Abstract
Cytotoxic-T-lymphocyte (CTL) escape mutations undermine the durability of effective human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T cell responses. The rate of CTL escape from a given response is largely governed by the net of all escape-associated viral fitness costs and benefits. The observation that CTL escape mutations can carry an associated fitness cost in terms of reduced virus replication capacity (RC) suggests a fitness cost-benefit trade-off that could delay CTL escape and thereby prolong CD8 response effectiveness. However, our understanding of this potential fitness trade-off is limited by the small number of CTL escape mutations for which a fitness cost has been quantified. Here, we quantified the fitness cost of the 29 most common HIV-1B Gag CTL escape mutations using an in vitro RC assay. The majority (20/29) of mutations reduced RC by more than the benchmark M184V antiretroviral drug resistance mutation, with impacts ranging from 8% to 69%. Notably, the reduction in RC was significantly greater for CTL escape mutations associated with protective HLA class I alleles than for those associated with nonprotective alleles. To speed the future evaluation of CTL escape costs, we also developed an in silico approach for inferring the relative impact of a mutation on RC based on its computed impact on protein thermodynamic stability. These data illustrate that the magnitude of CTL escape-associated fitness costs, and thus the barrier to CTL escape, varies widely even in the conserved Gag proteins and suggest that differential escape costs may contribute to the relative efficacy of CD8 responses.
INTRODUCTION
Cytotoxic-CD8+-T-lymphocyte (CTL) responses play an important role in the control of viremia during human immunodeficiency virus type 1 (HIV-1) infection (1–6), modulating important predictors of disease progression such as set-point viral load and the rate of CD4 T cell loss (7–9). Although in most HIV-1 infections the CTL response is only partially effective, the association of particular “protective” HLA class I alleles with sustained control of viremia (10) and delayed progression to AIDS (11–13) supports the potential of this arm of the immune response to potently suppress HIV-1. Moreover, recent studies in the rhesus macaque model demonstrate that vaccine-induced effector memory CD8+ T cells may be capable of conferring resistance to infection following mucosal exposure to pathogenic simian immunodeficiency virus (SIV) SIVmac239 (14, 15). Therefore, the induction of robust HIV-1-specific CTL responses remains a central goal of the effort to develop an effective vaccine against HIV-1 (16). However, the capacity of HIV-1 to “escape” from the suppression effected by some CTL responses (17–21), including those induced by vaccination in both human and macaque studies (22, 23), highlights the need to consider the potential for such CTL escape when designing an optimal HIV-1 vaccine immunogen.
HIV-1 escapes from specific CTL responses as the result of the expression of amino acid mutations that disrupt the normal processing (24, 25), presentation (26–28), or recognition (29–31) of the targeted CTL epitopes. The dynamics and kinetics of CTL escape are in large part governed by the impact of an escape mutation on virus relative fitness, a composite parameter comprising the net of all mutation-associated fitness costs and benefits. Interestingly, although virus expression of a CTL escape mutation confers a clear fitness benefit by facilitating evasion of CTL-mediated killing of infected cells, the same escape mutation can also carry a concomitant fitness cost as evidenced by reversion of CTL escape mutations following transmission to individuals that do not mount the selecting CTL responses (28, 32) and by CTL escape mutation-associated reductions in virus replication capacity (RC) (32–38). The quantification of such CTL escape-associated fitness costs is integral to determining the differential susceptibility of CTL responses to escape and by extension their potential to effectively suppress HIV-1. To date, however, the relatively small number of escape mutations for which RC has been quantified, the focus on CTL escape mutations associated with protective HLA class I alleles, e.g., HLA-B*27 and HLA-B*57, and the assay-to-assay variation which precludes quantitative comparison of RC results across studies all combine to limit our understanding of the generality of escape-associated fitness costs and their contribution to differential CTL effectiveness.
Accumulating evidence indicates that CTL targeting of the HIV-1 Gag protein is central to effective CTL-mediated control of HIV-1, including the immunodominant targeting of CD8+ T cell epitopes in Gag during the acute phase of infection (39). Indeed, the most protective HLA class I alleles, e.g., HLA-B*27 and HLA-B*57, are associated with early CTL targeting of Gag (40–42) and the consistent selection of CTL escape mutations in Gag (43). The structural HIV-1 Gag proteins are among the most conserved in the virus proteome (44), suggesting substantial functional constraints on amino acid substitution. Indeed, the mature HIV-1 capsid is composed of approximately 1,500 copies of Gag p24, multimerized as both hexamers and pentamers, which assemble into a highly coordinated conical fullerene lattice (45, 46). The implication is that the higher relative functional constraint on the Gag proteins imposes a higher fitness cost to CTL escape and thereby increases the relative effectiveness of Gag-specific CTL responses by reducing their susceptibility to loss from viral escape and by attenuating virus replication if escape does occur. However, the relative susceptibility of CTL responses to escape, both in Gag and across the rest of the proteome, remains unclear.
Here, we characterized the fitness cost of 29 commonly selected HIV-1 Gag CTL escape mutations (47, 48) by quantifying their impact on in vitro virus replication capacity (RC). We observed a broad range of clinically significant reductions in RC, with the mutations associated with protective HLA class I alleles carrying the largest escape-associated fitness costs. Although we observed no relationship between the impact of a mutation on replication capacity and the degree of genetic conservation (Shannon entropy) at the variant position, we found that escape-associated reductions in replication capacity were correlated with the computationally predicted impact of the escape mutation on protein structural stability. These data reveal that the fitness cost of CTL escape varies widely, even within the relatively conserved Gag proteins, and provide important insight into the relationship between the magnitude of the fitness cost of CTL escape and the protective value of particular CD8+ T cell responses. Additionally, the implementation of a computational approach for predicting high-cost CTL escape mutations will help to identify attractive CTL escape-refractory targets, which will facilitate construction of an HIV-1 immunogen that induces maximally effective and durable immune responses against HIV-1.
MATERIALS AND METHODS
HIV-1B recombinant plasmid clones.
The pNL4-3 HIV-1 subtype B (HIV-1B) infectious molecular clone (NIH AIDS Research and Reference Reagent Program) served as the basis for the recombinant viruses used in this study. A pNL4-3 gag subclone was constructed for use in mutagenesis reactions by restriction digest of pNL4-3 with SacI and SbfI followed by ligation of the resultant 2.3-kb fragment encompassing NL4-3 gag into the pUC19 vector (New England BioLabs). A pNL4-3 pol subclone was constructed for use in mutagenesis reactions by restriction digest of pNL4-3 with SbfI and EcoRI followed by ligation of the 2.9-kb fragment into the pUC19 vector. Subclone and full-length plasmids were propagated in TOP10 (Invitrogen) or STBLII (Invitrogen) chemically competent cells, respectively, per the manufacturer's protocol and were purified using Qiagen Miniprep and Maxiprep kits per the manufacturer's protocol.
Mutations of interest were generated by GeneTailor (Invitrogen) site-directed mutagenesis per the manufacturer's protocol. Double mutants were produced by successive rounds of mutagenesis. Mutagenesis was confirmed by sequencing of the entire subclone fragment both before and after cloning to produce the full-length mutant molecular clone. Prior to construction of the escape variant viruses, the native pNL4-3 was modified to restore HIV-1B consensus amino acid residues at two positions in Gag (Q28K and D93E). This modified pNL4-3 (pNL4-3**) served as the parental backbone for all variant viruses in the panel and is thus considered the “wild-type” virus in the analyses performed in this study. In vitro replication of NL4-3 and that of NL4-3** were equivalent (data not shown).
HIV-1B recombinant virus stocks.
Infectious virus stocks were produced by transfection of HEK293T cells (ATCC) with plasmid DNA for full-length infectious molecular clones using Polyfect transfection reagent (Qiagen) according to a modified manufacturer's protocol. Briefly, at 1 day prior to transfection, 2.8 × 106 HEK293 cells were seeded in a T75 flask in Dulbecco modified Eagle medium without glutamine (DMEM) (Sigma) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS) (Sigma), 1% (vol/vol) penicillin-streptomycin (Gibco), and glutamine (Gibco) and incubated overnight at 37°C with 5% CO2. For the transfection, 15 μg of plasmid DNA purified by Qiagen Maxiprep (Qiagen), at a minimum concentration of 1 μg/μl, was diluted to a 150-μl final volume in DMEM (without supplements), 115 μl of Polyfect reagent was added, and the solution was mixed by gentle pipetting and incubated for 10 min at room temperature. During the incubation, medium was removed from the cells to be transfected, they were washed once in cold phosphate-buffered saline (PBS), and 7.0 ml of fresh medium was added. Following the incubation, the transfection mixture was transferred to the cells, swirled gently to mix, and incubated at 37°C with 5% CO2. After 3 h, the medium was removed and discarded, the cells were washed once with PBS to remove excess plasmid DNA, and 7 ml of fresh medium was added before returning the cells to the incubator. Transfection supernatant was harvested after 72 h, filtered through a 0.45-μm filter, and stored in aliquots at −80°C.
Virus stocks were first screened by p24 enzyme-linked immunosorbent assay (ELISA) (Perkin-Elmer) per the manufacturer's protocol; stocks that contained less than 10 ng of p24 per ml were discarded. Next, the gag and pol sequences of the stock virus were confirmed. Finally, the endpoint virus titer of the stock was determined by 50% tissue culture infective dose (TCID50) per a standard protocol (NIH-NIAID-DAIDS). Briefly, CD8+ T cell-depleted peripheral blood mononuclear cells (CD8-depleted PBMC) were isolated from buffy coats from anonymous HIV-negative donors (Partners IRB protocol 2005-P001218) using CD8-specific RosetteSep reagent (Stem Cell Technologies) per the manufacturer's protocol. The CD8-depleted PBMC were washed once in cold 1× phosphate-buffered saline (PBS) supplemented with 1% (vol/vol) FBS, resuspended in freeze medium consisting of RPMI (Invitrogen) supplemented with 20% (vol/vol) FBS and 10% (vol/vol) dimethyl sulfoxide (DMSO), and frozen in aliquots overnight at −80°C prior to long-term storage in LiN2. For the TCID50 assay, CD8-depleted PBMC were thawed by gentle incubation in a 37°C water bath, washed three times in 50 ml PBS supplemented with 1% FBS, plated into 24 wells of a 96-well plate at 1 × 105 cells per well, and stimulated for 3 days in RPMI supplemented with 20% (vol/vol) FBS, 1% (vol/vol) penicillin-streptomycin, 5 μg/ml phytohemagglutinin (PHA) (Sigma-Aldrich), and 20 U/ml recombinant human interleukin-2 (rhIL-2) (Roche Applied Sciences, Indianapolis, IN) at 37°C with 5% CO2. After 3 days, the medium was removed and replaced with RPMI supplemented with 20% FBS, 1% penicillin-streptomycin, and 20 U/ml rhIL-2 and the mixture was inoculated in triplicate with 4-fold serial dilutions of stock virus ranging from 4−6 to 4−13. On day 4 postinfection (p.i.), 100 μl of medium was removed and replaced. On day 7 p.i., wells were scored for infection by p24 ELISA per the manufacturer protocol's and TCID50 was calculated by the Spearman-Karber method.
