TEXT
The COMPASS family, which functions in the regulation of developmental gene expression, is a group of histone H3 lysine 4 (H3K4) methylases that is evolutionarily conserved from Saccharomyces cerevisiae (yeast) to human (1). Although there is only one Set1/COMPASS in yeast, Drosophila cells possess three yeast Set1-related proteins: dSet1, Trithorax (Trx), and Trithorax-related (Trr), all found within COMPASS-like compositions (1). Mammalian cells possess two representatives for each of the three subclasses found in Drosophila for a total of six COMPASS family members: SET1A and SET1B (related to dSet1); MLL1 and MLL2 (related to Trx); and MLL3 and MLL4 (related to Trr). Expansion of this family over evolutionary time implies a diversification in the function of H3K4 methylation, and studies into the distinct roles of the different branches of the COMPASS family support this notion. Drosophila and mammalian Set1 complexes mediate the bulk of genomic H3K4 di- and trimethylation (2–4). In contrast, the Trx/MLL1/2 complexes act in a highly gene-specific manner, in particular, controlling expression of distinct homeotic genes, including those within the Hox gene clusters (1, 5). MLL1 has been extensively studied in mouse models and human cells, as MLL1 translocations cause aggressive infant leukemias (6–8). Trr/MLL3/4 complexes are involved in nuclear hormone receptor signaling in both Drosophila and mammals (9, 10), and inactivating mutations have recently been implicated in human cancer (11–16). Mammalian MLL3/4 are large proteins (approximately 5,000 amino acids), whereas Drosophila Trr is homologous to the carboxy-terminal PHD, FYRN, FYRC, and SET domain of MLL3/4. A separate gene, LPT (Lost PHDs of Trr), encodes a protein homologous to the MLL3/4 amino terminus (3, 17). Moreover, Trr and LPT associate in the same complex, suggesting that a gene fission event had occurred in an ancestral gene in the Drosophila lineage (3).
Set1/COMPASS in yeast is unique in its ability to mono-, di-, and trimethylate its nucleosomal substrate (1, 18). The pattern of localization of histone H3K4 trimethylation (H3K4me3) and COMPASS on chromatin was first demonstrated to strongly correlate with transcriptionally active promoters in yeast (19), and this role of H3K4me3 in marking actively transcribed genes is highly conserved across the eukaryotes and is indeed used as a landmark for finding active promoters (20, 21). In contrast to H3K4me3, H3K4 monomethylation (H3K4me1) is found on poised and/or active enhancers (22, 23). Given that there are six COMPASS family members in mammalian cells, it was not clear until recently which COMPASS family member is involved in implementing H3K4me1 on enhancers. Recent work has now uncovered an unexpected role for Trr/MLL3/4 in gene regulation through enhancer-promoter communication. It was demonstrated that Trr functions as a major H3K4 monomethylase targeting enhancers in Drosophila (24). Moreover, loss of Trr impairs long-range enhancer function during Drosophila wing development. Given the strong association of H3K4me1 with enhancers (22) and the emerging connections between MLL3/4 and human disease, the relationship between Trr/MLL3/4 methylase activity and gene regulation is an area of burgeoning interest.
In this issue, Kanda and coworkers from the Hariharan laboratory (25) report the use of elegant genetic tools in Drosophila to shed light on Trr function during development and draw a striking parallel between Drosophila Trr and MLL3/4 mutations in human cancer. Using genetic mosaics, Kanda et al. demonstrate that during Drosophila eye development, cells lacking Trr have a clonal growth advantage over their wild-type counterparts. In agreement with recent work identifying Trr as a major H3K4 monomethylase involved in enhancer function (24), they observed a dramatic loss of H3K4me1 in trr mutant tissue accompanied by altered activity of key developmental signaling pathways, namely, Notch, Dpp/BMP, and receptor tyrosine kinases (RTK). In stark contrast to the growth advantage conferred by Trr deficiency, Trx mutant clones fail to proliferate and display increased apoptosis, mirroring the phenotypes observed in mammalian Mll1/2 loss-of-function studies (26, 27).
