agroinfiltrated leaves were incubated on water and SA. After floating, proteins were extracted from leaf tissue, and GUS reporter activity was determined. In other experiments, we have infiltrated the two halves of a single leaf with Agrobacterium strains harboring different constructs in order to allow an even more direct comparison between effects exerted by the respective NIMIN proteins. Consistent with what has been described for NIMIN1 overexpression in transgenic Arabidopsis plants, agroinfiltration of 35SPro::NIMIN1 bacteria suppressed SA-mediated PR-1a promoter activation to nearly background levels as compared to GUS levels observed in GFP expressing leaf disks floated on water (Figures 3A and 4A). Quite surprisingly, NIMIN3 overexpression, too, clearly repressed GUS reporter gene induction from the Nt PR-1a promoter in N. benthamiana (Figures 3A and 4A). Repression with NIMIN3 was, however, weaker than with NIMIN1 (Figures 3A and 4A). The presence of NIMIN1 and NIMIN3 proteins in infiltrated N. benthamiana leaf tissue was monitored by immunodetection using specific antisera. NIMIN3 accumulated to high levels. The protein was readily detected in extracts from SA-floated leaf disks and also in extracts from agroinfiltrated tissue without SA induction (Figure 3B and data not shown). In contrast, we were not able to detect NIMIN1 expression in extracts from SA-treated leaf tissue. We therefore performed time course experiments monitoring NIMIN1 accumulation in twofold concentrated extracts from 1 to 4 days after agroinfiltration. Whereas GFP accumulated to high levels at 3 and 4 days post-inoculation (dpi; Figure 3D), NIMIN1 protein was detected only faintly (Figure 3C). The inability to detect high amounts of NIMIN1 in agroinfiltrated plant tissue is, however, not due to a low sensitivity of the anti-NIMIN1 serum we used. Detection of NIMIN1 and NIMIN3-Gal4 DNA binding domain (GBD) fusion proteins, which are expressed to similar levels in yeast (Weigel et al., 2001), was similar for both NIMIN3 and NIMIN1 with the specific antisera (Figure 3E).
NIMIN2 DOES NOT SIGNIFICANTLY AFFECT SALICYLIC ACID-INDUCED EXPRESSION OF TOBACCO PR-1 GENES
Likewise surprisingly, agroinfiltration of the N. benthamiana reporter line with 35SPro::NIMIN2 harboring bacteria did not repress SA-mediated induction of the PR-1aPro::GUS transgene (Figures 4A,B). Expression of NIMIN2 in N. benthamiana leaf tissue was demonstrated by immunodetection using a specific antiserum directed against Nt NIMIN2a-maltose binding protein (MBP) which exhibits cross-reactivity with Arabidopsis NIMIN2 (Figure 4C). Thus, albeit similar to each other and possessing similar NPR1 interaction motifs, NIMIN2 and NIMIN1 seem to fulfill different, even opposing, functions in the SA signal transduction pathway.
We also tested whether transient expression of At NIMIN genes in N. benthamiana is able to suppress induction of endogenous PR-1 genes. N. benthamiana (Nb) carries a gene for a basic PR-1 protein. The amino acid sequence for the basic PR-1 protein is co-linear with N. tabacum acidic PR-1 proteins except for a 19 amino acid-long extension at the C-terminus of Nb PR-1. In the co-linear region, the identity (similarity) between the basic Nb PR-1 protein and Nt PR-1a is 64% (87%). Consequently, using an antiserum raised against Nt PR-1a, we were able to detect a PR-1-related protein exhibiting a slightly higher molecular weight than the acidic Nt PR-1 proteins in extracts from N. benthamiana leaf disks floated on 1 mM SA (data not shown). SA induction of this protein was clearly suppressed in N. benthamiana tissue overexpressing NIMIN1 or NIMIN3, but not in tissue overexpressing NIMIN2 (Figure 4D).
Arabidopsis NIMIN PROTEINS CANNOT BIND SIMULTANEOUSLY TO NPR1 IN YEAST
Differential regulation of NIMIN genes and differential effects of NIMIN proteins on PR-1 induction strongly suggested that NIMINs serve unique functions at specific time points during the SAR response in Arabidopsis, an assumption fully consistent with our previous observation that NIMIN3 and NIMIN1/NIMIN2 bind to physically separate regions of At NPR1 (Weigel et al., 2001). Therefore, it was of interest to test whether NIMIN proteins are able to bind simultaneously to NPR1, or whether their binding excludes each other. To address this question, we made use of a yeast three-hybrid (Y3H) system. In this assay, interaction of two proteins can be monitored at different concentrations of a third protein whose expression level is controlled by methionine (Met) in the growth medium (Tirode et al., 1997). Previously, we have demonstrated that NIMIN proteins are able to interact with TGA transcription factors in presence of NPR1 (Figures 5C and 6B;Weigel et al., 2001), showing that NIMINs and TGA factors possess independent binding sites on NPR1 which can be occupied at the same time. The same assay was used for monitoring binding of two different NIMIN proteins to NPR1. To this end, we used partial NIMIN cDNA clones which we had isolated in a yeast two-hybrid (Y2H) screen with the At NPR1 bait (Weigel et al., 2001).
