Abstract
Dengue virus (DENV) causes dengue fever, a major health concern worldwide. We identified an amphipathic helix (AH) in the N-terminal region of the viral nonstructural protein 4A (NS4A). Disruption of its amphipathic nature using mutagenesis reduced homo-oligomerization and abolished viral replication. These data emphasize the significance of NS4A in the life cycle of the dengue virus and demarcate it as a target for the design of novel antiviral therapy.
TEXT
Dengue virus (DENV) infection is a growing public health threat, with more than one-third of the world population at risk (1). DENV is a positive single-strand RNA virus. Its genome is translated into a single polyprotein, which is cleaved to produce structural (components of the mature virus) and nonstructural (NS) proteins. In addition, the NS proteins generate the viral replication complexes (RC) (2). DENV replicates its RNA genome in association with modified intracellular membranes; the details of the assembly of these complexes are incompletely understood.
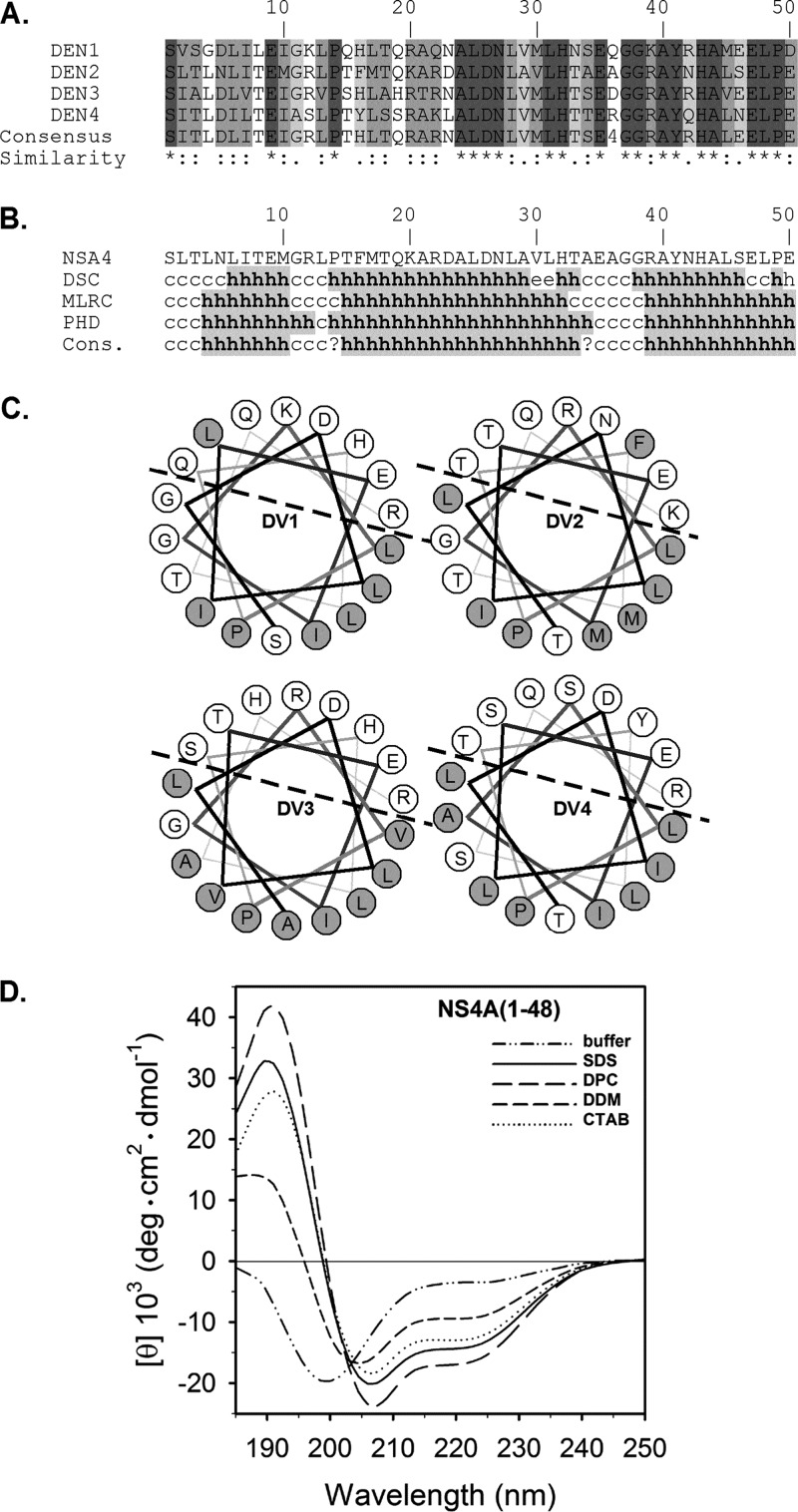
NS4A, a transmembrane endoplasmic reticulum (ER) resident protein, is thought to induce the host membrane modifications that harbor the viral RC (3). A similar function for NS4A was reported in other flaviviruses (4, 5). To further understand the role of NS4A, we analyzed its cytosolic N-terminal region (amino acids 1 to 48) using sequence alignment of the four DENV serotypes. Within this sequence, amino acids that differed in their identity maintained their biochemical properties, suggesting the presence of a conserved structural motif with a potential functional significance (Fig. 1A). Secondary structure algorithms (6) indicated that this segment is predicted to fold into an α-helix (Fig. 1B). Helical wheel projections of amino acids 3 to 20 indicated a conserved polar-nonpolar asymmetry indicative of an amphipathic helix (AH) (Fig. 1C). To experimentally examine the conformation of the NS4A N terminus, a recombinant peptide comprising amino acids 1 to 48 was prepared. Codon-optimized DENV2 NS4A 1-48 was cloned into pGEV2 (7) with an N-terminal fusion to the immunoglobulin binding domain of streptococcal protein G (GB1). A tobacco etch virus (TEV) protease cleavage site (ENLYFQ) was introduced into the beginning of the NS4A coding sequence. Due to difficulties in separating NS4A from GB1 after TEV protease cleavage, an N-terminal glutathione S-transferase (GST) affinity tag was added to GB1-TEV-NS4A(1-48) by cloning it into pGEX4T-2. Protein expression and purification of the GST-GB1-NS4A(1-48) fusion were performed as described previously (8), except that an on-column cleavage was performed by adding TEV protease. The flowthrough was concentrated and subjected