RC.
For each infection, 1 × 105 CD8-depleted PBMC were seeded per well of a 96-well plate and were stimulated for 72 h in RPMI supplemented with 20% FBS, 1% penicillin-streptomycin, 5 μg/ml PHA, and 20 U/ml rhIL-2 at 37C with 5% CO2. After 72 h, the stimulation medium was gently removed, the cells in each well were inoculated in at least triplicate for each virus at a multiplicity of infection (MOI) of 0.001, and the final volume in the well was brought to 200 μl with RPMI supplemented with 20% FBS, 1% penicillin-streptomycin, and 20 U/ml rhIL-2. RC was quantified for each mutation in at least two independent infection assays using CD8-depleted PBMC from at least two different donors. Each independent infection assay included triplicate wells inoculated at an MOI of 0.001 with the parental (NL4-3**) virus to control for nonspecific differences in HIV replication from infection to infection. Following overnight incubation, the inoculum was carefully removed and replaced with 200 μl of fresh RPMI supplemented with 20% FBS, 1% penicillin-streptomycin, and 20 U/ml rhIL-2. The following day (day 2 postinfection), and every day thereafter for 5 days (up to day 7 postinfection), 50 μl of supernatant was sampled with replacement and stored at −80°C.
Viral RNA (vRNA) was isolated from each 50-μl supernatant sample using the QiaXtractor (Qiagen) automated extraction platform per the manufacturer's protocol with the exception of the off-deck addition of VXL buffer to infectious supernatant and the addition of a 30-min, room-temperature, on-column DNase (Qiagen) treatment sandwiched between two VXW buffer wash steps. Viral RNA was eluted with 70 μl of VXE buffer and stored at −80°C prior to vRNA copy number quantification by quantitative reverse transcription-PCR (qRT-PCR) on a Roche LC480 system using the QuantiFAST SYBR green qRT-PCR kit (Qiagen) per the manufacturer's protocol using HIV-1 gag SK145 (AGTGGGGGGACATCAAGCAGCCATGCAAAT) and SK431 (TGCTATGTCACTTCCCCTTGGTTCTCT) primers.
Replication capacity (copies of vRNA per day) was derived from the slope of the log-linear phase of replication in each infected well. The log-linear phase was determined by plotting the viral RNA copy number versus time on a log scale and selecting the minimum number of earliest data points that could support a linear regression line with an r2 value of ≥0.98. The impact of an escape mutation on replication capacity (RC) was calculated by dividing the log-linear slope of replication of the escape virus (virus expressing the escape variant amino acid) by the log-linear slope of replication of the susceptible virus (virus expressing the susceptible amino acid). In most cases, this consisted of dividing the log-linear slope of the mutant virus construct by that of the parental NL4-3** virus. However, for three mutations (Gag amino acid positions 81, 260, and 312), the escape variant amino acid was expressed by the parental NL4-3** virus and the mutant construct expressing the susceptible amino acid was generated by PCR mutagenesis as described above. As a result, the log-linear slope of NL4-3** (the escape virus) comprised the numerator and that of the mutant construct (the susceptible virus) comprised the denominator. For each mutant, there were multiple independent infection assays comprising multiple infected wells. The RC in each replicate well in a given infection assay was calculated by normalization of the log-linear slope in that well to the mean log-linear slope from all replicate wells infected with the parental (NL4-3**) virus in that same infection assay. This normalization to parental (NL4-3**) RC data generated within each independent infection assay controlled for nonspecific variation in virus replication from assay to assay, e.g., donor-specific target cell differences, tissue culture conditions, etc. The RC for each independent infection assay was calculated as the average of the replicate RC values generated in that assay. This addressed the nonindependence of the RC data generated from each replicate well in a given infection assay. The final reported RC values reflect the mean and 95% confidence interval (CI) of the average of the RC values derived from each independent infection assay. For each mutation, the null hypothesis that RCescape = RCsusceptible was evaluated by calculation of the 95% CI of the mean RC; nonoverlap of the 95% CI with RCsusceptible (equal to 1.0 by design) resulted in rejection of the null hypothesis at P < 0.05. The null hypothesis was also evaluated for each mutation by calculation of the 99.8% CI to reflect the more stringent P < 0.002 threshold of statistical significance indicated by Bonferroni correction for multiple hypothesis testing. Again, nonoverlap of the 99.8% CI with RCsusceptible (equal to 1.0 by design) resulted in rejection of the null hypothesis at P < 0.002.
Shannon entropy.
To calculate the HIV-1B amino acid Shannon entropy for each of the Gag CTL escape positions, 1,700 HIV-1B Gag sequences were downloaded from the Los Alamos National Laboratories HIV sequence database (http://www.hiv.lanl.gov/), aligned with MUSCLE (49), and manually edited. The Shannon entropy was then calculated using an in-house script according to the equation , where n is the number of variant amino acid residues and p is the frequency of each variant.
Predicted structural stability.
The thermodynamic stability changes caused by CTL escape mutations in the Gag p17 and Gag p24 proteins were calculated using the FoldX software package (http://foldx.crg.es/) and a five-step procedure. First, the FoldX optimization procedure and probability-based rotamer library were used to remove steric clashes and other estimation errors and to reconstruct missing side chain atoms in the published atomic structures of p17 (Protein Data Bank [PDB] code: 2GOL) (50) and the p24 hexamer (PDB: 3H4E) (45) and pentamer (PDB: 3P0A) (46). Second, the amino acid sequence of the published structure was edited to match that of the parental (NL4-3**) backbone used in this study to construct the escape mutant viruses. Third, FoldX was used to in silico “mutate” amino acid side chains in the crystal structure using a probability-based rotamer library while exploring alternative conformations of the proximal side chains. For each mutant, five optimized structural models were generated from random restart. Fourth, the structural stability of the parental (NL4-3**) model and each of the five mutant models was calculated as the Gibbs free energy (ΔG) of folding as estimated using the FOLDEF empirical energy function (51) as implemented in FoldX: ΔG = ΔGvdw + ΔGsolvH + ΔGsolvP + ΔGhbond + ΔGwb + ΔGel + ΔSmc + ΔSsc. Finally, the predicted impact of the escape mutation on the thermodynamic stability of the structure was calculated by taking the average of the absolute value of the difference in ΔG (|ΔΔG|) between the parental (NL4-3**) model and each of the five mutant models and then normalizing for the number of subunits in the multimer, i.e., 3 for the p17 trimer, 5 for the p24 pentamer, and 6 for the p24 hexamer. Additionally, the potential of p24 mutations to impact RC through effects on the pentameric and/or the hexameric forms of the protein was captured by averaging the |ΔΔG| derived from the two structures.
RESULTS
Comprehensive panel of HIV-1 Gag CTL escape mutations.
The identification of HLA class I-associated amino acid polymorphisms in HIV-1 at the population level provides an efficient method for identifying CTL escape mutations (47, 52–54). For this study, we first considered Gag mutations that were identified as statistically significant (following correction for multiple comparisons) HLA class I-associated amino acid polymorphisms in large phylogenetically corrected analyses of HIV-1 subtype B (HIV-1B) (47, 48). We then filtered on location within, or immediately adjacent to, defined CTL epitopes (55) that are restricted by the same HLA class I allele that was identified in population-level association with the polymorphism. Application of these two criteria identified 29 mutations located in 25 CTL epitopes that were distributed across 5 of the 6 constituent Gag proteins and were restricted by 17 different HLA class I alleles (5 HLA-A, 10 HLA-B, 2 HLA-C) (Table 1). The mutations predominantly comprised classical CTL escape mutations in that the mutation is a nonconsensus amino acid variant that is infrequently expressed by circulating viruses. However, 3 of the 29 mutations (A81T, D260E, and D312E; susceptible amino acid listed first, escape variant listed last) were atypical in that the identified escape mutation was actually the amino acid residue most frequently expressed by circulating HIV-1B viruses, i.e., the consensus amino acid. Such “negatope” escape mutations may be indicative of historically targeted CTL epitopes for which continuous CTL selection pressure resulted in the accumulation over time of the escape mutation to high frequency, resulting in an epitope that is “preescaped” in the majority of circulating viruses (27, 57, 58). The construction of NL4-3-derived recombinant viruses expressing each of the 29 mutations individually provided the opportunity to comprehensively quantify the fitness cost of HIV-1 CTL escape in Gag without a priori focus on any particular HLA class I allele, CTL epitope, or Gag protein.
Table 1.
