Abstract
Far-upstream element-binding protein 2 (FBP2) is an internal ribosomal entry site (IRES) trans-acting factor (ITAF) that negatively regulates enterovirus 71 (EV71) translation. This study shows that EV71 infection cleaved FBP2. Live EV71 and the EV71 replicon (but not UV-inactivated virus particles) induced FBP2 cleavage, suggesting that viral replication results in FBP2 cleavage. The results also showed that virus-induced proteasome, autophagy, and caspase activity co-contribute to EV71-induced FBP2 cleavage. Using FLAG-fused FBP2, we mapped the potential cleavage fragments of FBP2 in infected cells. We also found that FBP2 altered its function when its carboxyl terminus was cleaved. This study presents a mechanism for virus-induced cellular events to cleave a negative regulator for viral IRES-driven translation.
INTRODUCTION
Enterovirus 71 (EV71), an RNA virus that belongs to the family Picornaviridae, has caused several outbreaks worldwide and often results in severe neurological complications and high mortality in patients (1–10). Picornavirus infection can affect host mechanisms, such as host cap-dependent translation (11, 12), transcription (13, 14), and immune responses (15–17). For example, viral protease 2Apro can cleave eukaryotic initiation factor 4G (eIF4G) (18–20), poly(A)-binding protein (PABP) (21, 22), beta interferon (IFN-β) promoter stimulator-1 (IPS-1) (17), and 62-kDa nucleoporin (Nup62) (23). Viral protease 3Cpro can cleave PABP (24), polypyrimidine tract-binding protein (PTB) (25), cleavage stimulation factor 64 (CstF-64) (26), IPS-1 (17), retinoic acid-inducible gene I (RIG-I) (16), Toll-interleukin 1 receptor (TIR) domain-containing adaptor inducing IFN-β (TRIF) (27), and the 68-kDa Src-associated substrate during mitosis (Sam68) (28). Racaniello and colleagues have demonstrated that poliovirus infection-induced cleavage of MDA-5 (melanoma differentiation-associated gene 5) is associated with virus-induced proteasome and caspase activities (15). This shows that, in addition to viral proteases, virus infection also cleaves proteins through other pathways.
The translation of EV71 is driven by the internal ribosomal entry site (IRES) on the viral genome (20). Proteins that bind to the IRES and regulate translation by affecting ribosome recruitment or by changing the structure of the IRES are called IRES trans-acting factors (ITAFs). PTB (29, 30), poly(rC)-binding protein 1 (PCBP1), PCBP2 (31), autoantigen La (32), upstream N-ras protein (Unr) (33), far-upstream element (FUSE) binding protein 1 (FBP1) (34), and FBP2 (35) are all ITAFs of picornaviruses (36).
FBP2, also called the KH-type splicing regulatory protein (KSRP), was first identified as a single-strand DNA-binding protein (37). FBP2 positively regulates c-myc transcription (37), enhances the splicing of the neuron-specific c-src N1 exon (38, 39), edits apolipoprotein B (apoB) mRNA (40), and is associated with mRNA decay (41, 42). FBP2 is also a component of the Dicer and Drosha complexes and regulates let-7 microRNA (miRNA) biogenesis (43–45). A previous study has shown that FBP2 binds to the EV71 5′ untranslated region (UTR), which contains an IRES, and downregulates IRES activity by competing with other, positive ITAFs, such as PTB (35).
In this study, we show that FBP2, a negative ITAF of EV71, is cleaved in EV71-infected cells. We also demonstrate that EV71-induced caspase activity, autophagic activity, and proteasome activity are involved in virus-induced cleavage of FBP2. Moreover, we found a cleavage product, FBP21-503, that loses its carboxyl terminus and positively regulates the IRES activity of EV71.
MATERIALS AND METHODS
Cell lines and virus infection.
Human embryonal rhabdomyosarcoma (RD) and HeLa cells were maintained in Dulbecco's modified Eagle medium (DMEM) (GIBCO) containing 10% fetal bovine serum (FBS; GIBCO) at 37°C. RD cells were grown to 80% to 90% confluence and were infected with enterovirus 71 strain Tainan/4643/98 at a multiplicity of infection (MOI) of 40 PFU per cell in serum-free DMEM. Virus was adsorbed at 37°C for 1 h. After adsorption, the cells were washed with phosphate-buffered saline (PBS) and were incubated with a medium containing 2% FBS. In several experiments, RD cells were infected with EV71 for 1 h, washed with PBS, and incubated with a medium containing 2% FBS and various inhibitors. UV-inactivated EV71 was prepared by following a method described elsewhere (46). In other experiments, RD cells were transfected with autophagy protein 5 (ATG5) small interfering RNA (siRNA) before infection. The sequence of ATG5 siRNA is 5′-AAUUCGUCCAAACCACACAUCUCG (GeneDireX), and the sequence of negative-control (NC) siRNA is 5′-UUCUCCGAACGUGUCACGUdTdT.
Reagents.
The proteasome inhibitor MG132 and the lysosome inhibitor NH4Cl were purchased from Sigma. The general caspase inhibitor QVD-OPh was purchased from MP Biomedicals. The apoptosis inducer staurosporine (STS) was purchased from Cell Signaling.
Plasmid construction and expression.
The pT7-EV71 5′ UTR and pGL3-EV71 5′ UTR-FLuc plasmids were constructed as described previously (34). The DNA of FBP2 was provided by Douglas L. Black (University of California, Los Angeles). Nucleotides 1 to 890 of FBP2 were optimized using GeneART to decrease GC content and increase protein expression without changing the amino acids. The cDNA of FBP2 was amplified using primers 5′-ACCGAATTCGCCACCATGAGCGACTACAGCAC and 5′-CTCGAGTCATTGAGCCTGCTGCTGTCCCTGCT and was inserted into the EcoRI and XhoI sites of the pCMV-Tag 2B vector (N-FLAG-FBP2). The cDNA of FBP2 was also amplified using primers 5′-ACCGAATTCGCCACCATGAGCGACTACAGCAC and 5′-GTACCGATATCAGTTGAGCCTGCTGCTGTCCCT and was inserted into the EcoRI and EcoRV sites of the C-terminal pFLAG-CMV-5.1 vector (C-FLAG-FBP2). The cDNA corresponding to amino acids 1 to 503 of FBP2 was amplified using PCR with primers 5′-ACCGAATTCGCCACCATGAGCGACTACAGCAC and 5′-CCCCTCGAGTCATCCAACTGGGCAGAG and was inserted into the EcoRI and XhoI sites of the pCMV-Tag 2B vector (N-FLAG-FBP21-503). The cDNA corresponding to amino acids 190 to 711 of FBP2 was amplified using PCR with primers 5′-CAGGAATTCGCCACCATGAGATCCGTGTCTCTGACT and 5′-GTACCGATATCAGTTGAGCCTGCTGCTGTCCCT and was inserted into the EcoRI and EcoRV sites of the C-terminal pFLAG-CMV-5.1 vector (C-FLAG-FBP2190-711). The cDNAs of viral proteins 2B, 2C, 2BC, 3A, and 3AB were amplified from a full-length infectious cDNA clone of EV71 by PCR (47). PCR products were inserted into the p3XFLAG-Myc-CMV vector using EcoRI and EcoRV. The primers for 2B were 5′-GAATTCAGGCGTGTCTGATTACATTAA and 5′-GATATCCTACTGCTTCTGAGCCATCGGTA. The primers for 2C were 5′-GAATTCAAGTGCCTCTTGGTTAAAGAA and 5′-GATATCCTATTGGAAAAGAGCTTCAATGG. The primers for 3A were 5′-GAATTCAGGACCCCCTAAATTTAGACC and 5′-GATATCCTATTGAAAACCGGCGAACAACT. The primers for 3AB were 5′-GAATTCAGGACCCCCTAAATTTAGACC and 5′-ATATCCTACTGCACAGTTGCCGTGCGCA. Plasmid N-FLAG-FBP2502-504AAA was constructed in 2 steps. The cDNA corresponding to amino acids 1 to 504 (residues 502 to 504 were mutated from VGP to AAA) of FBP2 was amplified using PCR with primers 5′-ACCGAATTCGCCACCATGAGCGACTACAGCAC and 5′-GGGCCTGGGCCACCTGGGCCTGCTGCAGCTGGGCAGAGAGGACCCTCG, and cDNA corresponding to amino acids 502 to 711 (residues 502 to 504 were mutated from VGP to AAA) of FBP2 was amplified using PCR with primers 5′-CGAGGGTCCTCTCTGCCCAGCTGCAGCAGGCCCAGGTGGCCCAGGCCC and 5′-CTCGAGTCATTGAGCCTGCTGCTGTCCCTGCT. The cDNA of FBP2502-504AAA was amplified using PCR with primers 5′-ACCGAATTCGCCACCATGAGCGACTACAGCAC and 5′-CTCGAGTCATTGAGCCTGCTGCTGTCCCTGCT and with fragments containing amino acids 1 to 504 and 502 to 711 as templates. The cDNA of FBP2502-504AAA was inserted into the EcoRI and XhoI sites of the pCMV-Tag 2B vector (N-FLAG-FBP2502-504AAA). The cDNA corresponding to amino acids 1 to 497 of FBP2 was amplified using PCR with primers 5′-ACCGAATTCGCCACCATGAGCGACTACAGCAC and 5′-GGCCAGCAGGGCCTGGGCCACCCTCGATCTTTTCCTCG, and cDNA corresponding to amino acids 508 to 711 of FBP2 was amplified using PCR with primers 5′-CGAGGAAAAGATCGAGGGTGGCCCAGGCCCTGCTGGCC and 5′-CTCGAGTCATTGAGCCTGCTGCTGTCCCTGCT. The cDNA of FBP2Δ498-507 was amplified using PCR with primers 5′-ACCGAATTCGCCACCATGAGCGACTACAGCAC and 5′-CTCGAGTCATTGAGCCTGCTGCTGTCCCTGCT and with fragments containing amino acids 1 to 497 and 508 to 711 (which lost cDNA corresponding to residues 498 to 507) as templates. The cDNA of FBP2Δ498-507 was inserted into the EcoRI and XhoI sites of the pCMV-Tag 2B vector (N-FLAG-FBP2Δ498-507).
Construction and expression of the EV71 replicon.