to size exclusion chromatography (HiLoad 16/60 Superdex 75) yielding pure NS4A(1-48). The resulting peptide was characterized using far-UV circular dichroism (CD) (Fig. 1D). In aqueous buffer, NS4A(1-48) showed limited solubility and a CD spectrum typical of a random coil conformation with an α-helix content below 10%. Addition of various membrane-mimicking, micelle-forming detergents significantly increased the amount of α-helical content. This α-helical content varied from 30 to 40% in the presence of charged micelles (cetyl trimethylammonium bromide [CTAB], sodium dodecyl sulfate [SDS], and dodecyl phosphocholine [DPC]). In the presence of uncharged detergent (n-dodecyl-β-d-maltoside [DDM]) micelles, the helix content was significantly increased compared to detergent-free buffer, but was lower than other micelles. Together, these data support the existence of a conserved AH in the N terminus of NS4A.
Fig 1.
Secondary structure analysis of the N-terminal of NS4A. (A) ClustalW (23) multiple alignments of NS4A from the 4 DENV serotypes (accession numbers NP733810, NP739588 YP001531173, and NP740322). Similarities are shown as follows: asterisk, invariant amino acids; colon, highly similar; and period, different amino acids that are somewhat similar. The degree of similarity is also color coded in a gradient of gray color where the darkest gray is the most similar. (B) Secondary structure prediction (6). Structures are shown as helical (h), extended (e), turn (t), or undetermined (c, coil). (C) A helical wheel plot of NS4A amino acids 3 to 20. Hydrophilic residues are labeled in gray. Dashed line separates the putative polar and nonpolar faces of the helix. (D) Far-UV CD analysis of recombinant wild-type NS4A(1–48) peptide in 50 mM sodium phosphate buffer (pH 7.0) (dashed dotted line) and in the presence of 100 mM sodium dodecyl sulfate (SDS; solid line), n-dodecyl phosphocholine (DPC; long dashed line), n-dodecyl-β-d-maltoside (DDM; short dashed line), respectively, or in the presence of 1 mM cetyl trimethylammonium bromide (CTAB; dotted line). Spectra were recorded on a Jasco J810 spectrometer (Jasco, Groß-Umstadt, Germany) at 298 K in a 0.1-cm path-length quartz cuvette (Hellma) with 30 μM peptide. Data collection was done at 260 to 185 nm, with a 0.5-increment and a bandwidth of 1 nm. Spectra were baseline corrected and smoothed by a local smoothing technique with bisquare weighting and polynomial regression (SigmaPlot 11; Systat Software, Inc.). Units are given as the mean residue ellipticity ([θ]) by using peptide concentrations based on UV light absorption at 280 nm. The α-helical content was estimated with various deconvolution methods by the use of CDPro software (http://lamar.colostate.edu/∼sreeram/CDPro/main.html).
