Abstract
Activation of a host DNA damage response (DDR) is essential for DNA replication of minute virus of canines (MVC), a member of the genus Bocavirus of the Parvoviridae family; however, the mechanism by which DDR contributes to viral DNA replication is unknown. In the current study, we demonstrate that MVC infection triggers the intra-S-phase arrest to slow down host cellular DNA replication and to recruit cellular DNA replication factors for viral DNA replication. The intra-S-phase arrest is regulated by ATM (ataxia telangiectasia-mutated kinase) signaling in a p53-independent manner. Moreover, we demonstrate that SMC1 (structural maintenance of chromosomes 1) is the key regulator of the intra-S-phase arrest induced during infection. Either knockdown of SMC1 or complementation with a dominant negative SMC1 mutant blocks both the intra-S-phase arrest and viral DNA replication. Finally, we show that the intra-S-phase arrest induced during MVC infection was caused neither by damaged host cellular DNA nor by viral proteins but by replicating viral genomes physically associated with the DNA damage sensor, the Mre11-Rad50-Nbs1 (MRN) complex. In conclusion, the feedback loop between MVC DNA replication and the intra-S-phase arrest is mediated by ATM-SMC1 signaling and plays a critical role in MVC DNA replication. Thus, our findings unravel the mechanism underlying DDR signaling-facilitated MVC DNA replication and demonstrate a novel strategy of DNA virus-host interaction.
INTRODUCTION
Parvoviruses are small, nonenveloped and single-stranded DNA (ssDNA) viruses and cause highly contagious diseases that are sometimes fatal in humans and animals (1, 2). The viral genome of parvoviruses is 5 to 6 kb and flanked by two terminal hairpin structures. Adeno-associated viruses (AAVs), in the genus Dependovirus of the family Parvoviridae, require helper virus for replication, whereas autonomous parvoviruses, such as minute virus of mice (MVM) and minute virus of canines (MVC), in the genera Parvovirus and Bocavirus, respectively, replicate autonomously in host cells. Because of its well-characterized reverse genetics system and efficient infection system, MVC has been used as a model to study the DNA replication mechanism of autonomous parvoviruses as well as the pathogenesis of bocavirus infection (3–6). During infection of Walter Reed/3873D (WRD) canine cells (7), MVC induces a gradual cell cycle arrest, from S phase in early infection to G2/M phase at a later stage, and mitochondrion-mediated apoptosis (3). Additionally, MVC hijacks the cellular DNA damage response (DDR) machinery to facilitate viral DNA replication (4). The MVC genome shares 50 to 60% identity with the genome of human bocavirus type 1 (HBoV1) (6, 8, 9), a newly identified human pathogen that causes acute respiratory tract infections in children worldwide (8, 10–14). Therefore, MVC has been used as a model for studying bocavirus replication.
Infections of many DNA viruses are able to subvert the cellular DDR machinery (15–18), a safeguarding system triggered by damaged cellular DNA structures such as ssDNA breaks (SSBs), double-stranded DNA (dsDNA) breaks (DSBs), and stalled replication forks (19, 20). The central role of the DDR is to protect genome stability and integrity through a cascade of phosphorylation events initiated by three phosphatidylinositol 3-kinase-like kinases (PI3Ks): ATM (ataxia telangiectasia-mutated kinase), ATR (ATM- and Rad3-related kinase), and DNA-PKcs (DNA-dependent protein kinase catalytic subunit) (21, 22). In the presence of damaged DNA structures, these three kinases are recruited and autophosphorylated and further recruit a number of effector proteins to coordinate cell cycle arrest, DNA repair, and apoptosis. ATM signaling has been reported to be coopted by the autonomous parvoviruses MVC and MVM (4, 23, 24) to help their productive infections. However, the beneficial effects of ATM signaling on parvovirus DNA replication have not been well understood.
In replicating cells in S phase, one of the most important outcomes of the DDR is the intra-S-phase arrest (25–27). Intra-S-phase arrest plays a crucial role in preventing damaged DNA from entering mitosis by slowing the rate of S-phase progression and stabilizing stalled replication forks (25, 28). The signaling proteins involved in the intra-S-phase arrest include a large number of checkpoint proteins and DNA repair factors. Intra-S-phase checkpoint proteins are activated to slow down cellular DNA replication through degradation of replication proteins or regulator factors such as Cdc25A (27, 29–32), while DDR signaling recruits repair factors to the damaged DNA foci for the rapid resumption of replication following DNA repair (33). ATM signaling plays a central role in regulating DSB-induced intra-S-phase arrest. Damaged DNA is first recognized by the Mre11-Rad50-Nbs1 (MRN) complex sensor and further recruits ATM kinase. Following ATM autophosphorylation, several proteins, such as Chk2 (checkpoint protein 2), BRCA1 (breast cancer type 1 susceptibility protein), and SMC1 (structural maintenance of chromosomes 1), are phosphorylated and recruited as checkpoint proteins (25). SMC1 was originally identified as a subunit of the cohesion complex that ensures proper segregation of sister chromatids (34). Further studies confirmed that it is an intra-S-phase checkpoint protein that is phosphorylated at serines 957 and 966 by ATM kinase (35–39). However, it is not clear how SMC1 interferes with cellular DNA replication proteins or regulator factors through its checkpoint function. Although replication of many DNA viruses occurs during the S phase of host cells and induces a DDR, the link between viral infection-induced DDR and the intra-S-phase arrest has not been well established.
Modulation of the host cellular environment through cell cycle control is an important strategy for replication of DNA viruses. By arresting cells in S phase, viral DNA synthesis is facilitated by the cellular DNA replication machinery; however, many DNA viruses also block cellular DNA synthesis for productive infection (40–42). Autonomous parvovirus MVM has been reported to inhibit host cell growth through p53-dependent inhibition of cyclin A, and the large nonstructural protein NS1 plays a key role in inhibiting host cell DNA synthesis (43–45). However, we found that expression of the nonstructural proteins of MVC, NS1 and NP1, failed to interfere with host cell cycle regulation (3), indicating that a mechanism without a direct involvement of viral proteins is involved in MVC-induced cell cycle arrest.
In this study, we aimed to determine whether DDR signaling and cell cycle modulation coordinate to facilitate MVC DNA replication. Our results confirm that MVC infection triggers the intra-S-phase arrest that is mediated by the ATM-SMC1 pathway and facilitates viral DNA replication. Moreover, our results provide direct evidence that MVC infection-induced DDR is elicited by the MRN complex that senses replicating viral genomes. These findings reveal a novel strategy by which MVC exploits cellular DNA replication and DDR machineries for its own DNA replication and provide new insights in the mechanisms of DNA virus-host interaction that directly contribute to viral DNA replication.
MATERIALS AND METHODS
Cell culture and virus infection.
WRD cells (7) were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum (FCS) in 5% CO2 at 37°C. MVC (GA3 strain) was cultured and titrated as previously described (3, 4, 6). WRD cells were infected with MVC at a multiplicity of infection (MOI) of 3. Both WRD cells and MVC were gifts from Colin Parrish at Cornell University.
Chemicals and treatment.
ATM kinase inhibitor KU55933 (ATMi) (Tocris Bioscience, Bristol, United Kingdom) was prepared in dimethyl sulfoxide (DMSO) as a stock solution at 10 mM. Bromodeoxyuridine (BrdU) (Sigma) was diluted in deionized water as a stock solution at 10 mM. WRD cells were seeded on 60-mm dishes 1 day prior to chemical treatment. KU55933 was applied to cells at a final concentration of 10 μM upon virus infection.
Antibodies used.
The rat anti-MVC NS1 polyclonal antibody was developed previously (6). All the other antibodies used in this study were purchased from companies listed as follows: anti-BrdU (clone B44) and anti-proliferating cell nuclear antigen (PCNA) antibodies (BD Biosciences, San Jose, CA); anti-γH2AX antibody (Novus, Littleton, CO); anti-Rad50 (Epitomics, Burlingame, CA); anti-p53 (Ser15) and anti-Flag epitope (Cell Signaling, Danvers, MA); anti-β-actin (Sigma); anti-cyclin A, anti-RFC1, anti-polymerase (pol) δ, and anti-Mre11 antibodies (Santa Cruz Biotechnology, Santa Cruz, CA); anti-p-Nbs1(Ser343) and anti-p-SMC1 (Ser957) antibodies (Genscript, Piscataway, NJ); and anti-SMC1 antibody (Genetex, Irvine, CA). All the secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA).
siRNA, plasmids, and transfection.
