Abstract
Several novel anti-influenza compounds are in various phases of clinical development. One of these, T-705 (favipiravir), has a mechanism of action that is not fully understood but is suggested to target influenza virus RNA-dependent RNA polymerase. We investigated the mechanism of T-705 activity against influenza A (H1N1) viruses by applying selective drug pressure over multiple sequential passages in MDCK cells. We found that T-705 treatment did not select specific mutations in potential target proteins, including PB1, PB2, PA, and NP. Phenotypic assays based on cell viability confirmed that no T-705-resistant variants were selected. In the presence of T-705, titers of infectious virus decreased significantly (P < 0.0001) during serial passage in MDCK cells inoculated with seasonal influenza A (H1N1) viruses at a low multiplicity of infection (MOI; 0.0001 PFU/cell) or with 2009 pandemic H1N1 viruses at a high MOI (10 PFU/cell). There was no corresponding decrease in the number of viral RNA copies; therefore, specific virus infectivity (the ratio of infectious virus yield to viral RNA copy number) was reduced. Sequence analysis showed enrichment of G→A and C→T transversion mutations, increased mutation frequency, and a shift of the nucleotide profiles of individual NP gene clones under drug selection pressure. Our results demonstrate that T-705 induces a high rate of mutation that generates a nonviable viral phenotype and that lethal mutagenesis is a key antiviral mechanism of T-705. Our findings also explain the broad spectrum of activity of T-705 against viruses of multiple families.
INTRODUCTION
Influenza is one of the most significant acute upper respiratory tract infections for which therapeutic options are limited. The available influenza-specific drugs are limited to two classes: M2 ion channel blockers (amantadine and rimantadine) and neuraminidase (NA) inhibitors (oseltamivir and zanamivir) (1). However, influenza viruses evolve under drug selection pressure to generate strains resistant to these drugs. Resistance to M2 ion channel inhibitors is now so widespread that the Centers for Disease Control and Prevention recommends only the NA inhibitors for human use in the United States (2). Progress in the development of influenza therapeutics accelerated after the emergence of highly pathogenic H5N1 influenza viruses in 1997 and the novel H1N1 pandemic influenza strain in 2009 (3, 4). New strategies for the use of the currently licensed agents, including alternative forms of delivery and combination therapy with other drugs, are being explored (5). In addition, several novel antiviral compounds with different anti-influenza mechanisms are in various clinical phases of development.
Among the promising investigational agents is a substituted pyrazine compound, T-705 (6-fluoro-3-hydroxy-2-pyrazinecarboxamide [favipiravir]) developed by Toyama Chemical (6). T-705 demonstrated inhibitory activity against viruses of different families that utilize viral RNA-dependent RNA polymerase (RdRP) for genomic replication (7). The agent showed potent activity in vitro against flaviviruses (8), bunyaviruses (9), arenaviruses (9, 10), noroviruses (11), seasonal influenza A (H1N1, H2N2, and H3N2), B, and C viruses (6), and influenza A (H5N1) viruses (12, 13). Importantly, viruses resistant to both oseltamivir and amantadine were susceptible to T-705 (6, 14). Orally administered T-705 at a dose of ≥30 mg/kg of body weight/day prevented death, inhibited lung consolidation, and reduced lung virus titers in BALB/c mice lethally challenged with H5N1- and H3N2-subtype viruses (12, 13). T-705 is currently undergoing clinical evaluation in Japan and may soon be available there for clinical use.
T-705 acts as a prodrug, exerting its broad antiviral activity primarily through its active metabolite, T-705 ribofuranosyltriphosphate (T-705RTP); the active drug is generated intracellularly via phosphorylation by various kinases (15). In an enzyme-based assay, T-705RTP inhibited influenza virus RdRP in a dose-dependent manner (15). However, the mechanism by which this inhibition occurs is not completely understood, and the viral proteins targeted by T-705 are not defined. Similarly, the mechanism of ribavirin, a well-known nucleoside analog with potential effects against influenza viruses, is undefined. After its synthesis in 1972, several mechanisms of action were proposed, including inhibition of influenza virus RdRp (16–19). However, the high cytotoxicity of ribavirin in cell culture and significant side effects in humans eliminated this drug from clinical use as an anti-influenza agent (20). Uncertainty about ribavirin's anti-influenza mechanism of action has been an obstacle to understanding RdRP inhibitors' specific target(s) in the influenza virus genome and developing more efficacious nucleoside analogs for clinical use.
Like all RNA viruses, influenza viruses have a rapid mutation rate and naturally exist as mutant spectra termed “viral quasispecies,” which include subpopulations with point mutations, deletions in the viral genes, and defective interfering particles (21, 22). This high mutation rate is thought to favor adaptation of influenza viruses to the changing environment; however, this rate is close to the error threshold, and therefore even a modest increase in the mutation rate can negatively affect viral fitness, eventually driving the virus population to extinction (21). This concept currently serves as the basis for a new approach to antiviral intervention (23). The compounds that exert antiviral activity via lethal mutagenesis are nucleoside analogs, whose incorporation into the viral genome by virus polymerase leads to their accumulation and virus population collapse.
Currently available anti-influenza drugs are nonmutagenic influenza virus inhibitors that target specific virus proteins: NA inhibitors target the NA protein, and adamantanes target the transmembrane domain of the M2 protein (24). Influenza viruses have been shown to develop resistance to both NA inhibitors and adamantanes when serially passaged in the presence of the drugs in vitro (25, 26). The drug-resistant viruses are viable, demonstrate decreased susceptibility to the drug in phenotypic assays, and possess mutations in the targeted protein (in the catalytic and/or framework residues of the NA protein after passage with NA inhibitors and in the transmembrane region of M2 protein after serial passage with adamantanes). These characteristics reflect the accumulation of drug-resistant gain-of-fitness mutations during repeated replication under drug pressure. In this case, the drug is the selective agent that drives the viral fitness gradient. Very few mutations (in some cases, only one) are required for virus adaptation to the nonmutagenic inhibitors; furthermore, these mutations do not inflict a high fitness cost and can be incorporated into an evolving virus population.
Because T-705 is a nucleoside analog, its antiviral activity may be attributable primarily to its incorporation into viral RNA and a resulting increase in the virus's mutation frequency. Here we examined T-705 toxicity to MDCK and A549 cells, its activity against seasonal and pandemic A (H1N1) influenza viruses in vitro, and whether its mechanism of action is lethal mutagenesis. When influenza A virus was cultured with increasing concentrations of T-705, we observed a loss of specific virus infectivity, an increase in mutation frequency, and a shift in the nucleotide profile. These findings demonstrate that lethal mutagenesis is the predominant mechanism by which T-705 inhibits influenza virus replication in vitro.
MATERIALS AND METHODS
Cells, viruses, and compounds.
Madin-Darby canine kidney (MDCK) and human lung carcinoma (A549) cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained as previously described (27, 28). Seasonal influenza A (H1N1) viruses A/Brisbane/59/2007 and A/New Jersey/15/2007 and 2009 pandemic H1N1 viruses A/Denmark/524/2009 and A/Denmark/528/2009 were obtained from the St. Jude Children's Research Hospital influenza virus repository. Virus stocks were grown in MDCK cells at 37°C for 72 h. Compound T-705 (6-fluoro-3-hydroxy-2-pyrazinecarboxamide) and the NA inhibitors oseltamivir carboxylate ([3R,4R,5S]-4-acetamido-5-amino-3-[1-ethylpropoxy]-1-cyclohexene-1-carboxylic acid) and zanamivir (4-guanidino-Neu5Ac2en) were provided by Hoffmann-La Roche, Ltd. (Nutley, NJ). T-705 was prepared as 10 mM stocks in 10% dimethyl sulfoxide (DMSO) and stored in aliquots at −20°C. T-705 working solution was freshly prepared in infection medium for each experiment. Stocks of oseltamivir carboxylate and zanamivir were prepared in distillated water, filter sterilized, and stored in aliquots at −20°C.
Infectivity of influenza A (H1N1) viruses.
The infectivity of influenza A (H1N1) viruses was determined by plaque assay in MDCK cells. Briefly, confluent MDCK cell monolayers were incubated at 37°C for 1 h with 1 ml of 10-fold serial dilutions of virus. The cells were than washed to remove unbound viral particles and overlaid with 3 ml of Dulbecco's modified Eagle's medium (DMEM) containing 0.3% bovine serum albumin and 0.45% immunodiffusion-grade agarose (MP Biomedicals, Solon, OH) and with 1 μg/ml l-tosylamido 2-phenylethyl chloromethyl ketone (TPCK)-treated trypsin (Worthington, Lakewood, NJ). After 3 days of incubation at 37°C and 5% CO2, cells were stained with 1% crystal violet in 10% formaldehyde. The diameters of 20 randomly selected plaques were measured by using a Finescale Comparator (Los Angeles, CA).
Antiviral activity and cytotoxicity of T-705.
The viability of MDCK and A549 cells was tested after 2 and 24 h of incubation with T-705 by using the CellTiterGlo cell viability assay (Promega, Madison, WI) and the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay (Sigma-Aldrich, St. Louis, MO) according to the manufacturer's instructions. Luminescence and absorbance were measured by using a Synergy 2 multimode microplate reader (BioTek Instruments, Winooski, VT). The mean value of the negative control in each plate was set at 100% luminescence, and the percentage of luminescence of each compound-containing well was determined in relation to this internal control. The 50% cellular cytotoxicity (CC50) and 50% effective concentration (EC50) were determined by using the 4-parameter logistic nonlinear regression model equation in GraphPad Prism 5 software (GraphPad Software, La Jolla, CA).