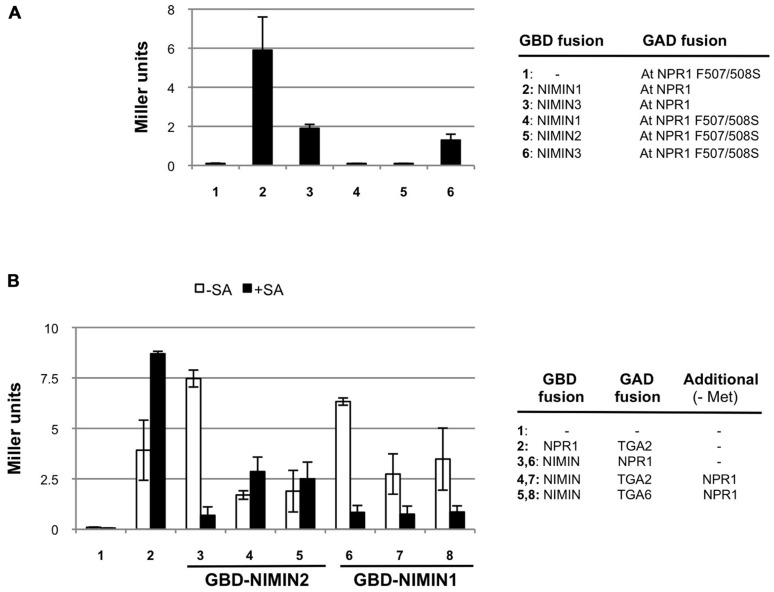
FIGURE 5.
Arabidopsis NIMIN1, NIMIN2, and NIMIN3 do not bind simultaneously to At NPR1 in yeast. (A) Yeast two-hybrid interaction of At NPR1-Gal4 DNA binding domain (GBD) and NIMIN1-Gal4 activation domain (GAD) fusion proteins in absence and presence of NIMIN2. NIMIN2 was expressed from the Met25 promoter which is repressed in presence and de-repressed in absence of methionine. (B) Immunodetection of NIMIN2 in yeast. Yeast cells analyzed for lacZ reporter gene expression in Figure 5A were probed for accumulation of NIMIN2 protein. N2, NIMIN2. (C) Yeast three-hybrid interaction of At NPR1 with NIMIN1 and NIMIN3 or with NIMIN2 and NIMIN3. NIMIN1, NIMIN2, and NIMIN3 were expressed as fusions with the GBD or GAD. Simultaneous interaction of At NPR1 with GAD-TGA2 and GBD-NIMIN3 serves as positive control for formation of a ternary protein complex.
FIGURE 6.
Arabidopsis NIMIN1 and NIMIN2 interact differentially with At NPR1 in yeast. (A) Yeast two-hybrid interaction of NIMIN1, NIMIN2 or NIMIN3 expressed as GBD fusions with the mutant protein At NPR1 F507/508S expressed as GAD fusion. Interactions of GAD-At NPR1 with GBD-NIMIN1 and GBD-NIMIN3 serve as positive controls. (B) Effect of SA on formation of ternary protein complexes comprising GBD-NIMIN1 or GBD-NIMIN2, GAD-At TGA2 or GAD-At TGA6 and At NPR1. The binary interactions of At NPR1 with NIMIN1 or NIMIN2 or TGA2 serve as controls for effects of SA (concentration 0.3 mM) on the NPR1–NIMIN1/2 interaction.
Initially, we tested whether NIMIN1 and NIMIN2 can bind together to NPR1 in Y3H assays. Both proteins possess similar NPR1 interaction motifs by which they bind to the C-terminus of NPR1 (Weigel et al., 2001; Figure 6A). Truncated NIMIN1 or NIMIN2 including their NPR1 interaction motif were expressed as fusions with the Gal4 transcription activation domain (GAD), and full-length NIMIN1 or NIMIN2 were expressed from the Met25 promoter, which is repressed in presence and de-repressed in absence of methionine. NPR1 was expressed as GBD fusion. The interactions of NIMIN1 or NIMIN2 with NPR1 were disrupted in presence of NIMIN2 or NIMIN1, respectively (Figure 5A and data not shown). Furthermore, complex formation between NPR1 and NIMIN1 was clearly dependent on the concentration of NIMIN2 (Figures 5A,B). Together, the data suggest that NIMIN1 and NIMIN2 may compete for the same binding site on NPR1.
Next, we asked whether NIMIN1 or NIMIN2 can bind to NPR1 in presence of NIMIN3 which interacts with NPR1 via a site distant from the NIMIN1/NIMIN2 binding site (Weigel et al., 2001). NIMIN1 and NIMIN3 or NIMIN2 and NIMIN3 were expressed as GBD or GAD fusions, while NPR1 was expressed from the de-repressed Met25 promoter. Surprisingly, the interaction between NIMIN1 and NPR1 and between NIMIN2 and NPR1 was disrupted in presence of NIMIN3 (Figure 5C). Hence, NIMIN3 binding to NPR1 seems to inhibit NIMIN1/NIMIN2 interaction, and simultaneous binding of NIMIN1, NIMIN2, and NIMIN3 to NPR1 may exclude each other.
NIMIN1 AND NIMIN2 INTERACT DIFFERENTIALLY WITH NPR1
In tobacco NPR1, binding of NIMIN2 proteins occurs in the region from amino acids 494 to 510 (Maier et al., 2011). The domain is highly conserved in NPR1 proteins from many plant species, including Arabidopsis, regarding both the sequence and its position within the amino acid chain (for At NPR1 94% identity, 100% similarity, from amino acids 496 to 512). To test whether both NIMIN1 and NIMIN2 bind to this region in At NPR1 and whether binding occurs in a similar fashion, we introduced mutations F507S and F508S into At NPR1. Nt NPR1 F505/506S is no longer able to interact with Nt NIMIN2a or Nt NIMIN2c (Maier et al., 2011). Similarly, mutation of F507/508S completely abolishes binding of NIMIN1 and NIMIN2 to At NPR1, but not binding of NIMIN3 (Figure 6A).
