Abstract
The human MLL genes (MLL1 to MLL4) and their Drosophila orthologs, trithorax (trx) and trithorax related (trr), encode proteins capable of methylating histone H3 on lysine 4. MLL1 and MLL2 are most similar to trx, while MLL3 and MLL4 are more closely related to trr. Several MLL genes are mutated in human cancers, but how these proteins regulate cell proliferation is not known. Here we show that trr mutant cells have a growth advantage over their wild-type neighbors and display changes in the levels of multiple proteins that regulate growth and cell division, including Notch, Capicua, and cyclin B. trr mutant clones display markedly reduced levels of H3K4 monomethylation without obvious changes in the levels of H3K4 di- and trimethylation. The trr mutant phenotype resembles that of Utx, which encodes a H3K27 demethylase, consistent with the observation that Trr and Utx are found in the same protein complex. In contrast to the overgrowth displayed by trr mutant tissue, trx clones are underrepresented, express low levels of the antiapoptotic protein Diap1, and exhibit only modest changes in global levels of H3K4 methylation. Thus, in Drosophila eye imaginal discs, Trr, likely functioning together with Utx, restricts tissue growth. In contrast, Trx appears to promote cell survival.
INTRODUCTION
In multicellular organisms, tissue growth and cell proliferation are regulated by diffusible growth factors and by cell-cell interactions. Extracellular signals eventually regulate gene expression by modulating the activity of proteins that bind to specific DNA sequences and influence the recruitment of the transcriptional machinery to specific promoters. A second layer of regulation is provided by the simultaneous recruitment of proteins that influence chromatin structure and thereby determine the accessibility of DNA for transcription (1, 2). While transcription factors have been studied intensively in the context of growth control, the role of chromatin-modifying proteins is less well understood.
Studies in Drosophila led to the discovery of two of the main classes of genes that regulate gene expression by influencing chromatin structure, namely, the trithorax group (TrG) and polycomb group (PcG) (3, 4). While specific mutations in TrG and PcG genes were initially found to alter the expression of Hox genes during development, it is now known that both PcG and TrG genes can have more widespread effects on gene expression. Both classes include members that encode proteins capable of regulating the methylation status of specific lysine residues on histone H3 (reviewed in references 4, 5, and 6). Activity of PcG proteins is associated with the methylation of lysine 27 on histone H3 (H3K27), typically a marker of genes that are repressed. In contrast, some members of the TrG are capable of methylating lysine 4 (H3K4), a modification often found in transcriptionally active genes.
The Drosophila genome encodes at least three genes encoding H3K4 methyl transferases, Set1 (7), trithorax (trx) (8–10), and trithorax related (trr) (11, 12). The catalytic portion of these proteins is the Su(var)3-9/Enhancer of zeste/Trithorax (SET) domain. The Set1, Trx, and Trr proteins are each incorporated into a multiprotein complex that has similarities to the COMPASS (complex of proteins associated with Set1) complex found in yeast (13). Recent studies (7, 13, 14) suggested that these three COMPASS-like complexes differ in their effects on global levels of H3K4 methylation. The Set1-containing complex appears to account for the majority of trimethylation of H3K4. Trx appears to account for very little H3K4 methylation in cell culture or in imaginal discs. Conflicting results have been reported for Trr; using Western blotting, two studies have reported decreased levels of H3K4 trimethylation in mutant embryos and prepupae (12, 15) whereas a recent study of knockdown in S2 cells showed that the main effect was a decrease in H3K4 monomethylation (7).
Our knowledge of the biological function of these Drosophila genes and their mammalian orthologs is extremely limited. Drosophila trx and its mammalian orthologs MLL1 and MLL2 appear to regulate the expression of homeotic genes (16, 17). The Trr protein, whose mammalian orthologs are MLL3 and MLL4, has been shown to modulate the function of the Ecdysone receptor (EcR) (12, 15); ecdysone is the steroid hormone that promotes important developmental transitions in Drosophila. Similarly, the MLL4 protein in mammals is capable of interacting with the estrogen receptor (ER) (18). Since Trx and Trr have been shown to bind to many sites in the genome (12, 13, 19), it is likely that they also regulate the expression of genes other than those characterized in these studies.
Several MLL-family members have been implicated in mammalian cancer. MLL1, a mammalian ortholog of Trx, is translocated in several types of leukemia (20–22). In the course of these translocations, the N terminus of MLL1 is fused to a variety of proteins, generating a chimeric protein that lacks the SET domain. Interestingly, a property shared by these chimeric proteins is that they are incorporated into a complex that regulates transcriptional elongation (5). While this observation provides an important mechanistic advance in our understanding of the pathogenesis of MLL-induced leukemias, the links between possible perturbations in transcriptional elongation and changes in cell proliferation are still not known. Both orthologs of trr, MLL3 and MLL4, have been implicated in mammalian cancer (5). MLL4 is mutated in 32% of diffuse B-cell lymphomas and 89% of follicular lymphomas (23). Currently, little is known about how these mutations contribute to lymphoma development. Thus, understanding how MLL-family proteins regulate cell proliferation is crucial to unraveling the pathogenesis of these types of leukemias and lymphomas.
Here we show that mutations in the Drosophila trr gene provide mutant cells with a growth advantage over their wild-type neighbors. Mutant tissue is characterized by a strong reduction in H3K4 monomethylation and alterations in multiple growth-promoting pathways. In contrast, trx mutations result in a decrease in tissue mass as a result of increased levels of apoptosis. Thus, inactivation of each of these two H3K4 methyltransferases, which are orthologs of MLL3 and -4 and MLL1 and -2, respectively, can have very different functions with respect to cell proliferation and survival in vivo.
MATERIALS AND METHODS
Fly stocks.
All four trr alleles were generated by ethylmethanesulfonate (EMS) mutagenesis of w1118 sn3 FRT19A (24). y w Ubi-GFP FRT19A; eyFLP (II) (25), w1118 sn FRT19A; eyFLP, y w l (1)cl 8.7 P[m-w+arm-lacZ] FRT19A/FM7a; eyFLP (II) (26), y w eyFLP; FRT82B P[mini-w+, armLacZ] (24, 27), and y w eyFLP glass-lacZ; FRT82B w+ cl3R3/TM6B, y+ (28) flies were used to generate mitotic clones. y; FRT82B trxE2/TM6C (29) was obtained from the Bloomington Stock Center. UAStrrRNAi was obtained from the Bloomington Stock Center (stock number 29563) and expressed using eyGal4.
Quantification of imaginal disc clone size.
Eye imaginal discs were imaged using a TCS confocal microscope (Leica). Images were edited in Adobe Photoshop, and areas were measured using ImageJ software.
Immunohistochemistry.