Susceptibility to NA inhibitors.
A modified fluorometric assay using the fluorogenic substrate 4-methylumbelliferyl-a-d-N-acetylneuraminic acid (MUNANA; Sigma-Aldrich) (final concentration, 100 μM) was used to measure the inhibition of NA activity by oseltamivir carboxylate and zanamivir. H1N1 viruses were preincubated for 30 min at 37°C in the presence of various drug concentrations (0.05 to 500 nM), and the concentration required to inhibit 50% of the NA enzymatic activity (IC50) was determined by nonlinear regression using Graph Pad Prism 5 software. The mean of 2 to 3 independent determinations was calculated.
Serial passage in the presence and absence of T-705.
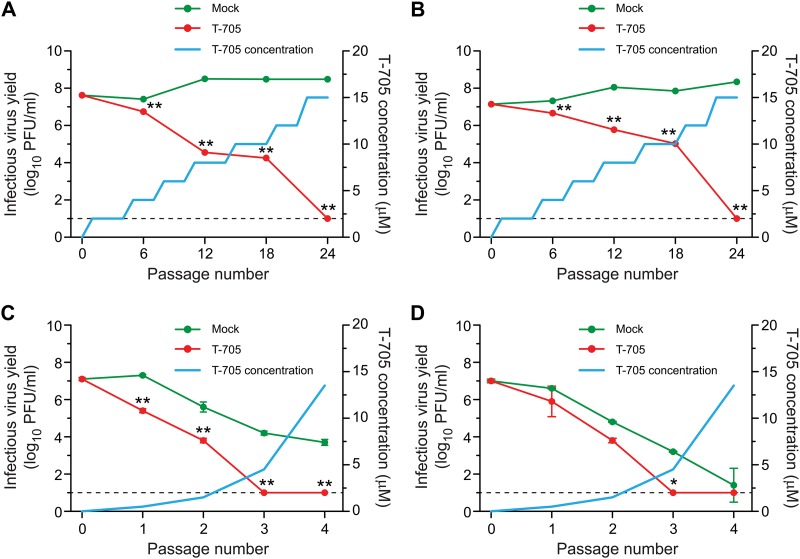
Two passaging approaches (using low and high infectious virus doses) were employed to assess the biological and genetic changes caused by T-705, including the emergence of viruses resistant to polymerase inhibitor. In the low-infectious-dose approach, confluent monolayers of MDCK cells (∼1.0 × 106 cells/well in 6-well plates) were infected with the seasonal influenza virus A/Brisbane/59/2007 or A/New Jersey/15/2007 at a multiplicity of infection (MOI) of 0.0001 PFU/cell. In each passage, the cultures were maintained for 72 h in 3 ml DMEM at 37°C and 5% CO2, in the presence or absence of T-705. The first passage was performed at a T-705 concentration of 0.5 μM, which was 1/30 the EC50. Supernatants from T-705-treated and mock-treated infected cells were harvested, clarified by centrifugation at 1800 rpm for 5 min at 4°C, and stored at −80°C. Virus was titrated by plaque assay in MDCK cells after each passage. For the second and subsequent passages, cells were infected by using supernatant from the previous passage containing an MOI of 0.001 PFU/cell. The two viruses were passaged 24 times at increasing T-705 concentrations (0.5 to 16 μM) (blue curve and scale at right in Fig. 1A and B). The parental viruses (MOI of 0.001 PFU/cell) were passaged in parallel in the absence of T-705.
Fig 1.
Infectivity of influenza A (H1N1) viruses during serial passage with T-705 in MDCK cells. Cells were inoculated with a low dose (MOI of 0.0001 PFU/cell) of influenza A/Brisbane/59/2007 (A) and A/New Jersey/15/2007 (B) viruses and a high dose (starting MOI of 10 PFU/cell) of A/Denmark/524/2009 (C) and A/Denmark/528/2009 (D) viruses. The T-705 concentration at each passage is indicated by the blue curve. After each passage, the infectivity (log10 PFU/ml) of progeny viruses was measured by plaque assay in MDCK cells. Values are the means ± standard deviations (SD) of two independent experiments. The dotted line indicates the assay's limit of detection (1.0 log10 PFU/ml). Virus titers in supernatants were compared by analysis of variance (ANOVA). *, P < 0.01; **, P < 0.0001.
In the high-infectious-dose approach, confluent monolayers of MDCK cells (48-well plates, ∼5.0 × 104 cells/well) were infected with 2009 pandemic influenza viruses (A/Denmark/524/2009 or A/Denmark/528/2009) at a starting MOI of 10 PFU/cell. In the first selection passage, the infected cells were treated with 0.5 μM T-705. After the appearance of cytopathic effects, 100 μl of the conditioned medium was used to infect fresh MDCK cells for the second selection passage in medium containing 1.5 μM T-705 (29). The third and fourth selection passages used 4.5 and 6.0 μM T-705, respectively. The T-705 doses used for each passage are shown as the blue curve (right-hand scale) in Fig. 1C and D. The parental viruses were passaged in parallel in the absence of T-705.
Cell-based influenza virus polymerase inhibition assay.
The inhibition of viral polymerase activity by T-705 was assessed in A549 cells by using a modified polymerase inhibition assay that utilizes whole virus (30). Reporter pPolI-358Luc was kindly provided by Megan Shaw (Mount Sinai School of Medicine, New York, NY). Each plate contained mock-infected cells (positive controls) and infected but untreated cells (negative controls). Luminescence was measured for 0.1 s/well in a Synergy 2 multimode microplate reader (BioTek Instruments, Winooski, VT). The mean value of the negative controls in each plate was set at 100% luminescence, and the percentage of luminescence of each compound-containing well was determined in relation to this internal control. The 50% inhibitory concentration (IC50) was determined by using the 4-parameter logistic nonlinear regression model equation in GraphPad Prism 5 software (GraphPad Software, La Jolla, CA).
Fixation of mutations in the viral genome.
RNA was extracted from the culture supernatants by using an RNeasy minikit (Qiagen, Germantown, MD) as directed by the manufacturer. RNA samples were amplified by reverse transcription-PCR (RT-PCR) by using the SuperScript III one-step RT-PCR system (Invitrogen). The list of primers used for amplification and sequencing is available upon request. The whole genomes of the influenza viruses were sequenced by the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Research Hospital, using Sanger sequencing-based technologies. The DNA template was sequenced by using rhodamine or dichlororhodamine dye terminator cycle sequencing ready reaction kits with AmpliTaq DNA polymerase FS (PerkinElmer Applied Biosystems, Waltham, MA). Samples were analyzed in a Perkin-Elmer Applied Biosystems DNA sequencer (model 3730 or 3770). DNA sequences were completed and edited by using the Lasergene sequence analysis software package (DNASTAR). The sequences were aligned and compared with the virus consensus sequence as a reference, and each new point mutation was recorded.
Mutation frequency with and without T-705.
MDCK cells were seeded into 48-well plates at 5 × 104 cells per well and incubated for 18 h at 37°C, 5% CO2, and 95% humidity. The cells were washed with Dulbecco's phosphate-buffered saline (DPBS) (Gibco), and the medium was replaced with DMEM containing mock compound (infection medium) or the compound of interest at the indicated concentrations. After 6 h of incubation, the cells were infected with influenza A/Denmark/524/2009 (H1N1) virus at an MOI of 0.001 PFU/cell and incubated for 1 h. Compounds were absent during the 1-h virus incubation but were present in the combination of DMEM infection medium plus 1 μg/ml TPCK-treated trypsin (Worthington, Lakewood, NJ). The infected cells were incubated at 37°C for 48 h, and cell supernatants were harvested and stored at −80°C. Cell monolayers were washed extensively with DPBS, incubated for 5 min at 37°C with enzyme-free PBS-based cell dissociation buffer (Gibco), and centrifuged for 5 min at 400 × g. Cell pellets were resuspended in 0.1 ml DMEM containing 5% fetal bovine serum (FBS) and underwent three freeze/thaw cycles in liquid nitrogen and a 37°C water bath. Samples were then centrifuged at 10,000 × g for 10 min at 4°C to remove cell debris. Extracellular and intracellular infectivity of influenza A/Denmark/524/2009 (H1N1) virus was determined by plaque assay, as described above, and virus RNA copies were determined by quantitative real-time RT-PCR assay, as described below, using total viral RNA from supernatants of infected mock-treated and T-705-treated (10 and 100 μM) MDCK cells. Samples treated with zanamivir (10 and 100 μM) were included for comparison. The nucleoprotein (NP) gene was amplified by RT-PCR, using the primer sets E01 (TATTCGTCTCAGGGAGCAAAAGCAGGGTA) and E02 (ATATCGTCTCGTATTAGTAGAAACAAGGGTATTTTTC). PCR products were gel purified (Qiagen, Germantown, MD) and TopoTA cloned (Invitrogen). Blue/white screening was used on single-colony transformants, and positive clones were sequenced. Sequence data were analyzed using the Lasergene sequence analysis software package (DNASTAR). For statistical purposes, we retained sequence data only over the region for which every clone was represented (1,513 nucleotides). The number of mutations per 104 nucleotides sequenced was determined as the total number of mutations identified per population over the total number of nucleotides sequenced for that population, multiplied by 104. For each population, 47 to 94 clones were sequenced (71,000 to 142,000 nucleotides per sample).
