Abstract
Yju2 is an essential splicing factor required for the first catalytic step after the action of Prp2. We dissected the structure of Yju2 and found that the amino (Yju2-N) and carboxyl (Yju2-C) halves of the protein can be separated and reconstituted for Yju2 function both in vivo and in vitro. Yju2-N has a weak affinity for the spliceosome but functions in promoting the first reaction, with the second reaction being severely impeded. The association of Yju2-N with the spliceosome is stabilized by the presence of Yju2-C at both the precatalytic and postcatalytic stages. Strikingly, Yju2-N supported a low level of the second reaction even in the absence of Prp16. Prp16 is known to mediate destabilization of Yju2 and Cwc25 after the first reaction to allow progression of the second reaction. We propose that in the absence of the C domain, Yju2-N is not stably associated with the spliceosome after lariat formation, and thus bypasses the need for Prp16. We also showed, by UV cross-linking, that Yju2 directly contacts U2 snRNA primarily in the helix II region both pre- and postcatalytically and in the branch-binding region only at the precatalytic stage, suggesting a possible role for Yju2 in positioning the branch point during the first reaction.
INTRODUCTION
Introns are removed from precursor mRNA via two steps of transesterification reactions that form lariat intermediates and products. The reactions are catalyzed by a large ribonucleoprotein complex, the spliceosome, which consists of five small nuclear RNAs (snRNAs), U1, U2, U4, U5, and U6, in the form of small nuclear ribonucleoprotein particles (snRNPs), and numerous protein factors (for reviews, see references 1 to 4). The spliceosome is a highly dynamic structure, assembled by sequential binding of the snRNAs in the order U1, U2, and then U4/U6.U5 as a preformed tri-snRNP. U1 and U2 play roles in mediating the recognition of the 5′ splice site and the branch site, respectively, through base pairing between the snRNAs and the intron sequences. Following binding of the tri-snRNP, the spliceosome undergoes a major structural rearrangement, releasing U1 and U4 and forming new base pairs between U6 and the 5′ splice site and between U2 and U6. A protein complex associated with Prp19, called NTC (nineteen complex), is added to the spliceosome to stabilize the association of U5 and U6 with the spliceosome (5–7), and this proceeds with catalytic activation of the spliceosome. The NTC remains associated with the spliceosome until completion of the reaction.
The spliceosome undergoes extensive remodeling throughout the splicing pathway (3, 8). Eight DEXD/H-box ATPases are required to mediate structural changes of the spliceosome during the splicing process (9). Prp2 and Prp16 are required for the first and the second catalytic step, respectively. Several other protein factors are required to promote the catalytic reactions following their actions. Prp2 has recently been shown to function in destabilizing the U2 components SF3a and SF3b (8, 10, 11). SF3b is known to interact with the branch site, presumably in stabilizing base pair interactions between U2 and the branch site during spliceosome formation. SAP155 and its yeast homologue Hsh155, a subunit of SF3b, have both been shown to cross-link to the intron sequence flanking the branch site (12, 13). Destabilization of SF3b likely leads to exposure of the branch point to initiate the catalytic reaction. At least two proteins, Yju2 and Cwc25, are required to promote the reaction independently of ATP, possibly for positioning the branch point and the 5′ splice site (14, 15). Yju2 and Cwc25 bind tightly to the spliceosome after the first reaction and need to be destabilized for repositioning of splice sites. Prp16 is required for destabilization of Yju2 and Cwc25 (16) and for the binding of Slu7, Prp18, and Prp22, to promote the second reaction in an ATP-independent manner (17–20). A schematic of the spliceosome pathway is shown in Fig. 1.
Fig 1.
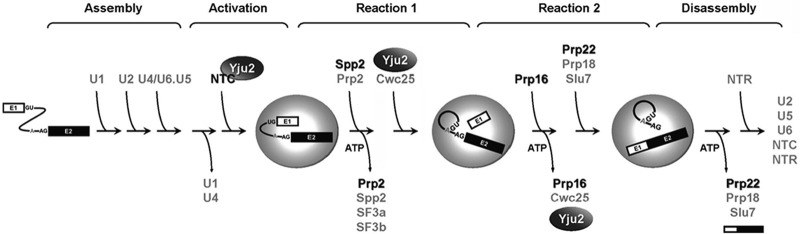
Schematic of the spliceosome pathway showing remodeling of the spliceosome during catalytic steps.
How DEXD/H-box proteins mediate remodeling of the spliceosome is an enigma. Prp16, Prp22, Prp43, and Brr2 have been demonstrated to unwind RNA duplexes in vitro (21–25), but whether the unwinding activity is associated with their function on the spliceosome is not clear. DEXD/H-box proteins are also known to displace proteins from RNA in vitro in simple RNA-protein complexes (26–28). In splicing, Sub2, Prp5, and Prp28 have been implicated in such a role (29–31). Prp22, which is required for the release of spliced mRNA from the spliceosome (32), is among the best-characterized splicing DEXD/H-box proteins with regard to their functions. Prp22 has been shown to bind to the pre-mRNA in a region upstream from the 3′ splice site during the second catalytic step (13). After exon ligation, Prp22 is placed on the downstream exon, where it translocates along the mRNA in a 3′-to-5′ direction to disrupt contacts with U5 snRNP (33). Whether Prp22 disrupts the interaction of mRNA with U5 snRNP components or unwinds U5 mRNA base pairing was not investigated. We recently showed that Prp2 shares a similar mechanism in displacing SF3a/b. Prp2 is first recruited to the spliceosome via interaction with Brr2, is translocated to the intron downstream of the branch site, and moves in the 3′-to-5′ direction to dislodge SF3a/b (34). The mechanism underlying Prp16-mediated displacement of Yju2 and Cwc25 has not been elucidated.
Studies of U2/U6 helix I in the spliceosome catalytic core have revealed genetic interactions between PRP16 and U2/U6 helix I (35). Mutations that weaken U2/U6 helix I were found to suppress the PRP16 cold-sensitive prp16-302 mutation. This led to the suggestion that U2/U6 helix I undergoes dynamic structural changes during catalytic steps and that Prp16 may be involved in destabilization of helix I. In view of Prp16 being responsible for destabilization of Yju2 and Cwc25, it is also possible that the Prp16-U2/U6 helix I interaction is mediated through Yju2 and/or Cwc25.
Yju2 has been shown to interact with the NTC components Ntc90 and Ntc77 and can be recruited to the spliceosome prior to or after the action of Prp2 (14, 36). In contrast, Cwc25 binds to the spliceosome only after the action of Prp2 and is dependent on the presence of Yju2 (15). Cwc25 can cross-link to the intron sequence near the branch site, and its association with the spliceosome is affected by branch point mutations, suggesting a role for Cwc25 in positioning the branch point for lariat formation (16, 34). How Yju2 functions in the first reaction is not known. Conceivably, its binding to the spliceosome may set the spliceosome in a proper conformation for the binding of Cwc25.
