Abstract
Macroautophagy is a cellular pathway that degrades intracellular pathogens and contributes to antigen presentation. Herpes simplex virus 1 (HSV-1) infection triggers both macroautophagy and an additional form of autophagy that uses the nuclear envelope as a source of membrane. The present study constitutes the first in-depth analysis of nuclear envelope-derived autophagy (NEDA). We established LC3a as a marker that allowed us to distinguish between NEDA and macroautophagy in both immunofluorescence and flow cytometry. NEDA was observed in many different cell types, indicating that it is a general response to HSV-1 infection. This autophagic pathway is known to depend on the viral protein γ34.5, which can inhibit macroautophagy via binding to beclin-1. Using mutant viruses, we were able to show that binding of beclin-1 by γ34.5 had no effect on NEDA, demonstrating that NEDA is regulated differently than macroautophagy. Instead, NEDA was triggered in response to γ34.5 binding to protein phosphatase 1α, an interaction used by the virus to prevent host cells from shutting off protein translation. NEDA was not triggered when late viral protein production was inhibited with acyclovir or hippuristanol, indicating that the accumulation of these proteins might stress infected cells. Interestingly, expression of the late viral protein gH was sufficient to rescue NEDA in the context of infection with a virus that otherwise does not support strong late viral protein expression. We argue that NEDA is a cellular stress response triggered late during HSV-1 infection and might compensate for the viral alteration of the macroautophagic response.
INTRODUCTION
Macroautophagy, the mechanism by which cells literally eat some of their own components, plays an essential role in cell homeostasis and nutrient recycling (1, 2). This process involves the de novo formation of a vesicle that engulfs cytosolic protein aggregates or organelles. To this end, a two-layered membrane cup forms around cytoplasmic cargo. This cup expands and closes into a vesicle, the autophagosome, which can then fuse with lysosomes or early endosomes for degradation of its content. The initiation of autophagosome formation requires the protein beclin-1 (3), while membrane extension is driven by two ubiquitin-like protein conjugation systems, one of which includes LC3b. The accumulation of membrane-bound LC3b as punctate patterns representing autophagic vesicles is a well-established hallmark of macroautophagy (4).
In addition to nutrient recycling, macroautophagy also serves as a defense mechanism against intracellular pathogens like viruses (5, 6). Macroautophagosomes can directly engulf pathogens in the cytosol and promote their degradation. This process is referred to as xenophagy. Furthermore, recent evidence shows that macroautophagy stimulates the presentation of intracellular viral antigens on major histocompatibility complexes I and II (7, 8), thus alerting the adaptive branch of the immune system to help clear the viral infection. Herpes simplex virus 1 (HSV-1) triggers macroautophagy very early in infection, prior to the production of viral proteins (9, 10). Later in infection, HSV-1 can inhibit both macroautophagosome formation and fusion of macroautophagosomes with lysosomes (11, 12).
Recently, we discovered a novel form of autophagy that involves vesicle formation at the nuclear envelope during HSV-1 infection in mouse macrophages (13). This process is characterized by the formation of 4-membrane-layered LC3b-positive structures made by coiling of the inner and outer nuclear membrane and is referred to here as nuclear envelope-derived autophagy (NEDA). We have shown that the γ34.5 viral protein is required for the induction of this autophagic pathway, as an HSV-1 mutant lacking this molecule is unable to induce NEDA. Other than the requirement of γ34.5, the molecular mechanisms governing NEDA were unknown.
The HSV-1 protein γ34.5 has multiple functions. It can interfere with macroautophagy induction by binding beclin-1 (12). Second, γ34.5 promotes translation of viral proteins by counteracting cellular translational shutoff, an antiviral defense mechanism. This translational shutoff is mediated by phosphorylation and inactivation of the α subunit of eukaryotic initiation factor 2 (eIF2α). HSV-1 γ34.5 binds both the protein phosphatase 1α (PP1α) and eIF2α, thus facilitating eIF2α dephosphorylation and active translation (14, 15). In the current study, we set out to analyze the function of HSV-1 γ34.5 in order to better understand the molecular requirements for NEDA.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
The BMA3.1A7 macrophage cell line was derived from C56/BL6 mice as described previously (16). BMA3.1A7, BHK-21 (ATCC CCL-10), MEF (provided by Gilbert Arthur, University of Manitoba, Canada), HeLa (ATCC CCL-2), Vero (ATCC CCL-81), Vero-gH CR1 (kindly provided by Tony Minson and Helena Browne, University of Cambridge, United Kingdom) (17, 18), Vero-ICP4 E5 (19), and MeWo (ATCC HTB-65, kindly provided by Bruce Banfield, Queens University, Kingston, Canada) were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% (vol/vol) fetal calf serum (FCS) and 2 mM glutamine.
The HSV-1 17+ Δγ34.5 and HSV-1 17+ Δbeclin-1 (Δaa68-87) viruses have been previously described (12, 13, 20, 21). The HSV-1 17+ ΔPP1α virus that carries Val178Glu and Phe180Leu substitutions was constructed by amplifying nucleotides 463 to 1088 of HSV-1 by PCR from wild-type (wt) strain 17syn+ infectious DNA with primers 5′-CAGACCACCAGGTGGCGCACCCGGACGTGGGGCGATAAGCGCTCCCGCGCGGGGGTC-3′ and 5′-GCACATGCTTGCCTGTCAAACTCT-3′. The amplified PCR fragment with nucleotide changes (underlined) was inserted into pCR-BluntII-TOPO (Invitrogen, Carlsbad, CA), resulting in pDA14, which was sequenced to verify that the plasmid contained the desired mutations. A 477-bp PflMI/BstEII fragment of pDA14 containing two nucleotide changes was ligated into PflMI/BstEII-digested pDA04, generating pDA13. pDA13 was cotransfected into Vero cells with infectious viral DNA from HSV-1 strain 17syn+ using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Viruses from this transfection were plaque purified and screened for incorporation of the nucleotide substitution by PCR using the following primers: 5′-GCACATGCTTGCCTGTCAAACTCT-3′ and 5′-TGTAACGTTAGACCGAGTTCGCCG-3′. These primers generate a PCR product that spans the mutation, which generates an AfeI site. Wild-type and PP1α templates generate 808-bp PCR products. Following digestion with AfeI, PP1α PCR products were cut into 588- and 216-bp fragments, but wild-type DNA was uncut. Plaques containing the AfeI restriction site were plaque purified three times prior to isolation of viral DNA and Southern blot analysis that was performed as previously described (22). Following DNA digestion of viral stocks with AfeI, an 812-bp BspEI/NcoI fragment corresponding to nucleotides 511 to 1323 of the HSV-1 genome was labeled with 32P-dCTP by random priming. This fragment was used as a probe to confirm the presence of the PP1α binding mutation (data not shown). HSV-1 17+ wt was kindly provided by Beate Sodeik (Medizinische Hochschule Hannover, Germany). HSV-1 stocks were propagated in BHK-21, and titers were determined on Vero cells as previously described (23, 24).