CTL escape mutations in HIV-1B Gag
| Escape mutationa | Gag protein | Associated HLA class I alleleb | CTL epitopec | Epitope sequenced |
|---|---|---|---|---|
| K26R | p17 | B*08 (nonprotective) | B*08-GK9 | GGKKKYKLK |
| K28R | p17 | A*03 (nonprotective) | A*03-RK9 | RLRPGGKKK |
| K28Q | p17 | A*03 (nonprotective) | A*03-RK9 | RLRPGGKKK |
| Y79F | p17 | A*01(nonprotective) | A*01-GY9 | GSEELRSLY |
| Y79H | p17 | Cw14 (protective) | Cw14-LL8 | LYNTVATL |
| A81T* | p17 | B*08 (nonprotective) | B*08-EV8 | ELRSLYNTV |
| A83V | p17 | A*11 (nonprotective) | A*11-TI9 | [A]TLYCVHQR |
| R91G | p17 | A*11 (nonprotective) | A*11-TK8 | TLYCVHQR |
| A146P | p24 | B*57 (protective) | B*57-ISW9 | [A]ISPRTLNAW |
| I147L | p24 | A*25 (protective) | A*25-QW11 | QAISPRTLNAW |
| A163G | p24 | B*57 (protective) | B*57-KF11 | KAFSPEVIPMF |
| Q182T | p24 | B*42 (undetermined) | B*42-TL9 | TPQDLNTML |
| T186M | p24 | Cw08 (protective) | Cw08-TL9 | TPQDLNTML |
| E211D | p24 | A*25 (protective) | A*25-EW10 | ETINEEAAEW |
| T242N | p24 | B*57 (protective) | B*57-TW10 | TSTLQEQIGW |
| G248A | p24 | B*57 (protective) | B*57-TW10 | TSTLQEQIGW |
| D260E* | p24 | B*35 (nonprotective) | B*35-NY10 | NPPIPVGEIY |
| R264K | p24 | B*27 (protective) | B*27-KK10 | KRWIILGLNK |
| L268M | p24 | B*27 (protective) | B*27-KK10 | KRWIILGLNK |
| T280S | p24 | B*52 (protective) | B*52-RI8 | RMYSPTSI |
| K302R | p24 | B*14 (protective) | B*14-DA9 | DRFYKTLRA |
| T303V | p24 | B*14 (protective) | B*14-DA9 | DRFYKTLRA |
| D312E* | p24 | B*44 (nonprotective) | B*44-AW11 | AEQASQEVKNW |
| K331R | p24 | B*08 (nonprotective) | B*08-DL9 | DCKTILKAL |
| G357S | p24 | A*11 (nonprotective) | A*11-AK11 | ACQGVGGPGHK |
| T427N | p7 | B*40 (protective) | B*40-TL8 | TERQANFL |
| K436R | p1 | B*13 (protective) | B*13-RI9 | RQANFLGKI |
| I437L | p1 | B*13 (protective) | B*13-RI9 | RQANFLGKI |
| E482D | p6 | B*40 (protective) | B*40-KL9 | KELYPLTSL |
CTL escape mutation denoted as susceptible residue followed by position (HXB2 Gag numbering) followed by escape residue; an asterisk denotes a putative negatope escape mutation.
HLA class I allele associated with mutation expression in HIV-1B (56) and its protective value in HIV-1 infection (10).
Defined CTL epitope spanning or immediately adjacent to the listed escape mutation.
HIV-1B consensus sequence for each epitope; position of escape mutation underlined; brackets denote mutation adjacent to epitope.
Benchmarking the in vitro RC assay.
To characterize the fitness cost of HIV-1 CTL escape in Gag, we quantified the impact of each of the mutations in our panel on virus replication capacity. The in vitro RC assay as used here calculated the rate of progeny virion production by qRT-PCR during the exponential growth phase of a short-term (≤7-day) infection of stimulated, heterologous, peripheral blood mononuclear cells (PBMC). To benchmark the in vivo relevance of the RC values calculated using this assay, we first quantified the RC of the M184V antiretroviral (ARV) drug resistance mutation in reverse transcriptase (RT) (Fig. 1A). This mutation causes a defined reduction in virus replication in vitro (35, 59) and is associated with a clinically significant reduction in viral load in vivo (60–62). The statistically significant 6% reduction in replication caused by the M184V mutation (RCRT M184V = 0.94) in our assay is consistent with the 7 to 14% reduction reported by prior studies (35, 59) and provides a benchmark of clinical relevance against which the RC results for the Gag CTL escape mutations could be compared.
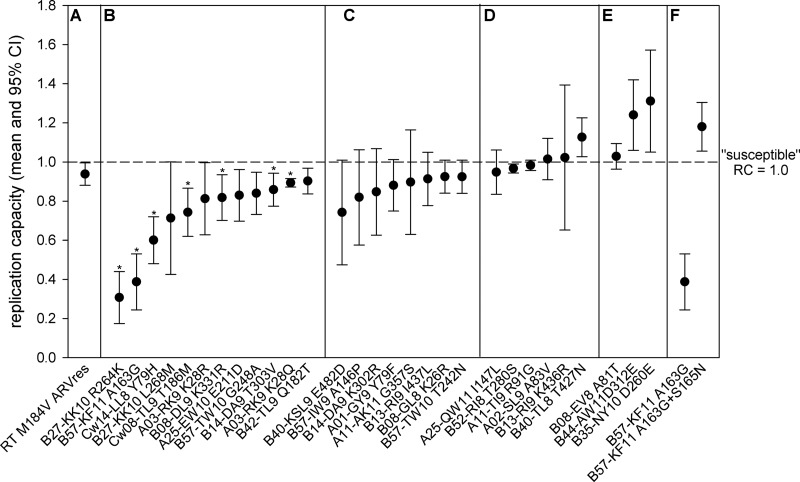
Fig 1.
The impact on replication capacity (RC) of CTL escape mutations in HIV-1B Gag. The mean replication capacity (RC; dots) with 95% confidence interval (CI; whiskers) is shown for the benchmark RT M184V mutation (A), the 12 CTL escape mutations associated with a statistically significant reduction in RC (P ≤ 0.05) (B), the 8 escape mutations associated with reduction in RC that exceeds that caused by the benchmark M184V mutation (C), the 6 escape mutations that had minimal impact on RC and the 1 that increased it (D), the 3 CTL negatope escape mutations (E), and the A163G mutation in the absence and presence of the S165N compensatory mutation (F). For all mutations, the susceptible amino acid residue is listed first and the escape residue is listed last. The null hypothesis that RCescape = RCsusceptible is rejected at P < 0.05 if the 95% CI of RCescape does not overlap RCsusceptible (horizontal dashed line), which is equal to 1.0 by definition. The mutations that withstand Bonferroni correction for multiple comparisons (P ≤ 0.002) are denoted by an asterisk.
HIV-1 Gag CTL escape mutations frequently impair viral RC.
We next quantified the RC associated with each of the 26 classical CTL escape mutations. Overall, 77% (20/26) of the classical escape mutations caused a reduction in RC in excess of that caused by the M184V benchmark, with 12 of the 20 exhibiting a statistically significant (P < 0.05) reduction in replication (RC, 0.31 to 0.90) and 7 of those withstanding Bonferroni correction for multiple comparisons (P < 0.002) (Fig. 1B). Within the 12 statistically significant mutations, 2 mutations (R264K and A163G) reduced virus replication by more than 50% (RCR264K = 0.31; RCA163G = 0.39). Notably, these mutations are selected by immunodominant CTL responses restricted by the protective HLA-B*27 and -B*57 alleles, respectively (9). Three additional mutations (Y79H, L268M, and T186M) impaired replication by >25% (RCY79H = 0.60; RCL268M = 0.71; RCT186M = 0.74), and all three of these mutations are again associated with protective HLA class I alleles (HLA-Cw14, -B*27, and -Cw08) (Table 1). The 7 remaining mutations that had a statistically significant impact caused more moderate reductions of 10 to 19% (RC, 0.81 to 0.90) that still exceeded that of the M184V benchmark of clinical relevance (RCRT M184V = 0.94).
In addition to the 12 mutations that exhibited a statistically significant reduction in RC, there were 8 mutations for which the reduction in virus replication exceeded the clinical relevance benchmark (RCRT M184V = 0.94) but did not achieve statistical significance (Fig. 1C). These included the well-described T242N escape mutation in the HLA-B*57 TW10 epitope that caused a reduction of 8% (RCT242N = 0.92), consistent with the previously reported in vivo (28) and in vitro (35, 63) fitness cost of this mutation. As shown in Fig. 1D, five of the final six classical HIV-1 Gag CTL escape mutations (I147L, T280S, R91G, A83V, and K436R) exhibited a minimal impact on virus replication (RC, 0.95 to 1.02), with the last mutation (T427N) causing a statistically significant 14% increase in virus replication (RC = 1.13). These data reveal that the majority of HIV-1 CTL escape mutations in Gag cause a reduction in virus replication capacity and that the magnitude of this escape-associated fitness cost is sufficient to be clinically relevant.
HIV-1 Gag negatope escape mutations and compensatory mutations increase RC.
Next, we examined the effect on replication of the 3 CTL negatope escape mutations (A81T, D312E, and D260E) in which the mutation conferring escape (listed second) is also the HIV-1B consensus amino acid residue. Here, we observed that the D312E and D260E mutations significantly increased virus replication by 26% and 31%, respectively (RCD312E = 1.24; RCD260E = 1.31) (Fig. 1E). These data represent the first quantification of the impact of CTL negatope escape mutations on virus replication and suggest that such mutations may enhance virus replication capacity. The observation that two of these negatope escape mutations (D260E and A81T) are associated with the hazardous HLA-B*35 and -B*08 alleles, respectively, suggests a possible contribution of the neutral, or even enhanced, escape-associated replication to ineffective control of HIV-1.
Finally, we quantified the impact on replication of the S165N mutation, which was previously described to partially compensate for the fitness cost of the A163G escape mutation in the HLA-B*57 KF11 epitope in HIV-1 subtype C (37). The 61% reduction in replication caused by the A163G mutation (RCA163G = 0.39) was completely rescued by coexpression of the S165N mutation (RCA163G+S165N = 1.18) (Fig. 1F). Taken together, these data support the idea that some HLA class I-associated mutations in Gag can restore, and even increase, viral replication capacity, which may influence the effects of specific HLA alleles and their restricted CTL responses on immune control of HIV-1.
Higher HIV-1 Gag CTL escape costs in epitopes restricted by protective HLA class I alleles.
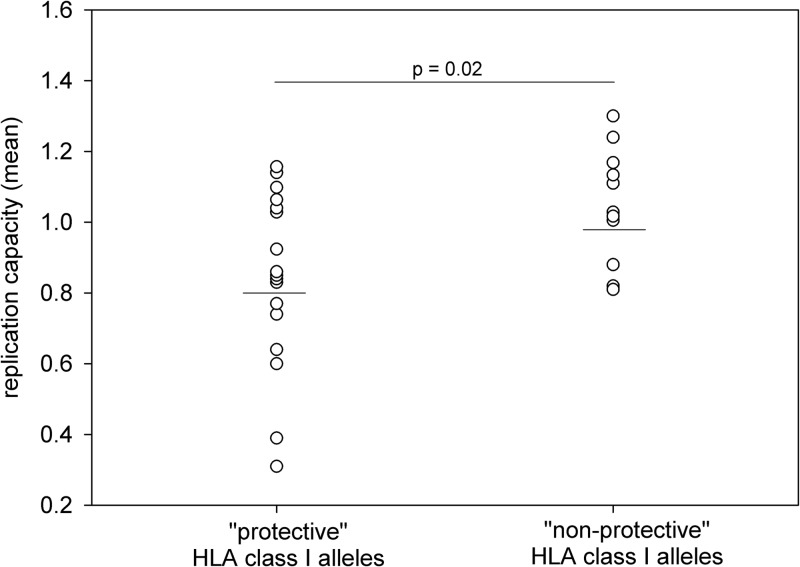
The protective benefit associated with a subset of HLA class I alleles is well established (10, 13), but the immunological basis for the effect remains less clear. The observed range of HIV-1 Gag CTL escape-associated fitness costs suggested the possibility that differences in the cost of escape from specific CTL responses may contribute to CTL effectiveness and thus the protective value attributed to a given HLA class I allele. We investigated the relationship between the fitness cost of CTL escape and the protective benefit of HLA class I alleles by sorting the 29 CTL escape mutations into two groups based on associated HLA alleles. One group comprised 17 mutations that confer escape from CTL responses restricted by “protective” HLA class I alleles, defined as those alleles that were enriched in a cohort of HIV-1 controllers (10), and the second group comprised 11 mutations that confer escape from CTL responses restricted by HLA class I alleles that are not protective (not enriched in HIV-1 controllers). The Q182T mutation could not be grouped because the protective value of the associated HLA-B*42 allele was not determined in that study. We observed that the mutations conferring escape from CTL responses restricted by protective alleles caused a significantly greater reduction in virus replication than did those conferring escape from CTL responses restricted by nonprotective alleles (mean RCprotective = 0.80; mean RCnonprotective = 0.98; P = 0.02; Student's t test) (Fig. 2). This difference in mean RC between the two groups remained significant even when we included in the comparison only the mutations exhibiting a statistically significant impact on replication capacity (data not shown). These data support the concept that immune targeting of CD8+ T cell epitopes that are escape refractory, by virtue of high escape-associated fitness costs, may contribute to the differential control of HIV-1 associated with many protective HLA class I alleles.
