Abstract
Glucocorticoids are among the most potent and effective agents for treating inflammatory diseases and hematological cancers. However, subpopulations of patients are often resistant to steroid therapy, and determining the molecular mechanisms that contribute to glucocorticoid resistance is thus critical to addressing this clinical problem affecting patients with chronic inflammatory disorders. Since the cellular level of the glucocorticoid receptor (GR) is a critical determinant of glucocorticoid sensitivity and resistance, we investigated the molecular mechanisms mediating repression of glucocorticoid receptor gene expression. We show here that glucocorticoid-induced repression of GR gene expression is mediated by inhibition of transcription initiation. This process is orchestrated by the recruitment of agonist-bound GR to exon 6, followed by the assembly of a GR-NCoR1-histone deacetylase 3-containing repression complex at the transcriptional start site of the GR gene. A functional negative glucocorticoid response element (nGRE) in exon 6 of the GR gene and a long-range interaction occurring between this intragenic response element and the transcription start site appear to be instrumental in this repression. This autoregulatory mechanism of repression implies that the GR concentration can coordinate repression with excess ligand, regardless of the combinatorial associations of tissue-specific transcription factors. Consequently, the chronic nature of inflammatory conditions involving long-term glucocorticoid administration may lead to constitutive repression of GR gene transcription and thus to glucocorticoid resistance.
INTRODUCTION
Glucocorticoids regulate diverse physiological processes, including development, metabolism, homeostasis, inflammation, and responses to both physiological and psychological stresses (1, 2). Clinically, synthetic glucocorticoids are employed for the treatment of numerous inflammatory conditions, including ectopia, dermatitis, allergies and asthma, osteoarthritis, adrenal insufficiency, autoimmune disorders, and organ rejection (3). Unfortunately, long-term and chronic exposure to glucocorticoids leads to unwanted side effects, such as osteoporosis and glucocorticoid resistance (4–6). Glucocorticoid resistance in patients is a major challenge for the treatment of these diseases, and thus an understanding of the molecular determinants of glucocorticoid resistance is of critical importance.
The actions of glucocorticoids are mediated through the glucocorticoid receptor (GR), a steroid hormone-regulated transcription factor that belongs to the nuclear hormone receptor superfamily (7). GR regulates gene expression by either transcriptional activation or transcriptional repression (transrepression). To mediate transcriptional activation, GR binds glucocorticoid response elements (GREs) and activates target gene transcription (8, 9). To initiate transrepression, GR is thought to physically interact with other transcription factors (such as NF-κB or activating protein 1 [AP-1]) and repress transcription of their downstream target genes (10, 11). Additionally, glucocorticoids may mediate anti-inflammatory effects via direct binding of GR to evolutionarily conserved negative glucocorticoid response elements (nGREs) (12). The transrepression activity of GR is often considered the major basis for the anti-inflammatory and immunosuppressive effects of glucocorticoids (10).
A major determinant of cellular responsiveness to glucocorticoids is the cellular concentration of GR protein, and the magnitude of transcription is proportional to receptor expression within a cell (13, 14). Thus, GR is a limiting factor for responsiveness, and small changes in receptor levels can affect cellular sensitivity to glucocorticoids (15). Ligand binding induces the repression of GR gene expression and ultimately leads to desensitization to further steroid administration (16), and this process may well contribute to glucocorticoid resistance.
Cellular levels of GR in cells are, however, dynamic and are regulated in a cell type-specific manner by the concentration of ligand, in addition to other factors (17, 18). In numerous cell lines and tissues, administration of GR agonists results in repression of the GR gene (18–24). The mechanism of glucocorticoid-induced repression of GR gene expression has been attributed to a decrease in GR gene expression as well as a decreased stability of the GR mRNA and protein (25–27). Both promoter-dependent (28) and -independent DNA elements have been proposed to be involved in the repression of GR mRNA expression (29–31). Furthermore, proteasome-mediated degradation contributes to increased turnover of the GR protein (32). However, a precise mechanism responsible for transcriptional repression of the GR gene remains elusive.
In this study, we investigated the molecular mechanism responsible for repression of GR gene transcription by glucocorticoids. Time course studies following glucocorticoid treatment revealed a rapid decrease in nascent GR RNA in human, mouse, and rat cell lines as well as in various mouse tissues. Maximal repression of nascent GR RNA was sustained for 6 to 8 h. Chromatin immunoprecipitation (ChIP) analysis indicated that GR is recruited to exons 6 and 8 of the GR gene, in a ligand-dependent manner, and that the ligand induces prolonged receptor occupancy at these GR coding regions. Our data also identified the existence of a functional nGRE in exon 6 of the GR gene that is instrumental for repression of the GR gene. Furthermore, we demonstrated that ligand-induced repression of GR gene transcription is mediated by assembly of a GR-NCoR1-histone deacetylase 3 (HDAC3)-containing repression complex at the proximal promoter region of the GR gene. This repression complex involves a long-range interaction between the intragenic response elements (exons 6 and 8) and the transcriptional start site (TSS) of the GR gene. This autoregulatory mechanism of repression implies that the GR concentration can coordinate downregulation with excess ligand, regardless of the combinatorial associations of tissue-specific transcription factors.
MATERIALS AND METHODS
Cell culture.
U-2 OS, A549, and HTC cells (ATCC, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium (DMEM)–F-12 medium supplemented with 10% (vol/vol) fetal calf serum (FCS) and 100 μg/ml penicillin-streptomycin (Invitrogen). U-2 OS cells stably expressing wild-type (WT) hGRα were maintained in 0.2 mg/ml Geneticin- and 0.2 mg/ml hygromycin (Invitrogen)-containing medium as described previously (33). 3T3L mouse fibroblast cells were maintained in DMEM supplemented with 10% (vol/vol) FCS and 100 μg/ml penicillin-streptomycin. All cells were maintained in a humidified 5% CO2 atmosphere at 37°C.
Animal experiments.
All animals were maintained in accordance with the National Institutes of Health directives for the care and use of laboratory animals, and the NIEHS Animal Care and Use Committee approved all experimental procedures. Adrenalectomized C57BL/6 male mice were purchased from Jackson Laboratory (Bar Harbor, ME). Adrenalectomized mice were treated with 1 mg of dexamethasone/kg of body weight or with vehicle intraperitoneally (i.p.) for different periods; after the specified periods, the mice were euthanized by cervical dislocation, and organs were excised and preserved in RNAlater (Qiagen) for RNA isolation.
RNA extraction and real-time quantitative PCR.
Following dexamethasone treatment, RNAs were extracted from cell lines and mouse tissues by use of a Qiagen RNeasy minikit, and real-time PCR was performed using a model 7900HT sequence detection system with predesigned primer-probe sets available from Applied Biosystems (Foster City, CA). The signal obtained from each gene transcript was normalized to the transcript of the unregulated cyclophilin B housekeeping gene and expressed relative to the transcript level in the control sample.
RNA interference.
NCoR1 and nontarget control (NTC) siRNA-SMART pools were purchased from Thermo Scientific. For each RNA interference experiment, cells were plated in 6-well plates at approximately 70% confluence 1 day prior to transfection. The cells were transfected with small interfering RNA (siRNA) at 50 nM, using DharmaFECT transfection reagent (Thermo Scientific) following the manufacturer's instructions. After 48 h of siRNA treatment, cells were induced with 100 nM dexamethasone or vehicle for the specified time points, and the cells were harvested for RNA and protein extraction.
ChIP assay.
