Abstract
The reprogramming of differentiated cells to pluripotent cells (induced pluripotent stem (iPS) cells) is known to be an inefficient process. We recently reported that cells with short telomeres cannot be reprogrammed to iPS cells despite their normal proliferation rates1, 2, probably reflecting the existence of ‘reprogramming barriers’ that abort the reprogramming of cells with uncapped telomeres. Here we show that p53 (also known as Trp53 in mice and TP53 in humans) is critically involved in preventing the reprogramming of cells carrying various types of DNA damage, including short telomeres, DNA repair deficiencies, or exogenously inflicted DNA damage. Reprogramming in the presence of pre-existing, but tolerated, DNA damage is aborted by the activation of a DNA damage response and p53-dependent apoptosis. Abrogation of p53 allows efficient reprogramming in the face of DNA damage and the generation of iPS cells carrying persistent DNA damage and chromosomal aberrations. These observations indicate that during reprogramming cells increase their intolerance to different types of DNA damage and that p53 is critical in preventing the generation of human and mouse pluripotent cells from suboptimal parental cells.
Nuclear reprogramming of differentiated cells into pluripotent stem cells is thought to be a possible source of patient-specific cells for transplantation therapies. Several strategies have been used to achieve nuclear reprogramming, including nuclear transplantation and cellular fusion3. Recently, the generation of iPS cells from differentiated cells was achieved by the overexpression of four transcription factors4, 5, 6. These factors are Oct4 (also known as Pou5f1), Sox2, Klf4 and c-Myc, although some of them are dispensable to reprogram certain cell types4, 5, 6, 7, 8, 9, 10, 11.
Notably, only a small proportion of cells become pluripotent iPS cells (less than 1% (ref. 3); Supplementary Table 1). Several models have been proposed to explain the low efficiency of reprogramming3. Here we reasoned that a further explanation may be related to the presence of DNA damage in the cells undergoing reprogramming.
This notion is on the basis of our observations that murine fibroblasts with increased DNA damage owing to the presence of critically short telomeres (third generation (G3) telomerase-deficient mouse embryonic fibroblasts (MEFs), or G3 Terc−/− MEFs) cannot be reprogrammed to iPS cells1, 2, despite the fact that the cells have normal proliferation rates12 (Supplementary Fig. 1) and are able to spontaneously immortalize and be transformed by oncogenes12. iPS cell generation from G3 Terc−/− MEFs can be restored after re-elongation of the shortest telomeres by telomerase1, 2, indicating that damaged/uncapped telomeres are responsible for their failure to reprogram. These observations suggest that an important impediment for reprogramming is the existence of reprogramming barriers that abort reprogramming of DNA-damaged cells, such as those with uncapped telomeres.
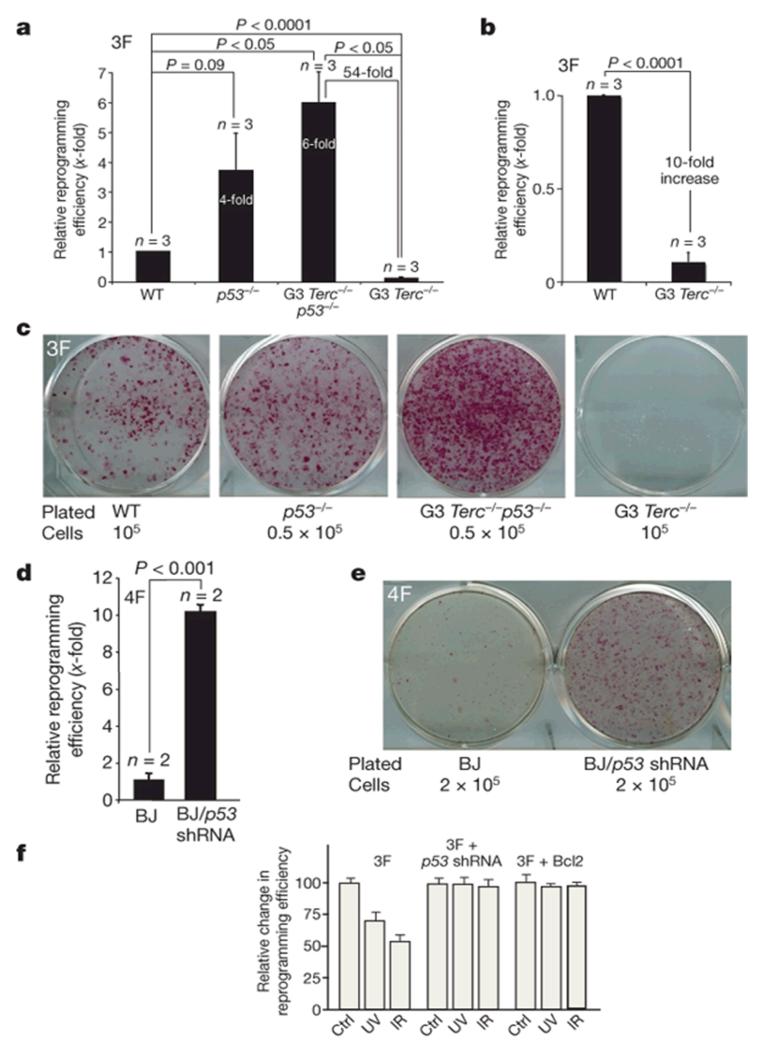