We then analyzed the relations of NPR1 with NIMIN1 and NIMIN2 in ternary protein complexes including TGA transcription factors. We have shown previously that SA administered to growth medium impairs formation of NPR1–NIMIN1 and NPR1–NIMIN2 complexes in Y2H assays, and that the sensitivity of loss of protein–protein interaction is very similar for both NIMIN1 and NIMIN2 (IC50 ≈ 20 μM SA;Maier et al., 2011). Here, we monitored effects of SA on NPR1–NIMIN1 and NPR1–NIMIN2 interactions in presence of TGA2 or TGA6. Interaction of Arabidopsis NPR1 with TGA factors is not diminished with SA (Figure 6B;Maier et al., 2011). Ternary complexes comprising NIMIN1 were sensitive to SA as observed before for the NIMIN1–NPR1 binary interaction (Figure 6B). Quite surprisingly, however, ternary complexes comprising NIMIN2 proved to be stable in presence of SA (Figure 6B). Thus, although possessing similar NPR1 interaction motifs and binding to the same site in the C-terminus of NPR1, NIMIN1, and NIMIN2 can form complexes with NPR1 and TGA factors exhibiting differential sensitivity to SA, implying that these two NIMIN proteins interact differentially with NPR1 in transcription complexes on PR gene promoters.
DISCUSSION
NIM1-INTERACTING proteins have been identified through a Y2H screen with Arabidopsis NPR1 as bait. Although of rather small molecular weight, the proteins share several conserved regions with each other which are likely of functional relevance. Thus, all NIMIN proteins encompass an LxLxL/EAR (ethylene-responsive element binding factor-associated amphiphilic repression) motif at their C-terminus, and NIMIN1 and NIMIN2 possess a common motif for interaction with a domain in the C-terminus of Arabidopsis and tobacco NPR1. On the other hand, NIMIN1 and NIMIN3 have been reported to share a conserved PA/SFQPEDF signature (Weigel et al., 2001), suggesting that NIMIN1 and NIMIN3, albeit binding to different regions of NPR1, may exert similar activities. To understand the action of related, yet distinct, NIMIN proteins on NPR1, we have performed a comparative analysis of Arabidopsis NIMIN1, NIMIN2, and NIMIN3. We have studied the expression profiles of NIMIN genes, the effects of NIMIN proteins on SA induction of the SAR marker PR-1 and their interaction with NPR1. Our results suggest that the Arabidopsis NIMIN proteins exert unique and complementary functions on NPR1 at different stages of the SAR response.
NIMIN3 REPRESSES PR-1 IN UNCHALLENGED PLANTS
As opposed to NIMIN1 and NIMIN2, which are clearly responsive to SA, NIMIN3 is expressed constitutively at a low level in Arabidopsis leaf tissue. In our current work, we have not found any indications for enhancement of NIMIN3 expression by SA or other plant defense hormones. Most importantly, the NIMIN3 promoter is weakly active in leaf tissue and does not respond to the SAR signal molecule SA. Hence, NIMIN3 is likely to function on a constitutive basis in unchallenged plants before the induction of SAR. This idea is consistent with our previous finding that NIMIN3 does not possess the interaction site by which NIMIN1 and NIMIN2 bind to the SA-sensitive NPR1 C-terminus (Weigel et al., 2001;Maier et al., 2011). When transiently overexpressed in the N. benthamiana -1533PR-1a::GUS reporter line created by us, NIMIN3, like NIMIN1, is able to suppress SA-induced activation of the reporter. Similarly, NIMIN3, like NIMIN1, also suppresses induced expression of an endogenous PR-1 gene in N. benthamiana. Altogether, repression effects exerted by NIMIN3 in N. benthamiana seem moderate, when compared to effects observed with NIMIN1. On the other side, we did not expect suppression of PR-1 gene induction to occur at all by NIMIN3 in Nicotiana species. First, a true NIMIN3 homolog has not been identified to date from tobacco or tomato. Furthermore, NPR1 family members from tobacco, Nt NPR1 and Nt NPR3, have not been found to interact with NIMIN3 in Y2H assays (Zwicker et al., 2007;Maier et al., 2011), whereas NIMIN3 clearly interacts with Arabidopsis NPR1 (Weigel et al., 2001). Thus, the biochemical basis of NIMIN3-mediated suppression of PR-1 in N. benthamiana is not clear. However, we have noted previously that NIMIN3 and NIMIN1 share the conserved amino acid signature PA/SFQPEDF (from here on termed EDF motif;Weigel et al., 2001). This signature is also present in the rice (Os) NIMIN homolog NRR and some of its paralogs (consensus sequence WRP-F-W/MEDF;Chern et al., 2012). Mutations of NRR and its paralogs in this region have uncovered the motif as domain for strong interaction with rice NH1/NPR1 causing repression of transcription activity of Os NH1/NPR1 in a rice transient assay system. In contrast, the motif mediates only very weak interaction between NRR and Arabidopsis NPR1 (Chern et al., 2012). We have introduced mutations in the EDF motifs of NIMIN3 and NIMIN1 (E63A D64V in NIMIN3; E94A D95V in NIMIN1), and tested activities of the mutant proteins in Y2H assays with Gal4 AD-At NPR1 and in the N. benthamiana transient