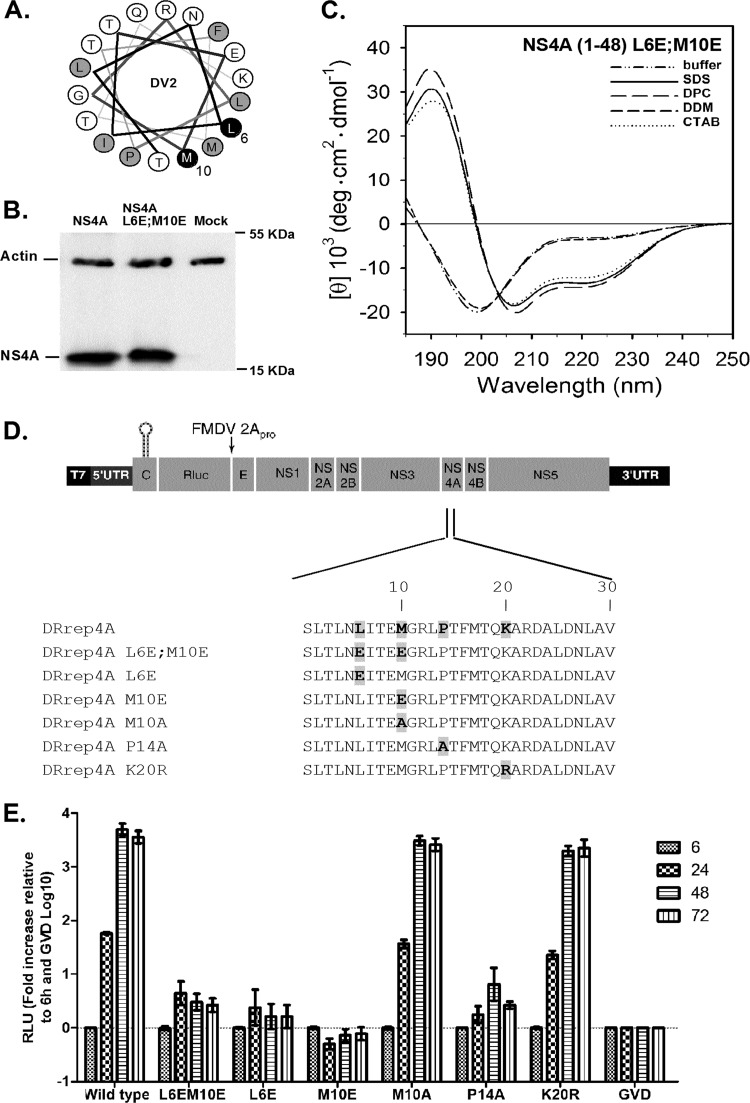
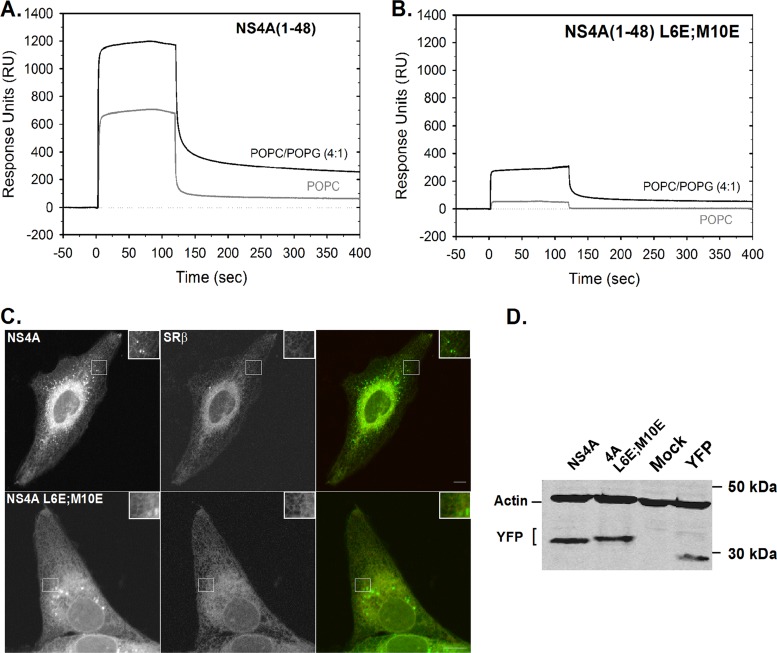
To test the relevance of this AH to the life cycle of DENV, the hydrophobic face of the helix was genetically disrupted by introduction of charged amino acids (L6E, M10E, or a combination designated L6E;M10E) (Fig. 2A) into the nonpolar face of the helix. A Western blot with comparable expression levels of full-length NS4A (in its mature form lacking the 2K signal sequence) (3, 9) and the L6E;M10E mutant is shown in Fig. 2B. A recombinant NS4A(1-48, L6E;M10E) mutant peptide was prepared using the plasmid described above and tail-to-tail mutagenesis to insert the L6E;M10E mutations (10). A CD analysis of the mutant peptide in the presence of charged micelles (SDS, DPC, and CTAB) did not lead to significant changes of the α-helix content compared to the wild-type peptide (Fig. 2C). In contrast to the wild type, the NS4A(1-48, L6E;M10E) mutant has lost its helix-forming capability in the presence of uncharged detergent (DDM) micelles (Fig. 2C). This result indicates that although the mutations did not change the helix propensity of NS4A(1-48), the wild-type and mutant peptides differ in their properties. The mutations were inserted into a luciferase reporter replicon of DENV2 (DRrep strain 16681) (11). RNA from mutated replicons was transcribed in vitro and transfected into BHK21 cells. Luciferase activity was measured as shown in Fig. 2E. DRrep with a lethal mutation (GVD) (12) in the RNA-dependent RNA polymerase served as a negative control. All of the AH mutants showed a severe replication defect displaying low levels of luciferase activity comparable to the GVD negative control. This result indicated that NS4A AH is essential for DENV replication. As a