p53 has a crucial involvement in preventing the propagation of DNA-damaged cells, including those containing short/dysfunctional telomeres13, 14. We began to test the role of p53 by reprogramming cells with critically short telomeres (G3 Terc−/− MEFs) in the presence or absence of p53. We used three reprogramming factors (Oct4, Klf4 and Sox2) following methods previously shown to reprogram wild-type MEFs into bona fide pluripotent iPS cells1. The efficiency of reprogramming of wild-type MEF cultures was 0.72% ± 0.05% (mean ± s.e.m) (Supplementary Table 1)1, 9. p53-null MEFs were reprogrammed with an efficiency that was on average fourfold higher than the wild-type MEFs (Fig. 1a-c and Supplementary Table 1), suggesting that p53 limits reprogramming of wild-type MEFs. We extended these findings to the reprogramming of BJ human foreskin fibroblasts with four factors (Oct4, Sox2, Klf4 and c-Myc). The simultaneous infection with a retrovirus expressing short hairpin RNA (shRNA) against p53 (p53 shRNA) resulted in a tenfold increase in reprogramming efficiency compared to BJ fibroblast controls (Fig. 1d, e). Furthermore, p53-null iPS cell colonies appeared 3 days earlier than in the p53-proficient controls. G3 Terc−/− MEFs showed a tenfold decrease in reprogramming efficiency compared to wild-type MEFs (Fig. 1a-c and Supplementary Table 1)1, and G3 Terc−/− iPS cells colonies appeared 2-3 days later. Notably, p53 abrogation in G3 Terc−/− p53−/− restored their reprogramming efficiency to similar levels seen in p53-null cells, which represented a 54-fold increase in reprogramming efficiency compared with the G3 Terc−/− controls, and a six fold increase compared with wild-type cells (Fig. 1a-c and Supplementary Table 1). We extended these results to other sources of DNA damage, such as low doses of γ-irradiation and ultraviolet light. In both cases, irradiated MEFs showed lower reprogramming efficiencies than the non-irradiated controls (Fig. 1f). Co-infection of wild-type MEFs with the three factors together with a retrovirus expressing p53 shRNA or the anti-apoptotic protein Bcl2, rendered the reprogramming of these cells essentially insensitive to these doses of DNA damage (Fig. 1f). Together, these findings indicate that p53 limits reprogramming of mouse and human cells by restricting conversion of DNA-damaged cells into iPS cells, probably by the elimination of these cells by apoptosis13, 15. This effect is exacerbated in cells containing a higher proportion of DNA-damaged cells owing to their dysfunctional telomeres or after infliction of exogenous DNA damage.
Figure 1. p53-deficiency allows reprogramming of MEFs with short telomeres.
a, b, Relative reprogramming efficiencies are shown, with the fold changes indicated. Student’s t-test (two-tailed) is used for statistics. Error bars, standard error. n = experiments with independent MEFs. 3F, three factors; WT, wild type. c, Reprogramming plates stained with alkaline phosphatase. The number of parental cells used is indicated. d, Reprogramming of BJ human fibroblasts with four factors (4F) together with a human shRNA against the human p53 gene. Fold changes relative to BJ + 4F (BJ) are indicated. e, Reprogramming plates stained with alkaline phosphatase. f, Relative reprogramming efficiencies of wild-type MEFs exposed to UVC (UV) or ionizing radiation (IR), and expressing three factors together with a retrovirus expressing mouse p53 shRNA or Bcl2. Error bars, standard deviation. Ctrl, control.