ChIP assays were performed in A549 cells as described previously (34), with minor modifications. For ChIP assay, A549 cells were plated on 150-mm dishes in 30 ml medium supplemented with 10% dextran-coated charcoal-stripped fetal calf serum and grown to 90% confluence, followed by dexamethasone or vehicle treatment for specified periods. Cells were then scraped, resuspended in cell lysis buffer (50 mM HEPES-KOH at pH 8, 1 mM EDTA, 140 mM NaCl, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, protease inhibitor cocktail), and subjected to nutation for 30 min at 4°C. The crude nuclei were collected by centrifugation (600 × g for 5 min at 4°C), resuspended in 600 μl of shearing buffer (10 mM Tris-HCl at pH 8, 1 mM EDTA, 140 mM NaCl, 1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, protease inhibitor cocktail), and sonicated using a Branson Sonifier 150 at setting 4. Chromatin extracts (100 μl) were precleared and immunoprecipitated with 2 μg of desired antibody (anti-GR [5], anti-NCoR1 [ab24552; Abcam], anti-SMRT [ab24551; Abcam], anti-polymerase II [anti-Pol II] [MMS-134R; Covance], anti-SRC1 [sc-6098; Santa Cruz], anti-HDAC2 [ab7029; Abcam], or anti-HDAC3 [sc-11417; Santa Cruz] antibody), and the immunoprecipitated DNA complex was pulled down using Magna ChIP protein A magnetic beads (Millipore). The amount of immunoprecipitated DNA was then quantified using real-time quantitative PCR with corresponding primer-probe sets custom ordered from Integrated DNA Technologies (Coralville, IA).
Western blot analysis.
Following the indicated treatments, cells were washed in phosphate-buffered saline (PBS) and lysed in ice-cold RIPA buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.5% sodium deoxycholate, 1% Igepal CA-630, and 0.1% sodium dodecyl sulfate) supplemented with protease inhibitor cocktail tablets (Roche). The cell extracts were cleared by centrifugation at 12,000 × g for 10 min at 4°C, and the protein concentration was quantified using a Bio-Rad protein assay kit. The cell lysates were resolved in 4 to 20% Tris-glycine Ready gels (Bio-Rad, Hercules, CA) and transferred onto nitrocellulose membranes. Subsequently, the membranes were blocked with 5% nonfat dry milk in Tris-buffered saline (20 mM Tris base [pH 7.5] and 150 mM NaCl) containing 0.05% Tween 20 (TBS-T) and then probed with the primary antibody diluted in 1% nonfat milk in TBS-T. After incubation in TBS-T, the membranes were incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies (GE Healthcare, Piscataway, NJ), and blots were visualized by chemiluminescence (GE Healthcare, Piscataway, NJ). To determine the amount of protein, chemiluminescence signals were quantified by densitometry using NIH ImageJ analysis software and normalized to the beta-actin signal for each band.
Luciferase plasmid constructs, transient transfection, and luciferase assays.
To create a pGL3 luciferase reporter plasmid with exons 6 and 8 of the GR gene at the promoter region, the pGL3 promoter plasmid was digested with BglII, and exon 6 or 8 was inserted. The potential nGRE in exon 6 was mutated by site-directed mutagenesis (12). A549 cells were plated in 6-well plates at approximately 80% confluence 1 day prior to transfection. The cells were then transiently transfected with the control pGL3 luciferase plasmid or the pGL3 luciferase construct with exon 6 or 8 and with pRL (Renilla luciferase) as a control for transfection, using TransIT-LT1 reagent (Mirus Bio LLC, Madison, WI). Following 18 to 24 h, the transfection medium was removed and replaced with fresh medium supplemented with FCS charcoal stripped of endogenous glucocorticoids for 8 h. Subsequently, the cells were treated with dexamethasone (Steraloids, Wilton, NH) or vehicle (H2O). Six hours after treatment, cells were harvested for RNA isolation, and the level of luciferase gene transcripts was measured using real-time quantitative PCR with corresponding primer-probe sets custom ordered from Integrated DNA Technologies (Coralville, IA). The luciferase mRNA was normalized to that of the unregulated cyclophilin B housekeeping gene and corrected to the Renilla luciferase level.
3C assay.
Chromosome conformation capture (3C) assay was performed with A549 cells as described previously (35), with minor modifications. A549 cells were formaldehyde cross-linked after treatment with vehicle or dexamethasone, resuspended in lysis buffer (50 mM HEPES-KOH at pH 8, 1 mM EDTA, 140 mM NaCl, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, protease inhibitor cocktail), and incubated for 30 min on ice, followed by centrifugation (400 × g, 5 min) at 4°C. The nuclear pellet was resuspended in 500 μl digestion buffer (1.2× BglII restriction enzyme digestion buffer [NEB] plus 0.3% [wt/vol] SDS) and incubated at 37°C for 1 h on a rocker. Triton X-100 (1.8% [vol/vol]) was then added to the samples and incubated for 1 h under similar conditions. BglII (400 units; NEB) was added and incubated at 37°C overnight. The restriction enzyme was inactivated by addition of SDS (1.6% [vol/vol] final concentration) and incubation at 65°C for 20 min. SDS was quenched by addition of Triton X-100 (1% [vol/vol] final concentration) and 1 h of incubation at 37°C. Individual samples were then diluted 12 times in ligase buffer. Ligation was done for 4 h at 16°C, using 100 units T4 DNA ligase (Promega), followed by 30 min of incubation at room temperature. Proteinase K (final amount, 300 μg) was added to the samples, which were incubated overnight at 65°C. Next, 30 μl of 10 mg/ml RNase (final amount, 300 μg) was added to the samples, which were incubated for 30 min at 37°C, and DNA was purified by phenol-chloroform extraction. The DNA concentration was estimated by spectrophotometry, and equal amounts of DNA were used for PCR amplification. Primers flanking either side of the putative interaction sites (restriction sites) were used to amplify a specific hybrid product by reverse transcription-PCR (RT-PCR) after ligation. As a control to ensure that equal amounts of DNA were used for PCR amplification of each sample, an aliquot of sample was amplified using a pair of primers that amplifies the GILZ promoter region, which is devoid of BglII sites. Equal amplification was obtained in all samples (data not shown). Primers flanking either side of the putative ligation sites (interaction sites), at the TSS and kb +122 (downstream of the TSS) of exon 6 and exon 8, were used to amplify a specific hybrid product by RT-PCR as a negative control. Each 3C assay result is expressed as the cross-linking frequency (y-axis label), which is a measure of the frequency of interaction between two DNA loci. The cross-linking frequency of any two DNA fragments is determined by the intensity of the PCR signal of a ligation product. The cross-linking frequency depends on the relative proximity of the two DNA fragments to each other at a given time point. The cross-linking frequency was calculated as described by Hagege et al. (35).
Statistical analysis.
Data are presented as means ± standard deviations (SD). Statistical significance was determined by analysis of variance (ANOVA) with Tukey's post hoc analysis.
RESULTS
Glucocorticoid-induced repression of the GR gene is mediated by inhibition of transcription initiation.