The following primary antibodies were used to detect cellular proteins: rabbit polyclonal antibody to cleaved LC3a (AP1805a; Abgent), rabbit polyclonal to cleaved LC3b (AP1806a; Abgent), mouse monoclonal antibody against total LC3 (M115-3; MBL), rabbit monoclonal anti-P-eIF2α antibody (D9G8; Cell Signaling), mouse monoclonal anti-eIF2α antibody (L57A5; Cell Signaling), mouse monoclonal anti-nuclear pore complex/p62 antibody (MAb414; Covance), mouse monoclonal anti-γ-tubulin antibody (GTU-88; Sigma), and rabbit polyclonal anti-calnexin antibody (kindly provided by John Bergeron, McGill University, Montreal, Canada). Viral proteins were detected with a rabbit polyclonal anti-HSV-1 antibody (RB-1425-A; Neomarkers), rabbit polyclonal anti-HSV-1 VP5 antibody (H1.4; Acris), rabbit polyclonal anti-HSV-1 γ34.5 antibody (25) (kindly provided by Ian Mohr, New York University), mouse monoclonal anti-HSV-1 gH antibody (BBH1; Abcam), mouse monoclonal anti-HSV-1 gC antibody (MBS609783; Virussys), mouse monoclonal anti-HSV-1/2 gB antibody (M612449; Fitzgerald), mouse monoclonal anti-HSV-1/2 gB antibody (clone 10B7, ab6506; Abcam), mouse monoclonal anti-HSV-1/2 ICP27 antibody (10-H44; Fitzgerald), mouse monoclonal anti-HSV-1 ICP0 antibody (clone 5H7, ab6513; Abcam), mouse monoclonal anti-HSV-1 ICP4 antibody (clone 10F1, ab6514; Abcam), and rabbit polyclonal anti-HSV-1 pUL36 147 antibody (26) (kindly provided by Ari Helenius, ETH Zürich, Switzerland, and Beate Sodeik, Medizinische Hochschule Hannover, Germany).
Infection and drug treatment.
Cells were inoculated at a multiplicity of infection (MOI) of 5 on a rocking platform at 37°C for 30 min. The inoculum was aspirated and replaced by cell culture medium, and infected cells were incubated until 8 h postinfection (hpi) unless indicated otherwise. Cycloheximide and acyclovir were purchased from Sigma-Aldrich (Oakville, Canada) and added directly after inoculation for 8 h at final concentrations of 0.5 mM and 200 to 400 μM, respectively. The translation inhibitor hippuristanol (27, 28) was kindly provided by Jerry Pelletier (McGill University, Montreal, Canada), used at a 1 μM final concentration, and added for the times indicated.
Immunofluorescence and FACS labeling.
For immunofluorescence analysis and fluorescence-activated cell sorting (FACS) labeling of intracellular antigens, cells were fixed and permeabilized with the Cytofix/Cytoperm kit according to the manufacturer's instructions (BD Biosciences, Mississauga, Canada). Proteins of interest were labeled with primary antibodies and the corresponding secondary antibodies coupled to Alexa 488, Alexa 568, or Alexa 647 (Invitrogen, Carlsbad). Cells were then either analyzed by flow cytometry with a FACSCalibur (Becton Dickinson, Mississauga, Canada) or embedded with ProLong Gold antifade reagent (Invitrogen, Carlsbad) and analyzed with a TCS confocal laser-scanning microscope (Leica Microsystems, Concord, Canada). Flow cytometry data were analyzed with FlowJo 8.7. Contrast adjustments of immunofluorescence images were performed equally for all images within each experiment with Adobe Photoshop CS3.
Electron microscopy.
For morphological analysis, infected cells were fixed in 2.5% (vol/vol) glutaraldehyde (Canemco), embedded in Epon (Mecalab), and thin sectioned as described previously (29). For immunogold labeling, cells were fixed in 1% (vol/vol) glutaraldehyde and embedded at −20°C in Lowicryl (Canemco). Thin sections were incubated overnight with the polyclonal anti-LC3a antibody at a dilution of 1:10 and for 1 h with protein A-gold.
SDS-PAGE and immunoblot assay.
Protein samples were separated by linear 4 to 15% SDS-PAGE (Bio-Rad) and transferred onto nitrocellulose membrane (Pall Corporation). Proteins of interest were detected using the primary antibodies described above, followed by secondary antibodies coupled to horseradish peroxidase (Jackson ImmunoResearch, West Grove, PA) for enhanced chemiluminescence (ECL) detection (PerkinElmer, Waltham, MA). If required, membranes were stripped by incubation with 2% SDS, 100 mM β-mercaptoethanol, and 50 mM Tris (pH 6.8) at 56°C for 15 min and reprobed with other antibodies.
RESULTS
NEDA is a general mechanism during HSV-1 infection.
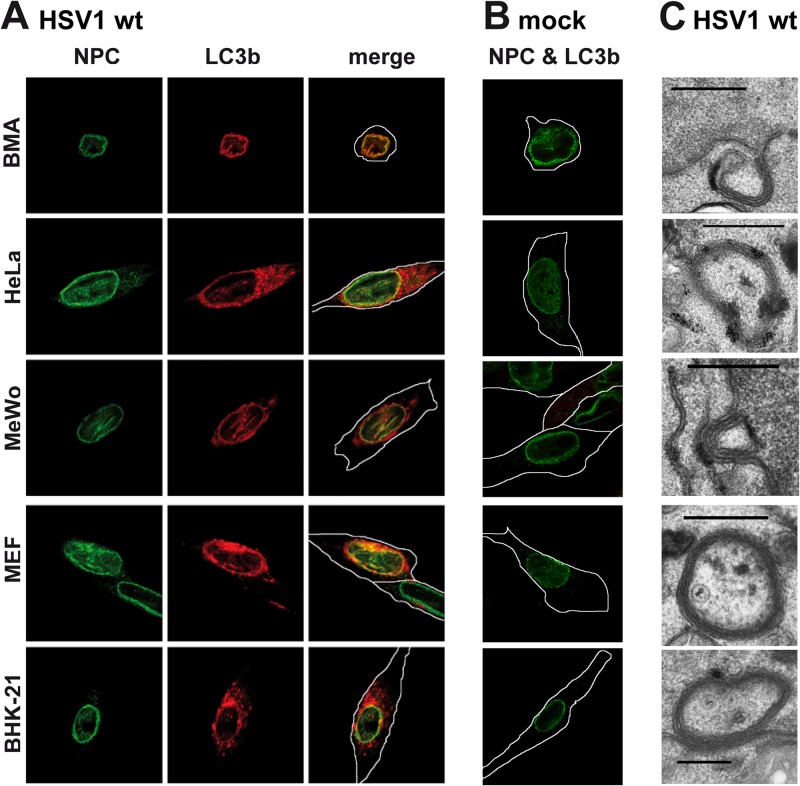
NEDA is a novel form of autophagy triggered during HSV-1 infection in mouse macrophages (13). Here, we show that this process is triggered in multiple cell types, including human cells, during HSV-1 infection (Fig. 1A). Immunofluorescence analyses with the autophagosome marker LC3b clearly show the accumulation of this protein on the nuclear envelope, the hallmark of NEDA, and its colocalization with the nuclear pore complex protein p62. Uninfected cells show no accumulation of LC3b around the nucleus (Fig. 1B). Electron microscopy provides direct evidence that NEDA generates 4-membrane-layered autophagosomes from the coiling of the nuclear envelope in all cell types analyzed (Fig. 1C). This autophagic process results in the sequestration of cytoplasmic components, and viral particles are often observed inside resulting vesicles. We conclude that NEDA is a general response to HSV-1 infection in macrophages, fibroblasts, and epithelial cells of human as well as rodent origin.