Fig 2.
CTL escape in HIV-1 Gag epitopes restricted by protective HLA class I alleles is associated with a greater reduction in replication capacity. The mean RCs of the 29 CTL escape mutations are shown grouped according to whether the associated HLA class I allele is enriched among HIV controllers (protective) or not (nonprotective). The horizontal bars denote the mean RC of mutations in epitopes restricted by protective alleles (0.80) and nonprotective alleles (0.98). The data are normally distributed (P = 0.445, Shapiro-Wilk test), and the difference in mean RC between the two groups is statistically significant (P = 0.02, Student's t test).
HIV-1 Gag CTL escape costs do not correlate with genetic conservation.
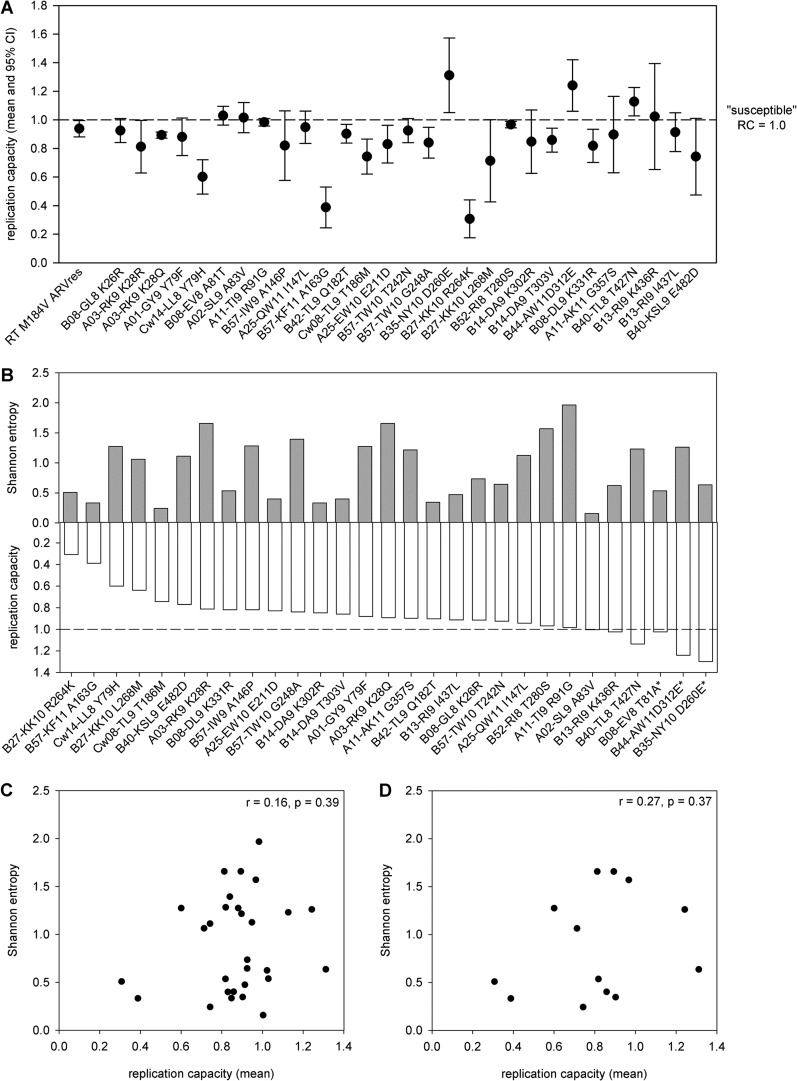
Quantification of the impact of a large number of Gag CTL escape mutations on viral replication capacity provided the opportunity to determine whether specific protein sequence or structural characteristics correlate with the impact of CTL escape mutations on virus replication capacity. First, we evaluated the RC data for evidence of gross protein-specific differences in mutational impact by reordering the RC data according to the linear position of the mutation within the Gag polyprotein, but we observed no obvious pattern (Fig. 3A). Shannon entropy provides an effective means of quantifying the population-scale genetic variability at any given residue across HIV-1. Therefore, we next calculated the HIV-1B amino acid Shannon entropy at the position of each of the mutations in our panel using a public set of 1,700 HIV-1B Gag sequences and then compared the Shannon entropy to the in vitro RC (Fig. 3B). We observed no correlation between Shannon entropy and impact on replication capacity regardless of whether we considered all mutations (r = 0.16, P = 0.39; Pearson product moment test) (Fig. 3C) or only those that caused a statistically significant impact on replication capacity (r = 0.27, P = 0.37; Pearson product moment test) (Fig. 3D). Although Shannon entropy has been used to predict which amino acid position within a given epitope is most likely to escape (64), the multiple factors influencing HIV-1 genetic diversity, e.g., HLA class I allele frequency, CTL immunodominance patterns (9), and the presence or absence of compensatory mutations, may have obscured any correlation with replication capacity. These data suggest that the fitness cost of CTL escape at a given amino acid position, and by extension the value of targeting the associated CTL epitope, cannot be accurately predicted based on population-scale genetic diversity at that position.
Fig 3.
The impact on replication capacity (RC) of CTL escape mutation in HIV-1 Gag is not correlated with position or genetic conservation (Shannon entropy). (A) The mean replication capacity (RC; dots) with 95% confidence interval (CI; whiskers) is shown for the benchmark RT M184V mutation and for the 29 individual HIV-1B Gag CTL escape mutations in order of their linear position in the Gag polyprotein. For all mutations, the susceptible residue is listed first and the escape residue is listed last. The horizontal dashed line denotes RCsusceptible, which is equal to 1.0 by definition. (B) The CTL escape mutations are plotted from greatest to least reduction in replication (lowest to highest RC value) (white bars) opposite the Shannon entropy of the mutant position (gray bars). (C and D) The lack of correlation between Shannon entropy and RC is shown for all mutations tested (r = 0.16, P = 0.39; Pearson product moment test) (C) and for only those mutations that caused a statistically significant impact on RC (r = 0.27, P = 0.37; Pearson product moment test) (D).
CTL escape costs correlate with predicted impact on protein structural stability.
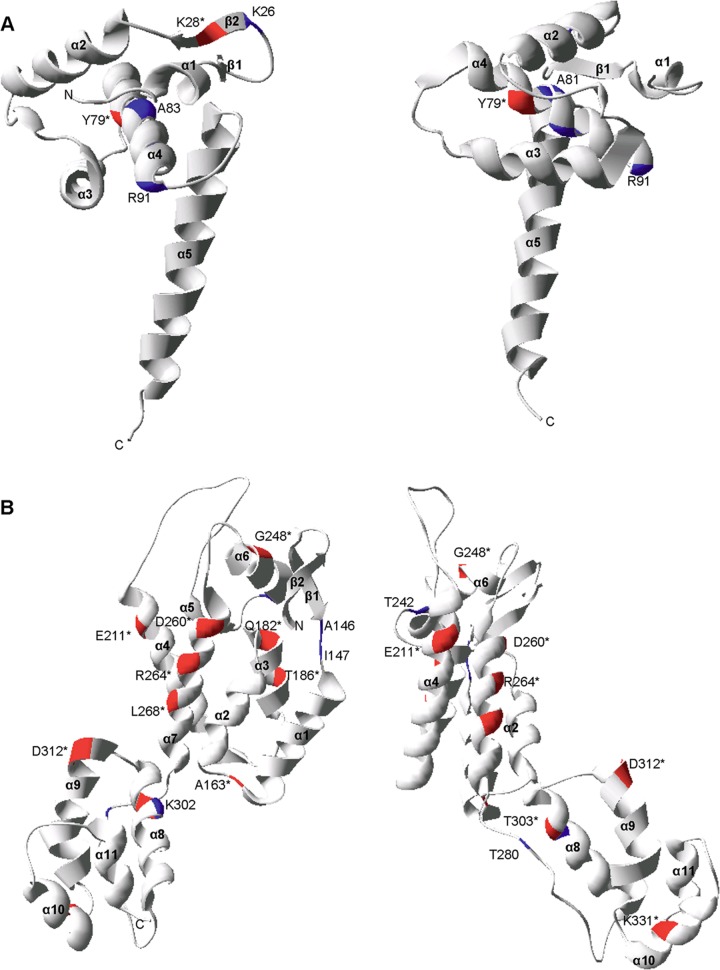
We next set out to determine whether there may be a protein structural basis for the differential impact of CTL escape mutations on viral replication. This analysis was facilitated by the availability of high-resolution, three-dimensional crystal structures for the Gag p17 trimer (PDB: 1HIW) (65) and p24 hexamer (PDB: 3GV2) (45) and of nuclear magnetic resonance (NMR) structures for the p7 (PDB: 1F6U) (66) and p6 (PDB: 2C55) (67) monomers. We first mapped the location of the CTL escape mutations in our panel onto the p17 and p24 structures (Fig. 4A and B, respectively) and the p7 and p6 structures (data not shown) to distinguish between mutations located at positions within protein secondary structure, e.g., alpha helices and beta sheets, and those located in nonstructured regions. Overall, 26 mutations could be mapped onto the available structures (p24 position 357 is not in the structure; there is no structure for p1) with the two different mutations each at positions 28 and 79 in p17 counted separately. We observed that an impact on in vitro viral replication capacity was associated with the location of the mutation in protein secondary structure, with 13/14 mutations that caused a statistically significant impact on RC located within secondary structure versus 5/12 mutations that did not have a significant impact on RC (P = 0.009; two-tailed Fisher's exact test).
