Abstract
The Kaposi's sarcoma-associated herpesvirus (KSHV) open reading frame 57 (ORF57)-encoded protein (Mta) is a multifunctional regulator of viral gene expression. ORF57 is essential for viral replication, so elucidation of its molecular mechanisms is important for understanding KSHV infection. ORF57 has been implicated in nearly every aspect of viral gene expression, including transcription, RNA stability, splicing, export, and translation. Here we demonstrate that ORF57 interacts with the KSHV K-bZIP protein in vitro and in cell extracts from lytically reactivated infected cells. To further test the biological relevance of the interaction, we performed a chromatin immunoprecipitation and microarray (ChIP-chip) analysis using anti-ORF57 antibodies and a KSHV tiling array. The results revealed four specific areas of enrichment, including the ORF4 and K8 (K-bZIP) promoters, as well as oriLyt, all of which interact with K-bZIP. In addition, ORF57 associated with DNA corresponding to the PAN RNA transcribed region, a known posttranscriptional target of ORF57. All of the peaks were RNase insensitive, demonstrating that ORF57 association with the viral genome is unlikely to be mediated exclusively by an RNA tether. Our data demonstrate that ORF57 associates with the viral genome by using at least two modes of recruitment, and they suggest that ORF57 and K-bZIP coregulate viral gene expression during lytic infection.
INTRODUCTION
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) is an oncogenic double-stranded DNA virus that causes KS, as well as the lymphoproliferative disorders primary effusion lymphoma (PEL) and multicentric Castleman's disease (1–4). Like the life cycle of all herpesviruses, that of KSHV includes both latent and lytic phases. During latency, viral replication is absent and few viral genes are expressed (5–7). Entry into the lytic phase launches a complex, temporally regulated pattern of viral gene expression that ultimately produces infectious virions. While the mechanism of viral transformation remains obscure, the expression of both latent- and lytic-phase genes has been implicated in viral pathogenesis and oncogenesis (1–4, 8). Thus, an understanding of the mechanisms that control KSHV gene expression is essential to understanding both the life cycle and the pathogenesis of KSHV.
In order to achieve the intricate regulation of gene expression necessary for viral replication and survival in its host, KSHV encodes factors that modulate gene expression at nearly every level (9–14). One such regulator of gene expression is the multifunctional 51-kDa open reading frame 57 (ORF57)-encoded protein Mta (KS-SM). ORF57 encodes a member of a family of proteins conserved throughout the Herpesviridae, and it is essential for KSHV replication (15–21). ORF57 has been reported to affect nearly every posttranscriptional stage of gene expression, including RNA stability, pre-mRNA splicing, RNA export, and translation (11, 13, 16, 22). Consistent with its role as a posttranscriptional regulator, ORF57 binds RNA directly (23–27). ORF57 binds specific RNA sequences preferentially, and this interaction promotes ORF57-mediated regulation of the transcript. However, ORF57 also binds and stimulates the expression of many reporter transcripts with no biological relevance for the host or virus, so it likely functions in a nonspecific fashion as well.
In addition to its posttranscriptional activities, several observations suggest that ORF57 regulates viral transcription. ORF57 interacts with the KSHV lytic transactivator Rta (replication and transcription activator; ORF50) (28–30), a transcription factor that is both necessary and sufficient to drive lytic reactivation of the virus (10, 31). Furthermore, viral gene expression from Rta-responsive promoters is synergistically upregulated in the presence of ORF57, and the effect is cell and promoter specific (28, 29, 32). ORF57 contains putative AT-hook and leucine zipper domains (16, 28, 33), both of which are commonly found in transcription factors. Indeed, ORF57 associates with DNA, as determined by gel shift and chromatin immunoprecipitation (ChIP) assays (28, 29). However, how these activities relate to ORF57 function during viral reactivation is unknown, as are the mechanistic details of ORF57 interactions with genes and transcription factors.
K-bZIP is a leucine zipper-containing protein that is a positional homolog of the Epstein-Barr virus (EBV) Zta (ZEBRA, BZLF1) protein (14). While Zta is involved in EBV DNA replication and the transcriptional activation of specific viral genes, the role of K-bZIP in KSHV lytic reactivation is less clear (14). Two independent studies showed that K-bZIP is not essential for lytic reactivation in KSHV BACmid systems (34, 35), but it was reported to be essential for virus production in infected PEL cells (36). K-bZIP associates with oriLyt (37–40) and is essential for oriLyt-dependent DNA replication in a plasmid-based system (41), although its absence can be complemented by high levels of Rta expression (38). In addition, K-bZIP appears to act at an early stage of viral replication, perhaps by modulating the activities of the latency protein LANA (38) or by promoting DNA replication early in latent viral infection (35). Additionally, K-bZIP interacts with Rta and represses transcription from some Rta-dependent viral promoters (42, 43) and from the cellular beta interferon promoter, suggesting a role as an immune response modulator (44). In addition to its role as a transcription repressor, K-bZIP activates the transcription of specific KSHV (45) and cellular (40) promoters. Thus, K-bZIP is a multifunctional KSHV protein involved in viral DNA replication and transcriptional control.
A high-throughput approach identified K-bZIP as an ORF57-interacting protein (30), but no validation of this putative interaction has been reported. Here we show that ORF57 and K-bZIP interact in cell extracts from both transfected cells and lytically reactivated PEL cells. Furthermore, we show that ORF57 and K-bZIP interact in vitro and identify a minimal region in ORF57 necessary for this binding. Using a ChIP and tiling microarray approach (ChIP-chip), we demonstrate that ORF57 binds to the KSHV genome at several of the same sites as K-bZIP during lytic reactivation. In addition to these regions, the ORF57 ChIP-chip experiments show interactions with the PAN RNA transcribed region, which encodes a noncoding RNA that binds ORF57. Interestingly, all of the interactions between ORF57 and the KSHV genome are insensitive to RNase, demonstrating that while RNA may recruit ORF57 to the PAN RNA gene, ORF57 makes additional contacts with protein factors at the genome. Taken together, our data support the conclusions that ORF57 interacts with K-bZIP and that ORF57 is recruited to the viral genome during lytic reactivation by at least two distinct mechanisms. We propose that one mechanism results from ORF57 binding to nascent transcripts and association with the transcription machinery. In the alternative mechanism, the viral transcription factor K-bZIP recruits ORF57 to specific viral promoters.
MATERIALS AND METHODS
Cell culture, transfection, and Northern blotting.
TREx BCBL1-Rta cells (46) were passaged in RPMI 1640 medium (Sigma) supplemented with 10% tetracycline-free fetal bovine serum (Clontech), penicillin-streptomycin (Sigma), 2 mM l-glutamate, and 100 μg/ml hygromycin (Sigma). Lytic reactivation was achieved by adding 1 μg/ml doxycycline to the medium. HEK293 cells were grown and transfected as previously described (27).
Plasmids.
For bacterial expression of glutathione S-transferase (GST) and GST–K-bZIP, the parallel series of vectors was used (47). The K-bZIP coding region was transferred from pcK8 (gift of Pinghui Feng, University of Southern California) into a GST-coupled parallel expression vector by using EcoRI and XhoI with standard molecular biology techniques. To generate an intronless version of ORF57, we amplified a cDNA version of the 5′ end of the ORF57 mRNA by using primers NC497 (5′ TGGGATTCCCTGGGCCGACT 3′) and NC495 (5′ ATTAGCGGATTCATGGTACAAGCAATGATAGAC 3′). This fragment was inserted into pcFl-ORF57II (27) by using BamHI and KpnI digestion to generate pcFl-ORF57IIΔi. The first 52 amino acids of ORF57 were deleted from pcFl-ORF57 to generate pcFl-ORF57IIΔ1-52 by using gene splicing by overlap extension PCR (48).
Microarray design.
With Roche NimbleGen, we developed a tiling microarray that covers the 138-kbp KSHV unique sequence using U75698 as a reference. The mean and median probe spacing was 14 bp, and the probe sizes ranged from 50 to 70 bp with a mean size of 51 bp. Repeat sequences were excluded, which generated a few apparent gaps in the data. For example, inspection of the ChIP-chip data around the oriLyt/PAN RNA locus shows three gaps, two in the oriLyt region and one immediately following the PAN RNA gene. All probes were unique to the KSHV genome and were checked against the human genome to minimize the possibility of cross-hybridization with human DNA. After these exclusions, 133,780 bp of the 137,508-bp KSHV genome was covered by 8,838 probes, all of which were present in both the forward and reverse orientations. The array had the probe set printed three times, which controls for random variation during the hybridization and washing steps. While only one replicate for each experiment is shown, complete data sets for the experiments, including biological and independent printing replicates, were evaluated during the manuscript review process.
ChIP.