Small interfering RNA (siRNA) oligonucleotides were synthesized via Dicer substrate RNA interference (RNAi) at Integrated DNA Technologies (IDT, Coralville, IA). The following siRNA sequences were chosen for targeting the genes of interest: siRNA specific to canine ATM (siATM), 5′-GUA CUA GUU GCU UGU GUA ACU GUA-3′; siRNA specific to canine SMC1A (siSMC1), 5′-CUC UCC CAA UCU CUG GAU AUU UGG-3′; siRNA specific to canine p53 (sip53), 5′-CCA CCA UCC CUA AAC UAA UGT G-3′. The following scrambled RNA was used as an siRNA control: 5′-CUU CCU CUC UUU CUC UCC CUU GUG A-3′. Transfection of all siRNAs was performed using Hiperfect reagent (Qiagen, Valencia, CA) following the manufacturer's instructions. At 48 h posttransfection, the cells were fed with fresh medium and infected with MVC.
Plasmids pcDNA3-5′cMyc-SMC1wt and pcDNA3-5′cMyc-SMC1(S957A/S966A) (36), expressing wild-type human hSMC1wt and the hSMC1(S957A/S966A) mutant, respectively, were purchased from Addgene (Cambridge, MA). MVC plasmids pIMVC, pIMVCNP1(−), pIMVCVP1/2(−), and pMVCNSCap and control vector pBB have been described previously (6). Nucleofection was used to transfect plasmid DNA using an AMAXIA Nucleofector (Lonza Inc.) with program T030.
Immunofluorescence assay.
Immunofluorescence staining was performed as previously described (4, 46). Briefly, cells were fixed in 3.7% paraformaldehyde and permeabilized in 0.1% Triton X-100, except for the staining with an anti-PCNA antibody, in which 90% methanol was used for permeabilization. Images were taken at a magnification of ×100 or ×40 under a confocal microscope (Eclipse C1 Plus; Nikon) with Nikon EZ-C1 software.
BrdU-based dot blot analysis of DNA replication.
WRD cells were mock or MVC infected. At 12 h, 18 h, 24 h, and 48 h postinfection (p.i.), the infected cells were collected. Half of the cells were used to purify total DNA (both cellular DNA and viral DNA) using the DNeasy blood and tissue kit (Qiagen). This kit is optimized for purification of the total DNA from various sources, including viruses, and has a recovery rate of over 90% for parvoviral DNA (data not shown). The other half of the infected cells were used to extract low-molecular-weight DNA (viral DNA) using the Hirt DNA extraction method (47, 48). Extracted DNA was diluted in 100 μl of deionized water. The BrdU-based dot blot assay was performed as previously described (49). Briefly, to expose the BrdU epitopes in cellular DNA, the DNA samples were denatured by heating at 95°C for 5 min and immediately kept on ice; 5 μl of the DNA samples was pipetted onto a nitrocellulose membrane. The DNA on the membrane was cross-linked by UV treatment at a dose of 700 mJ/cm2 in a Hoefer UVC 500 UV cross-linker (Hoefer, Inc., Holliston, MA). The membrane was then blocked in 5% nonfat milk in TBST (Tris-buffered saline, pH 7.4, with 0.1% Tween 20) at room temperature, after which a Western blotting procedure was followed.
BrdU incorporation and BrdU pulsing assays.
For the BrdU incorporation assay (50), BrdU was added to the cell culture medium at a final concentration of 30 μM and incubated for 1 h. After BrdU incorporation, cells were collected, fixed in 3.7% paraformaldehyde for 30 min, and permeabilized with 0.1% Triton X-100 for another 30 min. After permeabilization, two procedures were followed to differentiate the cell cycle (cellular DNA replication) from viral DNA replication. For the detection of the cell cycle, cells were treated with 1 M HCl for 30 min to denature chromosome DNA for the binding of the BrdU epitopes with an anti-BrdU antibody (clone B44) (51). For parvovirus DNA replication analysis (52, 53), the HCl treatment step was skipped, since parvovirus DNA replication generates ssDNA viral genome and replication intermediates that contain partial ssDNA (54). The cells were costained with anti-BrdU and anti-MVC NS1 antibodies followed by secondary antibodies and DAPI (4′,6-diamidino-2-phenylindole) to analyze the cell cycle and the percentage of NS1-positive (NS+) cells, respectively.
For the BrdU pulsing assay, at 18 h p.i., mock- or MVC-infected cells were incubated with BrdU at 30 μM for 20 min. Incubated cells were collected immediately after BrdU labeling and every hour thereafter. Collected cells were fixed, permeabilized, and treated with 1 M HCl as described above. Treated cells were then costained with DAPI and anti-BrdU and anti-MVC NS1 antibodies and were assessed by flow cytometry. Mock- and MVC-infected cells were gated according to NS1 staining, and the change in DNA content in the BrdU-labeled cells was monitored by DAPI staining.
Flow cytometry analysis.
The stained cell samples described above were analyzed on a three-laser flow cytometer (LSR II; BD Biosciences) at the Flow Cytometry Core of the University of Kansas Medical Center. All flow cytometry data were analyzed using FACSDiva software (BD Biosciences).
Western blot and Southern blot analyses.
Western blotting was performed as previously described (3, 4). For Southern blotting, low-molecular-weight (Hirt) DNA was extracted from infected cells (47, 48) and analyzed by Southern or dot blotting using an MVC NSCap probe as described previously (3, 4, 6).
Comet assay.
A Comet assay kit was purchased from Cell Biolabs Inc. (San Diego, CA) and used according to the manufacturer's instructions. Briefly, at 18 h p.i., mock- or MVC-infected cells were trypsinized and diluted in phosphate-buffered saline (PBS). Mock-infected cells were treated with 100 μM H2O2 at 4°C for 20 min as positive controls. Mock- and MVC-infected and H2O2-treated cells were mixed with 1% low-melting-point agarose and used to coat slides. Then, the slides were treated in an alkaline condition, electrophoresed, and stained with VISTA green dye. Stained slides were visualized under a confocal microscope (Eclipse C1 Plus, Nikon, Melville, NY) with Nikon EZ-C1 software. Images were taken at a magnification of ×40.
BrdU-based IP assay.
At 18 h p.i., mock- or MVC-infected cells were pulsed with BrdU at 100 μM for 1 h and collected. Immunoprecipitation (IP) was performed using the Pierce cross-link IP kit (Thermo Scientific, Rockford, IL). Briefly, treated cells were lysed and centrifuged; the supernatant that contained viral DNA was incubated with protein A/G-coated resins preincubated with an anti-BrdU antibody. The resins were then rinsed and diluted in protein loading buffer followed by Western blotting using an anti-Mre11 antibody.
RESULTS
MVC DNA replication arrests host cells in S phase during early infection.