assay system. Unfortunately, the mutant proteins did not accumulate to detectable levels, neither in yeast nor in plant tissue, and therefore, the significance of the EDF domain for NIMIN3 and NIMIN1 could not be assessed (Masroor and Pfitzner, unpublished data). It is of interest, however, to note that binding of At NPR1 to NIMIN3 occurs within the 60 amino acid-long C-terminal half including the EDF motif (Weigel et al., 2001). Given the conservation of the amino acid sequence in NPR1 interactors from multiple plant species and the clear results in the rice system reported byChern et al. (2012), we infer that the EDF signature is functional in Arabidopsis NIMINs, and that the domain is involved in regulation of PR genes via the NIMIN–NPR1 complex. The significance of the EDF domain for PR gene induction may, however, vary among different plant species. In this line, suppression of PR-1 induction in N. benthamiana may be mediated via the EDF domain in NIMIN3 and NIMIN1, and suppression by NIMIN1 would be stronger because NIMIN1, unlike NIMIN3, can interact via a second domain with the NPR1 C-terminus. Of note, several cDNAs from N. tabacum and N. benthamiana coding for NIMIN proteins with the EDF motif (consensus WNL/PA/TF/L-T/PEDF) have been described in the databanks, underscoring our assumption that the EDF domain may have functional relevance also in tobacco. The mechanism by which the EDF domain in NIMIN proteins could suppress PR-1 gene induction remains, however, elusive. Alternatively, suppression of PR-1 induction in N. benthamiana by NIMIN3 and NIMIN1 may occur via the C-terminal LxLxL/EAR motif which has been implicated in recruiting the transcriptional co-repressor TOPLESS (Arabidopsis Interactome Mapping Consortium, 2011). In summary, our data would support the view that NIMIN3 can target the NPR1 complex in tobacco, and that NIMIN3 is a repressor of inadvertent PR-1 gene expression in unchallenged Arabidopsis leaf tissue.
NIMIN2 DOES NOT AFFECT SALICYLIC ACID INDUCTION OF PR-1
We have noted previously that NIMIN2 is responsive to SA (Weigel et al., 2001;Glocova et al., 2005). Here, using RT-PCR analyses, we show that NIMIN2 mRNA accumulates very early after treatment of plants with SA, and, in several cases, NIMIN2 mRNA was already detectable in plant tissue without exposure to chemicals at all. From our observations, we conclude that NIMIN2 is more readily induced than NIMIN1 or PR-1, consistent with the finding that NIMIN2 expression, as opposed to NIMIN1 and PR-1 expression, is independent from an intact NPR1 gene requiring activation by SA. Surprisingly, overexpression of NIMIN2 in the N. benthamiana -1533PR-1a::GUS reporter line does not appear to have an effect on SA-induced PR-1 gene expression. This finding is consistent with our previous observation showing that overexpression of a NIMIN2 homolog, Nt NIMIN2a, in transgenic tobacco plants did not result in massive PR-1 repression as reported in similar experiments for At NIMIN1 and Os NRR overexpression (Chern et al., 2005, 2008;Weigel et al., 2005). Hence, NIMIN2 is likely to play a role at the very onset of SAR and is unlikely to be involved in repression of PR-1 gene induction.
NIMIN1 CONTROLS EXPRESSION OF LATE SAR-INDUCED PR-1
NIMIN1 is an early SA-activated and NPR1-dependent gene which is induced after NIMIN2, but clearly before PR-1. NIMIN1 is expressed only transiently, and the NIMIN1 protein does not appear to accumulate to high levels. These features are compatible with a role of NIMIN1 as regulator of late SAR genes, e.g., PR-1, preventing their premature activation. The repression effect exerted by NIMIN1 in the N. benthamiana -1533PR-1a::GUS reporter line is very strong. Above, we have argued that PR-1 repression may be mediated via the EDF domain in NIMIN1, although we were not able to provide direct proof for this assumption. It is important to note, however, that NIMIN2 does not possess the EDF motif and does not repress PR-1 gene induction in our system. Curiously, although not accumulating to substantial levels in agroinfiltrated N. benthamiana leaf tissue, NIMIN1 executes strong effects raising the question how NIMIN1 could suppress PR-1 gene expression in near physical absence? Different scenarios seem conceivable. For example, NIMIN1 could be stable and exert its function only in direct association with NPR1. Any excess NIMIN1 protein would immediately be degraded. In this scenario, NIMIN1 could act to prohibit contact of NPR1 to downstream transcription factors either by sterical hindrance or, in imitation to the action of a chaperone, by imposing a non-productive bent on NPR1. Together, our data support a view where NIMIN1 acts only later during the SAR response, after NIMIN2, keeping tight control over PR-1 by promoting its repression. Notably, we were not able to detect simultaneous binding of NIMIN3, NIMIN2, or NIMIN1 to NPR1, and we found that NIMIN1 and NIMIN2 bind differentially to NPR1 in ternary protein complexes including TGA transcription factors.