In this study, we dissected the structure of Yju2 for functional studies. We found that Yju2 can be separated into two functional domains. The amino half (N) of the protein is evolutionarily conserved, but the carboxyl (C) half is not. The two domains can be reconstituted in vivo for cellular growth and in vitro for its function in the splicing reaction. The conserved N domain is partially functional in splicing and has a low affinity for the spliceosome. The C domain binds the spliceosome more tightly, and its presence stabilizes the association of the N domain with the spliceosome. Strikingly, the N domain alone promoted a low level of the second reaction in the absence of Prp16, due to “self-destabilization” from the spliceosome. This finding not only provides direct evidence that structural changes in the catalytic core of the spliceosome mediated by Prp16 occur via regulation of the interaction of Yju2 and Cwc25 with the spliceosome but also suggests a role for Yju2, specifically the C domain, in the second step of splicing. We also showed that Yju2 directly contacts U2 snRNA prior to and after the first reaction by UV-cross-linking analysis. While most cross-links were mapped to the helix II region, one was mapped to the branch site-binding region, but only at the precatalytic stage. These results suggest that Yju2 may play a role in stabilizing the structure of the spliceosome catalytic core and positioning of the branch point during the first catalytic step.
MATERIALS AND METHODS
Preparation of total yeast cell lysates by glass beads.
Yeast cell pellets (3 mg) were mixed with 50 μl of 2× lysis buffer (0.25 M Tris-HCl [pH 6.8], 4% SDS, 0.28 M β-mercaptoethanol) and 0.1 ml of acid-washed glass beads. The mixture was shaken vigorously with a Vortex mixer for 5 min, boiled at 100°C for 2 min, and shaken again for another 3 min. After centrifugation in an Eppendorf centrifuge at top speed for 1 min, the supernatant was collected.
Splicing extracts, substrates, and splicing assays.
Yeast whole-cell extracts were prepared according to Cheng et al. (37). Actin precursors were synthesized in vitro using SP6 RNA polymerase, and splicing assays were carried out according to Cheng and Abelson (38). The splicing efficiency of the first and second reactions was calculated as the ratio of L + M to P + L + M (where L is the molar amount of lariat-IVS-E2, P is that of pre-mRNA, and M is that of mRNA) for the first step and the ratio of M to P + L + M for the second step. The amount of each RNA species was measured using a phosphorimager and normalized to the number of uridine residues for each RNA species.
Immunoprecipitation and immunodepletion.
Immunoprecipitation of the spliceosome with anti-Ntc20, antihemagglutinin (anti-HA), or anti-V5 antibody was performed as described by Liu et al. (14). For each 20 μl of the reaction mixture, 2 μl of anti-Ntc20 antibody, 20 μl of anti-HA antibody, and 2 μl of anti-V5 antibody were used.
UV cross-linking of Yju2 to snRNA.
Splicing reactions were carried out with 2 nM substrate RNA in Cwc25- or Prp16-depleted Yju2-HA extracts for 20 min. The reaction mixtures were spread onto a tissue culture plate placed on ice, irradiated with 254-nm UV at 0.8 J/cm2 (Stratalinker 1800), and then collected into a microcentrifuge tube. After addition to a mixture containing (final concentrations) 1% SDS, 1% Triton X-100, and 0.1 M dithiothreitol (DTT), the mixture was boiled for 2 min and allowed to cool to room temperature in a water bath. The mixtures were diluted 10-fold with a buffer containing 50 mM Tris-HCl (pH 7.5), 300 mM NaCl, 1 mM EDTA, 0.05% NP-40, and 0.2 mg/ml tRNA and subjected to immunoprecipitation with anti-HA antibody. The precipitates were washed with the same buffer without tRNA and then digested with proteinase K. RNA was extracted for Northern blotting or primer extension analysis. For Northern blotting, RNA was isolated from 1 ml of splicing reaction mixture after UV irradiation and fractionated on 8 M urea–5% (29:1) polyacrylamide gels. For primer extension analysis, RNA was isolated from 0.5 ml of splicing reaction mixture after UV irradiation. Each RNA sample was mixed with 2 × 105 cpm of 5′-, 32P-labeled primer U2-A or U2-B, denatured by heating at 70°C for 5 min, and quickly chilled on ice. Primer extension reactions were carried out with avian myeloblastosis virus (AMV) reverse transcriptase (Promega) at 42°C for 1.5 h. Reaction products were analyzed on 8 M urea–12% (19:1) polyacrylamide gels and visualized by autoradiography.
RESULTS
Dissection of Yju2 N and C domains for interactions with NTC components.
Sequence alignment of Yju2 orthologs has revealed a stretch of 21 amino acid residues in the middle region of the protein (amino acid residues 125 to 145) that are found only in Saccharomyces cerevisiae and not in other species, suggesting that the amino and carboxyl halves of the protein may form distinct modules. While the N-terminal domain of Yju2 is evolutionarily conserved, the C-terminal domain is highly divergent (see Fig. S1 in the supplemental material). Yju2 was thus divided into two halves, consisting of amino acids 1 to 130 (Yju2-N) and 125 to 278 (Yju2-C), for functional characterization.
We previously showed that Yju2 interacts with two NTC components, Ntc90/Syf1 and Ntc77/Syf3/Clf1 (14), and that Ntc90 is required for the recruitment of Yju2 to the spliceosome (36). In this study, we first examined whether Yju2 interacts with these two proteins via specific domains by yeast two-hybrid assays. Full-length Yju2, Yju2-N, and Yju2-C were individually fused to the GAL4 activation domain, and Ntc90 and Ntc77 were fused to the GAL4-DNA binding domain. The interaction within each pair of fusions was assayed by the activation of Ade2 expression. Figure 2A shows that Yju2-C interacted strongly with Ntc77 to nearly the same level as full-length Yju2 but did not interact with Ntc90. In contrast, Yju2-N showed weak interaction with Ntc90 but did not interact with Ntc77. The N and C domains did not interact with each other in trans (see Fig. S2 in the supplemental material). These results suggest that the Yju2 N domain interacts with Ntc90 and the Yju2 C domain interacts with Ntc77 of the NTC.
Fig 2.
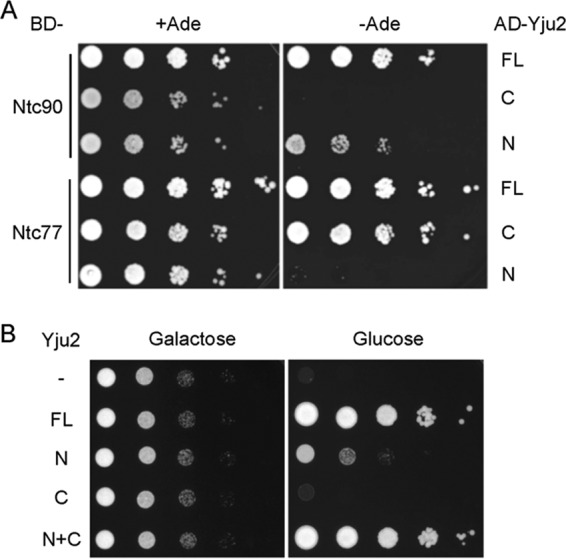
Analyses of Yju2 N and C domains. (A) Two-hybrid interactions of Yju2 domains with Ntc90 and Ntc77. The N and C domains of Yju2 and full-length Yju2 were fused to the GAL4-activation domain (AD), and Ntc77 and Nt90 were fused to the GAL4-DNA binding domain (BD). The interactions were assayed in terms of the activation of ADE2 under the control of the GAL2 promoter. FL, full-length; C, C domain; N, N domain. (B) Growth analysis. TC2 cells were transformed with pRS414 and pRS415 vectors alone (−), with pRS414.YJU2 and pRS415 (FL), with pRS414.YJU2-N and pRS415 (N), with pRS414.YJU2-C and pRS415 (C), or with pRS414.YJU2-N and pRS415.YJU2-C (N+C). Cells were cultured in raffinose-containing synthetic complete medium and then spotted onto galactose- or glucose-containing plates in 10-fold serial dilutions. FL, full-length; N, N domain; C, C domain; N+C, combination of N and C domains.