control for the mutagenesis experiment, we prepared two additional mutants, K20R and M10A; these replacements had no apparent effect on viral replication (Fig. 2E). Nevertheless, mutation of P14 to alanine abolished viral replication, indicating that this proline has a critical role in viral replication (Fig. 2E). AHs are typically known to associate with membranes, where their polar side faces the aqueous phase and the hydrophobic face is immersed in the membrane (13). Membrane binding of the AH was studied using surface plasmon resonance (SPR) (Fig. 3A and B). NS4A(1-48) and mutant NS4A(1-48, L6E;M10E) peptides were tested for their ability to bind to a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer or to a negatively charged POPC/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol (POPG; 4:1) bilayer. The sensogram in Fig. 3A shows that NS4A(1-48) binds strongly to chips coated with both bilayer types. Interestingly, the signal does not return to baseline levels during buffer washes. Moreover, repeated pulses of 100 mM sodium hydroxide or hydrochloric acid were not sufficient to restore the signal to the baseline (data not shown). This result indicates that once bound, the peptide remains tightly bound to the membranes, particularly to the negatively charged POPC/POPG bilayer. In contrast, injection of the NS4A(1-48, L6E;M10E) mutant resulted in a considerably low signal. Moreover, the mutant curve returned to baseline (POPC) or close to the baseline levels (POPC/POPG) following the termination of the peptide injection. These results indicate that the AH of NS4A has a high affinity for membranes in vitro and that the hydrophobic patch necessary for this membrane binding ability is disrupted in the L6E;M10E mutant.
Fig 2.