The EV71 replicon contained the 5′ UTR, luciferase, the P2 and P3 regions, and the 3′ UTR. The EV71 replicon was constructed as follows. The 5′ UTR of EV71 and luciferase were amplified using PCR from pRHF-EV71-5′ UTR (35) with primers 5′-TTAAAACAGCCTGTGGGTTGC and 5′-ACGCGTCACGGCGATCTTTCC. The PCR product was inserted into the yTA vector using TA cloning and was named yTA-5′ UTR-Luc. The cDNA of the viral P2 and P3 regions and of the 3′ UTR was amplified using PCR from a full-length infectious cDNA clone of EV71 with primers 5′-CACCACGCGTCGCACAGCAATCACCACC and 5′-CGAATAGTCGACAGCTCGGATCCACAAGTAC. The PCR product was inserted downstream of luciferase into a yTA-5′ UTR-Luc plasmid with MluI and SalI sites and was named the EV71 replicon. The EV71 replicon was linearized by SalI as the template for in vitro transcription. The EV71 replicon was transcribed to RNA and was transfected by Lipofectamine 2000 (LF2000) into RD cells. After 20 h of transfection, the cells were washed with PBS and were lysed with a CA630 lysis buffer (150 mM NaCl, 1% CA630, 50 mM Tris-base [pH 8.0]) for 30 min on ice. Cell lysates were collected by centrifugation at 10,000 × g for 10 min at 4°C for further analysis.
Construction and purification of recombinant protein.
Plasmids pET-23-EV71-3C (3C) and pET-23-EV71-m3C-C147S 3C (3CC147S) were constructed as described previously (26). pET-23-EV71-3C (3C) and pET-23-EV71-m3C-C147S 3C (3CC147S) were purified using a method described elsewhere (26). Plasmid pGEX-5X-1-EV71-2A (2A) was constructed as follows. EV71 2A cDNA was amplified from a full-length infectious cDNA clone of EV71 by PCR. The PCR product was inserted into pGEX-5X-1 using EcoRI and XhoI sites. The primers for 2A were 5′-CCGGAATTCGGGAAATTTGGACAG and 5′-CCGCTCGAGCTGCTCCATGGCTT. The DNA of pGEX-5X-1-EV71-2A (2A) was used as the template for the generation of plasmid pGEX-5X-1-EV71-m2A-C110S (2AC110S). The primers for pGEX-5X-1-EV71-m2A-C110S (2AC110S) were 5′-GAACCAGGGGATTCCGGTGGCATTCTTAG and 5′-CTAAGAATGCCACCGGAATCCCCTGGTTC. The PCR and plasmid construction were conducted using the QuikChange site-directed mutagenesis kit (Stratagene). pGEX-5X-1-EV71-2A (2A) and pGEX-5X-1-EV71-m2A-C110S (2AC110S) were purified using a glutathione S-transferase kit (Pharmacia). pBacPAK8-MTEGFP-His-FBP2 (FBP2), pBacPAK8-MTEGFP-His-FBP2190-711 (FBP2190-711), and pFastBac-His-FBP21-503 (FBP21-503) were used to express His-tagged recombinant FBP2 using a baculovirus expression system. The cDNA of full-length FBP2 was amplified using PCR with primers 5′-CCGCTCGAGGCCACCATGAGCGACTACAGCACAGGCGGA and 5′ (CCGGAATTCTCA ATGATGATGATGATGGTGTTGAGCCTGCTGCTGTCC. The cDNA corresponding to amino acids 190 to 711 of FBP2 was amplified using PCR with primers 5′-CCGCTCGAGGCCACCATGAGATCCGTGTCTCTGACT and 5′-CCGGAATTCTCAATGATGATGATGATGGTGTTGAGCCTGCTGCTGTCC. The reverse primers contained 6 His sequences. PCR products were inserted into the pBacPAK8-MTEGFP vector (Tsu-An Hsu, National Health Research Institute, MiaoLi, Taiwan) using XhoI and EcoRI sites (34). The cDNA corresponding to amino acids 1 to 503 of FBP2 was amplified using PCR with primers 5′-GAATTCGCCACCATGAGCGACTACAGCACAGGCGGA and 5′-CCCCTCGAGTCAATGATGATGATGATGGTGTCCAACTGGGCAGAG; the reverse primer contained 6 His sequences. The PCR product was inserted into the EcoRI and XhoI sites of the pFastBac vector (Invitrogen) (pFastBac-His-FBP21-503). His-tagged recombinant proteins FBP2, FBP21-503, and FBP2190-711 were produced using the baculovirus expression system. pBacPAK8-MTEGFP-His-FBP2 or pBacPAK8-MTEGFP-His-FBP2190-711 plasmid DNA was cotransfected with baculovirus DNA into Sf9 cells using a BaculoGold transfection kit (BD Biosciences). Transfected Sf9 cells were maintained in Grace's insect medium (Caisson) containing 10% FBS. The supernatants were collected after 4 days of infection. Infected cells were seeded in a 96-well plate in order to select a single virus clone. The pFastBac-His-FBP21-503 plasmid DNA was transformed into Escherichia coli DH10Bac cells for recombination. After 4 days, a single colony containing a recombinant bacmid was cultured to isolate the recombinant bacmid DNA. The recombinant bacmid DNA was transfected into Sf9 cells using the Cellfectin II reagent (Invitrogen). The baculovirus-containing supernatant was collected for further infection. Cell extracts were collected and were analyzed by Western blotting to assess the expression of FBP2. For large-scale amplification of FBP2 recombinant proteins, Sf9 cells (1 × 106/ml) were infected at an MOI of 1 with recombinant virus in 400 ml of Grace's insect medium and were cultured at 27°C for 4 days. The His-tagged FBP2 recombinant proteins were purified using a HiTrap kit (Pharmacia).
Western blotting.
Samples were loaded onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. After electrophoresis, the proteins in the sample were separated and were transferred to a polyvinylidene difluoride (PVDF) membrane. The PVDF membrane was blocked with Tris-buffered saline–0.1% (vol/vol) Tween 20 containing 5% nonfat dry milk and was probed with the antibodies. The anti-KSRP monoclonal antibody 5 (Ab5) was raised against the full length of the KSRP protein (dilution, 1:3,000; Douglas L. Black) (38), and the monoclonal antibody against EV71 3Cpro was generated from recombinant full-length 3Cpro in the laboratory (1:50) (26). Antibodies against CstF-64 (1:200; Clinton C. MacDonald), FLAG (F3156; 1:2,000; Sigma), poly(ADP-ribose) polymerase (PARP) (sc7150; 1:2,500; Cell Signaling), actin (A5316; 1:4,000; Sigma), eIF4G (sc11373; 1:200; Santa Cruz), microtubule-associated protein light chain 3 (LC3) (2775S; 1:500; Cell Signaling), and ATG5 (GTX62601; 1:5,000; GeneTex) were used. After washing, the membrane was incubated with a horseradish peroxidase (HRP)-conjugated anti-mouse or HRP-conjugated anti-rabbit antibody (1:2,000). HRP was detected using the Western Lightning chemiluminescence reagent (Amersham Pharmacia).
[35S]methionine-cysteine labeling assay.
RD cells were infected with EV71 (MOI, 40) or were treated with cycloheximide (CHX). After 1 h of adsorption, the cells were washed with PBS and were cultured in DMEM containing 2% FBS at 37°C. The medium was replaced by methionine- and cysteine-free DMEM 1 h before labeling with [35S]methionine-cysteine. Subsequently, the medium was replaced by a medium containing [35S]methionine-cysteine (50 μCi/ml). After 1 h of labeling, the cells were washed with PBS and were lysed with a CA630 lysis buffer for 30 min over ice. Cell lysates were collected by centrifugation at 10,000 × g for 10 min at 4°C. Radiolabeled proteins were analyzed using SDS-PAGE, transferred to a PVDF membrane, and detected using autoradiography. The same membrane was used for Western blotting. In numerous experiments, the RD cells were transfected with FBP2 siRNA or FLAG-FBP2 expression plasmids, and the transfected RD cells were reseeded in a 12-well plate after 48 h posttransfection. The cells were subsequently infected with EV71, after incubation for 24 h. The sequence of FBP2 siRNA is 5′-CACAUUCGUAUUCUGAGAUCCGUCC (KHSRP-HSS112554; Invitrogen), and the sequence of NC siRNA is 5′-AACUGGGUAAGCGGGCGCAAAUU.
In vitro protease cleavage assay.
The RD cells were washed with PBS and were collected using a CA630 lysis buffer without protease inhibitors. Three micrograms of the recombinant viral protease (pET-23-EV71-3C [3C], pET-23-EV71-m3C-C147S 3C [3CC147S], pGEX-5X-1-EV71-2A [2A], or pGEX-5X-1-EV71-m2A-C110S [2AC110S]) was incubated with RD cell lysates containing 40 μg of protein at 37°C for 4 h. The reactions were analyzed using Western blotting.
In vivo ubiquitination assay.
The RD cells were cotransfected with the pUI-myc-ubiquitin (myc-ub) (supplied by Michael M. C. Lai, Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan) (48) and N-FLAG-FBP2 expression plasmids. At 48 h posttransfection, the cells were either left untreated or treated with the proteasome inhibitor MG132 for 2 h. The cells were lysed in a FLAG-IP-lysis buffer (50 mM Tris HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100) over ice for 30 min. Cell lysates were collected using centrifugation at 10,000 × g for 10 min at 4°C and were incubated with an anti-FLAG M2 affinity gel (Sigma) at 4°C for 4 h. The anti-FLAG M2 affinity gel was washed 5 times in a wash buffer (0.5 M Tris HCl [pH 7.4], 1.5 M NaCl). The immunoprecipitation complex was eluted by competition with a 3× FLAG peptide. Bound proteins were analyzed using SDS-PAGE and Western blotting.
Preparation of cell extracts.
The RD cells were transfected with various FLAG-tagged FBP2 expression plasmids. At 48 h posttransfection, the cells were washed using PBS and were lysed with a 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) lysis buffer (10 mM Tris-HCl [pH 7.4], 1 mM MgCl2, 1 mM EGTA, 0.5% CHAPS, 10% glycerol, 0.1 mM phenylmethylsulfonyl fluoride [PMSF], 5 mM β-mercaptoethanol) over ice for 30 min. Cell lysates were collected using centrifugation at 10,000 × g for 10 min at 4°C. Cell lysates were stored at −80°C for further analysis.
In vitro transcription.
The EV71 5′ UTR fragment was cut from the pT7-EV71 5′ UTR plasmid using EcoRI as the template for transcription. To generate EV71-IRES-FLuc reporter RNA, the pGL3-EV71-5′ UTR-Fluc plasmid was linearized by XhoI as the template for in vitro transcription. To generate Cap-RLuc reporter RNA, the DNA fragment containing the T7 promoter and the Renilla luciferase coding sequence was amplified from the pRH plasmid using primers 5′-TAATACGACTCACTATAGGCTAGCCACCATGACTTCGAAAGTTTATGATC-3′ and 5′-TTATTGTTCATTTTTGAGAACT-3′ and was used as the template for in vitro transcription. A cap structure on the 5′ end of the Renilla reporter RNA was added using a vaccinia virus capping system (New England BioLabs). RNA transcripts were synthesized using the MEGAscript T7 kit (Ambion). The biotinylated RNA probe was synthesized in a 20-μl MEGAscript transcript reaction mixture containing 1.25 μl of 20 mM biotinylated UTP and biotin-16-UTP (Roche). The synthesized RNA products were purified using an RNeasy Protect minikit (Nobel).
