Abstract
Methylation of H3K79 is associated with chromatin at expressed genes, though it is unclear if this histone modification is required for transcription of all genes. Recent studies suggest that Wnt-responsive genes depend particularly on H3K79 methylation, which is catalyzed by the methyltransferase DOT1L. Human leukemias carrying MLL gene rearrangements show DOT1L-mediated H3K79 methylation and aberrant expression of leukemogenic genes. DOT1L inhibitors reverse these effects, but their clinical use is potentially limited by toxicity in Wnt-dependent tissues such as intestinal epithelium. Genome-wide positioning of the H3K79me2 mark in Lgr5+ mouse intestinal stem cells and mature intestinal villus epithelium correlated with expression levels of all transcripts and not with Wnt-responsive genes per se. Selective Dot1l disruption in Lgr5+ stem cells or in whole intestinal epithelium eliminated H3K79me2 from the respective compartments, allowing genetic evaluation of DOT1L requirements. The absence of methylated H3K79 did not impair health, intestinal homeostasis, or expression of Wnt target genes in crypt epithelium for up to 4 months, despite increased crypt cell apoptosis. Global transcript profiles in Dot1l-null cells were barely altered. Thus, H3K79 methylation is not essential for transcription of Wnt-responsive or other intestinal genes, and intestinal toxicity is not imperative when DOT1L is rendered inactive in vivo.
INTRODUCTION
Covalent histone modifications influence chromatin structure and diverse nuclear functions, including gene regulation (1, 2). Expressed genes are associated with di- or trimethylated H3K4, H3K36, and H3K79 and monomethylated H3K9 and H4K20, whereas repressed genes are enriched for trimethylated H3K9, H3K27, and H4K20 (2–4); various lysine methyltransferases (KMTs) place these marks. H3K79me2 denotes active gene transcription in Saccharomyces cerevisiae, Drosophila, and mammalian cells (5–8). Unlike other modified histone N-terminal “tail” residues, H3K79 is exposed on the nucleosome surface, may be methylated at both heterochromatin and euchromatin (5, 9), and is aberrantly methylated in human leukemias that carry MLL1 gene rearrangements (10, 11).
Disruption of Dot1 in yeast or its fly (grappa) and mammalian (Dot1l) homologs eliminates H3K79 methylation, revealing these as the only enzymes capable of H3K79 mono-, di-, and trimethylation (8, 12–15). Dot1 and DOT1L/KMT4 differ from other KMTs in possessing an arginine methyltransferase-like domain instead of a canonical SET domain (12), and H3K79 lacks known demethylases (9). Dot1-dependent H3K79 methylation is associated with telomere silencing and meiotic checkpoint controls (16), DNA repair (17), and modulation of constitutive heterochromatin (15), but its role in transcriptional control has drawn particular attention. Fruit fly grappa mutants dysregulate developmental genes and show embryonic defects (14). Dot1l-null mouse embryos are stunted and die in midgestation of restricted cardiovascular defects (15, 18) that seem incompatible with a global requirement for DOT1L activity in transcription. Indeed, recent studies implicate DOT1L-dependent H3K79 methylation specifically in the transcriptional output of Wnt signaling, which depends on T-cell factor (TCF) transcription factors and the coactivator β-catenin and is crucial for intestinal homeostasis (19).
The self-renewing gut mucosa requires the activity of Wnt-dependent intestinal stem cells (ISCs) and progenitor cells that reside in the crypts of Lieberkühn (20–22). Although there are probably two or more distinct ISC populations (23–25), a key “workhorse” population consists of 10 to 15 crypt base columnar cells (CBCs) that express the cell surface protein LGR5, require Wnt signals, repopulate adjacent intestinal villi for months, produce clonal “organoids” ex vivo, and serve as the cell of colorectal cancer origin (26–28). Mahmoudi et al. identified both DOT1L and its partner AF10 within β-catenin-dependent TCF4 complexes in mouse crypt and human colon cancer cells (29). AF10 depletion in cell lines impaired DOT1L recruitment to TCF4/β-catenin target genes; AF10 and DOT1L were both required for elongating Wnt-responsive transcripts and to express a Wnt-reporter transgene and maintain crypt cell replication in zebrafish embryos (29). In a reciprocal approach, Mohan et al. detected β-catenin, the transcriptional effector of canonical Wnt signals, within DOT1L-containing protein complexes and identified a specific requirement for H3K79me3 in regulating Wnt target genes. Depletion of dDot1 or AF10 in Drosophila embryos reduced expression of canonical Wnt targets, especially the high-threshold target senseless (30). Lastly, a human DOT1L polymorphism is associated with joint space width and reduced risk of osteoarthritis, and DOT1L knockdown in articular chondrocytes inhibited Wnt-dependent chondrogenesis (31). Each of these studies had certain limitations. First, as the investigators interrogated few known Wnt target genes, DOT1L/H3K79me specificity for the Wnt pathway was inconclusive. Second, biologic effects were studied in fly and morpholino-treated zebrafish embryos or in a cell line, not in native adult mammalian tissues. Nevertheless, the findings have important implications for H3K79me2 and me3 specificity in transcriptional regulation and, because Wnt signaling underlies gut epithelial homeostasis (20, 21), for possible intestinal toxicity if DOT1L activity is compromised in therapy.
The H3K4-specific KMT gene MLL1 is a universal target of gene rearrangement in a distinct subset of acute leukemias that accounts for 70% of cases in infants and 10% of adult cases (32, 33). In this disease subset, MLL1 is fused in frame to one of numerous different translocation partners, such as AF4, AF9, AF10, and ENL, which interact with DOT1L in complexes that promote transcriptional elongation (34, 35). Thus, MLL1 fusion proteins replace the native KMT domain for H3K4 with domains that recruit DOT1L, altering the balance between chromatin H3K79 and H3K4 methylation; the resulting ectopic H3K79 methylation is associated with increased expression of leukemogenic genes, such as HOXA9 and MEIS1 (10, 11). As DOT1L is necessary for MLL1 fusion proteins' oncogenic activities, including target gene overexpression (36–38), it is an appealing molecular target for treatment of leukemias that carry MLL1 rearrangements. EPZ004777, recently developed as a potent and selective small-molecule inhibitor of DOT1L KMT activity, reverses the gene signature of MLL1 translocation and kills MLL1-rearranged leukemic cells (39). Such results raise both the promise for DOT1L inhibitors in the clinic and the importance of anticipating mechanism-based toxicities. For example, DOT1L requirements in postnatal hematopoiesis (37, 40) predict possible but manageable myelotoxicity, and mice treated with EPZ004777 for 2 weeks remained overtly healthy and preserved bone marrow cellularity (39). Long-term effects on blood and other tissues are unknown, and the association of DOT1L with Wnt pathway activity (29–31) raises particular concern about gut toxicity. We therefore used whole-genome chromatin immunoprecipitation (ChIP) assays to map H3K79me-marked chromatin in different intestinal cell populations and gene disruption in mice to study DOT1L and H3K79me2 requirements in Lgr5+ ISCs and intestinal homeostasis.