Quantitative real-time RT-PCR.
The primers, probe, and amplification protocol targeting the matrix (M) gene of influenza A viruses were described previously (31). For quantification of virus, the viral RNA copy number was derived from a standard curve generated using known concentrations of plasmid DNA containing the target gene. Conversion to an equivalent genome copy number to obtain the standard curve was done using the formula gene copy = [DNA amount (ng) × 6.022 × 1023 (Avogadro's no.)]/[length of template (bp) × 109 × 650 (weight/bp)].
Serial log dilutions of the DNA standard were used to generate a standard curve ranging from 1 × 103 to 1 × 1013 genome copies/ml of viral RNA. The standard curve is the linear regression line through the data points on a plot of threshold cycle (CT) versus the logarithm of standard sample concentration. The points at which the test sample values crossed the standard curve were determined from the regression line, and the corresponding concentrations were calculated.
Statistical analysis.
The titers of passaged viruses, number of RNA copies, and number of nucleotide mutations were compared by using the unpaired t test or analysis of variance (ANOVA). For comparisons of mutation frequency, the χ2 test was used to compare the total number of mutations in a population to the total number of nucleotides sequenced.
RESULTS
Antiviral activity and toxicity of T-705 in vitro.
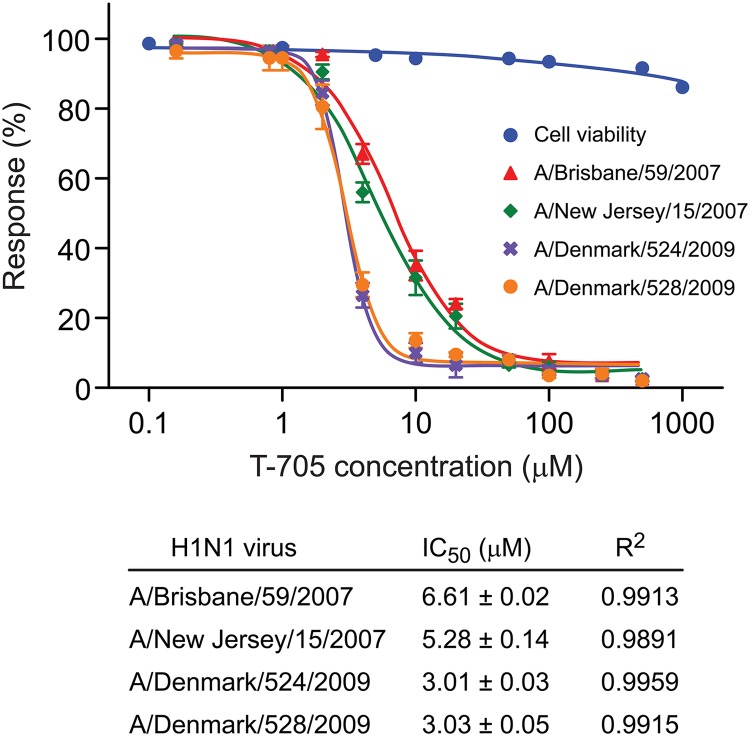
To select the initial T-705 concentrations and the cell line to be used in serial passaging studies, we assayed the drug's activity (IC50) against four influenza A (H1N1) viruses in A549 cells, using a polymerase activity assay. We used two pairs of recent human seasonal and pandemic 2009 H1N1 viruses (Table 1). One virus of each pair was oseltamivir resistant, and one was oseltamivir susceptible, to allow us to assess the influence of T-705 treatment on susceptibility to currently available antiviral drugs. The IC50s were 6.6 and 5.3 μM for seasonal influenza A/Brisbane/59/2007 and A/New Jersey/15/2007 viruses and 3.0 and 3.0 μM for the pandemic A (H1N1) viruses (A/Denmark/524/2009 and A/Denmark/528/2009) (Fig. 2). T-705 also appeared to be effective against all four viruses in MDCK cells inoculated at a low dose, with EC50s of 17.05 and 15.07 μM for the seasonal influenza viruses and 15.54 and 11.36 μM for the pandemic viruses (Table 1).
Table 1.
Susceptibility of influenza A (H1N1) viruses to T-705 and NA inhibitors before and after serial passage in MDCK cells in the presence and absence of T-705
| H1N1 virus | Passage | T-705 present | Susceptibility (mean ± SD) to: |
||
|---|---|---|---|---|---|
| T-705 (EC50 [μM])a | NA inhibitor (IC50 [nM])b |
||||
| Oseltamivir carboxylate | Zanamivir | ||||
| Seasonal viruses | |||||
| A/Brisbane/59/2007 | 0 | No | 17.05 ± 0.71 | 0.58 ± 0.00 | 0.38 ± 0.02 |
| 23 | No | 17.28 ± 0.78 | 0.55 ± 0.09 | 0.37 ± 0.04 | |
| 23 | Yes | 17.82 ± 0.27 | 0.51 ± 0.03 | 0.37 ± 0.00 | |
| A/New Jersey/15/2007 | 0 | No | 15.07 ± 0.14 | 296.85 ± 8.41 | 0.44 ± 0.00 |
| 23 | No | 12.50 ± 0.69 | 309.60 ± 6.16 | 0.49 ± 0.02 | |
| 23 | Yes | 12.95 ± 0.73 | 319.75 ± 0.35 | 0.32 ± 0.04 | |
| 2009 Pandemic viruses | |||||
| A/Denmark/524/2009 | 0 | No | 15.54 ± 0.56 | 0.42 ± 0.01 | 0.23 ± 0.03 |
| 3 | No | 14.76 ± 0.10 | 0.40 ± 0.01 | 0.28 ± 0.01 | |
| 3 | Yes | NDc | ND | ND | |
| A/Denmark/528/2009 | 0 | No | 11.36 ± 0.84 | 172.30 ± 3.11 | 0.33 ± 0.01 |
| 3 | No | 10.53 ± 0.73 | 186.10 ± 3.13 | 0.38 ± 0.00 | |
| 3 | Yes | ND | ND | ND | |
The concentration of T-705 required to protect 50% of MDCK cells (EC50) was determined by plotting the percentage of inhibition of cytotoxicity as a function of T-705 concentration and calculating values from the inhibitor-response curve by using GraphPad Prism 5 software. Values reflect three independent experiments.
The concentration required to inhibit 50% of NA enzymatic activity (IC50) as determined by using MUNANA substrate (100 μM final concentration). Values reflect three independent experiments.
ND, not determined due to low virus infectivity. The limit of detection was 1.0 log10 PFU/ml.
Fig 2.
Antiviral activity and toxicity of T-705 in vitro. A549 cells were transfected with the reporter pPolI-358Luc and infected with influenza A (H1N1) viruses (MOI of 2 PFU/cell) in the presence of increasing concentrations of T-705 (0.1 to 1,000 μM). At 24 h p.i., the results were assessed using the Bright-Glo luciferase reagent (Promega Corp., Madison, WI), following the manufacturer's instructions. The mean value of the negative controls in each plate was set at 100% luminescence, and the percentage of luminescence of each compound-containing well was determined in relation to it. The 50% inhibitory concentration (IC50) was determined by using the 4-parameter logistic nonlinear regression model equation in GraphPad Prism 5 software. Cell viability (CC50) was determined independently in mock-inoculated cells after 24 h of incubation using the CellTiterGlow kit, following the manufacturer's instructions. Values are means ± SD of three replicates.
As the observed antiviral activity might be attributable to metabolic inhibition of the host cells, we evaluated the effect of T-705 on the viability of uninfected MDCK cells. The CC50 value was >1,000 μM after 24 h of incubation (Fig. 2). Therefore, the selective index (SI = CC50/EC50) was >1, indicating that T-705 exerted strong anti-influenza activity but was not toxic to MDCK cells at the concentrations tested. To exclude T-705-induced mitochondrial toxicity, we used an MTT assay that assesses cellular mitochondrial metabolism. The results, like those reported by Furuta et al. (6), demonstrated no mitochondrial toxicity. We found no cytotoxicity in either MDCK or A549 cells at a concentration as high as 1,000 μM (data not shown).
On the basis of the antiviral activity and cytotoxicity of T-705, we selected T-705 concentrations of 2 μM and 0.5 μM for the initial passages of the seasonal and pandemic H1N1 viruses, respectively. To increase the likelihood of cumulative mutation, we used a 72-h incubation period for each passage. We also assessed the EC50s of the viruses in A549 cells; however, as the standard deviation of EC50s was relatively high in these cells after 72 h (data not shown), we used the MDCK cell line in subsequent studies.
Serial passage of influenza A viruses under T-705 pressure in MDCK cells.