Pulldown assay of streptavidin beads and biotinylated RNA.
The reaction mixtures contained RD cell extracts containing 120 μg protein, 12.5 pmol of the biotinylated EV71 5′ UTR, and an RNA mobility buffer (5 mM HEPES [pH 7.1], 40 mM KCl, 0.1 mM EDTA, 2 mM MgCl2, 2 mM dithiothreitol [DTT], 1 U RNasin, and 0.25 mg/ml heparin) at a final volume of 100 μl. The mixture was incubated at 30°C for 15 min. A total of 400 μl of streptavidin MagneSphere paramagnetic particles (Promega) was added to the mixture for 10 min at room temperature to pull down biotinylated RNA and interacting proteins. The protein-RNA complexes were washed 5 times in an RNA mobility buffer without heparin. After washing, 30 μl of SDS sample buffer was added to the streptavidin beads at room temperature for 10 min in order to dissociate the proteins from the RNA. The SDS sample buffers containing interacting proteins were further analyzed using SDS-PAGE and Western blotting.
Preparation of HeLa cell translation extracts.
Approximately 4.5 × 105 HeLa cells/ml were maintained in DMEM with 10% FBS at 37°C. Two liters of HeLa cells was trypsinized after washing with PBS and was centrifuged at 2,800 × g for 10 min at 4°C. The cells were washed 3 times with cold PBS, and the cell pellets were resuspended in 1.5 volume of hypotonic lysis buffer {10 mM HEPES-KOH [pH 7.6], 10 mM potassium acetate [KOAc], 2.5 mM magnesium acetate [Mg(OAc)2], 2 mM DTT} on ice for 10 min. The cells were subsequently homogenized with approximately 100 strokes of a 25-gauge 3/8-in needle. The percentage of cell lysis (>90%) was verified using trypan blue staining. Cell extracts were centrifuged at 10,400 × g for 20 min at 4°C. The supernatant was frozen in liquid nitrogen and was stored at −80°C for further analysis.
In vitro translation assay.
An in vitro translation assay for IRES activity was carried out by mixing 0.25 μg of EV71 5′ UTR-FLuc RNA, various amounts of recombinant proteins, 80 μg of HeLa cell translation extracts, and 20% rabbit reticulocyte lysates (RRL) (Promega) in a final volume of 25 μl. An in vitro translation assay for cap-dependent translation was carried out by mixing 0.25 μg of Cap-RLuc reporter RNA, various amounts of recombinant proteins, HeLa cell translation extracts containing 80 μg protein, and 20% RRL in a final volume of 25 μl. The mixtures were incubated at 30°C for 90 min. Firefly luciferase and Renilla luciferase activities were measured using the Luciferase Assay System (Promega).
Sucrose gradient fractionation.
The RD cells were transfected with the plasmid DNA of FLAG-FBP2 or FLAG-FBP21–503; 48 h after transfection, the cells were infected with EV71 at an MOI of 40. At 6 h postinfection, the cells were incubated with 0.1 mg/ml cycloheximide for 5 min at 37°C; subsequently, they were washed twice with ice-cold PBS containing 0.1 mg/ml cycloheximide. Cells were lysed with a 0.2-ml polysomal extraction buffer (20 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 100 mM KCl, 1% Triton X-100, 0.1 mg/ml cycloheximide) on ice for 30 min. Cell lysates were collected after centrifugation at 15,000 × g for 10 min. The cell extracts were layered onto 9.6-ml 7%-to-47% sucrose gradients (composed of 20 mM Tris-HCl [pH 7.], 5 mM MgCl2, and 100 mM KCl) and were centrifuged at 35,000 rpm for 150 min in a Beckman SW41-Ti rotor at 4°C. The gradients were fractionated with the Isco fractionator by pumping a 60% sucrose solution into the bottom of the tube, and fractions were collected from the top with concomitant measurement of the optical density at 254 nm (OD254). Ribosome subunits were detected in each fraction using Western blotting with an antibody against the S6 (2271; dilution, 1:1,000; Cell Signaling) and P0 (NBP1-57528; 1:50; Novus) ribosomal proteins.
RESULTS
EV71 infection induces FBP2 truncation.
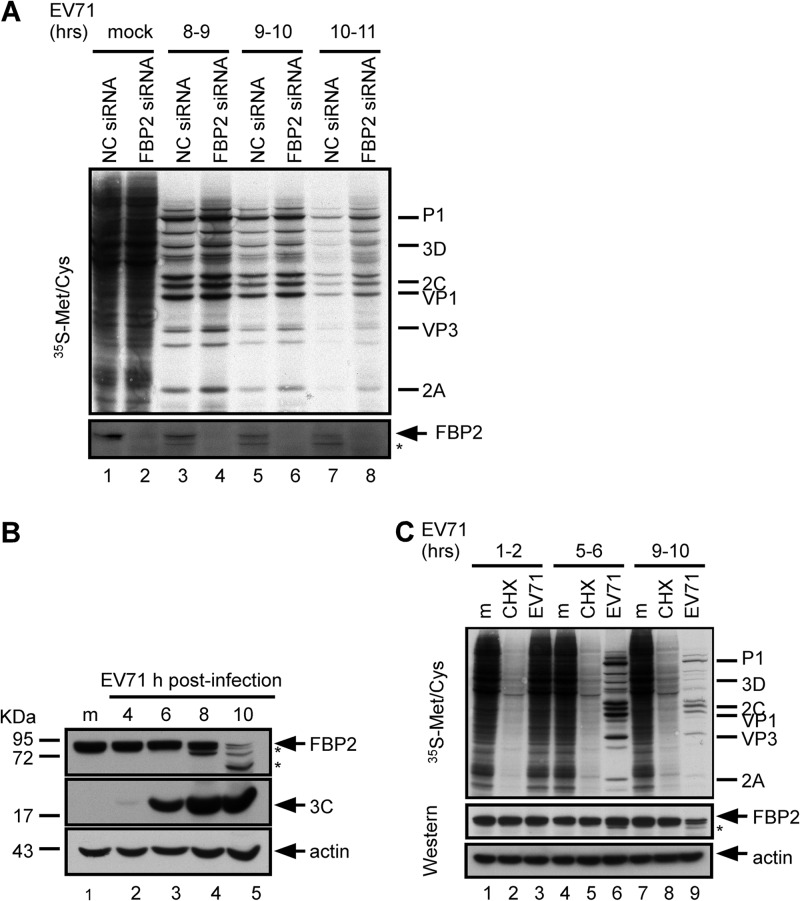
Lin et al. showed that FBP2 is a negative regulator and that levels of viral protein synthesis and IRES activity (but not viral RNA synthesis) increased in FBP2-knockdown cells (35). A [35S]methionine-cysteine labeling assay was performed to monitor newly synthesized proteins in EV71-infected cells with FBP2 depletion. In agreement with previous results (35), levels of viral protein synthesis in FBP2 siRNA-transfected cells (Fig. 1A, lanes 4, 6, and 8) were higher than those in NC siRNA-transfected cells (Fig. 1A, lanes 3, 5, and 7). FBP2 was truncated during EV71 infection (Fig. 1A, bottom). To further confirm FBP2 truncation, RD cells were infected with EV71; the cell lysates were collected at the indicated time points (8 to 9, 9 to 10, and 10 to 11 h) after infection occurred. The level of FBP2 in infected cells declined with the time of infection (Fig. 1B, lanes 2 to 5). The detection of FBP2 showed truncation products in EV71-infected cells; one of the dominant truncation products observed at 10 h postinfection was approximately 60 kDa (Fig. 1B, lane 5). The detection of viral protein 3C was used as the index of virus replication. Because the picornavirus infection stops host translation, the decrease in the level of FBP2 in infected cells may be caused by protein instability and the lack of newly synthesized protein. To examine the stability of FBP2, we used CHX to inhibit cellular protein synthesis and to detect FBP2 levels at various time points. RD cells were treated with CHX or were infected with EV71, and the newly synthesized proteins were labeled with [35S]methionine-cysteine for 1 h at 1 to 2, 5 to 6, and 9 to 10 h postinfection, as indicated in Fig. 1C. The inhibitory effect of CHX on new protein synthesis was verified (Fig. 1C, lanes 2, 5, and 8). Equivalent levels of FBP2 appeared in CHX-treated cells and mock-treated cells at various intervals (Fig. 1C, lanes 1, 2, 4, 5, 7, and 8), indicating that FBP2 was stable for as long as 10 h after CHX treatment. Compared with the levels of FBP2 expression in CHX-treated cells and EV71-infected cells, FBP2 levels decreased in cells at 10 h postinfection (Fig. 1C, lane 9); however, FBP2 levels did not decrease in CHX-treated cells (Fig. 1C, lane 8), indicating that the FBP2 reduction in EV71-infected cells is not caused by protein instability.
Fig 1.
Function and truncation of FBP2 in EV71-infected cells. (A) Effect of FBP2 knockdown on viral translation. Control RNA (NC siRNA)- or FBP2 siRNA-transfected RD cells were mock infected or infected with EV71. Protein synthesis in these cells was examined at 8 to 9, 9 to 10, and 10 to 11 h postinfection by the [35S]methionine-cysteine labeling assay. Viral proteins P1, 3D, 2C, VP1, and 2A are labeled according to protein size. The FBP2 expression level was detected by Western blotting. The truncated form of FBP2 is indicated by an asterisk. (B) FBP2 truncation in EV71-infected cells. FBP2 in mock (m)- or EV71-infected cells at 4, 6, 8, and 10 h postinfection was detected by Western blotting. Viral 3C protein in these infected cells was used as a viral marker. Truncated forms of FBP2 are indicated by asterisks. (C) FBP2 stability test. FBP2 in mock-infected, CHX (20 μM)-treated, or EV71-infected cells was detected by Western blotting. Protein synthesis was detected using the [35S]methionine-cysteine labeling assay at the intervals indicated. The levels of FBP2 in these cells were detected by Western blotting.
Truncation of FBP2 is associated with EV71 replication.