Quite remarkably, these distinct Trx (growth-promoting) versus Trr (growth-suppressing) functions may be conserved in mammals. Mll1 knockout mice lack hematopoetic stem cells and display embryonic proliferation defects, whereas gain-of-function Mll1 fusions cause aggressive leukemia (6, 27, 28). Similarly, Mll2 mutant embryos are severely growth retarded at early developmental stages and display widespread apoptosis (26). In contrast, mice lacking the Mll3 SET domain are viable but develop ureteric tumors, demonstrating a tumor suppressor function (29). Moreover, a series of genome-wide studies have identified loss-of-function mutations in MLL3 and MLL4 and in their cofactor, UTX, in diverse human cancers (11–16). Consistent with this, Drosophila Utx mutant clones also display an overgrowth phenotype (30). As for many of the Drosophila trr alleles characterized by the Hariharan laboratory, many cancer-associated MLL3 and MLL4 mutations result in truncation of point mutations in the catalytic SET domain (Fig. 1). Intriguingly, chromatin profiling in human cancer suggests a key role for H3K4me1. The genome-wide distribution of H3K4me1 undergoes a consistent alteration in colon cancer, often resulting in the loss of intestinal crypt-specific H3K4me1 marks (31). Collectively, these data provide evidence that Trr/MLL3/4-catalyzed H3K4 monomethylation functions to suppress tumorigenesis in specific contexts. The present work from the Hariharan laboratory is particularly significant and suggests that Drosophila eye mosaics could provide an ideal platform for dissecting the molecular mechanisms underlying MLL3/4 mutations in human cancer.
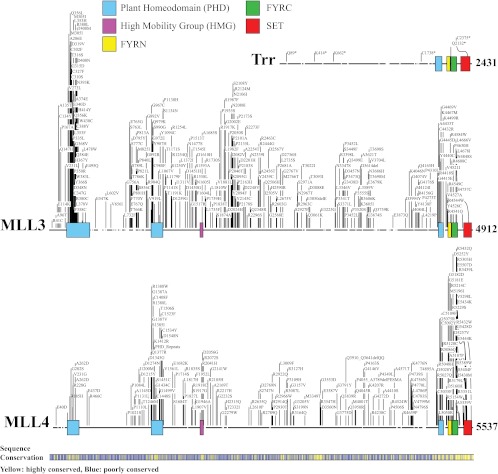
Fig 1.
Mutations of Trr and MLL3/4. Amino acid alignment of Trr, MLL3, and MLL4 was generated using CLC Sequence Viewer 6. Known protein domains are indicated. Sequence conservation between Trr, MLL3, and MLL4 is shown beneath the alignment. Yellow represents highly conserved regions, whereas blue indicates regions of poor sequence conservation between the 3 related proteins. Reported nonsense mutations of Trr, including those reported by Kanda et al. (25), are shown. Missense mutations of MLL3 and MLL4 were obtained from the Catalogue of Somatic Mutations in Cancer (COSMIC) database (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/) (35). Note that the COSMIC website reports sites of MLL4 mutations relative to an alternatively spliced transcript encoding a shortened 5,268-amino-acid protein. In the figure presented here, these positions were adjusted to match the 5,537-amino-acid protein that is most commonly reported in the literature.
Many important questions remain regarding Trr/MLL3/4 function. We do not understand what the precise mechanisms are that lead to overproliferation of trr mutant clones and MLL3/4 mutant cancer cells. What are the genome-wide targets affected by loss of Trr and MLL3/4, and are any of these targets conserved between Drosophila and mammals? What are the factors that recruit Trr/Mll3/4 to enhancer sequences, and do mammalian MLL3 and MLL4 share overlapping targets? Recent work suggests that direct enhancer-promoter interactions via cohesin complexes may organize the chromatin of the interphase nucleus (32–34). Could loss of the H3K4me1 and/or Trr/MLL3/MLL4 COMPASS-like complexes at enhancers grossly disrupt genome packaging and lead to genetic instability? The current work from the Hariharan laboratory firmly establishes Drosophila as a powerful genetic and biochemical model system to complement mammalian genetics and high-throughput sequencing of human cancer for the studies of Trr/MLL3/4 COMPASS-like complexes in development and disease.