WORKING MODEL FOR THE CONSECUTIVE ACTION OF Arabidopsis NIMIN PROTEINS IN THE COURSE OF SAR
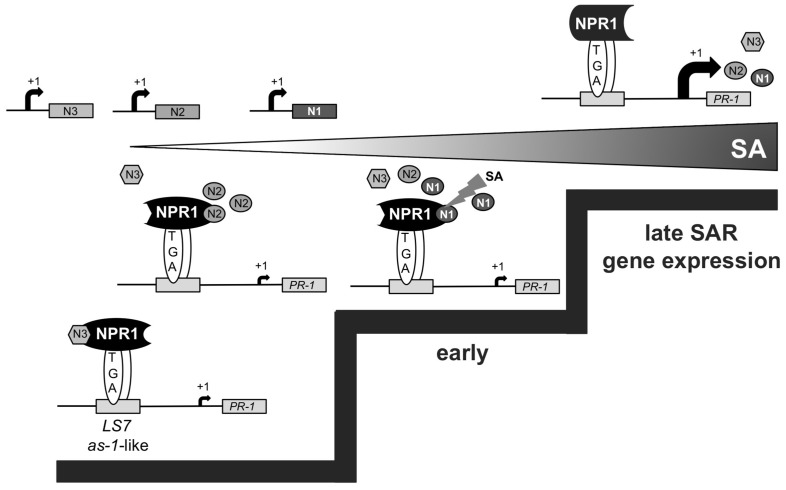
Based on our findings, we propose sequential formation of different NIMIN–NPR1 complexes to promote defense gene activation at distinct stages of SAR (Figure 7). While NIMIN3 represses inadvertent PR gene activation in unchallenged plants, NIMIN2 is induced at low tissue levels of SA to relieve NIMIN3 repression by binding to the NPR1 C-terminus. This process may allow activation of early SA- and NPR1-dependent genes, e.g., NIMIN1. Interaction of NIMIN2 with the NPR1 C-terminus does not, however, appear to be sufficient to activate substantial expression of the late SAR gene PR-1. NIMIN2 action on NPR1 is transient and is followed by NIMIN1 replacing NIMIN2. NIMIN1 suppresses activation of NPR1-dependent SAR genes. NIMIN1 action on NPR1 seems even more transient than NIMIN2 action, and instability of NIMIN1 protein would be a crucial prerequisite for relief of PR-1 gene repression. In this scenario, late SAR genes would be activated through direct action of SA on NPR1 (Maier et al., 2011;Fu et al., 2012;Wu et al., 2012) causing removal of repressing NIMIN1 from the NPR1 complex (Maier et al., 2011). In conclusion, consecutive action of NIMIN proteins with different biochemical capacities on the central SAR regulator NPR1 is needed to ensure sudden, strong and coordinate expression of defense genes to successfully combat invading pathogens. In this line, the NIMIN–NPR1 connection may constitute a molecular device to monitor ambient SA levels in diseased plants, enabling the plant to translate a steadily increasing gradient of the defense hormone SA into two clear decision steps, early and late SAR gene expression.
FIGURE 7.
Working model for the consecutive action of Arabidopsis NIMIN proteins in the course of SAR. The model implies sequential interaction between diverse NIMIN proteins and NPR1 to form regulatory complexes with differential biochemical capacities in the course of SAR. The model also suggests that sensing of ambient SA levels in diseased plants may occur through the various NIMIN–NPR1 complexes, enabling activation of PR genes at distinct threshold levels of SA (indicated by steps). In this scenario, the defense gene PR-1 is induced late during SAR by direct action of SA on the NIMIN1–NPR1 regulatory complex.
MATERIALS AND METHODS
DNA CONSTRUCTS
For transient gene expression assays, the coding regions from NIMIN1, NIMIN2, and NIMIN3 were inserted as BamHI/SacI fragments into pBin19/35S Pro::GUS (Jefferson et al., 1987) from which the GUS reporter gene had been excised. The coding regions were amplified from the respective pGBT9 plasmids (Weigel et al., 2001) using C-terminal primers with the native stop codons and a SacI restriction endonuclease site added 3′ to the stop codons.
The NIMIN3Pro::GUS reporter gene was constructed in analogy to the NIMIN1Pro::GUS and NIMIN2Pro::GUS chimeric genes (Glocova et al., 2005). The NIMIN3 promoter sequence was amplified from Arabidopsis thaliana (L.) Heynh. Col-0 genomic DNA using primers N3-P2 (5′-TTAAGCTTATACGGGACATAGTGCACAGCC) and N3-P1 (5′-AAGGATCCTGAACCGCTCTCTCTTCCTTCC). N3-P1 primes immediately upstream of the ATG translation start codon of NIMIN3. The resulting 1.4 kb fragment was ligated to HindIII/BamHI cleaved pBin19/35SPro::GUS from which the 35S RNA promoter had been removed.
To map the NIMIN1/NIMIN2 binding site in At NPR1, Phe-507 and Phe-508 were mutated to Ser using overlap extension PCR (Ho et al., 1989). The primers for mutagenesis were AtNPR1-14 (5′-CTCGGGAAACGAAGCAGCCCGCGCTGTTC) and AtNPR1-15 (5′-GAACAGCGCGGGCTGCTTCGTTTCCCGAG). The mutations were inserted in a C-terminal fragment of At NPR1. To this clone, the N-terminal At NPR1 sequence was added as a 1.4 kb BamHI/DraIII fragment, and the complete mutant sequence was ligated to BamHI/SalI cleaved pGBT9 and pGAD424.
All clones generated by PCR amplification were verified by DNA sequence analysis.
RNA ISOLATION AND RT-PCR ANALYSES
RNA isolation and RT-PCR analyses were performed as described byZwicker et al. (2007). The primer combinations and control plasmids used for the different gene fragments are listed in Table 1. For the time course experiment shown in Figure 2B, RT-PCR assays were conducted to give approximately equal amounts of reaction products in order to enable direct comparison of NIMIN1, NIMIN2, and PR-1 transcript accumulation at different time points after treatment of Arabidopsis with SA. To this end, RNAs were diluted 1:20 for RT-PCR amplification of NIMIN2 transcripts.