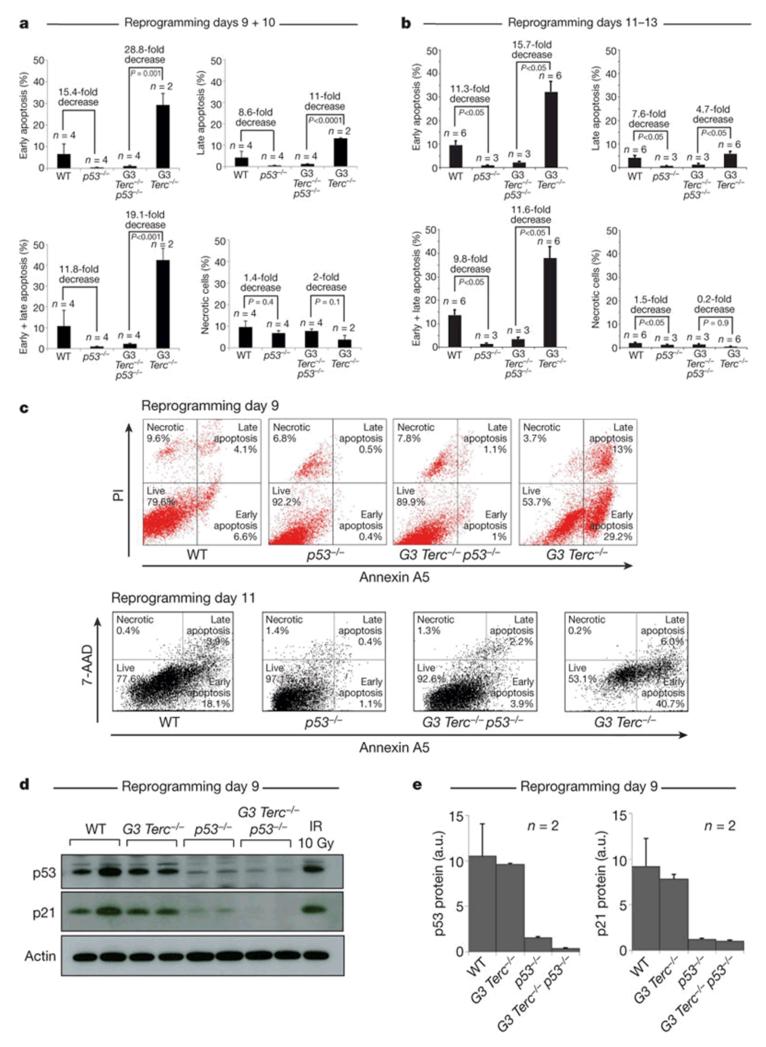
Our data indicates that p53 becomes sensitized to DNA damage once cells engage the iPS cell program. We sought to find direct evidence of p53 activity at early times during iPS cell formation before the appearance of typical iPS cell colonies. For this, we quantified the percentage of cells undergoing apoptosis at days 9-10 and 11-13 after infection, at the time of induction of pluripotency8, 9. A significant proportion (10% and 15%) of wild-type cells undergo apoptosis at days 9-10 and 11-13 after infection, respectively (Fig. 2a-c). This is further increased to 40% in G3 Terc−/− cultures at 9-13 days after infection (Fig. 2a-c), in agreement with their lower iPS cell yields (Fig. 1a-c). Notably, apoptosis was essentially abrogated in p53−/− and G3 Terc−/− p53−/− cells at 9-13 days after infection (Fig. 2a-c), whereas no differences in necrosis were observed (Fig. 2a-c). The increased apoptosis in wild-type and G3 Terc−/− cultures at day 9 of reprogramming was accompanied by p53 and p21 (also referred as Cdkn1a) protein levels similar to those of γ-irradiated wild-type MEFs, while this was not observed in the p53-null cultures (Fig. 2d, e). These results indicate that p53 limits reprogramming by inducing apoptosis of suboptimal cells at the time of pluripotency induction, in agreement with Bcl2-overexpression allowing normal reprogramming of cells with exogenously inflicted DNA damage (Fig. 1f). The activity of p53 restricting reprogramming of suboptimal cells is readily observed in wild-type cells and it is further exacerbated in G3 Terc−/− cells, which contain a higher proportion of damaged cells1.
Figure 2. p53-deficiency abrogates apoptosis at the onset of iPS cell formation.
a, b, Apoptosis and necrosis are determined on days 9 and 10 (a) and 11-13 (b) post-infection. Data are mean and s.e.m. A Student’s t-test is used for statistics. c, Representative FACS profiles at day 9 of reprogramming. PI, propidium iodide. 7-AAD, 7-amino-actinomycin D. d, Western blots of p53 and p21 protein levels at day 9 post-infection. As a control, wild-type MEFs were γ-irradiated (10 Gy). Two reprogramming experiments per genotype are shown. e, Quantification of p53 and p21 westerns shown in d. Values are in arbitrary units (a.u.). Error bars, standard error.