An intact N-terminal NS4A AH is required for DENV replication. (A) A helical wheel plot of NS4A amino acids 3 to 20 showing the location of the inserted mutations (black, L6E and M10E). (B) Comparable expression levels of full-length wild-type and L6E;M10E mutant NS4A as determined by Western blot analysis of FLAG-tagged wild-type or mutant NS4A expression levels using an anti-FLAG antibody. Monoclonal mouse anti-actin antibody was used as a loading control. (C) Spectra of the purified mutant peptide was recorded in 10 mM sodium phosphate buffer (pH 7.0) (dashed dotted line) or in the presence of 100 mM sodium dodecyl sulfate (SDS; solid line), n-dodecyl phosphocholine (DPC; long dashed line), n-dodecyl-β-d-maltoside (DDM; short dashed line), or 1 mM cetyl trimethylammonium bromide (CTAB; dotted line). (D) Schematic diagram of the DENV2 luciferase reporter replicon (pDRrep) used (11). Amino acids 1 to 30 of NS4A containing the AH region are shown for the wild-type and mutant replicons. Amino acid changes are in bold. (E) Replication assay for NS4A-AH mutated dengue replicons. Wild-type and mutated replicons were transfected into BHK21 cells. Luciferase activity was measured at 6, 24, 48, and 72 h posttransfection. Luciferase activity (relative light units [RLU]) is plotted for each time point. The data were normalized to the 6-h values that reflect transfection efficiency and to luciferase levels in replicon with a mutated polymerase. The mean values of at least two independent experiments performed in triplicates are shown. Error bars indicate standard errors of the means.
Fig 3.
The N-terminal NS4A AH binds membranes in vitro. (A) Binding of recombinant wild-type NS4A(1–48) peptide to lipid bilayers. Association of NS4A(1–48) to an L1 chip (GE Healthcare, Piscataway, NJ) covered with a POPC bilayer (gray) or a negatively charged bilayer containing 20% of POPG and 80% of POPC [POPC/POPG (4:1), black], prepared as described elsewhere (24). Sensograms were recorded with a Biacore X device (GE Healthcare) using a flow rate of 20 μl/min, an injection time of 1 min, and a peptide concentration of 50 μM. (B) Binding behavior of mutant NS4A(1–48, L6E;M10E) to lipid bilayers. Conditions are as in panel A, and the data shown are representative of three independent experiments. (C) Subcellular localization of NS4A-YFP fusion constructs. U2OS cells were cotransfected with wild-type NS4A-YFP or a similar construct with the AH mutations (M6E;M10E; see Fig. 2 for details) and SRβ-CFP. Images were captured 24 h posttransfection using a confocal microscope. Inserts show an enlarged portion of the cell (white square). Bar = 10 μm. (D) Comparable expression levels of the constructs shown in panel C as determined by Western blotting using an anti-GFP antibody. Monoclonal mouse anti-actin antibody was used as a loading control.
NS4A was previously implicated in the induction of the membrane domains that harbor the viral RCs (3). To create these rearrangements, NS4A most likely induces membrane curvature by an unknown mechanism (3). Oligomerization of membrane proteins can form a scaffold that can locally bend the membrane (14–16). A similar mechanism is thought to mediate membrane alterations induced by HCV NS4B (17, 18). To this end, we analyzed the effect of the above-mentioned mutations on the intracellular localization of NS4A.
Figure 3C shows live U2OS cells expressing the yellow fluorescent protein (YFP)-tagged full-length NS4A wild type or the L6E;M10E NS4A mutant and the β-subunit of the ER resident signal recognition particle (SRP) receptor (SRβ) fused to cyan fluorescent protein (CFP). Both NS4A proteins demonstrated localization to ER membranes and widely spread bright foci across the cell. Interestingly, although these NS4A-induced foci were previously shown to colocalize with a different ER marker (calreticulin) (3), SRβ was generally excluded from these foci. Photobleaching experiments demonstrated that NS4A was dynamically exchanged between ER membranes and the foci, which possibly represent densely packed, high-curvature membranes (data not shown). Both the wild-type and mutant NS4A YFP fusions showed similar expression levels (Fig. 3D).