GENERATION AND CULTIVATION OF TRANSGENIC PLANTS
Transformation of tobacco (N. tabacum L. cv. Samsun NN) by Agrobacterium tumefaciens was performed according toGrüner et al. (2003). Tobacco lines with PR-1aPro::GUS, 35SPro::GUS, NIMIN1Pro::GUS, and NIMIN2Pro::GUS have been described earlier (Grüner et al., 2003;Glocova et al., 2005). For localization of GUS enzyme activity in situ (Figure 1B) and for determination of SA-induced GUS activity in time course experiments (Figure 2C), seeds from transgenic tobacco were sown on MS medium with 400 μg ml-1 kanamycin or on selective medium supplemented with 0.3 mM SA.
Agrobacterium-MEDIATED TRANSIENT GENE EXPRESSION IN NICOTIANA BENTHAMIANA
The -1533PR-1aPro::GUS gene construct (Grüner et al., 2003) was integrated via Agrobacterium-mediated transformation into the genome of N. benthamiana Domin. All primary transformants exhibited strong and stringent induction of the GUS reporter gene in response to SA. A line with an intermediate GUS enzyme activity was propagated by selfing, and plants of the T2 generation were used for agroinfiltration experiments.
The pBin19 gene constructs were transferred by triparental mating to Agrobacterium tumefaciens strain LBA4404. Recombinant Agrobacterium strains were grown at 30°C in minimal medium supplemented with 50 μg ml-1 kanamycin and 50 μg ml-1 rifampicin to stationary phase. Cells were collected by centrifugation and resuspended in 10 mM MgCl2 and 150 μM acetosyringone to give an optical density (OD600) of 0.5 for all strains. Agrobacteria were incubated for 2–3 h at room temperature before agroinfiltration. To suppress post-transcriptional gene silencing, the bacterial suspensions were mixed with an equal volume of a strain carrying the p19 suppressor from Tomato bushy stunt virus (Voinnet et al., 2003). Four to six week-old greenhouse-grown N. benthamiana plants with integrated -1533PR-1aPro::GUS were agroinfiltrated in the abaxial air spaces. To allow for a direct comparison between effects produced by different NIMIN strains, leaves at the same position on the axis of different plants or the two halves of the same leaf were injected. In each experiment, three independent plants were infiltrated with the same Agrobacterium suspension, and plants infiltrated with a strain containing 35SPro::mGFP4 (Haseloff et al., 1997) were used to control gene expression levels in leaf tissue. Expression of GFP was monitored under UV light. GFP fluorescence remained always strictly confined to infiltrated leaf areas. Agroinfiltrated tissue was processed 4 or 5 days post-infiltration (dpi), when strong GFP fluorescence was observed. At this point of time, bacterial titers were similar in leaf tissue agroinfiltrated with strains 35SPro::mGFP4, 35SPro::NIMIN1, or 35SPro::NIMIN2 (Wöhrle and Pfitzner, unpublished data). Furthermore, co-overexpression of mGFP4 and NIMIN1 produced the same levels of GFP fluorescence and of GFP protein accumulation as overexpression of mGFP4 alone (Masroor and Pfitzner, unpublished data).
GUS REPORTER GENE ASSAYS AND IMMUNODETECTION OF PROTEIN ACCUMULATION
Determination of GUS enzyme activity and histochemical localization of GUS activity in situ were performed as described previously (Weigel et al., 2001;Glocova et al., 2005). GUS activity is given in units (1 unit = 1 nmol 4-MU per hour per mg protein). For the time course experiment shown in Figure 2C, GUS enzyme activities were determined from pools of 10 seedlings for each data point. The same extracts were used for immunodetection of endogenous PR-1 proteins. Equal amounts of protein were loaded in each lane of the sodium dodecyl sulfate (SDS) gels.
To determine GUS enzyme activity after transient expression of NIMIN genes in N. benthamiana, two leaf disks each were punched out from non-infiltrated control or from agroinfiltrated leaf tissue at 4 or 5 dpi. Disks were floated for 2 days on water or on 1 mM SA and thereafter extracted with 150 μl GUS lysis buffer. The SA-induced reporter gene expression from the PR-1a promoter was compared in non-agroinfiltrated leaf tissue and in tissue infiltrated with 35SPro::mGFP4 and 35SPro::NIMIN chimeric genes. The same extracts were used for immunodetection of protein accumulation.
Immunodetection of proteins separated by SDS gel electrophoresis was performed as described earlier (Zwicker et al., 2007). Specific antisera were raised in rabbits immunized with E. coli expressed and purified proteins NIMIN1-GST, Nt NIMIN2a-MBP, and NIMIN3 according to standard procedures. PR-1 protein accumulation in N. benthamiana was detected with a specific antiserum against Nt PR-1a. For detection of GFP and GUS proteins, rabbit polyclonal antisera were used as recommended by the manufacturers (Santa Cruz Biotechnology and Abcam, respectively). To analyze accumulation of NIMIN1 at different times after agroinfiltration (Figure 3C), four leaf disks were harvested directly from each infiltrated tissue and extracted with 150 μl GUS lysis buffer yielding twofold concentrated extracts. SA induction of the GUS reporter protein and of an endogenous N. benthamiana PR-1 protein was compared in tissue infiltrated with 35SPro::mGFP4 and 35SPro::NIMIN chimeric genes. Equal extract volumes were loaded in each lane of an SDS gel. The loading of SDS gels for immunodetection of protein accumulation was checked by staining the nitrocellulose filters with Ponceau S (0.1% in 5% acetic acid). Alternatively, unspecific bands reacting with the antisera used are marked for demonstration of equal gel loading.