For each sample, 5 × 106 TREx BCBL1-Rta cells in 20 ml medium were induced by adding 1 μg/ml doxycycline to the medium for 18 to 22 h. ChIP was performed as previously described (27), except that 12 μg of an affinity-purified polyclonal ORF57 antibody or Ig purified from serum from the same rabbit taken prior to inoculation (prebleeding) was used for immunoprecipitation (24). For the RNase experiments, the pellets were washed once with radioimmunoprecipitation assay buffer, equilibrated in TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA), and incubated for 30 min at room temperature with TE plus 0.1 mg/ml RNase A. Subsequently, the standard ChIP wash steps were performed (27). We also performed an experiment in which the RNase was added to the lysates during immunoprecipitation and obtained similar results (data not shown). To verify the RNase activity, the samples were removed at the steps in the procedure described in the legend to Fig. 5D. To harvest the RNA, we treated the samples with proteinase K for 60 min at 37°C and reversed the cross-links at 65°C for only 45 min to minimize hydrolysis. Northern blot assays were performed by using standard protocols, and 5′-end labeling was performed with T4 polynucleotide kinase (PNK) by using standard protocols. We note that the RNA was not decapped prior to PNK labeling, but we reasoned that this is suitable because our goal was to monitor RNA decay and not specifically examine mRNAs.
Fig 5.
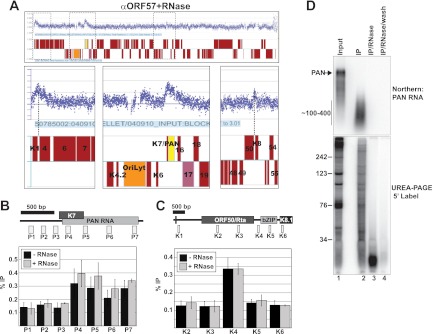
ORF57 association with KSHV genes is RNase insensitive. (A) ChIP-chip data from RNase-treated samples. Details are as described in the legend to Fig. 3. An independent biological replicate yielded the same results (B, C) ChIP analysis of the RNase-treated samples; the details are as described in the legend to Fig. 4. The untreated samples (black) are the same as those in Fig. 2 and are displayed here for comparison to the RNase-treated samples (gray). (D) Verification of efficient RNase treatment. RNA was harvested from a chip experiment before immunoprecipitation (Input, 1%), after immunoprecipitation (IP), immediately following RNase treatment, or after RNase treatment and bead washing. The samples were analyzed by Northern blot assay for PAN RNA (top) or by 5′-end radiolabeling and urea-PAGE (bottom). The mobility of the DNA markers (in nucleotides) is shown on the left. The distinct bands in the input lanes observed in the urea-PAGE are likely abundant small RNAs (e.g., tRNAs, snRNAs, etc.).
For the ChIP-chip assays, the DNA was amplified using the GenomePlex Complete WGA kit (WGA2; Sigma). In each case, we verified that the amplification was in the linear range by removing aliquots in the final two cycles of the PCR and visually inspecting the amplified DNA by agarose gel electrophoresis. If the amount of DNA did not change, we repeated the procedure with fewer cycles. However, if the DNA content doubled, we concluded that the PCR was still in the linear range. Generally, we used 13 or 14 cycles of amplification. WGA2-amplified input and pellet DNA was hybridized to the KSHV microarray, detected, and quantified by technicians at Roche-NimbleGen. For our confirmatory studies, we used real-time PCR for detection and quantification.
Real-time PCR.
Real-time PCR was performed by using iTAQ Fast SYBR green Supermix with ROX (Bio-Rad) with a final primer concentration of 100 nM. The conditions were 40 cycles of 95°C for 3 s and 60°C for 30 s with a 7500 Fast real-time PCR system (Applied Biosystems). Primers and amplification efficiencies (49) are given in Table 1.
Table 1.
Primers used in this study
| Primer use and amplicon or construct | Name | Orientation | Efficiency | Sequence (5′ to 3′) |
|---|---|---|---|---|
| Quantitative PCR | ||||
| K1 | NC710 | Forward | 1.89 | CAGAGTTCGCCAAGCTCTACGT |
| K1 | NC711 | Reverse | 1.89 | TTTGGTCAAGTACACCGAACACTTAA |
| K2 | NC712 | Forward | 1.89 | ACCGGTCTACCTTCCGAGGAT |
| K2 | NC713 | Reverse | 1.89 | GGCGAGCTGACCCTTGGT |
| K3 | NC714 | Forward | 1.90 | AGGTGTGCCGTGTAGAGATTCAA |
| K3 | NC715 | Reverse | 1.90 | TCTGCTTTCGTTTGGGTGTTG |
| K4 | NC716 | Forward | 1.90 | CGAATCTGGTTGATTGTGACTATTTG |
| K4 | NC717 | Reverse | 1.90 | TTGGCAGGGTTACACGTTTACAC |
| K5 | NC718 | Forward | 1.92 | GCTTCCAACTCGCAGATCCA |
| K5 | NC719 | Reverse | 1.92 | CCCTGTTTGGCCTTAGTGCAT |
| K6 | NC720 | Forward | 1.91 | GGCGAGATTGGAGGCACATA |
| K6 | NC721 | Reverse | 1.91 | GCTGGCTGCTGGCACATT |
| P1 | NC698 | Forward | 1.86 | CACGTCTTCGGCCAATGC |
| P1 | NC699 | Reverse | 1.86 | TTGTCGGACTCGTCTACCTATCAGT |
| P2 | NC700 | Forward | 1.90 | CCGACTGGTTGCGGAAGTATT |
| P2 | NC701 | Reverse | 1.90 | ACCCCCCGCCCAGTTATT |
| P3 | NC722 | Forward | 1.65 | TTGAAGGATGATGTTAATGACATAAAGG |
| P3 | NC723 | Reverse | 1.65 | ATCTCCAGTGTTCCCATATTTTGG |
| P4 | NC702 | Forward | 1.93 | GCTCGCTGCTTGCCTTCTT |
| P4 | NC703 | Reverse | 1.93 | CCAAAAGCGACGCAATCAA |
| P5 | NC704 | Forward | 1.92 | CTTGCGGGTTATTGCATTGG |
| P5 | NC705 | Reverse | 1.92 | GACACGTTAAGTATCCTCGCATATCA |
| P6 | NC706 | Forward | 1.81 | TTTTCCAGTGTAAGCAAGTCGATTT |
| P6 | NC707 | Reverse | 1.81 | TGTTCTTACACGACTTTGAAACTTCTG |
| P7 | NC708 | Forward | 1.85 | TTAACGTGCCTAGAGCTCAAATTAAAC |
| P7 | NC709 | Reverse | 1.85 | TTGACCTTTATTTATGTTGTAAGTTGCATTA |
| Generation of in vitro translation templates | ||||
| FL | NC487 | Forward | ATATAAGCAGAGCTCTCTGG | |
| FL | NC496 | Reverse | AAAAGGCTCGAGTTAAGAAAGTGGATAAAAGAATAAACCC | |
| 1-370 | NC861 | Reverse | TTAGAAGTTAAGCTTGACCTCGCC | |
| 1-341 | NC862 | Reverse | TTATGCTAGCTCGCTCTGCTTGTC | |
| 1-300 | NC956 | Reverse | TTACCCGTGGCGCACAACCGTCTG | |
| 1-260 | NC957 | Reverse | TTAGCGGCTGGTCACAAAAGACAG | |
| 1-220 | NC863 | Reverse | TTACCTGTCCGTAAACACCTCCGG | |
| 1-157 | NC864 | Reverse | TTACTGGCAATTGACCTGCGG | |
| 96-455 | NC958 | Forward | TAATACGACTCACTATAGGGAGACCCAAGCTGCCACCATGGACTACAAGGACGACGATGACAAGTCACCAGTAAACAGGTACGGT | |
| 152-455 | NC873 | Forward | TAATACGACTCACTATAGGGAGACCCAAGCTGCCACCATGGACTACAAGGACGACGATGACAAGCCGCAGGTCAATTGCCAG |
Recombinant and in vitro translated proteins.
Recombinant GST and GST–K-bZIP were expressed in Rosetta (DE3) pLysS cells (Novagen). Transformed bacteria were grown to mid-log phase in Luria broth containing 15 μg/ml chloramphenicol and 100 μg/ml ampicillin. Isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added to 1 mM, and induction was allowed to proceed for 2 h at 30°C. Bacteria were pelleted, frozen at −80°C, and lysed in lysis buffer (150 mM NaCl, 50 mM Tris HCl [pH 7.5], 1% Triton X-100) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF), 1× protease inhibitors (cocktail V; Calbiochem), and 1 mg/ml lysozyme for 30 min on ice. Subsequently, the lysate was sonicated and clarified by centrifugation for 15 min at 10,000 × g at 4°C. The clarified extract was bound to glutathione Sepharose beads (Pierce) in batch, and the beads were washed extensively with ice-cold phosphate-buffered saline (PBS) supplemented with 1% Triton X-100. Bound proteins were eluted in 50 mM Tris HCl (pH 8.0)–10 mM glutathione–10% glycerol. Elution buffer was replaced with exchange buffer (20 mM Tris HCl [pH 8.0], 1 mM dithiothreitol [DTT], 10% glycerol, 50 mM NaCl) by using an Amicon Ultra-4 centrifugal filter unit (Millipore) with a molecular mass cutoff of 3,000 Da.