Third-instar imaginal discs were dissected and fixed in 4% paraformaldehyde–phosphate-buffered saline (PBS). Washes were performed with 0.1% Triton X-100–PBS. Primary antibodies used included rabbit anti-cleaved caspase 3 (Cell Signaling) (1:200), mouse anti-cyclin B (Developmental Studies Hybridoma Bank [DSHB]) (F2F4; 1:5), mouse anti-Diap1 (from Bruce Hay) (1:200), guinea pig anti-Capicua (anti-Cic) (1:300) (30), rabbit anti-phospho-Mad (Cell Signaling) (1:100), mouse anti-Notch intercellular domain (ICD) (DSHB) (9C6; 1:200), mouse anti-Dlg (DHSB) (4F3; 1:100), mouse anti-Crumbs (anti-Crb) (Cq4; 1:50) (31), guinea pig anti-cyclin E (from Terry Orr-Weaver), mouse anti-cyclin D (from Wei Du and Nick Dyson), mouse anti-Myc (from Robert Eisenman), rabbit anti-green fluorescent protein (anti-GFP) (Torrey Pines Biolabs Inc.) (1:500; TP401), mouse anti-GFP (Roche) (1:500), mouse anti-β-galactosidase (Sigma) (1:500), and rabbit anti-β-galactosidase (Promega) (1:500). Secondary antibodies used were anti-rabbit antibody–Alexa Fluor 555, anti-mouse antibody–Alexa Fluor 555, anti-rabbit antibody–Alexa Fluor 488, anti-mouse antibody-Alexa Fluor 488 (all from Invitrogen), and anti-guinea pig antibody–Alexa Fluor 555 (Molecular Probes) at 1:500.
Reverse transcription-quantitative PCR (RT-qPCR).
Fifty third-instar eye imaginal discs were dissected and placed in RNAlater (Qiagen). The discs used had trr or trx clones or clones from the parent chromosome generated by mitotic recombination with a chromosome bearing a recessive cell-lethal mutation such that the twin spots were eliminated, leaving the mutant clones and some heterozygous tissue. Total RNA was extracted using TRIzol (Invitrogen) and then an RNAeasy minikit (Qiagen). cDNA synthesis was performed with a Transcriptor First-Strand Synthesis kit (Roche) using the oligo(dT) priming method. Samples were diluted (1:20 to 1:40) and amplified by quantitative PCR (qPCR) using SYBR GreenER SuperMix (Invitrogen) and an Applied Biosystems StepOnePlus real-time PCR system. Reactions were run in triplicate using the standard curve method. Melting curve analysis and conventional PCR were used to confirm primer specificity. Eight potential endogenous reference genes were analyzed, and actin5C was selected as the normalization transcript. Three biological replicates of each genotype were tested. A two-tailed Student's t test was used for statistical analysis.
RESULTS
In order to identify genes that regulate cell proliferation in Drosophila imaginal discs, we have screened the five main chromosome arms for mutations that allow mutant cells to outgrow their wild-type neighbors. By generating mosaic eyes that contained clones of mutant cells (marked white) and sister clones (marked red), we were able to screen for mutations that resulted in an increase in the relative representation of mutant versus wild-type tissue (27). In our screen of the X chromosome, we recovered a lethal complementation group consisting of four members (Fig. 1A to E) where each had a nonsense mutation in the trithorax related (trr) gene.
Fig 1.
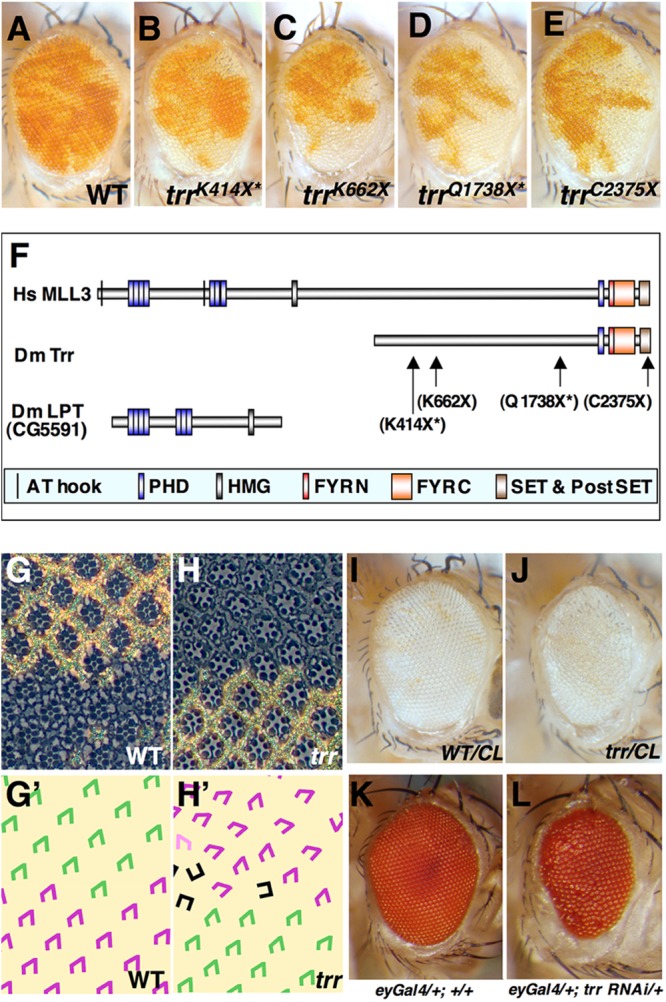
Mutations in trr result in a relative overrepresentation of mutant tissue. (A to E) Mosaic adult eyes generated by mitotic recombination, where the tissue homozygous for the parent chromosome (A) or four different trr alleles (B to E) appears white. The trr mutant tissue is overrepresented (B to E) compared to the relative amounts of nonmutant tissue (red) and mutant tissue (white) found in mosaic eyes generated using the isogenic parent chromosome (A). The alleles that are no longer being maintained are indicated with an asterisk. Genotypes are w1118 sn3 FRT19A/yw P[w+Ubi-GFP] FRT19A; eyFLP/+ for panel A and trr w1118 sn3 FRT19A/yw P[w+Ubi-GFP] FRT19A; eyFLP/+for panels B to E. WT, wild type. (F) Schematic representation of the Trr protein of Drosophila melanogaster (Dm Trr) as well as its human ortholog MLL3 (Hs MLL3). In Drosophila melanogaster and some other insects, a portion of the protein orthologous to MLL3 is found in a separate gene Lost PHD domains of Trr (LPT). The major domains found in the proteins are shown, as are the changes caused by two of the trr mutations characterized. (G and H) Sections of adult eyes showing the arrangement of tissue homozygous for the FRT19A parent chromosome (G) or the chromosome bearing the trrK662X mutation (H). (G′ and H′) The trapezoidal arrangement of photoreceptor cells in the ommatidia in panels G and H is shown in panels G′ and H′. Ommatidia from the mutant portion of the retina (shown in purple) show defects in rotation and sometimes have a rectangular rather than trapezoidal arrangement (black) suggestive of defects in the specification of the R3 and R4 photoreceptor cells. (I and J) Generation of eyes composed mostly of tissue homozygous for the FRT19A parent chromosome (I) or the trrK662X chromosome (J) using eyFLP-induced mitotic recombination with a chromosome bearing a recessive cell-lethal (CL) mutation that eliminates the wild-type twin spots. The genotypes were w1118 sn3 FRT19A/y w l (1)cl 8.7 P[m-w+arm-lacZ] FRT19A; eyFLP/+ (I) and w1118 sn3 trrK662 FRT19A/y w l (1)cl 8.7 P[m-w+arm-lacZ] FRT19A; eyFLP/+ (J). (K and L) Expression of an RNAi transgene directed at trr in the eye primordium using the eyGAL4 driver reduces eye size and causes an irregularity in the arrangement of ommatidia. The genotypes were y w/+; eyGAL4/+ for panel K and y w/+; eyGAL4/+; UAS-trrTRiP_JF03242/+ for panel L.