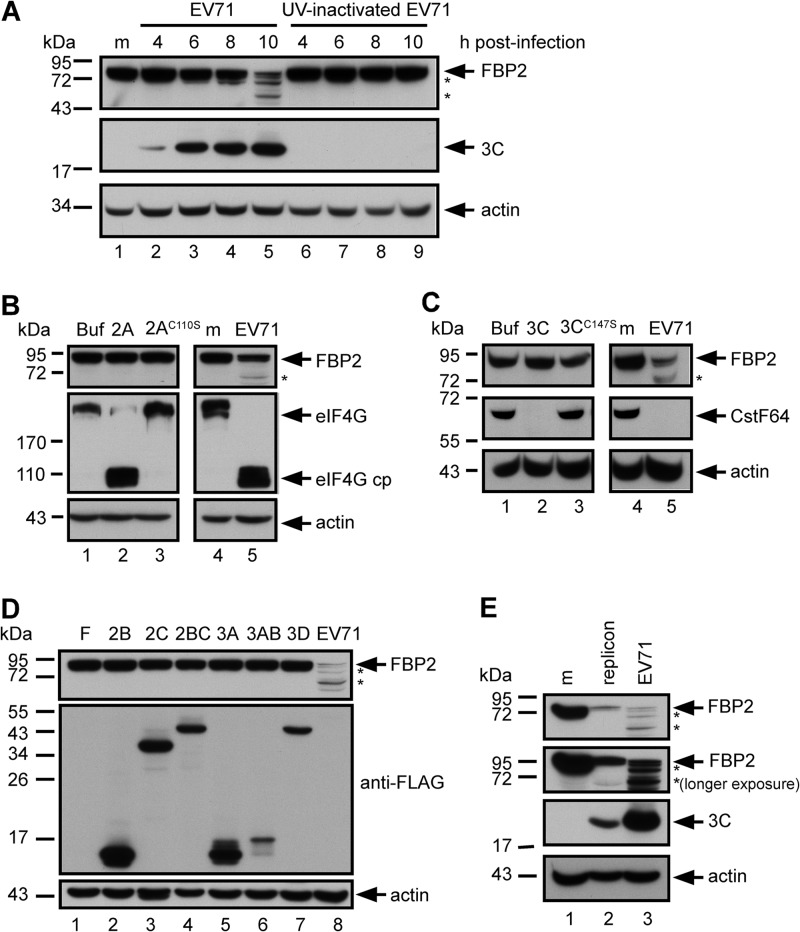
The results showed that FBP2 was truncated in EV71-infected cells (Fig. 1B). Subsequently, we investigated the manner in which a virus induces FBP2 truncation. To determine whether virus replication is essential for triggering FBP2 truncation, we used either EV71 or UV-inactivated EV71 to infect RD cells, and we examined FBP2 in cell lysates at various time points after infection. The results show that FBP2 was truncated in EV71-infected cells (Fig. 2A, lanes 2 to 5). In contrast, infection with the replication-incompetent virus (UV-inactivated EV71) did not induce FBP2 truncation (Fig. 2A, lanes 6 to 9). These results also indicate that the process of viral replication or the presence of viral nonstructural proteins, which are produced during viral replication, may be essential for triggering FBP2 truncation.
Fig 2.
Truncation of FBP2 was associated with EV71 replication. (A) FBP2 in RD cells infected with EV71 or a UV-inactivated virus at 4, 6, 8, and 10 hours postinfection. Truncated FBP2 forms are indicated by asterisks. The levels of viral 3C protein in these cells were used as viral replication indicators. (B) FBP2 was detected in RD cell extracts incubated with wild-type 2Apro (2A) or mutant 2Apro (2AC110S) for 4 h at 37°C and in mock-infected (m) or EV71-infected cells. eIF4G and its cleavage product (eIF4G cp) were detected in these lysates to demonstrate the activity of 2Apro. (C) FBP2 in RD cell extracts incubated with wild-type 3Cpro (3C) or mutant 3Cpro (3CC147S) were analyzed by Western blotting. Western blotting was used for detection of CstF-64 in these lysates and in mock-infected (m) or EV71-infected cells in order to demonstrate the activity of 3Cpro. (D) FBP2 levels in RD cells transfected with FLAG-tagged 2B, 2C, 2BC, 3A, 3AB, and 3D plasmids were analyzed by Western blotting. FBP2 levels in EV71-infected cell lysates were also analyzed (lane 8). Truncated forms of FBP2 are indicated by asterisks. (E) FBP2 in mock-infected (m), EV71 replicon-transfected (20 h posttransfection), or virus-infected (EV71) RD cells was detected. The second panel shows the longer-exposure result of Western blotting. Truncated forms of FBP2 are indicated by asterisks. The viral 3C protein was detected as an indicator of viral protein expression.
Two viral nonstructural proteins (proteases 2A and 3C) cleave viral polypeptide to become functional viral proteins; they also cleave numerous cellular proteins. Purified recombinant proteins (EV71 2A and 3C) were used to determine whether FBP2 truncation was caused by the catalytic activity of 2A or 3C. RD cell lysates were incubated with recombinant wild-type 2A or mutant 2A protease (2AC110S) for 4 h and were analyzed using Western blotting. The catalytic activity of recombinant 2A was confirmed by detecting a known 2A substrate, eIF4G, and its cleavage product in the reaction mixture (18–20). 2A cleaved eIF4G, yielding a cleavage product (Fig. 2B, lane 2). The catalytically defective mutant 2AC110S failed to cleave eIF4G (Fig. 2B, lane 3). The results show that wild-type 2A did not cleave FBP2 in the cell extract (Fig. 2B, lane 2). The same experiment was conducted to assay the catalytic activity of 3C cleaving FBP2. The results show that wild-type 3C cleaved a known substrate, CstF-64 (26), but did not cleave FBP2 (Fig. 2C, lane 2). This indicates that FBP2 was not a direct substrate of viral proteases 2A and 3C. To determine whether other EV71 nonstructural proteins contribute to FBP2 truncation, we analyzed FBP2 in cells overexpressing FLAG-tagged 2B, 2C, 2BC, 3A, 3AB, and 3D. The results show that none of these cells overexpressing viral proteins induced FBP2 truncation (Fig. 2D).
FBP2 in EV71 replicon-transfected cells was analyzed using Western blotting to confirm the effect of virus replication on FBP2 truncation. The EV71 replicons induced FBP2 truncation (Fig. 2E, lane 2). The results show that virus replication is an essential process in inducing FBP2 truncation.
Involvement of virus-induced autophagy, lysosomes, and proteasomes in FBP2 turnover.
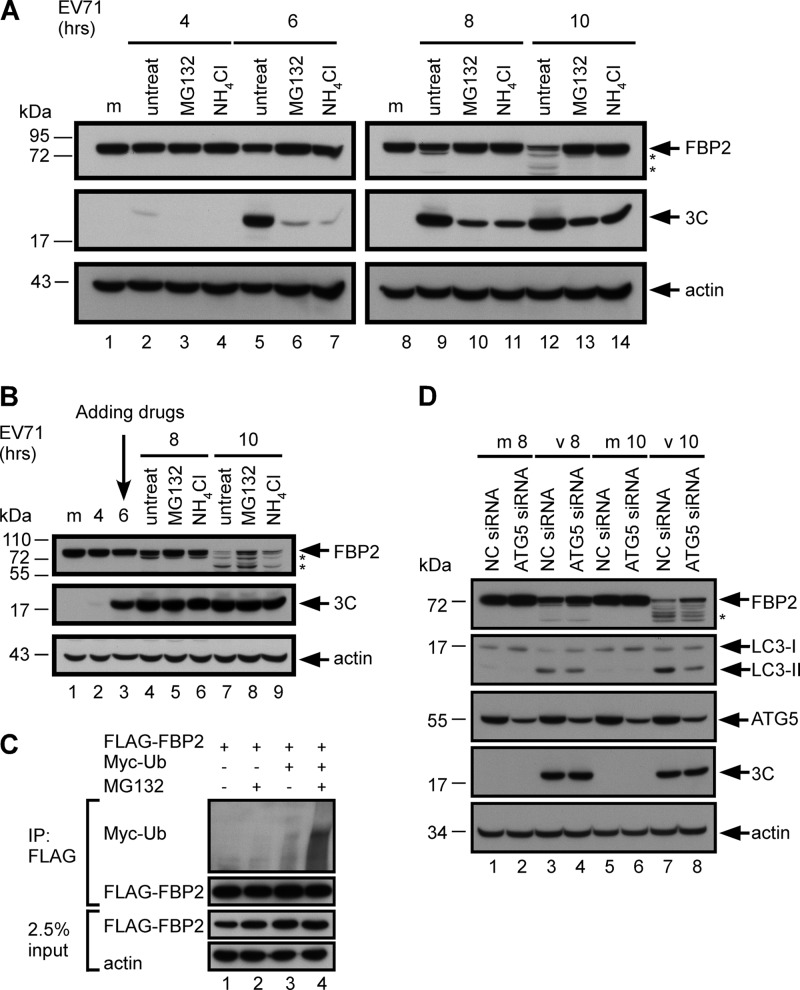
Because FBP2 truncation in infected cells was associated with viral replication, we determined whether pathways induced by EV71 were involved in this truncation. Virus infection triggers pathways that are involved in protein degradation (15, 17, 49–51). Two major degradation pathways are involved in cytoplasmic protein degradation: proteasome-dependent and lysosome-dependent pathways (52). To determine whether proteasomes and lysosomes are involved in FBP2 turnover, we examined FBP2 in EV71-infected cells after treatment with proteasome and lysosome inhibitors. RD cells were infected with EV71 and were treated with either the proteasome inhibitor MG132 or the lysosome inhibitor NH4Cl after virus adsorption (Fig. 3A). The cell lysates were subsequently collected and analyzed using Western blotting. Truncation of FBP2 was not obviously detected at 10 h postinfection in MG132- or NH4Cl-treated cells (Fig. 3A, lanes 13 and 14 versus lane 12). Expression of 3C in MG132- or NH4Cl-treated cells decreased from that in untreated cells, which indicated that the inhibitors may also delay viral replication (Fig. 3A, lanes 10 and 11 versus lane 9; lanes 13 and 14 versus lane 12). To avoid sabotaging viral replication, inhibitors were added at 6 h postinfection (Fig. 3B). Comparable levels of viral protein 3C were produced at 8 and 10 h postinfection in untreated and inhibitor-treated cells (Fig. 3B, lanes 5 and 6 versus lane 4; lanes 8 and 9 versus lane 7). The levels of FBP2 in infected cells treated with MG132 or NH4Cl were increased (Fig. 3B, lanes 8 and 9) over those in infected cells without treatment (Fig. 3B, lane 7). Because ubiquitination is required for protein degradation by proteasomes, we performed an in vivo ubiquitination experiment to further validate the involvement of proteasomes in FBP2 truncation. RD cells were transfected with the FLAG-FBP2 plasmid or cotransfected with FLAG-FBP2 and pUI-myc-ubiquitin (myc-ub) plasmids. FLAG-FBP2 was immunoprecipitated by an anti-FLAG antibody, and the ubiquitinated FBP2 was detected using an anti-Myc antibody. MG132 was used to enrich the ubiquitinated protein in the cells. FBP2 in the cells was ubiquitinated at a basal level when coexpressed with myc-Ub (Fig. 3C, lane 3), and the ubiquitinated FBP2 was enriched in cells treated with MG132 (Fig. 3C, lane 4). These results indicate that proteasomes and lysosomes participate in EV71-induced FBP2 truncation.