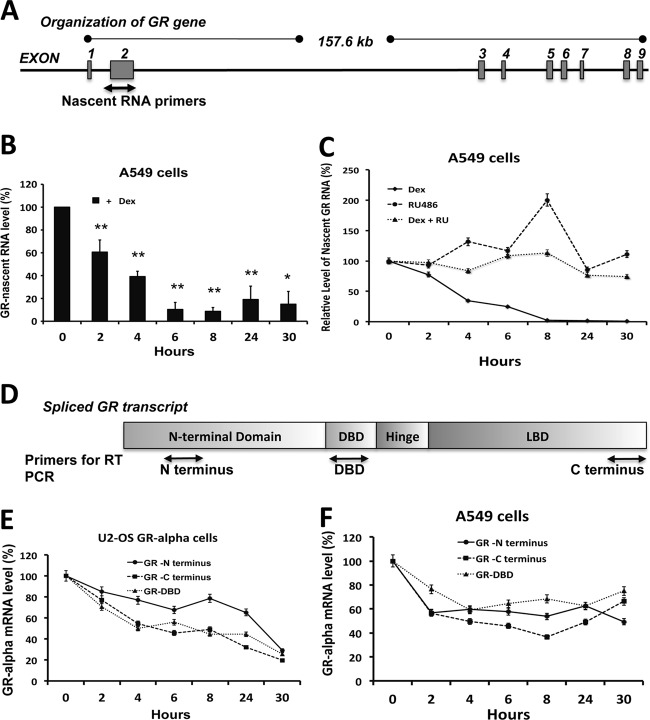
Studies from our laboratory and others have demonstrated that glucocorticoid treatment can repress the expression of mature GR mRNA by 50 to 80% in different tissues of humans, mice, and rats (18–24). However, the molecular mechanisms responsible for these effects remain poorly understood. To define these mechanisms and trans-acting factors necessary for repression of GR gene expression in the presence of ligands that bind to the glucocorticoid receptor, we initiated studies to analyze GR primary transcript (nascent RNA) expression in response to glucocorticoid treatment. To evaluate the effects of dexamethasone and the antagonist RU486 (36) on GR primary transcripts, RT-PCR was performed on A549 cells, which contain endogenous glucocorticoid receptors and their regulatory elements. Using primers and a probe spanning exon 2 and intron 2, as shown in Fig. 1A, a 50 to 90% decrease in newly synthesized nascent GR RNA was observed with glucocorticoid treatment (Fig. 1B). The glucocorticoid receptor antagonist RU486 failed to downregulate nascent GR RNA and was able to block glucocorticoid-induced downregulation of GR primary transcripts (Fig. 1C).
Fig 1.
Glucocorticoid-induced repression of the GR gene is mediated by inhibition of transcription. (A) Schematic showing the GR gene with the relative positions of the primers for detection of nascent GR RNA. (B) Nascent GR RNA levels were examined by RT-PCR analysis of A549 cells following dexamethasone treatment for 2, 4, 6, 8, 24, and 30 h. The data represent the means ± SD for five independent experiments. **, P < 0.01; *, P < 0.05. (C) Nascent GR RNA levels were examined by RT-PCR analysis of A549 cells treated with dexamethasone, RU486, or dexamethasone plus RU486 for 2, 4, 6, 8, 24, and 30 h. Each assay result was normalized to the cyclophilin B housekeeping gene's nascent RNA level. The data represent the means ± SD for three independent experiments. (D) Schematic representation of the GR protein showing the relative positions of the primers for different domains of GR for RT-PCR analysis. (E) GR mRNA expression was examined by RT-PCR analysis of U-2 OS cells stably expressing GRα following dexamethasone treatment for 2, 4, 6, 8, 24, and 30 h, using primers and probes for the different regions of GR. (F) GR mRNA expression was examined by RT-PCR analysis of A549 cells following dexamethasone treatment for 2, 4, 6, 8, 24, and 30 h, using primers and probes for the different regions of GR.
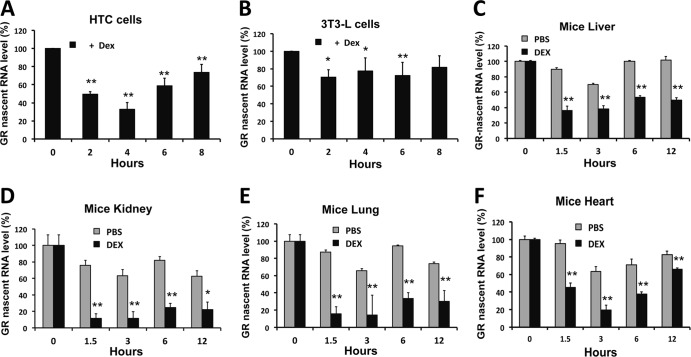
Since dexamethasone has been reported to cause repression of GR mRNA in various cell types and in a variety of mouse tissues in vivo, we also investigated whether this glucocorticoid induced a decrease in GR primary transcript expression in HTC cells, 3T3L cells, and a variety of mouse tissues (liver, kidney, lung, and heart). Figure 2A shows that dexamethasone induced a significant downregulation of GR primary transcripts, within hours, in HTC cells. Inhibition of GR gene transcription, measured by a decrease in primary transcript expression, was also observed in 3T3L cells but was more limited than that seen in the other cell types (Fig. 2B). Maximum repression of GR primary transcript expression in vivo was observed in the kidney and lung (Fig. 2C, D, E, and F). Together, these results indicate that glucocorticoids impede the transcription of the glucocorticoid receptor gene in a fashion largely independent of cell or tissue specificity.
Fig 2.
Glucocorticoids induce repression of the GR gene by inhibiting transcription in mice and rats. (A and B) Primary GR transcript levels were measured in HTC cells and 3T3L cells, using primers and probes specific for nascent GR transcripts in rats and mice, respectively. (C, D, E, and F) Adrenalectomized mice were treated with dexamethasone or PBS for different intervals, and then the mice were euthanized, followed by extraction of the liver, kidney, lung, and heart for RNA isolation. Quantitative RT-PCR was then performed to determine the levels of mGR primary transcript. The data represent the means ± SD for three independent experiments. **, P < 0.01; *, P < 0.05.
In order to elucidate if glucocorticoid-mediated downregulation of GR mRNA is mediated only by impeding transcription initiation or also by RNA Pol II pausing during transcription elongation, we analyzed GR mRNA levels in A549 cells and U-2 OS hGRα cells, using primers and probes designed to evaluate different regions of the mature GR mRNA, as shown in Fig. 1D. Quantification of GR mRNA measured with primers and probes specific to different regions of the mRNA revealed a kinetics similar to that with the C-terminal primers (Fig. 1E and F), indicating that repression of GR mRNA is mediated by inhibition of transcription initiation rather than by RNA Pol II pausing or attenuation of elongation. Thus, our results support the notion that glucocorticoid-mediated repression of GR mRNA expression is mediated specifically by blocking transcription.
The glucocorticoid receptor binds to the 3′ end of its own gene.