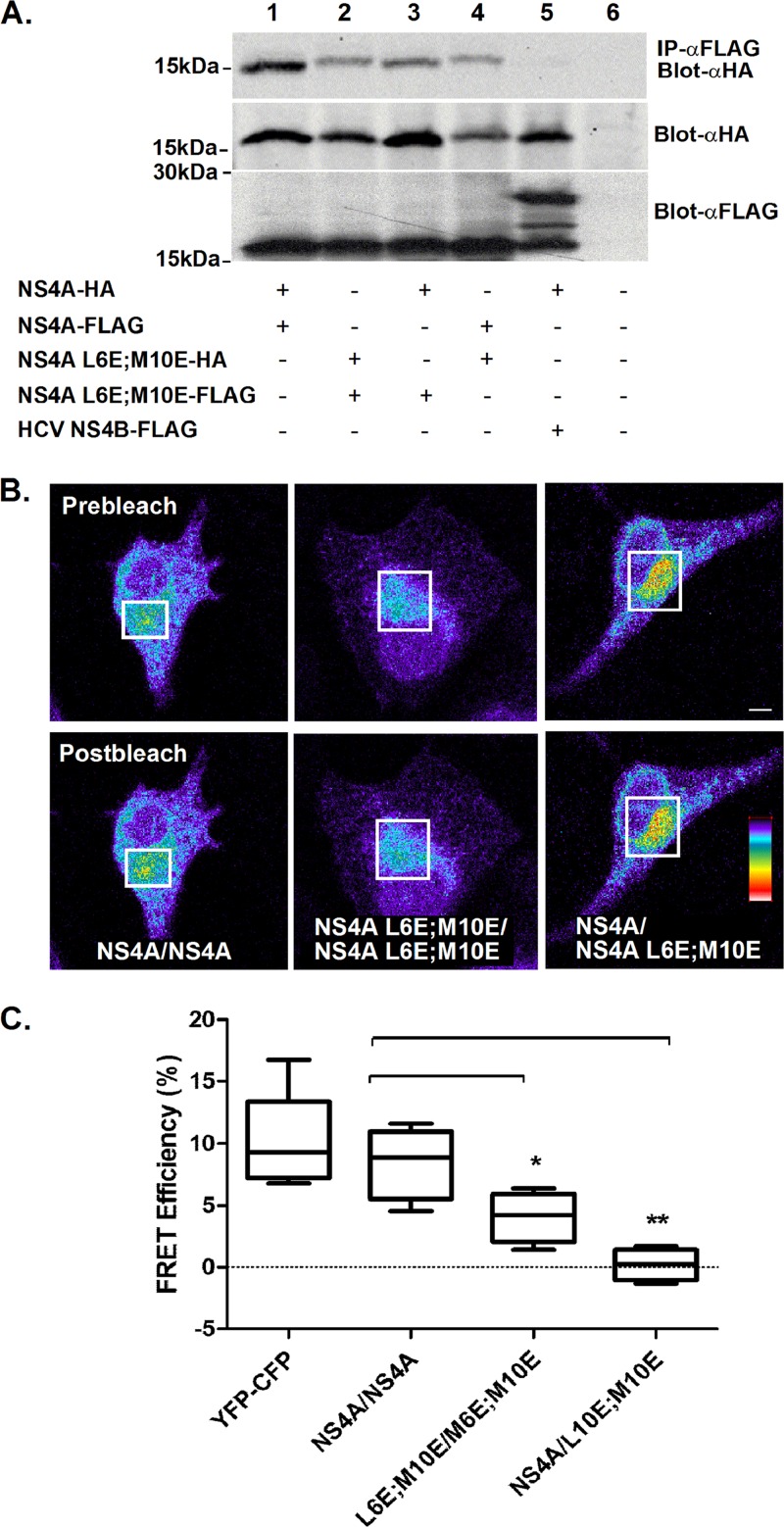
To test if NS4A can self-oligomerize, immunoprecipitation was performed. A modified vaccinia virus expressing T7 polymerase (19) was used to obtain increased levels of NS4A expression. Cells were infected with the recombinant vaccinia virus and cotransfected with full-length wild-type or mutant NS4A, each with a C-terminal FLAG or hemagglutinin (HA) tag. Cells coexpressing NS4A and HCV NS4B were used as a negative control. Lysates from cells expressing all combinations of these plasmids were subjected to immunoprecipitation using FLAG beads. Figure 4A shows that NS4A forms oligomers (lane 1). The ability of NS4A to form oligomers was not dramatically changed by the presence of the AH mutations (lanes 2 to 4). Since immunoprecipitation tests protein interactions in solubilized membranes, we verified NS4A self-association using an experimental approach that allows analysis of the proteins in their native intact membranes. On this premise, we analyzed the fluorescence resonance energy transfer (FRET) using acceptor photobleaching (20). Full-length NS4A or NS4A with AH mutants were fused to CFP or YFP which were coexpressed in U2OS cells. The cells were fixed 24 h posttransfection, and the NS4A-YFP (acceptor) was repeatedly bleached in a region of interest (ROI). The changes in NS4A-CFP (donor) and photobleached acceptor fluorescence were measured. FRET efficiency (EF) from at least 10 different cells was calculated using the equation EF = (Dpost − Dpre)/Dpost, where Dpost is the fluorescence intensity of the donor after acceptor photobleaching and Dpre is the fluorescence intensity of the donor before acceptor photobleaching. Figure 4B shows typical cells coexpressing CFP and YFP fusions of wild-type NS4A or NS4A mutants before (top panel) and after (bottom panel) photobleaching of the acceptor within the ROI (white square). Images are pseudocolored for fluorescence intensity; colors reflect the fluorescence intensity from lowest (black) to highest (red). The increase in donor fluorescence after acceptor photobleaching can be observed in the cell coexpressing a pair of wild-type NS4A (Fig. 4B). Figure 4C shows quantification of the fluorescence intensities within the labeled ROI. FRET was observed in cells coexpressing YFP- and CFP-tagged wild-type NS4A (mean EF, 8.5% ± 2.9%). Intermediate FRET efficiency was detected in cells coexpressing the AH mutants. FRET was not observed in cells (mean EF, 4% ± 2%) coexpressing