The N and C domains of Yju2 can function in trans for cellular growth and for splicing.
The results above showing that the C domain of Yju2 interacts with Ntc77 nearly as strongly as the full-length protein despite the sequence not being evolutionarily conserved and that the conserved N domain interacts with Ntc90 less strongly than the full-length protein were unexpected. Therefore, we next examined the requirement of the N and C domains of Yju2 for growth. A yeast strain, TC2, was constructed such that the YJU2 gene is under the control of the inducible GAL1 promoter. YJU2 is essential for cellular viability, and repression of the GAL1 promoter results in cellular lethality. TC2 was transformed separately with a centromeric plasmid vector alone, with a plasmid carrying full-length YJU2, YJU2-N, or YJU2-C, or with two plasmids each carrying YJU2-N and YJU2-C, respectively, all under the control of the authentic YJU2 promoter. Cells grown in glucose-containing medium expressed Yju2 proteins only from the plasmid. Figure 2B shows that cells expressing only the N domain of Yju2 were viable but grew more slowly than those expressing the full-length protein, suggesting that the C domain of Yju2 is dispensable but plays an auxiliary role. Cells expressing the C domain alone, however, were inviable. Strikingly, expression of both N and C domains of Yju2 recovered full growth of YJU2-repressed cells. Thus, although the C domain of Yju2 by itself is not functional, it can act together with the N domain to alleviate the growth defect caused by truncation of the C domain. This suggests that the N domain and C domain each form separate functional modules that act in concert to assume the function of Yju2.
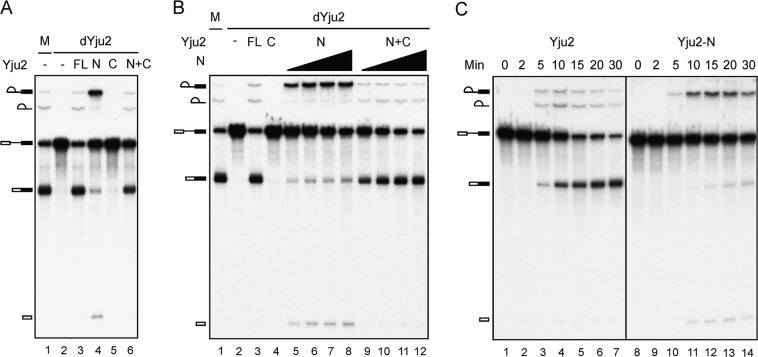
To correlate the growth phenotype with the function of Yju2 in splicing, we set up an in vitro complementation assay for the splicing reaction. Yju2 was depleted from extracts using anti-Yju2 antibody, and the extract was complemented with purified recombinant Yju2 proteins. Depletion of Yju2 completely abolished the splicing activity (Fig. 3A, lane 2), but addition of recombinant full-length Yju2 efficiently restored the splicing activity of the Yju2-depleted extract (lane 3). Complementation with the N domain of Yju2 alone partly restored the splicing activity, with the second reaction being severely impeded, resulting in the accumulation of splicing intermediates (Fig. 3A, lane 4). This result correlates with the growth defect seen in YJU2-N cells. The C domain alone consistently failed to complement the Yju2-depleted extracts (Fig. 3A, lane 5); nevertheless, the splicing activity was recovered in the presence of both the N and C domains of Yju2 (lane 6), suggesting that the N and C domains of Yju2 can act together to function in the splicing reaction as the full-length Yju2 protein, regardless of being covalently unlinked.
Fig 3.
Complementation of Yju2-depleted extracts. (A) Splicing reactions were carried out for 30 min in mock-depleted (lane 1) or Yju2-depleted extracts without addition (lane 2) or with the addition of 0.3 μM Yju2-FL (lane 3), 1.5 μM Yju2-N (lane 4), 0.4 μM Yju2-C (lane 5), or a combination of 1.5 μM Yju2-N and 0.4 μM Yju2-C (lane 6). (B) Splicing was carried out for 30 min in mock-depleted (lane 1) or Yju2-depleted extracts without addition (lane 2) or with the addition of 0.3 μM Yju2-FL (lane 3), 0.4 μM Yju2-C (lane 4), or 0.4 μM (lanes 5 and 9), 0.8 μM (lanes 6 and 10), 1.5 μM (lanes 7 and 11), or 3.0 μM (lanes 8 and 12) Yju2-N without Yju2-C addition (lanes 5 to 8) or with the addition of 0.4 μM Yju2-C (lanes 9 to 12). (C) Splicing was carried out in Yju2-depleted extracts with the addition of 0.2 μM Yju2-FL (lanes 1 to 7) or Yju2-N (lanes 8 to 14) for 0 to 30 min. dYju2, Yju2 depletion; M, mock treatment; FL, full length; N, Yju2-N; C, Yju2-C.
We previously showed that Yju2 is required for the first catalytic reaction after the action of Prp2 and then undergoes Prp16-mediated destabilization from the spliceosome to allow the binding of the second-step factors Slu7, Prp22, and Prp18 (16). It is interesting that Yju2-N is capable of promoting the first catalytic reaction but results in a retarded second step, implying a function for Yju2 in the second step. To more carefully characterize the effect of Yju2-N on the two catalytic steps, we titrated the amount of Yju2-N for complementation of Yju2-depleted extracts (Fig. 3B). Quantification of the complementation activity revealed that while 0.3 μM full-length Yju2 supported splicing to nearly 80% completion of both steps (Fig. 3B, lane 3), Yju2-N supported splicing only at higher concentrations. Both steps of splicing increased as amounts of Yju2-N increased from 0.4 to 3.0 μM, attaining up to 60% completion of the first-step reaction and 30% of the second step (Fig. 3B, lane 8), indicating a defect in Yju2-N in both steps but a more severe defect in the second step. In the presence of 0.4 μM Yju2-C, splicing was achieved at near 80% completion in both steps when Yju2-N was added at 3.0 μM (Fig. 3B, lane 12), suggesting that the C domain has a function in the second step. A kinetics study also revealed serious impairment of the second step with Yju2-N protein (Fig. 3C). While the second reaction progressed in concert with the first reaction with full-length Yju2, it was notably retarded with the Yju2-N protein. Extracts depleted of Yju2 could not be complemented by Yju2-C protein alone at up to 4.0 μM the protein (see Fig. S3 in the supplemental material). These results demonstrate that the C domain of Yju2 has auxiliary roles in Yju2 function, primarily in facilitating the second step and also moderately enhancing the first reaction, acting in concert with Yju2-N. The N domain alone could promote the first reaction, but not as efficiently as the full-length protein. Addition of increasing amounts of Yju2-N gradually increased the level of the first reaction, but the level did not reach that of the full-length protein even with a 10-fold excess of full-length Yju2. In agreement with this result, slight growth improvement was observed when cells overexpressed the Yju2-N protein (see Fig. S4 in the supplemental material).
The C domain of Yju2 stabilizes the binding of the N domain to the spliceosome.