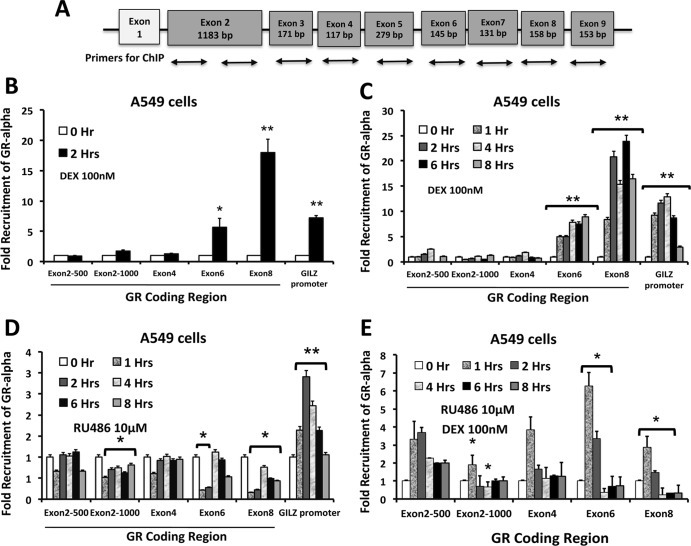
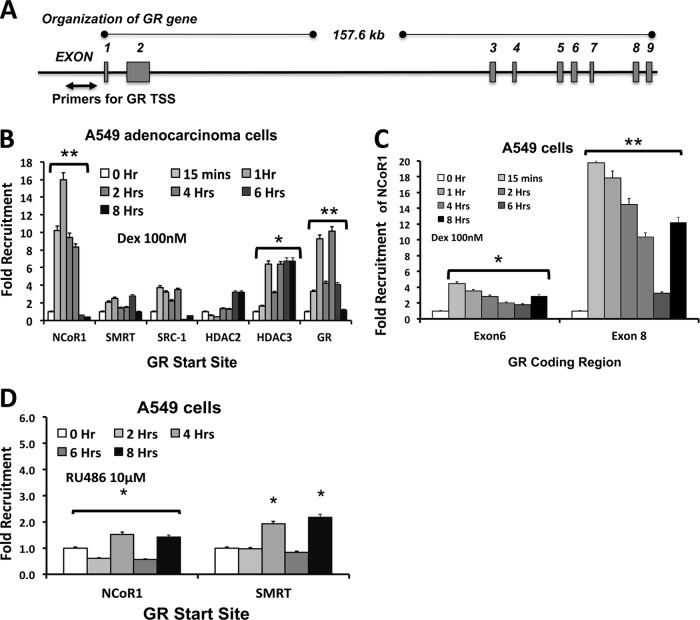
Our results from U-2 OS cells stably transfected with hGRα cDNA with a cytomegalovirus (CMV) promoter (Fig. 1E) suggest that the GR cDNA contains a sequence sufficient for glucocorticoid-induced downregulation of GR mRNA (30). Furthermore, studies using transcriptional and translational inhibitors indicated that the glucocorticoid receptor is directly responsible for hormone-mediated downregulation (30, 37). Thus, we wished to determine if GR binds its own coding region in a ligand-dependent manner and, if so, where precisely it binds. In order to address this question, A549 cells were treated with ligand or vehicle, and the entire exonic region of the GR gene was scanned for GR recruitment by ChIP assays. Figure 3A depicts how primers were designed to scan the entire exonic region of the hGR gene. Consistent with previous findings for transiently transfected Cos-1 cells, our data indicate that GR is recruited to the 3′ end of the GRα coding region distinctive to exons 6 and 8, in a ligand-dependent manner (Fig. 3B). For evaluation of the kinetics of GR recruitment to this region, ChIP scanning experiments with A549 cells were performed at different time points after glucocorticoid treatment. These experiments revealed that the ligand induced prolonged GR occupancy at exons 6 and 8 of the GR gene for up to 8 h after dexamethasone treatment (Fig. 3C).
Fig 3.
The 3′ end of the GRα coding region recruits GR in a ligand-dependent manner. (A) Schematic representation of the GR gene showing the relative positions of the primers for ChIP scanning of the exons. (B) GR recruitment to all the exons of the GR gene was assessed by ChIP assay of A549 cells treated with dexamethasone for 2 h. The fold change was calculated from the percentage of input for each pulldown and by comparison to vehicle-treated controls for 5 independent experiments. Recruitment of GR to the GILZ gene promoter (positions −2220 to −2133) was used as a positive control. (C) GR recruitment to all the exons of the GR gene was assessed by ChIP assay of A549 cells treated with dexamethasone for different times. (D) Recruitment of GR to all the exons of the GR gene was assessed by ChIP assay of A549 cells treated with RU486 for different times. (E) GR recruitment to all the exons of the GR gene was assessed by ChIP assay of A549 cells treated with RU486 and dexamethasone for different times. The data represent the means ± SD for three independent experiments. **, P < 0.01; *, P < 0.05.
We also investigated whether the recruitment of GR to this exonic region is unique to agonist treatment. ChIP scanning assays were performed on A549 cells following treatment with either RU486 or dexamethasone plus RU486. Treatment with RU486 alone failed to recruit GR to exons 6 and 8 (Fig. 3D). Cotreatment of cells with dexamethasone and RU486 showed moderate recruitment of GR to exons 6 and 8 (Fig. 3E). These data indicate that the ligand induces prolonged occupancy of GR at exons 6 and 8 of the GR gene. Furthermore, RU486 inhibits the binding of antagonist-bound GR to these regions of DNA.
Ligand-induced repression of GR is mediated by GR binding to an nGRE in exon 6 of the GR gene.
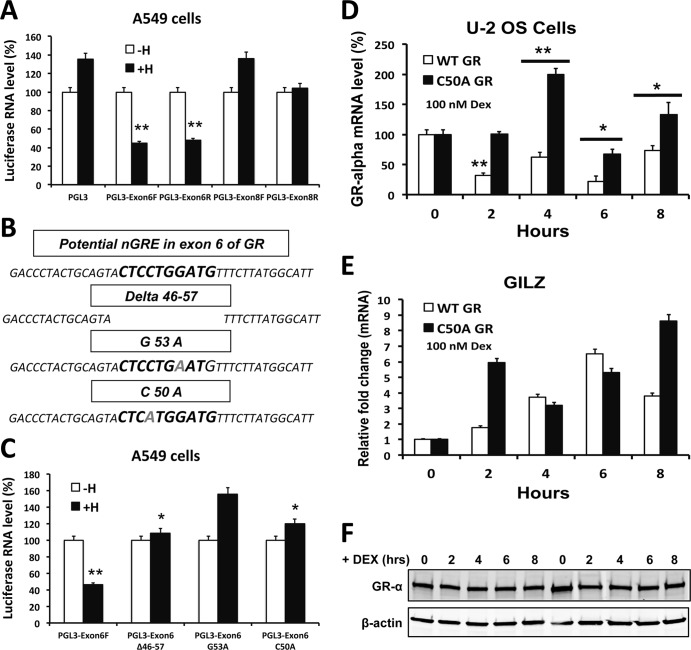
Moreover, to determine if a putative DNA element in exons 6 and 8 is required for eliciting GR gene transcriptional repression, we cloned exon 6 or exon 8 of GR upstream of the enhancerless simian virus 40 (SV40) early promoter, located 5′ of the luciferase coding sequence of the pGL3 vector, and transfected it into A549 cells, followed by addition of dexamethasone or vehicle. Glucocorticoid treatment did not affect luciferase expression from the plasmid containing exon 8. However, with the plasmid containing exon 6, dexamethasone treatment resulted in a 60% decrease in luciferase expression compared to basal expression (Fig. 4A). Thus, we analyzed the exon 6 DNA sequence for potential nGREs. Interestingly, exon 6 of the GR gene harbors a putative nGRE described by Surjit et al. (12), and this putative nGRE is conserved in humans, rats, and mice. We next assayed the recruitment of GR to exon 6 in the stably integrated GR construct in the U-2 OS cells. As shown in Fig. S4 in the supplemental material, there was significant recruitment of GR to exon 6 in these cells, which shows that the putative nGRE in exon 6 is functional in the context of stably integrated cDNA and actively recruits GR. To determine the functionality of this putative nGRE in repression, we mutated critical bases necessary for receptor binding or deleted the whole nGRE (Fig. 4B) and measured luciferase expression in reporter assays. Mutation of the critical bases and deletion of the nGRE resulted in the elimination of the repression function (Fig. 4C). Together, these observations suggest that the nGRE in exon 6 is critical for glucocorticoid-induced repression of the GR gene.