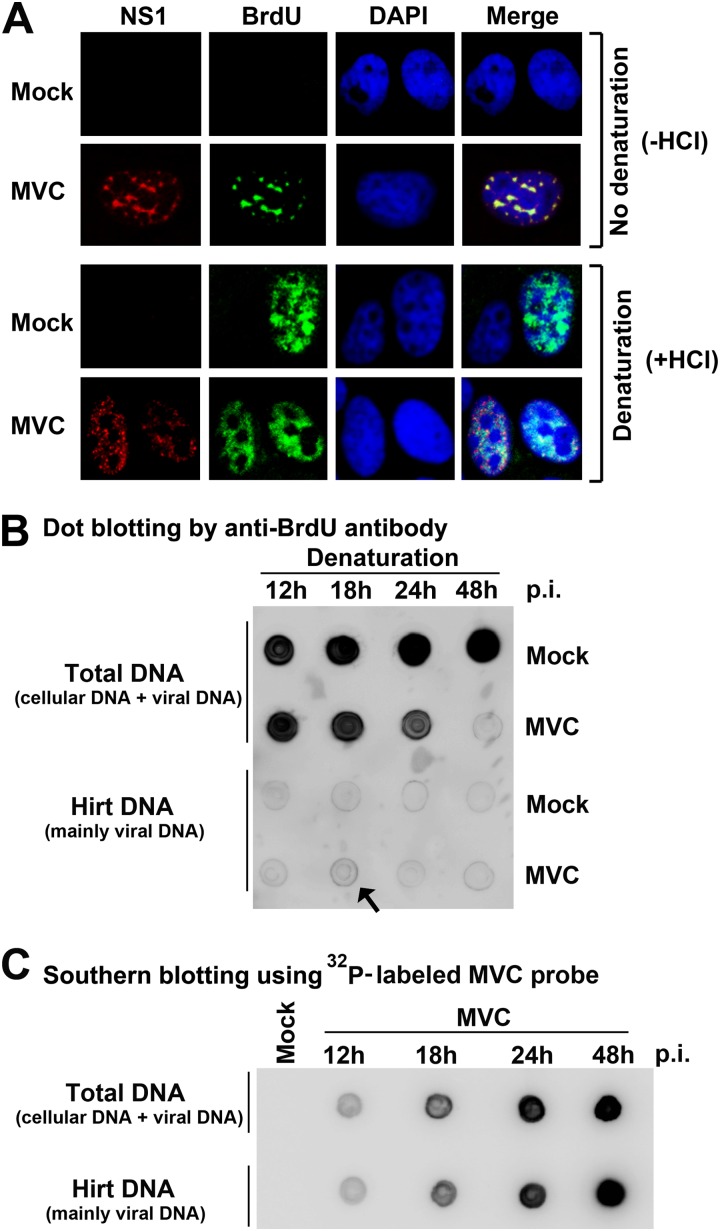
We have shown previously that MVC infection induces a host cell cycle change from S phase in early infection to G2/M phase in later infection (3); however, whether such a change is related to viral DNA replication is unknown. To determine the relationship between viral and cellular DNA replication, we performed a BrdU pulse-labeling assay. BrdU is a thymidine analog that can be incorporated into replicating DNA. For the detection of cellular DNA replication by BrdU incorporation, a denaturation process, such as treatment with hydrochloride (HCl), is necessary because the BrdU epitopes are detectable only in the context of the ssDNA form (51, 55). In contrast, for detection of parvovirus DNA replication by BrdU incorporation, the denaturation step is not required (52, 53). As anticipated, without denaturation, incorporated BrdU was undetectable in mock-infected cells (Fig. 1A, −HCl). MVC-infected cells showed punctate foci of anti-BrdU staining that colocalized with the foci stained for MVC NS1 (Fig. 1A, −HCl), which represent active viral DNA replication centers. Notably, with denaturation, both mock- and MVC-infected cells showed a much broader distribution of BrdU-incorporated foci, which presumably contained both newly synthesized cellular DNA and both dsDNA and ssDNA forms of viral DNA (Fig. 1A, +HCl).
Fig 1.
Cellular DNA replication decreases but still prevails over viral DNA replication during early infection of MVC. (A) Immunofluorescence analysis of DNA replication. WRD cells were seeded on chamber slides 24 h prior to MVC infection. At 18 h p.i., cells were incubated with BrdU for 1 h. The cells on slides were fixed and treated with (+HCl) or without (−HCl) HCl as indicated. Fixed cells were costained with anti-MVC NS1 and anti-BrdU antibodies and DAPI. Confocal images were taken at a magnification of ×100. (B) Dot blot analysis of viral and cellular DNA replication. At the indicated times p.i., mock- or MVC-infected cells were incubated with BrdU for 1 h. BrdU-labeled cells were collected and extracted for total DNA and Hirt DNA (lower-molecular-weight DNA), respectively. The DNA samples were denatured, dot blotted, and immunostained with an anti-BrdU antibody. (C) Southern blot analysis of viral DNA in preparations of total DNA and Hirt DNA. The total DNA and Hirt DNA samples extracted from MVC-infected cells were denatured, dot-blotted, and hybridized with a 32P-labeled MVC NSCap probe (6).
To determine the relative levels of incorporated BrdU in cellular DNA versus viral DNA under the denaturation condition, total DNA and low-molecular-weight DNA (Hirt DNA) of infected cells were extracted from equal numbers of cells and were analyzed by a dot blot assay. Incorporated BrdU in Hirt DNA of MVC-infected cells was detected at a background level, as was that seen in Hirt DNA of mock-infected cells (Fig. 1B, Hirt DNA), except for the Hirt DNA prepared from MVC-infected cells at 18 h (Fig. 1B, arrow), suggesting a peak of viral DNA replication. At this peak, the incorporation of BrdU into Hirt DNA was over 20 times lower than that into cellular DNA (Fig. 1B, 18 h). At 48 h p.i., BrdU incorporation into cellular DNA abruptly dropped (Fig. 1B, 48 h/MVC/Total DNA), which was due to the G2/M arrest of infected cells (Fig. 2A) (3). Notably, the Hirt DNA samples contained nearly all the viral DNA in purified total DNA of MVC-infected cells (Fig. 1C) and were contaminated only with a very low level (<5% of) cellular DNA (Fig. 1B, 48 h/Mock). Hence, these results confirm that the majority of incorporated BrdU signaling resulted from cellular DNA replication in infected cells. In subsequent studies, denaturation of infected cells was used to differentiate cellular DNA replication from viral DNA replication.
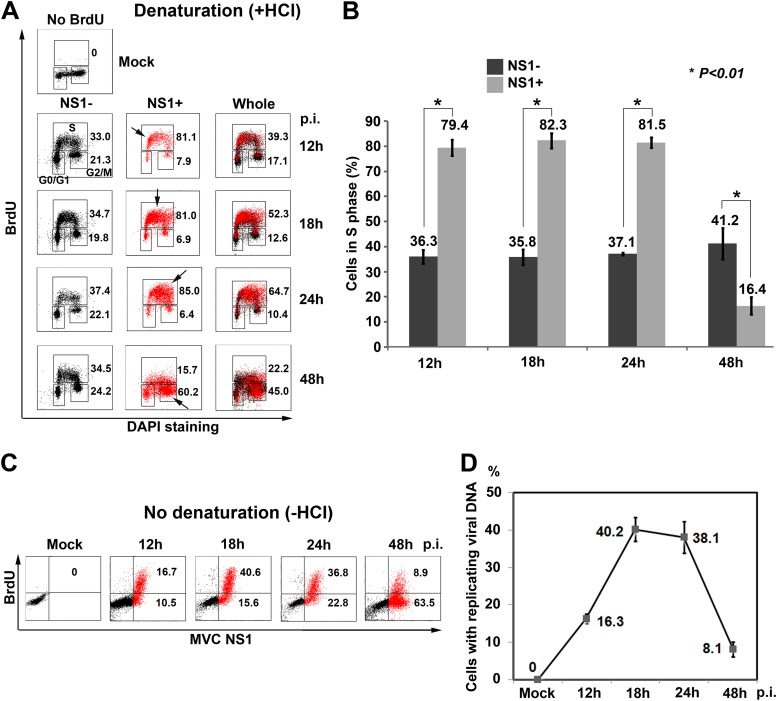
Fig 2.
MVC DNA replication arrests host cells in S phase. (A and B) Flow cytometry analysis of the cell cycle. (A) At the indicated times p.i., mock- or MVC-infected cells were BrdU labeled, treated with HCl (+HCl), stained, and analyzed by flow cytometry. Unlabeled mock-infected cells were used as a negative control for anti-BrdU staining. Both the NS1-positive (NS1+, marked in red) and -negative (NS1−, marked in black, bystander of whole population) cell populations in infected cells were gated. The percentage of cells in each phase was gated in the NS1+ and NS1− cell populations and in the whole population (whole) based on the intensity of BrdU staining and DNA content. Numbers show percentages of cells in S phase (upper) and G2/M phase (lower) in each histogram. Arrows indicate the most concentrated cell population. (B) The percentages of cells in S phase are shown as means (numbers) and standard deviations (error bars) and were generated from at least three independent experiments. P values were determined using Student's t test. (C and D) Flow cytometry analysis of viral DNA replication. (C) At the indicated times p.i., mock- or MVC-infected cells were incubated with BrdU for 1 h. The cells were costained with anti-NS1 and anti-BrdU antibodies and DAPI and analyzed by flow cytometry. NS1+ cells were marked in red. Numbers shown in the histograms indicate percentages of cells with BrdU incorporation from one representative experiment. (D) The percentages of BrdU-positive (BrdU+) cells are plotted to the time points p.i. Averages and standard deviations are shown at each time point and were obtained from at least three independent experiments.