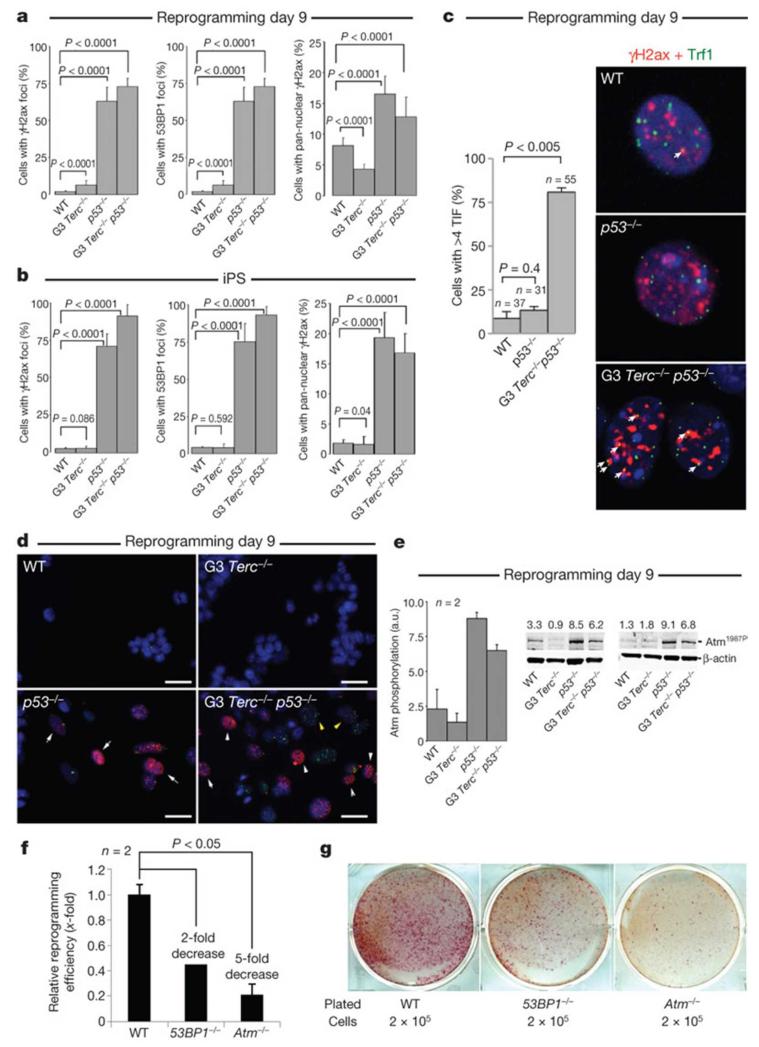
If p53-deficiency is allowing the conversion of DNA-damaged cells into iPS cells, we should expect to see persistent activation of the DNA damage response (DDR) in the p53-null genotypes, both during reprogramming and in the resulting iPS cell clones, while this should be less apparent in p53-proficient cells in which damaged cells are being eliminated by apoptosis. DDR activation was evidenced by the presence of γH2ax (also known as γH2afx) and 53BP1 (also known as Trp53bp1) foci at day 9 after infection in p53−/− and G3 Terc−/− p53−/− cultures (Fig. 3a), which persisted in the isolated iPS cell clones (Fig. 3b) and the teratomas derived from them (Supplementary Fig. 4d). γH2ax and 53BP1 foci showed frequent co-localization, indicating a robust DDR activation (Fig. 3d, yellow arrows). Furthermore, in the G3 Terc−/− p53−/− cultures, a significant proportion of cells with γH2ax foci (>75%) showed co-localization with the Trf1 (also known as Terf1) telomeric protein forming the so-called telomere-induced DNA damage foci (TIF) (Fig. 3c), indicative of telomere dysfunction. Cells with pan-nuclear γH2ax staining were also increased in the p53-null cultures at day 9 after infection and in the corresponding iPS cell clones (Fig. 3a, b and white arrows in Fig. 3d). Pan-nuclear γH2ax staining is associated with replication-induced DNA damage16, suggesting generation of this type of endogenous DNA damage during the reprogramming process16. Finally, we detected Atm phosphorylation in the p53-null cultures at day 9 after infection (Fig. 3e), a hallmark of Atm activation17 and further indicating DDR activation during reprogramming of these cells.
Figure 3. DDR activation during reprogramming.
a, b, γH2ax and 53BP1 foci 9 days after infection (a) and in iPS cell clones (b). Results are the mean of two experiments. Two-hundred cells were analysed per genotype/experiment. Error bars, s.d. c, Left, telomere-induced DNA damage foci (TIF) 9 days after infection. n = cells with γH2ax foci analysed. Values correspond to two reprogramming experiments. Error bars, standard error. Student’s t-test was used for statistics. Right, representative images of γH2ax (red) and Trf1 (green) staining. Arrows indicate co-localization events (yellow). Original magnification, ×63. d, Representative images that are quantified in a. White arrows, pan-nuclear γH2ax; yellow arrows, co-localization of γH2ax and 53BP1. Scale bars, 10 μm. e, Western blot (right) showing Atm phosphorylation 9 days after infection. Atm1987P denotes Atm protein phosphorylated at Ser 1987. Numbers above each lane represent the relative quantification of Atm levels. Two western blots were used for quantification (left). Error bars, standard deviation. f, Relative reprogramming efficiencies of 53BP1−/− and Atm−/− MEFs compared to wild-type MEFs. Fold changes are indicated. Student’s t-test is used for statistics. Error bars, standard error. n = experiments performed with independent MEF cultures. g, Reprogramming plates stained with alkaline phosphatase.
To test the unique role of p53 in the elimination of DNA-damaged cells during reprogramming, we considered the effect of Atm and 53BP1 deficiencies on reprogramming. These two proteins participate in the repair of DNA breaks and, consequently, their absence increases the endogenous levels of DNA damage18, 19. Furthermore, the two proteins participate in the signalling of DNA damage to p53 (refs 18, 19). MEFs deficient in either Atm or 53BP1 showed a decreased reprogramming efficiency compared to wild-type controls (Fig. 3f, g), which might reflect increased endogenous DNA damage in these cultures18, 19. These results are in agreement with the central role of p53 as the integrator of several and redundant DNA damage signalling pathways, whereas other upstream components of the DDR, such as Atm or 53BP1, are not essential to prevent the reprogramming of DNA-damaged cells.