Fig 3.
Involvement of the proteasome, the lysosome, and autophagy in FBP2 turnover. (A) FBP2 in mock-infected (m) or EV71-infected RD cells treated with MG132 (20 μM) or NH4Cl (20 mM) at 0 h postinfection was analyzed. (B) FBP2 in mock-infected (m) or EV71-infected RD cells treated with MG132 (20 μM) or NH4Cl (20 mM) at 6 h postinfection was also analyzed. Viral 3C protein in these cells was used as an indicator for viral replication. (C) RD cells overexpressing FLAG-FBP2 were cotransfected with Myc-ubiquitin. After 48 h posttransfection, cell lysates were either left untreated or treated with MG132 (20 μM) for 2 h. The FLAG-fused proteins in these lysates were immunoprecipitated by an anti-FLAG M2 affinity gel. The ubiquitinated proteins were detected by the anti-Myc antibody. In total, 2.5% of the lysates was used as the input control. (D) FBP2 in mock-infected (m) or EV71-infected (v) RD cells transfected with NC siRNA or ATG5 siRNA at 8 (v 8) and 10 (v 10) h postinfection was analyzed. FBP2, LC3-I (precursor) and LC3-II (autophagy activation marker), ATG5, 3C, and actin were detected by Western blotting. Truncated forms of FBP2 are indicated by asterisks.
EV71 infection induces autophagy (53). Autophagosomes can fuse with lysosomes to form autolysosomes, which can degrade organelles or proteins (54, 55). Here we used an siRNA target for ATG5 to determine whether autophagy is involved in FBP2 truncation. ATG5 is an essential component in assembling autophagosomes. Therefore, we examined FBP2 truncation in infected cells after ATG5 siRNA transfection. RD cells were infected with EV71 after transfection with ATG5 siRNA for 48 h. Cell extracts were collected at 8 and 10 h postinfection and were analyzed using Western blotting. The expression level of microtubule-associated protein light chain 3 II (LC3-II) was used to demonstrate the level of autophagy in the cells. The result shows that LC3-II (autophagic activity) was induced in EV71-infected cells (Fig. 3D, lanes 3 and 7). The level of virus-induced LC3-II decreased in ATG5 siRNA-transfected cells (Fig. 3D, lanes 4 and 8) from that in NC siRNA-transfected cells (Fig. 3D, lanes 3 and 7). The expression level of viral protein 3C in ATG5 siRNA-transfected cell extracts was equivalent to that in NC siRNA-transfected cell extracts, whereas the intact form of FBP2 was more abundant in ATG5 siRNA-transfected cells than in NC siRNA-transfected cells (Fig. 3D, lane 4 versus lane 3; lane 8 versus lane 7). These results show that proteasomal and lysosomal degradation may regulate FBP2 protein turnover.
Virus-induced caspase activity cleaves FBP2.
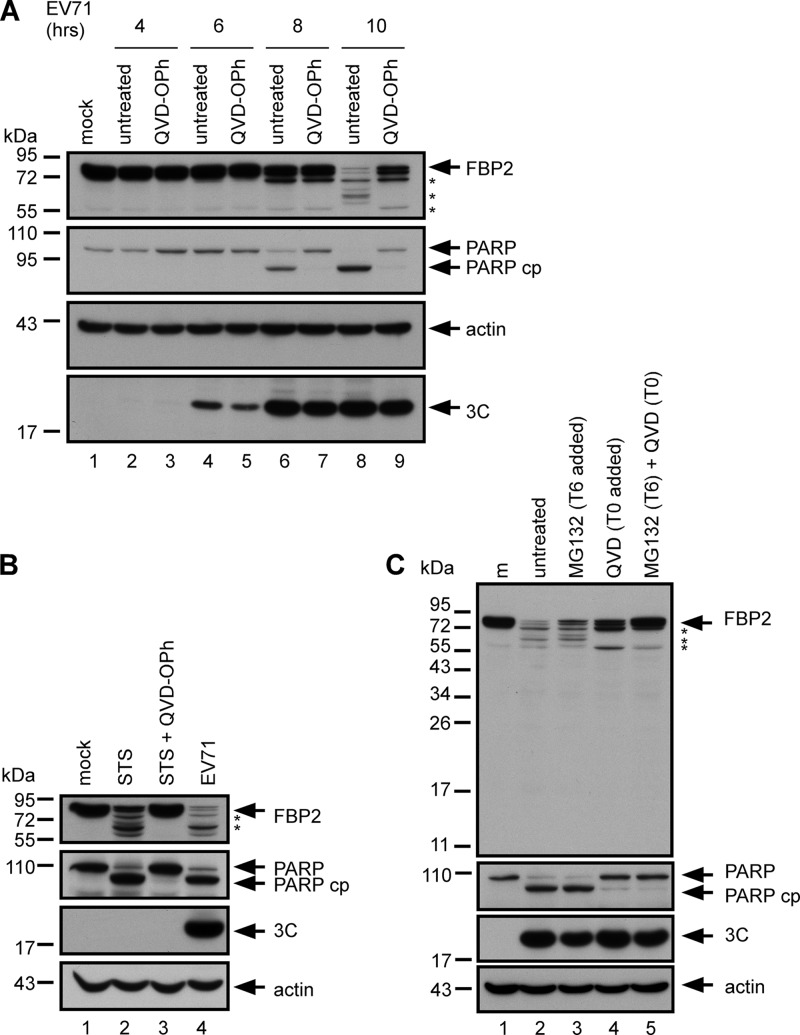
FBP2 is a substrate of caspase (56). EV71 infection induces caspase activation (46, 57). A pan-caspase inhibitor, QVD-OPh, was used to determine whether EV71-induced caspase activity is involved in FBP2 truncation. The results show that PARP, a substrate of caspase 3, was cleaved in EV71-infected cells (Fig. 4A, lanes 6 and 8) and that the cleavage of PARP was inhibited by QVD-OPh treatment (Fig. 4A, lanes 7 and 9). Treatment with QVD-OPh increased the intact form of FBP2 and decreased the cleavage products in infected cells (Fig. 4A, lane 9 versus lane 8). Viral 3C protein expression was not affected by QVD-OPh treatment (Fig. 4A, lane 7 versus lanes 6; lane 9 versus lane 8). Several caspases were activated by EV71-induced apoptosis (57, 58). To determine whether apoptosis-induced caspase activity is involved in FBP2 cleavage, RD cells were treated with the apoptosis inducer STS and the pan-caspase inhibitor QVD-OPh and were subsequently collected and analyzed using Western blotting. The cleavage of PARP confirmed STS-induced and EV71-induced caspase activation (Fig. 4B, lanes 2 and 4). The STS-treated cells exhibited FBP2 cleavage (Fig. 4B, lane 2) and generated cleavage products similar to those in infected cells (Fig. 4B, lane 4). Treating cells with QVD-OPh completely inhibited STS-induced caspase activation and FBP2 cleavage (Fig. 4A, lane 3). Cells treated with caspase (Fig. 4A) or proteasome inhibitors (Fig. 3B) had more intact FBP2 than untreated cells in EV71 infection. However, both inhibitors only partially restored the intact form of FBP2, indicating the possible presence of multiple pathways in EV71-induced FBP2 cleavage. Therefore, cells were treated with QVD-OPh and MG132. The cells treated with both inhibitors had more intact FBP2 than did cells treated with only one inhibitor (Fig. 4C, lane 5 versus lanes 3 and 4). These results showed that multiple factors are involved in EV71-induced FBP2 cleavage, including caspase activation, proteasomes, and autophagy.
Fig 4.
Involvement of caspase activity in FBP2 cleavage. (A) FBP2 in mock-infected (m) or EV71-infected RD cells treated with a pan-caspase inhibitor, QVD-OPh (20 μM), at 4, 6, 8, and 10 h postinfection were analyzed. Truncated forms of FBP2 are indicated by asterisks. Detection of PARP and its cleavage product (PARP cp) were used to demonstrate virus-induced caspase activity. (B) FBP2 in RD cells treated with an apoptosis inducer, STS (2 μM), or treated with STS combined with a pan-caspase inhibitor (STS + QVD-OPh), was analyzed. Truncated forms of FBP2 are indicated by asterisks. (C) RD cells were mock infected (m) or infected with EV71. They were treated either with MG132 alone at 6 h postinfection (T6 added), with QVD-OPh alone at 0 h postinfection (T0 added), or with combined QVD-OPh and MG132. FBP2, PARP, PARP cp, and viral 3C in these cells were analyzed by Western blotting. Truncated forms of FBP2 are indicated by asterisks.
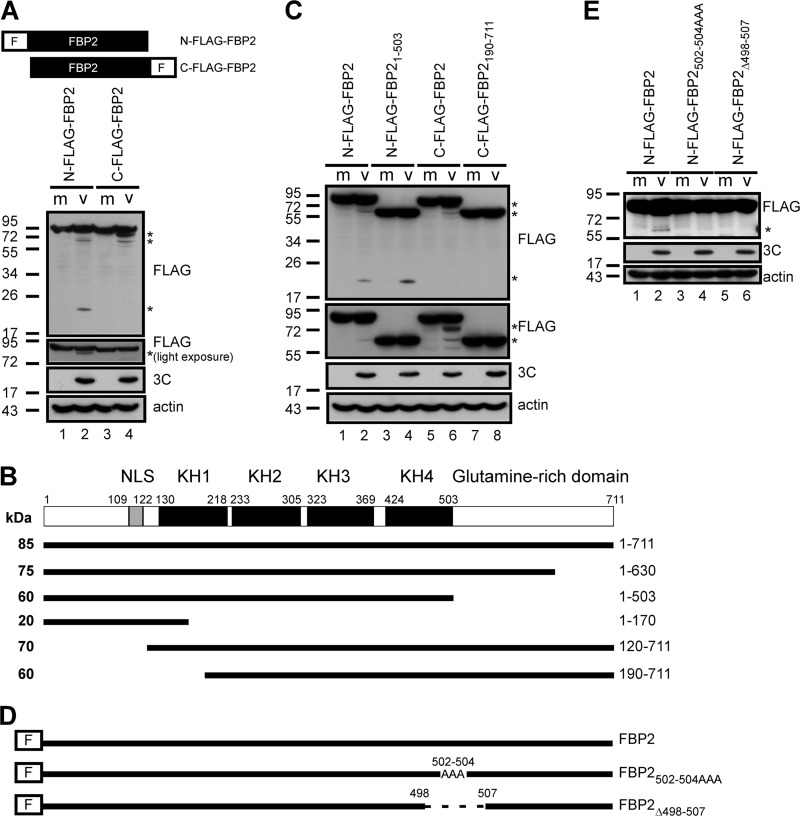
FBP2 is cleaved from both the amino (N) and the carboxyl (C) terminus.