The cell cycle change was then examined in MVC-infected cells. At all the time points p.i., approximately 36% of NS1-negative cells (NS1−) in infected cells were in S phase (Fig. 2A, NS1−). At 12 h p.i., NS1-positive cells (NS1+) in infected cells showed 81% in S phase. The majority of these NS1+ cells were actually in early S phase as shown by a lower DNA content (Fig. 2A, NS1+/12 h). NS1+ cells progressed to mid-S phase at 18 h p.i. and late S phase at 24 h p.i. (Fig. 2A, NS1+/18 h and 24 h). At 48 h p.i., viral DNA replication slowed down; only 16% of NS1+ cells were in S phase, and the majority (∼60%) of NS1+ cells had moved to G2/M phase (Fig. 2A, NS1+/48 h). A statistical analysis of the cell cycle over the course of MVC infection was summarized (Fig. 2B). Overall, MVC infection induced 80% of NS1+ cells in S phase from 12 h to 24 h p.i. but only 33 to 37% of NS1− cells in MVC-infected cells.
We next detected BrdU incorporation in infected cells without denaturation to probe viral DNA replication. Approximately 40% of the total cell population produced a significant level of BrdU-positive signal, which suggests active parvoviral DNA replication (52), at 18 h and 24 h p.i. At 48 h p.i. viral DNA replication had slowed down to 9% of the total cell population, although most of the cells (approximately 72%) expressed NS1, as shown by NS1+ staining (Fig. 2C). These results indicate that active viral DNA replication occurs from 18 h to 24 h p.i. (Fig. 2D).
Taken together, these results show that MVC infection induces accumulation of infected cells in S phase during early infection, which supports active viral DNA replication. Notably, we observed that at early infection (18 h to 24 h p.i.), cellular DNA replication was active but at a lower rate (Fig. 1B, compare dots in lines between Mock and MVC for Total DNA), indicating that S-phase progression was perturbed during early infection.
MVC DNA replication prolongs S phase.
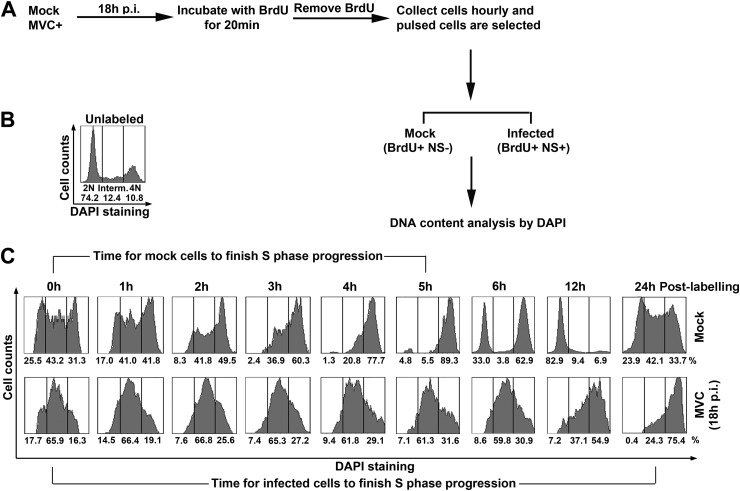
To further investigate whether S-phase accumulation is due to a prolonged S-phase progression, we performed a BrdU pulsing assay to analyze the rate of S-phase progression. Immediately after labeling (0 h postlabeling), infected cells (at 18 h p.i.) with 2N, intermediate (Interm.), and 4N DNA contents were all labeled in the mock group for cellular DNA synthesis in early, mid, and late S phase, respectively (Fig. 3C, Mock/0 h). In contrast, the majority of the labeled infected cells had an intermediate DNA content immediately after labeling (Fig. 3C, MVC/0 h), which was consistent with the cell cycle arrest in mid-S phase at 18 h p.i. (Fig. 2A). Labeled mock-infected cells were able to synthesize DNA smoothly, as evidenced by the fast increase in cells with a 4N DNA content every hour postlabeling. At 5 h postlabeling, approximately 90% of the cells had a DNA content of 4N. At 6 h postlabeling, a large portion of cells finished mitosis and became 2N cells, indicating that those cells had finished one round of replication and the daughter cells had entered G1 phase (Fig. 3C, Mock). In contrast, MVC-infected cells synthesized DNA slowly as the increase in 4N cells was much slower than in the mock-infected group. At 12 h postlabeling, only 55% of the labeled cells had a DNA content of 4N. None of the labeled cells were able to pass G2/M phase even after 24 h postlabeling (approximately 42 p.i.). It took at least 12 h for infected cells to move from early S to late S phase, suggesting that the S phase of MVC-infected cells is prolonged.
Fig 3.
MVC replication delays host cell S-phase progression. (A) Diagram of BrdU pulsing assay. WRD cells were infected with MVC or mock infected. At 18 h p.i., infected cells were incubated with BrdU for 20 min. After BrdU was removed, cells were taken every hour as indicated in panel C. The cells were treated with HCl and then costained with anti-NS1 and anti-BrdU antibodies and DAPI for flow cytometry analysis. (B and C) DNA content analysis. DNA content was gated as 2N, 4N, and intermediate (Interm.; between 2N and 4N) in unlabeled cells (B) based on DAPI staining, which was used as a reference to gate labeled cells (C) with 2N, 4N, and intermediate DNA content. The numbers under each histogram show percentages of the cell population in each gate.
Collectively, these results confirm that MVC DNA replication induces S-phase arrest. Moreover, cellular DNA synthesis in MVC-infected cells is still active but slower, which is consistent with the fact that BrdU was less incorporated in MVC-infected cells than in mock-infected cells (Fig. 1B). Thus, we hypothesized that MVC infection creates a prolonged S phase to block cellular DNA replication and to facilitate viral DNA replication.
Host cellular DNA replication factors are associated with the viral replication centers.
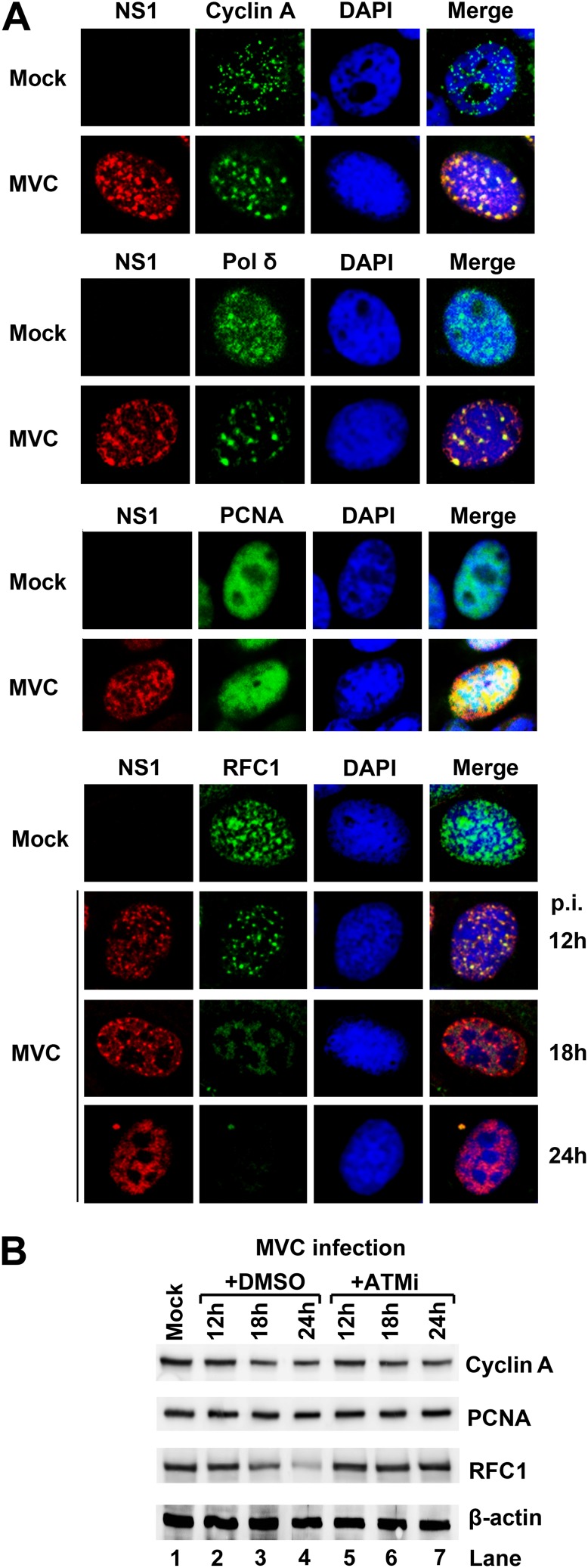
S phase is critical for parvoviruses to hijack the cellular replication machinery (56–58). Previous studies on MVM and H-1 parvovirus have shown that the DNA replication factors PCNA (proliferating cell nuclear antigen), RFC1 (replication factor C1), cyclin A, pol α, and pol δ were recruited into the viral DNA replication compartments during infection (59, 60). Since MVC infection delays S-phase progression, we assessed the localization of these DNA replication factors in the nuclei of infected cells. During MVC infection, at 18 h p.i., RFC1, pol δ, and cyclin A were strongly associated with MVC NS1 (Fig. 4A), and PCNA was distributed in a pan-nuclear pattern in MVC-infected cells, suggesting that these replication factors were abundantly associated with the viral replication centers in the environment of prolonged S phase. Notably, the level of RFC1, a component of the clamp loader RFC complex that drives PCNA and polymerase loading onto the replication fork, disappeared gradually in the viral DNA replication centers during infection (Fig. 4A, RFC1). Western blot analysis confirmed that the total RFC1 level was significantly reduced at 24 h p.i. (Fig. 4B, RFC1/+DMSO). The levels of other replication factors, such as cyclin A and PCNA, were not significantly decreased during early infection.