In vitro-translated proteins were generated by using the TNT T7 Coupled Reticulocyte Lysate System (Promega) according to the manufacturer's instructions. The products were radiolabeled by the incorporation of radiolabeled [35S]methionine. DNA fragments used for coupled transcription-translation reactions were generated by PCR using pcFl-ORF57IIΔi as a PCR template, except for fragments 96-260Δ181-208 and 53-455, which used pcFl-ORF57IIΔReBD (50) and pcFl-ORF57Δ1-52 as PCR templates, respectively. The sequences of the primers used are given in Table 1. All of the proteins generated that include the natural translation start site were generated by using primer NC487, and those with the natural termination codon used NC496 as a reverse primer. Each of proteins produced contains an N-terminal Flag tag and Kozak consensus sequences, which were derived from pcFl-ORF57Δi or from the primers used for amplification (NC958, NC873).
GST–K-bZIP pull-down reactions.
For binding reactions, 12 μg of GST or GST-K8 (final concentrations, ∼10 and 5 μM, respectively) in exchange buffer was mixed with 10 μl in vitro-translated ORF57 fragment in binding buffer (20 mM Tris HCl [pH 7.5], 100 mM NaCl, 0.1% Igepal, 1 mM DTT) supplemented with 1 mM PMSF, 1× protease inhibitors (cocktail V; Calbiochem), and 0.1 mg/ml RNase A in a total volume of 50 μl. The reaction mixtures were incubated for 15 min at 30°C and then added to glutathione beads for 1 h of incubation at room temperature. The beads were washed five times in 1 ml binding buffer. Input and bound fractions were subjected to SDS-PAGE and detected with a PhosphorImager (Molecular Dynamics). For quantification, the signals from the bound samples were divided by the signals from the inputs and expressed relative to a full-length ORF57 sample, which was repeated in every binding experiment to control for experimental variation.
Antibodies.
The ORF57 antibodies used were previously described (24). Prebleed antibodies were derived from serum from the same rabbit that generated the ORF57 antibodies but were taken prior to inoculation. The prebleed antibodies were purified from serum by using protein A agarose (Pierce). Anti-Flag antibodies and agarose were obtained from Sigma. K-bZIP polyclonal antibodies were a generous gift from Pinghui Feng (University of Southern California).
Coimmunoprecipitation experiments.
For coimmunoprecipitation experiments, HEK293 cell transfections were conducted with 60-mm dishes and a total of 5 μg of plasmid DNA. For the BCBL1 experiments, a total of 5 × 106 TREx BCBL1-Rta cells per condition were collected at 18 to 20 h postinduction and washed in 10 ml of ice-cold PBS (Sigma) prior to use.
Cells were collected by centrifugation at 700 × g for 3 min at 4°C and then lysed by incubation on ice for 5 min in RSB100-I buffer (100 mM NaCl, 2.5 mM MgCl2, 0.5% Igepal, 10 mM Tris [pH 7.5]) plus 0.1 mg/ml RNase A, protease inhibitors (cocktail V; Calbiochem), and 1 mM PMSF (Sigma). Lysate was clarified by centrifugation at 16,000 × g for 10 min at 4°C. For HEK293 experiments, the soluble protein extracts were incubated with 20 μl of washed anti-Flag M2 affinity beads (Sigma) for a total of 2 h at 4°C. Immunoprecipitation from TREx BCBL1-Rta cell lysates was carried out by nutation for 1 h with antibody and then for an additional hour with washed protein A agarose. Beads were washed five times with 500 μl RSB100-I buffer. Proteins were eluted by boiling in SDS gel loading buffer and analyzed by 10 or 12% SDS-PAGE.
Western blot assays.
Western blot assays were performed by using standard procedures, except for the TREx BCBL1-Rta cell immunoprecipitations, which utilized the Clean Blot IP detection reagent in accordance with the manufacturer's instructions.
RESULTS
ORF57 coimmunoprecipitates with K-bZIP.
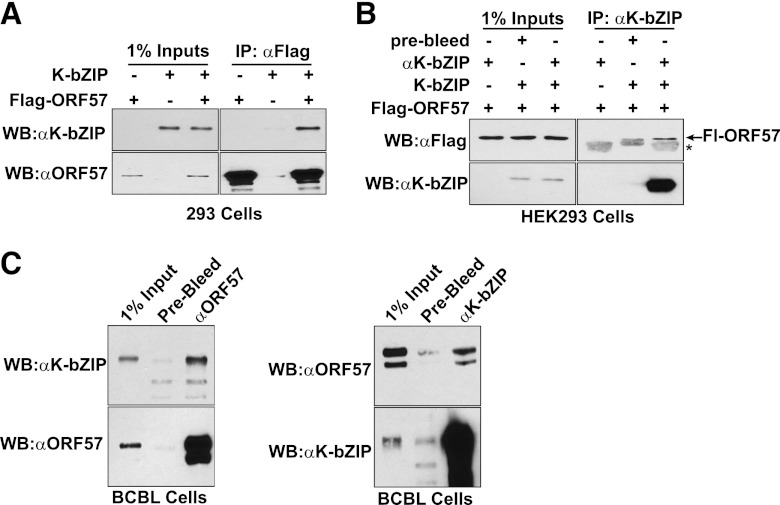
High-throughput screening showed that ORF57 interacts with K-bZIP in a yeast two-hybrid assay (30). To validate this interaction in a biologically relevant context, we performed coimmunoprecipitation experiments. Initially, we transfected HEK293 cells with Flag-tagged ORF57 (Fl-ORF57) and a K-bZIP expression vector. We immunoprecipitated Fl-ORF57 with immobilized anti-Flag antibodies and observed coimmunoprecipitation of K-bZIP (Fig. 1A). As expected, no coimmunoprecipitation was observed when Fl-ORF57 was not coexpressed. In addition, we performed the reciprocal coimmunoprecipitation using an anti-K-bZIP antibody (Fig. 1B). Fl-ORF57 was immunoprecipitated by these antibodies but only when K-bZIP was expressed, further supporting an interaction between K-bZIP and ORF57. We extended the coimmunoprecipitation experiments by assaying coimmunoprecipitation from extracts generated from a lytically reactivated PEL cell line (46). Immunoprecipitation with affinity-purified anti-ORF57 polyclonal antibodies led to coimmunoprecipitation of K-bZIP (Fig. 1C, left). Conversely, ORF57 coimmunoprecipitated with K-bZIP antibodies (Fig. 1C, right). Importantly, minimal coimmunoprecipitation was observed with prebleed control antibodies. These data demonstrate that ORF57 and K-bZIP interact in cell lysate and imply an in vivo interaction between these two regulators of KSHV gene expression.
Fig 1.
ORF57 and K-bZIP coimmunoprecipitate. (A) Cell extracts from HEK293 cells transfected with Fl-ORF57 and/or K-bZIP were precipitated with anti-Flag antibody. The immunoprecipitated proteins were assayed by Western blotting with either K-bZIP (top) or ORF57 (bottom) antibodies. As negative controls, we immunoprecipitated by using extracts from cells not expressing Fl-ORF57 or K-bZIP, as indicated. The panels on the left are Western blot assays of 1% of the total lysate used for immunoprecipitation. (B) Immunoprecipitations were performed as in panel A, except that anti-K-bZIP antibodies were used. Proteins were detected by Western blot (WB) assay with anti-Flag (top) or anti-K-bZIP (bottom) antibodies. We substituted mouse IgG for the anti-K-bZIP antibody in the no-antibody control. The asterisk marks cross-reaction with the antibody heavy chain. (C) Immunoprecipitations (IP) were performed with extracts from lytically reactivated TREx BCBL1-Rta cells. Immunoprecipitation was performed with anti-ORF57 (left), anti-K-bZIP (right), or prebleed negative-control antibodies, as indicated. The total cell lysate used in each experiment is also shown (1% Input). Immunoprecipitated proteins were detected by anti-K-bZIP or anti-ORF57 antibodies, as indicated.
Amino acids 96 to 260 are sufficient for ORF57–K-bZIP interaction in vitro.