The trr gene encodes two protein isoforms that differ by 21 amino acids near the N terminus, the larger being 2,431 amino acids long (11) (Fig. 1F). Both isoforms include a C-terminal SET domain that accounts for its histone lysine methyltransferase activity. The human orthologs of Trr are MLL3 and MLL4. However, in some insects, including Drosophila, the domains found in the N-terminal portion of human MLL3 or -4 are found in a separate protein, Lost PHDs of Trr (LPT) (also known as Cara Mitad [Cmi]), that remains physically associated with Trr (13, 15) (Fig. 1F). Of the original four alleles recovered in the screen, only trrK662X and trrK2375X were still being maintained (Fig. 1C and E). Data pertaining to the other two alleles (trrK414X and trrC1738X) are therefore marked with an asterisk.
For all four alleles, mosaic eyes containing trr mutant clones exhibit a relative overrepresentation of mutant over wild-type tissue, indicating that the mutant tissue had a growth advantage. This disparity is more evident in the anterior part of the eye, likely resulting from the more prolonged period of cell proliferation that occurs in the anterior part of the eye disc. However, for two of the alleles, trrK662X or trrK414X, each of which is predicted to generate an extremely truncated protein, two additional phenotypic abnormalities are observed. First, the mutant portion of the eye appears rough, which is indicative of irregularities in the size or organization of the individual facets (ommatidia) of the compound eye. Indeed, sections of adult retinas reveal occasional ommatidia with missing photoreceptor cells as well as abnormalities in the orientation of individual ommatidia that often reflect defects in planar cell polarity (Fig. 1G, G′, H, and H′). Second, despite the relative growth advantage displayed by mutant clones, there is a small decrease in the overall size of the mosaic eyes. Additionally, when the wild-type cells are eliminated by a cell-lethal mutation (Fig. 1I and J) or when trr gene function is uniformly reduced by expression of a hairpin RNA interference (RNAi) transgene (Fig. 1K and L), the resulting eye is slightly reduced in size and roughness is observed similar to that observed with the strongest alleles of trr. All of the phenotypic abnormalities found in mutant clones are rescued by a transgene containing a wild-type copy of the trr gene (data not shown). These findings suggest that disruption of the C-terminal portion of the protein, which has the histone methyltransferase activity, is sufficient to give mutant cells a slight growth advantage over their neighbors. Elimination of functions involving regions nearer the N terminus would be predicted to disrupt the interaction of Trr with the Ecdysone receptor (12) and thus impair progression of the morphogenetic furrow, as has previously been shown by others (12, 32, 33), which could explain the reduction in eye size. Importantly, loss of trr function does not appear to prevent photoreceptor differentiation per se, as assessed by the presence of rhabdomeres in trr mutant photoreceptor cells in sections from adult eyes.
trr mutant clones have a growth advantage as well as alterations in cell survival.
To determine whether the increased representation of mutant tissues in adult eyes reflects a growth advantage, we examined imaginal discs from third-instar larvae. The Trr protein is expressed throughout the eye disc and cannot be detected in mutant clones of the trrK662X allele (data not shown). Clones of the trr mutant occupied a significantly larger portion of the disc than clones homozygous for the isogenic parent chromosome (Fig. 2A and B; quantified in Fig. 2C). Despite their growth advantage, mutant cells appear to exit the cell cycle at the appropriate time since we did not observe ectopic bromodeoxyuridine (BrdU) incorporation posterior to the second mitotic wave as occurs in cells mutant for several negative regulators of cell proliferation such as components of the Hippo pathway (Fig. 2F and F′). Thus, trr mutant cells outgrow their wild-type neighbors but still exit the cell cycle at the appropriate time.
Fig 2.
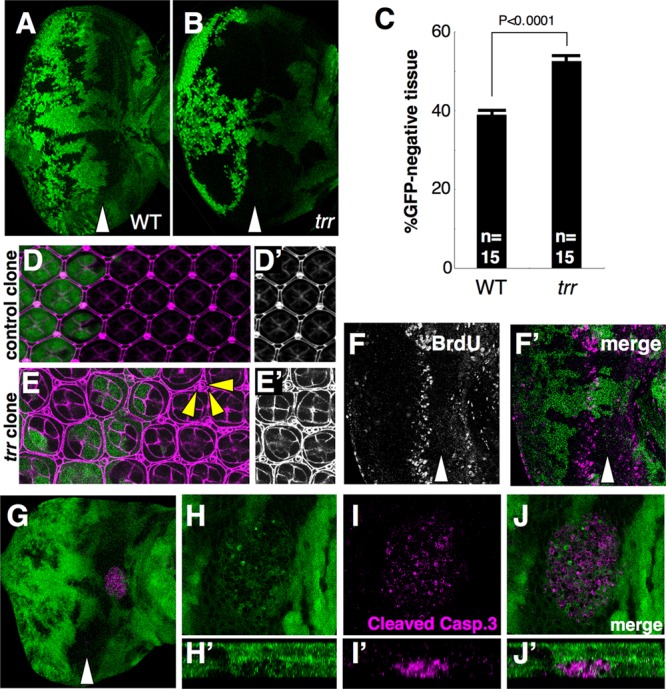
Mutations in trr result in increased growth and alterations in the pattern of apoptosis in the eye imaginal disc. (A to C) The ratio of mutant (black) to nonmutant (green) tissue in the eye is increased in trrK662X mutants (B) compared to the FRT19A parent chromosome (A) as quantified in panel C. The data were compared using a 2-tailed t test (n = 15 eye discs each). P = 1.4e-5. Error bars indicate the standard errors of the means (SEM). (D and E) Mosaic pupal retinas showing a single layer of interommatidial cells in tissue homozygous for the FRT19A chromosome. In contrast, there are additional interommatidial cells in the trrK662X tissue. Anti-Dlg staining is shown in magenta. GFP (green) indicates the presence of the wild-type chromosome. Yellow arrowheads indicate some examples of extra interommatidial cells. (F) Incorporation of BrdU in trrK662X mosaic larval discs shows no differences between the wild-type and mutant portions of the disc. (G to J) A clump of tissue expressing activated caspase is observed basal to the epithelium near the junction of the eye and antennal portions of the disc in discs bearing trr clones. Mutant tissue does not express GFP. Z projections shown in panels H′ to J′ show that the tissue expressing activated caspase (Casp.) is basal to the epithelium of the disc proper. The genotypes were w1118 sn3 FRT19A/yw Ubi-GFP FRT19A; eyFLP/+ for panels A, D, and D′ and trrK662X w1118 sn3 FRT19A/yw Ubi-GFP FRT19A; eyFLP/+ for panels B and E to J.
We also observed changes in the pattern of cell death in discs containing mutant clones. In pupal retinas, supernumerary cells are normally eliminated by a wave of apoptosis, leaving a regular array of cells with specified fates (Fig. 2D and D′). In contrast, in trr clones, additional interommatidial cells were observed, indicating that this wave of cell death may not have proceeded to completion (34) (Fig. 2E and E′). Additionally, in the larval disc, we observed clumps of cells expressing activated caspase basal to the disc but seldom observed these cells in the disc epithelium itself (Fig. 2G to J), suggesting that cells undergoing apoptosis are rapidly extruded from the epithelium. The mass of activated caspase 3-positive cells included material that was GFP positive, suggesting that at least some wild-type cells were present. These wild-type cells could have derived from the disc epithelium or, alternatively, could have been hemocytes that were engulfing the corpses of dead cells. Thus, while decreased trr function seems to protect cells from the normal developmental apoptosis that occurs at the pupal stage, it appears to promote some apoptosis of either mutant cells or their wild-type neighbors at an earlier stage.
trr mutations have markedly reduced levels of H3K4 monomethylation.