YEAST TWO-HYBRID AND THREE-HYBRID ASSAYS
Yeast two-hybrid and yeast three-hybrid analyses in absence and presence of SA were conducted as reported earlier (Weigel et al., 2001;Maier et al., 2011). LacZ reporter gene activities are given in Miller units. Most plasmids used in the protein–protein interaction assays have been described (Weigel et al., 2001). pGAD10/NIMIN1 35/142, pGAD10/NIMIN2 20/122, and pGAD10/NIMIN3 13/112 encode NIMIN proteins truncated at their N-terminus. The plasmids were isolated in a Y2H screen with the At NPR1 bait (Weigel et al., 2001).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Sylvia Zwicker for communication of Y2H data, Xinnian Dong, Duke University, USA, and Jane Glazebrook, University of Minnesota, USA, for npr1 mutant seeds, Jim Haseloff, University of Cambridge, England, for the generous gift of pBIN 35S-mGFP4, and Klaus Harter, Universität Tübingen, Germany, for providing the Agrobacterium strain with the p19 silencing suppressor. Thanks also to David Neeley for advice on the English language. Ashir Masroor was supported by a fellowship from Katholischer Akademischer Ausländer-Dienst (KAAD).
REFERENCES
- Arabidopsis Interactome Mapping Consortium (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco F., Salinas P., Cecchini N. M., Jordana X., Van Hummelen P., Alvarez M. E., et al. (2009). Early genomic responses to salicylic acid in Arabidopsis. Plant Mol. Biol. 70 79–102 [DOI] [PubMed] [Google Scholar]
- Canet J. V., Dobón A., Roig A., Tornero P. (2010). Structure–function analysis of npr1 alleles in Arabidopsis reveals a role for its paralogs in the perception of salicylic acid. Plant Cell Environ. 33 1911–1922 [DOI] [PubMed] [Google Scholar]
- Cao H., Bowling S. A., Gordon A. S., Dong X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Glazebrook J., Clarke J. D., Volko S., Dong X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 57–63 [DOI] [PubMed] [Google Scholar]
- Cao H., Li X., Dong X. (1998). Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. U.S.A. 95 6531–6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern M., Bai W., Sze-To W. H., Canlas P. E., Bartley L. E., Ronald P. C. (2012). A rice transient assay system identifies a novel domain in NRR required for interaction with NH1/OsNPR1 and inhibition of NH1-mediated transcriptional activation. Plant Methods 8 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern M., Canlas P. E., Fitzgerald H. A., Ronald P. C. (2005). Rice NRR, a negative regulator of disease resistance, interacts with Arabidopsis NPR1 and rice NH1. Plant J. 43 623–635 [DOI] [PubMed] [Google Scholar]
- Chern M., Canlas P. E., Ronald P. C. (2008). Strong suppression of systemic acquired resistance in Arabidopsis by NRR is dependent on its ability to interact with NPR1 and its putative repression domain. Mol. Plant 1 552–559 [DOI] [PubMed] [Google Scholar]
- Després C., DeLong C., Glaze S., Liu E., Fobert P. R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12 279–290 [PMC free article] [PubMed] [Google Scholar]
- Fonseca J. P., Menossi M., Thibaud-Nissen F., Town C. D. (2010). Functional analysis of a TGA factor-binding site located in the promoter region controlling salicylic acid-induced NIMIN-1 expression in Arabidopsis. Genet. Mol. Res. 9 167–175 [DOI] [PubMed] [Google Scholar]
- Friedrich L., Lawton K., Dietrich R., Willits M., Cade R., Ryals J. (2001). NIM1 overexpression in Arabidopsis potentiates plant disease resistance and results in enhanced effectiveness of fungicides. Mol. Plant Microbe Interact. 14 1114–1124 [DOI] [PubMed] [Google Scholar]
- Friedrich L., Lawton K., Ruess W., Masner P., Specker N., Gut Rella M., et al. (1996). A benzothiadiazole derivative induces systemic acquired resistance in tobacco. Plant J. 10 61–70 [Google Scholar]
- Fu Z. Q., Yan S., Saleh A., Wang W., Ruble J., Oka N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Rogers E. E., Ausubel F. M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glocova I., Thor K., Roth B., Babbick M., Pfitzner A. J. P., Pfitzner U. M. (2005). Salicylic acid (SA)-dependent gene activation can be uncoupled from cell death-mediated gene activation: the SA-inducible NIMIN-1 and NIMIN-2 promoters, unlike the PR-1a promoter, do not respond to cell death signals in tobacco. Mol. Plant Pathol. 6 299–314 [DOI] [PubMed] [Google Scholar]
- Grüner R., Pfitzner U. M. (1994). The upstream region of the gene for the pathogenesis-related protein 1a from tobacco responds to environmental as well as to developmental signals in transgenic plants. Eur. J. Biochem. 220 247–255 [DOI] [PubMed] [Google Scholar]
- Grüner R., Strompen G., Pfitzner A. J. P., Pfitzner U. M. (2003). Salicylic acid and the hypersensitive response initiate distinct signal transduction pathways in tobacco that converge on the as-1-like element of the PR-1a promoter. Eur. J. Biochem. 270 4876–4886 [DOI] [PubMed] [Google Scholar]
- Haseloff J., Siemering K. R., Prasher D. C., Hodge S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. U.S.A. 94 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77 51–59 [DOI] [PubMed] [Google Scholar]