ACKNOWLEDGMENTS
We thank Hans-Martin Herz and Edwin Smith for insightful discussions and for the critical reading of the manuscript and Laura Shilatifard for editorial assistance.
Footnotes
Published ahead of print 4 March 2013
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1. Shilatifard A. 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81: 65– 95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T. 2011. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 30: 2817– 2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, Shilatifard A. 2011. The COMPASS family of H3K4 methylases in Drosophila. Mol. Cell. Biol. 31: 4310– 4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. 2008. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol. Cell. Biol. 28: 7337– 7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, Ge K, Krumlauf R, Shilatifard A. 2009. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol. Cell. Biol. 29: 6074– 6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Daser A, Rabbitts TH. 2004. Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Genes Dev. 18: 965– 974 [DOI] [PubMed] [Google Scholar]
- 7. Mohan M, Lin C, Guest E, Shilatifard A. 2010. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat. Rev. Cancer 10: 721– 728 [DOI] [PubMed] [Google Scholar]
- 8. Rowley JD. 1993. Rearrangements involving chromosome band 11Q23 in acute leukaemia. Semin. Cancer Biol. 4: 377– 385 [PubMed] [Google Scholar]
- 9. Lee S, Kim DH, Goo YH, Lee YC, Lee SK, Lee JW. 2009. Crucial roles for interactions between MLL3/4 and INI1 in nuclear receptor transactivation. Mol. Endocrinol. 23: 610– 619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sedkov Y, Cho E, Petruk S, Cherbas L, Smith ST, Jones RS, Cherbas P, Canaani E, Jaynes JB, Mazo A. 2003. Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature 426: 78– 83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, Asangani IA, Ateeq B, Chun SY, Siddiqui J, Sam L, Anstett M, Mehra R, Prensner JR, Palanisamy N, Ryslik GA, Vandin F, Raphael BJ, Kunju LP, Rhodes DR, Pienta KJ, Chinnaiyan AM, Tomlins SA. 2012. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487: 239– 243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stütz AM, Rausch T, Warnatz HJ, Ryzhova M, Bender S, Sturm D, Pleier S, Cin H, Pfaff E, Sieber L, Wittmann A, Remke M, Witt H, Hutter S, Tzaridis T, Weischenfeldt J, Raeder B, Avci M, Amstislavskiy V, Zapatka M, Weber UD, Wang Q, Lasitschka B, Bartholomae CC, Schmidt M, von Kalle C, Ast V, Lawerenz C, Eils J, Kabbe R, Benes V, van Sluis P, Koster J, Volckmann R, Shih D, Betts MJ, Russell RB, Coco S, Tonini GP, Schüller U, et al. 2012. Dissecting the genomic complexity underlying medulloblastoma. Nature 488: 100– 105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, Jackman S, Krzywinski M, Scott DW, Trinh DL, Tamura-Wells J, Li S, Firme MR, Rogic S, Griffith M, Chan S, Yakovenko O, Meyer IM, Zhao EY, Smailus D, Moksa M, Chittaranjan S, Rimsza L, Brooks-Wilson A, Spinelli JJ, Ben-Neriah S, Meissner B, Woolcock B, Boyle M, McDonald H, Tam A, Zhao Y, Delaney A, Zeng T, Tse K, Butterfield Y, Birol I, Holt R, Schein J, Horsman DE, Moore R, Jones SJ, Connors JM, Hirst M, Gascoyne RD, Marra MA. 2011. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476: 298– 303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, Gallia GL, Jallo GI, Binder ZA, Nikolsky Y, Hartigan J, Smith DR, Gerhard DS, Fults DW, VandenBerg S, Berger MS, Marie SK, Shinjo SM, Clara C, Phillips PC, Minturn JE, Biegel JA, Judkins AR, Resnick AC, Storm PB, Curran T, He Y, Rasheed BA, Friedman HS, Keir ST, McLendon R, Northcott PA, Taylor MD, Burger PC, Riggins GJ, Karchin R, Parmigiani G, Bigner DD, Yan H, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. 