The Trr protein belongs to a family of proteins that contain SET domains that are capable of methylating the N-terminal tail of histone H3 on the lysine residue at position 4 (H3K4). In Drosophila cells, Trr proteins, along with two other SET domain-containing proteins, Set1 and Trithorax (Trx), assemble into multiprotein complexes that share several of their components (13). Of these, Trr alone forms a complex that includes Utx, a protein that is capable of removing methyl groups attached to the lysine residue at position 27 on histone H3 (H3K27). Interestingly, Utx mutants were previously isolated from our screen based on the phenotype of a relative overrepresentation of mutant tissue (35).
Mutations in Drosophila trr have been described previously and were shown to modulate the function of the receptor for the steroid hormone ecdysone (12, 15), a function similar to that described for its human ortholog, MLL4, which has been shown to interact with the estrogen receptor (18). The authors (12) also reported that the levels of dimethylation and trimethylation of H3K4 were much reduced in trr mutant embryos. Another study, also using Western blotting, documented decreased levels of H3K4 trimethylation in both embryos and prepupae (15). However, two recent studies using RNAi-based approaches have reached a different conclusion. In one study, knockdown of Trr function in wing imaginal discs resulted in only a marginal decrease in di- and trimethylation of H3K4 (13). In another, knockdown of Trr in cell culture primarily caused a decrease in H3K4 monomethylation and the Trr protein showed robust H3K4 monomethylation activity in vitro (7).
We therefore examined H3K4 mono-, di-, and trimethylation levels in mosaic eye imaginal discs, where the adjacent wild-type cells provide an ideal reference for comparison. Consistent with the recent findings in Drosophila cell culture (7), we found no detectable change in the levels of di- or trimethylation of H3K4 (Fig. 3A to A″ and B to B″). However, monomethylation of H3K4 was dramatically reduced (Fig. 3C to C″). These findings are also similar to those observed in Utx mutants (35) and are consistent with the finding that Utx and Trr are found in the same protein complex. Thus, reducing trr or Utx function seems to primarily impair H3K4 monomethylation, a posttranslational modification that appears to be associated with enhancers (36–38). We did observe one difference between Utx and trr mutants. In Utx clones, a slight elevation of H3K27 trimethylation that is consistent with the ability of the Utx protein to demethylate H3K27 has been observed (35). In trr clones, H3K27 trimethylation levels appeared unchanged (Fig. 3D to D″). Thus, even though Trr and Utx are in the same complex, loss of either protein results in reduced H3K4 monomethylation. However, loss of Utx function but not Trr function results in slightly increased H3K27 trimethylation. These results suggest that the decrease in H3K4 monomethylation in either trr or Utx mutants is not a consequence of altered H3K27 trimethylation but is more likely the result of a direct loss of H3K4 monomethylation activity of the complex. Additionally, loss of Trr function may not compromise the H3K27 demethylase activity of the complex. Alternatively, the Utx protein could make only a minor contribution to the overall levels of H3K27 demethylation and a slight increase might not be obvious using immunofluorescence.
Fig 3.
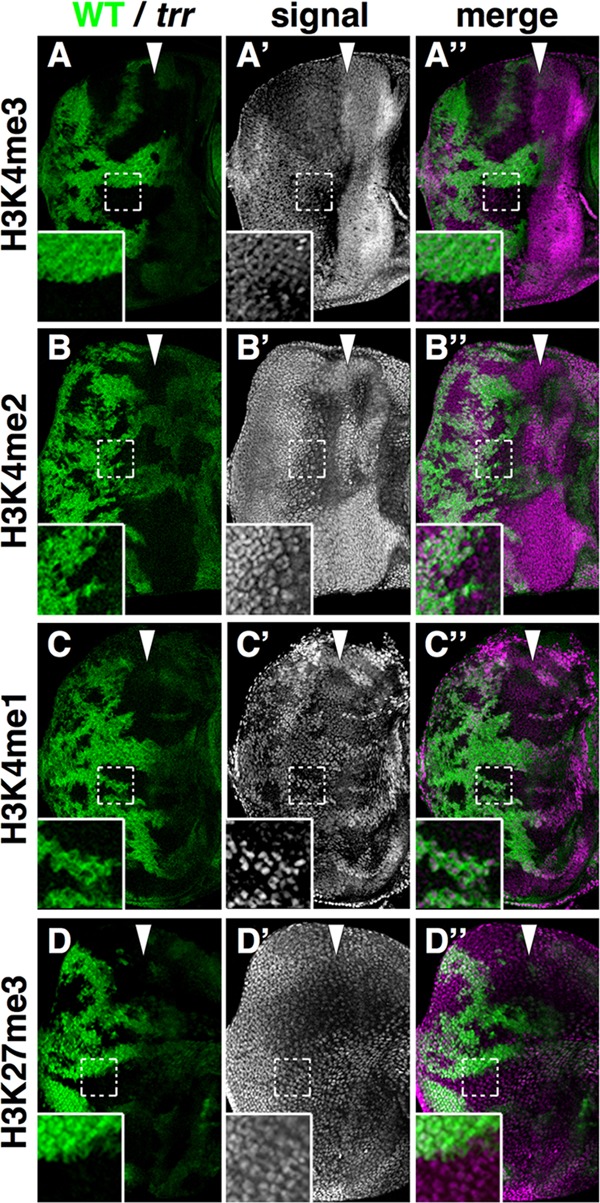
trr mutant cells have decreased levels of H3K4 monomethylation. (A to D) Mosaic eye imaginal discs stained with antibodies that detect methylation of H3K4 (A to C) and H3K27 (D). (A to C) Mutant tissue does not express GFP (green). trr mutant cells express greatly diminished levels of H3K4 monomethylation (H3K4me1) (C) but not di- or trimethylation (A and B). (D) Trimethylation of H3K27 was not detectably changed. The genotypes were trrK662X w1118 sn3 FRT19A/y w P[wUbi-GFP] FRT19A; eyFLP/+ for panels A to D.
Indeed, while this work was being prepared for submission, another study used both immunohistochemistry and genome-wide chromatin immunoprecipitation (ChIP) studies to demonstrate that decreasing trr function reduces levels of both H3K4 monomethylation and H3K27 acetylation (which is normally a mark of enhancer elements) (39). Moreover, the absence of Trr results in diminished levels of Utx, likely by destabilizing the Utx protein.
trr mutations impact Notch signaling as well as levels of other growth regulators.
Mutations in Drosophila Utx augment Notch signaling, as shown by both an increase in the expression level of genes downstream of Notch and the ability of Utx mutants to dominantly suppress phenotypes resulting from decreased Notch signaling (35). To determine whether this aspect of Utx function is shared by trr, we examined the ability of trr mutations to modify the wing-notching phenotype of Notch (N) and Serrate (Ser) mutants (Fig. 4). As with Utx mutants, trr mutants demonstrated some suppression of both phenotypes. We also examined the level of Notch protein in mutant clones and observed markedly increased staining with an antibody directed against the intracellular domain of Notch (Fig. 5A to A″).