Fig 1.
NEDA is triggered by HSV-1 in different cell types. BMA, HeLa, MeWo, MEF, and BHK-21 cells were infected with HSV-1 wt at a multiplicity of infection (MOI) of 5 for 8 h (A) or mock infected (B), and NEDA onset was analyzed by labeling with antibodies against cleaved LC3b and colocalization with the nuclear pore marker p62 (NPC). (C) HSV-1 wt-infected cells were also analyzed by electron microscopy. Scale bar, 500 nm. White lines denote outlines of cells as observed in transilluminated images in this and the following figures.
LC3a is detected on NEDA but not macroautophagosomes.
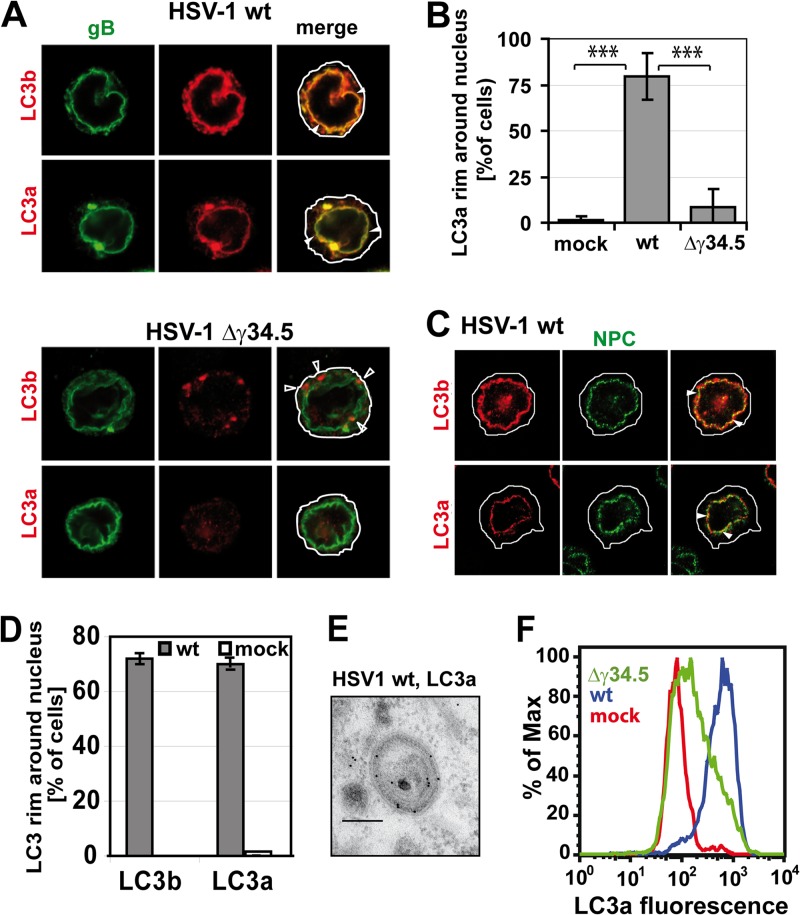
So far, no exclusive marker associated with NEDA has been identified, hindering a more thorough characterization of the molecular mechanisms regulating it. As LC3b is a marker of both macroautophagy and NEDA (13), we sought to verify whether an isoform of this protein, LC3a, might also label autophagosomes and/or the nuclear membrane during NEDA. Accordingly, we performed immunofluorescence analyses to localize LC3a and LC3b in cells infected with wt HSV-1 where both NEDA and macroautophagy can occur or with HSV-1 lacking γ34.5 (the HSV-1 Δγ34.5 virus) to induce macroautophagy only. In HSV-1 wt-infected cells, both LC3a and LC3b colocalized with gB, a viral protein present on the nuclear envelope (Fig. 2A, closed arrowheads). In contrast, upon infection with the HSV-1 Δγ34.5 virus, only LC3b-positive cytoplasmic punctae were detected (Fig. 2A, bottom). There were no discernible LC3a-positive structures and very little LC3a on the nuclear envelope (Fig. 2B). The nuclear membrane was formally identified by labeling with the nuclear pore complex protein p62, which localized very closely to LC3a in about 80% of wt HSV-1-infected cells but not in uninfected cells (Fig. 2C, D). Immunogold labeling and electron microscopy demonstrated that LC3a localized to the 4-membrane-bound vesicles that are characteristic of NEDA (Fig. 2E). These data indicate that LC3a can be used to monitor the occurrence of NEDA during HSV-1 infection and that LC3a does not occur on macroautophagosomes. To facilitate the detection of NEDA and quantify its occurrence, we then went on to develop a flow cytometric assay based on the measurement of the fluorescent signal for LC3a. (Fig. 2F). Establishing LC3a as a means to distinguish NEDA from macroautophagy allowed us to initiate the characterization of this pathway in more detail and address the particular role played by the viral protein γ34.5 in its initiation.
Fig 2.
LC3a localizes to NEDA but not macroautophagosomes. (A) Immunofluorescence analysis of BMA cells infected with the HSV-1 wt or Δγ34.5 virus, MOI 5, 8 hpi. Antibodies against both forms of LC3 labeled a perinuclear rim (closed arrowheads) in the presence but not in the absence of γ34.5. The antibody against cleaved LC3b also labeled punctae in the cytosol in the absence of γ34.5 (open arrowheads). (B) LC3a colocalization with the nuclear envelope in mock-infected cells or cells infected with the HSV-1 wt or Δγ34.5 virus. Quantification of three immunofluorescence experiments, 150 cells per condition. Error bars, standard deviations (SD); ***, P < 0.001 in a two-sided Student t test. (C) Immunofluorescence of cells infected with HSV-1 wt, MOI 5, 8 hpi. Nuclear pore complexes (NPC) were labeled with an antibody against p62, and LC3 was detected with isoform-specific antibodies against the cleaved forms of LC3b and LC3a. (D) Quantification of immunofluorescence data with HSV-1 wt-infected and mock-infected cells. About 100 cells for each condition were evaluated; error bars, SD. (E) Immunogold labeling and electron microscopy confirmed the presence of LC3a on four membrane-bound vesicles in HSV-1 wt-infected cells. Scale bar, 250 nm. (F) Flow cytometry analysis of LC3a labeling intensity in BMA cells infected with the HSV-1 wt or Δγ34.5 virus or in uninfected cells.