We used serial passaging to increase the complexity of the mutant viral spectra (quasispecies) generated under T-705 pressure. Two different strategies were utilized: a low-dose virus inoculum (MOI of 0.0001 PFU/cell) and a high-dose virus inoculum (MOI of 10 PFU/cell). First, we serially passaged low-dose A/Brisbane/59/2007 and A/New Jersey/15/2007 viruses in MDCK cells in the presence of increasing concentrations of T-705 to allow the accumulation of mutant subpopulations and possibly select for a resistant phenotype. As influenza virus replication is prone to error and naturally generates defective interfering particles during serial passage (32), identical influenza viruses not treated with T-705 (mock-treated viruses) were serially passaged in parallel.
The titers of the two seasonal influenza viruses were approximately equal in T-705-treated and mock-treated cultures during passages 1 to 5 (Fig. 1A and B) but were significantly lower in treated cultures during passages 6 to 23 (P < 0.0001). After 24 passages, neither virus was detected by plaque assay in the presence of 15 μM T-705, while viruses in control (mock-treated) cells grew to high titers (>108 PFU/ml).
To assess the effect of the high-dose MOI on the accumulation of viral genomic defects under T-705 pressure, we conducted four serial passages of the 2009 pandemic H1N1 influenza viruses in MDCK cells. The T-705 concentration was increased by a factor of 3 in each successive passage (T-705 concentrations of 0.5 to 13.5 μM). Extinction of the virus population was observed after three passages in T-705-treated cells (no viable viruses were detected in plaque assays) (Fig. 1C and D).
By utilizing the serial passaging approach, we demonstrated a progressive decline of viral infectivity ultimately leading to viral extinction (loss of infectivity) during the passages in T-705-treated cells inoculated with either a low or high virus dose.
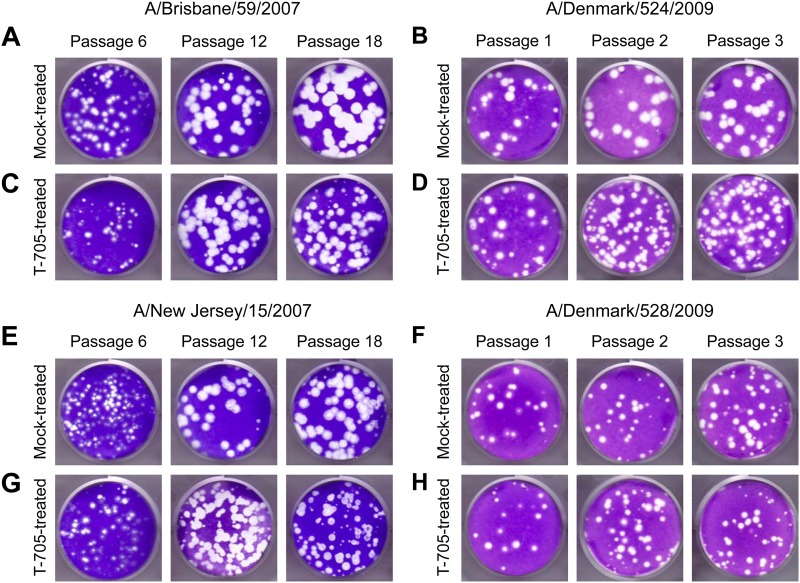
The plaque size of A/Brisbane/59/2007 and A/New Jersey/15/2007 viruses was significantly decreased (diameters of 0.26 ± 0.06 and 0.29 ± 0.06 mm, respectively, versus 0.5 ± 0.07 and 0.4 ± 0.09 mm in mock-treated cultures; P < 0.05) after passage 18 in T-705-treated MDCK cells (Fig. 3). After three sequential passages in T-705-treated MDCK cells, the plaque size of A/Denmark/524/2009 and A/Denmark/528/2009 influenza viruses was also decreased (diameters of 0.13 ± 0.07 and 0.16 ± 0.08 mm, respectively, versus 0.21 ± 0.04 and 0.29 ± 0.08 mm in mock-treated cultures) (Fig. 3). These results suggested that the changes observed in virus properties reflected T-705 selection of viral genetic composition.
Fig 3.
Plaque morphology of influenza A (H1N1) viruses during serial passage with T-705 in MDCK cells. Panels show plaques from cultures inoculated with low (A, C, E, and G) and high (B, D, F, and H) infectious doses of influenza virus: (A) A/Brisbane/59/2007, mock treated; (B) A/Denmark/524/2009, mock treated; (C) A/Brisbane/59/2007, T-705 treated; (D) A/Denmark/524/2009, T-705 treated; (E) A/New Jersey/15/2007, mock treated; (F) A/Denmark/528/2009, mock treated; (G) A/New Jersey/15/2007, T-705 treated; (H) A/Denmark/528/2009, T-705 treated. After the indicated serial passage, MDCK cells were inoculated with supernatant containing the specified virus and overlaid with 0.45% immunodiffusion-grade agarose (MP Biomedicals, Solon, OH) containing 1 μg/ml TPCK-treated trypsin. After 3 days of incubation at 37°C and 5% CO2, the cells were stained with 1% crystal violet in 10% formaldehyde.
Susceptibility of passage-generated variants to T-705.
To investigate the possible emergence of a T-705-resistant phenotype during serial passage under drug pressure, we compared T-705 EC50s after 6, 12, and 18 passages in T-705-treated and mock-treated cultures. In T-705-treated cells inoculated with high or low virus doses, the mean EC50s remained stable during passages 0 to 23 (A/Brisbane/59/2007, 17.82 ± 0.27 μM; A/New Jersey/15/2007, 12.95 ± 0.73 μM) and did not differ significantly from those in mock-treated cultures (Table 1). Importantly, the susceptibility of the seasonal and pandemic H1N1 viruses to NA inhibitors (oseltamivir carboxylate and zanamivir) was not altered by serial passage with or without T-705 (Table 1). Thus, by using both low and high virus infectious doses, we demonstrated the absence of phenotypically resistant viruses after multiple rounds of replication in vitro with T-705. This finding indicates that the selection of drug-specific resistance mutations is unlikely, at least in vitro.
Fixation of mutations into the virus population.
To screen for possible mutations in the viral genome during sequential passages, we sequenced the eight genes of the A/Brisbane/59/2007 and A/New Jersey/15/2007 viruses after 12 and 24 serial passages and those of A/Denmark/524/2009 and A/Denmark/528/2009 viruses after 4 serial passages, in T-705-treated and mock-treated MDCK cells. We observed two amino acid substitutions in the influenza A/Brisbane/59/2007 virus, at residue 203 (I203N) of the HA protein and at residue 377 (P377L) of the NA protein. These mutations were present in both T-705-treated and mock-treated cells, suggesting that they were host-selected mutations acquired during serial passage. Only a single amino acid substitution, at residue 34 of NA (A34T), was specific for viruses from T-705-treated cells. In the influenza A/New/Jersey/15/2007 virus, we observed an amino acid substitution at NA residue 377 (P377L) that was acquired during passage in MDCK cells, and we found two changes in the NA protein (I53T and N248D) that were specific for T-705-treated cells. In the two pandemic viruses, we found a single host-selected D103N amino acid substitution in the HA protein that was present in both T-705-treated and mock-treated viruses.
Interestingly, the mock-treated A/Brisbane/59/2007 and A/New/Jersey/15/2007 clones contained 11 and 7 single-nucleotide substitutions, respectively, among the 7,749 sequenced nucleotides. In contrast, the T-705-treated A/Brisbane/59/2007 and A/New Jersey/15/2007 viruses had 30 and 24 single-nucleotide substitutions, respectively. These changes showed a significant increase in G→A and C→T nucleotide mutations in both of the T-705-treated seasonal viruses. A single G→A nucleotide mutation was observed in 2009 H1N1 pandemic viruses after passage 4 with T-705 (Table 2). These observations indicated an increase in fixed nucleotide mutations in treated versus untreated viruses, suggesting that T-705RTP (the active metabolite of T-705) may induce G→A mutations during positive-strand RNA synthesis, whereas C→T mutations are predicted to result from incorporation of T-705RTP during negative-strand RNA synthesis.
Table 2.
Number of mutations observed in influenza A (H1N1) viruses after serial passage in MDCK cells treated with T-705
| H1N1 virus | Passage | No. of nucleotide changes ina: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T-705 treated |
Mock treated |
||||||||||||||
| G→A | C→T | A→G | T→C | T→A | C→A | G→C | G→A | C→T | A→G | T→C | T→A | C→A | G→C | ||
| Seasonal viruses | |||||||||||||||
| A/Brisbane/59/2007 | 12 | 2 | − | − | − | 2 | − | − | − | 2 | − | − | 2 | − | − |
| 24 | 9* | 8* | 2 | 1 | 2 | 1 | 1 | 1 | 2 | − | 1 | 2 | 1 | − | |
| A/New Jersey/15/2007 | 12 | − | 2 | 2 | 2 | − | − | − | − | − | − | 1 | − | − | − |
| 24 | 5* | 5* | 4 | 3 | − | 1 | − | 1 | − | 2 | 3 | − | − | − | |
| 2009 Pandemic viruses | |||||||||||||||
| A/Denmark/524/2009 | 4 | 1 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| A/Denmark/528/2009 | 4 | 1 | − | − | − | − | − | − | − | − | − | − | − | − | − |
Whole virus genomes were sequenced. Shown is the number of nucleotide changes in a total of 7,749 nucleotide sequences in viruses from the supernatants of T-705-treated MDCK cells compared to those in mock-treated MDCK cells. −, no nucleotide changes detected; *, P < 0.05 compared to mock-treated samples (unpaired two-tailed t test).