Fig 4.
Host DNA replication factors are associated with the viral replication centers. (A) Immunofluorescence staining of cellular DNA replication factors. At 18 h p.i., mock- and MVC-infected cells were fixed and stained with the indicated antibodies and DAPI. For RFC1 staining, infected cells were also analyzed at 12 h and 24 h p.i. (B) Western blot analysis of cellular DNA replication factors. Mock- and MVC-infected cells were either treated with ATM inhibitor KU55933 (ATMi) or DMSO control. At the indicated times p.i., the cells were collected and analyzed by Western blotting using antibodies against proteins as indicated.
Since ATM signaling-mediated intra-S-phase arrest has been reported to be involved in inhibition of cellular DNA replication during S phase (35–39), and since ATM signaling is also required for MVC replication (4), we assessed the protein levels of these replication factors in ATM-inactivated infected cells. The reduction in the total RFC1 level was obviously diminished by an ATM-specific inhibitor, KU55933 (61), at 24 h p.i. (Fig. 4B, RFC1/+ATMi), indicating that the reduction is dependent on ATM signaling. Although ATM inhibitor treatment of MVC-infected cells significantly reduced the percentage of NS1+ cells, it did not change the colocalization patterns of these DNA replication factors with the viral replication centers (data not shown).
Taken together, these results suggest that cellular DNA replication factors are associated with the MVC replication centers and that RFC1 is one of the replication factors to mediate slowing down cellular DNA replication in the intra-S-phase arrest. MVC infection not only creates a prolonged S-phase environment for hijacking cellular DNA replication factors but also reduces the overall level of cellular DNA replication factor RFC1 to inhibit cellular DNA synthesis. Since the reduction in RFC1 was blocked by an ATM-specific inhibitor, we hypothesized that ATM signaling may play a critical role in the inhibition of cellular DNA synthesis that contributes to the delay in S-phase progression and to the intra-S-phase arrest (25, 27).
ATM signaling regulates MVC infection-induced intra-S-phase arrest.
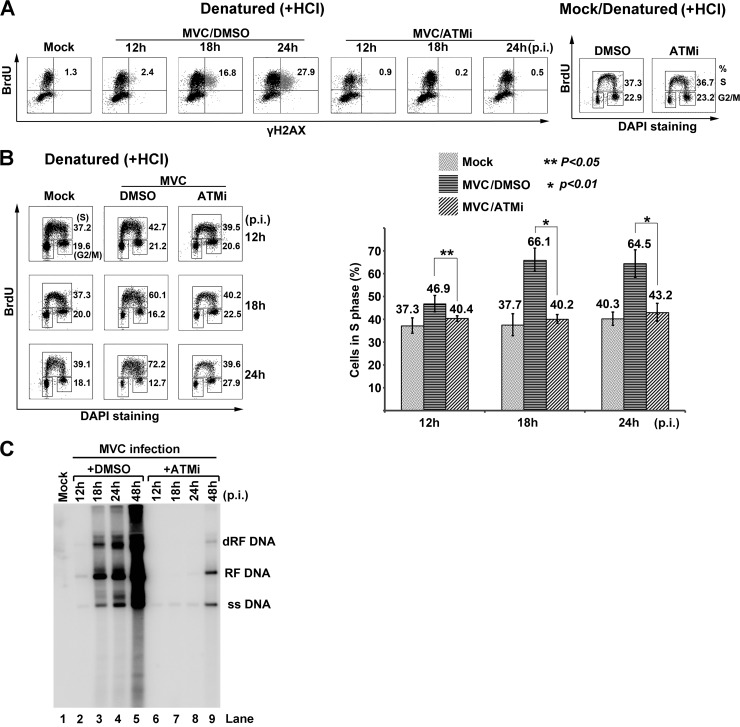
To further examine the correlation between S-phase arrest and ATM signaling, we pulse-chased infected cells with BrdU and analyzed BrdU-labeled cells under denaturation for expression of γH2AX, which is induced by ATM activation during MVC infection (4). As shown in Fig. 5A, nearly all γH2AX-positive cells were also BrdU positive, suggesting that ATM activation correlates with the infection-induced S-phase arrest, whereas treatment with an ATM-specific inhibitor blocked this correlation (Fig. 5A, MVC/ATMi). As a control, treatment with the ATM inhibitor did not change the cell cycle pattern in mock-infected cells (Fig. 5A, Mock/ATMi).
Fig 5.
MVC infection-induced S-phase arrest is regulated by ATM signaling. (A and B) Flow cytometry analysis of DDR. WRD cells were infected with MVC or mock infected and were treated with ATMi or DMSO. At the indicated times p.i., the cells were incubated with BrdU for 1 h. (A) Labeled cells were treated with HCl and then costained with anti-γH2AX and anti-BrdU antibodies for flow cytometry analysis. Numbers in each histogram show percentages of both BrdU- and γH2AX-positive cells. A cell cycle analysis of ATMi-treated mock-infected cells is shown on the right side. Treated cells were costained with an anti-BrdU antibody and DAPI. Numbers show percentages of cells in S phase (upper) and G2/M phase (lower), respectively. (B) Labeled cells were treated with HCl and then costained with an anti-BrdU antibody and DAPI for cell cycle analysis. The whole-cell population in S phase, either treated with ATMi or DMSO, and mock-infected cells were quantified at the indicated times p.i., and data are shown as means ± standard deviations. P values were determined using Student's t test. (C) Southern blot analysis of viral DNA replication. MVC-infected cells treated with either DMSO control or ATMi were collected for preparation of Hirt DNA at the indicated times p.i. Samples were analyzed by Southern blotting. Mock-infected cells were used as a negative control. RF DNA, replicative form DNA; dRF DNA, double RF DNA; ss DNA, single-stranded DNA.
To define the function of ATM signaling in the MVC infection-induced intra-S-phase arrest better, we examined the cell cycle status of infected cells treated with the ATM inhibitor or DMSO (as a control). In ATM inhibitor-treated groups, the population of the cells in S phase was almost reduced to the level of mock-infected cells (Fig. 5B). Thus, inhibition of ATM signaling significantly blocked the infection-induced S-phase arrest. These results strongly suggest that the S-phase arrest, which occurs in replicating cells, is ATM activation dependent. Hence, we conclude that the MVC infection-induced S-phase arrest mimics the intra-S-phase arrest elicited by cellular DSBs. Inhibition of the S-phase arrest by the ATM inhibitor significantly blocked viral DNA replication (Fig. 5C), which is consistent with our previous observations (4).