MATERIALS AND METHODS
Isolation of Lgr5hi ISCs, villus cells, and enterocytes.
Lgr5hi ISCs were isolated from crypts in the duodenum and proximal jejunum of Lgr5GFP-IRES-CreER mice. Intestines were washed with cold phosphate-buffered saline (PBS), and villi were scraped using glass slides. Intestinal tissue was incubated in 5 mM EDTA in PBS for 45 min with occasional gentle shaking, and crypt epithelium was depleted of contaminating villi by passage through 70-μm filters. Crypt epithelial cells were disaggregated by treatment with 3.5× TrypLE (Invitrogen) at 37°C for 40 min. The cell suspension was washed in PBS, stained with Live/Dead cell viability dye (Invitrogen), and sorted using a MoFlo instrument (Beckman Coulter) to collect GFPhi cells. To collect villus cells or enterocytes from tamoxifen-treated wild-type or Villin-CreER(T2); Atoh1fl/fl mice, intestines were incubated in 5 mM EDTA in PBS for 20 to 30 min (41) and the villus epithelium was trapped on 70-μm filters.
Immunofluorescence and immunohistochemistry.
Tissues were fixed overnight at 4°C in 4% paraformaldehyde. For cryosections, fixed tissues were further incubated in 30% sucrose in PBS overnight at 4°C and embedded in optimal cutting temperature compound (OCT; Sakura). Tissue sections were permeabilized with 0.5% Triton X-100 overnight at 4°C and sequentially incubated with rabbit H3K79me2 antibody (Ab) (Abcam) in 0.5% Triton X-100 for 12 h and Cy3-conjugated anti-rabbit IgG (Millipore). Staining was visualized using a Nikon E800 fluorescence microscope. For paraffin sections, fixed tissues were dehydrated in 70% ethanol, embedded in paraffin, and cut into 5-μm sections. Staining with hematoxylin and eosin (H&E stain), alcian blue, and alkaline phosphatase used routine methods (42). For immunohistochemistry, 10 mM sodium citrate buffer (pH 6) was used to retrieve antigens and endogenous peroxidase activity was inhibited in methanol containing 3% H2O2. Tissues were blocked with 5% fetal bovine serum (FBS; Gibco) or 10% bovine serum albumin and incubated overnight at 4°C with one of the following Abs: rabbit lysozyme (1:50; Invitrogen), Ki67 (clone MM1, 1:1,000; Vector Laboratories), chromogranin A (1:500; Immunostar), active caspase 3 (1:20; Cell Signaling), PCNA (1:200; Neomarkers), H3K79me2 (1:500; Abcam), and H3K79me3 (1:400; Abcam). Sections were washed in PBS and treated with biotin-conjugated anti-mouse or anti-rabbit IgG (1:300; Vector Laboratories). Color reactions were developed using diaminobenzidine substrate (DAB; Sigma-Aldrich) and Vectastain avidin-biotin complex (ABC kit; Vector Laboratories).
Conditional mutant mice and tamoxifen administration.
All mice were at least 4 weeks old. To deplete Dot1l in Lgr5hi ISCs or whole intestinal epithelium, Dot1l fl/fl mice carrying Lgr5GFP-CreER or Villin-CreER(T2) alleles were injected with 2 mg tamoxifen daily for 5 days and tissues were harvested 4 or more days later. Control littermates were treated identically. To deplete Atoh1 to retrieve purified villus enterocytes, 1 mg tamoxifen was injected daily for 5 days and cells were collected on day 9. For lineage tracing of Dot1-null ISCs, Dot1lfl/fl; Lgr5GFP-CreER; Rosa26-YFP mice were injected with 2 mg tamoxifen daily for 5 days.
Histone extraction and immunoblotting.
Intestinal epithelial cells, harvested as described above, were suspended in extraction buffer containing 0.5% (vol/vol) Triton X-100, 0.02% (vol/vol) NaN3, and 2 mM phenylmethylsulfonyl fluoride (PMSF). Histones were extracted overnight at 4°C in 0.2 N HCl (43), resolved by SDS-PAGE, and immunoblotted with the following rabbit Abs: histone H3 (ab1791), H3K79me2 (ab3594), H3K4me3 (ab8580), H3K27Ac (ab4729), H4K20me (ab9051), H3K9me3 (ab8898), H3K36me3 (ab9050), and H3K9me2 (ab1220), all from Abcam, and H3K4me2 (07-030) and H3K27me3 (07-449) from Millipore.
Gene expression analyses.
Total RNA was extracted using TRIzol (Invitrogen) and RNeasy kits (Qiagen). For quantitative reverse transcription-PCR (RT-PCR) analysis, RNA was reverse transcribed with QuantiTect reverse transcription kit (Qiagen) and analyzed using SYBR green (Applied Biosystems). CT values were normalized with respect to β-actin and expressed as transcript ratios in mutant versus control tissues. For global profiling using 430 A2.0 arrays (Affymetrix), RNA was prepared and labeled as recommended by the manufacturer and data were analyzed using dChIP and Gene Pattern 3.2.3 software (44).
ChIP-seq and RNA-seq.
For ChIP sequencing (ChIP-seq), epithelial cell chromatin was cross-linked in 1% formaldehyde at ambient temperature for 25 min, washed in PBS, and sonicated to generate 200- to 500-bp fragments. H3K79me2, H4K20me, and H3K36me3 chromatin immunoprecipitation was performed as described previously (42), and immunoprecipitates were tested for enrichment of expected fragments. DNA from H3K79me2-marked chromatin was amplified to generate libraries using the ChIP-Seq DNA sample prep kit (Illumina) and sequenced using Illumina Hi-seq 2000. Fragments were mapped to the Mus musculus reference genome mm9, build 37. For RNA sequencing (RNA-seq), mRNA was isolated from total RNA using oligo(dT)25 magnetic beads (New England BioLabs). cDNA was synthesized, sonicated, amplified using the Encore Complete RNA-Seq Library System (Nugen), and sequenced using Illumina Hi-seq 2000. Sequence tags were mapped to the mouse reference genome, build mm9, and differences in transcript levels were determined using Tophat and Cufflinks software (45) at a false-discovery rate (FDR) of 0.001.
Microarray data accession number.
Data are deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under record number GSE41543.
RESULTS AND DISCUSSION
Study objectives and design.