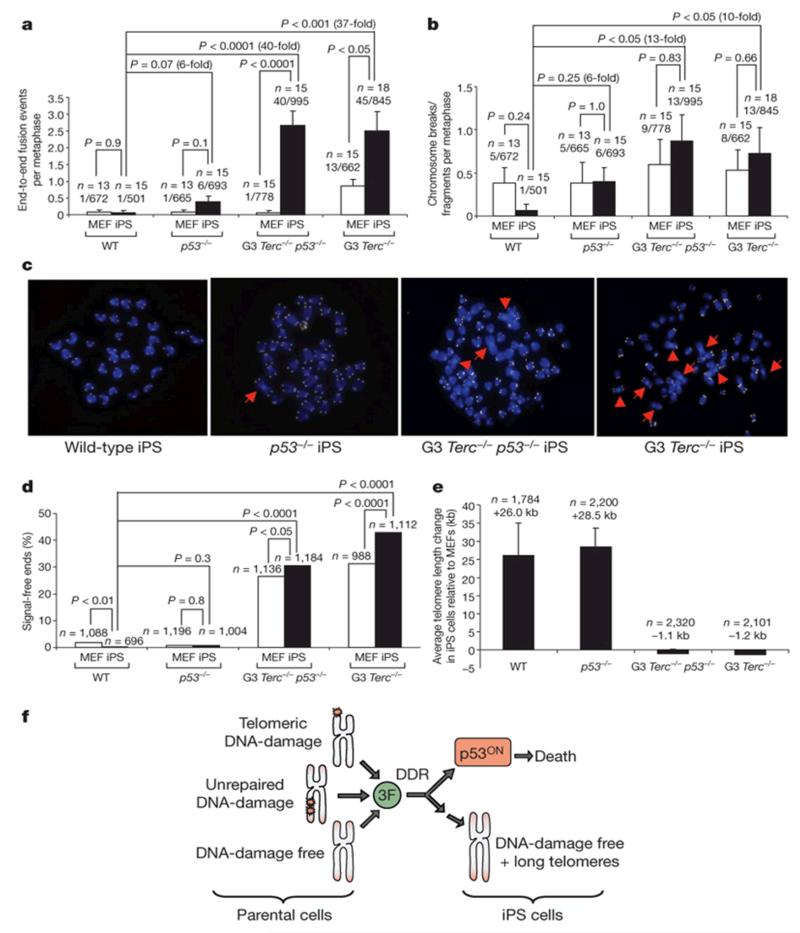
We wondered whether reprogramming of p53-null cultures in the face of DNA damage was accompanied by increased chromosomal damage. Short/uncapped telomeres are known to lead to chromosome end-to-end fusions12. p53−/− iPS cells showed sixfold more end-to-end fusions than wild-type iPS cells (Fig. 4a, c), and this was increased by 40- and 37-fold in G3 Terc−/− p53−/− and G3 Terc−/− iPS cells, respectively (Fig. 4a, c). Of note, G3 Terc−/− iPS cells represent rare reprogramming events (Fig. 1a) after widespread apoptotic death (Fig. 2a-c), which may explain the high levels of fusions in the face of functional p53. Chromosomal breaks/fragments were also increased in p53−/− iPS cells compared to wild-type iPS cells (sixfold), and further increased in G3 Terc−/− and G3 Terc−/− p53−/− iPS cells (13- and 10-fold, respectively) (Fig. 4b). Increased fusions in G3 Terc−/− p53−/− and G3 Terc−/− iPS cells were coincidental with a high percentage (30% and 42%, respectively) of chromosome ends with undetectable telomere signals or ‘signal-free ends’, compared to only 0.29% of signal-free ends in wild-type iPS cells (Fig. 4d).
Figure 4. p53-null iPS cells show chromosomal instability.
a, b, Frequency of end-to-end fusions (a) and breaks/fragments (b) in the indicated cells. n = metaphase number. The number of aberrations out of the chromosomes scored is indicated. Student’s t-test was used for statistics. Error bars, s.e.m. c, Representative metaphases. Red arrows, end-to-end fusions. Original magnification, ×100. d, Percentage of signal-free ends. n = telomeres used for the analysis. Chi-square test is used for statistics. e, Average telomere elongation (kilobase (kb)) in iPS cell clones compared to parental MEFs. n = telomeres used for the analysis. At least two independent iPS cell clones were used per genotype. MEF passage number = 3; iPS cell passage number = 4-6. Error bars, s.e.m. f, Summary illustrating that p53 constitutes a main barrier to reprogramming of cells with increased DNA damage by preventing that they become iPS cells.
Telomeres undergo telomerase-dependent elongation during reprogramming—a process that continues after reprogramming until iPS cells clones acquire the typically long telomeres of embryonic stem (ES) cells1, 2. Telomeres were similarly elongated in wild-type and p53-null iPS cells compared with the parental MEFs, but suffered further shortening in the telomerase-deficient G3 Terc−/− and G3 Terc−/− p53−/− iPS cells (Fig. 4e). These results indicate that the p53-deficiency allows reprogramming of G3 Terc−/− cells independently of telomere length.