NEDA requires the PP1α-binding domain of γ34.5 but not the beclin-1-binding domain.
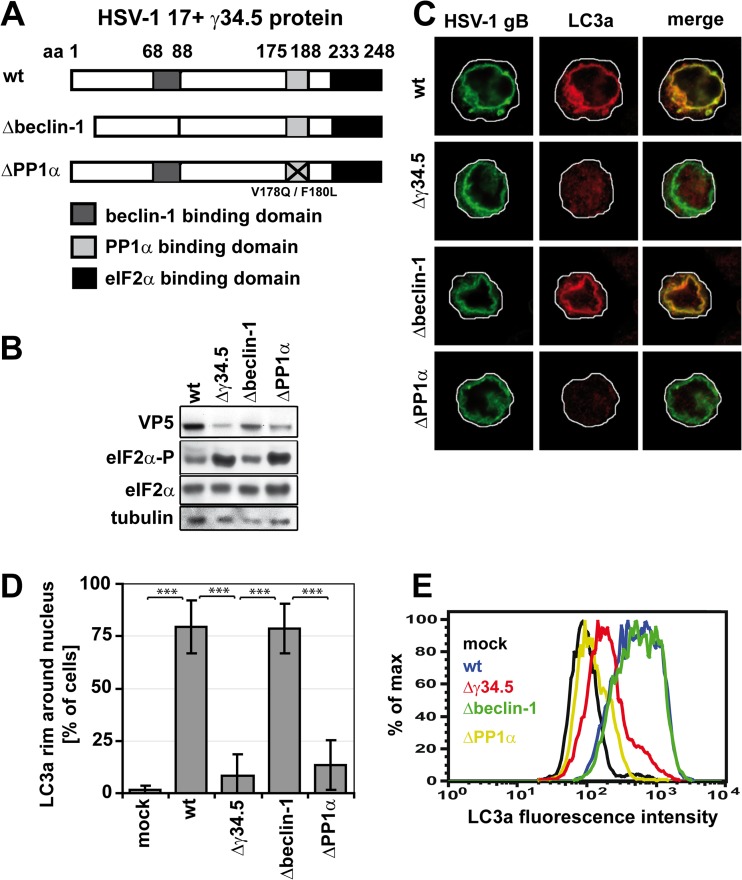
We have shown that the expression of the viral protein γ34.5 during HSV-1 infection is required for NEDA to occur (13), suggesting that this autophagic process is engaged in response to a specific effect of γ34.5 on the host cell. This viral protein interferes with at least two distinct host defense mechanisms. First, it partially inhibits macroautophagy by binding beclin-1 through its beclin-1-binding domain (12, 20). Thus, NEDA could be an alternative autophagic process triggered to counteract the inhibition of macroautophagy. Second, γ34.5 also binds and bridges two host proteins, PP1α and eIF2α, that are part of the machinery involved in shutting off protein translation in response to HSV-1 infection (15). Binding of these proteins by γ34.5 interferes with the ability of infected cells to shut off protein translation. Thus, we hypothesized that infected cells could respond with NEDA when γ34.5 prevents shutoff of protein translation. To determine whether NEDA is caused as a response to the effect of γ34.5 on macroautophagy and/or protein translation, we used two mutant viruses displaying specific mutations in the γ34.5 protein. The first one lacks the beclin-1-binding domain (the Δbeclin-1 virus) (12, 13, 20, 21), while the second one contains two point mutations interfering with PP1α binding (the ΔPP1α virus) (Fig. 3A). These two point mutations were sufficient to increase the phosphorylation of eIF2α (eIF2α-P) to levels similar to those observed with a mutant virus lacking the complete γ34.5 protein (Fig. 3B), indicating that this mutant is appropriate to study the effect of translation on NEDA.
Fig 3.
The PP1α but not the beclin-1-binding domain of γ34.5 is required for NEDA. (A) HSV-1 γ34.5 mutants. The γ34.5 protein contains a beclin-1-binding domain (amino acids [aa] 68 to 87), a PP1α binding domain (aa 175 to 188), and an eIF2α binding domain (aa 233 to 248). Aa 68 to 87 are deleted in the HSV-1 Δbeclin-1 virus, and in the HSV-1 ΔPP1α virus, two point mutations inhibit PP1α binding. (B to E) BMA cells were infected with the HSV-1 wt, Δγ34.5, Δbeclin-1, or ΔPP1α virus for 8 h at an MOI of 5. (B) Immunoblot analysis with antibodies against total eIF2α and phosphorylated eIF2α (eIF2α-P). (C) Immunofluorescence with an antibody against cleaved LC3a. (D) Quantification of the number of cells in which LC3a colocalized with the nuclear envelope in immunofluorescence. About 150 cells per condition in three experiments. Error bars, SD; ***, P < 0.001 in a two-sided Student t test. (E) Flow cytometry analysis of LC3a labeling intensity in cells infected with the four viruses.
A strong signal for LC3a on the nuclear envelope, indicative of NEDA, was observed in about 75% of cells infected either with the HSV-1 wt or HSV-1 Δbeclin-1 virus (Fig. 3C and D), suggesting that the inhibition of macroautophagy through the binding of γ34.5 to beclin is not required for the occurrence of NEDA. In contrast, no significant recruitment of LC3a to the nuclear membrane was observed in cells infected with the HSV-1 Δγ34.5 and HSV-1 ΔPP1α viruses. Similar results were obtained when observing LC3b localization to the nuclear envelope (data not shown). Quantitative reverse transcription (RT)-PCR demonstrated that the expression of LC3a and LC3b was not altered by these viruses (data not shown). The accumulation of cleaved LC3a after infection with the HSV-1 wt and HSV-1 Δbeclin-1 viruses, but not with the Δγ34.5 and ΔPP1α viruses, was confirmed with the LC3a-specific flow cytometric assay (Fig. 3E). Due to accumulation of LC3a in cytosolic clusters in some cells infected with the HSV-1 Δγ34.5 virus but not the HSV-1 ΔPP1α virus (not shown), overall LC3a signals appeared slightly higher with the HSV-1 Δγ34.5 virus in the flow cytometry assay. Together, these results strongly suggest that the occurrence of NEDA is linked to elevated levels of eIFα phosphorylation, which in turn is known to regulate translation of both viral and cellular proteins.
NEDA induction requires protein translation.