Fig 4.
Glucocorticoid-induced repression of the GR gene is mediated by GR binding to a negative GRE in exon 6 of the GR gene. (A) A549 cells were transfected with luciferase reporter constructs with exon 6 in the forward direction (exon 6F), exon 6 in the reverse direction (exon 6R), exon 8 in the forward direction (exon 8F), or exon 8 in the reverse direction (exon 8R). Reporter activity was measured by RT-PCR, using primers and a probe specific for firefly luciferase, following 6 h of treatment with dexamethasone. Empty pGL3 was used as a negative control. (B) Schematic representation of the site-directed mutagenesis of the nGRE in exon 6 of the GR gene. (C) Luciferase assay performed on A549 cells transfected with the pGL3-exon6F reporter construct with different mutations at the putative nGRE. Cells were treated with vehicle or dexamethasone for 6 h, and the reporter activity was measured by RT-PCR using primers and a probe specific for the luciferase gene (n = 3). **, P < 0.01; *, P < 0.05. (D) U-2 OS cells were transfected with either WT GR or C50A mutant GR expression vector, and GR mRNA was measured by RT-PCR after dexamethasone treatment for 0, 2, 4, 6, and 8 h (n = 3). **, P < 0.01; *, P < 0.05. (E) GILZ mRNA levels in U-2 OS cells transfected with either WT GR or C50A mutant GR expression vector and treated with dexamethasone for 0, 2, 4, 6, and 8 h (n = 3). **, P < 0.01; *, P < 0.05. (F) Immunoblot of GR protein in U-2 OS cells transfected with either WT GR or C50A mutant GR expression vector and treated with dexamethasone for 0, 2, 4, 6, and 8 h.
To ascertain if mutation of a key residue in the putative nGRE in exon 6 would inhibit ligand-induced repression of the GR gene, we mutated the C residue at position 50 in exon 6 without altering the function of the GR protein, as this C residue is key for the nGRE to be functional. We transiently transfected U-2 OS cells with either WT GR or C50A mutant GR (Fig. 4F) and examined the ligand-induced repression of GR mRNA. As shown in Fig. 4D, cells transfected with the C50A nGRE mutant were unable to repress GR mRNA compared to cells transfected with WT GR. In order to validate if the C50A GR mutant was functional, we also analyzed the expression of the GILZ gene, a classic GR target gene (Fig. 4E). These data suggest that C50A mutant GR is functional in terms of transactivation. Together, our results imply that the putative nGRE in exon 6 is critical for ligand-induced transrepression of the GR gene.
Binding of ligand-bound GR to its exonic region enables the formation of a repression complex near the transcriptional start site.
We next investigated how recruitment of GR to the 3′ end of the GR gene causes repression of GR transcription by formation of a repression complex at the promoter of the GR gene by using a ChIP assay (Fig. 5A). We observed significant recruitment of NCoR1, occurring within 15 min of glucocorticoid treatment and maintained for up to 4 h, whereas SMRT and SRC1 were weakly bound at the promoter-proximal region (Fig. 5B). Since NCoR1 is known to recruit histone deacetylases (HDACs) to repression complexes, we also examined the recruitment of HDAC2 and HDAC3 to the GR start site in the presence of dexamethasone. While HDAC2 was weakly bound to the GR start site, the glucocorticoid strongly enhanced the recruitment of HDAC3 to the GR proximal promoter (Fig. 5B), further supporting the idea that ligand-induced binding of GR to the intragenic region generates a repression complex. ChIP analysis revealed the recruitment of GR to the promoter-proximal region of the GR gene (Fig. 5B) and NCoR1 to exons 6 and 8 (Fig. 5C). In keeping with the above data (Fig. 3D), RU486 treatment precluded the formation of this repression complex at the GR start site (Fig. 5D). Recruitment of these coregulators to the GILZ gene TSS was also analyzed as a control, and there was no significant recruitment of these coregulators to the GILZ gene start site. This finding suggests that NCoR1-mediated transcriptional repression is a potential mechanism by which ligand influences GR downregulation.
Fig 5.
Binding of ligand-bound GR to the exonic GR gene region enables the formation of a repression complex at the TSS of the GR gene. (A) Schematic representation of the GR gene showing the relative positions of the primers for the transcriptional start site for ChIP analysis. (B) Recruitment of GR, coregulators (NCoR1, SMRT, and SRC1), and HDACs (HDAC2 and HDAC3) was assessed by ChIP assay following treatment with dexamethasone for different times. (C) Recruitment of NCoR1 to exons 6 and 8 of the GR gene was assessed by ChIP assay following treatment with dexamethasone for different times. (D) Recruitment of corepressors to the GR gene start site was examined by ChIP assay following treatment with RU486 for different times.
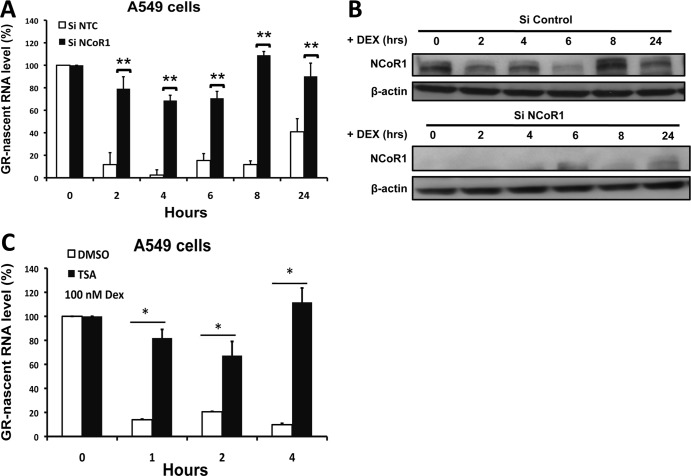
The data described above suggest that NCoR1 is the primary coregulator protein involved in this repression process. Thus, to evaluate the role of NCoR1 in glucocorticoid-induced repression, we knocked down the expression of NCoR1 in A549 cells by using selective siRNA and examined the nascent GR RNA levels after dexamethasone treatment. Knockdown of NCoR1 prevented the repression of nascent GR RNA (Fig. 6A and B). These results indicate that NCoR1 is the key corepressor protein involved in the formation of this repression complex. We also evaluated the role of SMRT in the formation of this repression complex by knocking down SMRT in A549 cells by use of selective siRNA (see Fig. S2A and B in the supplemental material) and examining nascent GR RNA levels after dexamethasone treatment. Knockdown of SMRT had no significant effect on the repression of nascent GR RNA (see Fig. S1 in the supplemental material). These results indicate that SMRT might not be involved in the formation of this repression complex.
Fig 6.
NCoR1 and HDAC3 are instrumental in the repression of GR gene transcription. (A) A549 cells were transfected with siRNA for NCoR1 or a control siRNA, and regulation of nascent GR RNA by dexamethasone was analyzed by quantitative RT-PCR. (B) NCoR1 protein levels were determined by Western blotting of A549 cells transfected with siNCoR1 or siNTC and treated with dexamethasone for different times. (C) Nascent GR RNA levels were examined by RT-PCR analysis of A549 cells after treatment with 2 μM TSA and dexamethasone for 0, 1, 2, and 4 h. The data represent the means ± SD for three independent experiments. **, P < 0.01; *, P < 0.05.