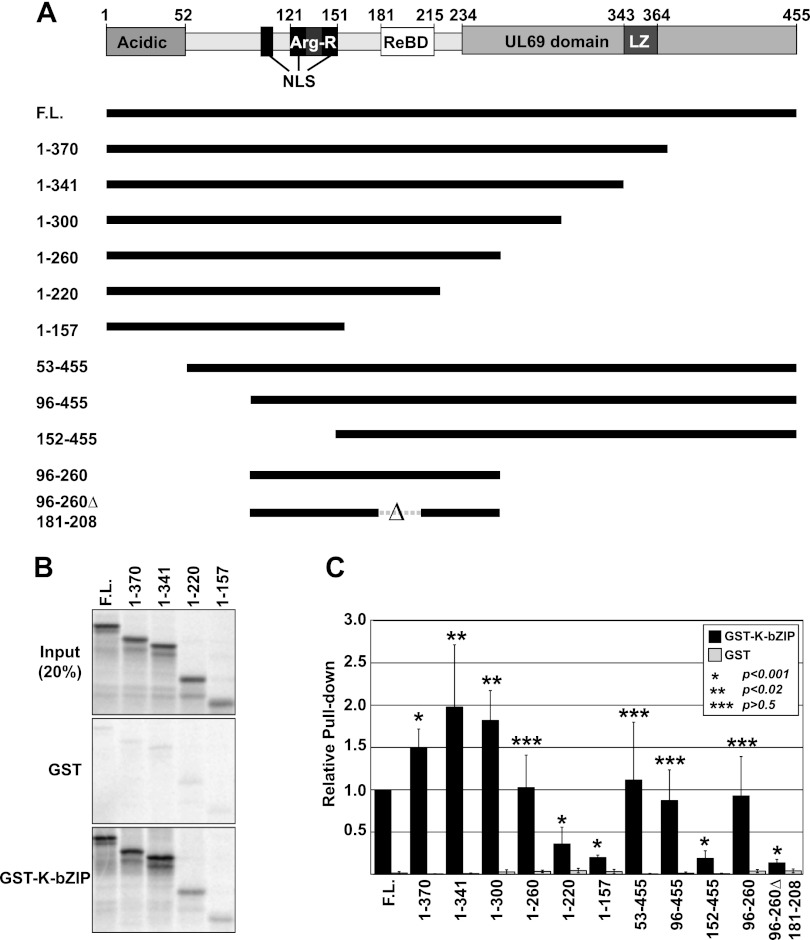
We next used an in vitro pulldown assay to define the amino acids in ORF57 necessary for interaction with K-bZIP. Figure 2A highlights several ORF57 protein domains, including three nuclear localization signals (NLS) (51), the REF/Aly interaction domain (ReBD) (52), an Arg-rich motif (Arg-R), a putative leucine zipper (LZ), and the C-terminal UL69 domain, a motif of unknown function conserved among ORF57 homologs. We expressed and purified the GST and GST–K-bZIP proteins and determined their ability to pull down radiolabeled, in vitro-translated fragments of ORF57. Representative results are shown in Fig. 2B. The top panel shows the proteins prior to binding (20% input), the middle and bottom panels show the bound fractions from GST alone and GST–K-bZIP, respectively. To quantify the results across several different experiments, each replicate included a full-length ORF57 control and the bound fraction of each fragment was determined relative to this control (Fig. 2C). Because both ORF57 and K-bZIP have putative leucine zipper motifs, we hypothesized that they interacted in this domain. To test this, we first compared two C-terminal deletions, 1 to 370 and 1 to 341, that maintain and delete the putative leucine zipper, respectively. Deletion of the leucine zipper motif did not decrease the binding of ORF57 to K-bZIP. In fact, both of these truncations resulted in a statistically significant increase in ORF57 binding to K-bZIP compared to full-length ORF57 (Fig. 2C). The reason for this increase is unknown, but it may result from more efficient protein folding when this region is lost or it may be due to a regulatory domain at the C terminus of the protein. We conclude that the interaction between ORF57 and K-bZIP is not the result of a direct interaction between the putative leucine zipper regions.
Fig 2.
ORF57 amino acids 96 to 260 are sufficient for K-bZIP interaction in vitro. (A, top) Diagram of ORF57 highlighting several domains, including an acidic N terminus, NLS (black boxes), an arginine-rich domain (Arg-R, overlaps two NLS), the REF/Aly binding domain (ReBD), a predicted leucine zipper (LZ), and a C-terminal UL-69 domain. (A, bottom) ORF57 protein derivatives used in GST–K-bZIP pulldown experiments. (B) Representative data from a pulldown experiment. The top panel shows 20% of the input in vitro-translated 35S-labeled ORF57 fragments. The middle panel shows fragments bound to GST, while the bottom panel shows the fragments bound to GST–K-bZIP. (C) Quantification of pull-down data for GST–K-bZIP (black) and GST alone (gray). First, pellet-to-input ratios were determined and then each value was normalized to the full-length control from the same experiment. Bar graphs show average values, and error bars represent standard deviations. Each fragment was tested a minimum of three times but up to seven times. P values are shown for each data set relative to the full-length ORF57. P values were determined with a two-tailed unpaired Student t test.
To determine which region is necessary for the interaction, we tested additional truncations of the C terminus of ORF57 (Fig. 2A). The first 300 amino acids of ORF57 bound to GST–K-bZIP slightly more efficiently than the wild type, and the first 260 amino acids were indistinguishable from those of the wild type (P > 0.5) (Fig. 2C). In contrast, proteins consisting of the N-terminal 220 or the N-terminal 157 amino acids bound significantly less efficiently to GST–K-bZIP (Fig. 2C). Three truncations of the N terminus that delete the first 52, 95, and 151 amino acids were also tested. Among these truncations, only deletion of the first 151 amino acids produced a significant decrease in association with K-bZIP. We then tested the binding of minimal ORF57 amino acids 96 to 260, and this region binds K-bZIP with an efficiency similar to that of the full-length protein. Finally, we tested an ORF57 fragment that deletes the REF/Aly interaction, ΔReBD (50), in the context of the minimal binding fragment (Fig. 2A, 96-260Δ181-208). This deletion reduced binding to near background levels, demonstrating that the amino acids between 181 and 208 contribute to the ORF57–K-bZIP interaction. In addition, this observation demonstrates that the Arg-R region is not sufficient for binding, thereby ruling out the hypothesis that the ORF57 interaction with K-bZIP is due exclusively to a highly charged basic patch in the protein. Interestingly, coimmunoprecipitation assays showed that Flag-ORF57ΔReBD maintained its interaction with K-bZIP (data not shown). Therefore, regions of full-length ORF57 outside amino acids 96 to 260 may contribute to its interaction with K-bZIP in vivo. Alternatively, cellular proteins not present in the in vitro assay may contribute to K-bZIP-ORF57 binding in vivo. Regardless, we can conclude that ORF57 amino acids 96 to 260 are sufficient for the interaction with K-bZIP in vitro. Taken with the coimmunoprecipitation experiments, our results strongly support the idea that ORF57 and K-bZIP bind each other directly and specifically and they suggest that the proteins interact in the context of viral reactivation.
ChIP-chip analysis reveals ORF57 association with transcribed regions and promoters.
A ChIP-chip assay with K-bZIP antibodies revealed that the genomic regions with the highest occupancy of K-bZIP were promoters for ORF4, K8, ORF57, ORF75, and K15 (45). In addition, K-bZIP binds oriLyt (37–40). If ORF57 interacts with K-bZIP in the context of viral replication, we reasoned that the interaction may occur at one or more of these promoters. To test this idea, we performed ChIP-chip analysis with ORF57 antibodies. We induced TREx BCBL1-Rta cells (46) with doxycycline for ∼20 h. This cell line is latently infected by KSHV but has a copy of the gene for Rta under the control of a tetracycline-responsive promoter stably integrated into the host genome. Addition of doxycycline to the medium induces Rta expression, resulting in KSHV lytic reactivation. We avoided the use of sodium butyrate for reactivation, as its deacetylase inhibition activity could alter epigenetic marks and thereby alter transcription factor association with the viral genome. We performed ChIP-chip with affinity-purified anti-ORF57 antibodies or with prebleed control antibodies (24). Input DNA and pellet samples were used for hybridization to a tiling array that covers the 138-kb KSHV genome, excluding the terminal repeats. The probes are ∼50- to 70-base overlapping oligonucleotides specific for the KSHV genome with an average offset of 14 bp (see Materials and Methods). A trace of the pellet/input ratios for one such experiment is shown in Fig. 3A. While the prebleed control showed no consistent peaks, four peaks were reproducibly observed with the ORF57 antibodies in three biological replicates (Fig. 3A and data not shown). Proceeding from left to right on the genome, the first peak was observed near the genes encoding ORF K1 and ORF4. The center of the peak aligned very closely with the small promoter region immediately downstream of the K1 gene that drives ORF4 expression (Fig. 3B, dotted line). The second observed peak is disrupted by a repeat sequence that was absent from the tiling array, generating the observed gaps in the histogram. There are no coding genes in this region, but the lytic origin of replication is in this area (orange, Fig. 3C). The third peak was distributed over the transcribed region of PAN RNA (yellow, Fig. 3C), and the fourth peak was centered at the K8 promoter immediately downstream of the ORF50 coding region (Fig. 3D). Thus, three of our observed ORF57 peaks, the ORF4 promoter, the K8 promoter, and oriLyt, correlate with K-bZIP binding sites. These data support the hypothesis that ORF57 interacts with K-bZIP in specific regions of the KSHV genome in lytically reactivated cells.