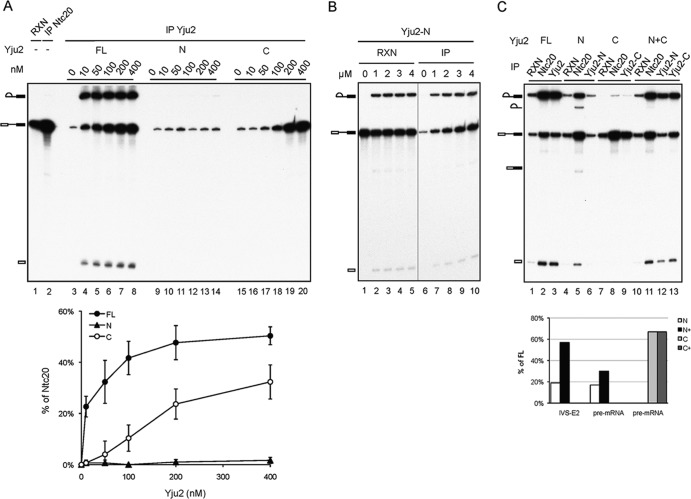
To investigate how the N and C domains of Yju2 coordinate to promote the first reaction, we next examined whether each domain alone can bind the spliceosome. The splicing reaction was carried out in Yju2-depleted extracts for 20 min, and the reaction mixture was then depleted of ATP. Recombinant HA-tagged full-length Yju2, V5-tagged N domain, or HA-tagged C domain was added to the reaction mixture at concentrations of 0 to 400 nM, and the Yju2-associated spliceosome was precipitated with anti-HA or anti-V5 antibody. Prior to the addition of Yju2 proteins, the reaction mixture was also precipitated with anti-Ntc20 antibody for postactivation spliceosomes. At 10 nM, full-length Yju2 was able to promote efficient splicing, and the amount of Yju2 associated with the precatalytic spliceosome increased with increasing concentrations of Yju2 added (Fig. 4A, lanes 3 to 8). However, the amount of Yju2 associated with splicing intermediates remained unchanged, indicating that some other component required for catalysis was limiting. The amount of Yju2-N binding to the spliceosome was near the background level at concentrations up to 400 nM (Fig. 4A, lanes 9 to 14), and only a tiny amount of splicing intermediates was detected at 400 nM (lane 14). Binding of Yju2-C to the spliceosome was observed significantly beyond 100 nM and increased with increasing concentrations of Yju2-C (Fig. 4A, lanes 15 to 20). Although different antibodies may precipitate the spliceosome with different efficiencies, we observed approximately the same precipitation efficiency of the spliceosome with anti-HA and anti-V5 antibodies for Yju2 tagged with HA and V5, respectively (data not shown). The binding affinities for the spliceosome of Yju2, Yju2-N, and Yju2-C are shown on the graph in Fig. 4A, in which the fractions of Yju2-containing activated spliceosome, calculated as the ratio of the total amount of Yju2-bound spliceosome to that of the Ntc20-associated spliceosome, are plotted against the concentration of Yju2 added. The association of Yju2-N with the spliceosome was more pronounced when the amount of Yju2-N was increased to the micromolar range (Fig. 4B). These results indicate that both the N and C domains of Yju2 can individually bind to the spliceosome, possibly via interaction with Ntc90 and Ntc77, respectively. The N domain, although it has a lower affinity for the spliceosome, can partially function in promoting the first reaction, as its binding to the spliceosome allows subsequent binding of Cwc25 (see Fig. S5 in the supplemental material). In contrast, the C domain has a higher affinity for the spliceosome, but its association with the spliceosome is inadequate for Cwc25 recruitment (see Fig. S5) and consequently fails to promote the reaction.
Fig 4.
Binding affinities of Yju2 domains to the spliceosome. (A) (Top) Splicing reactions were carried out in Yju2-depleted extracts for 20 min, and ATP was then depleted by addition of 10 mM glucose and incubation for 5 min. Following the addition of recombinant HA-tagged Yju2, V5-tagged Yju2-N, or HA-tagged Yju2-C at final concentrations of 0, 10, 50 100, 200, and 400 nM, the reaction mixtures were incubated for 10 min and precipitated with anti-HA (lanes 3 to 8 and 15 to 20) or anti-V5 (lanes 9 to 14) antibody. (Bottom) Binding affinities for the spliceosome of Yju2, Yju2-N, and Yju2-C. The fractions of Yju2-containing activated spliceosome, calculated as the ratio of the total amount of Yju2-bound spliceosome to that of the Ntc20-associated spliceosome, are plotted against the concentration of Yju2 added. (B) Splicing reactions were carried out in Yju2- and Prp16-depleted extracts supplemented with 0, 1, 2, 3, and 4 μM V5-tagged Yju2-N (lanes 1 to 5), and each reaction mixture was precipitated with anti-V5 antibody. (C) Splicing reactions were carried out in Yju2- and Prp16-depleted extracts supplemented with 0.3 μM HA-tagged full-length Yju2 (lanes 1 to 3), 1.5 μM V5-tagged Yju2-N (lanes 4 to 6), 0.4 μM HA-tagged Yju2-C (lanes 7 to 9), or 1.5 μM V5-tagged Yju2-N and 0.4 μM HA-tagged Yju2-C (lanes 10 to 13), followed by immunoprecipitation with anti-Ntc20 (lanes 2, 5, 8, and 11), anti-HA (lanes 3, 9, and 13) or anti-V5 (lanes 6 and 12) antibody. RXN, 1/10 of the reaction mixture used for immunoprecipitation.
The above results showed that the C domain could complement the deficiency of the N domain in the second step, so we next examined whether the C domain also functions in stabilizing the binding of the N domain to the spliceosome. Splicing extracts were depleted of Yju2 and supplemented with full-length Yju2, N domain, C domain, or both N and C domains. Prp16 was also depleted from the extract so that Yju2 could be retained on the spliceosome for analysis. Splicing reaction mixtures were precipitated with anti-HA or anti-V5 antibody, as well as with anti-Ntc20 antibody as an indication of activated spliceosomes. With full-length Yju2, over 80% of the splicing intermediates and 70% of pre-mRNA precipitated by anti-Ntc20 antibody were precipitated by anti-HA antibody (Fig. 4C, lanes 2 and 3). With Yju2-N, less than 20% of both splicing intermediates and pre-mRNA precipitated by anti-Ntc20 antibody were precipitated by anti-V5 antibody (Fig. 4C, lanes 5 and 6). With Yju2-C, around 50% of the precatalytic spliceosome (pre-mRNA precipitated by anti-Ntc20 antibody) was precipitated by anti-HA antibody (Fig. 4C, lanes 8 and 9), which accounts for ∼70% of the efficiency of full-length Yju2. In the presence of both the N and C domains, the precipitation efficiency of Yju2-N increased to a level of near 60% of that of full-length Yju2 for splicing intermediates and 30% for pre-mRNA (Fig. 4C, lanes 11 and 12), indicating stabilization of the association of Yju2-N with the spliceosome by Yju2-C. The precipitation efficiency of Yju2-C was not affected in the presence of Yju2-N, being retained at a level of around 70% of the level of full-length Yju2 (Fig. 4C, lanes 11 and 13). This result suggests that the presence of the C domain stabilizes the association of the N domain with the spliceosome at both the pre- and postcatalytic stages, but more pronouncedly at the postcatalytic stage.