Furthermore, we tested whether p53-abrogation in p53−/− and G3 Terc−/− p53−/− iPS cells had an affect on their ability to contribute to mouse chimaerism and teratomas. Although some of the p53−/− and G3 Terc−/− p53−/− iPS cell clones lost the typical rounded iPS cell morphology after expansion (Supplementary Fig. 2), we were able to obtain chimaeras from p53−/− and G3 Terc−/− p53−/− iPS cells by picking individual iPS cell colonies with robust Nanog and Oct4 expression (Supplementary Table 2 and Supplementary Figs 3 and 4a). p53−/− chimaeras were able to contribute to the germ line, however, the only G3 Terc−/− p53−/− chimaera obtained died at 14 days of age with severe intestinal atrophy (Supplementary Table 2). All of the p53−/− and G3 Terc−/− p53−/− iPS cell clones tested were able to form teratomas (Supplementary Fig. 4b, c). Notably, teratomas derived from G3 Terc−/− p53−/− iPS cells showed abundant γH2ax staining and anaphase bridges concomitant with lower p21 and apoptosis levels than wild-type teratomas (Supplementary Fig. 4d), indicating that DDR activation persists during the differentiation of p53-null iPS cell clones.
In summary, these data indicate that p53 constitutes a main barrier to reprogramming of wild-type cells, which is exacerbated in cells with pre-existing DNA damage (that is, cells with short telomeres or deficient for Atm and 53BP1) and in cells in which DNA damage has been exogenously inflicted (irradiated cells). In this manner, suboptimal cells carrying DNA damage are eliminated by p53-dependent apoptosis and prevented from becoming iPS cells (Fig. 4f). These results agree with previous findings showing that p53 downregulation improves reprogramming efficiency20. A p53-dependent counterselection of DNA-damaged cells during reprogramming is shown by increased Atm phosphorylation and increased DNA damage foci both in p53−/− and G3 Terc−/− p53−/− cultures. Given that some of the reprogramming factors promote tumorigenesis in vivo21, it is tempting to propose that the observed DDR in p53−/− cultures might be equivalent to the oncogene-induced DDR reported in the context of malignant transformation22, 23. In both models, reprogramming and transformation, p53 is critical to control the spreading of damaged cells.
Online Methods
Mice, cells and culture conditions
Terc+/− and p53+/− mice were first intercrossed to generate Terc+/− p53+/− double heterozygous mice and then mated to generate first generation (G1) Terc−/− p53+/− littermates. G1 Terc−/− p53+/− littermates were interbred for successive generations to obtain late generation G3 Terc−/− p53−/− double mutant mice, as well as their G3 Terc−/− littermate controls, as previously described14, 24. The genetic background for all genotypes was a pure C57BL/6 background.
Primary MEFs (passage 2) were obtained from embryos of the indicated Terc and p53 genotypes as described previously14, 24. Atm−/− and 53BP1−/− MEFs and their corresponding wild-type controls were obtained from the null-mouse strains described before19, 25.
MEFs were cultured in standard DMEM medium with 10% FBS (Gibco). ES and iPS cells were cultured in DMEM (high glucose) supplemented with serum replacement (KSR, Invitrogen), Lif (1,000 U ml−1), non-essential amino acids, glutamax and β-mercaptoethanol. C57BL/6 ES cells were derived at the Transgenic Mice Unit of the CNIO from C57BL/6 blastocysts.
Generation of mouse iPS cells
Reprogramming of primary (passage 2-4) MEFs was performed as previously described1 following modifications of a previous protocol26. In brief, primary MEFs of the indicated genotypes in a pure C57BL/6 background were seeded in 6-well plates (0.25-1 × 105 cells per well). They were infected four times in the next two days with a cocktail of the retroviral constructs pMXsOct3/4, pMXsKlf4 and pMXsSox2 as described1. Reprogramming was assessed 2 weeks after infection by counting alkaline-phosphatase-positive colonies. Alkaline phosphatase staining was performed according to manufacturer’s instructions (Alkaline Phosphatase Detection kit; Millipore). The results were normalized to the respective efficiencies of retroviral transduction as assessed by transducing with the three retroviruses pMXsOct3/4, pMXsKlf4 and pMXsSox2, plus a retrovirus expressing GFP. Absolute reprogramming efficiencies are shown in Supplementary Table 1. The efficiency of reprogramming was also calculated as the relative change compared to that of wild-type MEFs (Fig. 1). Colonies were picked after 2 weeks and expanded on feeder fibroblasts using standard procedures.
Generation of human iPS cells
Reprogramming of BJ human foreskin fibroblasts (passage 15; obtained from the American Type Culture Collection (ATCC)) was done as previously described5, 27. In brief, retroviral supernatants were produced in HEK-293T cells (5 × 106 cells per 100-mm-diameter dish) transfected with the ecotropic packaging plasmid pCL-Ampho (4 μg) together with one of the following retroviral constructs (4 μg): pMXs-hKlf4, pMXs-hSox2 or pMXs-hOct4 (obtained from Addgene and previously described)5. The retroviral construct expressing human shRNA against p53, pRETRO-Super, was provided by R. Bernards. Transfections and infections were performed the same as the mouse iPS cell reprogramming described earlier. Twenty-thousand BJ fibroblasts had been seeded the previous day (2 × 105 cells per 60-mm-diameter gelatin-coated dish) and received 1.5 ml of each of the corresponding retroviral supernatants (a total of either three or four—the fourth being the p53 shRNA retroviral supernatant). This procedure was repeated every 12 h for 2 days (a total of four additions). The day after infection was completed, media was replaced by human fibroblast media, and kept for a further 2 days. At day 8, cells were trypsinized and reseeded on feeder plate. At day 9, media was changed to human ES cell media. Cultures were maintained in the absence of drug selection with daily medium changes. At day 20, colonies with ES-like morphology became visible at the microscope. Colonies were picked after 3 weeks and expanded on feeder fibroblasts using standard procedures.