Fig 3.
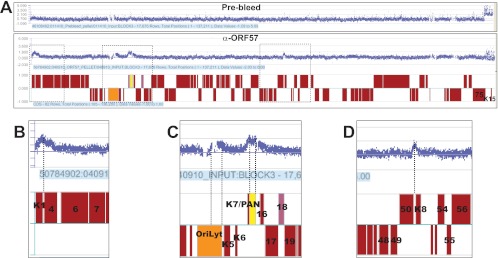
ChIP-chip analysis with ORF57. (A) Traces from a ChIP-chip analysis with antibodies against ORF57 or a prebleed control. TREx BCBL1-Rta cells were induced with doxycycline for 20 h, and ChIP-chip was performed with DNA immunoprecipitated by anti-ORF57 antibodies (bottom) or a prebleed control (top). The red blocks below the data represent the ORFs in the KSHV genome using the GenBank entry with accession no. U75698 as a reference. The upper ORFs are transcribed in the forward orientation (left to right as drawn), and the lower ORFs are in the reverse orientation. The positions of the oriLyt and PAN RNA gene are colored orange and yellow, respectively. Three independent experiments for the anti-ORF57 antibodies and two replicates of the prebleed antibodies yielded similar results (data not shown). (B to D) Enlargements of ChIP-chip peaks found at the ORF4 promoter, oriLyt, PAN RNA, and K8 promoter; the regions shown are boxed in panel A. Selected ORF numbers are shown as markers. The broken spaces in the data near the oriLyt region are due to repeated sequences excluded from the microarray probe set. Vertical dashed lines demonstrate the alignment between the peaks and the promoter regions of ORF4 (B) and K8 (D) or highlight the breadth of the peaks (C).
The observation that ORF57 binds to the PAN RNA transcribed region is consistent with data showing that ORF57 binds directly to PAN RNA and stabilizes the transcript (23–25, 27, 32). It is important to note that RNA-binding proteins that associate with nascent transcripts can often be detected by ChIP analysis (for example, see reference 53). Such interactions are observed because formaldehyde efficiently cross-links proteins with other proteins or nucleic acids (54). As a result, cross-linking networks between the nascent RNA, the RNA-binding protein, RNA polymerase II (pol II), and DNA may be detected by ChIP. Thus, our data show that ORF57 binds to PAN RNA prior to its release from the transcription site, most likely as the nascent transcript emerges from pol II.
Validation of ChIP-chip results with the PAN RNA transcribed region and K8 promoter.
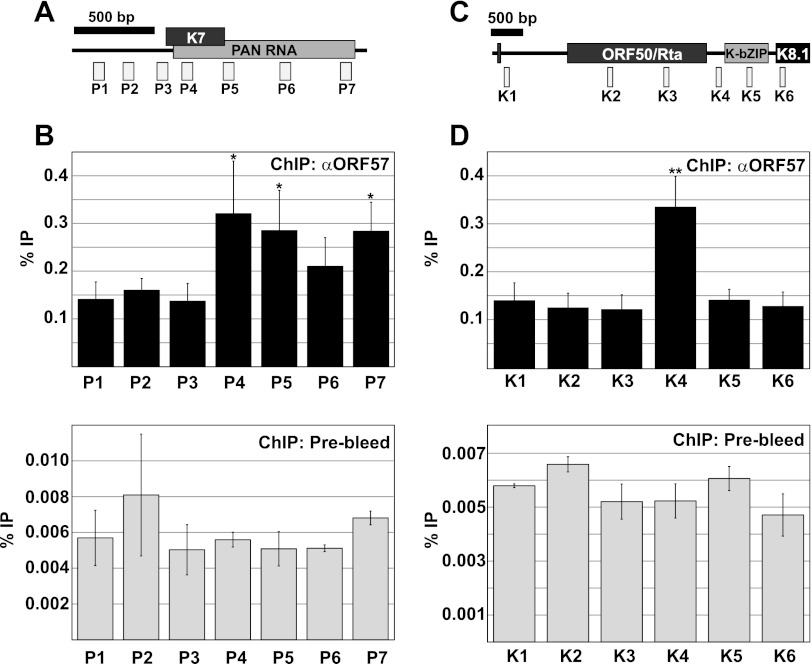
We next confirmed the ChIP-chip results by using a conventional ChIP assay with real-time PCR as the readout. To do this, we designed real-time PCR oligonucleotide sets across the PAN RNA and ORF50-K8 regions. We chose these two genes because they show different patterns of ORF57 binding to the locus. For PAN RNA, the ChIP peak was seen broadly across the transcribed region whereas the K8 peak was sharp and restricted to the K8 promoter region (Fig. 3). To test these ChIP patterns, we designed multiple primer pairs that were upstream or downstream of or overlapped the expected ChIP peak (Fig. 4A and C). The PAN RNA transcribed region was precipitated with anti-ORF57 antibodies, but the promoter region was not (compare the P1 to P3 signals with the P4, P5, and P7 signals). This change occurs relatively sharply in the transcribed region, as the P3 and P4 amplicons (Fig. 4A) were separated by only 78 bp of intervening sequence. This result confirms the data observed in the ChIP-chip assay, and it also is consistent with reports demonstrating that ORF57 binds to a 5′ element in PAN RNA (24, 25, 27). Interestingly, we saw a consistent decrease in the ChIP signal when using the P6 primer pair and a subtle M-shaped pattern in the PAN RNA ChIP-chip peak (Fig. 3C). While the relevance of this is uncertain, we speculate that this may be the result of gene or RNA looping during transcription bringing the 5′ and 3′ ends together. Next, we verified the ORF57 ChIP results at the ORF50-K8 locus. Once again, the data from the ChIP-chip were confirmed. The primer set at the K8 promoter showed a significant enhancement of ORF57, while all five other amplicons were not enhanced (Fig. 4C and D). We also tested the same amplicons with prebleed antibodies and saw no enhancement of the ChIP signal at any of the primer sets (Fig. 4B and D, bottom). Interestingly, the percent immunoprecipitation was over 10-fold lower with the prebleed sample at each of the loci, which could be due to nonspecific interactions of ORF57 with the viral genome-associated proteins or nonspecific interactions between the antibodies and the genome. In either case, these results validate our ChIP-chip approach and demonstrate that ORF57 associates with the transcribed region of the PAN RNA gene, as well as the K8 promoter region.
Fig 4.
Validation of ChIP-chip results. (A) Schematic diagram of the K7-PAN RNA genomic locus. The K7 protein-coding mRNA overlaps the PAN RNA but has an independent upstream transcription start site. Each primer set, corresponding to amplicons P1 to P7 as shown, was tested to determine the amplification efficiency prior to its use for ChIP. The diagram is drawn to scale, with the heavy black bar representing 500 bp. The P3 amplicon includes the Rta-responsive element (59, 60). (B) ChIP data from the PAN RNA locus. Top, ChIP data with anti-ORF57 antibodies analyzed using primers described in panel A. Each value is the average percent immunoprecipitation (% IP) from four independent experiments, and the error bars are standard deviations. An asterisk indicates a P value of <0.05 compared to the P1 data set in a two-tailed unpaired Student t test. Bottom, ChIP data from prebleed controls (n = 3). Note the difference in scale from the top panel. (C) Schematic diagram of the ORF50-K8(K-bZIP)-K8.1 genomic locus. This locus has a complex transcription profile with multiple splice isoforms and transcription start sites (61). Each primer set, corresponding to amplicons K1 to K6 as shown, was tested to determine the amplification efficiency prior to its use for ChIP. The diagram is drawn to scale; the heavy black bar corresponds to 500 bp. (D) ChIP data from the ORF50–K-bZIP locus. Top, ChIP data analyzed using ORF50-K8 locus primers shown in panel C. Details are as in panel B. An asterisk indicates a P value of <0.001 compared to the K1 data set. Bottom, ChIP data from prebleed controls. Details are as in the top, except that only three replicates were performed. Note the difference in scale from the top panel. P values were determined using a two-tailed unpaired Student t test.