The fact that Yju2-N also has a low affinity for lariat intermediate indicates that it may not be stably associated with the spliceosome after catalysis in the absence of the C domain. In light of the strict requirement for DEAH-box protein Prp16 to promote the second catalytic reaction, we were surprised to consistently observe a small amount of spliced product coprecipitated with Ntc20 when Yju2-N alone was used for splicing in Prp16-depleted extracts (Fig. 4C, lane 5) but not when full-length Yju2 (lane 2) or both Yju2-N and -C (lane 11) were used. Since Prp16 is required for destabilization of Yju2 and Cwc25 before the second reaction, we speculated that Prp16 might become dispensable if Yju2-N is self-destabilized from the spliceosome.
The requirement for Prp16 in the second step is compensated for by the absence of the Yju2 C domain.
To further investigate the requirement for Prp16 in the absence of the Yju2 C domain, the splicing reaction was performed in extracts depleted of both Yju2 and Prp16 and complemented with recombinant Yju2 proteins with or without further addition of Prp16 (Fig. 5A). As expected, a small amount of mature mRNA was produced when the reaction mixture was incubated with Yju2-N (Fig. 5A, lane 6), but splicing was less efficient than with full-length Yju2 (lane 4) or the combination of Yju2-N and Yju2-C (lane 10). The addition of Prp16 did not further enhance the second reaction (Fig. 5A, lane 7), indicating that Prp16 does not function in the second reaction with Yju2-N. In contrast, the addition of Prp16 efficiently promoted the second reaction when complemented with full-length Yju2 (Fig. 5A, lane 5) or with the combination of Yju2-N and Yju2-C (lane 11). These results support the notion that Prp16 is required for destabilization of Yju2 due to its high affinity for the spliceosome. Truncation of the C-terminal half of the protein thus decreases the affinity of Yju2 for the spliceosome and relieves its dependency on Prp16 for destabilization.
Fig 5.
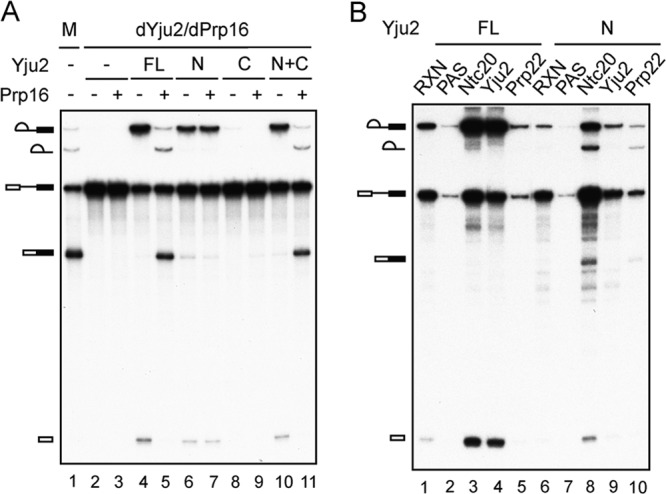
Yju2-N can bypass the requirement for Prp16 function in the second step. (A) Splicing reactions were carried out in mock-depleted (lane 1) and Yju2- and Prp16-depleted extracts (lanes 2 to 11) with no addition (lanes 2 and 3) or with the addition of 0.3 μM Yju2 (lanes 4 and 5), 1.5 μM Yju2-N (lanes 6 and 7), 0.4 μM Yju2-C (lanes 8 and 9), or 1.5 μM Yju2-N and 0.4 μM Yju2-C (lanes 10 and 11), in the presence (lanes 3, 5, 7, 9, and 11) or absence (lanes 2, 4, 6, 8, and 10) of 80 nM Prp16. dYju2/dPrp16, depletion of both Yju2 and Prp16; M, mock-depleted extracts; FL, full-length Yju2; N, the N domain of Yju2; C, the C domain of Yju2; N+C, both N and C domains of Yju2. (B) Splicing reactions were carried out in Yju2- and Prp16-depleted extracts with the addition of recombinant 130 nM 4×V5-tagged Prp22D603A (lanes 1 to 10), 0.3 μM HA-tagged full-length Yju2 (lanes 1 to 5), or 1.5 μM Yju2-N (lanes 6 to 10) for 25 min, followed by immunoprecipitation without antibody (lanes 2 and 7) or with anti-Ntc20 (lanes 3 and 8), anti-HA (lane 4), anti-Yju2 (lane 9), or anti-V5 (lanes 5 and 10) antibody. RXN, 1/8 of the reaction mixture used for immunoprecipitation; PAS, protein A-Sepharose; FL, full-length Yju2; N, Yju2-N.
Destabilization of Yju2 and Cwc25 allows the binding of Slu7, Prp22, and Prp18 to the spliceosome to promote the second reaction. Conceivably, these step 2 factors can bind to a fraction of the spliceosome formed with Yju2-N without the need for Prp16. This hypothesis was investigated by immunoprecipitation of the spliceosome to examine the association of Prp22. Splicing extracts were depleted of Yju2 and Prp16 and complemented with either HA-tagged full-length Yju2 or Yju2-N. A V5-tagged Prp22 mutant protein, with the D603A mutation in the DEAH motif, was also added to the reaction mixture. The Prp22D603A protein is able to bind to the spliceosome and promote the second reaction but is unable to hydrolyze ATP to catalyze mRNA release (39) and is retained on the spliceosome after exon ligation. After splicing, the reaction mixtures were precipitated with anti-V5 antibody for Prp22, anti-HA antibody for full-length Yju2, and anti-Yju2 antibody for Yju2-N (Fig. 5B). Indeed, a small amount of the second-step products coprecipitated with Prp22 when Yju2-N was used (Fig. 5B, lane 10) but not when full-length Yju2 was used (lane 5). Furthermore, Yju2-N is associated with splicing intermediates but not with spliced products (Fig. 5B, lane 9). These results confirmed that the step 2 factors could bind to the spliceosome after self-destabilization of Yju2-N to catalyze exon ligation in the absence of Prp16.
Direct interaction of Yju2 with U2 within the catalytic spliceosome.