Effect of T-705 on virus infectivity in MDCK cells.
To investigate the mechanism by which T-705 leads to viral extinction in vitro, we first compared the infectivity of T-705-treated viruses to that of viruses mock treated with T-705 after 6, 12, 18, and 24 passages in MDCK cells inoculated with low virus doses. Titers of the seasonal influenza viruses remained stable in mock-treated cells over passages 6 to 24, ranging from 7.4 to 8.5 log10 PFU/ml (A/Brisbane/59/2007) and from 7.3 to 8.3 log10 PFU/ml (A/New Jersey/15/2007) (Table 3). In contrast, the mean infectious titers of both viruses decreased significantly under T-705 selection (from 6.7 log10 PFU/ml to undetectable; P < 0.0001) during passages 6 to 24.
Table 3.
Effect of T-705 on infectious virus yield and viral RNA copy number during serial passage in MDCK cells inoculated with a low dose of seasonal influenza A (H1N1) virusa
| H1N1 virus | Passage | Infectious virus yield (log10 PFU/ml)b |
Viral RNA copy no. (log10 genome copies/ml)c |
Specific infectivity (infectious virus yield/viral RNA copy no.)d |
|||
|---|---|---|---|---|---|---|---|
| Mock treatment | T-705 treatment | Mock treatment | T-705 treatment | Mock treatment | T-705 treatment | ||
| A/Brisbane/59/2007 | 0 | 7.6 ± 0.18 | NAe | 9.3 ± 0.33 | NA | 0.8 ± 0.26 | NA |
| 6 | 7.4 ± 0.10 | 6.7 ± 0.05** | 9.2 ± 0.01 | 9.0 ± 0.09 | 0.8 ± 0.06 | 0.7 ± 0.07 | |
| 12 | 8.5 ± 0.02 | 4.6 ± 0.02** | 10.1 ± 0.05 | 8.4 ± 0.14* | 0.8 ± 0.04 | 0.5 ± 0.08 | |
| 18 | 8.5 ± 0.16 | 4.3 ± 0.17** | 10.2 ± 0.09 | 8.7 ± 0.04* | 0.8 ± 0.13 | 0.5 ± 0.11 | |
| 24 | 8.5 ± 0.03 | <1.0** | 8.5 ± 0.69 | 4.3 ± 0.17** | 1.0 ± 0.36 | 0.2 ± 0.17** | |
| A/New Jersey/15/2007 | 0 | 7.1 ± 0.06 | NA | 9.7 ± 0.03 | NA | 0.7 ± 0.05 | NA |
| 6 | 7.3 ± 0.06 | 6.7 ± 0.04** | 9.7 ± 0.04 | 9.6 ± 0.01 | 0.8 ± 0.05 | 0.7 ± 0.03 | |
| 12 | 8.0 ±0.09 | 5.8 ± 0.04** | 9.9 ± 0.03 | 9.0 ± 0.12** | 0.8 ± 0.06 | 0.6 ± 0.08 | |
| 18 | 7.9 ± 0.08 | 5.0 ± 0.01** | 9.9 ± 0.09 | 9.4 ± 0.02 | 0.8 ± 0.09 | 0.5 ± 0.02 | |
| 24 | 8.3 ± 0.13 | <1.0** | 8.7 ± 0.68 | 3.8 ± 0.13** | 1.0 ± 0.41 | 0.3 ± 0.13** | |
*, P < 0.01, or **, P < 0.0001, significant differences with respect to mock-treated samples by two-way ANOVA.
Infectious virus was titrated by plaque assay in MDCK cells. Values are means ± SD from three independent experiments.
Virus RNA copy number was determined by quantitative real-time RT-PCR of the matrix gene. Values are means ± SD from three independent experiments.
Infectious virus yield × 10−3/genome copy number, determined 24 h p.i.
NA, not applicable.
To distinguish between naturally occurring defective virus particles and T-705 activity as the cause of virus extinction, we used the high-dose inoculation approach. Titers of mock-treated viruses were 7.3 to 3.7 log10 PFU/ml (A/Denmark/524/2009) and 6.6 to 1.4 log10 PFU/ml (A/Denmark/528/2009) during passages 1 to 4 (Table 4). In the presence of T-705, however, no infectious virus of either type was identified after 3 serial passages.
Table 4.
Effect of T-705 on infectious virus yield and viral RNA copy number during serial passage in MDCK cells inoculated with a high dose of 2009 pandemic influenza A (H1N1) virusa
| H1N1 virus | Passage | Infectious virus yield (log10 PFU/ml)b |
Virus RNA copy no. (log10 genome copies/ml)c |
Specific infectivity (infectious virus yield/viral RNA copy no.)d |
|||
|---|---|---|---|---|---|---|---|
| Mock treatment | T-705 treatment | Mock treatment | T-705 treatment | Mock treatment | T-705 treatment | ||
| A/Denmark/524/2009 | 0 | 7.1 ± 0.09 | NAe | 9.4 ± 0.07 | NA | 0.8 ± 0.08 | NA |
| 1 | 7.3 ± 0.05 | 5.4 ± 0.11** | 8.3 ± 0.64 | 8.2 ± 0.64 | 0.9 ± 0.35 | 0.7 ± 0.75 | |
| 2 | 5.6 ± 0.27 | 3.8 ± 0.12** | 6.0 ± 0.43 | 5.1 ± 0.30 | 0.9 ± 0.35 | 0.7 ± 0.21 | |
| 3 | 4.2 ± 0.11 | <1.0** | 5.9 ± 0.39 | 3.5 ± 0.12* | 0.7 ± 0.25 | 0.3 ± 0.12 | |
| 4 | 3.7 ± 0.16 | <1.0** | 5.5 ± 0.37 | <3.0** | 0.7 ± 0.27 | <0.3 | |
| A/Denmark/528/2009 | 0 | 7.0 ± 0.10 | NA | 9.2 ± 0.24 | NA | 0.8 ± 0.17 | NA |
| 1 | 6.6 ± 0.03 | 5.9 ± 0.82 | 7.8 ± 0.61 | 7.8 ± 0.63 | 0.8 ± 0.32 | 0.8 ± 0.73 | |
| 2 | 4.8 ± 0.03 | 3.8 ± 0.12 | 5.6 ± 0.44 | 5.5 ± 0.37 | 0.9 ± 0.24 | 0.7 ± 0.25 | |
| 3 | 3.2 ± 0.03 | <1.0* | 4.7 ± 0.22 | 3.0 ± 0.10* | 0.7 ± 0.13 | 0.3 ± 0.10 | |
| 4 | 1.4 ± 0.91 | <1.0 | 4.2 ± 0.5 | <3.0** | 0.3 ± 0.71 | <0.3 | |
*, P < 0.01, or **, P < 0.0001, significant differences with respect to mock-treated samples by two-way ANOVA.
Infectious virus was titrated by plaque assay in MDCK cells. Values are means ± SD from three independent experiments.
Virus RNA copy number was determined by quantitative real-time PCR of the matrix gene. Values are means ± SD from three independent experiments.
Infectious virus yield × 10−3/genome copy number, determined 24 h p.i.
NA, not applicable.
These findings showed that extinction of the virus population under T-705 pressure paralleled the loss of virus infectivity at both the low and high virus doses. Therefore, T-705-induced mutagenesis was the likely cause of influenza virus extinction.
Comparison of RNA load with infectivity.
Our next step in investigating the mechanism of T-705-induced viral extinction was to quantify viral RNA at each passage by quantitative real-time RT-PCR. In mock-treated cells inoculated with a low virus dose, the viral RNA load did not change significantly, ranging from 9.2 to 10.2 log10 genome copies/ml (A/Brisbane/59/2007 virus) and from 9.7 to 9.9 log10 genome copies/ml (A/New Jersey/15/2007 virus) (Table 3). In T-705-treated cells, the viral RNA load remained unchanged during 18 passages (9.0 to 8.7 log10 genome copies/ml for A/Brisbane/59/2007 virus and 9.6 to 9.4 log10 genome copies/ml for A/New Jersey/15/2007 virus), despite the dramatic decline in infectivity.
In cells inoculated at a high MOI, passage of influenza viruses resulted in a decreased viral RNA load, from 8.3 to 5.5 log10 genome copies/ml (A/Denmark/524/2009) and from 7.8 to 4.2 log10 genome copies/ml (A/Denmark/528/2009) in mock-treated cells. This observation is consistent with the defective interfering particle theory, which holds that viruses are prone to generate defective genomes during their natural infectious cycle. Importantly, when the same viruses were passaged in the presence of T-705, the number of viral RNA copies declined dramatically (from 8.2 log10 genome copies/ml of A/Denmark/524/2009 and 7.8 log10 genome copies/ml of A/Denmark/528/2009 after passage 1 to undetectable levels of both viruses after passage 4) (Table 4).
These results demonstrated that T-705 treatment induced interfering activity greater than that observed in mock-treated cells (33). The reduction of virus infectivity was disproportionately greater than the reduction of viral RNA copy number (Table 4). This finding supports the hypothesis that the antiviral effect of T-705 is caused by lethal mutagenesis of the internal genes of influenza viruses.
Effect of T-705 on specific viral infectivity.