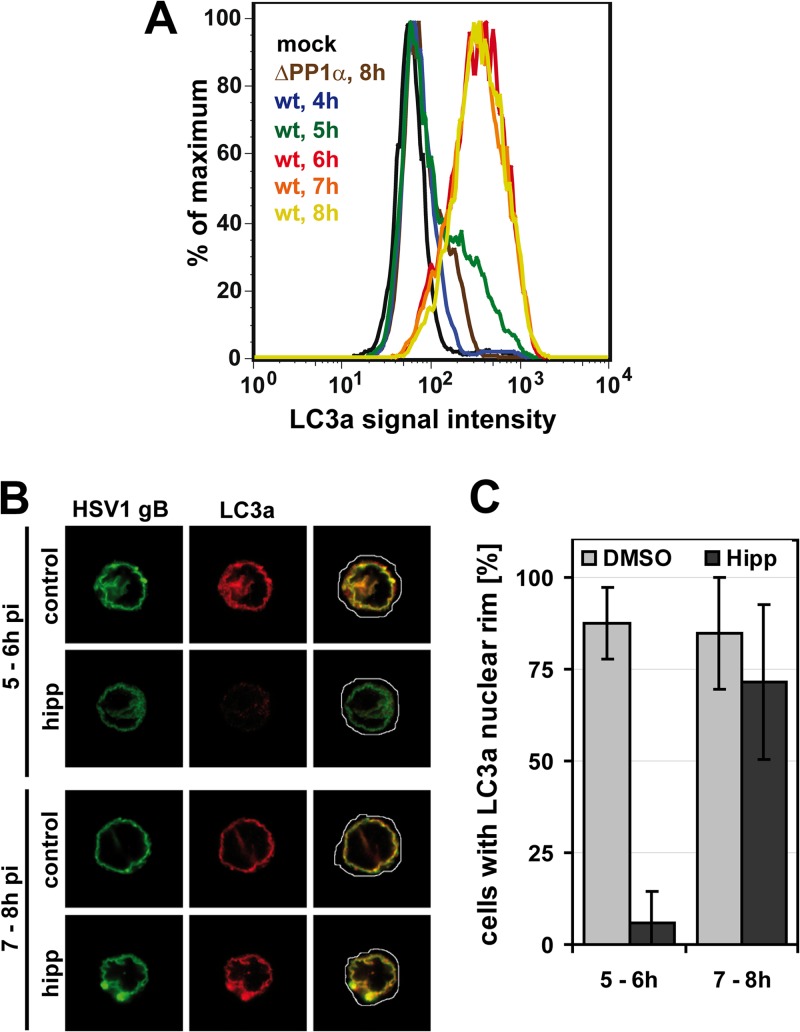
To further investigate whether NEDA is engaged during changes in protein translation, we used a pharmacological approach to alter host and viral protein translation at various times after HSV-1 infection. A detailed kinetic study indicated that NEDA is first observed sometime between 5 and 6 hpi in HSV-1-infected macrophages (Fig. 4A). Therefore, we focused on that window of time and inhibited protein translation at 5 hpi by treating cells with hippuristanol, a compound isolated from corals that interferes strongly and very rapidly with translation by inhibiting eukaryotic initiation factor 4 (28). Remarkably, while hippuristanol treatment inhibited the recruitment of LC3a to the nuclear membrane when added between 5 to 6 hpi, this drug had no effect on the nuclear localization of this protein when added at 7 to 8 hpi (Fig. 4B and C). These results indicate that proteins translated between 5 to 6 hpi are required to induce NEDA and that continuous translation is not needed to maintain the pathway once induced.
Fig 4.
NEDA is triggered in response to translation in between 5 to 6 h of infection. (A) Cells infected with HSV-1 wt for 4 to 8 h or the HSV-1 ΔPP1α virus for 8 h were analyzed for LC3a signal intensity by flow cytometry. Mock-infected and HSV-1 ΔPP1α virus-infected cells served as negative controls. (B and C) Macrophages were infected with HSV-1 wt and treated with dimethyl sulfoxide (DMSO, control) or 1 μM hippuristanol (hipp) for 1 h. NEDA induction was analyzed at 6 or 8 hpi in immunofluorescence with an antibody against LC3a. The occurrence of an LC3a nuclear rim was analyzed in about 50 cells per condition. Error bars, SD.
Inhibition of late viral protein production interferes with NEDA induction, even in the presence of γ34.5.
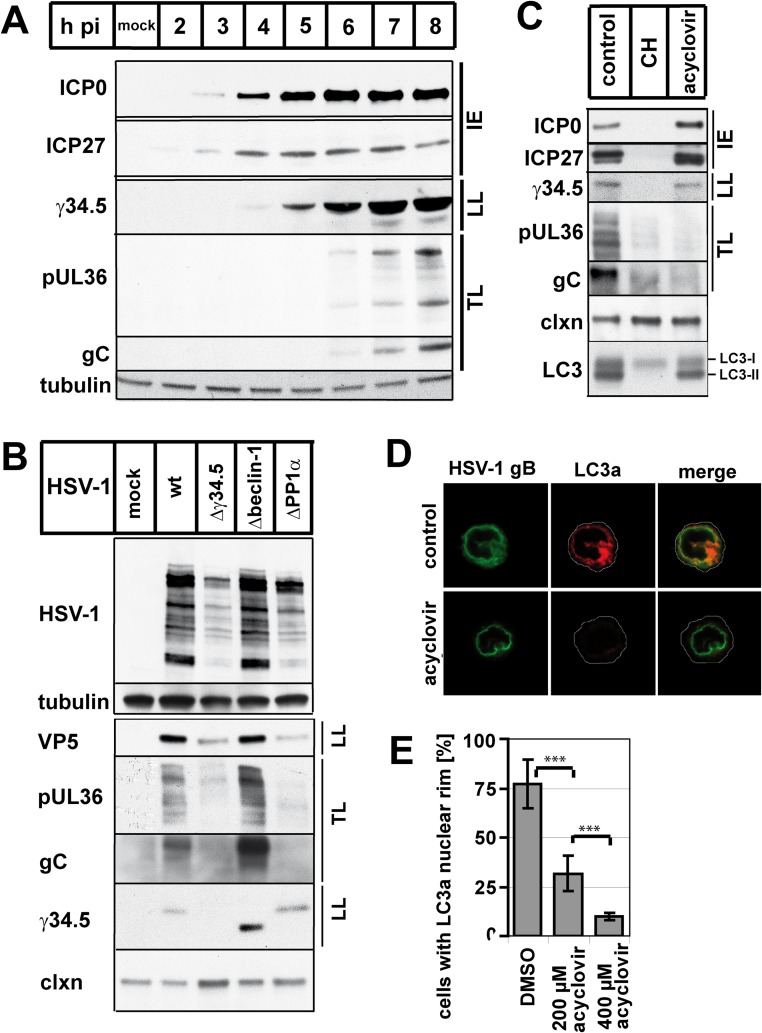
In the window of time between 5 to 6 h of HSV-1 infection, the cell produces mostly viral proteins. Therefore, we wanted to know whether NEDA occurrence was linked to translation of viral proteins. HSV-1 proteins are produced in three waves (30). Proteins referred to as immediate early proteins are the first proteins expressed during infection. These proteins initiate the production of the next wave of so-called early proteins (31, 32). After viral DNA replication has been initiated, the third and largest wave of viral protein production ensues. These late proteins are mostly structural and allow the production of progeny viral particles. However, this separation in three groups is not absolute. Some late proteins, referred to as leaky late, are produced in small quantities already prior to DNA replication, and their production increases later on (31). Other so-called true late proteins are produced only after the onset of viral DNA replication.