Fig 4.
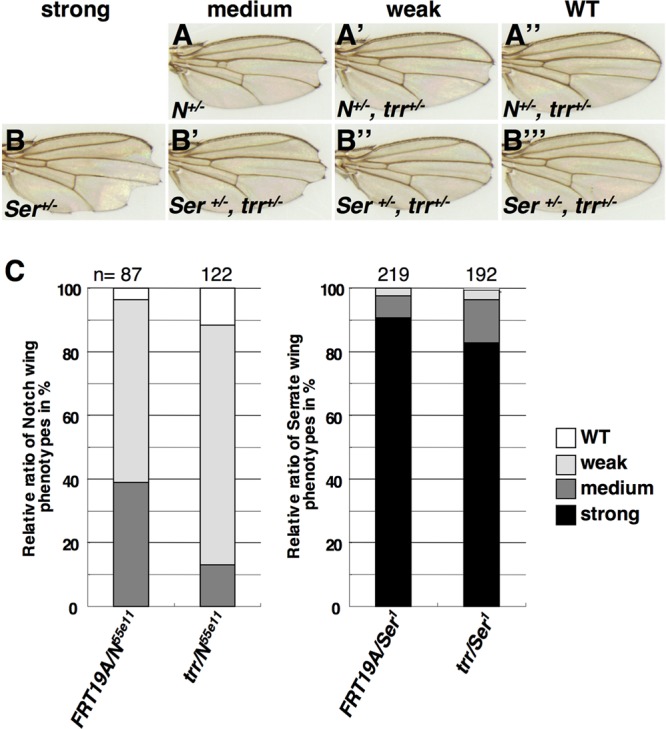
trr mutations suppress the dominant wing-notching phenotypes of Notch (N) and Serrate (Ser) mutants. The frequencies of different classes of notched wings were compared between flies heterozygous for the parent chromosome (FRT19A) and a chromosome bearing the trrK662X mutation. The genotypes were N55e11 FRT19A/w1118 sn3 FRT19A for panel A, N55e11 FRT19A/trrK662X w1118 sn3 FRT19A for panels A′ and A″), w1118 sn3 FRT19A/+; TM3, Ser/+ for panel B, and trrK662X w1118 sn3 FRT19A/+; TM3, Ser/+ for panels B′ to B‴.
Fig 5.
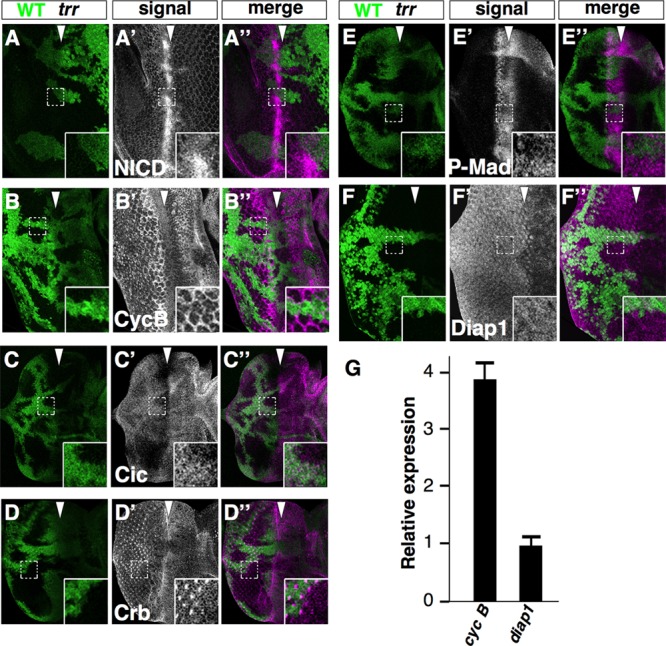
trr cells have altered levels of multiple regulators of tissue growth and cell survival. (A to F) Eye imaginal discs mosaic for clones homozygous for trrK662X stained with antibodies to the intracellular domain of Notch (NICD; A to A″), cyclin B (CycB; B to B″), Capicua (Cic; C to C″), Crumbs (Crb; D to D″), phospho-Mad (P-Mad; E to E″), and Diap1 (F to F″). Mutant tissue did not express GFP (green). (G) Relative levels of cyclin B and diap1 RNA as assessed by RT-qPCR. Error bars indicate the SEM of the results determined with three biological replicates. The genotypes were trrK662X w1118 sn3 FRT19A/yw Ubi-GFP FRT19A; eyFLP/+ for panels A to F″.
Since mutations in epigenetic regulators such as trr are likely to affect multiple genes and signaling pathways, we surveyed a number of regulators of growth and cell cycle progression. We found a decrease in the levels of the corepressor Capicua (Cic) (Fig. 5C to C″), which is indicative of increased receptor tyrosine kinase (RTK) signaling (40), and an increase in phospho-Mad levels, which is indicative of increased Dpp signaling (Fig. 5E to E″). Additionally, levels of the cell-surface protein Crumbs were decreased (Fig. 5D to D″). Complete inactivation of crumbs (crb) results in a modest upregulation of Yorkie-regulated genes such as diap1 (41–44). However, there does not appear to have been an increase in diap1 expression (Fig. 5F), suggesting that alterations in Hippo pathway signaling were perturbed even less than they are in crb mutant clones. The level of the mitotic cyclin, cyclin B, was also elevated (Fig. 5B to B″). However, the levels of the G1 cyclins, cyclin D and cyclin E, as well as of the Myc protein which promotes growth and cell cycle progression were unchanged (data not shown). We tested whether these changes in the levels of growth and cell cycle regulators were the result of changes in RNA levels by RT-PCR. For the genes examined, only cyclin B showed a 4-fold increase in RNA levels (Fig. 5G); others showed little change. Thus, the overgrowth phenotype cannot easily be explained by a simple effect on a single pathway but likely results from a complex interplay of transcriptional and posttranscriptional changes involving multiple genes that eventually result in altered activity of multiple growth-promoting pathways.
Mutations in trithorax (trx) affect tissue growth and gene expression in ways that differ from those seen with trithorax related (trr).
Drosophila has three SET domain-containing proteins that assemble into COMPASS-like complexes (13). The Set1-containing complex accounts for most of the H3K4 trimethylation observed. The complexes containing the MLL-family proteins Trr and Trx appear to contribute very little to the trimethylation (7, 13). Inhibiting trr function reduces H3K4 monomethylation in cell culture (7, 13) and also in imaginal disc cells (Fig. 3) (39). In contrast, reducing trx function did not lead to marked changes in the global levels of H3K4 monomethylation in imaginal discs (Fig. 6G and G′). Moreover, levels of H3K4 di- or trimethylation were also not different (data not shown). When we generated mosaic eyes containing trx clones, the mutant tissue in adult eyes was greatly underrepresented (Fig. 6A and B). Indeed, a trx allele was previously identified in a mosaic screen for mutant clones that reduced eye size or caused scarring (45). This was in marked contrast to the mild overgrowth phenotype displayed by trr mutant clones (Fig. 1A to E). When mitotic recombination was induced with a chromosome bearing a recessive cell-lethal mutation such that most of the eye was mutant for trx, the eye was markedly reduced in size (Fig. 6C). These results indicate that trx cells either had a severe growth defect or had been eliminated during development.