To determine the FBP2 cleavage product that is induced by EV71 infection, plasmids containing N- or C-terminally FLAG tagged FBP2 were transfected into RD cells, and the cells were then infected with EV71. Cell lysates were analyzed using Western blotting with an anti-FLAG antibody. Five cleavage products from N- and C-terminally FLAG tagged FBP2 were generated in infected cells. The N-terminally FLAG tagged FBP2 in EV71-infected cells yielded 3 major cleaved products of approximately 75, 60, and 20 kDa (Fig. 5A, lane 2). However, C-terminally FLAG tagged FBP2 yielded 2 major cleaved products of approximately 70 and 60 kDa (Fig. 5A, lane 4). Therefore, the potential cleavage sites can be predicted based on the sizes of the cleavage products. This was roughly estimated according to the size of the truncated proteins: the size of full-length FBP2 was approximately 85 kDa according to our protein marker. Therefore, we estimated that each kDa of FBP2 contains, on average, approximately 8.4 amino acids. The potential cleavage sites were near the amino acid positions 120, 170, 190, 503, and 630 in FBP2 (Fig. 5B). Figure 1B shows that one of the dominant cleavage products was approximately 60 kDa, a size similar to those of the predicted cleaved products (residues 190 to 711 of FBP2 [FBP2190-711] and residues 1 to 503 of FBP2 [FBP21-503]) (Fig. 5B). The RD cells transfected with plasmids containing FLAG-FBP2190-711 and FLAG-FBP21-503 were infected with EV71 at 48 h posttransfection. Cell lysates were analyzed using Western blotting. The sizes of FBP21-503 and FBP2190-711 were approximately 60 kDa and were similar to that of EV71-cleaved FBP2 (Fig. 5C, lane 3 versus lane 2; lane 7 versus lane 6), indicating that cleaved FBP2 at approximately 60 kDa may correspond to multiple cospecies. We chose the amino acid position 503 for FBP2 to confirm our prediction. Plasmids encoding N-terminally FLAG fused FBP2 with either a mutation of residues VGP at positions 502 to 504 to AAA (N-FLAG-FBP2502-504AAA) or a 10-amino-acid deletion from positions 498 to 507 (N-FLAG-FBP2Δ498-507) were generated to confirm the prediction of the cleavage sites (Fig. 5D). The cells were transfected with plasmids containing N-FLAG-FBP2, N-FLAG-FBP2502-504AAA, or N-FLAG-FBP2Δ498-507 and were infected with EV71. Cell lysates were analyzed using SDS-PAGE and Western blotting with an anti-FLAG antibody. The results showed that cells expressing the N-FLAG-FBP2 plasmid produced a 60-kDa cleavage product after virus infection (Fig. 5E, lane 2), and cells expressing the N-FLAG-FBP2502-504AAA plasmid produced a small amount of the 60-kDa cleavage product (Fig. 5E, lane 4). The 60-kDa cleavage product was not detected in cells that expressed the N-FLAG-FBP2Δ498-507 plasmid after virus infection (Fig. 5E, lane 6). These data suggested that our prediction of the cleavage site on residue 503 of FBP2 correlated with the actual cleavage site.
Fig 5.
FBP2 was cleaved from both the N and the C terminus. (A) Cells were transfected with N-terminally or C-terminally FLAG tagged-FBP2 plasmids. Following transfection, cells were either mock infected (m) or infected with EV71 (v). FLAG-tagged FBP2 in these cells was detected by Western blotting. The second panel shows the light-exposure result of Western blotting. Cleavage products containing the FLAG tag are indicated by asterisks. (B) FBP2 contains a nuclear localization signal (NLS), KH1 to KH4, and a glutamine-rich domain. A schematic diagram of the potential cleavage products of FBP2 (solid lines) induced by EV71 infection is shown. The predicted cleavage products are from amino acids 1 to 630, 1 to 503, 1 to 170, 120 to 711, and 190 to 711 of FBP2. The predicted sizes of these products are also indicated. (C) RD cells overexpressing full-length FLAG-tagged FBP2 (N-FLAG-FBP2 and C-FLAG-FBP2) or truncated FLAG-tagged FBP2 (N-FLAG-FBP21–503 and C-FLAG-FBP2190–711) were mock infected or infected with EV71. The lysate was analyzed with 12% SDS-PAGE (top panel) and with 8% SDS-PAGE (second panel). FLAG-FBP2 or cleavage products in these lysates were detected by Western blotting. Detection of 3C was used to demonstrate viral protein expression. The detection of actin was used as a loading control. (D) Plasmids encoding N-terminally FLAG fused FBP2 with residues VGP at positions 502 to 504 mutated to AAA (N-FLAG-FBP2502-504AAA) or with a 10-amino-acid deletion from position 498 to 507 (FLAG-FBP2Δ498–507) were constructed. (E) RD cells overexpressing wild-type (N-FLAG-FBP2) and mutant (N-FLAG-FBP2502-504AAA and N-FLAG-FBP2Δ498–507) FBP2 were mock infected or infected with EV71. FLAG-FBP2 or its cleavage products in these lysates were detected by Western blotting. Truncated FLAG-FBP2 was detected and is indicated by an asterisk. The detection of viral protein 3C was used as an indicator for virus infection.
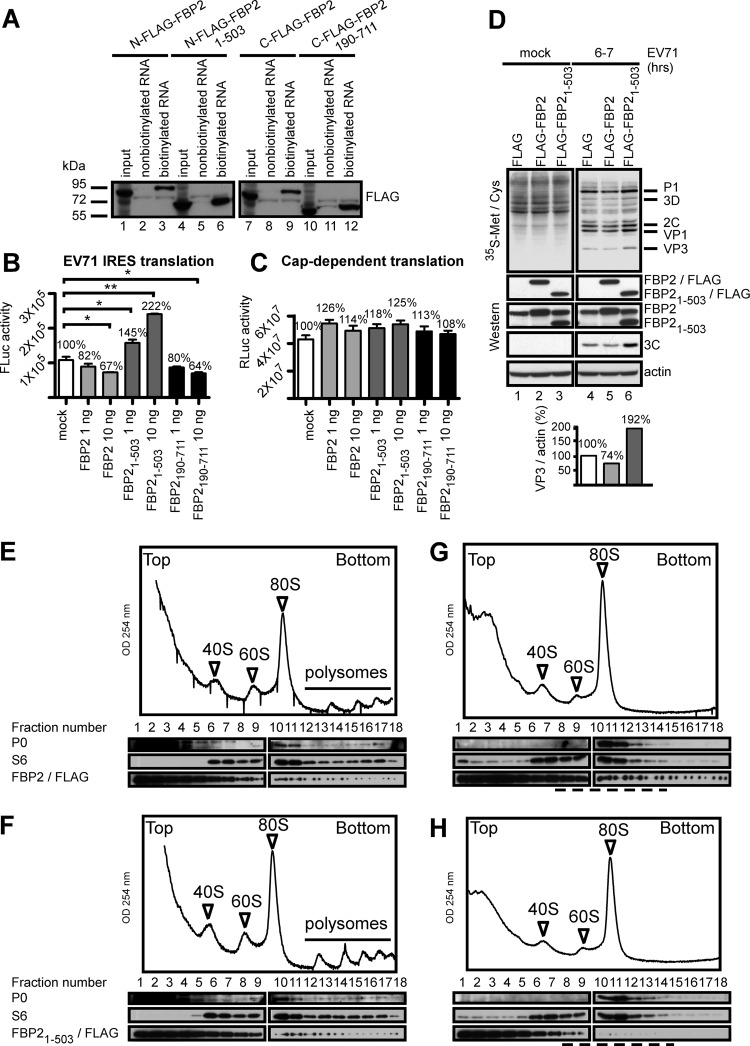
FBP21–503 positively regulated EV71 IRES activity.
The results show that the N- and C-terminal domains of FBP2 may be cleaved in EV71-infected cells (Fig. 5). To determine whether cleavage of the N- or C-terminal domains of FBP2 affected its functions, we overexpressed plasmids encoding FLAG-tagged-FBP2, FBP21–503, and FBP2190–711 in cells and collected the cell lysates. We chose FBP21–503 and FBP2190–711 for further study because the sizes of FBP21–503 and FBP2190–711 are similar to that of primary cleaved FBP2 (60 kDa) in virus-infected cells (Fig. 1B, lane 5). Previous studies have shown that FBP2 binds to the EV71 5′ UTR and negatively regulates IRES activity by competing with other, positive ITAFs, such as PTB (35). We tested the binding activity of cleaved FBP2 using an RNA-protein pulldown assay. Biotinylated EV71 5′ UTR RNA containing EV71 IRES was incubated with lysates transfected with plasmids containing FLAG-FBP2, FLAG-FBP21–503, or FLAG-FBP2190–711. Streptavidin beads were used to pull down the biotinylated EV71 5′ UTR and its associated proteins. The pulled down FLAG-tagged FBP2 proteins were detected using Western blotting with anti-FLAG antibodies. The results showed that FBP2 and the truncated forms FBP21–503 and FBP2190–711 bound to the EV71 5′ UTR (Fig. 6A, lanes 3, 6, 9, and 12). FBP2 is a negative ITAF that inhibits EV71 IRES activity; therefore, we further examined the effects of FBP2 or truncated FBP2 (FBP2190–711 and FBP21–503) recombinant proteins on IRES activity using an in vitro translation assay. Like full-length FBP2, truncated FBP2190–711 decreased IRES activity in vitro, whereas FBP21–503 increased EV71 IRES activity (Fig. 6B). We also performed a similar assay on cap-dependent translation. No significant effect of full-length or truncated FBP2 on cap-dependent translation was found in this assay (Fig. 6C). These data indicated the specificity of FBP2 regulation of IRES activity. To determine the effect of FBP21–503 on viral translation, we performed a [35S]methionine-cysteine labeling assay to detected newly synthesized proteins in EV71-infected cells. In the [35S]methionine-cysteine labeling assay, viral protein synthesis was lower in cells expressing FLAG-FBP2 (Fig. 6D, lane 5) than in cells expressing only a FLAG vector (Fig. 6D, lane 4). Viral protein synthesis was higher in cells expressing FLAG-FBP21–503 (Fig. 6D, lane 6) than in cells expressing only the FLAG vector (Fig. 6D, lane 4). In addition, we used quantification software (Multi Gaugel Fujifilm) to quantify the ratio of viral protein VP3 to actin. The VP3/actin ratio decreased (74%) in cells expressing FLAG-FBP2 relative to that in cells expressing FLAG (100%), and the VP3/actin ratio increased (192%) in cells expressing FLAG-FBP21–503 relative to that in cells expressing FLAG (100%) (Fig. 6D). The contrasting effects of FBP2 and FBP21–503 were also observed in viral 3C protein levels, which were detected using a Western blotting assay. Viral protein 3C expression increased in cells expressing FLAG-FBP21–503 (Fig. 6D, lane 6) and decreased in cells expressing FLAG-FBP2 (Fig. 6D, lane 5) relative to that in cells expressing the FLAG vector (Fig. 6D, lane 4). The levels of accumulation of 3C are consistent with the viral protein labeling results, which also verified that FBP21–503 increases IRES-driven translation. The [35S]methionine-cysteine labeling assay showed equal amounts of cellular protein synthesis in FLAG-, FLAG-FBP2-, and FLAG-FBP21–503-expressing cells (Fig. 6D, lanes 1 to 3), a finding consistent with the results shown in Fig. 6C. To examine the effects of the intact or truncated forms of FBP2 on ribosome profiling, RD cells were transfected with plasmids containing FLAG-FBP2 or FLAG-FBP21–503. The cells were mock infected (Fig. 6E and F) or infected with EV71 (Fig. 6G and H) at 48 h posttransfection. Ribosome profiles were assayed by a sucrose gradient using extracts from EV71-infected cells at 6 h postinfection and were analyzed using a Western blotting assay. Fractions 12 to 18 show polysomes in mock-infected cells (Fig. 6E and F). After virus infection, the amount of polysomes decreased (Fig. 6G and H, fractions 12 to 18), which is consistent with certain published studies (59, 60). The result presents the differing cosedimentation for ribosomal subunit proteins for FBP2 (Fig. 6G) and FBP21–503 (Fig. 6H) in EV71-infected cells. FBP2 appeared in the sediments that contained the small ribosomal subunit protein S6 (Fig. 6G, fractions 8 to 14). However, FBP21–503 occurred only in the parts of sediments that contained S6 (Fig. 6H, fractions 8 to 14). The results suggest that FBP2 and FBP21–503 are associated with various components in the translation mechanisms, which may cause various regulatory effects on EV71 IRES activity. Overall, these results show that FBP21–503 binds to the EV71 5′ UTR and positively regulates IRES activity and viral protein synthesis.