Our next step in investigating the mechanism of T-705-induced viral extinction was to explore the kinetics of the loss of specific infectivity of influenza H1N1 virus under various T-705 concentrations by analyzing both the extracellular and intracellular pools of A/Denmark/524/2009 (H1N1) virus. For comparison, samples treated with the NA inhibitor zanamivir were included. Zanamivir (10 and 100 μM) did not significantly alter either infectious virus yield or RNA load as determined 48 h postinfection (p.i.). Treatment with T-705 decreased the infectious virus yield in a dose-dependent manner, and extracellular virus was below the limit of detection in samples treated with 100 μM T-705 (Table 5). The viral RNA load did not change significantly at 10 μM T-705, but 100 μM T-705 significantly reduced RNA copy number in the extracellular pool. These results demonstrated a significant reduction of specific viral infectivity (P < 0.01) under the pressure of T-705.
Table 5.
Specific infectivity of influenza A/Denmark/524/2009 (H1N1) virus in the presence and absence of T-705 and zanamivira
| Drug | Concn (μM) | Infectious virus yield (log10 PFU/ml)b |
Virus RNA copy no. (log10 genome copies/ml)c |
Specific infectivity (infectious virus yield × 10−3/genome copy no.) |
|||
|---|---|---|---|---|---|---|---|
| Extracellular | Intracellular | Extracellular | Intracellular | Extracellular | Intracellular | ||
| None (mock treated) | 0 | 6.71 ± 0.03 | 6.84 ± 0.15 | 10.04 ± 0.14 | 10.06 ± 0.64 | 0.490 ± 0.175 | 0.714 ± 0.600 |
| T-705 | 10 | 4.68 ± 0.08* | 5.29 ± 0.08 | 9.40 ± 0.02 | 9.70 ± 0.01 | 0.019 ± 0.004* | 0.039 ± 0.01* |
| 100 | <1.0* | 1.24 ± 0.3* | 5.83 ± 0.15* | 7.19 ± 0.05 | 0.001 ± 0.000* | 0.001 ± 0.00* | |
| Zanamivir | 10 | 5.17 ± 0.22 | 5.89 ± 0.04 | 8.80 ± 0.11 | 10.30 ± 0.15 | 0.224 ± 0.084 | 0.128 ± 0.110 |
| 100 | 5.73 ± 0.08 | 5.72 ± 0.05 | 8.46 ± 0.11 | 9.57 ± 0.14 | 0.184 ± 0.013 | 0.148 ± 0.050 | |
*, P < 0.01 in comparison to mock-treated samples (two-way ANOVA).
Infectious virus in supernatants and cellular lysates was titrated by plaque assay in MDCK cells. Values are means ± SD from two independent experiments.
Virus RNA copy number was determined by quantitative real-time PCR of the matrix gene. Values are means ± SD from three independent experiments.
Mutation frequency with and without T-705.
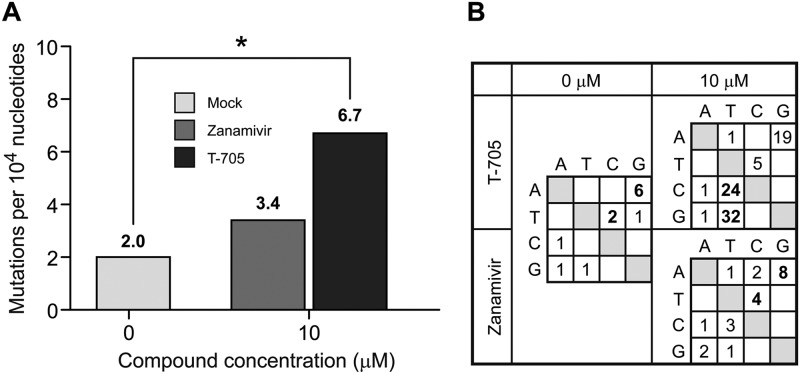
To demonstrate that T-705 exerts mutagenic activity on the influenza virus genome, we investigated mutation frequency by analyzing individual clones of influenza A/Denmark/524/2009 obtained from infected MDCK cells treated with T-705 (10 and 100 μM) or mock treated and by sequencing their NP genes. For each population, between 47 and 94 clones were sequenced, representing 71,000 to 142,000 nucleotides per sample (Fig. 4). The mock-treated samples and samples treated with zanamivir were used as controls. Zanamivir was chosen as a control because it targets the NA gene and prevents the release of influenza virions from an infected cell but appears unlikely to significantly increase mutation frequency. Ribavirin is a well-known virus mutagen but was not used as a control because no mutagenic activity has been demonstrated in influenza viruses. Our T-705-treated (10 μM) samples showed a significantly higher mutation frequency (P < 0.0001) than mock-treated samples (Fig. 4). Interestingly, two attempts to sequence 200 individual clones from samples treated with 100 μM T-705 failed to obtain sufficient base pairs to calculate the mutation rate per 10,000 nucleotides. Although both T-705-treated and zanamivir-treated clones were selected by blue-white screening, only those obtained from samples treated with 100 μM T-705 contained small insertions of the NP genes into the plasmid, suggesting that the drug adversely affected virus genetic information.
Fig 4.
Mutation frequency and profile of nucleotide changes in influenza A/Denmark/524/2009 (H1N1) virus. (A) MDCK cells were mock treated or treated with T-705 (10 μM) or zanamivir (10 μM) and inoculated with influenza A/Denmark/524/2009 (H1N1) virus at an MOI of 0.001 PFU/cell. Virus was extracted from the supernatant after a 48-h incubation at 37°C. A 1,523-kb region of the NP gene was RT-PCR amplified and subcloned. Individual clones were sequenced to obtain the observed mutation frequencies. The numbers of mutations per 10,000 nucleotides differed significantly between T-705-treated and mock-treated viruses (P < 0.001). (B) Mutation profiles of influenza A/Denmark/524/2009 (H1N1) viruses grown in the presence of mock-treated, T-705-treated (10 μM), or zanamivir-treated (10 μM) MDCK cells. Bold numerals indicate the most frequent nucleotide changes.
We then examined whether T-705 treatment biased the mutation profile toward specific nucleotide mutations, as RNA mutagens are reported to do (21, 23, 34). In untreated viruses, the transition mutations (A→G and T→C) were most common (Fig. 4B). Treatment with T-705 resulted in accumulation of G→T and C→T transition mutations that increased linearly with the T-705 concentration, suggesting a dose-dependent effect. Treatment with zanamivir slightly increased the number of nucleotide changes in a statistically insignificant manner but did not bias the natural mutation profile (Fig. 4B).
DISCUSSION
Taken together, our results provide the first evidence that T-705 exerts mutagenic activity in influenza viruses and that lethal mutagenesis is either wholly or in part the anti-influenza A virus mechanism of T-705 in vitro. During serial passage, treatment with T-705 resulted in a disproportionate decrease in infectious virus without a corresponding decrease in RNA copy number, thereby reducing virus infectivity (the ratio of PFU to viral RNA copies). It also increased the mutation frequency and shifted the nucleotide profiles of individual NP gene clones. The greater increase in the virus mutant spectrum observed after passage in the presence versus the absence of T-705 suggests that T-705RTP is incorporated into the viral genome, although we did not address this possibility or the possibility that T-705 indirectly affects nuclear transcription or translation. We hypothesize that the small increase observed in the rate of viral mutation leads to a disproportionately larger reduction of viral infectivity as T-705 promotes accumulation of mutations in numerous genes, ultimately causing the generation of nonviable progeny. T-705-enhanced mutagenesis was required for virus extinction, which was not observed in parallel mock-treated samples. The stability of IC50 and EC50s over the course of multiple passages in MDCK cells suggests the absence of phenotypic changes in susceptibility to T-705; therefore, drug-resistant variants are unlikely to emerge in vitro.
Lethal mutagenesis is a mechanism by which drug treatment increases the viral mutation rate sufficiently to overwhelm the virus population's ability to retain fitness (35). Our hypothesis that lethal mutagenesis is the antiviral mechanism of T-705 was based on available data about the antiviral mechanism of nucleoside analogs against poliovirus, vesicular stomatitis virus (36, 37), West Nile virus (34), foot-and-mouth disease virus (38), lymphatic choriomeningitis virus (39), and human immunodeficiency virus type 1 (35). Lethal mutagenesis is suggested when the percentage of noninfectious virus genomes is much higher after passaging with a nucleoside analog than after passaging in mock-treated cells; furthermore, incorporation of a nucleoside analog into the viral genome is known to induce hypermutation (34, 38, 39).
We used two approaches to assess the anti-influenza mechanism of T-705: serial passage with a low virus infectious dose and serial passage with a high virus infectious dose. If the antiviral activity of T-705 is caused by acceleration of the viral mutation rate, then serial passage after the low infectious dose would allow accumulation of the mutations, resulting in lethal mutagenesis. Indeed, after low-dose inoculation we observed virus extinction after 24 serial passages in the presence of T-705. Quantification and infectivity testing of RNA genomes after low- and high-dose virus inoculation and serial passage showed strikingly lower specific infectivity in viruses passaged in the presence of T-705 than in mock-treated viruses (e.g., seasonal influenza A viruses showed a >60% decrease in specific infectivity after 23 passages with T-705), suggesting that T-705 cased lethal mutagenesis.