RNase treatment does not alter the ORF57 ChIP pattern.
Because ORF57 binds to RNA, its association with genes may be due solely to interaction with nascent transcripts that, in turn, cross-link pol II and the gene. If this were true, removal of the RNA tether should eliminate the association of ORF57 with DNA. In contrast, ORF57 genome association mediated by direct DNA contacts or by direct interactions with DNA-binding proteins will be unaffected by RNase treatment. To test this, we determined the effects of RNase treatment on the association of ORF57 with KSHV genes. After immunoprecipitation but prior to washes, we resuspended the beads in buffer containing high levels of RNase A and incubated the mixture at room temperature to degrade RNA. We then washed away complexes under stringent conditions. First, we used the samples for a ChIP-chip experiment. As shown in Fig. 5A, the pattern of the ORF57 ChIP-chip was unaffected by the addition of RNase. The ORF4 promoter, PAN RNA, oriLyt, and K8 promoter peaks were all present after this treatment. To verify these results, we next performed confirmatory ChIP experiments at the PAN RNA and ORF50/K8 loci (Fig. 5B and C). Consistent with the ChIP-chip results, we saw no significant difference between the samples with RNase and those without RNase.
Given that ORF57 binds directly to PAN RNA, we were initially surprised that RNase treatment had no effect on the ChIP signal across the PAN RNA transcribed region. To verify that the RNase treatment worked, we followed PAN RNA (Fig. 5D, top) or bulk RNA (Fig. 5D, bottom) through the ChIP protocol (Fig. 5D). We harvested RNA from samples prior to immunoprecipitation (lane 1), after immunoprecipitation but prior to RNase treatment (lane 2), immediately following RNase treatment (lane 3), and after washes (lane 4). The ChIP protocols were not optimized to maintain RNA integrity (e.g., we added no RNase inhibitor to these assays), so some RNA degradation was observed in the input samples and this was exacerbated during the long immunoprecipitation step (lanes 1 and 2, top). However, PAN RNA was detectable in both samples and it remained relatively long (∼100 to 400 nucleotides [nt]), even after immunoprecipitation. Most importantly, the PAN RNA signal was completely lost after RNase treatment. We further examined bulk RNA with 5′-end labeling and urea-PAGE. After RNase treatment, the average RNA sizes were <30 nt (lane 3, bottom). Moreover, these fragments were nearly completely lost from the beads after washing (lane 4, bottom). Thus, the RNase treatment step degraded PAN RNA and the overwhelming majority of viral and cellular RNA.
One interpretation of these results is that ORF57 association with the viral genome is exclusively driven by ORF57-protein or ORF57-DNA interactions. However, the RNase insensitivity of the ChIP signal does not necessarily indicate that ORF57 is recruited by DNA or proteins. It is formally possible that the small amount of ORF57-associated nascent PAN RNA is resistant to RNase but is not abundant enough to be detected in our assays. Alternatively, the small RNA fragments remaining after RNase digestion (Fig. 5D) may be sufficient to maintain ORF57 at the gene independent of other proteins. However, if such a small fragment were sufficient, it seems unlikely that ORF57 would not be contacting other gene-associated proteins and we would expect to observe at least some diminution of the ChIP signal. Given the effects of PAN RNA sequences on ORF57 binding and activity (24, 25, 27), we favor the model in which ORF57 is initially recruited by PAN RNA elements but then contacts DNA or DNA-associated proteins. Taken with the results above, our data suggest that ORF57 associates with the KSHV genome by using two modes of recruitment. In one case, ORF57 is recruited to PAN RNA through RNA, while the interactions at the K8 and ORF4 promoters and oriLyt are mediated by interactions with K-bZIP. These models are described in more detail in the Discussion.
DISCUSSION
ORF57 functions as a viral RNA-binding protein and has been proposed to modulate multiple posttranscriptional events in mRNA metabolism, including RNA stability, splicing, export, and translation (11, 13, 16, 22). In addition, ORF57 has been implicated in transcription (28–30). Here we have shown that ORF57 interacts with the KSHV transcription factor K-bZIP in vitro and in extracts from both transfected and infected cells (Fig. 1 and 2). In addition, our data show that ORF57 associates with the viral genome at oriLyt and the ORF4 and K-bZIP promoters, consistent with locations of K-bZIP binding (Fig. 3). In addition to these peaks, K-bZIP was reported to bind the ORF75, K15, and ORF57 promoters (45), but these were not found in our ORF57 ChIP-chip. However, our data for the ORF75 and K15 promoters were inconclusive because of the high background level, so ORF57 may indeed co-occupy these sites (Fig. 3A, right side of the genome). Moreover, the interaction of K-bZIP with the ORF57 promoter suggests the existence of a potential regulatory loop involving K-bZIP and ORF57 (see below). The interaction between K-bZIP and ORF57 and the association with similar regions in the viral genome during lytic reactivation support the model in which ORF57 interacts with K-bZIP on the viral genome to regulate viral gene expression.
Interactions between ORF57 and the viral genome were also observed at the PAN RNA transcribed region. ChIP-quantitative PCR analysis confirmed the existence of a broad association across the PAN RNA transcribed region, as predicted if it associates with the nascent RNA. Even so, extensive RNase treatment does not diminish the ORF57 association with KSHV DNA, demonstrating that ORF57 does not exclusively contact the viral genome through an RNA tether. Thus, it seems likely that ORF57 contacts components of the transcriptional machinery cotranscriptionally, but in this case, the RNA is likely responsible for the initial interaction (24, 25, 27) (see below). Given the broad role of ORF57 as a posttranscriptional modulator of gene expression, we were surprised that other transcribed regions were not enriched in the ORF57 ChIP-chip experiments. We suspect that we can detect a signal for the PAN RNA transcribed region because it is, by far, the most highly expressed transcript in reactivated cells. Even so, the ChIP signal is only about as strong as the promoter peaks, even for this highly transcribed transcript. Therefore, we think it is likely that other KSHV nascent transcripts associate with ORF57 but the interactions do not exceed the limits of detection of our assay.
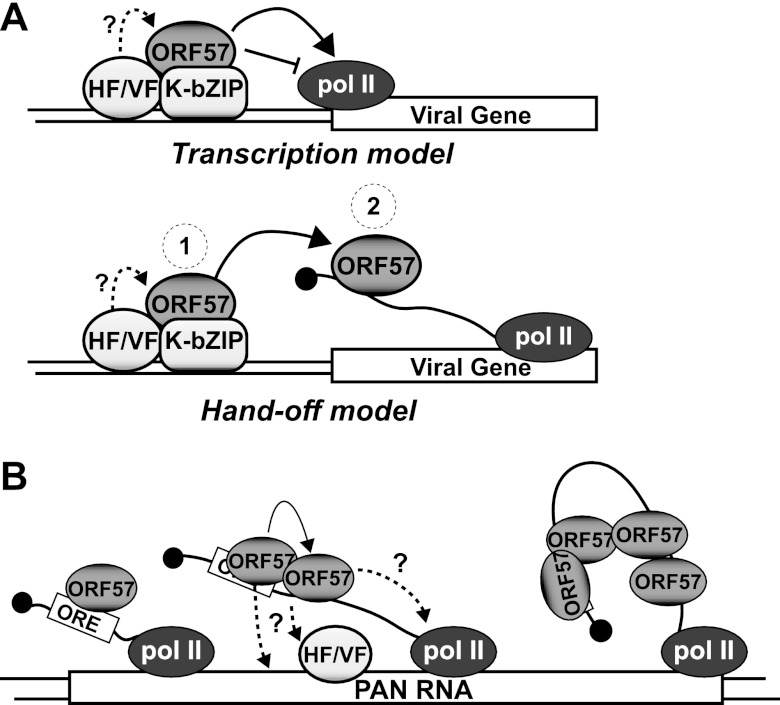
We propose that ORF57 associates with the KSHV genome by using at least two distinct molecular mechanisms (Fig. 6). In one of these mechanisms, the K-bZIP protein, perhaps in concert with other viral or host factors, recruits ORF57 to the viral promoters (Fig. 6A). Upon the recruitment of ORF57, the complex may be involved in the negative or positive regulation of transcription (Fig. 6A, transcription model). Alternatively, it is possible that it is recruited to these promoters and is subsequently transferred to the emerging nascent transcript (Fig. 6A, hand-off model) to enhance ORF57-mediated posttranscriptional activity. The second mode of ORF57 association with the genome is observed with PAN RNA (Fig. 6B). After pol II initiates transcription elongation, the 5′-proximal ORF57-responsive element (ORE) emerges from the transcript (24, 25, 27) and is bound by ORF57. Next, ORF57 multimerization and nonspecific RNA binding activity drive the association of ORF57 to lower-affinity sites on the RNA. The nonspecific binding of ORF57 may be strongest at the sites adjacent to the ORE and to the 3′ end of the transcript (Fig. 3 and 4) (24, 25), suggesting a potential RNA looping mechanism (Fig. 6B). While bound to the RNA, ORF57 makes additional contacts with the polymerase, with DNA-bound factors, or directly with the DNA (dashed arrows, Fig. 6B). Indeed, the ORF57 homolog in herpes simplex virus, ICP27, binds pol II (55). While our data strongly support the existence of two modes of ORF57 genome recruitment, significant gaps remain in the definition of the molecular mechanisms of ORF57-mediated gene regulation. Future studies will focus on the validation of these models and the testing of their molecular details.