Fig 4.
Structural locations of the CTL escape mutations in HIV-1 Gag p17 and p24. The CTL escape mutations mapped onto two rotations of the ribbon representation of the three-dimensional monomer structures of p17 (PDB: 1HIW) (A) and p24 (PDB: 3GV2) (B). The locations of CTL escape mutations are denoted by the consensus amino acid and the Gag amino acid position. Mutations that caused a statistically significant impact on virus replication are denoted by an asterisk and by red highlighting of the ribbon. All other mutations are denoted by blue highlighting.
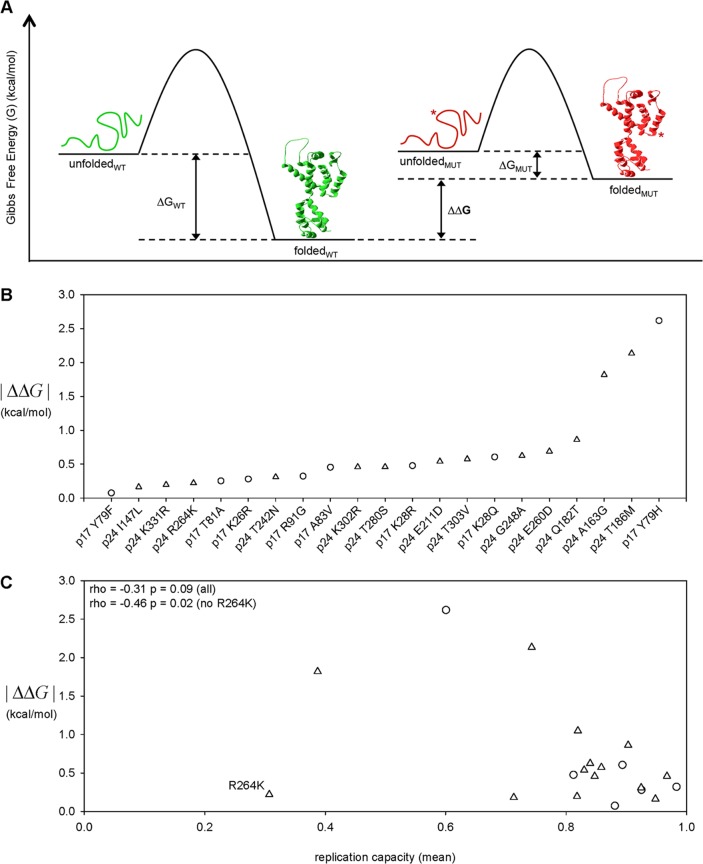
Next, we refined our analysis of the structural basis of CTL escape-associated fitness costs by utilizing a well-described protein folding energy algorithm, FOLDEF (for FOLD-X Energy Function) (51), which can estimate the Gibbs free energy of folding (ΔG) and of protein complex formation by imputing fundamental thermodynamic parameters, e.g., van der Waals forces, side chain solvation, entropy changes, etc., from the atomic coordinates of the three-dimensional protein structure (see Materials and Methods). We used FOLDEF to estimate the impact of the 21 classical CTL escape mutations located in Gag p17 and p24 on protein thermodynamic stability by calculating the change in the ΔG caused by each mutation (ΔΔG) (Fig. 5A). We reasoned that the p17 and p24 three-dimensional structures occupy narrowly defined stability optima and that change in either direction, i.e., both stability increases and decreases, could reduce function and thus replication capacity. We therefore considered the magnitude of thermodynamic stability changes without regard for the direction of change by taking the absolute value (|ΔΔG|). We accounted for the different number of monomers present in the structures of p17 (trimer) and p24 (pentamer and hexamer) by normalizing the computed |ΔΔG| for a given mutation for the number of monomers present in the structure. In addition, we accounted for the possibility that a mutation in p24 could impact the structural stability of the distinct pentameric and hexameric forms of the protein by taking the average of the |ΔΔG| values determined for each of the structures. The estimated impacts of the 21 mutations analyzed spanned a wide range of effects with the majority (18/21) following essentially a linear continuum from |ΔΔG| = 0.07 (p17 Y79F) to |ΔΔG| = 0.86, with the exception of three mutations (A163G, T186M, and Y79H in p24) that exhibited a substantially larger impact (Fig. 5B). We next assessed the relationship between estimated CTL escape-associated impact on protein stability and the observed reduction in replication capacity for the 21 classical CTL escape mutations by rank correlation to reduce the impact of either the single data point in the top left quadrant or the cluster of data points in the bottom right quadrant (Fig. 5C). Overall, the estimated |ΔΔG| and the replication capacity exhibited a moderate negative correlation (r = −0.31; P = 0.09; Spearman rank correlation). However, the outlier R264K mutation in p24 has been previously shown to alter normal virus interaction with the host protein cyclophilin A as indicated by increased RC in the presence of cyclosporine and in a cyclophilin A-deficient target cell (34). In contrast to R264K, none of the other 16 p24 CTL escape mutations in this study exhibited any increase in RC in the presence of cyclosporine (data not shown), suggesting that it impacts RC through a unique molecular mechanism. When the R264K mutation was excluded from the analysis on this basis, we observed that |ΔΔG| and RC were in fact negatively correlated (r = −0.46, P = 0.02, Spearman rank correlation).
Fig 5.
In silico estimation of the impact on protein stability of CTL escape mutations in HIV-1B Gag p17 and p24. (A) Protein stability is defined by the difference in Gibbs free energy (ΔG) between the unfolded and folded states. The FOLDEF energy function (51) was used to estimate the ΔG for the “wild-type” (WT) protein sequence and for each CTL escape mutant sequence (MUT), and the stability impact of a mutation was quantified as the absolute value of the difference between WT ΔG and MUT ΔG (|ΔΔG|). (B) The impact on protein stability (|ΔΔG|) was estimated for the 21 classical CTL escape mutations located within the Gag p17 trimer and p24 pentamer and hexamer crystal structures. (C) The CTL escape-associated impact on protein stability (|ΔΔG|) and reduction in replication capacity (RC) were negatively correlated when the outlier p24 R264K mutation was excluded from the analysis (rho = −0.46, P = 0.02; Spearman rank correlation). CTL escape mutations in Gag p17 and p24 are denoted by circles and triangles, respectively.
DISCUSSION
The capacity of HIV-1 to escape from cytotoxic-CD8+-T-lymphocyte (CTL) responses undermines the effectiveness of CTL-mediated control of virus replication and poses a significant challenge to the durability of vaccine-induced CTL responses and vaccine efficacy (22, 23). Importantly, however, CTL escape mutations not only confer a fitness benefit by increasing virus evasion of CTL-mediated killing but also can confer a concomitant fitness cost by reducing virus replication (32–38), suggesting a fitness trade-off associated with CTL escape. Given that the process of CTL escape is largely governed by the net balance of all fitness costs and benefits, the implications of such a trade-off are significant as it suggests that CTL escape may be delayed, or perhaps precluded entirely, if expression of the escape mutation is associated with viral attenuation of sufficient magnitude. Thus, a CTL response that specifically targets epitopes in which escape occurs with a high fitness cost (escape refractory) would be more durable and effective than one that targets epitopes in which escape occurs with little or no fitness cost (escape permissive). To begin to distinguish escape-refractory epitopes from escape-permissive epitopes to help inform HIV-1 vaccine design, we undertook an extensive quantification of CTL escape-associated fitness costs in the immunologically critical HIV-1 Gag protein.
The Gag CTL escape mutations in this study were selected for analysis on the basis of both their identification as statistically significant (following correction for multiple comparisons) HIV-1B HLA class I allele-associated amino acid polymorphisms in large phylogenetically corrected analyses (43, 47) and their location within experimentally defined CD8+ T cell epitopes (LANL “A List”) that are restricted by the associated HLA class I alleles, i.e., an HLA-B*14-associated amino acid polymorphism located within a defined HLA-B*14 CTL epitope. Although the majority of these mutations have not been formally characterized in immunological assays to confer loss of CD8+ T cell-mediated targeting and/or killing, we believe that satisfying the two abovementioned criteria provides a high level of confidence that these are indeed CTL escape mutations. We observed a wide range of impacts on in vitro replication capacity (RC), with some mutations reducing replication by as much as 69% while others had minimal impact, indicating substantial variation in structure/function plasticity across Gag despite its high genetic conservation relative to other HIV-1 proteins. Overall, approximately 75% (20/26) of the classical escape mutations tested carried an associated fitness cost sufficient to impact viral replication in vivo as measured against the RC of the RT M184V antiretroviral drug resistance mutation, which is clinically associated with reduced viral load (35, 59–62). This potential for CTL escape-associated fitness costs to attenuate viral replication in vivo is supported by observations of lower set-point viral loads in subjects with a greater number of deleterious CTL escape mutations (68–70). Our finding that the majority of the analyzed CTL escape mutations in Gag carry an associated fitness cost as indicated by reduced in vitro replication capacity may help to explain the association between Gag-specific CD8+ T cell responses and the relative level of control of HIV-1 (40, 41) because escape from Gag responses has a high likelihood of attenuating HIV-1. However, the range of escape-associated costs suggests that even against a well-conserved protein like Gag, not all CD8+ T cell responses will exploit this Achilles heel of the virus and will thus not all be equally effective at controlling HIV-1.
In addition to the 26 traditional CTL escape mutations, we examined 3 mutations (A81T, D260E, and D312E) that displayed a distinct HLA class I association in which the escape mutation (T81, E260, or E312) reflects the high-frequency HIV-1B consensus amino acid residue and the susceptible, or targetable, amino acid (A81, D260, or D312) is present at lower frequency. This phenomenon is hypothesized to result from continuous adaptation of HIV-1 to CTL pressure that has caused the escape mutation to increase in frequency among circulating viruses until it has become the consensus amino acid residue (27, 52, 54, 71, 72). As a result, the CD8 epitope becomes a preescaped CTL negatope that is no longer effectively targeted by the majority of individuals expressing the restricting HLA class I allele (27, 57). The increase in RC reported here for the D260E and D312E putative negatope escape mutations in the HLA-B*35 NY10 and HLA-B*44 AW11 epitopes, respectively, helps to explain how such negatopes can develop. For these mutations, there is not a fitness cost-benefit trade-off but rather dual fitness benefits in terms of both increased survival by immune evasion and increased replication capacity. The escape mutation thus continually accumulates in the circulating virus population over time, i.e., without reverting upon transmission to a nonselecting individual, and ultimately becomes the consensus residue. As a result, most contemporary HIV-1B infections of individuals expressing HLA-B*35 or -B*44 will consist of a transmitted virus that is already escaped in the NY10 or AW11 epitope, respectively, which may contribute to the lack of protective benefit associated with these alleles (10, 13). Interestingly, Matthews et al. recently reported that the HLA-B*3501 Gag NY10 epitope is intact and not a negatope (D260) in the HIV-1C consensus, that the resultant effective targeting of the NY10 epitope in the HIV-1C-infected population was the only significant difference in HLA-B*3501-restricted CTL responses observed between HIV-1B- and HIV-1C-infected cohorts, and that unlike in HIV-1B infections the HLA-B*3501 allele is not deleterious in populations infected with HIV-1C (58). Overall, these data highlight the idea that negatopes are likely to represent uniquely poor targets for CD8+ T cell responses.