We first examined if some cells in the mouse intestinal epithelium show disproportionately high DOT1L activity and if levels of H3K79 methylation at genes correspond to the levels of the transcripts. We asked, in particular, if Wnt target genes carry stronger methyl-H3K79 marks than other genes. To determine functions of H3K79 methylation in vivo, we used Lgr5EGFP-IRES-Cre(ER) (here called Lgr5GFP-Cre) mice (26) to induce Dot1l deletion in Lgr5+ ISCs and their progeny and, separately, Villin-CreER(T2) mice (46) to remove DOT1L from all intestinal epithelial cells. Lastly, we measured the expression of canonical Wnt targets and other genes and the status of other histone marks in Dot1l-null intestinal cells.
Levels of the H3K79me2 chromatin mark correlate better with gene expression in ISCs and differentiated cells than with Wnt target genes.
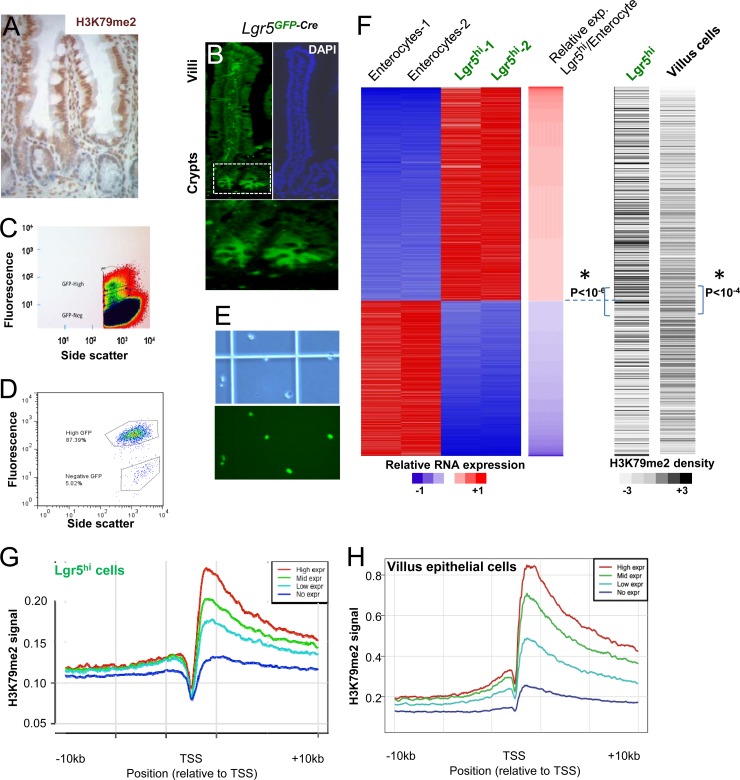
The intensity of H3K79me2 immunostaining is consistently higher in mature intestinal villus cell nuclei than in crypt progenitors (Fig. 1A). To examine the relationship between H3K79me2-marked chromatin and mRNA expression of individual genes in ISCs, we used flow cytometry to isolate green fluorescent protein (GFP)-expressing Lgr5+ CBCs from Lgr5GFP-Cre mice (26). Lgr5+/GFPhi stem cells were readily detected by fluorescence microscopy (Fig. 1B) and flow cytometry (Fig. 1C). Analysis of isolated GFPhi ISCs by flow cytometry (Fig. 1D) or microscopy (Fig. 1E) confirmed purity that approached or exceeded 90%. For comparison with these stem cells, we isolated either unfractionated villus cells from wild-type mouse duodenum, where enterocytes constitute 85% to 90% of the villus population (47), or pure enterocytes from Atoh1-null intestines, which lack all secretory cells (48). For the questions addressed in this study, unfractionated villus cells and purified enterocytes represent comparable populations. We used microarrays to profile transcripts and ChIP-seq with H3K79me2 Ab to profile this chromatin mark in intestinal stem and differentiated cells. Although H3K79me2 and H3K79me3 marks may occur on different genes in yeast (49), they colocalize in mammalian cells (9), and because we and others find that available H3K79me3 Abs cross-react with H3K79me2 (8), we focused on H3K79me2 as a marker of DOT1L enzyme activity.
Fig 1.
Distributions of the H3K79me2 chromatin mark in Lgr5+ crypt stem cells and mature intestinal villus cells. (A) Structure of mammalian crypt-villus units, showing stronger H3K79me2 immunostaining in mature villus epithelial cells than in undifferentiated crypt cells. (B) Crypt base columnar (CBC) intestinal stem cells (ISCs) are exclusively marked by GFP expression in Lgr5GFP-Cre mice. The boxed area in the left panel is magnified below to highlight CBCs. DAPI, 4′,6-diamidino-2-phenylindole. (C) Flow-cytometric isolation of Lgr5+/GFPhi ISCs. The highest gate in the scatter plot identifies cells with the highest GFP expression, which we isolated for this study. (D and E) Flow cytometry (D) and microscopic analyses (E; top, phase-contrast; bottom, fluorescence) confirm ∼90% purity of isolated Lgr5+ ISCs. (F) Left, heat map representation of genes showing at least 4-fold differential expression between Lgr5+ CBCs and purified villus enterocytes. Red and blue indicate high and low mRNA levels, respectively. Right, corresponding heat maps of H3K79me2 ChIP-seq tag density in reads per kilobase per million reads mapped (RPKM) in villus and Lgr5+ stem cells. Shades of gray are proportional to the global H3K79me2 signal over the gene body, and P values refer to the difference in H3K79me2 levels between the gene groups above and below the dotted lines (*, independent t test). (G and H) Correlation between gene expression and H3K79me2 marks in Lgr5+ ISCs (G) and villus cells (H), shown in aggregate for unexpressed genes (blue curve) and the top (red), middle (green), and bottom (cyan) tertiles of all expressed genes.
Approximately 1,200 genes are expressed selectively in Lgr5+ ISCs, and approximately 870 genes are active only in mature enterocytes (≥4-fold difference, P < 0.05) (Fig. 1F, left, red = high expression); this differential gene expression is similar to that reported by others (50). H3K79me2 marks were generally higher in mature villus cells than in ISCs (compare the y axes in Fig. 1G and H), consistent with stronger immunostaining in villi than in crypts (Fig. 1A). However, both cell populations showed the expected distribution of H3K79me2-marked nucleosomes, across the bodies of active genes, with the highest density near transcription start sites (TSSs) (Fig. 1G and H), and normalization of the two ChIP-seq libraries allowed reliable comparisons. Genes expressed at higher levels in Lgr5+ cells tended to carry higher H3K79me2 marking in these than in villus cells, and genes expressed at higher levels in enterocytes tended to give higher H3K79me2 signals in villus cells than in Lgr5+ CBCs; the differences were statistically highly significant (Fig. 1F, right panel). Even beyond differentially expressed genes, the H3K79me2 signal correlated well with gene expression levels in both ISCs (Fig. 1G) and villus cells (Fig. 1H), as other groups have reported occurs in other cell types (5–8).