Fig 6.
Two modes of ORF57 recruitment to the KSHV genome. (A) K-bZIP recruitment models. The details are discussed in the text. HF/VF ovals represent undefined host or viral factors, respectively (e.g., Rta or RBPJκ). Note that we are not proposing any specific molecular architecture of the complex, except for the existence of an ORF57–K-bZIP interaction. For example, we do not know which component(s) directly contacts DNA. In the transcription model (top), recruitment of ORF57 to the promoter regulates transcription efficiency. In the “hand-off” model, ORF57 recruitment at gene promoters leads to higher local concentrations that promote ORF57 binding to nascent transcripts. (B) ORF57 recruitment to the PAN RNA gene. See the text for details.
ORF57 interacts with the transcription factor Rta, the primary latency-to-lytic switch protein (28–30), but we do not see a correlation between ORF57 ChIP and Rta-responsive promoters. For example, the Rta-responsive PAN RNA promoter does not interact with ORF57 in our ChIP-chip or conventional ChIP assays (Fig. 3 and 4). We interpret these negative results cautiously and cannot conclude that ORF57 does not interact with Rta at viral promoters for several reasons. First, our ChIP assays were all performed at approximately the same time following lytic reactivation (∼18 to 22 h after doxycycline addition), so it remains possible that the interaction between ORF57 and Rta occurs at a different stage of viral reactivation. Second, the ORF57 epitopes may be buried in the context of an ORF57-Rta complex but remain exposed in other ORF57 complexes. Finally, it is important to note that the K8 promoter is Rta responsive (56–58), so it is possible that the ORF57 association at the K8 promoter reflects the importance of both the ORF57-Rta and ORF57–K-bZIP interactions. Thus, while our data do not lend direct support to the existence of a functional interaction between ORF57 and Rta at viral promoters, they do not exclude the model in which ORF57 and Rta functionally interact at promoters during viral reactivation.
Three viral gene regulatory proteins, Rta, ORF57, and K-bZIP, appear to function interdependently to regulate KSHV gene expression. ORF57 binds to K-bZIP and to Rta (Fig. 1 and 2) (28–30) and binds to the same promoters as K-bZIP (Fig. 3 and 4) (45). K-bZIP binds Rta and inhibits its transcriptional activation function, but in some cases, it coactivates Rta-dependent promoters (42, 43, 45). Rta activates the ORF57 promoter, and K-bZIP also binds the ORF57 promoter, presumably to regulate ORF57 expression (45). We do not know how these interactions affect KSHV gene regulation. Unfortunately, attempts to use reporter constructs in HeLa or HEK293 cell lines to assess the functional consequences of ORF57–K-bZIP interaction have failed thus far (unpublished observations). In addition, the dissection of the molecular mechanism is complicated by several factors. First, transcriptional activation is cell line dependent and the K8 promoter specifically behaves differently in distinct cell lines (57). Second, other viral components could be necessary for regulation. Third, distinguishing a mechanism that entails synergistic posttranscriptional and transcriptional activities from a mechanism in which the transcriptional and posttranscriptional activities are independent can be difficult. For example, we can imagine that a particular gene depends on Rta for transcription activation and on ORF57 for posttranscriptional activation. In this case, ORF57 posttranscriptional stimulation strictly depends on the presence of Rta, giving the appearance of synergy. However, the mechanism is indirect and not the result of a bona fide mechanistic coupling between ORF57 and Rta. Empirical distinction between this type of activation and true mechanistic coupling is challenging. Fourth, ORF57 acts nonspecifically, so care must be taken to discern whether stimulation of Rta or K-bZIP activity is due to molecular interactions or simply due to ORF57 upregulation of Rta or K-bZIP. Indeed, <2-fold differences in the expression of any of these factors can alter the gene expression of transiently expressed K8 promoter-driven constructs (unpublished observations). These experimental challenges require rigorous approaches to address the complex regulatory interplay among ORF57, K-bZIP, and Rta. However, an understanding of these mechanisms is necessary to elucidate the molecular mechanisms of KSHV gene regulation.
ACKNOWLEDGMENTS
We thank Pinghui Feng (University of Southern California) for reagents and Ivan D'Orso and Julie K. Pfeiffer for critical reviews of the manuscript.
This work was funded by NIH-NIAID grant AI081710. N.K.C. is a Southwestern Medical Foundation Scholar in Biomedical Research. E.S. is supported by NIH-NIAID grant AI081710-02S1.
Footnotes
Published ahead of print 30 January 2013
REFERENCES
- 1. Ganem D. 2006. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu. Rev. Pathol. 1:273–296 [DOI] [PubMed] [Google Scholar]
- 2. Nicholas J. 2007. Human herpesvirus 8-encoded proteins with potential roles in virus-associated neoplasia. Front. Biosci. 12:265–281 [DOI] [PubMed] [Google Scholar]
- 3. Dourmishev LA, Dourmishev AL, Palmeri D, Schwartz RA, Lukac DM. 2003. Molecular genetics of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) epidemiology and pathogenesis. Microbiol. Mol. Biol. Rev. 67:175–212, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Greene W, Kuhne K, Ye F, Chen J, Zhou F, Lei X, Gao SJ. 2007. Molecular biology of KSHV in relation to AIDS-associated oncogenesis. Cancer Treat. Res. 133:69–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sarid R, Flore O, Bohenzky RA, Chang Y, Moore PS. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun R, Lin SF, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Staudt MR, Dittmer DP. 2003. Viral latent proteins as targets for Kaposi's sarcoma and Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) induced lymphoma. Curr. Drug Targets Infect. Disord. 3:129–135 [DOI] [PubMed] [Google Scholar]
- 9. Deng H, Liang Y, Sun R. 2007. Regulation of KSHV lytic gene expression. Curr. Top. Microbiol. Immunol. 312:157–183 [DOI] [PubMed] [Google Scholar]
- 10. Staudt MR, Dittmer DP. 2007. The Rta/Orf50 transactivator proteins of the gamma-herpesviridae. Curr. Top. Microbiol. Immunol. 312:71–100 [DOI] [PubMed] [Google Scholar]
- 11. Conrad NK. 2009. Posttranscriptional gene regulation in Kaposi's sarcoma-associated herpesvirus. Adv. Appl. Microbiol. 68:241–261 [DOI] [PubMed] [Google Scholar]
- 12. Glaunsinger BA, Ganem DE. 2006. Messenger RNA turnover and its regulation in herpesviral infection. Adv. Virus Res. 66:337–394 [DOI] [PubMed] [Google Scholar]
- 13. Swaminathan S. 2005. Post-transcriptional gene regulation by gamma herpesviruses. J. Cell. Biochem. 95:698–711 [DOI] [PubMed] [Google Scholar]
- 14. Sinclair AJ. 2003. bZIP proteins of human gammaherpesviruses. J. Gen. Virol. 84:1941–1949 [DOI] [PubMed] [Google Scholar]
- 15. Boyne JR, Whitehouse A. 2006. Gamma-2 herpes virus post-transcriptional gene regulation. Clin. Microbiol. Infect. 12:110–117 [DOI] [PubMed] [Google Scholar]
- 16. Majerciak V, Zheng ZM. 2009. Kaposi's sarcoma-associated herpesvirus ORF57 in viral RNA processing. Front. Biosci. 14:1516–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Han Z, Swaminathan S. 2006. Kaposi's sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J. Virol. 80:5251–5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Majerciak V, Pripuzova N, McCoy JP, Gao SJ, Zheng ZM. 2007. Targeted disruption of Kaposi's sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8alpha, and K8.1 and the production of infectious virus. J. Virol. 81:1062–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ote I, Piette J, Sadzot-Delvaux C. 2010. The varicella-zoster virus IE4 protein: a conserved member of the herpesviral mRNA export factors family and a potential alternative target in antiherpetic therapies. Biochem. Pharmacol. 80:1973–1980 [DOI] [PubMed] [Google Scholar]
- 20. Sandri-Goldin RM. 2008. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front. Biosci. 13:5241–5256 [DOI] [PubMed] [Google Scholar]
- 21. Toth Z, Stamminger T. 2008. The human cytomegalovirus regulatory protein UL69 and its effect on mRNA export. Front. Biosci. 13:2939–2949 [DOI] [PubMed] [Google Scholar]
- 22. Boyne JR, Jackson BR, Whitehouse A. 2010. ORF57: master regulator of KSHV mRNA biogenesis. Cell Cycle 9:2702–2703 [DOI] [PubMed] [Google Scholar]