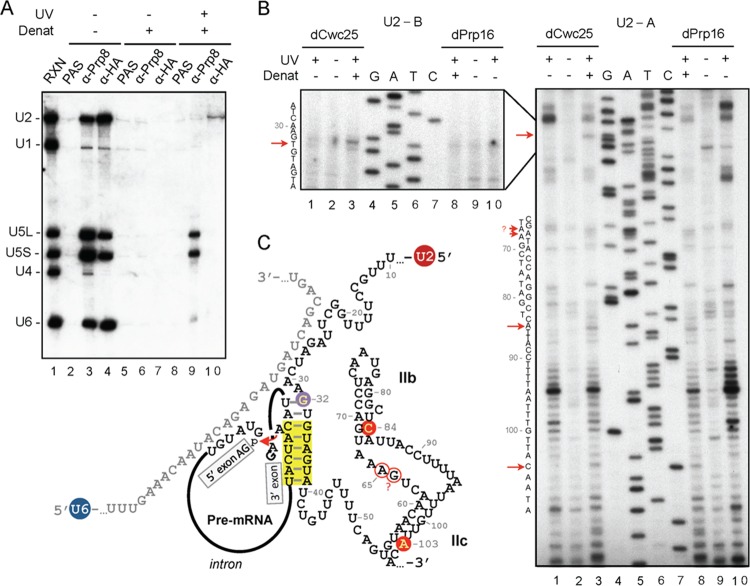
We showed previously that although Yju2 can be recruited to the spliceosome via interaction with NTC components, it is not required for either NTC-mediated spliceosome activation or Prp2-mediated remodeling of the spliceosome (14). Instead, Yju2 is required after the action of Prp2 to promote the first catalytic reaction in an ATP-independent manner. Furthermore, the recruitment of Cwc25 to the spliceosome requires prior association of Yju2 with the spliceosome (15). These findings suggest that Yju2 may play a crucial role in orchestrating the catalytic core of the spliceosome after Prp2 action. To gain insights into how Yju2 may interact with spliceosomal RNAs in the catalytic core, UV cross-linking was performed to examine whether Yju2 directly contacts substrate pre-mRNA or snRNAs. Spliceosomes were assembled in Cwc25-depleted Yju2-HA extracts to arrest the spliceosome prior to the first reaction, and the reaction mixture was irradiated with 254-nm UV to generate protein-RNA cross-links. After treatment with denaturant to disrupt noncovalent interactions, the mixture was precipitated with anti-HA antibody. While pre-mRNA was not found to cross-link to Yju2 (data not shown), U2 snRNA was found to cross-link to Yju2, as revealed by Northern blotting (Fig. 6A, lane 10). A control experiment with anti-Prp8 antibody revealed cross-linking of Prp8 primarily with U5 snRNA and with a small amount of U6 (Fig. 6A, lane 9) as previously reported (40–43). Without UV irradiation and treatment with denaturant, Yju2 was able to coprecipitate the activated spliceosome, containing U2, U5, and U6 snRNAs (Fig. 6A, lane 4). Without UV irradiation, no RNA was coprecipitated with Prp8 or Yju2 under denaturation condition (Fig. 6A, lanes 6 and 7), precluding possible nonspecific backgrounds from noncovalent RNA-protein interactions. Cross-linking of Yju2 to U2 was also observed on the spliceosome that was arrested after the first reaction using Prp16-depleted extracts (see Fig. S6 in the supplemental material).
Fig 6.
UV cross-linking of Yju2 to U2 snRNA within the activated spliceosome. (A) A splicing reaction was performed in Cwc25-depleted Yju2-HA extracts with a pre-mRNA substrate of low radioactivity. The reaction mixture was subjected to immunoprecipitation (lanes 2 to 4) or either irradiated (lanes 8 to 10) with 254-nm UV or not irradiated (lanes 5 to 7); following denaturation, mixtures were immunoprecipitated without (lanes 2, 5, and 8) or with anti-Prp8 (lanes 3, 6, and 9) or anti-HA (lanes 4, 7, and 10) antibody. RNA was extracted and analyzed by Northern blotting. RXN, 1/10 of the reaction mixture used for immunoprecipitation; PAS, protein A-Sepharose; UV, UV irradiation; Denat, denaturation. (B) Primer extension analysis of U2 cross-link sites. The splicing reactions were carried out in Cwc25-depleted (lanes 1 to 3) or Prp16-depleted (lanes 8 to 10) Yju2-HA extracts, followed by UV irradiation (lanes 1, 3, 7, and 10) or no UV treatment (lanes 2 and 9). With (lanes 3 and 8) or without (lanes 1, 2, 9, and 10) treatment with denaturant, the reaction mixtures were immunoprecipitated with anti-HA antibody, and the extracted RNA was subjected to primer extension analysis using U2-A (right) or U2-B (left) primers. dCwc25, depletion of Cwc25; dPrp16, depletion of Prp16; UV, UV irradiation; Denat, denaturation. (C) Schematic representation of the secondary structure of U2/U6/pre-mRNA in the catalytic center of the spliceosome in the first step of the reaction. The branch point region helix is highlighted in yellow, and the attack of the phosphate in the 5′ splice junction by branch point A in the step 1 reaction is marked with a red arrow. Sites in U2 snRNA cross-linked to Yju2 are indicated by open and filled circles, representing weak and strong cross-links, respectively. The purple filled circle indicates the cross-link seen in the precatalytic spliceosome formed in dCwc25 extracts but not in the postcatalytic spliceosome formed in dPrp16 extracts. The positions of U2 snRNA are numbered. The question mark at 64G indicates the ambiguous result.
Prp2 was previously shown to play a role in destabilizing U2 components SF3a/b from the spliceosome, presumably to allow the access of Yju2 and Cwc25 to the catalytic center to promote the first reaction (16). To investigate whether Yju2 interacts with U2 at the sites important for U2 function, we mapped the Yju2-U2 cross-linked sites by primer extension using reverse transcriptase (RT) (Fig. 6B). To identify specific Yju2-U2 cross-links, we compared the RT stops from samples that had been treated with denaturants prior to immunoprecipitation (Fig. 6B, lanes 3 and 8) or not treated (lanes 1 and 10). Only those enriched in denaturant-treated samples were assigned as Yju2-U2 cross-links. Figure 6C shows many strong RT stops from both complexes after UV irradiation without denaturant treatment (lanes 1 and 10), representing cross-links of U2 to spliceosomal components. They show similar patterns in the precatalytic and postcatalytic spliceosomes, except for three strong stops from the region around positions 20 to 50, corresponding to the region that was proposed to form the branch point-interacting stem-loop (BSL) structure during formation of the prespliceosome (44). A change in the cross-linking pattern suggests a structural change in the catalytic core of the spliceosome after the reaction that leads to changes in the contact of splicing factors with U2. Nevertheless, these cross-links were not associated with Yju2, as they were not observed after denaturant treatment, and therefore were not further investigated.
Several potential cross-linking sites for Yju2 were identified (Fig. 6B, lanes 3 and 8). Weak cross-links at 84C (in the stem of the U2 IIb) and 103A (in the stem of the U2 IIc) and an even weaker cross-link at 65A (located just upstream of the U2 IIb) were observed in both complexes, suggesting that Yju2 may be involved in modulating the conformation of U2 helix II. Intriguingly, a cross-link at U2-32G, located just upstream of the branch point interacting sequence, was observed in the precatalytic spliceosome but not in the postcatalytic spliceosome, suggesting that Yju2 may mediate positioning of the branch point.
DISCUSSION
Yju2 is essential for the first catalytic step of splicing, and functions after the action of Prp2. We previously showed that Yju2 interacts with two NTC components, Ntc90 and Ntc77, and can be recruited to the spliceosome either before or after Prp2 action (14). Here, we showed that Yju2 can be physically separated into two distinct domains, and each domain interacts with one component of the NTC. Two-hybrid analysis revealed that the N domain interacts with Ntc90 but not with Ntc77, whereas the C domain interacts with Ntc77 but not with Ntc90. The interaction between Yju2-C and Ntc77 is more robust than that between the Yju2-N and Ntc90.
The N domain of Yju2 is evolutionarily conserved, but the C domain is highly diverged, implying the functional significance of the N domain. Our analysis indeed found that Yju2-N is sufficient for cellular growth and for the in vitro splicing reaction. Yju2-N could promote the first reaction to a level of around 75% of that promoted by full-length Yju2 given sufficient amounts of the protein. But Yju2-N has a very low affinity for the spliceosome and can bind the spliceosome to significant levels only at micromolar concentrations. Yju2-C has no function in splicing itself but can bind the spliceosome much more tightly than Yju2-N, in agreement with the relative strength of their two-hybrid interactions. Although the affinity of Yju2-C for the spliceosome is substantially lower than that of the full-length protein, the amount of Yju2-C binding to the spliceosome could reach nearly 70% of the level of the full-length Yju2 with large amounts of the protein present.