Fig 6.
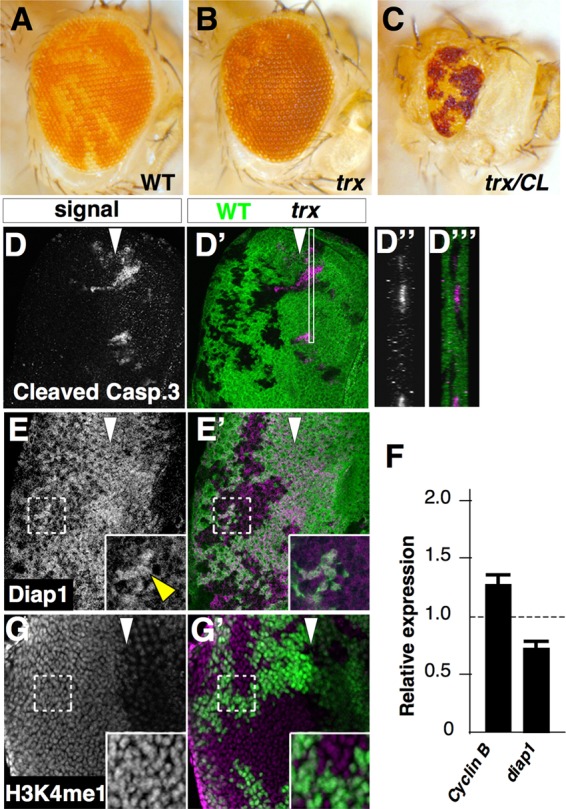
Mutations in trithorax (trx) elicit phenotypes that are qualitatively distinct from those observed in trr mutants. (A and B) Eyes mosaic for the FRT82B chromosome (A) or the trxE2 mutation (B) show a relative underrepresentation of trx mutant tissue. (C) Eyes composed mostly of trx tissue were generated by mitotic recombination with a chromosome bearing a recessive cell-lethal mutation and were much smaller. The genotypes were y w eyFLP; FRT82B P[mini-w+, armLacZ]/FRT82B for panel A, y w eyFLP; FRT82B P[mini-w+, armLacZ]/FRT82B trxE2 for panel B, and y w eyFLP glass-lacZ; FRT82B w+ cl3R3/FRT82B trxE2 for panel C. (D) Increased staining with an antibody that recognizes cleaved caspase is observed in trx clones. Z projections of the region boxed in panel D′ are shown in panels D″ and D‴. (E) Staining of mosaic discs with an antibody to Diap1 shows reduced staining in mutant clones compared to wild-type tissue (green). The yellow arrowhead indicates the expression of Diap1 in wild-type tissue. (F) Relative levels of cyclin B and diap1 RNA in discs containing trx clones as measured using RT-qPCR. Error bars indicate the SEM of the results determined with three biological replicates. (G) Staining with an antibody to the monomethylated form of H3K4 (H3K4me1) shows no change in the level of signal in mutant clones. The genotypes were y w eyFLP; FRT82B P[mini-w+, armLacZ]/FRT82B trxE2 for panels D to G.
An examination of imaginal discs containing trx clones demonstrated high levels of cell death, as assessed by elevated levels of activated caspase in the disc epithelium. Moreover, the activated caspase 3-positive material was found to have been extruded basally from the epithelium (Fig. 6D to D‴). Levels of the antiapoptotic protein Diap1 (46) were reduced in trx clones (Fig. 6E and E′). This is likely to have resulted, at least in part, from decreased transcription, since diap1 RNA levels were also reduced (Fig. 6F). Moreover, this measurement of diap1 RNA levels is likely to underestimate its reduction in trx mutant cells since the samples were prepared from discs that also contained heterozygous (trx/+) cells. In contrast, the levels of Notch, cyclin B, phospho-Mad, Capicua, and Crumbs, as assessed by antibody staining, were all unchanged in trx mutant clones compared to adjacent wild-type tissue (data not shown). Of these, there was a small increase in cyclin B RNA (Fig. 6F). A previous study demonstrated a reduction in the expression of eyeless and teashirt and increased expression of homothorax in trx clones that together resulted in a fate change from eye to head cuticle; sometimes these transformed clones appeared larger than those confined to the eye primordium (45).
Thus, trr and trx mutant cells differ considerably in their phenotypic characteristics. Consistent with the growth advantage exhibited by trr clones, several growth-promoting signaling pathways appear to function at higher levels whereas Diap1 RNA or protein levels were unchanged. In contrast, these same growth-promoting pathways were unchanged in trx clones whereas Diap1 RNA and protein levels were reduced. The reduction in Diap1 levels likely contributes to the apoptosis observed in trx clones. Additionally, trx promotes the specification of retinal fates in the eye disc; mutant cells can often fail to express eye-specific markers (45). In contrast, photoreceptor cell specification and differentiation appear to occur relatively normally in trr clones.
DISCUSSION
Several members of the MLL family of genes have been implicated in mammalian cancer. However, we still know very little about how these genes regulate cell proliferation. Study of the function of MLL-family genes is simplified in Drosophila because trr appears to correspond to both MLL3 and MLL4 whereas trx corresponds to MLL1 and MLL2. Here we have characterized the properties of each of these major subclasses of MLL genes. First, we show that mutations in trr promote tissue overgrowth without obviously interfering with differentiation. In contrast, inactivating mutations in trx cause increased levels of cell death. Second, we demonstrate that trr mutant clones, but not trx clones, are characterized by a marked decrease in H3K4 monomethylation. Third, we find that multiple pathways that promote growth and cell cycle progression are altered in trr cells, indicating that the phenotypic abnormalities are unlikely to result from the deregulation of a single pathway. Finally, we observe considerable similarities between the phenotypic abnormalities displayed by trr and Utx clones, consistent with the finding that the two proteins are present in the same complex.
In the past, studies of H3K4 methyltransferase complexes have emphasized their role in trimethylation. However, the main change observed in vivo with mutations in Utx or in trr is a decrease in H3K4 monomethylation. Recent work indicates that H3K4 monomethylation is characteristic of enhancer sequences and genome-wide surveys of H3K4 monomethylation can be used to predict the location of enhancers (36–38). Thus, it is likely that the function of a number of enhancers in the genome is altered in some way in trr mutants.
trr mutants and Utx mutants display remarkable similarities in their mutant phenotypes. Alleles of trr (this work) and alleles of Utx (35) were both identified in unbiased genetic screens for mutations that increased the relative overrepresentation of mutant tissue compared to wild-type tissue. Additionally, trr and Utx mutations display similar genetic interactions, with mutations in Notch (N) and Serrate (Ser). These findings indicate that these phenotypic consequences are likely to arise from a disruption of the complex that includes both proteins and that likely regulates the levels of H3K4 monomethylation at multiple enhancers.