The lethal mutagenesis theory emphasizes the role of a class of defective interfering particles, which can also be produced during the natural course of influenza virus infection. Therefore, we substantiated the role of lethal mutagenesis by demonstrating that the interfering activity generated by T-705 treatment was greater than the interfering activity naturally associated with influenza virus replication. For that purpose, we inoculated cells with a high dose of infectious virus to allow the generation of defective interfering particles during the course of serial passage. In cells inoculated with the high dose, no viable viruses were detected after 3 serial passages with T-705, although virus remained detectable after passage 4 in mock-treated cells. After 3 passages, specific infectivity decreased by approximately 60% in the presence of T-705 but only by 20% in mock-treated viruses. Therefore, virus extinction required T-705-induced mutagenesis.
Our data suggesting the incorporation of T-705RTP into viral RNA are consistent with the lethal mutagenesis theory. Specifically, we demonstrated a 9-fold increase (A/Brisbane/59/2007) and a 5-fold increase (A/New Jersey/15/2007) in G→A transitions. In cells inoculated with a low virus dose, virus-specific increases in RNA C→T transition were observed after serial passage in treated, but not mock-treated, cells. G→A transitions were also increased at high infectious doses, but not significantly, likely reflecting dose-related rapid virus extinction. A→G mutations within the coding viral genome cause the production of functionally impaired mutant proteins.
We utilized Sanger sequencing for quasispecies substructuring; this method is much less sensitive than ultradeep sequencing, and therefore we cannot completely rule out the selection of a very small population of T-705-resistant influenza variants. However, if such mutants were selected, their prevalence was less than ∼10% of the virus population and their fitness was severely impaired. Furthermore, the lethal mutagenesis theory suggests to us that lethal mutations in the viral genome would have likely prevented the survival of such mutants. Such a mechanism would add to the advantages of an antiviral strategy that targets the viral mutation rate rather than viral proteins.
We observed an increased number of G→A and C→T mutations, suggesting that T-705RTP base pairs with either cytosine or uracil. RNA viruses can also undergo an A→G/U→C mutation pattern caused by a large isoform of the RNA-specific adenosine deaminase (ADAR1-L); this pattern is usually induced by the innate immune response to viral infection (4). ADAR1-L can deaminate adenosine (A) to inosine (I) in the cellular cytoplasm, as demonstrated in the case of hepatitis C viruses (40). Inosine is then recognized as guanosine by decoding ribosomes and transcribing polymerases, leading to A→G transitions. However, we believe that the A→G mutation pattern observed in this study during passages with T-705 is attributable to viral polymerase errors caused by T-705, as no hypermutation was observed in mock-treated viruses. Moreover, the sites of the A→G mutations were not related to the presence of the A, C, or G nucleotides at positions −1, −2, +1, and +2 (data not shown), as would be observed in ADAR1-L-induced mutation (41). Taken together, our results suggest that T-705 increases the influenza virus mutation rate beyond the biological tolerance threshold, causing lethal mutagenesis.
An early understanding of potential resistance to an agent is crucial to guide the development of novel antivirals and maximize their usefulness. It has not been established whether resistance to mutagenic agents such as T-705 emerges as readily as resistance to nonmutagenic inhibitors such as NA inhibitors and adamantanes. At least two other groups have attempted to generate T-705-resistant influenza viruses (7, 42). In these studies, the viruses were extinguished within 20 serial passages in the presence of T-705. Importantly, neither study characterized the passaged variants or sought to determine the exact conditions (cell line, drug concentrations, viral MOI, method of susceptibility testing) required for generation of a T-705-resistant strain.
Of the nucleoside analogs, only ribavirin is demonstrated to have anti-influenza activity (16). Ribavirin and T-705 have analogous carboxamide groups. Multiple ribavirin-resistant mutants of many RNA viruses have been described (43–49). These mutants showed amino acid substitutions in RdRP that increase the general template-copying fidelity of the enzyme, thereby restricting the incorporation of ribavirin triphosphate into RNA during viral RNA synthesis. However, to the best of our knowledge, no ribavirin-resistant influenza viruses have been reported.
It is postulated that an influenza virus is resistant to currently licensed antiviral agents if the virus is (i) infectious in the presence of a drug (e.g., forms plaques on MDCK cells), (ii) is phenotypically less susceptible, and (iii) possesses mutation(s) in the target protein. In our study, the T-705 EC50s were 8 to 18 μM for both the seasonal and 2009 pandemic viruses. Notably, T-705 yielded comparable values for both oseltamivir-susceptible and oseltamivir-resistant viruses, suggesting that T-705 is effective against both. These data support previous reports of T-705 antiviral activity against oseltamivir-resistant and amantadine-resistant influenza viruses (6, 14). In addition, no specific amino acid substitutions in the polymerase gene were detected, and no viable viruses were detected after several serial passages. Thus, T-705-resistant mutants were not selected under our conditions.
Although a drug that increases the mutation rate of a virus may also increase the mutation rate of its host cells, the most recent generation of nucleoside analogs, including T-705, were designed to specifically target viral RdRP without significantly affecting human DNA and RNA synthesis (12, 15). Importantly, however, nucleoside analogs exert relatively high cytotoxicity. Ribavirin is the only known nucleoside analog with potent anti-influenza activity, and like all first-generation nucleoside analogs, it is cytotoxic (50). We found that T-705 was not toxic to MDCK or human lung A549 cells at concentrations of >1,000 μM. This value is at least 10 times the toxicity threshold of ribavirin (CC50 of 94 μM in MDCK cells) (6). The T-705 concentration sufficient to inhibit influenza virus replication in both human and canine cell lines was in the 10 μM range, resulting in a selective index (CC50/EC50) of >1. Furthermore, we observed neither early (1 h posttreatment) nor late (24 h posttreatment) cytotoxic effects. Furuta and colleagues demonstrated that T-705RTP was not significantly incorporated into MDCK-cell DNA at T-705 concentrations of <637 μM (15). Although the available evidence suggests that T-705 treatment would not severely affect cellular RNA and DNA synthesis in humans, further studies are warranted to investigate the long-term effect of T-705 on the human genome.
Overall, given the increased mutation frequency in the influenza virus genome during the T-705 treatment, the specific mutational bias, and the fact that this increase is dose dependent, this study demonstrated that T-705 is an influenza virus mutagen. This provides a novel and promising approach to influenza therapy. Further studies are required to clarify whether the increased mutation frequency in the influenza virus genome resulting from T-705 treatment is due to the direct incorporation of T-705 in the virus genome and/or indirectly due to the interaction with some unknown host factors that would lead to lowering the intracellular concentrations of nucleoside triphosphate pools.
ACKNOWLEDGMENTS
We are grateful to Kristi Prevost and Bindumadhav Marathe for technical support, Hassan Zaraket for helpful discussions, and Sharon Naron for excellent editing of the manuscript. We are highly appreciative of the comments and suggestions provided by the reviewers that helped us to improve the quality and clarity of our study.