Altogether, these results show that MVC infection-induced S-phase arrest is blocked by inhibition of ATM signaling, suggesting that ATM signaling induces the intra-S-phase arrest.
MVC infection-induced intra-S-phase arrest is p53 independent.
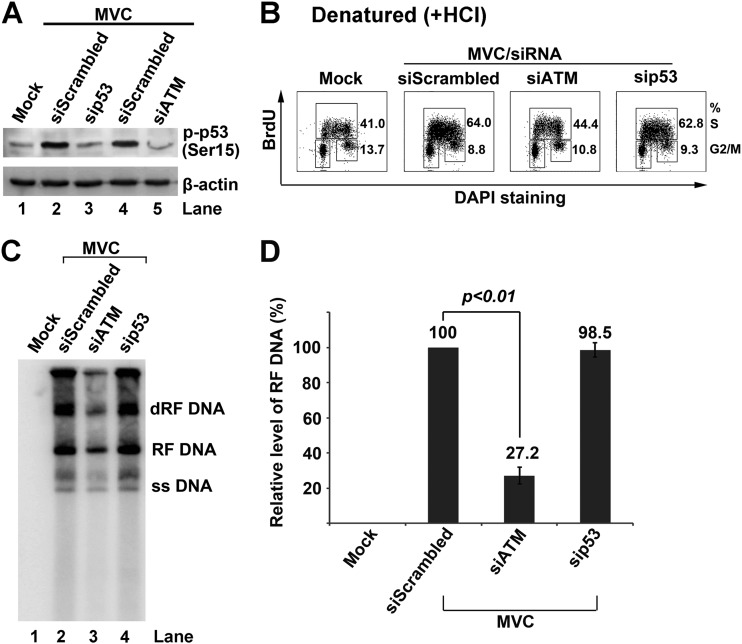
ATM-dependent accumulation of p53 plays a pivotal role in regulating G1 phase arrest to block cellular DNA synthesis following DNA damage (62). It has also been reported that p53 is involved in MVM NS1-mediated S-phase arrest (45). In addition, our previous study also showed that p53 was phosphorylated at serine 15 in the late stage of MVC infection (4). To explore whether p53 activation plays a role in MVC infection-induced intra-S-phase arrest, we assessed the cell cycle pattern and viral DNA replication of MVC-infected cells with knockdown of p53. We confirmed that p53 was phosphorylated at serine 15 at 18 h p.i. (Fig. 6A). Transfection of either p53 siRNA or ATM siRNA reduced p53 phosphorylation to the background level (Fig. 6A), indicating that p53 was phosphorylated by ATM signaling during MVC replication. However, while knockdown of ATM significantly reduced the cell population in S phase, knockdown of p53 did not change the cell population in S phase (Fig. 6B). In parallel, knockdown of ATM but not of p53 significantly blocked MVC DNA replication (Fig. 6C and D). The inhibition of MVC DNA replication by siATM was not as effective as by the ATM inhibitor (Fig. 5C) since siATM knocked down approximately 80% of the endogenous ATM (data not shown).
Fig 6.
p53 is dispensable for MVC infection-induced S-phase arrest. WRD cells were transfected with scrambled siRNA (siScrambled) control, ATM siRNA (siATM), and p53 siRNA (sip53), and then infected with MVC. (A) Western blot analysis of phosphorylated p53 (p-p53). At 18 h p.i., mock- and MVC-infected cells were collected and analyzed by Western blotting for p53 phosphorylated at serine 15, p-p53(Ser15). The same membrane was reprobed for β-actin. (B) Flow cytometry analysis of the cell cycle. At 18 h p.i., mock- or MVC-infected cells were incubated with BrdU for 1 h. Labeled cells were treated with HCl and costained with an anti-BrdU antibody and DAPI and then analyzed by flow cytometry. Numbers show percentages of cells in S phase and G2/M phase in each histogram. (C and D) Southern blot analysis of viral DNA replication. (C) At 18 h p.i., mock- or MVC-infected cells were collected at the indicated times p.i., and Hirt DNA was extracted for Southern blot analysis. (D) The levels of the MVC RF DNA on the blot were quantified using Image Quant TL software (GE Healthcare). The RF DNA level in the scrambled siRNA-treated group was arbitrarily set as 100%. Data are shown as means ± standard deviations from three independent experiments. The P value was determined using Student's t test.
Taken together, we conclude that p53 is not involved in MVC infection-induced intra-S-phase arrest. This result is also consistent with the notion that p53 is not associated with the intra-S-phase arrest induced by cellular DNA damage (25).
SMC1 plays a key role in MVC infection-induced intra-S-phase arrest.
As a well-established intra-S-phase checkpoint protein, SMC1 is a downstream effector of the ATM signaling pathway sensed by the MRN complex (37). We have shown previously that the MRN complex was recruited to the viral replication compartments and that SMC1 was phosphorylated at serine 957 during MVC infection (4). Therefore, we decided to assess the role of SMC1 in the MVC-induced intra-S-phase arrest.
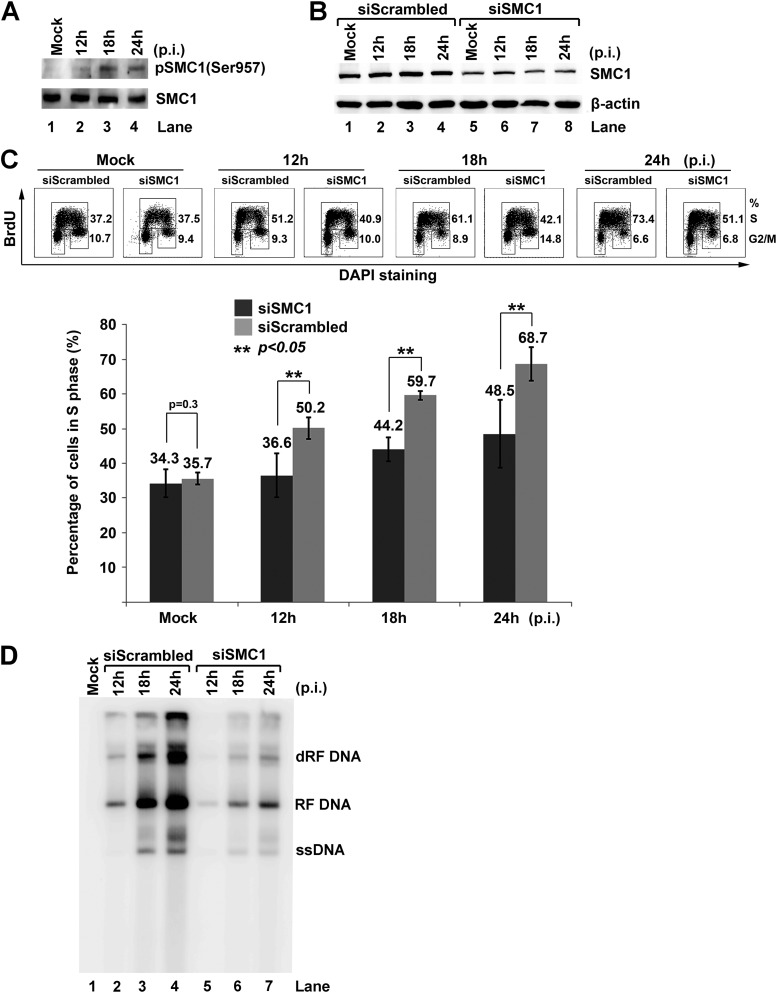
We observed that SMC1 was phosphorylated during early infection (Fig. 7A). Next, we knocked down approximately 60% of the endogenous SMC1 (Fig. 7B), at which level the regular cell cycle pattern was not altered (Fig. 7C, Mock). Notably, knockdown of SMC1 caused 14, 16, and 20% decreases in the cell population in S phase at 12 h, 18 h, and 24 h p.i., respectively (Fig. 7C). The cell cycle patterns of SMC1 siRNA (siSMC1)-treated groups at 12 h and 18 h p.i. were close to those of the mock groups. In addition, knockdown of SMC1 significantly blocked MVC DNA replication (Fig. 7D).
Fig 7.