In a survey of the main pathways that are known to regulate imaginal disc growth in Drosophila, we found multiple changes in trr mutants. Consistent with the genetic interactions with N and Ser, we found increased levels of the Notch protein in mutant clones. Thus, the increased activity of the Notch pathway in trr mutants could result from increased levels of the Notch protein itself. This would explain why this phenotype could suppress the wing-notching phenotype of N heterozygotes, where Notch protein levels are reduced. We also observed reduced levels of Capicua, which is indicative of increased RTK/Ras signaling (40), and increased levels of phospho-Mad, which is suggestive of increased signaling via Dpp receptors (47). Despite at least three growth-promoting pathways being activated at increased levels, the overgrowth phenotype of trr mutants is relatively subtle, suggesting that the extent of change in these pathways has a small impact on tissue growth or that there are also unknown changes in other pathways that counteract these effects.
Although trr and trx encode proteins of the same superfamily, their effects on tissue growth are completely different. Each of the growth-regulatory pathways that displayed alterations in trr mutant clones was unchanged in trx clones. In contrast, levels of the antiapoptotic protein Diap1 are reduced in trx clones and likely contribute to the increased cell death in trx mutants. Additionally, in screens of the right arm of the third chromosome conducted in our laboratory for adult eyes with an increased representation of mutant tissue, despite obtaining multiple alleles of several negative regulators of growth, including Tsc1, salvador, warts, and capicua, we never obtained any alleles of trx with an overgrowth phenotype. Thus, in contrast to trr, inactivating mutations in trx do not promote tissue growth while preserving the ability of cells to differentiate.
Mutations that are predicted to decrease the function of MLL3 and MLL4, the mammalian proteins most similar to Trr, are being found in an increasing number of solid tumors (see, for example, references 48, 49, and 50) in addition to the mutations described in lymphomas (23). This suggests that, as seen with trr, inactivation of MLL3 or MLL4 also promotes cell proliferation. In leukemias, however, MLL1 is disrupted by a variety of chromosomal translocations, each of which fuses the N terminus of MLL1, which lacks the SET domain, to a protein that functions in transcriptional elongation, resulting in the incorporation of this chimeric protein into a multiprotein complex that regulates transcriptional elongation (reviewed in reference 5). Thus, the effect of MLL1 on leukemic cell proliferation may not reflect a normal role for MLL1 as a regulator of cell proliferation. At least in Drosophila, inactivation of its ortholog trx does not seem to result in excess proliferation. Rather, trx mutant cells display increased levels of apoptosis as well as cell fate changes. Thus, these two major subclasses of MLL genes appear to have different types of effect on cell proliferation and survival.
While genome-wide changes in histone methylation caused by manipulation of genes encoding histone methyltransferases and demethylases are being increasingly documented, we still lack a basic understanding of how these changes relate to biological consequences. In yeast, inactivation of Set1 abolishes most if not all H3K4 methylation. Yet mutant strains are viable, albeit with alterations in growth and in morphology, and display abnormalities in gene silencing at specific loci (51–53). Thus far, it has been difficult to ascribe the phenotypic abnormalities to specific changes in transcription. The demonstration that mutations in the Drosophila genes trr and trx elicit specific and distinct phenotypic abnormalities pertaining to growth and cell survival will provide a useful starting point for dissecting those aspects of the biological functions of MLL-class genes that are most relevant to human cancers.
ACKNOWLEDGMENTS
We thank Wei Du, Nick Dyson, Robert Eisenman, Bruce Hay, Alexander Mazo, and Terry Orr-Weaver for antibodies and fly stocks as well as the following stock centers: VDRC, Bloomington Drosophila Stock Center, NIG-FLY, the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947), and the Drosophila Genome Resource Center at Kyoto. We are especially grateful to Brett Pellock for initiating the FLP/FRT screen of the X chromosome and for providing stocks and encouragement during this project and to current and former members of the Hariharan, Bilder, and Okano laboratories for discussions and advice. We thank Rieko Shimamura for technical support.
I.K.H. is funded by the NIH (R01 GM61672) and a Research Professor Award from the American Cancer Society (120366-RP-11-078-01-DDC). H.K. and H.O. received support from the Japanese Ministry of Education, Science, Sports, Culture and technology (H.K. and H.O.), the Japan Society for the Promotion of Science (H.K.), Keio Gijuku Academic Development Funds (H.K.), Keio University Grant-in-Aid for Encouragement of Young Medical Scientists (H.K.), the Strategic Research Foundation Grant-aided Project for Private Universities from MEXT(H.K.), and the Grant-in-Aid for the G-COE program from MEXT to Keio University (H.K. and H.O.).
H.K. designed and conducted the genetic screen with the assistance of L.C. and identified the mutants. A.N. identified trr as the gene responsible for the mutant phenotype and conducted its molecular characterization. H.K. and A.N. characterized the mutant phenotypes and prepared the figures. Data were interpreted by H.K., A.N., and I.K.H. The manuscript was written by I.K.H., H.K., and A.N. I.K.H. oversaw the work conducted at Berkeley. H.O. oversaw the experiments conducted by H.K. after he moved to Keio University.
Footnotes
Published ahead of print 4 March 2013
REFERENCES
- 1.Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78:273–304 [DOI] [PubMed] [Google Scholar]
- 2.Greer EL, Shi Y. 2012. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13:343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ringrose L, Paro R. 2004. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 38:413–443 [DOI] [PubMed] [Google Scholar]
- 4.Schuettengruber B, Cavalli G. 2009. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136:3531–3542 [DOI] [PubMed] [Google Scholar]
- 5.Shilatifard A. 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81:65–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuettengruber B, Martinez AM, Iovino N, Cavalli G. 2011. Trithorax group proteins: switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 12:799–814 [DOI] [PubMed] [Google Scholar]
- 7.Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T. 2011. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 30:2817–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mozer BA, Dawid IB. 1989. Cloning and molecular characterization of the trithorax locus of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 86:3738–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111:185–196 [DOI] [PubMed] [Google Scholar]
- 10.Smith ST, Petruk S, Sedkov Y, Cho E, Tillib S, Canaani E, Mazo A. 2004. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat. Cell Biol. 6:162–167 [DOI] [PubMed] [Google Scholar]
- 11.Sedkov Y, Benes JJ, Berger JR, Riker KM, Tillib S, Jones RS, Mazo A. 1999. Molecular genetic analysis of the Drosophila trithorax-related gene which encodes a novel SET domain protein. Mech. Dev. 82:171–179 [DOI] [PubMed] [Google Scholar]
- 12.Sedkov Y, Cho E, Petruk S, Cherbas L, Smith ST, Jones RS, Cherbas P, Canaani E, Jaynes JB, Mazo A. 2003. Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature 426:78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, Shilatifard A. 2011. The COMPASS family of H3K4 methylases in Drosophila. Mol. Cell. Biol. 31:4310–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hallson G, Hollebakken RE, Li T, Syrzycka M, Kim I, Cotsworth S, Fitzpatrick KA, Sinclair DA, Honda BM. 2012. dSet1 is the main H3K4 di- and tri-methyltransferase throughout Drosophila development. Genetics 190:91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chauhan C, Zraly CB, Parilla M, Diaz MO, Dingwall AK. 2012. Histone recognition and nuclear receptor co-activator functions of Drosophila cara mitad, a homolog of the N-terminal portion of mammalian MLL2 and MLL3. Development 139:1997–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingham P, Whittle R. 1980. Trithorax: a new homeotic mutation of Drosophila melanogaster causing transformations of abdominal and thoracic imaginal segments. Mol. Gen. Genet. 179:607–614 [Google Scholar]