both the wild type and the AH mutant. These results indicate that the AH has an important role in the self-oligomerization of NS4A. The intermediate FRET efficiency of the mutant pair might imply that additional elements within NS4A are required for oligomerization. The lack of FRET in the mixed wild-type–mutant pair might result from the alteration of the mutant's membrane binding capabilities; this may, in turn, induce a conformational change that dissociates the mutant NS4A from the wild type.
Fig 4.
NS4A forms AH-dependent oligomers. (A) Immunoprecipitation of wild-type NS4A and AH mutants. HEK293T cells were cotransfected with the indicated combinations. The cells were lysed 24 h posttransfection and subjected to immunoprecipitation using anti-FLAG beads followed by Western blotting with an anti-HA antibody. Direct Western blotting of the cell lysates with anti-FLAG and anti-HA antibodies are shown in the lower panels. Molecular mass markers are indicated on the left. (B) NS4A CFP and YFP fusion constructs and AH mutants thereof were transfected into U2OS cells as indicated. FRET was determined using acceptor photobleaching. Fluorescence of donor and acceptor was measured as a function of time during acceptor photobleaching. Intensities within the labeled (white square) region of interest were analyzed and are plotted in the graphs. Bar = 5 μm. Asterisks denote statistical significance in an up-paired Student t test. **, P < 0.05; ***, P < 0.0.005.
To summarize, we identified an N-terminal AH within NS4A that has a critical role in the DENV life cycle. As AHs were previously found to be amenable to pharmacological inhibition (21, 22), the data here merit consideration of the NS4A AH as a target for anti-DENV drugs. A more detailed study to investigate the exact structure of the AH in diverse membrane model systems and the role(s) of this motif in DENV replication is under way.
ACKNOWLEDGMENTS
This work was supported by a grant from the Sackler Faculty of Medicine V. Schreiber Fund (to E.H.S.) and by a grant from the Jürgen Manchot Foundation, Molecules of Infection Graduate School (to Y.-F.H.).
We thank Julian M. Glück for help with the SPR measurements and Simone König (Integrated Functional Genomics, University of Münster, Germany) for mass spectrometric analysis. Special thanks go to Koret Hirschberg (Tel Aviv University) and Bernd W. Koenig for critical reading of the manuscript and to Menashe Elazar (Stanford University) for helpful discussions.