Our data demonstrate that the magnitude of the CTL escape-associated fitness cost (reduction in RC) is significantly greater for mutations conferring escape in epitopes restricted by protective HLA class I alleles (mean RC = 0.80) than for those conferring escape in epitopes restricted by nonprotective alleles (mean RC = 0.98). These data support a potential critical role for CTL escape-associated fitness costs in determining the relative effectiveness of CD8+ T cell responses. This finding is consistent with our prior report illustrating that acute/early HIV-1 Gag sequences derived from individuals expressing protective HLA class I alleles were associated with significantly lower RC than were Gag sequences derived from individuals lacking those alleles (73) and suggests that the effect of RC on control may be imparted during the first few weeks after infection by impacting the acute-phase CD8+ T cell responses that determine primary viremia and set-point viral load (9, 74). Taken together, these data suggest that the protective effect associated with some HLA class I alleles may result from the acute immunodominant CTL targeting of epitopes that carry high escape-associated fitness costs.
In vitro RC assays allow for quantification of the fitness cost of CTL escape mutations but can be extremely laborious. As such, there is a substantial need to identify computational methods to more efficiently identify high-cost CTL escape mutations and thus susceptible regions of HIV-1. Shannon entropy failed to correlate with the impact of CTL escape mutations on RC. This is likely due to the fact that the Shannon entropy of an amino acid position calculated from a large data set of HIV-1B sequences is itself influenced by the frequencies of different HLA class I alleles and, by extension, the frequencies with which different epitopes are targeted, and mutations selected, in the host population. For example, although low Shannon entropy may be indicative of a position under high structure/function constraint and thus a high fitness cost of CTL escape, it may also simply indicate lack of frequent escape because the restricting HLA class I allele is rare in the population. Conversely, a high Shannon entropy may indicate either CTL escape with minimal or no associated fitness cost or CTL escape that carries an associated fitness cost but is driven to high frequency by a highly immunodominant CD8+ T cell response restricted by a common HLA class I allele or by a compensatory mutation that alleviates its fitness defect. In contrast to Shannon entropy, the estimation of the impact of CTL escape mutations on the Gibbs free energy of folding (ΔG), a measure of protein stability, using the FOLDEF energy function shows promise as an in silico tool for predicting mutations that carry an associated fitness cost. Specifically, we observed a correlation between in vitro RC and the estimated change in stability (|ΔΔG|) when we excluded an outlier mutation known to reduce RC through a stability-independent mechanism. These data suggest that the FOLDEF-based approach provides a valuable prescreening tool to identify CTL escape mutations that are likely to reduce RC as a result of their impact on protein stability. Importantly, this approach will nonetheless have a nonzero “false-negative” rate, i.e., will fail to identify some costly mutations, as it is not designed to identify mutations that impact RC through other molecular mechanisms such as host protein interaction. Indeed, the basis for excluding the R264K mutation when calculating the correlation between |ΔΔG| and RC is that there is evidence that the mutation exerts its impact through disruption of a p24 interaction with a host protein more than through disruption of p24-p24 interactions. Future development will include application of the method to three-dimensional structures of virus-host protein complexes in an attempt to capture such interactions as well as the simultaneous analysis of multiple mutations, e.g., escape and compensatory mutations, to extend the utility of the method. This and other in silico methods for predicting the impact of immune (CTL and antibody) escape mutations on HIV replication capacity will improve the speed and efficiency with which we can identify the most advantageous (escape-refractory) and deleterious (escape-permissive) targets for vaccine-induced immune responses.
The in vitro replication capacity (RC) assay provides an effective means of quantifying CTL escape-associated fitness costs, but as for any in vitro assay, one must be cognizant of the need to extrapolate the experimental results of such RC assays to HIV-1 infection in vivo. In an effort to address this limitation, we included in our assay the quantification of the RT M184V antiretroviral (ARV) drug resistance mutation to provide an internal benchmark of clinical relevance for the RC data produced by our assay. The RT M184V mutation is well suited as a reference because it reduces HIV-1 replication both in vitro (35, 59) and in vivo (60–62). The ability to compare RC data to such a benchmark is particularly valuable given that the relatively high well-to-well variation intrinsic to “traditional” single-well, primary-cell infection assays such as the one used here can limit the statistical power to detect small changes in RC. In addition to providing a point of reference for values derived within a particular assay, the inclusion of the RT M184V mutation in RC assays provides a mechanism for normalizing and comparing RC data generated across studies using different versions of the RC assay. The genetic context in which RC is quantified is another important point of consideration for in vitro RC assays. Here, we quantified the impact on RC that is specifically attributable to individual mutations by engineering them in isolation into a recombinant NL4-3 HIV-1B backbone. The use of a recombinant clone provides a consistent background in which to analyze each mutation, but it is possible that the specific genetic context of the backbone could differentially interact with the tested mutations, thereby perturbing the measured RC in a backbone-specific manner. For example, the impact on RC of the A163G mutation observed in this study in the context of the HIV-1B NL4-3 backbone is greater than that observed previously in the context of the HIV-1C MJ4 backbone (35), and Wright et al. have reported that the magnitude of the impact of the Gag T186S mutation also differed when assayed in the context of HIV-1B versus HIV-1C (38). The most well-described example of secondary genetic variation influencing the impact of a mutation on RC is the rescue of escape mutation-associated reductions in RC by compensatory mutations, which has been shown both in this study (A163G + S165N) and in others (35, 38); the presence of a such a compensatory mutation in the assay backbone (NL4-3 in this study) could mask the impact of a mutation of interest on RC. Although we found no evidence in the NL4-3 genome of known compensatory mutations for the escape mutations analyzed in this study, we cannot rule out the possibility that unknown genetic variants, compensatory or otherwise, native to the NL4-3 genome may have differentially impacted measured RC values. The large number of mutations and replicate infections in this study precluded recapitulation of the experiments in an additional genetic background, but future studies will seek to investigate the reproducibility of the impact on RC of CTL escape mutations by evaluating them in additional recombinant and clinically derived genetic backgrounds.
The potential of a CTL vaccine to induce potent protection against HIV-1 has been demonstrated recently by studies in the SIV/macaque model (15), highlighting the importance of inducing effective CD8+ T cell responses with any HIV-1 vaccine. This effort is challenged not only by the capacity of HIV-1 for CTL escape but also by the lack of clarity regarding exactly what constitutes an effective vaccine-induced CD8+ T cell response. Although effective, the RC assay is labor-intensive and the development of an in silico prescreen for the identification of CTL escape mutations with a high fitness cost could dramatically increase the rate at which escape-refractory, and thus higher-value, CD8+ T cell epitopes can be identified. The ability to distinguish between effective and ineffective targets of CD8+ T cell responses for the most common HLA class I alleles would allow for the development of novel HIV-1 vaccine immunogens designed to direct vaccine-induced responses against more protective, escape-refractory regions of the virus while simultaneously avoiding the priming of ineffective responses against escape-permissive “decoy” targets.
ACKNOWLEDGMENTS
This project was funded in part by the Bill and Melinda Gates Foundation (T.M.A.) and the National Institute of Allergy and Infectious Diseases (NIAID) under grants P01-AI074415 (T.M.A.), R01-AI090698 (T.M.A.), and T32-AI07245 (C.L.B.). Z.L.B. is the recipient of a New Investigator Award from the Canadian Institutes of Health Research and a Scholar Award from the Michael Smith Foundation for Health Research.
The pNL4-3 plasmid was obtained from Malcolm Martin through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. We thank Katherine Kane for assistance in the construction of recombinant pNL4-3-derived molecular clones.