Knockdown of SMC1 blocks MVC infection-induced intra-S-phase arrest. (A) Western blot analysis of SMC1 expression. WRD cells were infected with MVC. At the indicated times p.i., the cells were collected and analyzed for expression of SMC1 and SMC1 phosphorylated at serine 957, p-SMC1(Ser957). Mock-infected cells were used as a control. (B to D) Knockdown of SMC1 reduces cell population in S phase and viral DNA replication. WRD cells were transfected with siRNA control (siScrambled) or SMC1 siRNA (siSMC1). At 2 days posttransfection, the cells were mock or MVC infected. At the indicated times p.i., cells were analyzed as follows. (B) One-third of the cells were collected and analyzed for SMC1 by Western blotting. (C) One-third of the cells were incubated with BrdU for 1 h, denatured by HCl, and costained with an anti-BrdU antibody and DAPI for flow cytometry analysis. Numbers show percentages of the cell population in S phase and G2/M phase, respectively. The statistical analysis of the percentage of cells in S phase from three independent experiments is shown. Data are shown as means ± standard deviations. P values were determined using Student's t test. (D) One-third of the cells were collected for Hirt DNA extraction and Southern blot analysis.
Collectively, these results show that knockdown of SMC1 blocks MVC infection-induced S-phase arrest and represses MVC DNA replication, suggesting a key role of SMC1 in the intra-S-phase arrest (37).
Phosphorylation of SMC1 at serines 957 and 966 is essential for the intra-S-phase arrest induced during MVC infection.
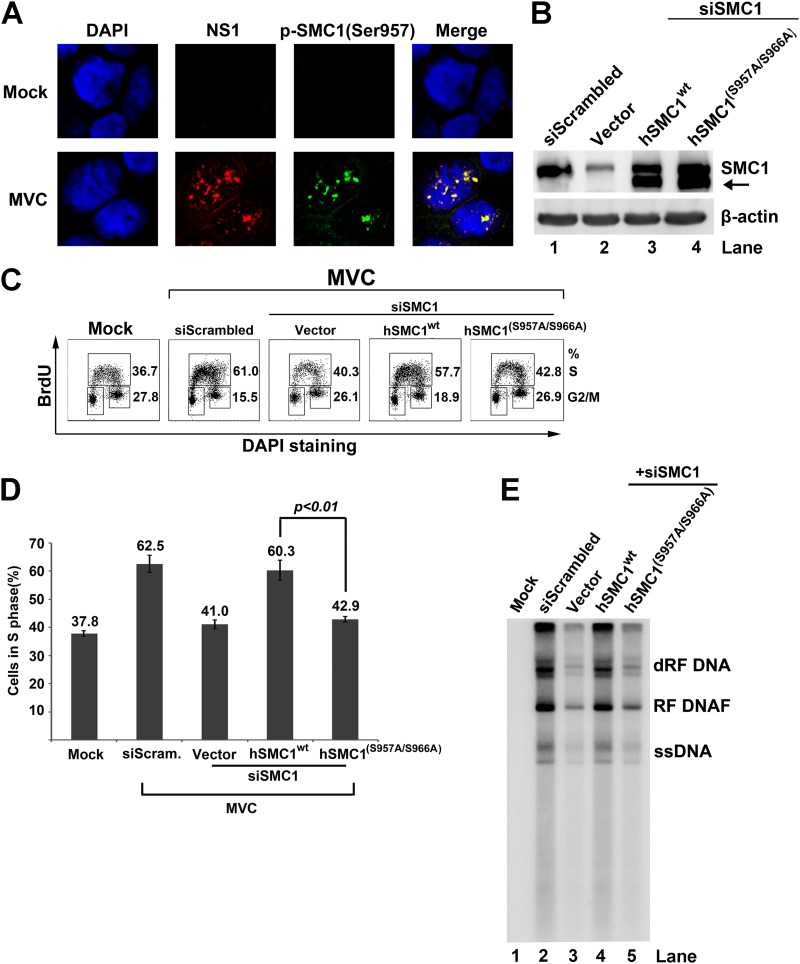
Since phosphorylation of SMC1 at serines 957 and 966 is important for the checkpoint function of SMC1 (36), we assessed their role in the intra-S-phase arrest induced during MVC infection. We observed that phosphorylated SMC1 colocalized with MVC NS1 during early infection (Fig. 8A). To test whether SMC1 phosphorylation at serines 957 and 966 is required for the intra-S-phase arrest, endogenous SMC1 was replaced by ectopic expression of a wild-type human SMC1 (hSMC1wt) or hSMC1 mutated at serines 957 and 966. Canine SMC1 mRNA (XM_538049.3) and human SMC1 (NM_006306.2) encode an identical SMC1 protein sequence (36). The majority of endogenous SMC1 was complemented by hSMCwt (Fig. 8B, hSMC1wt), which nearly fully restored the function of SMC1 in inducing the intra-S-phase arrest (Fig. 8C, hSMC1wt). A SMC1 protein band, shown by an arrow in Fig. 8B, at a position lower than the size of the endogenous SMC1 in transfected cells, is likely an isoform of the transfected human SMC1-encoding gene. Notably, while a dominant negative form of SMC1 (36), termed hSMC1(S957A/S966A), was used to complement the lack of endogenous SMC1 in SMC1 knockdown cells, it was able to block approximately 17% of the cell population in S phase (Fig. 8C and D), indicating that the intra-S-phase arrest is blocked by this dominant negative mutant. Consistently, overexpression of the wild-type hSMC1 but not the mutant hSMC1(S957A/S966A) was able to rescue MVC DNA replication in MVC-infected cells in which SMC1 was knocked down (Fig. 8E). Thus, these results suggest that the phosphorylation of SMC1 at serines 957 and 966 is necessary for the full function of SMC1 as an intra-S-phase checkpoint during MVC infection.
Fig 8.
Complementation of endogenous SMC1 with an SMC1 dominant negative mutant, hSMC1(S957A/S966A), rescues MVC infection-induced intra-S-phase arrest. (A) Immunofluorescence analysis of SMC1 phosphorylation. WRD cells were seeded on chamber slides 24 h prior to MVC infection. At 18 h p.i., cells were fixed and costained with anti-MVC NS1 and anti-p-SMC1(Ser957) antibodies and DAPI. Confocal images were taken at a magnification of ×100. Mock-infected cells were used as a negative control. (B to E) Analysis of SMC1 complementation with hSMC1(S957A/S966A) on the cell cycle. WRD cells were transfected twice with siScrambled or siSMC1 siRNA and subsequently transfected with an empty vector, a plasmid expressing wild-type human SMC1A (hSMC1wt) or dominant negative mutant hSMC1(S975A/S966A). (B) At 24 h posttransfection, cells were collected and analyzed by Western blotting using an anti-SMC1 antibody. The same membrane was reprobed for β-actin. The arrow shows a potential isoform of human SMC1. (C) At 24 h posttransfection, cells were infected with MVC or mock infected. Infected cells were incubated with BrdU for 1 h at 18 h p.i. Then cells were collected, treated with HCl, and costained with an anti-BrdU antibody and DAPI for cell cycle analysis by flow cytometry. Numbers shown in each histogram are percentages of the cell population in S phase and G2/M phase, respectively, as indicated. (D) The statistical analysis of the percentage of cells in S phase, performed from three independent experiments, is shown. Data are shown as means ± standard deviations. P value was determined using Student's t test. (E) At 24 h posttransfection, mock- or MVC-infected cells were collected for Hirt DNA extraction and Southern blot analysis.
Taken together, our results provide evidence that replication of MVC triggers SMC1 phosphorylation, which functions as a checkpoint protein to induce the intra-S-phase arrest of the host cells.
Replicating viral genome, but not damaged cellular DNA, induces intra-S-phase arrest during MVC infection.