Footnotes
Published ahead of print 16 January 2013
REFERENCES
- 1. Deen JL, Harris E, Wills B, Balmaseda A, Hammond SN, Rocha C, Dung NM, Hung NT, Hien TT, Farrar JJ. 2006. The WHO dengue classification and case definitions: time for a reassessment. Lancet 368:170–173 [DOI] [PubMed] [Google Scholar]
- 2. Lindenbach BD, Rice CM, Thiel HJ. 2007. Flaviviridae: the viruses and their replication, p 1101–1152 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed Lippincott, Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 3. Miller S, Kastner S, Krijnse-Locker J, Buhler S, Bartenschlager R. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 282:8873–8882 [DOI] [PubMed] [Google Scholar]
- 4. Mackenzie JM, Khromykh AA, Jones MK, Westaway EG. 1998. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245:203–215 [DOI] [PubMed] [Google Scholar]
- 5. Roosendaal J, Westaway EG, Khromykh A, Mackenzie JM. 2006. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J. Virol. 80:4623–4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Combet C, Blanchet C, Geourjon C, Deléage G. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25:147–150 [DOI] [PubMed] [Google Scholar]
- 7. Huth JR, Bewley CA, Jackson BM, Hinnebusch AG, Clore GM, Gronenborn AM. 1997. Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. 6:2359–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Feuerstein S, Solyom Z, Aladağ A, Hoffmann S, Willbold D, Brutscher B. 2011. 1H, 13C, and 15N resonance assignment of a 179 residue fragment of hepatitis C virus non-structural protein 5A. Biomol. NMR Assign. 5:241–243 [DOI] [PubMed] [Google Scholar]
- 9. Lin C, Amberg SM, Chambers TJ, Rice CM. 1993. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J. Virol. 67:2327–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hemsley A, Arnheim N, Toney MD, Cortopassi G, Galas DJ. 1989. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 17:6545–6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clyde K, Barrera J, Harris E. 2008. The capsid-coding region hairpin element (cHP) is a critical determinant of dengue virus and West Nile virus RNA synthesis. Virology 379:314–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guyatt KJ, Westaway EG, Khromykh AA. 2001. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J. Virol. Methods 92:37–44 [DOI] [PubMed] [Google Scholar]
- 13. Rees DC, DeAntonio L, Eisenberg D. 1989. Hydrophobic organization of membrane proteins. Science 245:510–513 [DOI] [PubMed] [Google Scholar]
- 14. McMahon HT, Gallop JL. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438:590–596 [DOI] [PubMed] [Google Scholar]
- 15. Miller S, Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yaffe Y, Shepshelovitch J, Nevo-Yassaf I, Yeheskel A, Shmerling H, Kwiatek JM, Gaus K, Pasmanik-Chor M, Hirschberg K. 2012. The MARVEL transmembrane motif of occludin mediates oligomerization and targeting to the basolateral surface in epithelia. J. Cell Sci. 125:3545–3556 [DOI] [PubMed] [Google Scholar]
- 17. Gouttenoire J, Roingeard P, Penin F, Moradpour D. 2010. Amphipathic alpha-helix AH2 is a major determinant for the oligomerization of hepatitis C virus nonstructural protein 4B. J. Virol. 84:12529–12537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu G-Y, Lee K-J, Gao L, Lai MMC. 2006. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J. Virol. 80:6013–6023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wyatt LS, Moss B, Rozenblatt S. 1995. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 210:202–205 [DOI] [PubMed] [Google Scholar]
- 20. Kenworthy AK. 2001. Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods 24:289–296 [DOI] [PubMed] [Google Scholar]
- 21. Cho NJ, Dvory-Sobol H, Lee C, Cho SJ, Bryson P, Masek M, Elazar M, Frank CW, Glenn JS. 2010. Identification of a class of HCV inhibitors directed against the nonstructural protein NS4B. Sci. Transl. Med. 2:15ra16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Elazar M, Cheong KH, Liu P, Greenberg HB, Rice CM, Glenn JS. 2003. Amphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replication. J. Virol. 77:6055–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cooper MA, Hansson A, Lofas S, Williams DH. 2000. A vesicle capture sensor chip for kinetic analysis of interactions with membrane-bound receptors. Anal. Biochem. 277:196–205 [DOI] [PubMed] [Google Scholar]