Considering that NEDA is triggered between 5 and 6 hpi, we wanted to determine whether this window of time corresponded to the production of a given group of viral proteins. In our system, immediate early and leaky late viral proteins were already produced prior to 5 hpi with wt HSV-1 (Fig. 5A). The production of true late proteins took place at 5 to 6 hpi, the precise time at which NEDA occurred. These analyses led us to propose that the production of true late viral proteins is required to induce NEDA. This hypothesis was supported by the finding that cells infected with the HSV-1 Δγ34.5 and ΔPP1α viruses that do not trigger NEDA also did not produce the true late viral proteins pUL36 and gC (Fig. 5B). Furthermore, cells infected with wt HSV-1 and treated with acyclovir, an inhibitor of DNA replication (33), did not produce true late viral proteins and displayed a dose-dependent reduction in NEDA (Fig. 5C to E). Under these conditions, both γ34.5 and LC3 were present. We conclude that NEDA is triggered as a response to true late viral protein expression.
Fig 5.
NEDA is triggered in response to true late viral protein expression. (A) Viral protein expression kinetics in macrophages infected with HSV-1 wt for 2 to 8 h or mock-infected cells, with antibodies for the immediate early proteins (IE) ICP0 and ICP27, the leaky late (LL) protein γ34.5, and the true late (TL) proteins gC and pUL36. Tubulin was used as a loading control. (B) Immunoblot analysis of macrophages infected with the HSV-1 wt, Δγ34.5, Δbeclin-1, or ΔPP1α virus, using a polyclonal anti-HSV-1 antibody and antibodies for VP5, γ34.5, gC, and pUL36. Calnexin and tubulin were used as loading controls. (C to E) Macrophages were infected with HSV-1 wt and treated with DMSO, cycloheximide (CH), or 200 to 400 μM acyclovir at 0 hpi and analyzed at 8 hpi. (C) Immunoblot with antibodies against ICP0, ICP27, γ34.5, pUL36, gC, calnexin (clxn), and LC3 (LC3-I uncleaved, LC3-II cleaved membrane-associated form). (D and E) Immunofluorescence analysis with an antibody against cleaved LC3a. Error bars, SD; ***, P < 0.001 in a two-sided Student t test.
Expression of one true late viral protein rescues NEDA.
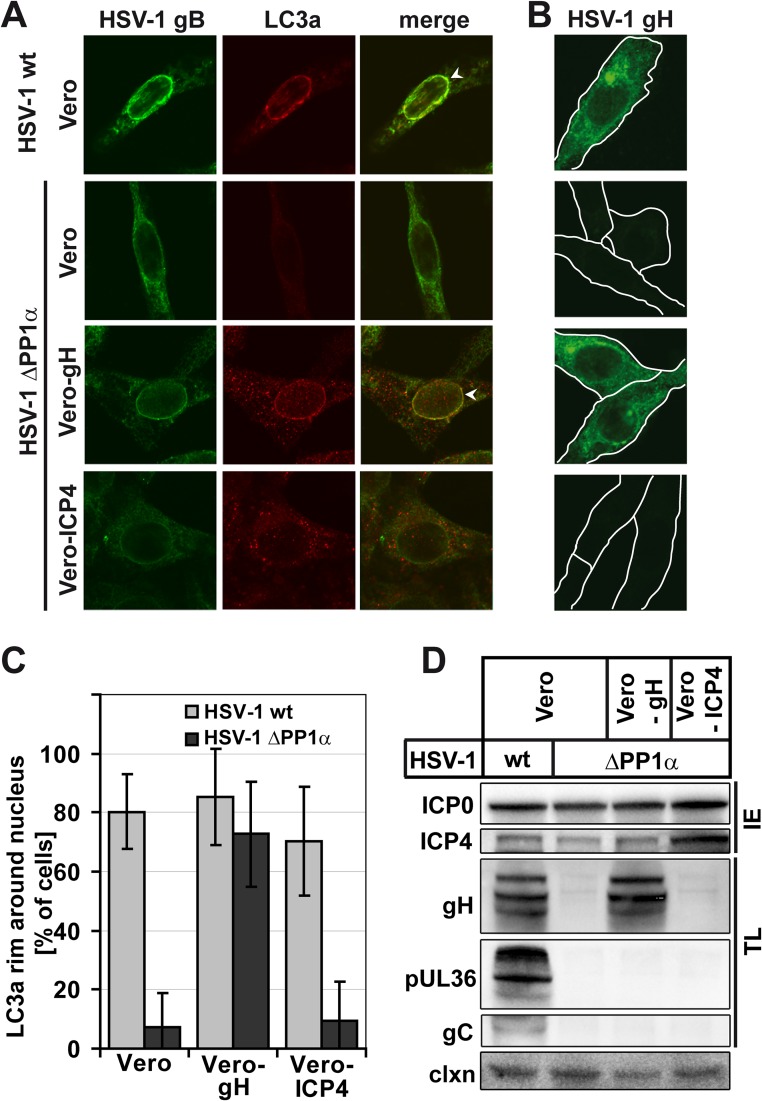
Next, we wanted to know if NEDA could be induced by expressing a true late protein in cells infected with a virus that otherwise fails to induce NEDA. To this end, we used Vero-gH (CR1) cells, which express the true late HSV-1 protein gH under the control of the promoter of an early-late HSV-1 protein, and thus only when infected with the virus (17, 18). Interestingly, the ΔPP1α virus induced NEDA in Vero-gH cells but not in control Vero cells or in a Vero cell line expressing the immediate early protein ICP4 (Vero-ICP4, E5) (Fig. 6A). The level of gH expression in Vero-gH cells infected with the ΔPP1α virus was similar to that during HSV-1 wt infection of control Vero cells (Fig. 6B). Quantification of NEDA in the three different Vero cell lines demonstrated that the occurrence of this autophagic pathway in the HSV-1 ΔPP1α virus-infected Vero-gH cells was similar to that observed during infection of any Vero cell line with the wt virus (Fig. 6C). This complete rescue of the phenotype can likely be attributed to the presence of the true late protein gH, since the expression levels of other late viral proteins did not differ between the three Vero cell lines (Fig. 6D). These results support our hypothesis that expression of true late viral proteins is required for NEDA and point to a possible function of gH in this pathway.
Fig 6.
NEDA is rescued by gH overexpression. Vero cells, Vero CR1 cells that express the late HSV-1 protein gH (Vero-gH), or Vero E5 cells that express the immediate early protein ICP4 (Vero-ICP4) were infected with the HSV-1 wt or HSV-1 ΔPP1α virus at an MOI of 5 for 8 h. (A) Infected cells were detected with an antibody against HSV-1 gB, and NEDA was monitored by LC3a labeling. HSV-1 ΔPP1α caused an accumulation of LC3a around the nucleus (white arrowheads) only in Vero-gH but not wt Vero or Vero-ICP4 cells. (B) Upon HSV-1 ΔPP1α virus infection, Vero-gH cells contained gH levels similar to those of Vero cells infected with HSV-1 wt. (C) Quantification of LC3a nuclear rims in immunofluorescence experiments with the three Vero cell lines in about 50 cells per condition; error bars, SD. (D) Immunoblot analysis of viral protein expression in different Vero cell lines with antibodies against the immediate early (IE) proteins HSV-1 ICP0, ICP4, and the true late (TL) proteins gH, pUL36, and gC. Calnexin (clxn) served as a loading control.