- Horvath D. M., Huang D. J., Chua N.-H. (1998). Four classes of salicylate-induced tobacco genes. Mol. Plant Microbe Interact. 11 895–905 [DOI] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D. G., Dangl J. L. (2006). The plant immune system. Nature 444 323–329 [DOI] [PubMed] [Google Scholar]
- Lawton K. A., Friedrich L., Hunt M., Weymann K., Delaney T., Kessmann H., et al. (1996). Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 10 71–82 [DOI] [PubMed] [Google Scholar]
- Lebel E., Heifetz P., Thorne L., Uknes S., Ryals J., Ward E. (1998). Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 16 223–233 [DOI] [PubMed] [Google Scholar]
- Maier F., Zwicker S., Hückelhoven A., Meissner M., Funk J., Pfitzner A. J. P., et al. (2011). NONEXPRESSOR OF PATHOGENESIS-RELATED PROTEINS1 (NPR1) and some NPR1-related proteins are sensitive to salicylic acid. Mol. Plant Pathol. 12 73–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J., Carr J. P., Klessig D. F., Raskin I. (1990). Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250 1002–1004 [DOI] [PubMed] [Google Scholar]
- Métraux J. P., Signer H., Ryals J., Ward E., Wyss-Benz M., Gaudin J., et al. (1990). Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250 1004–1006 [DOI] [PubMed] [Google Scholar]
- Mou Z., Fan W., Dong X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113 935–944 [DOI] [PubMed] [Google Scholar]
- Rochon A., Boyle P., Wignes T., Fobert P. R, Després C. (2006). The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell 18 3670–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A. F. (1961). Systemic acquired resistance induced by localized virus infections in plants. Virology 14 340–358 [DOI] [PubMed] [Google Scholar]
- Ryals J., Weymann K., Lawton K., Friedrich L., Ellis D., Steiner H. Y., et al. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell 9 425–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J., Tsui F., Klessig D. F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant Microbe Interact. 10 69–78 [DOI] [PubMed] [Google Scholar]
- Strompen G., Grüner R., Pfitzner U. M. (1998). An as-1-like motif controls the level of expression of the gene for the pathogenesis-related protein 1a from tobacco. Plant Mol. Biol. 37 871–883 [DOI] [PubMed] [Google Scholar]
- Tirode F., Malaguti C., Romero F., Attar R., Camonis J., Egly J. M. (1997). A conditionally expressed third partner stabilizes or prevents the formation of a transcriptional activator in a three-hybrid system. J. Biol. Chem. 272 22995–22999 [DOI] [PubMed] [Google Scholar]
- Uknes S., Mauch-Mani B., Moyer M., Potter S., Williams S., Dincher S., et al. (1992). Acquired resistance in Arabidopsis. Plant Cell 4 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes S., Winter A. M., Delaney T., Vernooij B., Morse A., Friedrich L., et al. (1993). Biological induction of systemic acquired resistance in Arabidopsis. Mol. Plant Microbe Interact. 6 692–698 [DOI] [PubMed] [Google Scholar]
- van Loon L. C., Rep M, Pieterse C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44 135–162 [DOI] [PubMed] [Google Scholar]
- van Loon L. C, van Kammen A. (1970). Polyacrylamide disc electrophoresis of the soluble leaf proteins from Nicotiana tabacum var. ‘Samsun’ and ‘Samsun NN’. II. Changes in protein constitution after infection with tobacco mosaic virus. Virology 40 199–211 [DOI] [PubMed] [Google Scholar]
- Vernooij B., Friedrich L., Ahl Goy P., Staub T., Kessmann H., Ryals J. (1995). 2,6-Dichloroisonicotinic acid-induced resistance to pathogens without the accumulation of salicylic acid. Mol. Plant Microbe Interact. 8 228–234 [Google Scholar]
- Vernooij B., Friedrich L., Morse A., Reist R., Kolditz-Jawhar R., Ward E., et al. (1994). Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell 6 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33 949–956 [DOI] [PubMed] [Google Scholar]
- Ward E. R., Uknes S. J., Williams S. C., Dincher S. S., Wiederhold D. L., Alexander D. C., et al. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel R. R., Bäuscher C., Pfitzner A. J. P., Pfitzner U. M. (2001). NIMIN-1, NIMIN-2 and NIMIN-3, members of a novel family of proteins from Arabidopsis that interact with NPR1/NIM1, a key regulator of systemic acquired resistance in plants. Plant Mol. Biol. 46 143–160 [DOI] [PubMed] [Google Scholar]
- Weigel R. R., Pfitzner U. M., Gatz C. (2005). Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 17 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. F. (1979). Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology 99 410–412 [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhang D., Chu J. Y., Boyle P., Wang Y., Brindle I. D., et al. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1 639–647 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Fan W., Kinkema M., Li X., Dong X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. U.S.A. 96 6523–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.-M., Trifa Y., Silva H., Pontier D., Lam E., Shah J., et al. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 13 191–202 [DOI] [PubMed] [Google Scholar]
- Zwicker S., Mast S., Stos V., Pfitzner A. J. P., Pfitzner U. M. (2007). Tobacco NIMIN2 proteins control PR gene induction through transient repression early in systemic acquired resistance. Mol. Plant Pathol. 8 385–400 [DOI] [PubMed] [Google Scholar]