Fig 6.
FBP21–503 positively regulated EV71 IRES activity. (A) Binding assay of full-length and truncated FBP2 on EV71 5′ UTR RNA. Lysates from RD cells overexpressing N-terminally FLAG tagged FBP2 (N-FLAG-FBP2), N-terminally FLAG tagged FBP21–503 (N-FLAG-FBP2 1–503), C-terminally FLAG tagged FBP2 (C-FLAG FBP2), or C-terminally FLAG tagged FBP2190–711 (C-FLAG FBP2 190–711) were collected and incubated with nonbiotinylated or biotinylated EV71 5′ UTR RNA. The protein-RNA complex was pulled down by streptavidin beads and was analyzed by SDS-PAGE. FLAG-tagged-FBP2 was detected by Western blotting. (B) Effects of FBP2 and the truncated FBP2 proteins on EV71 IRES activity in vitro. The in vitro EV71 IRES activity assay was conducted with 1 or 10 μg of full-length FBP2 or a truncated FBP2 (FBP21–503 or FBP2190–711) recombinant protein. EV71 IRES-driven luciferase expression (Fluc activity) is represented by bars (*, P < 0.05; **, P < 0.01; analyzed by the Student t test; n = 3). (C) The in vitro cap-dependent translation assay was conducted with 1 or 10 μg of full-length FBP2 or with a truncated FBP2 (FBP21–503 or FBP2190–711) recombinant protein. Cap-dependent Renilla luciferase expression (RLuc activity) is represented by bars. Error bars represent standard deviations (n = 3). (D) Effect of full-length or truncated FBP2 (FBP21–503) on viral protein synthesis. RD cells were transfected with plasmids containing FLAG, FLAG-FBP2, and FLAG-FBP21–503. Following transfection, RD cells were mock infected or infected with EV71. Protein synthesis in these cells was examined using the [35S]methionine-cysteine labeling assay at 6 to 7 h postinfection (within 1 h). Viral proteins, including P1, 3D, 2C, VP1, and VP3, are indicated according to protein size. The levels of 35S-labeled viral VP3 in the cells of each group were also quantified and normalized by the levels of actin detected by Western blotting (VP3/actin). The normalized VP3 levels are shown in the graph. The levels of viral 3C protein and FBP2 expression level were also detected by Western blotting. (E to H) Extracts were generated from mock-infected RD cells that overexpressed plasmids containing FLAG-FBP2 (E) or FLAG-FBP21–503 (F). Extracts were generated from EV71-infected RD cells that overexpressed plasmids containing FLAG-FBP2 (G) or FLAG-FBP21–503 (H). The extracts were sedimented through 7%-to-47% sucrose gradients and were fractionated. The fractions were assayed by Western blotting to detect polysome proteins P0, P6, and FLAG-FBP2. The broken lines indicate the differing cosedimentation with ribosomal subunit proteins for FBP2 and FBP21–503 in EV71-infected cells.
DISCUSSION
This study shows that EV71 infection induces FBP2 cleavage, which is associated with caspase, proteasome, and autophagic activities. The cleavage of FBP2 reduces the level of a negative regulator of viruses. In addition, this study also demonstrated that a truncated form of FBP2 without a C-terminal domain changed its role from that of a negative regulator to that of a positive regulator (Fig. 6). This datum may help further our understanding of the molecular functions of FBP2, which is molecularly significant for translation (35, 61), RNA stability (41, 42), and miRNA biogenesis (43–45).
The results show that treatment with the protease inhibitor MG132 and the pan-caspase inhibitor QVD-OPh can decrease FBP2 cleavage in infected cells (Fig. 3 and 4). These results are consistent with those of Racaniello and colleagues; that is, the cleavage of MDA-5 in poliovirus-infected cells is caspase and proteasome dependent (15). The cleavage patterns of FBP2 differ in MG132- and QVD-OPh-treated cells (Fig. 4C). MG132 treatment protects certain types of FBP2 from cleavage. However, cells treated with the caspase inhibitor QVD-OPh have only 2 detectable cleaved forms of FBP2 (Fig. 4C, lane 4), fewer than the number in untreated cells (lane 2) and MG132-treated cells (lane 3), indicating the role of caspases in virus-induced FBP2 cleavage. In addition to the proteasomes and caspases involved in FBP2 cleavage, we show that the mechanisms of virus-induced autophagy and lysosomes are also involved in FBP2 cleavage (Fig. 3D). Cleavage of FBP2 may be a by-product of all these cellular activities, which are induced by EV71 infection, and which can then provide an advantage to viruses.
We tagged FLAG on the N and C termini of FBP2 to examine the possible cleavage products during EV71 infection based on the size of the cleavage products (Fig. 5). Detecting cleavage products by tagging the FLAG peptide on the N or C terminus may cause us to lose the ability to detect a number of potential candidates, which lose both the N and C termini. In addition, we used mutant or deleted FBP2 to further validate the cleavage sites. FBP2Δ498–507 did not generate 60-kDa cleavage products after virus infection (Fig. 5E), indicating that residue 503 is correlated to the actual cleavage site.
FBP21–503, which loses the C-terminal glutamine-rich domain, can positively regulate viral translation. FBP2190–711, another potential cleavage product, remains at the C-terminal domain and, as an intact form of FBP2, is a negative ITAF. These results indicate the importance of the C-terminal domain of FBP2 for its negative regulation activity on EV71 IRES. The C-terminal glutamine-rich domain of FBP2 contains 4 degenerate copies involved in protein-protein interaction (62–64). It is crucial to examine the FBP2 interacting proteins through the C-terminal domain in order to further understand the manner in which FBP2 negatively regulates the EV71 IRES.
In conclusion, EV71 infection induces the cleavage of FBP2, a negative ITAF, through multiple mechanisms, including caspase activation, proteasome activity, and autophagy. One of the cleavage products, FBP21–503, becomes a positive regulator, which may provide insight into how a cellular protein fine-tunes viral translation. The information obtained may also help us to further understand the function of FBP2 and the manner in which FBP2 changes its function when it loses the C-terminal domain.
ACKNOWLEDGMENTS
We thank Douglas L. Black for providing the pEGFPC1-6×His-FLKSRP plasmid and the anti-KSRP monoclonal antibody Ab5. We thank Michael M. C. Lai for providing the pUI-myc-ubiquitin plasmid. We also thank Kai-Yin Lo for technical support with ribosome profiling.
This study was supported by The National Science Council of the Republic of China, Taiwan (NSC-101-2325-B-182-015) and by Chang Gung Memorial Hospital (CMRPD1A0671).