Intriguingly, the N and C domains in combination were able to reconstitute the function of Yju2 both in vivo for growth and in vitro for the splicing activity. Although Yju2-N has a low affinity for the spliceosome, its binding is stabilized by the presence of Yju2-C. Nevertheless, Yju2-N and Yju2-C do not interact with each other in two-hybrid assays or biochemical analysis. These results indicate that the N and the C domains of Yju2 each form a functional module, interacting with Ntc90 and Ntc77, respectively, but do not interact with each other in a stable manner. How Yju2-N and Yju2-C are functionally coupled to assume the activity of the full-length protein without stable interaction between themselves is an interesting question. In the absence of Yju2-C, the association of Yju2-N with the spliceosome is loose due to its weak interaction with Ntc90. The binding of Yju2-C may induce a local conformational change in the spliceosome to stabilize the interaction of Yju2-N or may simply impose steric hindrance to prevent Yju2-N from dissociation from the spliceosome.
Despite Yju2 only being required for the first reaction, Yju2-N promotes the second step to a limited extent, implying that the C domain of Yju2 may have a previously unidentified function in the second step. In addition to Yju2, the first step also requires Cwc25, which is recruited to the spliceosome only in the presence of Yju2. Although the C domain of Yju2 can bind to the spliceosome independently of the N domain, its binding is inadequate for Cwc25 recruitment (see Fig. S5 in the supplemental material) and consequently cannot promote the reaction. In contrast, Cwc25 can bind to the spliceosome in the presence of Yju2-N and remains associated after lariat formation without the C domain (see Fig. S5), suggesting that the binding of Yju2-N is necessary and sufficient to create an active binding site for Cwc25. To proceed to the second step, Yju2 and Cwc25 need to be destabilized and freed from the catalytic center of the spliceosome so that splice sites can be repositioned. Prp16 plays an essential role in destabilizing Yju2 and Cwc25. In the absence of Prp16, Yju2, and Cwc25 accumulate on the spliceosome after lariat formation. In the presence of Prp16, only small amounts of Yju2 and Cwc25 are retained on the spliceosome. Our results thus reveal dual roles of the Yju2 C domain in stabilizing the N domain of Yju2 to efficiently promote the catalytic reaction in the first step and in facilitating Prp16-mediated destabilization of Yju2 and Cwc25 in the second step. Although we did not find evidence that the C domain facilitates or stabilizes the binding of Prp16 to the spliceosome, it remains possible that the C domain stimulates the function of Prp16 in destabilizing Yju2 and Cwc25.
How Prp16 mediates destabilization of Yju2 and Cwc25 is not clear. Prp16 has been demonstrated to unwind RNA duplexes in vitro (45), so it may unwind RNA helices in the catalytic core of the spliceosome to disrupt the interactions of Yju2 and Cwc25 with the spliceosome, or it may directly displace Yju2 and Cwc25. Studies of U2/U6 helix I in the spliceosome catalytic core have revealed genetic interactions between PRP16 and U2/U6 helix I (35). Mutations that weaken U2/U6 helix I were found to suppress the cold-sensitive PRP16 prp16-302 mutation, and specific mutations that retain the helix I structure abolished suppression of the prp16-302 mutant (35). Based on these results, it was suggested that U2/U6 helix I undergoes dynamic structural changes during the catalytic steps, and Prp16 may be involved in destabilization of helix I directly or indirectly (35). In light of the fact that Prp16 is responsible for destabilization of Yju2 and Cwc25, it is also possible that the Prp16-U2/U6 helix I interaction is mediated through Yju2 and/or Cwc25.
In this study, we show that while Prp16 is strictly required for the second step with full-length Yju2, Yju2-N allowed progression of a small amount of the second reaction in the absence of Prp16, indicating that Prp16 can be dispensable under certain conditions. Since Yju2-N shows low affinity for the spliceosome, Yju2-N may be more easily dissociated from the spliceosome after lariat formation without requiring Prp16, and consequently, Cwc25 is also dissociated, thus allowing the binding of step 2 factors to promote the second reaction. Destabilization of Yju2 and Cwc25 likely leads to destabilization of RNA helices in the catalytic core for rearrangement of RNA-RNA or RNA-protein interactions. When Yju2-N is stabilized by Yju2-C on the spliceosome, Prp16 becomes necessary for the second step, suggesting an active role of Prp16 in efficiently displacing Yju2 from the spliceosome, hence releasing the catalytic core for Slu7/Prp18/Prp22. In this view, our results argue against direct unwinding of U2/U6 helix I by Prp16 to destabilize Yju2 and Cwc25. Although Yju2-N was able to bypass the requirement for Prp16 in vitro, attempts to recapitulate this effect in vivo were not successful (data not shown), possibly due to low read-through levels of the second-step reaction. Collectively, our results shed light on Prp16-mediated remodeling of the spliceosome in the second catalytic step of splicing. The detailed mechanism through which Prp16 acts to destabilize Yju2 and Cwc25 remains to be investigated.
Since Yju2 and Cwc25 are required to promote the first reaction after destabilization of SF3a/b, they presumably function in positioning the branch point at the 5′ splice site upon binding to the catalytic core of the spliceosome. We recently showed that Cwc25 directly interacts with substrate RNA in the form of lariat-intron-exon 2 but not the pre-mRNA and can cross-link to the intron sequence 3 bases downstream of the branch point (46). Yju2 was not found to cross-link to pre-mRNA (data not shown). Instead, Yju2 could cross-link to U2 snRNA likely in the helix II region, which has been reported to fluctuate between two different conformations along the spliceosome pathway (47, 48). Whether Yju2 plays a role in modulating U2 helix II structure during the first catalytic step remains to be studied. Yju2 could also cross-link to G32 of U2, located just two bases upstream of the region that form base pairs with the consensus branch site sequence. Interestingly, this cross-link was seen only in the precatalytic spliceosome and not in the postcatalytic spliceosome. This suggests that Yju2 may play a role in mediating positioning of the branch point to the 5′ splice site. Nevertheless, the first reaction cannot occur without Cwc25. Cwc25 is hardly detected on the precatalytic spliceosome, since the reaction occurs immediately upon its binding. How Cwc25 coordinates with Yju2 to promote the reaction is not known. Conceivably, a structural rearrangement may occur after lariat formation, disrupting the interaction of Yju2 with the branch point-interacting region of U2 snRNA. This may be necessary to allow close contact of Cwc25 with the intron sequence and stable association of Cwc25 with the spliceosome, since G32 of U2 is expected to be spatially very close to the Cwc25-cross-linked residue in the intron 3′ tail. Taken together, our results demonstrate that Yju2 closely interacts with the core of the precatalytic spliceosome, inducing an active architecture for the binding of Cwc25. These results provide insights into the formation of the catalytic center of the spliceosome and how protein factors coordinate with RNAs to promote the first-step reaction and prepare for the second-step reaction.
Supplementary Material
ACKNOWLEDGMENTS
We thank C.-K. Tseng for preparation of Prp22D603A protein, M. Loney for English editing, and members of the Cheng lab for helpful discussions.
This work was supported by a grant from the Academia Sinica and National Science Council (Taiwan), NSC100-2745-B-001-001-ASP.
Footnotes
Published ahead of print 25 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00035-13.