Reprogramming of irradiated cells
Primary wild-type MEFs were exposed to UVC (2 J m−2) or ionizing radiation (2 Gy), and 24 h later cells were infected with the three factors together with a fourth retrovirus expressing p53 shRNA or Bcl2, as indicated. Bcl2 was overexpressed using retroviral pBabe-PURO-Bcl2 (provided by A. Carnero). p53 shRNA against murine p53 was expressed using retroviral pRetro-SUPER (provided by R. Bernards).
Analysis of the DDR
γH2ax (Upstate Biotechnology) and 53BP1 (Novus Biologicals), as well as secondary antibodies conjugated with Alexa 488 or Alexa 594 (Molecular Probes), were used. Image acquisition was done with high-throughput-microscopy. In brief, cells were grown on gelatinized mCLEAR bottom 96-well dishes (Greiner Bio-One), and analysed on a BD Pathway 855 BioImager (Beckton Dickinson). All of the images used for quantitative analyses were acquired under non-saturating exposure conditions. Western analyses were performed on the LICOR platform (Biosciences) with β-actin (Sigma) and Atm1987P (gift from A. Nussenzweig) antibodies.
Analysis of telomere damage-induced foci
Reprogramming with the three factors was performed in the indicated MEFs. At days 9 and 10 after infection, cells were collected and plated in gelatinized coverslips. Once attached to the coverslips, cells were rinsed with PBS and incubated with Triton X-100 buffer (20 mM Tris-HCl, pH 8, 50 mM NaCl, 3 mM MgCl2, 0.5% Triton X-100 and 300 mM sucrose) at room temperature for 5 min. Cells were then fixed in PBS-buffered 4% paraformaldehyde for 10 min at room temperature, followed by permeabilization in PBS-0.1% Triton X-100 for 10 min. Cells were then blocked with 2% BSA (Sigma) for 1 h. The samples were incubated overnight at 4 °C with the primary antibodies. Phosphorylated γH2ax and Trf1 foci were detected using a mouse monoclonal anti-phospho-histone γH2ax (Ser 139) antibody (1:500; from Upstate Biotechnology) and an anti-Trf1 antibody (1:500). After washing with 0.1% Tween-20 buffer for 30 min at room temperature, the samples were incubated with Cy3-goat anti-mouse and Alexa 488-goat anti-rabbit antibody (1:400; Jackson ImmunoResearch Laboratories), for 1 h at room temperature. Slides were mounted in Vectashield with 4′,6-diamino-2-phenylindole (DAPI). Images were obtained using a confocal microscope (Leica TCS-SP5 (Acousto-optical beamsplitter)).
iPS cell chimaeras
The capacity of the p53−/− and G3 Terc−/− p53−/− iPS cell clones to generate chimaeras in vivo was tested by microinjection into C57BL/6J-Tyr(C-2J)/J (albino) blastocysts and the assessment of hair colour in the resulting progeny (Supplementary Table 2 and Supplementary Fig. 4a). Wild-type iPS cell clones were previously described to produce chimaeras and to contribute to the germ line1. Late generation Terc−/− iPS cells were previously described to fail to produce any viable chimaeras1.
Teratoma formation
The indicated number of mice (nu/nu) was subcutaneously injected with 1 × 106 cells of each iPS cell clone. All injected clones showed high expression of the pluripotency genes Nanog and Oct4 (Supplementary Fig. 3). Tumour growth was measured at the indicated days post-injection with a calibre, and tumour volume was calculated according to the formula: long diameter × (short diameter)2 × 0.51.
Apoptosis assay
Reprogramming with the three factors was performed in the indicated MEFs. At days 9 and 10 after infection, cells were collected, washed in PBS, resuspended in 1× annexin A5 binding buffer, and stained with annexin-A5–FITC (BD Pharmingen) and propidium iodide (Sigma). After a 15-min incubation in the dark at room temperature, annexin-A5-positive cells were quantified using a FACS Canto within 1 h. Similarly, at days 11, 12 and 13 after infection, cells were collected, washed in PBS, resuspended in 1× annexin A5 binding buffer (BD Biosciences), and stained with annexin-A5–APC (BD Biosciences) and the viability marker 7-amino-actinomycin D (BD Biosciences). After a 15-min incubation in the dark at room temperature, annexin-A5-positive cells were quantified using a FACS Canto within 1 h.