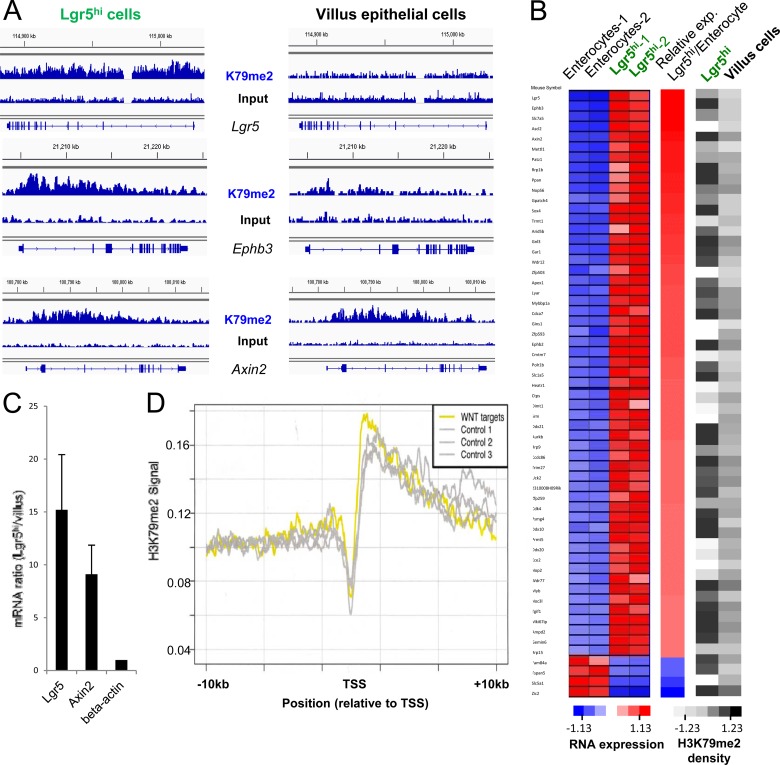
Because Dot1L activity and the H3K79me2 mark are postulated to mediate Wnt pathway activity (29–31), we assessed H3K79me2 marks on Wnt target genes. Well-validated intestinal Wnt target genes such as Lgr5 and EphB (26, 51) were indeed more highly expressed (Fig. 2C and data not shown) and marked more prominently with H3K79me2 (Fig. 2A) in ISCs than in villus cells. However, although the universal Wnt target Axin2 also showed higher expression in ISCs (Fig. 2C), this gene was marked similarly in the two populations (Fig. 2A), indicating that the relationship is not strict. Among the mouse homologs of 207 strong-candidate Wnt target genes identified in human intestinal cells (52), 55 genes were expressed at least 4-fold higher in Lgr5+ ISCs than in villus cells (Fig. 2B, left panel, where red indicates high expression). The H3K79me2 signal density over many of these Wnt target genes was higher in ISCs, but some target genes were minimally marked (Fig. 2B, right panel). To distinguish pathway-specific effects from those related to the level of gene expression, we assessed relative H3K79me2 levels at all 207 Wnt target genes and at 10 different random sets of 207 non-Wnt target genes whose average expression in microarray analysis was comparable to that of the group of Wnt targets. H3K79me2 levels were similarly distributed across Wnt target and nontarget genes that are expressed at similar levels (Fig. 2D). Thus, H3K79me2 abundance correlates with gene expression levels and not specifically with Wnt pathway target genes.
Fig 2.
Better correlation of H3K79me2 chromatin marks with gene expression than with Wnt target genes. (A) Distributions of H3K79me2-marked chromatin, as determined by ChIP-seq, in representative known Wnt target genes Lgr5, Ephb3, and Axin2 in Lgr5+ ISCs (left) and mature villus epithelial cells (right). Results of H3K79me2 ChIP are shown above those for input DNA. (B) Left, expression heat maps of 59 known intestinal Wnt target genes in Lgr5+ ISCs and enterocytes, ordered according to the fold difference in expression levels in the two populations. Right, corresponding heat maps of H3K79me2 density in Lgr5+ ISCs and villus cells. Shades of gray represent the H3K79me2 signal, expressed in ChIP-seq reads per kilobase per million reads mapped (RPKM). (C) Quantitative RT-PCR evidence that representative Wnt target genes express at higher levels in Lgr5+ ISCs than in villus cells. (D) Distribution of H3K79me2 marks, averaged for 207 known intestinal Wnt target genes (yellow), compared to groups of 207 non-Wnt target genes (gray) expressed at similar average levels in Lgr5+ ISCs. Results were similar for 7 other groups of non-Wnt target genes; for simplicity, only 3 groups are shown here.
H3K79me2 loss in Lgr5+ ISCs does not impede intestinal crypt cell growth or differentiation.