REFERENCES
- 1. Brow DA. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36:333–360 [DOI] [PubMed] [Google Scholar]
- 2. Wahl MC, Will CL, Lührmann RL. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136:701–718 [DOI] [PubMed] [Google Scholar]
- 3. Will CL, Lührmann R. 2011. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3:a003707 doi:10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen H-C, Cheng S-C. 2012. Functional roles of protein splicing factors. Biosci. Rep. 32:345–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tarn W-Y, Hsu C-H, Huang K-T, Chen H-R, Kao H-Y, Lee K-R, Cheng S-C. 1994. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 13:2421–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan S-P, Cheng S-C. 2005. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J. Biol. Chem. 280:31190–31199 [DOI] [PubMed] [Google Scholar]
- 7. Chan S-P, Kao D-I, Tsai W-Y, Cheng S-C. 2003. The Prp19p-associated complex in spliceosome activation. Science 302:279–282 [DOI] [PubMed] [Google Scholar]
- 8. Warkocki Z, Odenwälder P, Schmitzová J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Lührmann R. 2009. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat. Struct. Mol. Biol. 16:1237–1243 [DOI] [PubMed] [Google Scholar]
- 9. Staley JP, Guthrie C. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92:315–326 [DOI] [PubMed] [Google Scholar]
- 10. Lardelli RM, Thompson JX, Yates JR, III, Stevens SW. 2010. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16:516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ohrt T, Prior M, Dannenberg J, Odenwalder P, Dybkov O, Rasche N, Schmitzova J, Gregor I, Fabrizio P, Enderiein J, Lührmann R. 2012. Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed dcFCCS. RNA 18:1244–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gozani O, Potashkin J, Reed R. 1998. A potential role for U2AF SAP155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 18:4752–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McPheeters DS, Muhlenkamp P. 2003. Spatial organization of protein-RNA interactions in the branch site-3′ splice site region during pre-mRNA splicing in yeast. Mol. Cell. Biol. 23:4174–4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Y-C, Chen H-C, Wu N-Y, Cheng S-C. 2007. A novel splicing factor Yju2 is associated with NTC and acts after Prp2 in promoting the first catalytic reaction of pre-mRNA splicing. Mol. Cell. Biol. 27:5403–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chiu Y-F, Liu Y-C, Chiang T-W, Yeh T-C, Tseng C-K, Wu NY, Cheng S-C. 2009. Cwc25 is a novel splicing factor required after Prp2 and Yju2 to facilitate the first catalytic reaction. Mol. Cell. Biol. 29:5671–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tseng C-K, Liu H-L, Cheng S-C. 2011. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA 17:145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frank D, Guthrie C. 1992. An essential splicing factor, SLU7, mediates 3′ splice site choice in yeast. Genes Dev. 6:2112–2124 [DOI] [PubMed] [Google Scholar]
- 18. Ansari A, Schwer B. 1995. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J. 14:4001–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Horowitz DS, Abelson J. 1993. Stages in the second reaction of pre-mRNA splicing: the final step is ATP independent. Genes Dev. 7:320–329 [DOI] [PubMed] [Google Scholar]
- 20. Schwer B, Gross CH. 1998. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 17:2086–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Raghunathan PL, Guthrie C. 1998. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 8:847–855 [DOI] [PubMed] [Google Scholar]
- 22. Laggerbauer B, Achsel T, Lührmann R. 1998. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl. Acad. Sci. U. S. A. 95:4188–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wagner JDO, Jankowsky E, Company M, Pyle AM, Abelson JN. 1998. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 17:2926–2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Y, Guthrie C. 1998. PRP16, a DEAH-box RNA helicase, is recruited to the spliceosome primarily via its nonconserved N-terminal domain. RNA 4:1216–1229 [PMC free article] [PubMed] [Google Scholar]
- 25. Tanaka N, Schwer B. 2006. Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry 45:6510–6521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jankowsky E, Bowers H. 2006. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 34:4181–4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jankowsky E, Cross CH, Shuman S, Pyle AM. 2001. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science 291:121–125 [DOI] [PubMed] [Google Scholar]
- 28. Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. 2004. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science 304:730–734 [DOI] [PubMed] [Google Scholar]
- 29. Kistler AL, Guthrie C. 2001. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for Sub2, an essential spliceosomal ATPase. Genes Dev. 15:42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perriman R, Barta I, Voeltz GK, Abelson J, Ares M., Jr 2003. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc. Natl. Acad. Sci. U. S. A. 100:13857–13862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen JY-F, Stands L, Staley JP, Jackups RR, Jr, Latus LJ, Chang T-H. 2001. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell 7:227–232 [DOI] [PubMed] [Google Scholar]
- 32. Company M, Arenas J, Abelson J. 1991. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 349:487–493 [DOI] [PubMed] [Google Scholar]
- 33. Schwer B. 2008. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol. Cell 30:743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu H-L, Cheng S-C. 2012. The Interaction of Prp2 with a defined Region of the intron is required for the first splicing reaction. Mol. Cell. Biol. 32:5056–5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mefford MM, Staley JP. 2009. Evidence that U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps. RNA. 15:1386–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chang K-J, Chen H-C, Cheng S-C. 2009. Ntc90 is required for recruiting first step factor Yju2 but not for spliceosome activation. RNA 15:1729–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cheng S-C, Newman A, Lin R-J, McFarland GD, Abelson JN. 1990. Preparation and fractionation of yeast splicing extract. Methods Enzymol. 181:89–96 [DOI] [PubMed] [Google Scholar]
- 38. Cheng S-C, Abelson J. 1987. Spliceosome assembly in yeast. Genes Dev. 1:1014–1027 [DOI] [PubMed] [Google Scholar]
- 39. Schneider S, Hotz H-R, Schwer B. 2002. Characterization of dominant-negative mutants of the DEAH-box splicing factors Prp22 and Prp16. J. Biol. Chem. 277:15452–15458 [DOI] [PubMed] [Google Scholar]
- 40. Dix I, Russell CS, O'Keefe RT, Newman AJ, Beggs JD. 1998. Protein-RNA interactions in the U5 snRNP of Saccharomyces cerevisiae. RNA 4:1675–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grainger RJ, Beggs JD. 2005. Prp8 protein: At the heart of the spliceosome. RNA 11:533–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Urlaub H, Hatmuth K, Kostka S, Grelle G, Lührmann R. 2000. A general approach for identification of RNA-protein cross-linking sites within native human spliceosomal nuclear ribonucleoproteins (snRNPs). J. Biol. Chem. 275:41458–41468 [DOI] [PubMed] [Google Scholar]
- 43. Vidal VP, Verdone L, Mayes AE, Beggs JD. 1999. Characterization of U6 snRNA-protein interactions. RNA 5:470–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Perriman R, Ares M., Jr 2010. Invariant U2 snRNA nucleotides form a stem loop to recognize the intron early in splicing. Mol. Cell 38:416–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Y, Wagner JDO, Guthrie C. 1998. The DEAH-box splicing factor Prp16 unwinds RNA duplexes in vitro. Curr. Biol. 8:441–451 [DOI] [PubMed] [Google Scholar]
- 46. Chen H-C, Tseng C-K, Tsai R-T, Chung C-S, Cheng S-C. 2013. Link of NTR-mediated spliceosome disassembly with DEAH-box ATPases Prp2, Prp16 and Prp22. Mol. Cell. Biol. 33:514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Perriman R, Ares M., Jr 2007. Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev. 21:811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hilliker AK, Mefford MA, Staley JP. 2007. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev. 21:821–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.