Footnotes
Published ahead of print 30 January 2013
REFERENCES
- 1. Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103–6110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koup RA, Safrit JT, Cao Y, Andrews CA, McLeod G, Borkowsky W, Farthing C, Ho DD. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Safrit JT, Andrews CA, Zhu T, Ho DD, Koup RA. 1994. Characterization of human immunodeficiency virus type 1-specific cytotoxic T lymphocyte clones isolated during acute seroconversion: recognition of autologous virus sequences within a conserved immunodominant epitope. J. Exp. Med. 179:463–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goonetilleke N, Liu MK, Salazar-Gonzalez JF, Ferrari G, Giorgi E, Ganusov VV, Keele BF, Learn GH, Turnbull EL, Salazar MG, Weinhold KJ, Moore S, Letvin N, Haynes BF, Cohen MS, Hraber P, Bhattacharya T, Borrow P, Perelson AS, Hahn BH, Shaw GM, Korber BT, McMichael AJ. 2009. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 206:1253–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857–860 [DOI] [PubMed] [Google Scholar]
- 6. Jin X, Bauer DE, Tuttleton SE, Lewin S, Gettie A, Blanchard J, Irwin CE, Safrit JT, Mittler J, Weinberger L, Kostrikis LG, Zhang L, Perelson AS, Ho DD. 1999. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mellors JW, Rinaldo CR, Jr, Gupta P, White RM, Todd JA, Kingsley LA. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167–1170 [DOI] [PubMed] [Google Scholar]
- 8. Mellors JW, Margolick JB, Phair JP, Rinaldo CR, Detels R, Jacobson LP, Munoz A. 2007. Prognostic value of HIV-1 RNA, CD4 cell count, and CD4 cell count slope for progression to AIDS and death in untreated HIV-1 infection. JAMA 297:2349–2350 [DOI] [PubMed] [Google Scholar]
- 9. Streeck H, Jolin JS, Qi Y, Yassine-Diab B, Johnson RC, Kwon DS, Addo MM, Brumme C, Routy JP, Little S, Jessen HK, Kelleher AD, Hecht FM, Sekaly RP, Rosenberg ES, Walker BD, Carrington M, Altfeld M. 2009. Human immunodeficiency virus type 1-specific CD8+ T-cell responses during primary infection are major determinants of the viral set point and loss of CD4+ T cells. J. Virol. 83:7641–7648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O'Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, Alvino DM, Vine S, Addo MM, Allen TM, Altfeld M, Henn MR, Le Gall S, Streeck H, Haas DW, Kuritzkes DR, Robbins GK, Shafer RW, et al. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O'Brien SJ, Gao X, Carrington M. 2001. HLA and AIDS: a cautionary tale. Trends Mol. Med. 7:379–381 [DOI] [PubMed] [Google Scholar]
- 12. Carrington M, Nelson GW, Martin MP, Kissner T, Vlahov D, Goedert JJ, Kaslow R, Buchbinder S, Hoots K, O'Brien SJ. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748–1752 [DOI] [PubMed] [Google Scholar]
- 13. Carrington M, O'Brien SJ. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54:535–551 [DOI] [PubMed] [Google Scholar]
- 14. Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, Legasse AW, Chiuchiolo MJ, Parks CL, Axthelm MK, Nelson JA, Jarvis MA, Piatak M, Jr, Lifson JD, Picker LJ. 2011. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473:523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hansen SG, Vieville C, Whizin N, Coyne-Johnson L, Siess DC, Drummond DD, Legasse AW, Axthelm MK, Oswald K, Trubey CM, Piatak M, Jr, Lifson JD, Nelson JA, Jarvis MA, Picker LJ. 2009. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 15:293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koup RA, Douek DC. 2011. Vaccine design for CD8 T lymphocyte responses. Cold Spring Harbor Perspect. Med. 1:a007252 doi:10.1101/cshperspect.a007252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Price DA, Goulder PJ, Klenerman P, Sewell AK, Easterbrook PJ, Troop M, Bangham CR, Phillips RE. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. U. S. A. 94:1890–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bailey JR, Zhang H, Wegweiser BW, Yang HC, Herrera L, Ahonkhai A, Williams TM, Siliciano RF, Blankson JN. 2007. Evolution of HIV-1 in an HLA-B*57-positive patient during virologic escape. J. Infect. Dis. 196:50–55 [DOI] [PubMed] [Google Scholar]
- 19. Ammaranond P, van Bockel DJ, Petoumenos K, McMurchie M, Finlayson R, Middleton MG, Davenport MP, Venturi V, Suzuki K, Gelgor L, Kaldor JM, Cooper DA, Kelleher AD. 2011. HIV immune escape at an immunodominant epitope in HLA-B*27-positive individuals predicts viral load outcome. J. Immunol. 186:479–488 [DOI] [PubMed] [Google Scholar]
- 20. Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H, Nelson JA, Gairin JE, Hahn BH, Oldstone MB, Shaw GM. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205–211 [DOI] [PubMed] [Google Scholar]
- 21. Feeney ME, Tang Y, Roosevelt KA, Leslie AJ, McIntosh K, Karthas N, Walker BD, Goulder PJ. 2004. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J. Virol. 78:8927–8930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barouch DH, Kunstman J, Kuroda MJ, Schmitz JE, Santra S, Peyerl FW, Krivulka GR, Beaudry K, Lifton MA, Gorgone DA, Montefiori DC, Lewis MG, Wolinsky SM, Letvin NL. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335–339 [DOI] [PubMed] [Google Scholar]
- 23. Betts MR, Exley B, Price DA, Bansal A, Camacho ZT, Teaberry V, West SM, Ambrozak DR, Tomaras G, Roederer M, Kilby JM, Tartaglia J, Belshe R, Gao F, Douek DC, Weinhold KJ, Koup RA, Goepfert P, Ferrari G. 2005. Characterization of functional and phenotypic changes in anti-Gag vaccine-induced T cell responses and their role in protection after HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 102:4512–4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Allen TM, Altfeld M, Yu XG, O'Sullivan KM, Lichterfeld M, Le Gall S, John M, Mothe BR, Lee PK, Kalife ET, Cohen DE, Freedberg KA, Strick DA, Johnston MN, Sette A, Rosenberg ES, Mallal SA, Goulder PJ, Brander C, Walker BD. 2004. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J. Virol. 78:7069–7078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Draenert R, Le Gall S, Pfafferott KJ, Leslie AJ, Chetty P, Brander C, Holmes EC, Chang SC, Feeney ME, Addo MM, Ruiz L, Ramduth D, Jeena P, Altfeld M, Thomas S, Tang Y, Verrill CL, Dixon C, Prado JG, Kiepiela P, Martinez-Picado J, Walker BD, Goulder PJ. 2004. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J. Exp. Med. 199:905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kelleher AD, Long C, Holmes EC, Allen RL, Wilson J, Conlon C, Workman C, Shaunak S, Olson K, Goulder P, Brander C, Ogg G, Sullivan JS, Dyer W, Jones I, McMichael AJ, Rowland-Jones S, Phillips RE. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leslie A, Kavanagh D, Honeyborne I, Pfafferott K, Edwards C, Pillay T, Hilton L, Thobakgale C, Ramduth D, Draenert R, Le Gall S, Luzzi G, Edwards A, Brander C, Sewell AK, Moore S, Mullins J, Moore C, Mallal S, Bhardwaj N, Yusim K, Phillips R, Klenerman P, Korber B, Kiepiela P, Walker B, Goulder P. 2005. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J. Exp. Med. 201:891–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, Tang Y, Holmes EC, Allen T, Prado JG, Altfeld M, Brander C, Dixon C, Ramduth D, Jeena P, Thomas SA, St. John A, Roach TA, Kupfer B, Luzzi G, Edwards A, Taylor G, Lyall H, Tudor-Williams G, Novelli V, Martinez-Picado J, Kiepiela P, Walker BD, Goulder PJ. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282–289 [DOI] [PubMed] [Google Scholar]
- 29. Brander C, Hartman KE, Trocha AK, Jones NG, Johnson RP, Korber B, Wentworth P, Buchbinder SP, Wolinsky S, Walker BD, Kalams SA. 1998. Lack of strong immune selection pressure by the immunodominant, HLA-A*0201-restricted cytotoxic T lymphocyte response in chronic human immunodeficiency virus-1 infection. J. Clin. Invest. 101:2559–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sewell AK, Harcourt GC, Goulder PJ, Price DA, Phillips RE. 1997. Antagonism of cytotoxic T lymphocyte-mediated lysis by natural HIV-1 altered peptide ligands requires simultaneous presentation of agonist and antagonist peptides. Eur. J. Immunol. 27:2323–2329 [DOI] [PubMed] [Google Scholar]
- 31. Yang OO, Sarkis PT, Ali A, Harlow JD, Brander C, Kalams SA, Walker BD. 2003. Determinant of HIV-1 mutational escape from cytotoxic T lymphocytes. J. Exp. Med. 197:1365–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martinez-Picado J, Prado JG, Fry EE, Pfafferott K, Leslie A, Chetty S, Thobakgale C, Honeyborne I, Crawford H, Matthews P, Pillay T, Rousseau C, Mullins JI, Brander C, Walker BD, Stuart DI, Kiepiela P, Goulder P. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 80:3617–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schneidewind A, Brockman MA, Sidney J, Wang YE, Chen H, Suscovich TJ, Li B, Adam RI, Allgaier RL, Mothe BR, Kuntzen T, Oniangue-Ndza C, Trocha A, Yu XG, Brander C, Sette A, Walker BD, Allen TM. 2008. Structural and functional constraints limit options for cytotoxic T-lymphocyte escape in the immunodominant HLA-B27-restricted epitope in human immunodeficiency virus type 1 capsid. J. Virol. 82:5594–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schneidewind A, Brockman MA, Yang R, Adam RI, Li B, Le Gall S, Rinaldo CR, Craggs SL, Allgaier RL, Power KA, Kuntzen T, Tung CS, LaBute MX, Mueller SM, Harrer T, McMichael AJ, Goulder PJ, Aiken C, Brander C, Kelleher AD, Allen TM. 2007. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 81:12382–12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boutwell CL, Rowley CF, Essex M. 2009. Reduced viral replication capacity of human immunodeficiency virus type 1 subtype C caused by cytotoxic-T-lymphocyte escape mutations in HLA-B57 epitopes of capsid protein. J. Virol. 83:2460–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Prado JG, Honeyborne I, Brierley I, Puertas MC, Martinez-Picado J, Goulder PJ. 2009. Functional consequences of human immunodeficiency virus escape from an HLA-B*13-restricted CD8+ T-cell epitope in p1 Gag protein. J. Virol. 83:1018–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Crawford H, Prado JG, Leslie A, Hue S, Honeyborne I, Reddy S, van der Stok M, Mncube Z, Brander C, Rousseau C, Mullins JI, Kaslow R, Goepfert P, Allen S, Hunter E, Mulenga J, Kiepiela P, Walker BD, Goulder PJ. 2007. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J. Virol. 81:8346–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wright JK, Naidoo VL, Brumme ZL, Prince JL, Claiborne DT, Goulder PJ, Brockman MA, Hunter E, Ndung'u T. 2012. Impact of HLA-B*81-associated mutations in HIV-1 Gag on viral replication capacity. J. Virol. 86:3193–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Streeck H, Lichterfeld M, Alter G, Meier A, Teigen N, Yassine-Diab B, Sidhu HK, Little S, Kelleher A, Routy JP, Rosenberg ES, Sekaly RP, Walker BD, Altfeld M. 2007. Recognition of a defined region within p24 gag by CD8+ T cells during primary human immunodeficiency virus type 1 infection in individuals expressing protective HLA class I alleles. J. Virol. 81:7725–7731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, Bishop K, van der Stok M, Nair K, Khan N, Crawford H, Payne R, Leslie A, Prado J, Prendergast A, Frater J, McCarthy N, Brander C, Learn GH, Nickle D, Rousseau C, Coovadia H, Mullins JI, Heckerman D, Walker BD, Goulder P. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46–53 [DOI] [PubMed] [Google Scholar]