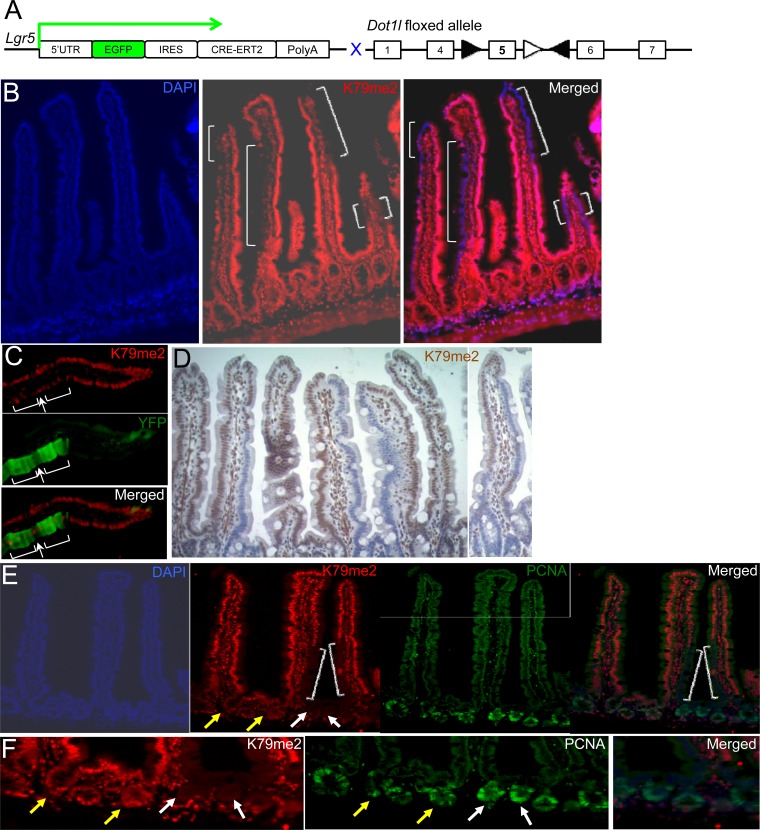
To determine H3K79me2 requirements in intestinal crypt cell function, we inactivated Dot1l, the only K79 methyltransferase gene (8, 15), in Lgr5+ ISCs, using mice with a Dot1l allele floxed at exon 5 (36) and the tamoxifen-inducible Cre recombinase gene inserted into the Lgr5 locus in Lgr5GFP-Cre mice (Fig. 3A). Lgr5GFP-Cre mice have variegated but clonal expression of the fusion insert. Thus, every Lgr5+ CBC in 30% to 50% of crypts expresses GFP-Cre, and all Lgr5+ CBCs in the remaining crypts do not, and mature intestinal villus cells derive from adjoining GFP-Cre+ and GFP/Cre-negative monoclonal crypts (26, 53). Accordingly, treatment of Lgr5GFP-Cre;Dot1lfl/fl mice with tamoxifen produced mosaic villi, with columns of H3K79me2-null cells interspersed among columns of wild-type cells (Fig. 3B), indicating that DOT1L and H3K79me2 loss in ISCs did not compromise their ability to replenish the epithelium. Lineage tracing with the Rosa26Fl-STOP-Fl-EYFP reporter allele (Rosa26YFP) (54) in tamoxifen-treated Lgr5GFP-Cre; Dot1lfl/fl; Rosa26YFP mice confirmed that H3K79me2-null villus cells originated in mutant Lgr5+ ISCs and not in alternative stem cells (Fig. 3C). Villi containing large fractions of H3K79me2-null cells were not small or dysmorphic (Fig. 3D), and cells lacking H3K79me2 were present 3 weeks after tamoxifen injection (Fig. 3B and C), indicating ISC activity over 5 or more renewal cycles. Consistent with this output, 3 weeks after Dot1l inactivation, Lgr5+ CBCs and their progeny in DOT1L-null crypts (white arrows in Fig. 3E and F) proliferated as robustly as their counterparts in neighboring DOT1L-proficient, H3K79me2+ crypts (yellow arrows). Taken together, these data indicate that intestinal crypts, which require continuous Wnt signaling (20, 21), do not depend on DOT1L-mediated methylation of H3K79.
Fig 3.
Elimination of the H3K79me2 chromatin mark in Lgr5+ ISCs and their progeny. (A) Strategy for conditional disruption of Dot1l in Lgr5+ CBCs. Lgr5GFP-Cre; Dot1lfl/fl mice are predicted to develop variegated loss of DOT1L function upon exposure to tamoxifen and Cre-mediated deletion of exon 5, which encodes the KMT domain. (B) H3K79me2 immunostaining (red) shows definitive loss of DOT1L activity in many tall columns (white brackets) of mature villus cells that must have originated in mutant ISCs. (C) Rosa26-YFP (yellow fluorescent protein) lineage tracing in Lgr5Cre; Dot1l f/f; Rosa26YFP mouse intestines demonstrates the origin of YFP-positive (green) villus cells in DOT1L-deficient ISCs, whose daughters lack H3K79me2 immunostaining (red). Villi are oligoclonal, and like other cells in this villus, the lone H3K79me2+ cell (arrow) amid unstained cells (brackets) must have originated in a DOT1L-proficient ISC. (D) Histology and H3K79me2 immunohistochemistry of villi in variegated Dot1l mutant intestines. (E and F). Robust crypt cell proliferation is preserved in the absence of H3K79me2. Dot1l-null crypts (white arrows) shown at low (E) and high (F) magnification spawn H3K79me2-deficient villus cells (white brackets in panel E) and show PCNA staining comparable to that of neighboring DOT1L-proficient crypts (yellow arrows). DAPI, 4′,6-diamidino-2-phenylindole.
Limited consequences of total intestinal epithelial loss of DOT1L function and H3K79me2.
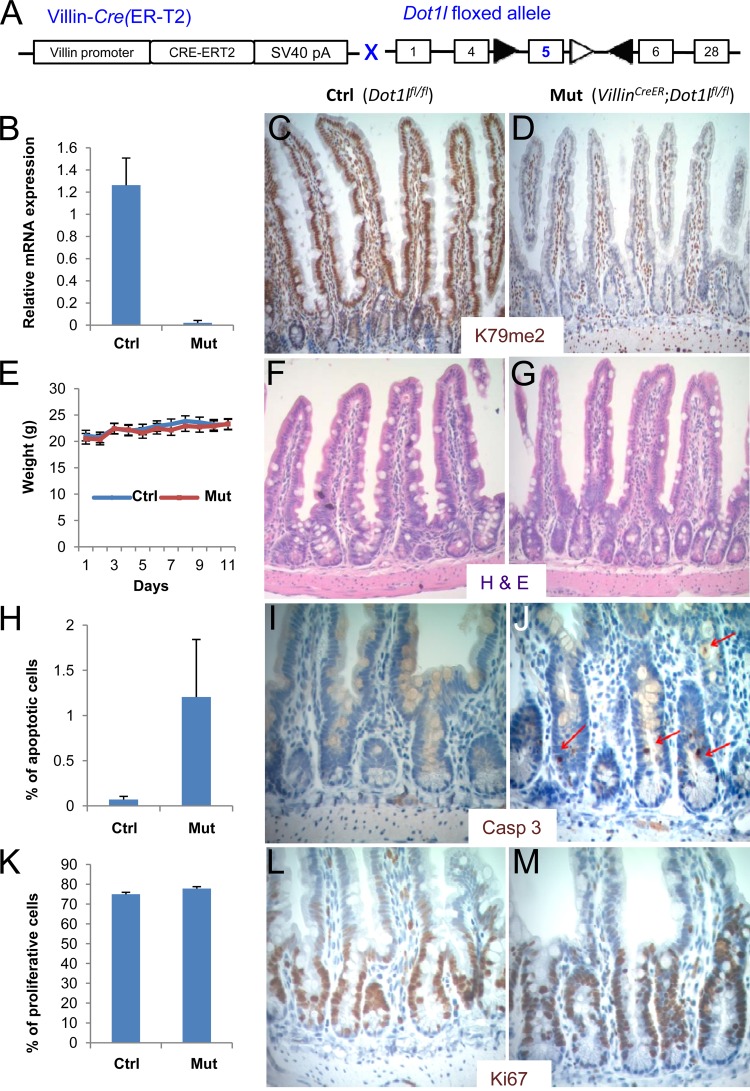
To bypass the limitations of variegated Cre expression in Lgr5GFP-Cre mice, we ablated DOT1L function simultaneously in all intestinal epithelial cells using Villin-CreER mice (Fig. 4A), which express inducible Cre recombinase throughout the epithelium (46). RT-PCR analysis confirmed that tamoxifen induced deletion of Dot1l exon 5 (Fig. 4B). Although RNA-seq analysis, described later in this report, revealed that transcripts containing other exons remained (see Fig. 6D), loss of exon 5 eliminated KMT activity, as expected, selectively in the surface epithelium, sparing the lamina propria and subepithelial smooth muscle (Fig. 4C and D). Both H3K79me2 (Fig. 4C and D) and H3K79me3 (see Fig. S1A in the supplemental material) were lost. Even with global loss of H3K79me2 from crypt and villus epithelium, mutant mice gained and maintained weight normally (Fig. 4E) and were not malnourished. Intestinal morphology remained intact, and the H3K79me2 mark was absent for periods ranging from 3 weeks to 4 months after administration of tamoxifen (Fig. 4F and G). The relative proportions of mucosal enterocytes and goblet, enteroendocrine, and Paneth cells were unaffected in Villin-CreER; Dot1lfl/fl intestines (see Fig. S1C to J in the supplemental material), but immunohistochemistry for cleaved caspase 3 revealed up to 20-fold increase in crypt cell apoptosis (Fig. 4H to J). This abnormality, evident in 5 mutant intestines harvested and processed exactly as were tamoxifen-treated Dot1l-proficient littermate controls, was reflected both in the number of crypts carrying at least one apoptotic body and in the number of apoptotic bodies per crypt. In contrast, Ki67 immunostaining was comparable in control and mutant intestines (Fig. 4K to M), indicating robust proliferation of ISCs and transit amplifying progenitors that could sustain H3K79me2-null tissue in the face of increased cell death. Apoptosis along intestinal villi was not increased. Thus, H3K79me2 loss subtly compromised intestinal crypt cell survival without impairing intestinal epithelial morphology or function, which is markedly different from phenotypes associated with Wnt pathway inactivation in the intestine (20, 21).
Fig 4.
Consequences of removing H3K79me2 from all intestinal epithelial cells. (A) Strategy for conditional Dot1l gene disruption in all intestinal epithelial cells in mice that carry a Villin-CreERT2 transgene, specifically expressed in intestinal epithelium, and the floxed Dot1l allele. (B) Relative expression of Dot1l exon 5 mRNA in intestinal epithelial cells isolated from Dot1lfl/lf (Ctrl) and VillinCreER; Dot1lfl/lf (Mut) mouse intestines, determined by quantitative RT-PCR. (C and D) H3K79me2 immunohistochemistry in control (C) and Dot1l-null (D) intestines, showing robust H3K79me2 signal in all control cells and selective loss in the mutant epithelium, with retention of the signal in subepithelial cells. (E) Body weights after administration of tamoxifen to VillinCreER; Dot1lfl/fl (Mut) and Dot1lfl/fl (Ctrl) mice. (F and G) Hematoxylin and eosin (H&E) staining of control (F) and Dot1l-null (G) mouse small intestines. (H to J) Immunohistochemistry (I and J) and quantitation, based on counting 1,000 crypts (H), of activated caspase 3 in control (I) and Dot1l-null (J) mouse intestinal crypts. Caspase 3-stained apoptotic bodies are indicated by red arrows. (K to M) Ki67 immunostain (L and M) and quantitation from counting 10,000 crypt cell nuclei (K) in control (L) and Dot1l-null (M) mouse intestinal crypts.
Fig 6.
RNA-seq analysis of VillinCreER;Dot1lfl/fl (Mut) and Dot1lfl/fl (Ctrl) villus epithelial cells. (A) Statistical analysis of autosomally encoded transcripts. Red dots denote the 189 genes with significantly altered (FDR = 0.001) expression in mutant cells. (B and C) Examples of RNA-seq data for genes that showed appreciably higher (B) or lower (C) signals in mutant cells. Sequence tags were mapped to a reference mouse genome and displayed in browser views (left panels) and by tag counts in fragments per kilobase of exon per million reads mapped (FPKM) (right panels). (D) Left panel, expression heat map of the 189 affected genes in mutant villus cells (red, increased expression; blue, reduced expression). Right panel, heat maps of H3K79me2 density at the corresponding genes in wild-type Lgr5+ ISCs and villus cells, in grayscale representation of FPKM. P values refer to the difference in H3K79me2 levels between the gene groups above and below the dotted lines (independent t test).
Wnt target gene activity in the absence of DOT1L-catalyzed H3K79me2.
Intact crypt cell proliferation and renewal in Villin-CreER; Dot1lfl/fl intestines suggested unperturbed Wnt pathway activity. It is possible, however, that DOT1L is necessary for optimal expression of Wnt target genes and that modestly reduced target gene levels do not overtly affect intestinal homeostasis. Quantitative RT-PCR analysis of isolated crypt epithelium revealed that selected known Wnt-dependent transcripts were present in DOT1L-deficient cells at levels similar to or slightly higher than the levels in control crypt cells (Fig. 5A).
Fig 5.
Wnt target gene activity and alternative activating histone marks in the absence of Dot1l enzyme activity. (A) Quantitative RT-PCR analysis of selected known intestinal Wnt target genes in isolated intestinal crypt epithelium. Average fold differences and standard deviations (±SD) from 3 replicate samples were calculated for each gene in mutant cells compared to controls and normalized to β-actin mRNA. (B) Total levels of known activation- and repression-associated chromatin marks in Dot1lfl/fl (Ctrl) and VillinCreER; Dot1lfl/fl (Mut) mouse intestinal epithelial cells, assessed by immunoblotting of histone extracts. Total H3 served as a loading control. (C) H3K79me2, H4k20me, and H3K36me3 ChIP at representative gene loci in crypt epithelial cells isolated from Dot1l-deficient (Mut) and control (Ctrl) mice. Immunoprecipitated DNA was analyzed by quantitative PCR, and results are represented in relation to the enrichment of a known unmarked fragment.