EdU proliferation assay
Proliferation assay was performed using a Click-it EdU Proliferation kit (Invitrogen) according to manufacturer’s instructions. In brief, MEFs were allowed to incorporate EdU (5-ethynyl-2′-deoxyuridine) overnight and then collected and fixed. Fixed cells were stained with Click-it EdU detection reagent, and EdU-positive cells were visualized using a FACS Canto.
Western blots
Cell extracts were prepared using RIPA buffer, resolved on NuPAGE 4-12% gradient Bis-Tris gels, transferred to nitrocellulose and hybridized using antibodies against Nanog (1:5,000; Chemicon), Oct4 (1:500; SantaCruz), p53 (1:500; Cell Signaling), p21 (1:500; Santa Cruz), actin (1:10,000; Sigma) and tubulin (1:10,000; Sigma).
Quantitative FISH analysis
We prepared metaphases and performed quantitative FISH (Q-FISH) hybridization as previously described28, 29. To correct for lamp intensity and alignment, images from fluorescent beads (Molecular Probes, Invitrogen) were analysed in parallel, using the TFL-Telo program (a gift from P. Lansdorp). Telomere fluorescence values were extrapolated from the telomere fluorescence of lymphoma cell lines LY-R (R cells) and LY-S (S cells) with known telomere lengths of 80 and 10 kb, respectively. There was a linear correlation (r2 = 0.999) between the fluorescence intensity of the R and S telomeres. We captured the images using a CCD camera (FK7512; COHU) on a fluorescence microscope (DMRB; Leica). We captured the images using Q-FISH software (Leica) in a linear acquisition mode to prevent the over-saturation of fluorescence intensity. TFL-Telo software30 was used to quantify the fluorescence intensity of telomeres from at least six metaphases for each data point.
Chromosomal aberrations
FISH hybridization was performed as described before28, 29. At least 15 metaphases per genotype and from at least two independent cultures per genotype were scored for chromosomal aberrations by superimposing the telomere image on the DAPI chromosomes image using the TFL-Telo software.
Histopathology and immunohistochemistry
After mice excision, the specimens were fixed in 10% buffered formalin (Sigma) and embedded in paraffin. For histopathological analysis of teratomas, tumours were serially sectioned (3 μm) to find the different germ-layer components, and every tenth section was stained with haematoxylin and eosin. Remaining sections were used for immunohistochemical studies with the primary antibodies mouse monoclonal anti-phospho-Histone H2A.X (Ser139) (clone JBW30) (1:350, Millipore), goat polyclonal anti-p21 (1:2,000; Santa Cruz Biotechnology), and TUNEL kit (ApopTag, Chemicon) following manufacturer instructions. Following incubation with the primary antibodies, positive cells were visualized using 3,3-diaminobenzidine tetrahydrochloride plus (DAB+) as a chromogen. Counterstaining was performed with nuclear haematoxylin. Images were captured with a DP-10 digital camera in an Olympus Vanox microscope at the indicated magnifications.
Anaphase bridges in teratomas
For detection of anaphase bridges in the teratomas, samples were fixed in 10% buffered formalin, dehydrated, and embedded in paraffin. Four-micrometre sections were deparaffinized and stained with DAPI. Images were captured using a CCD camera (FK7512; COHU) on a fluorescence microscope (DMRB; Leica).
Supplementary Material
Acknowledgements
We thank R. Serrano for mouse colony management, and M. Cañamero and the Comparative Pathology Unit at the CNIO for teratoma analysis. K.S. is recipient of a contract from the Spanish Association Against Cancer (AECC). Work in the laboratory of M.A.B. is funded by grants from the MICINN (CONSOLIDER), the Regional Government of Madrid, the European Union, the European Research Council (ERC), the AECC, and the Korber European Research Award.
References
- 1.Marion RM, et al. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 2.Davy P, Allsopp R. Balancing out the ends during iPSC nuclear reprogramming. Cell Stem Cell. 2009;4:95–96. doi: 10.1016/j.stem.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 7.Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nature Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- 10.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nature Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 11.Kim JB, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]; Blasco MA, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 12.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Chin L, et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 14.Vousden KH, Lane DP. p53 in health and disease. Nature Rev. Mol. Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 15.Toledo LI, Murga M, Gutierrez-Martinez P, Soria R, Fernandez-Capetillo O. ATR signaling can drive cells into senescence in the absence of DNA breaks. Genes Dev. 2008;22:297–302. doi: 10.1101/gad.452308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 17.Xu Y, et al. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 18.Ward IM, Minn K, van Deursen J, Chen J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 21.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 22.Di Micco R, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]; Herrera E, et al. Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J. 1999;18:2950–2960. doi: 10.1093/emboj/18.11.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barlow C, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 24.Blelloch R, Venere M, Yen J, Ramalho-Santos M. Generation of induced pluripotent stem cells in the absence of drug selection. Cell Stem Cell. 2007;1:245–247. doi: 10.1016/j.stem.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 26.Blasco MA, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 27.Samper E, Goytisolo FA, Slijepcevic P, van Buul PP, Blasco MA. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 2000;1:244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zijlmans JM, et al. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl Acad. Sci. USA. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.