Footnotes
Published ahead of print 23 January 2013
REFERENCES
- 1. AbuBakar S, Chee HY, Al-Kobaisi MF, Xiaoshan J, Chua KB, Lam SK. 1999. Identification of enterovirus 71 isolates from an outbreak of hand, foot and mouth disease (HFMD) with fatal cases of encephalomyelitis in Malaysia. Virus Res. 61:1–9 [DOI] [PubMed] [Google Scholar]
- 2. Alexander JP, Jr, Baden L, Pallansch MA, Anderson LJ. 1994. Enterovirus 71 infections and neurologic disease—United States, 1977–1991. J. Infect. Dis. 169:905–908 [DOI] [PubMed] [Google Scholar]
- 3. Brown BA, Pallansch MA. 1995. Complete nucleotide sequence of enterovirus 71 is distinct from poliovirus. Virus Res. 39:195–205 [DOI] [PubMed] [Google Scholar]
- 4. Fan X, Jiang J, Liu Y, Huang X, Wang P, Liu L, Wang J, Chen W, Wu W, Xu B. 2013. Detection of human enterovirus 71 and Coxsackievirus A16 in an outbreak of hand, foot, and mouth disease in Henan Province, China in 2009. Virus Genes 46:1–9 [DOI] [PubMed] [Google Scholar]
- 5. Gilbert GL, Dickson KE, Waters MJ, Kennett ML, Land SA, Sneddon M. 1988. Outbreak of enterovirus 71 infection in Victoria, Australia, with a high incidence of neurologic involvement. Pediatr. Infect. Dis. J. 7:484–488 [DOI] [PubMed] [Google Scholar]
- 6. Ho M, Chen ER, Hsu KH, Twu SJ, Chen KT, Tsai SF, Wang JR, Shih SR. 1999. An epidemic of enterovirus 71 infection in Taiwan. Taiwan Enterovirus Epidemic Working Group. N. Engl. J. Med. 341:929–935 [DOI] [PubMed] [Google Scholar]
- 7. Lee HS, Park LC, Lee EM, Lee SJ, Shin SH, Im H, Do KM, Kim EJ, Ye BJ, Song MK, Kim SH, Lee SM, Lee WS, Kim YS. 2012. Incidence rates and risk factors for vascular events in patients with essential thrombocythemia: a multicenter study from Korea. Clin. Lymphoma Myeloma Leuk. 12:70–75 [DOI] [PubMed] [Google Scholar]
- 8. McMinn P, Lindsay K, Perera D, Chan HM, Chan KP, Cardosa MJ. 2001. Phylogenetic analysis of enterovirus 71 strains isolated during linked epidemics in Malaysia, Singapore, and Western Australia. J. Virol. 75:7732–7738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McMinn PC. 2002. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol. Rev. 26:91–107 [DOI] [PubMed] [Google Scholar]
- 10. Weng KF, Chen LL, Huang PN, Shih SR. 2010. Neural pathogenesis of enterovirus 71 infection. Microbes Infect. 12:505–510 [DOI] [PubMed] [Google Scholar]
- 11. Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW. 1982. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 257:14806–14810 [PubMed] [Google Scholar]
- 12. Rose JK, Trachsel H, Leong K, Baltimore D. 1978. Inhibition of translation by poliovirus: inactivation of a specific initiation factor. Proc. Natl. Acad. Sci. U. S. A. 75:2732–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clark ME, Hammerle T, Wimmer E, Dasgupta A. 1991. Poliovirus proteinase 3C converts an active form of transcription factor IIIC to an inactive form: a mechanism for inhibition of host cell polymerase III transcription by poliovirus. EMBO J. 10:2941–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clark ME, Lieberman PM, Berk AJ, Dasgupta A. 1993. Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Mol. Cell. Biol. 13:1232–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barral PM, Morrison JM, Drahos J, Gupta P, Sarkar D, Fisher PB, Racaniello VR. 2007. MDA-5 is cleaved in poliovirus-infected cells. J. Virol. 81:3677–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barral PM, Sarkar D, Fisher PB, Racaniello VR. 2009. RIG-I is cleaved during picornavirus infection. Virology 391:171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Drahos J, Racaniello VR. 2009. Cleavage of IPS-1 in cells infected with human rhinovirus. J. Virol. 83:11581–11587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haghighat A, Svitkin Y, Novoa I, Kuechler E, Skern T, Sonenberg N. 1996. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J. Virol. 70:8444–8450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krausslich HG, Nicklin MJ, Toyoda H, Etchison D, Wimmer E. 1987. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. J. Virol. 61:2711–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thompson SR, Sarnow P. 2003. Enterovirus 71 contains a type I IRES element that functions when eukaryotic initiation factor eIF4G is cleaved. Virology 315:259–266 [DOI] [PubMed] [Google Scholar]
- 21. Joachims M, Van Breugel PC, Lloyd RE. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 73:718–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kerekatte V, Keiper BD, Badorff C, Cai A, Knowlton KU, Rhoads RE. 1999. Cleavage of poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J. Virol. 73:709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park N, Skern T, Gustin KE. 2010. Specific cleavage of the nuclear pore complex protein Nup62 by a viral protease. J. Biol. Chem. 285:28796–28805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuyumcu-Martinez NM, Joachims M, Lloyd RE. 2002. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. J. Virol. 76:2062–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Back SH, Kim YK, Kim WJ, Cho S, Oh HR, Kim JE, Jang SK. 2002. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3Cpro. J. Virol. 76:2529–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weng KF, Li ML, Hung CT, Shih SR. 2009. Enterovirus 71 3C protease cleaves a novel target CstF-64 and inhibits cellular polyadenylation. PLoS Pathog. 5:e1000593 doi:10.1371/journal.ppat.1000593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lei X, Sun Z, Liu X, Jin Q, He B, Wang J. 2011. Cleavage of the adaptor protein TRIF by enterovirus 71 3C inhibits antiviral responses mediated by Toll-like receptor 3. J. Virol. 85:8811–8818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lawrence P, Schafer EA, Rieder E. 2012. The nuclear protein Sam68 is cleaved by the FMDV 3C protease redistributing Sam68 to the cytoplasm during FMDV infection of host cells. Virology 425:40–52 [DOI] [PubMed] [Google Scholar]
- 29. Hellen CU, Witherell GW, Schmid M, Shin SH, Pestova TV, Gil A, Wimmer E. 1993. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. U. S. A. 90:7642–7646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Verma B, Bhattacharyya S, Das S. 2010. Polypyrimidine tract-binding protein interacts with coxsackievirus B3 RNA and influences its translation. J. Gen. Virol. 91:1245–1255 [DOI] [PubMed] [Google Scholar]
- 31. Blyn LB, Towner JS, Semler BL, Ehrenfeld E. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71:6243–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Costa-Mattioli M, Svitkin Y, Sonenberg N. 2004. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 24:6861–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boussadia O, Niepmann M, Creancier L, Prats AC, Dautry F, Jacquemin-Sablon H. 2003. Unr is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J. Virol. 77:3353–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang PN, Lin JY, Locker N, Kung YA, Hung CT, Huang HI, Li ML, Shih SR. 2011. Far upstream element binding protein 1 binds the internal ribosomal entry site of enterovirus 71 and enhances viral translation and viral growth. Nucleic Acids Res. 39:9633–9648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin JY, Li ML, Shih SR. 2009. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res. 37:47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shih SR, Stollar V, Li ML. 2011. Host factors in enterovirus 71 replication. J. Virol. 85:9658–9666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Davis-Smyth T, Duncan RC, Zheng T, Michelotti G, Levens D. 1996. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J. Biol. Chem. 271:31679–31687 [DOI] [PubMed] [Google Scholar]
- 38. Hall MP, Huang S, Black DL. 2004. Differentiation-induced colocalization of the KH-type splicing regulatory protein with polypyrimidine tract binding protein and the c-src pre-mRNA. Mol. Biol. Cell 15:774–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Min H, Turck CW, Nikolic JM, Black DL. 1997. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 11:1023–1036 [DOI] [PubMed] [Google Scholar]
- 40. Lellek H, Kirsten R, Diehl I, Apostel F, Buck F, Greeve J. 2000. Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J. Biol. Chem. 275:19848–19856 [DOI] [PubMed] [Google Scholar]
- 41. Chou CF, Mulky A, Maitra S, Lin WJ, Gherzi R, Kappes J, Chen CY. 2006. Tethering KSRP, a decay-promoting AU-rich element-binding protein, to mRNAs elicits mRNA decay. Mol. Cell. Biol. 26:3695–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. 2004. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 14:571–583 [DOI] [PubMed] [Google Scholar]
- 43. Briata P, Chen CY, Giovarelli M, Pasero M, Trabucchi M, Ramos A, Gherzi R. 2011. KSRP, many functions for a single protein. Front. Biosci. 16:1787–1796 [DOI] [PubMed] [Google Scholar]
- 44. Gherzi R, Chen CY, Trabucchi M, Ramos A, Briata P. 2010. The role of KSRP in mRNA decay and microRNA precursor maturation. Wiley Interdiscip. Rev. RNA 1:230–239 [DOI] [PubMed] [Google Scholar]
- 45. Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. 2009. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459:1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shih SR, Weng KF, Stollar V, Li ML. 2008. Viral protein synthesis is required for enterovirus 71 to induce apoptosis in human glioblastoma cells. J. Neurovirol. 14:53–61 [DOI] [PubMed] [Google Scholar]
- 47. Shih SR, Chiang C, Chen TC, Wu CN, Hsu JT, Lee JC, Hwang MJ, Li ML, Chen GW, Ho MS. 2004. Mutations at KFRDI and VGK domains of enterovirus 71 3C protease affect its RNA binding and proteolytic activities. J. Biomed. Sci. 11:239–248 [DOI] [PubMed] [Google Scholar]
- 48. Liao TL, Wu CY, Su WC, Jeng KS, Lai MM. 2010. Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication. EMBO J. 29:3879–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fliss PM, Jowers TP, Brinkmann MM, Holstermann B, Mack C, Dickinson P, Hohenberg H, Ghazal P, Brune W. 2012. Viral mediated redirection of NEMO/IKKγ to autophagosomes curtails the inflammatory cascade. PLoS Pathog. 8:e1002517 doi:10.1371/journal.ppat.1002517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shackelford J, Pagano JS. 2005. Targeting of host-cell ubiquitin pathways by viruses. Essays Biochem. 41:139–156 [DOI] [PubMed] [Google Scholar]
- 51. Thomas M, Pim D, Banks L. 1999. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene 18:7690–7700 [DOI] [PubMed] [Google Scholar]
- 52. Ciechanover A. 2005. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 6:79–87 [DOI] [PubMed] [Google Scholar]
- 53. Huang SC, Chang CL, Wang PS, Tsai Y, Liu HS. 2009. Enterovirus 71-induced autophagy detected in vitro and in vivo promotes viral replication. J. Med. Virol. 81:1241–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Klionsky DJ. 2007. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8:931–937 [DOI] [PubMed] [Google Scholar]
- 55. Klionsky DJ, Emr SD. 2000. Autophagy as a regulated pathway of cellular degradation. Science 290:1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Seok H, Cho J, Cheon M, Park IS. 2002. Biochemical characterization of apoptotic cleavage of KH-type splicing regulatory protein (KSRP)/far upstream element-binding protein 2 (FBP2). Protein Pept. Lett. 9:511–519 [DOI] [PubMed] [Google Scholar]
- 57. Chang SC, Lin JY, Lo LY, Li ML, Shih SR. 2004. Diverse apoptotic pathways in enterovirus 71-infected cells. J. Neurovirol. 10:338–349 [DOI] [PubMed] [Google Scholar]
- 58. Sun ZM, Xiao Y, Ren LL, Lei XB, Wang JW. 2011. Enterovirus 71 induces apoptosis in a Bax dependent manner. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 25:49–52 (In Chinese.) [PubMed] [Google Scholar]
- 59. Fitzgerald KD, Semler BL. 2011. Re-localization of cellular protein SRp20 during poliovirus infection: bridging a viral IRES to the host cell translation apparatus. PLoS Pathog. 7:e1002127 doi:10.1371/journal.ppat.1002127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Johannes G, Sarnow P. 1998. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA 4:1500–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dhamija S, Kuehne N, Winzen R, Doerrie A, Dittrich-Breiholz O, Thakur BK, Kracht M, Holtmann H. 2011. Interleukin-1 activates synthesis of interleukin-6 by interfering with a KH-type splicing regulatory protein (KSRP)-dependent translational silencing mechanism. J. Biol. Chem. 286:33279–33288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Duncan R, Collins I, Tomonaga T, Zhang T, Levens D. 1996. A unique transactivation sequence motif is found in the carboxyl-terminal domain of the single-strand-binding protein FBP. Mol. Cell. Biol. 16:2274–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Labourier E, Adams MD, Rio DC. 2001. Modulation of P-element pre-mRNA splicing by a direct interaction between PSI and U1 snRNP 70K protein. Mol. Cell 8:363–373 [DOI] [PubMed] [Google Scholar]
- 64. Siebel CW, Admon A, Rio DC. 1995. Soma-specific expression and cloning of PSI, a negative regulator of P element pre-mRNA splicing. Genes Dev. 9:269–283 [DOI] [PubMed] [Google Scholar]