2011. The genetic landscape of the childhood cancer medulloblastoma. Science 331: 435– 439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, Wells VA, Grunn A, Messina M, Elliot O, Chan J, Bhagat G, Chadburn A, Gaidano G, Mullighan CG, Rabadan R, Dalla-Favera R. 2011. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 43: 830– 837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, Greulich H, Lawrence MS, Lennon NJ, McKenna A, Meldrim J, Ramos AH, Ross MG, Russ C, Shefler E, Sivachenko A, Sogoloff B, Stojanov P, Tamayo P, Mesirov JP, Amani V, Teider N, Sengupta S, Francois JP, Northcott PA, Taylor MD, Yu F, Crabtree GR, Kautzman AG, Gabriel SB, Getz G, Jäger N, Jones DT, Lichter P, Pfister SM, Roberts TM, et al. 2012. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 488: 106– 110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith E, Lin C, Shilatifard A. 2011. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 25: 661– 672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schneider J, Wood A, Lee JS, Schuster R, Dueker J, Maguire C, Swanson SK, Florens L, Washburn MP, Shilatifard A. 2005. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol. Cell 19: 849– 856 [DOI] [PubMed] [Google Scholar]
- 19. Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11: 721– 729 [DOI] [PubMed] [Google Scholar]
- 20. Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77– 88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shilatifard A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75: 243– 269 [DOI] [PubMed] [Google Scholar]
- 22. Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108– 112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40: 897– 903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A. 2012. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 26: 2604– 2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanda H, Nguyen A, Chen L, Okano H, Hariharan IK. 2013. The Drosophila ortholog of MLL3 and MLL4, trithorax related, functions as a negative regulator of tissue growth. Mol. Cell. Biol. 33: 1702– 1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, Tufteland KR, Aasland R, Anastassiadis K, Ang SL, Stewart AF. 2006. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development 133: 1423– 1432 [DOI] [PubMed] [Google Scholar]
- 27. Yu BD, Hanson RD, Hess JL, Horning SE, Korsmeyer SJ. 1998. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc. Natl. Acad. Sci. U. S. A. 95: 10632– 10636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ. 2004. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev. Cell 6: 437– 443 [DOI] [PubMed] [Google Scholar]
- 29. Lee J, Kim DH, Lee S, Yang QH, Lee DK, Lee SK, Roeder RG, Lee JW. 2009. A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc. Natl. Acad. Sci. U. S. A. 106: 8513– 8518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Herz HM, Madden LD, Chen Z, Bolduc C, Buff E, Gupta R, Davuluri R, Shilatifard A, Hariharan IK, Bergmann A. 2010. The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol. Cell. Biol. 30: 2485– 2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, Myeroff L, Lutterbaugh J, Jarrar A, Kalady MF, Willis J, Moore JH, Tesar PJ, Laframboise T, Markowitz S, Lupien M, Scacheri PC. 2012. Epigenomic enhancer profiling defines a signature of colon cancer. Science 336: 736– 739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gause M, Schaaf CA, Dorsett D. 2008. Cohesin and CTCF: cooperating to control chromosome conformation? Bioessays 30: 715– 718 [DOI] [PubMed] [Google Scholar]
- 33. Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430– 435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sanyal A, Lajoie BR, Jain G, Dekker J. 2012. The long-range interaction landscape of gene promoters. Nature 489: 109– 113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, Teague JW, Stratton MR, Futreal PA. 2010. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 38: D652– D657 [DOI] [PMC free article] [PubMed] [Google Scholar]