Since significant recruitment of HDAC3 to the GR promoter was observed in our ChIP assay, we examined the effect of the HDAC3 inhibitor trichostatin A (TSA) on ligand-induced repression of the GR gene. Treatment of A549 cells with TSA and dexamethasone significantly blunted the ligand-induced repression of the nascent GR RNA (Fig. 6C). These data show that HDAC3 is the key HDAC protein in the repression of GR gene transcription.
Finally, 3T3L cells showed minimal repression of GR, so we examined the corepressor complement in 3T3L1 cells and compared it with that in A549 cells. In general, the levels of different corepressor proteins were lower in 3T3L cells than in A549 cells (see Fig. S3 in the supplemental material). Interestingly, the level of NCoR1 was negligible in 3T3L cells compared to the levels of the other corepressor proteins examined, suggesting that the minimal downregulation of GR upon ligand binding observed in these cells was due to the lack of NCoR1.
Glucocorticoid-induced formation of the repression complex is mediated by chromatin looping of the intragenic elements to the transcriptional start site of the GR gene.
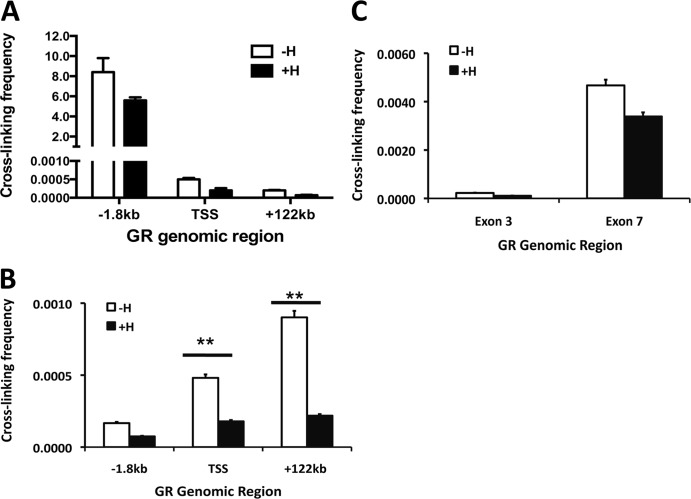
Our observation that GR is involved in the formation of an NCoR1-containing repression complex near the transcriptional start site of its own gene raises the important question of how distal intragenic chromatin regions are involved in the formation of this repression complex. Based on our data (Fig. 5B), we hypothesized that the repression complex is formed by looping of the intragenic chromatin region of the GR gene to the promoter-proximal region. To gain insight into the possible mechanism, we performed a chromosome conformation capture (3C) assay to evaluate the presence of a physical interaction between the intragenic chromatin region and the promoter-proximal region in A549 cells. One primer in the GR promoter-proximal region and one primer in the intragenic region flanking the putative ligation site were used to PCR amplify the potentially ligated fragments of DNA. The data (Fig. 7A) suggest that constitutive chromatin looping occurs between exon 6 and the promoter-proximal region of the GR gene, whereas no looping was observed with exon 8 (Fig. 7B). We also determined the looping frequency of exon 6/8 to the TSS and a region 122 kb downstream of the GR gene TSS as a control: the cross-linking frequency observed in Fig. 7A and B is nearly 5,000-fold lower than the cross-linking frequency between exon 6 and the promoter-proximal region at kb −1.8. Thus, our data demonstrate that looping is very limited between exon 6/8 and any of these GR genomic regions and that the observed cross-linking frequency is due to relatively infrequent interactions occurring between these DNA loci. As shown in Fig. 7C, the cross-linking frequency between exon 7 and the promoter-proximal region 1.8 kb upstream of the GR TSS was ∼1,300-fold lower than that between exon 6 and the promoter-proximal region at kb −1.8. These data clearly show that very little interaction is observed between exon 3/7 of the GR gene and the promoter-proximal regions. Thus, in the absence of ligand, a direct physical interaction between the intragenic region and the promoter-proximal region is present and transcription proceeds, suggesting that looping of the chromatin is inherent to the GR gene whether the gene is transcribed or not. However, in the presence of hormone, GR is recruited to the intragenic region and facilitates the formation of a repression complex at the transcriptional start site due to the physical interaction between the intragenic region and the transcriptional start site of the GR gene. This interaction of the distal intragenic GR (136 to 152 kb) binding site with the promoter of the GR gene confirms the authenticity of this distal intragenic chromatin region as a regulatory element for ligand-induced downregulation of the GR gene.
Fig 7.
Ligand-induced assembly of the repression complex is mediated by looping of the intragenic regulatory element to the TSS of the GR gene. A chromatin capture assay was performed after digesting fixed chromatin from vehicle- or dexamethasone-treated cells with the BglII restriction enzyme. Primers flanking either side of the putative interaction sites (restriction sites) were used to amplify a specific hybrid product by RT-PCR after ligation. (A) Interaction frequencies between exon 6 and different genomic loci on the GR gene. (B) Interaction frequencies between exon 8 and different genomic loci on the GR gene. (C) Interaction frequencies of exons 3 and 7 with the promoter (kb −1.8) of the GR gene (n = 3). **, P < 0.01; *, P < 0.05.
DISCUSSION
The regulation of hormone receptors is an important feature of physiologic control, since the sensitivity of target cells to a hormone is directly related to the receptor concentration. Several laboratories, including our own, have demonstrated that glucocorticoid receptor gene expression is repressed by exposure to its own ligand (19, 22, 24, 26). In addition to GR, homologous downregulation has also been documented for other steroid receptors. Repression of GR mRNA is time and dose dependent, and furthermore, the process is independent of ongoing protein synthesis, suggesting that downregulation of GR is mediated by the glucocorticoid receptor itself (29). Earlier studies using deletion analysis suggested that the coding region of the GR gene contains regulatory signals sufficient for downregulation of GR (30, 31). However, the molecular mechanisms responsible for this agonist-mediated repression of GR are not known. Since the biological effects of glucocorticoids are dependent on the presence of a functional receptor, this repression of the GR gene may also be associated with the tissue resistance that develops with chronic high-dose glucocorticoid treatment.
We have now investigated the molecular mechanism responsible for the repression of endogenous GR mRNA. Kinetic analysis following dexamethasone treatment showed a rapid decrease in the steady-state level of the GR primary transcript in several human, mouse, and rat cell lines as well as in various tissues. A 50 to 90% decrease in nascent GR RNA was observed, and the downregulation was sustained for 8 h after dexamethasone treatment (Fig. 1). Furthermore, our results show that glucocorticoid-mediated downregulation of GR transcripts is mediated by impeding transcription initiation, since a larger repression effect was seen with nascent RNA measurement. Additionally, we showed that repression depends on regulatory elements in the coding regions of the GR gene itself and is independent of the promoter, because the repression was also seen in U-2 OS cells, which have stably transfected hGR cDNA under the control of a CMV promoter. The similarities in repression between different cell lines and in various mouse tissues suggest that a conserved intragenic element is involved in the downregulation of the GR gene.
Functional GR is required for the rapid repression of GR mRNA, which confirms previous studies from our lab which showed that transcriptionally functional GR is required for this downregulation (29). ChIP analysis revealed that GR binds exons 6 and 8 of the GR gene in a ligand-dependent manner, and prolonged receptor occupancy was observed at both of these GR coding regions. The prolonged occupancy of the receptor at exons 6 and 8 indicated that the downregulation of GR mRNA might be mediated by RNA Pol II pausing. However, ChIP analysis showed that RNA Pol II is not enriched at exon 6 and/or exon 8, confirming that the repression of GR mRNA is not mediated by RNA Pol II pausing at these regions (Fig. 3). We also examined the existence of a functional nGRE in exon 6 of the GR gene that might be instrumental in active repression. This putative nGRE is similar to the IR1 nGRE described by Surjit et al. (12), with a tolerable change in one base, and this nGRE is conserved in humans, mice, and rats. In vitro investigation of the IR1 nGRE in exon 6 by site-directed mutagenesis showed that this nGRE is critical for glucocorticoid-induced repression of GR (Fig. 4). Interestingly, even though there was recruitment of GR and NCoR1 to exon 8, there were no putative GR binding sites observed in exon 8, and thus it had no repressive activity in the reporter assay. We believe that the recruitment of GR and NCoR1 to exon 8 might be through an alternative mechanism and have some other unknown function.