Although H3K79me2 and other chromatin marks are associated with active genes (2, 4, 8), it is unclear if such marks induce gene activity or if they are by-products of active transcription with other functions (55). Intact intestinal function and Wnt gene activity in Dot1l-null intestines argue against a requirement for H3K79me2 in gene transcription, but another activation mark could potentially compensate for its absence. The activation-associated mark H4K20me1 may be especially pertinent in this regard because, like H3K79me2, it is also implicated in Wnt target gene regulation (56). Immunoblots of histone extracts from Dot1l-null and control intestinal crypt epithelial cells showed equivalent levels of total H3K4me2, H3K4me3, H3K36me3, and H3K27ac, as well as the repressive marks H3K9me2, H3K9me3, and H3K27me3; total levels of only the H4K20me1 activation mark seemed modestly elevated (Fig. 5B). Within the constraints of using different Abs, quantitative PCR analysis of immunoprecipitated chromatin at randomly chosen, active, H3K79me2-marked loci suggested higher basal marking by H3K79me2 than by H4K20me1 in wild-type crypt cells (Fig. 5C). H4K20me1 levels at these genes were comparable in control and Dot1l mutant crypts, as were H3K36me3 levels (Fig. 5C). Thus, H3K79me2 loss is not overtly compensated for by the other histone modifications that we tested.
Global gene expression profiles in the absence of Dot1l function and H3K79me2.
On the one hand, strong and global association of H3K79me2 with active genes (6–8) suggests that transcription-related functions could be compromised in its absence. On the other hand, few genes were affected by loss or inhibition of DOT1L in mouse hematopoietic, cardiac, or induced pluripotent stem cells (18, 36, 57), and selected Wnt target genes were not affected in intestinal crypts (Fig. 5A). The H3K79me2 signal is stronger in villus than in crypt epithelium (Fig. 1A), and therefore, changes resulting from DOT1L loss may be most evident in intestinal villi. To quantify gene expression rigorously in DOT1L-deficient villus epithelial cells, we used RNA-seq to compare cells from tamoxifen-treated Dot1lfl/fl and Villin-CreER; Dot1lfl/fl mice. Because the gross phenotype of Dot1l mutant intestines suggested few differences and sexual dimorphism in intestinal gene expression is limited, we used mice of different genders in the two groups, and differential expression of sex-linked genes gave confidence that RNA-seq could reveal small and large changes in the mutant cells (see Fig. S2A and B in the supplemental material). Among 11,629 autosomal genes that yielded at least 20 sequenced fragments, fewer than 200 genes showed altered expression in mutant cells (false discovery rate, 0.001) (Fig. 6A). Examples of affected transcripts (Fig. 6B and C), coupled with evidence that sequence tags from Dot1l exon 5 were selectively diminished in mutant cells (see Fig. S2C in the supplemental material), demonstrate that the data are robust and reliable. Most affected transcripts, 151 of 189, were expressed at a higher level in mutant cells, and only 38 genes showed reduced expression, indicating minor gene activation in the absence of H3K79 methylation. The few affected genes were not enriched for functions in Wnt signaling or other pathways (see Fig. S2D in the supplemental material). Furthermore, altered gene expression in Dot1l-null villi showed no relation to the basal level of H3K79me2 marking at the corresponding loci in wild-type crypt or villus cells, and genes with the largest change were not among the most heavily marked (Fig. 6D). Taken together, these findings suggest that expression changes are unrelated to H3K79 methylation status per se.
Implications of the data for gene activity, Wnt pathway regulation, and clinical use of DOT1L inhibitors.
Our assessment of a Wnt-dependent tissue in genetic mouse models indicates that DOT1L-mediated H3K79 methylation does not have a special role in regulating intestinal Wnt-responsive genes and that increased apoptosis in Dot1l-null intestinal crypts does not affect the animals' health. The lack of a substantive effect of Dot1l gene disruption on gut function and gene expression cannot be attributed to conventional redundancy because H3K79me2 signals were completely lost (Fig. 4D and 5B), as expected from prior evidence that DOT1L is the only KMT for H3K79 (8, 12–15). Furthermore, in both intestinal crypt and villus epithelium, the signal from H3K79me2 ChIP correlated with overall gene expression and not with Wnt target genes per se. These findings improve prospects for developing DOT1L antagonists in targeted therapy of MLL-rearranged leukemias. An essential role in Wnt-responsive gene regulation would predict severe mechanism-based toxicity, possibly precluding safe drug development. The lack of DOT1L dependence in intestinal homeostasis suggests that intestinal toxicity is not imperative and, if it occurs, is likely unrelated to the primary mechanism of DOT1L inhibition.
Our observations have broad implications for transcriptional control because loss of DOT1L activity and methylated H3K79 had a negligible consequence on gene expression at large. This finding directly addresses a central question: whether histone modifications cue and instruct transcription or whether they merely correlate with gene activity and reflect ongoing transcript elongation (55). On the one hand, overexpressed leukemogenic genes in MLL-rearranged cells show hypermethylation of H3K79 (11), and the DOT1L inhibitor EPZ004777 reverses aberrant expression of these genes (39). In mice, deficiency of Menin, a component of H3K4 methylation complexes, specifically impairs H3K4 trimethylation at Hox loci and expression of most Hox genes (58). These findings support instructive roles for methylated H3K79 and H3K4 in disease or certain genetically engineered states. On the other hand, the data we present here indicate that H3K79me marks in particular are dispensable in a native tissue in vivo and may, in physiologic states, reflect a consequence rather than a requirement for transcription.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R01DK082889 (R.A.S.), R01CA140575 (S.A.A.), K01DK088868 (M.V.), RC2CA148222 (R.A.S. and S.A.A.), and P50CA127003.
We thank Sylvie Robine for gifting Villin-CreER transgenic mice and Myles Brown for helpful discussions.
Author contributions are as follows. L.-L.H. contributed to the study design, conducting of experiments, data analysis, and editing of the manuscript. A.S. and S.A.A. contributed to the study design, data analysis, and editing of the manuscript. M.V. and K.M.B. contributed to conducting of experiments. R.A.S. contributed to the study design and supervision, data analysis, and writing and editing of the manuscript.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 19 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01463-12.