DISCUSSION
NEDA is a cellular process induced in macrophages during HSV-1 infection. It differs from any of the known autophagic pathways by the use of the inner and outer nuclear membranes as the main source of material for the formation of 4-membrane-layered autophagic structures. Although these structures display LC3b, the hallmark marker of macroautophagosomes, we discovered that the related protein LC3a is recruited to the nuclear envelope but not macroautophagosomes during infection. This characteristic allowed us to develop a FACS-based assay to measure the occurrence of NEDA in various conditions. Using this assay and immunofluorescence microscopy, we were able to show that NEDA is triggered in a variety of cell types during HSV-1 infection, including human cells. These results clearly show that NEDA is a widely used host response to HSV-1 infection. The molecular mechanisms regulating this pathway are still poorly understood. In our first report, we observed that the viral protein γ34.5 is required for the occurrence of NEDA, since a mutant virus deleted for this protein could not induce this autophagic pathway (13). Since it is known that the binding of γ34.5 to beclin-1 interferes with macroautophagy (12), we wanted to know whether the binding of beclin-1 also affected NEDA. Therefore, we infected macrophages with a virus mutant that lacked the γ34.5 beclin-1-binding domain. Remarkably, this virus triggered NEDA as efficiently as the wt virus, demonstrating that in contrast to macroautophagy, NEDA was not affected by beclin-1 binding. This strongly suggests that NEDA is a beclin-1-independent pathway regulated by molecular mechanisms different than those associated with macroautophagy. Furthermore, it demonstrates that NEDA and macroautophagy are altered by γ34.5 through different mechanisms.
In addition to macroautophagy, γ34.5 can interfere with a second cellular response to HSV-1 infection—the shutoff of protein translation (34). This response limits the production of viral proteins and the assembly of viral particles. To interfere with this antiviral response, HSV-1 γ34.5 recruits both eIF2α and the phosphatase PP1α to enable eIF2α dephosphorylation (15). In order to investigate whether the activation of protein translation (by lifting the shutoff response) is required to trigger NEDA, we constructed a mutant virus that could no longer bind PP1α and thus could not interfere with host translational shutoff. Infection of macrophages with this virus did not trigger NEDA. We concluded that active protein translation was required for NEDA. To further investigate the link between active protein translation and NEDA, we used an approach where we infected cells with HSV-1 wt (a strong inducer of NEDA) and inhibited protein translation by using specific drugs. Inhibiting overall protein production with hippuristanol lifted the ability of HSV-1 wt to trigger NEDA, further supporting the link between protein translation and this autophagic pathway. Furthermore, we treated infected cells with acyclovir, a drug that inhibits the production of late viral proteins. This treatment effectively prevented the occurrence of NEDA, demonstrating that the production of late viral proteins is required for the triggering of this autophagic pathway. This explains why NEDA is observed only at late times of infection.
Late viral proteins are expressed rapidly and in large amounts, and some of them, like gH, localize to the nuclear envelope (35), where they might exert a stress due to protein accumulation. NEDA could counteract such a stress by clearing proteins from the nuclear envelope. This idea is supported by our previous finding that gB is present in NEDA-derived autophagosomes (13).
As mentioned before, NEDA is not observed in cells infected with the ΔPP1α mutant virus. Interestingly, expression of a single late viral protein (gH) in these cells was sufficient to trigger NEDA despite the host translational shutoff. This could be caused by an accumulation of large amounts of gH and resulting stress on the nuclear envelope. Alternatively, gH might fulfill a specific function in the NEDA pathway. This protein is known to contribute to membrane fusion events at the nuclear envelope (36). However, efficient fusion activity probably requires a concerted action of gH with the viral proteins gL and gD (37). In a quantitative mass spectrometry analysis of ΔPP1α virus-infected cells, we could not detect gL, while gD was expressed in smaller amounts than in wt-infected cells (C. Bell, K. Radtke, P. Thibault, and M. Desjardins, unpublished observations). Since gH expression in the context of ΔPP1α virus infection and thus without gL was sufficient to trigger NEDA, the fusion activity of this protein might not play a direct role in this autophagic pathway. Further experiments will be required to understand the role of gH in NEDA, either as a stress inducer or an active contributor to the pathway.
Macroautophagy is a cellular response triggered in stress conditions. It has been shown that HSV-1 is able to interfere in several ways with macroautophagy, a key antiviral host response involved in clearing of particles from the cytosol and the presentation of viral antigens (5, 7, 8, 11, 12, 20). Our results support a model where in reaction to the initial impairment of macroautophagy during HSV-1 infection, the production of high levels of viral proteins triggers a host response characterized by the induction of NEDA. This novel form of autophagy appears to be driven by molecular mechanisms different than those governing macroautophagy. Future research will be directed at identifying the viral and cellular machinery of NEDA and the confirmation of the role of NEDA in viral infection, viral protein degradation, and antigen presentation.
ACKNOWLEDGMENTS
This work was supported by the Canadian Institute of Health Research (CIHR) to M.D. and a postdoctoral fellowship from the German Research Foundation to K.R. (RA1608/4-1).
We thank Magali Chemali (Université de Montréal, Canada) for helpful discussions. We are grateful to Ian Mohr (New York University), Beate Sodeik (Medizinische Hochschule Hannover, Germany), and Ari Helenius (ETH Zürich, Switzerland) for sharing antibodies and to Jerry Pelletier (McGill University, Montreal, Canada), who generously provided us with hippuristanol. We are indebted to Tony Minson and Helena Browne (University of Cambridge, United Kingdom) for providing Vero CR1 cells and to Bruce Banfield (Queens University, Kingston, Canada) for providing MeWo cells.