- 17.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. 1995. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378:505–508 [DOI] [PubMed] [Google Scholar]
- 18.Mo R, Rao SM, Zhu YJ. 2006. Identification of the MLL2 complex as a coactivator for estrogen receptor alpha. J. Biol. Chem. 281:15714–15720 [DOI] [PubMed] [Google Scholar]
- 19.Chinwalla V, Jane EP, Harte PJ. 1995. The Drosophila trithorax protein binds to specific chromosomal sites and is co-localized with Polycomb at many sites. EMBO J. 14:2056–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Djabali M, Selleri L, Parry P, Bower M, Young BD, Evans GA. 1992. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nat. Genet. 2:113–118 [DOI] [PubMed] [Google Scholar]
- 21.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce CM, Canaani E. 1992. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell 71:701–708 [DOI] [PubMed] [Google Scholar]
- 22.Tkachuk DC, Kohler S, Cleary ML. 1992. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 71:691–700 [DOI] [PubMed] [Google Scholar]
- 23.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, Jackman S, Krzywinski M, Scott DW, Trinh DL, Tamura-Wells J, Li S, Firme MR, Rogic S, Griffith M, Chan S, Yakovenko O, Meyer IM, Zhao EY, Smailus D, Moksa M, Chittaranjan S, Rimsza L, Brooks-Wilson A, Spinelli JJ, Ben-Neriah S, Meissner B, Woolcock B, Boyle M, McDonald H, Tam A, Zhao Y, Delaney A, Zeng T, Tse K, Butterfield Y, Birol I, Holt R, Schein J, Horsman DE, Moore R, Jones SJ, Connors JM, Hirst M, Gascoyne RD, Marra MA. 2011. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476:298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu T, Rubin GM. 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117:1223–1237 [DOI] [PubMed] [Google Scholar]
- 25.Moon NS, Frolov MV, Kwon EJ, Di Stefano L, Dimova DK, Morris EJ, Taylor-Harding B, White K, Dyson NJ. 2005. Drosophila E2F1 has context-specific pro- and antiapoptotic properties during development. Dev. Cell 9:463–475 [DOI] [PubMed] [Google Scholar]
- 26.Fehon RG, Oren T, LaJeunesse DR, Melby TE, McCartney BM. 1997. Isolation of mutations in the Drosophila homologues of the human Neurofibromatosis 2 and yeast CDC42 genes using a simple and efficient reverse-genetic method. Genetics 146:245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK. 2001. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 105:345–355 [DOI] [PubMed] [Google Scholar]
- 28.Newsome TP, Asling B, Dickson BJ. 2000. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127:851–860 [DOI] [PubMed] [Google Scholar]
- 29.Gindhart JG, Jr, Kaufman TC. 1995. Identification of Polycomb and trithorax group responsive elements in the regulatory region of the Drosophila homeotic gene Sex combs reduced. Genetics 139:797–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tseng AS, Tapon N, Kanda H, Cigizoglu S, Edelmann L, Pellock B, White K, Hariharan IK. 2007. Capicua regulates cell proliferation downstream of the receptor tyrosine kinase/ras signaling pathway. Curr. Biol. 17:728–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tepass U, Knust E. 1993. Crumbs and stardust act in a genetic pathway that controls the organization of epithelia in Drosophila melanogaster. Dev. Biol. 159:311–326 [DOI] [PubMed] [Google Scholar]
- 32.Brennan CA, Ashburner M, Moses K. 1998. Ecdysone pathway is required for furrow progression in the developing Drosophila eye. Development 125:2653–2664 [DOI] [PubMed] [Google Scholar]
- 33.Zelhof AC, Ghbeish N, Tsai C, Evans RM, McKeown M. 1997. A role for ultraspiracle, the Drosophila RXR, in morphogenetic furrow movement and photoreceptor cluster formation. Development 124:2499–2506 [DOI] [PubMed] [Google Scholar]
- 34.Wolff T, Ready DF. 1991. Cell death in normal and rough eye mutants of Drosophila. Development 113:825–839 [DOI] [PubMed] [Google Scholar]
- 35.Herz HM, Madden LD, Chen Z, Bolduc C, Buff E, Gupta R, Davuluri R, Shilatifard A, Hariharan IK, Bergmann A. 2010. The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol. Cell. Biol. 30:2485–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459:108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38:576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, Ideker T, Glass CK, Murre C. 2010. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 11:635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A. 2012. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 26:2604–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roch F, Jimenez G, Casanova J. 2002. EGFR signalling inhibits Capicua-dependent repression during specification of Drosophila wing veins. Development 129:993–1002 [DOI] [PubMed] [Google Scholar]
- 41.Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, Halder G. 2010. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 107:15810–15815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. 2010. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr. Biol. 20:573–581 [DOI] [PubMed] [Google Scholar]
- 43.Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, Wu S, Pan D. 2010. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc. Natl. Acad. Sci. U. S. A. 107:10532–10537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson BS, Huang J, Hong Y, Moberg KH. 2010. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein expanded. Curr. Biol. 20:582–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janody F, Lee JD, Jahren N, Hazelett DJ, Benlali A, Miura GI, Draskovic I, Treisman JE. 2004. A mosaic genetic screen reveals distinct roles for trithorax and polycomb group genes in Drosophila eye development. Genetics 166:187–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hay BA, Wassarman DA, Rubin GM. 1995. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell 83:1253–1262 [DOI] [PubMed] [Google Scholar]
- 47.Dorfman R, Shilo BZ. 2001. Biphasic activation of the BMP pathway patterns the Drosophila embryonic dorsal region. Development 128:965–972 [DOI] [PubMed] [Google Scholar]
- 48.Ashktorab H, Schaffer AA, Daremipouran M, Smoot DT, Lee E, Brim H. 2010. Distinct genetic alterations in colorectal cancer. PLoS One 5:e8879 doi:10.1371/journal.pone.0008879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, Ng S, Lin L, Crowder R, Snider J, Ballman K, Weber J, Chen K, Koboldt DC, Kandoth C, Schierding WS, McMichael JF, Miller CA, Lu C, Harris CC, McLellan MD, Wendl MC, DeSchryver K, Allred DC, Esserman L, Unzeitig G, Margenthaler J, Babiera GV, Marcom PK, Guenther JM, Leitch M, Hunt K, Olson J, Tao Y, Maher CA, Fulton LL, Fulton RS, Harrison M, Oberkfell B, Du F, Demeter R, Vickery TL, Elhammali A, Piwnica-Worms H, et al. 2012. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486:353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, Lim KH, Ong CK, Huang D, Chin SY, Tan IB, Ng CC, Yu W, Wu Y, Lee M, Wu J, Poh D, Wan WK, Rha SY, So J, Salto-Tellez M, Yeoh KG, Wong WK, Zhu YJ, Futreal PA, Pang B, Ruan Y, Hillmer AM, Bertrand D, Nagarajan N, Rozen S, Teh BT, Tan P. 2012. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat. Genet. 44:570–574 [DOI] [PubMed] [Google Scholar]
- 51.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent Winston SYF, Allis CD. 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15:3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bryk M, Briggs SD, Strahl BD, Curcio MJ, Allis CD, Winston F. 2002. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 12:165–170 [DOI] [PubMed] [Google Scholar]
- 53.Nislow C, Ray E, Pillus L. 1997. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol. Biol. Cell 8:2421–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]