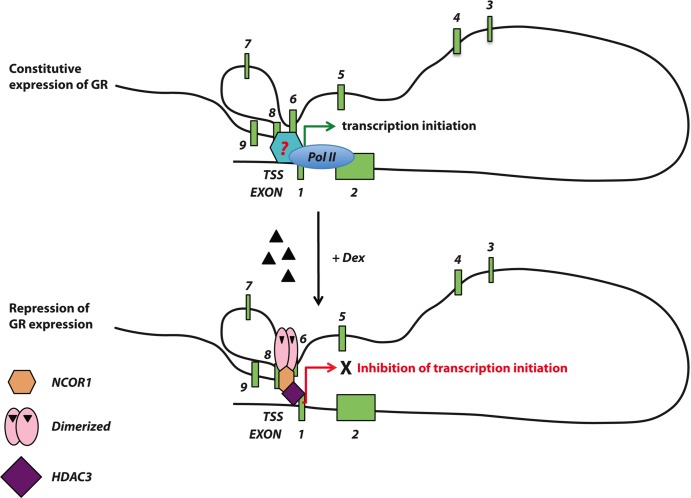
The data presented here indicate that the ligand-induced repression of the GR gene is mediated by transcriptional repression by promoting the assembly of a cis-acting GR-NCoR1-HDAC3 repression complex (Fig. 5). Glucocorticoid-induced GR repression was prevented by concomitant knockdown of NCoR1 but was not significantly affected by knockdown of SMRT. These findings demonstrate that NCoR1 is instrumental in glucocorticoid-induced nGRE-mediated repression (Fig. 6). Based on the literature, the variation in NCoR1 protein levels after dexamethasone treatment (Fig. 6B, top panel) might be due to turnover of the NCoR1 repression complex by the 26S proteasome (38, 39), but we have not directly investigated this issue in our studies. Intriguingly, ligand-induced repression of the GR gene was inhibited by the HDAC3 inhibitor TSA, suggesting that HDAC3 is a key player in the process (Fig. 6C). The formation of the repression complex requires the presence of a glucocorticoid agonist, which raises the important question of whether binding of GR to nGRE may induce an allosteric conformational change of GR enabling it to interact with corepressors in the presence of ligand. The recruitment of GR and NCoR1 to the distal intragenic chromatin regions combined with the ability of chromatin conformation capture assays to demonstrate that these distant sites are physically associated with promoter-proximal regions suggests that these intragenic chromatin regions play an important role in glucocorticoid-mediated downregulation of GR. Interestingly, the interaction between the two chromatin loci is not dependent on the presence of hormone. Furthermore, the addition of hormone maintains the looping conformation but results in the assembly and formation of a repression complex at the GR gene start site. These data suggest that active looping may be essential for constitutive expression of the GR gene in the absence of ligand (Fig. 8). Several studies have shown the existence of transcriptional regulatory sites at distal regions several kilobases removed from the transcriptional start site (40). For example, Hakim et al. have shown that the GRE proximal to the Lcn2 promoter apparently functions to regulate both the Lcn2 gene and the distal Ciz1 gene and that the interaction between the two loci is not dependent on the presence of glucocorticoid (41). Combined, these findings demonstrate that higher-order chromatin organization allows a DNA sequence to function both as a coding exon, in the absence of hormone, and as a regulatory element (42), in the presence of hormone. Since a cell type-specific loop structure is associated with transcription, the correlation between repression and the loop structure might play an important role in determining glucocorticoid sensitivity.
Fig 8.
Schematic representation of ligand-induced downregulation of the GR gene. In the absence of hormone, the higher-order chromatin structure of the GR gene is maintained, and the GR gene is constitutively expressed. Upon binding of glucocorticoids, GR binds the nGRE on exon 6; since the chromatin loop is still maintained, GR recruits NCoR1 and HDAC3 to the promoter-proximal region to form a repression complex and inhibit GR transcription.
In conclusion, our study demonstrates that repression of the GR gene is mediated by the agonist-bound GR complex via intragenic regulatory elements present in the 3′ region of the GR gene by looping of the chromatin between the intragenic regulatory element and the transcriptional start site, present several kilobases upstream. To our knowledge, this is the first functional demonstration that DNA sequences can act as a protein coding sequence in the absence of hormone but repress the expression of the same gene in the presence of hormone. This new mechanism of repression also implies that by acquiring an intragenic regulatory element for autoregulation, the GR concentration can be downregulated coordinately with excess ligand, regardless of the various combinatorial associations of tissue-specific transcription factors. While hormone-induced downregulation of GR represents a mechanism for maintaining glucocorticoid homeostasis in normal cells, it also has the potential to limit therapeutic responses to glucocorticoids under inflammatory and malignant conditions. Due to the chronic nature of inflammatory conditions, treatment paradigms involve long-term glucocorticoid administration, which may lead to constitutive repression of GR gene transcription leading to glucocorticoid resistance.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
Footnotes
Published ahead of print 19 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01151-12.