- 41. Novitsky V, Gilbert P, Peter T, McLane MF, Gaolekwe S, Rybak N, Thior I, Ndung'u T, Marlink R, Lee TH, Essex M. 2003. Association between virus-specific T-cell responses and plasma viral load in human immunodeficiency virus type 1 subtype C infection. J. Virol. 77:882–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zuniga R, Lucchetti A, Galvan P, Sanchez S, Sanchez C, Hernandez A, Sanchez H, Frahm N, Linde CH, Hewitt HS, Hildebrand W, Altfeld M, Allen TM, Walker BD, Korber BT, Leitner T, Sanchez J, Brander C. 2006. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J. Virol. 80:3122–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Carlson JM, Listgarten J, Pfeifer N, Tan V, Kadie C, Walker BD, Ndung'u T, Shapiro R, Frater J, Brumme ZL, Goulder PJ, Heckerman D. 2012. Widespread impact of HLA restriction on immune control and escape pathways of HIV-1. J. Virol. 86:5230–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li B, Gladden AD, Altfeld M, Kaldor JM, Cooper DA, Kelleher AD, Allen TM. 2007. Rapid reversion of sequence polymorphisms dominates early human immunodeficiency virus type 1 evolution. J. Virol. 81:193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. 2009. X-ray structures of the hexameric building block of the HIV capsid. Cell 137:1282–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pornillos O, Ganser-Pornillos BK, Yeager M. 2011. Atomic-level modelling of the HIV capsid. Nature 469:424–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brumme ZL, John M, Carlson JM, Brumme CJ, Chan D, Brockman MA, Swenson LC, Tao I, Szeto S, Rosato P, Sela J, Kadie CM, Frahm N, Brander C, Haas DW, Riddler SA, Haubrich R, Walker BD, Harrigan PR, Heckerman D, Mallal S. 2009. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PLoS One 4:e6687 doi:10.1371/journal.pone.0006687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang YE, Li B, Carlson JM, Streeck H, Gladden AD, Goodman R, Schneidewind A, Power KA, Toth I, Frahm N, Alter G, Brander C, Carrington M, Walker BD, Altfeld M, Heckerman D, Allen TM. 2009. Protective HLA class I alleles that restrict acute-phase CD8+ T-cell responses are associated with viral escape mutations located in highly conserved regions of human immunodeficiency virus type 1. J. Virol. 83:1845–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kelly BN, Howard BR, Wang H, Robinson H, Sundquist WI, Hill CP. 2006. Implications for viral capsid assembly from crystal structures of HIV-1 Gag(1–278) and CA(N)(133–278). Biochemistry 45:11257–11266 [DOI] [PubMed] [Google Scholar]
- 51. Guerois R, Nielsen JE, Serrano L. 2002. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 320:369–387 [DOI] [PubMed] [Google Scholar]
- 52. Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439–1443 [DOI] [PubMed] [Google Scholar]
- 53. Boutwell CL, Essex M. 2007. Identification of HLA class I-associated amino acid polymorphisms in the HIV-1C proteome. AIDS Res. Hum. Retroviruses 23:165–174 [DOI] [PubMed] [Google Scholar]
- 54. Bhattacharya T, Daniels M, Heckerman D, Foley B, Frahm N, Kadie C, Carlson J, Yusim K, McMahon B, Gaschen B, Mallal S, Mullins JI, Nickle DC, Herbeck J, Rousseau C, Learn GH, Miura T, Brander C, Walker B, Korber B. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 315:1583–1586 [DOI] [PubMed] [Google Scholar]
- 55. Llano A, Frahm N, Brander C. 2009. How to optimally define optimal cytotoxic T lymphocyte epitopes in HIV infection?, p I-3–5 In Yusim K, Korber B, Brander C, Haynes B, Koup R, Moore JP, Walker B, Watkins D. (ed), HIV molecular immunology 2009, LA-UR 09-05941. Los Alamos National Laboratory, Theoretical Biology and Biophysics, Los Alamos, NM. [Google Scholar]
- 56. Carlson JM, Brumme CJ, Martin E, Listgarten J, Brockman MA, Le AQ, Chui C, Cotton LA, Knapp DJ, Riddler SA, Haubrich R, Nelson G, Pfeifer N, Deziel CE, Heckerman D, Apps R, Carrington M, Mallal S, Harrigan PR, John M, Brumme ZL. 2012. Correlates of protective cellular immunity revealed by analysis of population-level immune escape pathways in HIV-1. J. Virol. 86:13202–13216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Altfeld M, Allen TM, Kalife ET, Frahm N, Addo MM, Mothe BR, Rathod A, Reyor LL, Harlow J, Yu XG, Perkins B, Robinson LK, Sidney J, Alter G, Lichterfeld M, Sette A, Rosenberg ES, Goulder PJ, Brander C, Walker BD. 2005. The majority of currently circulating human immunodeficiency virus type 1 clade B viruses fail to prime cytotoxic T-lymphocyte responses against an otherwise immunodominant HLA-A2-restricted epitope: implications for vaccine design. J. Virol. 79:5000–5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Matthews PC, Koyanagi M, Kloverpris HN, Harndahl M, Stryhn A, Akahoshi T, Gatanaga H, Oka S, Molina CJ, Ponce HV, Avila Rios S, Cole D, Carlson J, Payne RP, Ogwu A, Bere A, Ndung'u T, Gounder K, Chen F, Riddell L, Luzzi G, Shapiro R, Brander C, Walker B, Sewell AK, Reyes Teran G, Heckerman D, Hunter E, Buus S, Takiguchi M, Goulder PJ. 2012. Differential clade-specific HLA-B*3501 association with HIV-1 disease outcome is linked to immunogenicity of a single Gag epitope. J. Virol. 86:12643–12654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lu J, Kuritzkes DR. 2001. A novel recombinant marker virus assay for comparing the relative fitness of HIV-1 reverse transcriptase variants. J. Acquir. Immune Defic. Syndr. 27:7–13 [DOI] [PubMed] [Google Scholar]
- 60. Deeks SG, Wrin T, Liegler T, Hoh R, Hayden M, Barbour JD, Hellmann NS, Petropoulos CJ, McCune JM, Hellerstein MK, Grant RM. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472–480 [DOI] [PubMed] [Google Scholar]
- 61. Nijhuis M, Deeks S, Boucher C. 2001. Implications of antiretroviral resistance on viral fitness. Curr. Opin. Infect. Dis. 14:23–28 [DOI] [PubMed] [Google Scholar]
- 62. Wainberg MA, Hsu M, Gu Z, Borkow G, Parniak MA. 1996. Effectiveness of 3TC in HIV clinical trials may be due in part to the M184V substitution in 3TC-resistant HIV-1 reverse transcriptase. AIDS 10(Suppl 5):S3–S10 [DOI] [PubMed] [Google Scholar]
- 63. Brockman MA, Schneidewind A, Lahaie M, Schmidt A, Miura T, Desouza I, Ryvkin F, Derdeyn CA, Allen S, Hunter E, Mulenga J, Goepfert PA, Walker BD, Allen TM. 2007. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J. Virol. 81:12608–12618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Allen TM, Altfeld M, Geer SC, Kalife ET, Moore C, O'Sullivan KM, Desouza I, Feeney ME, Eldridge RL, Maier EL, Kaufmann DE, Lahaie MP, Reyor L, Tanzi G, Johnston MN, Brander C, Draenert R, Rockstroh JK, Jessen H, Rosenberg ES, Mallal SA, Walker BD. 2005. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J. Virol. 79:13239–13249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. U. S. A. 93:3099–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. 2000. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 301:491–511 [DOI] [PubMed] [Google Scholar]
- 67. Fossen T, Wray V, Bruns K, Rachmat J, Henklein P, Tessmer U, Maczurek A, Klinger P, Schubert U. 2005. Solution structure of the human immunodeficiency virus type 1 p6 protein. J. Biol. Chem. 280:42515–42527 [DOI] [PubMed] [Google Scholar]
- 68. Chopera DR, Woodman Z, Mlisana K, Mlotshwa M, Martin DP, Seoighe C, Treurnicht F, de Rosa DA, Hide W, Karim SA, Gray CM, Williamson C. 2008. Transmission of HIV-1 CTL escape variants provides HLA-mismatched recipients with a survival advantage. PLoS Pathog. 4:e1000033 doi:10.1371/journal.ppat.1000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Matthews PC, Prendergast A, Leslie A, Crawford H, Payne R, Rousseau C, Rolland M, Honeyborne I, Carlson J, Kadie C, Brander C, Bishop K, Mlotshwa N, Mullins JI, Coovadia H, Ndung'u T, Walker BD, Heckerman D, Goulder PJ. 2008. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J. Virol. 82:8548–8559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Goepfert PA, Lumm W, Farmer P, Matthews P, Prendergast A, Carlson JM, Derdeyn CA, Tang J, Kaslow RA, Bansal A, Yusim K, Heckerman D, Mulenga J, Allen S, Goulder PJ, Hunter E. 2008. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J. Exp. Med. 205:1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kawashima Y, Pfafferott K, Frater J, Matthews P, Payne R, Addo M, Gatanaga H, Fujiwara M, Hachiya A, Koizumi H, Kuse N, Oka S, Duda A, Prendergast A, Crawford H, Leslie A, Brumme Z, Brumme C, Allen T, Brander C, Kaslow R, Tang J, Hunter E, Allen S, Mulenga J, Branch S, Roach T, John M, Mallal S, Ogwu A, Shapiro R, Prado JG, Fidler S, Weber J, Pybus OG, Klenerman P, Ndung'u T, Phillips R, Heckerman D, Harrigan PR, Walker BD, Takiguchi M, Goulder P. 2009. Adaptation of HIV-1 to human leukocyte antigen class I. Nature 458:641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schellens IM, Navis M, van Deutekom HW, Boeser-Nunnink B, Berkhout B, Kootstra N, Miedema F, Kesmir C, Schuitemaker H, van Baarle D, Borghans JA. 2011. Loss of HIV-1-derived cytotoxic T lymphocyte epitopes restricted by protective HLA-B alleles during the HIV-1 epidemic. AIDS 25:1691–1700 [DOI] [PubMed] [Google Scholar]
- 73. Brockman MA, Brumme ZL, Brumme CJ, Miura T, Sela J, Rosato PC, Kadie CM, Carlson JM, Markle TJ, Streeck H, Kelleher AD, Markowitz M, Jessen H, Rosenberg E, Altfeld M, Harrigan PR, Heckerman D, Walker BD, Allen TM. 2010. Early selection in Gag by protective HLA alleles contributes to reduced HIV-1 replication capacity that may be largely compensated for in chronic infection. J. Virol. 84:11937–11949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Altfeld M, Kalife ET, Qi Y, Streeck H, Lichterfeld M, Johnston MN, Burgett N, Swartz ME, Yang A, Alter G, Yu XG, Meier A, Rockstroh JK, Allen TM, Jessen H, Rosenberg ES, Carrington M, Walker BD. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8(+) T cell response against HIV-1. PLoS Med. 3:e403 doi:10.1371/journal.pmed.0030403 [DOI] [PMC free article] [PubMed] [Google Scholar]