- 23. Nekorchuk M, Han Z, Hsieh TT, Swaminathan S. 2007. Kaposi's sarcoma-associated herpesvirus ORF57 protein enhances mRNA accumulation independently of effects on nuclear RNA export. J. Virol. 81:9990–9998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sei E, Conrad NK. 2011. Delineation of a core RNA element required for Kaposi's sarcoma-associated herpesvirus ORF57 binding and activity. Virology 419:107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Massimelli MJ, Kang JG, Majerciak V, Le SY, Liewehr DJ, Steinberg SM, Zheng ZM. 2011. Stability of a long noncoding viral RNA depends on a 9-nt core element at the RNA 5′ end to interact with viral ORF57 and cellular PABPC1. Int. J. Biol. Sci. 7:1145–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boyne JR, Colgan KJ, Whitehouse A. 2008. Recruitment of the complete hTREX complex is required for Kaposi's sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog. 4:e1000194 doi:10.1371/journal.ppat.1000194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sahin BB, Patel D, Conrad NK. 2010. Kaposi's sarcoma-associated herpesvirus ORF57 protein binds and protects a nuclear noncoding RNA from cellular RNA decay pathways. PLoS Pathog. 6:e1000799 doi:10.1371/journal.ppat.1000799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Palmeri D, Spadavecchia S, Carroll KD, Lukac DM. 2007. Promoter- and cell-specific transcriptional transactivation by the Kaposi's sarcoma-associated herpesvirus ORF57/Mta protein. J. Virol. 81:13299–13314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Malik P, Blackbourn DJ, Cheng MF, Hayward GS, Clements JB. 2004. Functional co-operation between the Kaposi's sarcoma-associated herpesvirus ORF57 and ORF50 regulatory proteins. J. Gen. Virol. 85:2155–2166 [DOI] [PubMed] [Google Scholar]
- 30. Uetz P, Dong YA, Zeretzke C, Atzler C, Baiker A, Berger B, Rajagopala SV, Roupelieva M, Rose D, Fossum E, Haas J. 2006. Herpesviral protein networks and their interaction with the human proteome. Science 311:239–242 [DOI] [PubMed] [Google Scholar]
- 31. West JT, Wood C. 2003. The role of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 regulator of transcription activation (RTA) in control of gene expression. Oncogene 22:5150–5163 [DOI] [PubMed] [Google Scholar]
- 32. Kirshner JR, Lukac DM, Chang J, Ganem D. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 74:3586–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aravind L, Landsman D. 1998. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 26:4413–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kato-Noah T, Xu Y, Rossetto CC, Colletti K, Papousková I, Pari GS. 2007. Overexpression of the Kaposi's sarcoma-associated herpesvirus transactivator K-Rta can complement a K-bZIP deletion BACmid and yields an enhanced growth phenotype. J. Virol. 81:13519–13532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Sathish N, Hollow C, Yuan Y. 2011. Functional characterization of Kaposi's sarcoma-associated herpesvirus open reading frame K8 by bacterial artificial chromosome-based mutagenesis. J. Virol. 85:1943–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lefort S, Flamand L. 2009. Kaposi's sarcoma-associated herpesvirus K-bZIP protein is necessary for lytic viral gene expression, DNA replication, and virion production in primary effusion lymphoma cell lines. J. Virol. 83:5869–5880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lin CL, Li H, Wang Y, Zhu FX, Kudchodkar S, Yuan Y. 2003. Kaposi's sarcoma-associated herpesvirus lytic origin (ori-Lyt)-dependent DNA replication: identification of the ori-Lyt and association of K8 bZip protein with the origin. J. Virol. 77:5578–5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rossetto C, Yamboliev I, Pari GS. 2009. Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 K-bZIP modulates latency-associated nuclear protein-mediated suppression of lytic origin-dependent DNA synthesis. J. Virol. 83:8492–8501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Izumiya Y, Ellison TJ, Yeh ET, Jung JU, Luciw PA, Kung HJ. 2005. Kaposi's sarcoma-associated herpesvirus K-bZIP represses gene transcription via SUMO modification. J. Virol. 79:9912–9925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu FY, Wang SE, Tang QQ, Fujimuro M, Chiou CJ, Zheng Q, Chen H, Hayward SD, Lane MD, Hayward GS. 2003. Cell cycle arrest by Kaposi's sarcoma-associated herpesvirus replication-associated protein is mediated at both the transcriptional and posttranslational levels by binding to CCAAT/enhancer-binding protein alpha and p21(CIP-1). J. Virol. 77:8893–8914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. AuCoin DP, Colletti KS, Cei SA, Papousková I, Tarrant M, Pari GS. 2004. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318:542–555 [DOI] [PubMed] [Google Scholar]
- 42. Izumiya Y, Lin SF, Ellison T, Chen LY, Izumiya C, Luciw P, Kung HJ. 2003. Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression. J. Virol. 77:1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liao W, Tang Y, Lin SF, Kung HJ, Giam CZ. 2003. K-bZIP of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (KSHV/HHV-8) binds KSHV/HHV-8 Rta and represses Rta-mediated transactivation. J. Virol. 77:3809–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lefort S, Soucy-Faulkner A, Grandvaux N, Flamand L. 2007. Binding of Kaposi's sarcoma-associated herpesvirus K-bZIP to interferon-responsive factor 3 elements modulates antiviral gene expression. J. Virol. 81:10950–10960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ellison TJ, Izumiya Y, Izumiya C, Luciw PA, Kung HJ. 2009. A comprehensive analysis of recruitment and transactivation potential of K-Rta and K-bZIP during reactivation of Kaposi's sarcoma-associated herpesvirus. Virology 387:76–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakamura H, Lu M, Gwack Y, Souvlis J, Zeichner SL, Jung JU. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77:4205–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sheffield P, Garrard S, Derewenda Z. 1999. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15:34–39 [DOI] [PubMed] [Google Scholar]
- 48. Horton RM. 1995. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol. Biotechnol. 3:93–99 [DOI] [PubMed] [Google Scholar]
- 49. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stubbs SH, Hunter OV, Hoover A, Conrad NK. 2012. Viral factors reveal a role for REF/Aly in nuclear RNA stability. Mol. Cell. Biol. 32:1260–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Majerciak V, Yamanegi K, Nie SH, Zheng ZM. 2006. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J. Biol. Chem. 281:28365–28378 [DOI] [PubMed] [Google Scholar]
- 52. Malik P, Blackbourn DJ, Clements JB. 2004. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 279:33001–33011 [DOI] [PubMed] [Google Scholar]
- 53. Sapra AK, Ankö ML, Grishina I, Lorenz M, Pabis M, Poser I, Rollins J, Weiland EM, Neugebauer KM. 2009. SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol. Cell 34:179–190 [DOI] [PubMed] [Google Scholar]
- 54.Conrad NK. 2008. Chapter 15. Co-immunoprecipitation techniques for assessing RNA-protein interactions in vivo. Methods Enzymol. 449:317–342 [DOI] [PubMed] [Google Scholar]
- 55. Dai-Ju JQ, Li L, Johnson LA, Sandri-Goldin RM. 2006. ICP27 interacts with the C-terminal domain of RNA polymerase II and facilitates its recruitment to herpes simplex virus 1 transcription sites, where it undergoes proteasomal degradation during infection. J. Virol. 80:3567–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lukac DM, Garibyan L, Kirshner JR, Palmeri D, Ganem D. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang Y, Chong OT, Yuan Y. 2004. Differential regulation of K8 gene expression in immediate-early and delayed-early stages of Kaposi's sarcoma-associated herpesvirus. Virology 325:149–163 [DOI] [PubMed] [Google Scholar]
- 58. Seaman WT, Quinlivan EB. 2003. Lytic switch protein (ORF50) response element in the Kaposi's sarcoma-associated herpesvirus K8 promoter is located within but does not require a palindromic structure. Virology 310:72–84 [DOI] [PubMed] [Google Scholar]
- 59. Song MJ, Brown HJ, Wu TT, Sun R. 2001. Transcription activation of polyadenylated nuclear rna by rta in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:3129–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chang PJ, Shedd D, Gradoville L, Cho MS, Chen LW, Chang J, Miller G. 2002. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J. Virol. 76:3168–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zheng ZM. 2003. Split genes and their expression in Kaposi's sarcoma-associated herpesvirus. Rev. Med. Virol. 13:173–184 [DOI] [PMC free article] [PubMed] [Google Scholar]