REFERENCES
- 1. Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature 403:41–45 [DOI] [PubMed] [Google Scholar]
- 2. Kouzarides T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12:198–209 [DOI] [PubMed] [Google Scholar]
- 3. Vakoc CR, Sachdeva MM, Wang H, Blobel GA. 2006. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol. Cell. Biol. 26:9185–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837 [DOI] [PubMed] [Google Scholar]
- 5. Ng HH, Ciccone DN, Morshead KB, Oettinger MA, Struhl K. 2003. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. U. S. A. 100:1820–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, Bell SP, Groudine M. 2004. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18:1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122:517–527 [DOI] [PubMed] [Google Scholar]
- 8. Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, Blobel GA, Vakoc CR. 2008. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol. Cell. Biol. 28:2825–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nguyen AT, Zhang Y. 2011. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 25:1345–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. 2005. hDOT1L links histone methylation to leukemogenesis. Cell 121:167–178 [DOI] [PubMed] [Google Scholar]
- 11. Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, Kutok JL, Kung AL, Armstrong SA. 2008. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 14:355–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. 2002. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12:1052–1058 [DOI] [PubMed] [Google Scholar]
- 13. van Leeuwen F, Gafken PR, Gottschling DE. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109:745–756 [DOI] [PubMed] [Google Scholar]
- 14. Shanower GA, Muller M, Blanton JL, Honti V, Gyurkovics H, Schedl P. 2005. Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics 169:173–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S, Baltus GA, Kadam S, Zhai H, Valdez R, Gonzalo S, Zhang Y, Li E, Chen T. 2008. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 4:e1000190 doi:10.1371/journal.pgen.1000190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16:1518–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432:406–411 [DOI] [PubMed] [Google Scholar]
- 18. Nguyen AT, Xiao B, Neppl RL, Kallin EM, Li J, Chen T, Wang DZ, Xiao X, Zhang Y. 2011. DOT1L regulates dystrophin expression and is critical for cardiac function. Genes Dev. 25:263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clevers H. 2006. Wnt/beta-catenin signaling in development and disease. Cell 127:469–480 [DOI] [PubMed] [Google Scholar]
- 20. Pinto D, Gregorieff A, Begthel H, Clevers H. 2003. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 17:1709–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuhnert F, Davis CR, Wang HT, Chu P, Lee M, Yuan J, Nusse R, Kuo CJ. 2004. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc. Natl. Acad. Sci. U. S. A. 101:266–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP, Batlle E, Simon-Assmann P, Clevers H, Nathke IS, Clarke AR, Winton DJ. 2004. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 18:1385–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sangiorgi E, Capecchi MR. 2008. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 40:915–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. 2011. Interconversion between intestinal stem cell populations in distinct niches. Science 334:1420–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. 2011. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478:255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449:1003–1007 [DOI] [PubMed] [Google Scholar]
- 27. Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. 2009. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457:608–611 [DOI] [PubMed] [Google Scholar]
- 28. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459:262–265 [DOI] [PubMed] [Google Scholar]
- 29. Mahmoudi T, Boj SF, Hatzis P, Li VS, Taouatas N, Vries RG, Teunissen H, Begthel H, Korving J, Mohammed S, Heck AJ, Clevers H. 2010. The leukemia-associated Mllt10/Af10-Dot1l are Tcf4/beta-catenin coactivators essential for intestinal homeostasis. PLoS Biol. 8:e1000539 doi:10.1371/journal.pbio.1000539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A. 2010. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes Dev. 24:574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Castano Betancourt MC, Cailotto F, Kerkhof HJ, Cornelis FM, Doherty SA, Hart DJ, Hofman A, Luyten FP, Maciewicz RA, Mangino M, Metrustry S, Muir K, Peters MJ, Rivadeneira F, Wheeler M, Zhang W, Arden N, Spector TD, Uitterlinden AG, Doherty M, Lories RJ, Valdes AM, van Meurs JB. 2012. Genome-wide association and functional studies identify the DOT1L gene to be involved in cartilage thickness and hip osteoarthritis. Proc. Natl. Acad. Sci. U. S. A. 109:8218–8223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krivtsov AV, Armstrong SA. 2007. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 7:823–833 [DOI] [PubMed] [Google Scholar]
- 33. Muntean AG, Hess JL. 2012. The pathogenesis of mixed-lineage leukemia. Annu. Rev. Pathol. 7:283–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bitoun E, Oliver PL, Davies KE. 2007. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum. Mol. Genet. 16:92–106 [DOI] [PubMed] [Google Scholar]
- 35. Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, Slany RK. 2007. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood 110:4445–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, Pollock RM, Richon VM, Kung AL, Armstrong SA. 2011. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20:66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. 2011. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood 117:4759–4768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nguyen AT, Taranova O, He J, Zhang Y. 2011. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood 117:6912–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, Jin L, Kuntz KW, Chesworth R, Moyer MP, Bernt KM, Tseng JC, Kung AL, Armstrong SA, Copeland RA, Richon VM, Pollock RM. 2011. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20:53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nguyen AT, He J, Taranova O, Zhang Y. 2011. Essential role of DOT1L in maintaining normal adult hematopoiesis. Cell Res. 21:1370–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weiser MM. 1973. Intestinal epithelial cell surface membrane glycoprotein synthesis. I. An indicator of cellular differentiation. J. Biol. Chem. 248:2536–2541 [PubMed] [Google Scholar]
- 42. Verzi MP, Shin H, Ho LL, Liu XS, Shivdasani RA. 2011. Essential and redundant functions of caudal family proteins in activating adult intestinal genes. Mol. Cell. Biol. 31:2026–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shechter D, Dormann HL, Allis CD, Hake SB. 2007. Extraction, purification and analysis of histones. Nat. Protoc. 2:1445–1457 [DOI] [PubMed] [Google Scholar]
- 44. Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. 2006. GenePattern 2.0. Nat. Genet. 38:500–501 [DOI] [PubMed] [Google Scholar]
- 45. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7:562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. 2004. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39:186–193 [DOI] [PubMed] [Google Scholar]
- 47. Bjerknes M, Cheng H. 2006. Intestinal epithelial stem cells and progenitors. Methods Enzymol. 419:337–383 [DOI] [PubMed] [Google Scholar]
- 48. Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. 2001. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science 294:2155–2158 [DOI] [PubMed] [Google Scholar]
- 49. Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, Haug J, Johnston M, Jaspersen SL, Kobor MS, Shilatifard A. 2009. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol. Cell 35:626–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Munoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S, Volckmann R, Kung KS, Koster J, Radulescu S, Myant K, Versteeg R, Sansom OJ, van Es JH, Barker N, van Oudenaarden A, Mohammed S, Heck AJ, Clevers H. 2012. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4′ cell markers. EMBO J. 31:3079–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Batlle E, Henderson JT, Beghtel H, van den Born MM, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering M, Pawson T, Clevers H. 2002. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell 111:251–263 [DOI] [PubMed] [Google Scholar]
- 52. van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, Tjon-Pon-Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H. 2002. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111:241–250 [DOI] [PubMed] [Google Scholar]
- 53. Kim TH, Escudero S, Shivdasani RA. 2012. Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc. Natl. Acad. Sci. U. S. A. 109:3932–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1:4 doi:10.1186/1471-213X-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Henikoff S, Shilatifard A. 2011. Histone modification: cause or cog? Trends Genet. 27:389–396 [DOI] [PubMed] [Google Scholar]
- 56. Li Z, Nie F, Wang S, Li L. 2011. Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc. Natl. Acad. Sci. U. S. A. 108:3116–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, Lander ES, Armstrong SA, Daley GQ. 2012. Chromatin-modifying enzymes as modulators of reprogramming. Nature 483:598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, Ge K, Krumlauf R, Shilatifard A. 2009. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol. Cell. Biol. 29:6074–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.