Footnotes
Published ahead of print 30 January 2013
REFERENCES
- 1. Yang Z, Klionsky DJ. 2009. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 335:1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Longatti A, Tooze SA. 2009. Vesicular trafficking and autophagosome formation. Cell Death Differ. 16:956–965 [DOI] [PubMed] [Google Scholar]
- 3. Yang Z, Klionsky DJ. 2010. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22:124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clave C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Droge W, Dron M, Dunn WA, Jr, Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fesus L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, Gonzalez-Estevez C, Gorski S, Gottlieb RA, Haussinger D, He YW, Heidenreich K, Hill JA, Hoyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jaattela M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovacs AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, Lopez-Otin C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Melendez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Munz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nurnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Talloczy Z, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcategui NL, van der Klei I, Vaquero EC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL. 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4:151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Levine B, Deretic V. 2007. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 7:767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deretic V, Levine B. 2009. Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5:527–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Münz C. 2010. Antigen processing via autophagy—not only for MHC class II presentation anymore? Curr. Opin. Immunol. 22:89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chemali M, Radtke K, Desjardins M, English L. 2011. Alternative pathways for MHC class I presentation: a new function for autophagy. Cell. Mol. Life Sci. 68:1533–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McFarlane S, Aitken J, Sutherland JS, Nicholl MJ, Preston VG, Preston CM. 2011. Early induction of autophagy in human fibroblasts after infection with human cytomegalovirus or herpes simplex virus 1. J. Virol. 85:4212–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rasmussen SB, Horan KA, Holm CK, Stranks AJ, Mettenleiter TC, Simon AK, Jensen SB, Rixon FJ, He B, Paludan SR. 2011. Activation of autophagy by α-herpesviruses in myeloid cells is mediated by cytoplasmic viral DNA through a mechanism dependent on stimulator of IFN genes J. Immunol. 187:5268–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Santana S, Bullido MJ, Recuero M, Valdivieso F, Aldudo J. 2012. Herpes simplex virus type I induces an incomplete autophagic response in human neuroblastoma cells. J. Alzheimers Dis. 30:815–831 [DOI] [PubMed] [Google Scholar]
- 12. Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. 2007. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1:23–35 [DOI] [PubMed] [Google Scholar]
- 13. English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, Alexander D, Leib D, Norbury C, Lippé R, Desjardins M. 2009. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat. Immunol. 10:480–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Verpooten D, Feng Z, Valyi-Nagy T, Ma Y, Jin H, Yan Z, Zhang C, Cao Y, He B. 2009. Dephosphorylation of eIF2alpha mediated by the gamma134.5 protein of herpes simplex virus 1 facilitates viral neuroinvasion. J. Virol. 83:12626–12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Y, Zhang C, Chen X, Yu J, Wang Y, Yang Y, Du M, Jin H, Ma Y, He B, Cao Y. 2011. ICP34.5 protein of herpes simplex virus facilitates the initiation of protein translation by bridging eukaryotic initiation factor 2alpha (eIF2alpha) and protein phosphatase 1. J. Biol. Chem. 286:24785–24792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kovacsovics-Bankowski M, Rock KL. 1995. A phagosome-to-cytosol pathway for exogenous antigens presented on MHC class I molecules. Science 267:243–246 [DOI] [PubMed] [Google Scholar]
- 17. Boursnell ME, Entwisle C, Blakeley D, Roberts C, Duncan IA, Chisholm SE, Martin GM, Jennings R, Ni CD, Sobek I, Inglis SC, McLean CS. 1997. A genetically inactivated herpes simplex virus type 2 (HSV-2) vaccine provides effective protection against primary and recurrent HSV-2 disease. J. Infect. Dis. 175:16–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang X, O'Shea H, Entwisle C, Boursnell M, Efstathiou S, Inglis S. 1998. An efficient selection system for packaging herpes simplex virus amplicons. J. Gen. Virol. 79:125–131 [DOI] [PubMed] [Google Scholar]
- 19. DeLuca NA, Schaffer PA. 1987. Activities of herpes simplex virus type 1 (HSV-1) ICP4 genes specifying nonsense peptides. Nucleic Acids Res. 15:4491–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alexander DE, Ward SL, Mizushima N, Levine B, Leib DA. 2007. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J. Virol. 81:12128–12134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ward SL, Scheuner D, Poppers J, Kaufman RJ, Mohr I, Leib DA. 2003. In vivo replication of an ICP34.5 second-site suppressor mutant following corneal infection correlates with in vitro regulation of eIF2 alpha phosphorylation. J. Virol. 77:4626–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rader KA, Ackland-Berglund CE, Miller JK, Pepose JS, Leib DA. 1993. In vivo characterization of site-directed mutations in the promoter of the herpes simplex virus type 1 latency-associated transcripts. J. Gen. Virol. 74:1859–1869 [DOI] [PubMed] [Google Scholar]
- 23. Döhner K, Radtke K, Schmidt S, Sodeik B. 2006. Eclipse phase of herpes simplex virus type 1 infection: efficient dynein-mediated capsid transport without the small capsid protein VP26. J. Virol. 80:8211–8224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sodeik B, Ebersold MW, Helenius A. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mulvey M, Poppers J, Sternberg D, Mohr I. 2003. Regulation of eIF2alpha phosphorylation by different functions that act during discrete phases in the herpes simplex virus type 1 life cycle. J. Virol. 77:10917–10928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wolfstein A, Nagel CH, Radtke K, Döhner K, Allan VJ, Sodeik B. 2006. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7:227–237 [DOI] [PubMed] [Google Scholar]
- 27. Dauber B, Pelletier J, Smiley JR. 2011. The herpes simplex virus 1 Vhs protein enhances translation of viral true late mRNAs and virus production in a cell type-dependent manner. J. Virol. 85:5363–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J. 2006. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2:213–220 [DOI] [PubMed] [Google Scholar]
- 29. Griffiths G, Quinn P, Warren G. 1983. Dissection of the Golgi complex. I. Monensin inhibits the transport of viral membrane proteins from medial to trans Golgi cisternae in baby hamster kidney cells infected with Semliki Forest virus. J. Cell Biol. 96:835–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rajčáni J, Andrea V, Ingeborg R. 2004. Peculiarities of herpes simplex virus (HSV) transcription: an overview. Virus Genes 28:293–310 [DOI] [PubMed] [Google Scholar]
- 31. Roizman B, Gu H, Mandel G. 2005. The first 30 minutes in the life of a virus: unREST in the nucleus. Cell Cycle 4:1019–1021 [DOI] [PubMed] [Google Scholar]
- 32. Kelly BJ, Fraefel C, Cunningham AL, Diefenbach RJ. 2009. Functional roles of the tegument proteins of herpes simplex virus type 1. Virus Res. 145:173–186 [DOI] [PubMed] [Google Scholar]
- 33. Furman PA, McGuirt PV. 1983. Effect of acyclovir on viral protein synthesis in cells infected with herpes simplex virus type 1. Antimicrob. Agents Chemother. 23:332–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Laurent AM, Madjar JJ, Greco A. 1998. Translational control of viral and host protein synthesis during the course of herpes simplex virus type 1 infection: evidence that initiation of translation is the limiting step. J. Gen. Virol. 79:2765–2775 [DOI] [PubMed] [Google Scholar]
- 35. Farnsworth A, Wisner TW, Webb M, Roller R, Cohen G, Eisenberg R, Johnson DC. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. U. S. A. 104:10187–10192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roizman B, Campadelli-Fiume G. 2007. Alphaherpes viral genes and their functions, p 2502–2601 In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K. (ed), Fundamental virology. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 37. Wright CC, Wisner TW, Hannah BP, Eisenberg RJ, Cohen GH, Johnson DC. 2009. Fusion between perinuclear virions and the outer nuclear membrane requires the fusogenic activity of herpes simplex virus gB. J. Virol. 83:11847–11856 [DOI] [PMC free article] [PubMed] [Google Scholar]