This work was supported in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under contract no. HHSN266200700005C, and by the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
Published ahead of print 16 January 2013
REFERENCES
- 1. Monto AS. 2003. The role of antivirals in the control of influenza. Vaccine 21:1796–1800 [DOI] [PubMed] [Google Scholar]
- 2. Fiore AE, Fry A, Shay D, Gubareva L, Bresee JS, Uyeki TM, Centers for Disease Control and Prevention 2011. Antiviral agents for the treatment and chemoprophylaxis of influenza—recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm. Rep. 60:1–24 [PubMed] [Google Scholar]
- 3. Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, Gubareva LV, Xu X, Bridges CB, Uyeki TM, Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360:2605–2615 [DOI] [PubMed] [Google Scholar]
- 4. Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393–396 [DOI] [PubMed] [Google Scholar]
- 5. Boltz DA, Aldridge JR, Webster RG, Govorkova EA. 2010. Drugs in development for influenza. Drugs 70:1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Furuta Y, Takahashi K, Fukuda Y, Kuno M, Kamiyama T, Kozaki K, Nomura N, Egawa H, Minami S, Watanabe Y, Narita H, Shiraki K. 2002. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob. Agents Chemother. 46:977–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Furuta Y, Takahashi K, Shiraki K, Sakamoto K, Smee DF, Barnard DL, Gowen BB, Julander JG, Morrey JD. 2009. T-705 (favipiravir) and related compounds: novel broad-spectrum inhibitors of RNA viral infections. Antiviral Res. 82:95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Julander JG, Shafer K, Smee DF, Morrey JD, Furuta Y. 2009. Activity of T-705 in a hamster model of yellow fever virus infection in comparison with that of a chemically related compound, T-1106. Antimicrob. Agents Chemother. 53:202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gowen BB, Wong MH, Jung KH, Sanders AB, Mendenhall M, Bailey KW, Furuta Y, Sidwell RW. 2007. In vitro and in vivo activities of T-705 against arenavirus and bunyavirus infections. Antimicrob. Agents Chemother. 51:3168–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mendenhall M, Russell A, Juelich T, Messina EL, Smee DF, Freiberg AN, Holbrook MR, Furuta Y, de la Torre JC, Nunberg JH, Gowen BB. 2011. T-705 (favipiravir) inhibition of arenavirus replication in cell culture. Antimicrob. Agents Chemother. 55:782–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rocha-Pereira J, Jochmans D, Dallmeier K, Leyssen P, Nascimento MS, Neyts J. 2012. Favipiravir (T-705) inhibits in vitro norovirus replication. Biochem. Biophys. Res. Commun. 424:777–780 [DOI] [PubMed] [Google Scholar]
- 12. Kiso M, Takahashi K, Sakai-Tagawa Y, Shinya K, Sakabe S, Le QM, Ozawa M, Furuta Y, Kawaoka Y. 2010. T-705 (favipiravir) activity against lethal H5N1 influenza A viruses. Proc. Natl. Acad. Sci. U. S. A. 107:882–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sidwell RW, Barnard DL, Day CW, Smee DF, Bailey KW, Wong MH, Morrey JD, Furuta Y. 2007. Efficacy of orally administered T-705 on lethal avian influenza A (H5N1) virus infections in mice. Antimicrob. Agents Chemother. 51:845–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sleeman K, Mishin VP, Deyde VM, Furuta Y, Klimov AI, Gubareva LV. 2010. In vitro antiviral activity of favipiravir (T-705) against drug-resistant influenza and 2009 A(H1N1) viruses. Antimicrob. Agents Chemother. 54:2517–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Furuta Y, Takahashi K, Kuno-Maekawa M, Sangawa H, Uehara S, Kozaki K, Nomura N, Egawa H, Shiraki K. 2005. Mechanism of action of T-705 against influenza virus. Antimicrob. Agents Chemother. 49:981–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eriksson B, Helgstrand E, Johansson NG, Larsson A, Misiorny A, Norén JO, Philipson L, Stenberg K, Stening G, Stridh S, Oberg B. 1977. Inhibition of influenza virus ribonucleic acid polymerase by ribavirin triphosphate. Antimicrob. Agents Chemother. 11:946–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miller JP, Kigwana LJ, Streeter DG, Robins RK, Simon LN, Roboz J. 1977. The relationship between the metabolism of ribavirin and its proposed mechanism of action. Ann. N. Y. Acad. Sci. 284:211–229 [DOI] [PubMed] [Google Scholar]
- 18. Sidwell RW, Huffman JH, Khare GP, Allen LB, Witkowski JT, Robins RK. 1972. Broad-spectrum antiviral activity of virazole: 1-beta-d-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science 177:705–706 [DOI] [PubMed] [Google Scholar]
- 19. Streeter DG, Witkowski JT, Khare GP, Sidwell RW, Bauer RJ, Robins RK, Simon LN. 1973. Mechanism of action of 1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide (virazole), a new broad-spectrum antiviral agent. Proc. Natl. Acad. Sci. U. S. A. 70:1174–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parker WB. 2005. Metabolism and antiviral activity of ribavirin. Virus Res. 107:165–171 [DOI] [PubMed] [Google Scholar]
- 21. Crotty S, Cameron CE, Andino R. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. U. S. A. 98:6895–6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Domingo E, Menéndez-Arias L, Holland JJ. 1997. RNA virus fitness. Rev. Med. Virol. 7:87–96 [DOI] [PubMed] [Google Scholar]
- 23. Perales C, Martín V, Domingo E. 2011. Lethal mutagenesis of viruses. Curr. Opin. Virol. 1:419–422 [DOI] [PubMed] [Google Scholar]
- 24. Das K, Aramini JM, Ma LC, Krug RM, Arnold E. 2010. Structures of influenza A proteins and insights into antiviral drug targets. Nat. Struct. Mol. Biol. 17:530–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brown AN, McSharry JJ, Weng Q, Driebe EM, Engelthaler DM, Sheff K, Keim PS, Nguyen J, Drusano GL. 2010. In vitro system for modeling influenza A virus resistance under drug pressure. Antimicrob. Agents Chemother. 54:3442–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ives JA, Carr JA, Mendel DB, Tai CY, Lambkin R, Kelly L, Oxford JS, Hayden FG, Roberts NA. 2002. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antiviral Res. 55:307–317 [DOI] [PubMed] [Google Scholar]
- 27. Hoffmann HH, Kunz A, Simon VA, Palese P, Shaw ML. 2011. Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 108:5777–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ilyushina NA, Bovin NV, Webster RG, Govorkova EA. 2006. Combination chemotherapy, a potential strategy for reducing the emergence of drug-resistant influenza A variants. Antiviral Res. 70:121–131 [DOI] [PubMed] [Google Scholar]
- 29. Su CY, Cheng TJ, Lin MI, Wang SY, Huang WI, Lin-Chu SY, Chen YH, Wu CY, Lai MM, Cheng WC, Wu YT, Tsai MD, Cheng YS, Wong CH. 2010. High-throughput identification of compounds targeting influenza RNA-dependent RNA polymerase activity. Proc. Natl. Acad. Sci. U. S. A. 107:19151–19156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoffmann HH, Palese P, Shaw ML. 2008. Modulation of influenza virus replication by alteration of sodium ion transport and protein kinase C activity. Antiviral Res. 80:124–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Centers for Disease Control and Prevention Accessed 27 July 2012 CDC protocol of realtime RTPCR for influenza A (H1N1). http://www.who.int/csr/resources/publications/swineflu/en/
- 32. Aggarwal S, Bradel-Tretheway B, Takimoto T, Dewhurst S, Kim B. 2010. Biochemical characterization of enzyme fidelity of influenza A virus RNA polymerase complex. PLoS One 5:e10372 doi:10.1371/journal.pone.0010372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marriott AC, Dimmock NJ. 2010. Defective interfering viruses and their potential as antiviral agents. Rev. Med. Virol. 20:51–62 [DOI] [PubMed] [Google Scholar]
- 34. Day CW, Smee DF, Julander JG, Yamshchikov VF, Sidwell RW, Morrey JD. 2005. Error-prone replication of West Nile virus caused by ribavirin. Antiviral Res. 67:38–45 [DOI] [PubMed] [Google Scholar]
- 35. Loeb LA, Essigmann JM, Kazazi F, Zhang J, Rose KD, Mullins JI. 1999. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc. Natl. Acad. Sci. U. S. A. 96:1492–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holland JJ, Domingo E, de la Torre JC, Steinhauer DA. 1990. Mutation frequencies at defined single codon sites in vesicular stomatitis virus and poliovirus can be increased only slightly by chemical mutagenesis. J. Virol. 64:3960–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vignuzzi M, Stone JK, Andino R. 2005. Ribavirin and lethal mutagenesis of poliovirus: molecular mechanisms, resistance and biological implications. Virus Res. 107:173–181 [DOI] [PubMed] [Google Scholar]
- 38. Sierra S, Dávila M, Lowenstein PR, Domingo E. 2000. Response of foot-and-mouth disease virus to increased mutagenesis: influence of viral load and fitness in loss of infectivity. J. Virol. 74:8316–8323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martín V, Abia D, Domingo E, Grande-Pérez A. 2010. An interfering activity against lymphocytic choriomeningitis virus replication associated with enhanced mutagenesis. J. Gen. Virol. 91:990–1003 [DOI] [PubMed] [Google Scholar]
- 40. Polson AG, Bass BL. 1994. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 13:5701–5711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zahn RC, Schelp I, Utermöhlen O, von Laer D. 2007. A-to-G hypermutation in the genome of lymphocytic choriomeningitis virus. J. Virol. 81:457–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smee DF, Hurst BL, Egawa H, Takahashi K, Kadota T, Furuta Y. 2009. Intracellular metabolism of favipiravir (T-705) in uninfected and influenza A (H5N1) virus-infected cells. J. Antimicrob. Chemother. 64:741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Coffey LL, Beeharry Y, Bordería AV, Blanc H, Vignuzzi M. 2011. Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc. Natl. Acad. Sci. U. S. A. 108:16038–16043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Feigelstock DA, Mihalik KB, Feinstone SM. 2011. Selection of hepatitis C virus resistant to ribavirin. Virol. J. 8:402 doi:10.1186/1743-422X-8-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Levi LI, Gnädig NF, Beaucourt S, McPherson MJ, Baron B, Arnold JJ, Vignuzzi M. 2010. Fidelity variants of RNA dependent RNA polymerases uncover an indirect, mutagenic activity of amiloride compounds. PLoS Pathog. 28:e1001163 doi:10.1371/journal.ppat.1001163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pfeiffer JK, Kirkegaard K. 2003. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl. Acad. Sci. U. S. A. 100:7289–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pfeiffer JK, Kirkegaard K. 2005. Ribavirin resistance in hepatitis C virus replicon-containing cell lines conferred by changes in the cell line or mutations in the replicon RNA. J. Virol. 79:2346–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Scheidel LM, Durbin RK, Stollar V. 1987. Sindbis virus mutants resistant to mycophenolic acid and ribavirin. Virology 158:1–7 [DOI] [PubMed] [Google Scholar]
- 49. Young KC, Lindsay KL, Lee KJ, Liu WC, He JW, Milstein SL, Lai MM. 2003. Identification of a ribavirin-resistant NS5B mutation of hepatitis C virus during ribavirin monotherapy. Hepatology 38:869–878 [DOI] [PubMed] [Google Scholar]
- 50. Fried MW. 2002. Side effects of therapy of hepatitis C and their management. Hepatology 36:S237–S244 [DOI] [PubMed] [Google Scholar]