REFERENCES
- 1. Barnes PJ. 1998. Anti-inflammatory actions of glucocorticoids: molecular mechanisms. Clin. Sci. (Lond.) 94:557–572 [DOI] [PubMed] [Google Scholar]
- 2. Sapolsky RM, Romero LM, Munck AU. 2000. How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocr. Rev. 21:55–89 [DOI] [PubMed] [Google Scholar]
- 3. Rhen T, Cidlowski JA. 2005. Antiinflammatory action of glucocorticoids—new mechanisms for old drugs. N. Engl. J. Med. 353:1711–1723 [DOI] [PubMed] [Google Scholar]
- 4. Schacke H, Docke WD, Asadullah K. 2002. Mechanisms involved in the side effects of glucocorticoids. Pharmacol. Ther. 96:23–43 [DOI] [PubMed] [Google Scholar]
- 5. Schmidt S, Rainer J, Ploner C, Presul E, Riml S, Kofler R. 2004. Glucocorticoid-induced apoptosis and glucocorticoid resistance: molecular mechanisms and clinical relevance. Cell Death Differ. 11(Suppl 1):S45–S55 [DOI] [PubMed] [Google Scholar]
- 6. Stanbury RM, Graham EM. 1998. Systemic corticosteroid therapy—side effects and their management. Br. J. Ophthalmol. 82:704–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evans RM. 1988. The steroid and thyroid hormone receptor superfamily. Science 240:889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beato M. 1989. Gene regulation by steroid hormones. Cell 56:335–344 [DOI] [PubMed] [Google Scholar]
- 9. Freedman LP. 1992. Anatomy of the steroid receptor zinc finger region. Endocr. Rev. 13:129–145 [DOI] [PubMed] [Google Scholar]
- 10. Necela BM, Cidlowski JA. 2004. Mechanisms of glucocorticoid receptor action in noninflammatory and inflammatory cells. Proc. Am. Thorac. Soc. 1:239–246 [DOI] [PubMed] [Google Scholar]
- 11. Yamamoto KR. 1985. Steroid receptor regulated transcription of specific genes and gene networks. Annu. Rev. Genet. 19:209–252 [DOI] [PubMed] [Google Scholar]
- 12. Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, Li M, Chambon P. 2011. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell 145:224–241 [DOI] [PubMed] [Google Scholar]
- 13. Bourgeois S, Newby RF. 1979. Correlation between glucocorticoid receptor and cytolytic response of murine lymphoid cell lines. Cancer Res. 39:4749–4751 [PubMed] [Google Scholar]
- 14. Vanderbilt JN, Miesfeld R, Maler BA, Yamamoto KR. 1987. Intracellular receptor concentration limits glucocorticoid-dependent enhancer activity. Mol. Endocrinol. 1:68–74 [DOI] [PubMed] [Google Scholar]
- 15. Bloomfield CD, Smith KA, Peterson BA, Munck A. 1981. Glucocorticoid receptors in adult acute lymphoblastic leukemia. Cancer Res. 41:4857–4860 [PubMed] [Google Scholar]
- 16. Silva CM, Powell-Oliver FE, Jewell CM, Sar M, Allgood VE, Cidlowski JA. 1994. Regulation of the human glucocorticoid receptor by long-term and chronic treatment with glucocorticoid. Steroids 59:436–442 [DOI] [PubMed] [Google Scholar]
- 17. Alarid ET. 2006. Lives and times of nuclear receptors. Mol. Endocrinol. 20:1972–1981 [DOI] [PubMed] [Google Scholar]
- 18. Hoeck W, Rusconi S, Groner B. 1989. Down-regulation and phosphorylation of glucocorticoid receptors in cultured cells. Investigations with a monospecific antiserum against a bacterially expressed receptor fragment. J. Biol. Chem. 264:14396–14402 [PubMed] [Google Scholar]
- 19. Cidlowski JA, Cidlowski NB. 1981. Regulation of glucocorticoid receptors by glucocorticoids in cultured HeLa S3 cells. Endocrinology 109:1975–1982 [DOI] [PubMed] [Google Scholar]
- 20. Danielsen M, Stallcup MR. 1984. Down-regulation of glucocorticoid receptors in mouse lymphoma cell variants. Mol. Cell. Biol. 4:449–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lacroix A, Bonnard GD, Lippman ME. 1984. Modulation of glucocorticoid receptors by mitogenic stimuli, glucocorticoids and retinoids in normal human cultured T cells. J. Steroid Biochem. 21:73–80 [DOI] [PubMed] [Google Scholar]
- 22. Schlechte JA, Ginsberg BH, Sherman BM. 1982. Regulation of the glucocorticoid receptor in human lymphocytes. J. Steroid Biochem. 16:69–74 [DOI] [PubMed] [Google Scholar]
- 23. Svec F, Rudis M. 1981. Glucocorticoids regulate the glucocorticoid receptor in the AtT-20 cell. J. Biol. Chem. 256:5984–5987 [PubMed] [Google Scholar]
- 24. Tornello S, Orti E, De Nicola AF, Rainbow TC, McEwen BS. 1982. Regulation of glucocorticoid receptors in brain by corticosterone treatment of adrenalectomized rats. Neuroendocrinology 35:411–417 [DOI] [PubMed] [Google Scholar]
- 25. Dong Y, Poellinger L, Gustafsson JA, Okret S. 1988. Regulation of glucocorticoid receptor expression: evidence for transcriptional and posttranslational mechanisms. Mol. Endocrinol. 2:1256–1264 [DOI] [PubMed] [Google Scholar]
- 26. Rosewicz S, McDonald AR, Maddux BA, Goldfine ID, Miesfeld RL, Logsdon CD. 1988. Mechanism of glucocorticoid receptor down-regulation by glucocorticoids. J. Biol. Chem. 263:2581–2584 [PubMed] [Google Scholar]
- 27. Vedeckis WV, Ali M, Allen HR. 1989. Regulation of glucocorticoid receptor protein and mRNA levels. Cancer Res. 49:2295s–2302s [PubMed] [Google Scholar]
- 28. Govindan MV, Pothier F, Leclerc S, Palaniswami R, Xie B. 1991. Human glucocorticoid receptor gene promoter-homologous down regulation. J. Steroid Biochem. Mol. Biol. 40:317–323 [DOI] [PubMed] [Google Scholar]
- 29. Burnstein KL, Jewell CM, Cidlowski JA. 1990. Human glucocorticoid receptor cDNA contains sequences sufficient for receptor down-regulation. J. Biol. Chem. 265:7284–7291 [PubMed] [Google Scholar]
- 30. Burnstein KL, Jewell CM, Sar M, Cidlowski JA. 1994. Intragenic sequences of the human glucocorticoid receptor complementary DNA mediate hormone-inducible receptor messenger RNA down-regulation through multiple mechanisms. Mol. Endocrinol. 8:1764–1773 [DOI] [PubMed] [Google Scholar]
- 31. Okret S, Poellinger L, Dong Y, Gustafsson JA. 1986. Down-regulation of glucocorticoid receptor mRNA by glucocorticoid hormones and recognition by the receptor of a specific binding sequence within a receptor cDNA clone. Proc. Natl. Acad. Sci. U. S. A. 83:5899–5903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wallace AD, Cidlowski JA. 2001. Proteasome-mediated glucocorticoid receptor degradation restricts transcriptional signaling by glucocorticoids. J. Biol. Chem. 276:42714–42721 [DOI] [PubMed] [Google Scholar]
- 33. Lu NZ, Cidlowski JA. 2005. Translational regulatory mechanisms generate N-terminal glucocorticoid receptor isoforms with unique transcriptional target genes. Mol. Cell 18:331–342 [DOI] [PubMed] [Google Scholar]
- 34. Nissen RM, Yamamoto KR. 2000. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 14:2314–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T. 2007. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2:1722–1733 [DOI] [PubMed] [Google Scholar]
- 36. Bamberger CM, Chrousos GP. 1995. The glucocorticoid receptor and RU 486 in man. Ann. N. Y. Acad. Sci. 761:296–310 [DOI] [PubMed] [Google Scholar]
- 37. Burnstein KL, Bellingham DL, Jewell CM, Powell-Oliver FE, Cidlowski JA. 1991. Autoregulation of glucocorticoid receptor gene expression. Steroids 56:52–58 [DOI] [PubMed] [Google Scholar]
- 38. Glass CK, Saijo K. 2010. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat. Rev. Immunol. 10:365–376 [DOI] [PubMed] [Google Scholar]
- 39. Perissi V, Jepsen K, Glass CK, Rosenfeld MG. 2010. Deconstructing repression: evolving models of co-repressor action. Nat. Rev. Genet. 11:109–123 [DOI] [PubMed] [Google Scholar]
- 40. Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38:1289–1297 [DOI] [PubMed] [Google Scholar]
- 41. Hakim O, John S, Ling JQ, Biddie SC, Hoffman AR, Hager GL. 2009. Glucocorticoid receptor activation of the Ciz1-Lcn2 locus by long range interactions. J. Biol. Chem. 284:6048–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Birnbaum RY, Clowney EJ, Agamy O, Kim MJ, Zhao J, Yamanaka T, Pappalardo Z, Clarke SL, Wenger AM, Nguyen L, Gurrieri F, Everman DB, Schwartz CE, Birk OS, Bejerano G, Lomvardas S, Ahituv N. 2012. Coding exons function as tissue-specific enhancers of nearby genes. Genome Res. 22:1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.