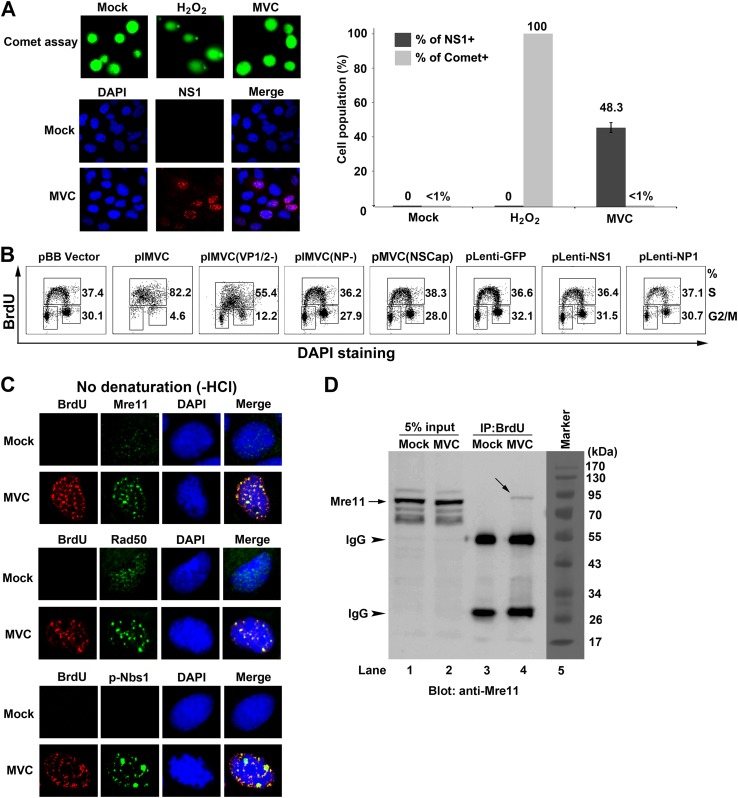
Previous studies of DDR induced by the autonomous parvoviruses MVC, MVM, and B19V have demonstrated that viral DNA replication, but not individual viral protein, triggers a DDR (4, 23, 63, 64). To further examine the cause of MVC infection-induced DDR, we asked whether virus infection is able to cause cellular DNA damage. To this end, we performed a Comet assay, which is commonly used for the detection of both DSBs and SSBs of chromosome DNA (65–68). H2O2 treatment, as a control, was able to cause severe damage to cellular DNA as shown by the fact that nearly all the cells were Comet positive (Comet+; DNA damaged); however, neither mock- nor MVC-infected cells contained Comet+ cells at a level of over 1% (Fig. 9A). These results suggest that the DDR signaling induced during MVC infection comes from viral DNA or its replicative intermediate molecules, rather than from cellular DNA.
Fig 9.
MVC infection-induced DDR and intra-S-phase arrest are dependent on replicating viral DNA. (A) Comet assay analysis. WRD cells were mock or MVC infected. At 18 h p.i., half of the cells were collected and analyzed for cells with damaged DNA (Comet+) by Comet assay; the other half of the cells were fixed and costained with an anti-NS1 antibody and DAPI to quantify the percentage of infected (NS1+) cells by immunofluorescence analysis. Confocal images were taken at a magnification of ×40. A statistical analysis of the percentage of cells with damaged DNA was performed from three independent Comet assays. Data are shown as means ± standard deviations. (B) Flow cytometry analysis of the cell cycle in transfected cells. WRD cells were transfected with the indicated plasmids for 18 h and then were incubated with BrdU for 1 h. The cells were collected, treated with HCl and costained with DAPI, anti-BrdU and anti-NS1 antibodies (for pIMVC and mutant derivatives), or an anti-Flag antibody (for pLenti-based plasmids) for flow cytometry analysis. (C and D) The MRN complex is associated with replicating viral DNA. Mock- or MVC-infected cells were incubated with BrdU for 1 h at 18 h p.i. (C) Immunofluorescence analysis of the MRN complex. The cells were fixed and costained with the indicated antibodies and DAPI. Confocal images were taken at a magnification of ×100. (D) Coimmunoprecipitation (Co-IP) with an anti-BrdU antibody. The BrdU-labeled cells were lysed and centrifuged. The supernatant containing BrdU-labeled viral ssDNA was immunoprecipitated with an anti-BrdU antibody. Immunoprecipitated samples were blotted with an anti-Mre11 antibody. An amount of lysate equal to 5% was used as an input control for each sample. Arrows show the Mre11 bands, whereas arrowheads show the immunoprecipitated light and heavy chains of the IgG antibody.
To determine whether viral DNA replication is required for the intra-S-phase arrest induced during early infection, we transfected cells with a wild-type infectious clone of MVC (pIMVC) and its derivative mutants, pMVC(NSCap) or pIMVC(NP−) and pIMVC(VP1/2−), which do not have the terminal hairpins or express NP1 and capsid proteins, respectively (6, 69). pMVC(NSCap) does not replicate but expresses all viral proteins, pIMVC(NP−) replicates very poorly (approximately 50-fold decrease compared with the wild type), and pIMVC(VP1/2−) replicates at an intermediate level without production of ssDNA (4, 6). In addition, NS1 and NP1 (Flag tagged) were expressed individually. Transfected cells were selected by either NS1 or Flag tagging and analyzed for the cell cycle pattern. Most of the cells transfected with pIMVC and pIMVC(VP1/2−), which replicate viral DNA, were accumulated in S phase, while cells transfected with other plasmids that were replication incompetent or inefficient (6) failed to arrest cells in S phase, though viral proteins were expressed (Fig. 9B). These results indicate that the replicating viral genome, but not viral proteins, was the cause of the intra-S-phase arrest.
The MRN complex is not only the sensor of DDR but also the initiator of the ATM-SMC1 signaling-induced intra-S-phase arrest (36, 37, 70). We hypothesized that the replicating viral DNA is likely sensed by the MRN complex as damaged DNA, which activates the DDR signaling and thereafter the intra-S-phase arrest. To prove this, we checked the localization of the MRN complex in the nuclei of MVC-infected cells. At 18 h p.i., a time point at which MVC DNA actively replicates, Mre11, Rad50, and phosphorylated Nbs1 (p-Nbs1) all colocalized within the viral DNA replication centers as shown with anti-BrdU staining (Fig. 9C). Furthermore, we performed a BrdU-IP assay to determine whether the newly synthesized viral ssDNA/intermediates was associated with the MRN complex. Notably, we were able to precipitate Mre11, the DNA binding component of the MRN complex (71), from BrdU-incorporated MVC-infected cells but not from mock-infected cells (Fig. 9D, lane 4 versus lane 3).
Collectively, we have provided evidence that cellular DNA is not damaged in MVC-infected cells and that the viral DNA replication process is critical for the intra-S-phase arrest induced during MVC infection. More importantly, the replicating viral DNA is able to mimic damaged cellular DNA, perhaps due to aberrant DNA structures, to recruit the MRN complex, which in turn activates ATM signaling and initiates the intra-S-phase arrest.
DISCUSSION
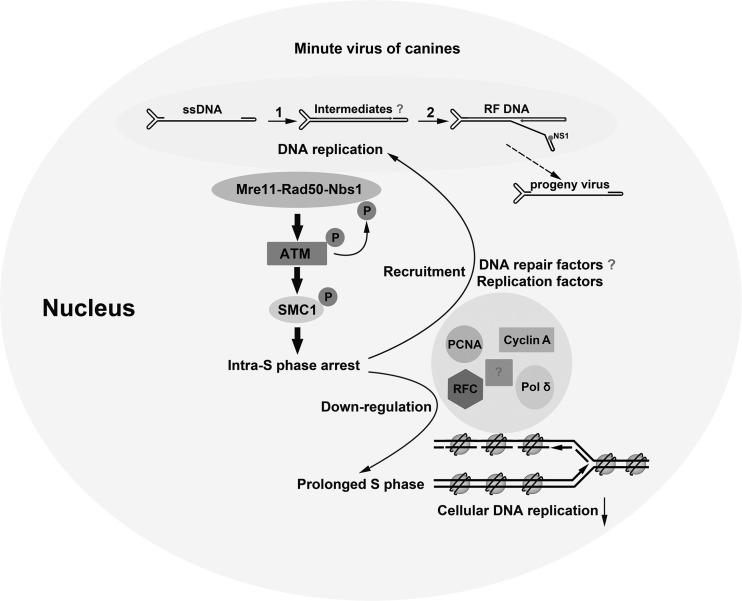
In this study, we demonstrated that infection of Bocavirus MVC induces the intra-S-phase arrest to delay S-phase progression and to hijack cellular DNA replication factors for viral DNA replication. The intra-S-phase arrest is mediated by ATM signaling through phosphorylation of SMC1. The study also provided evidence that the MVC infection-induced DDR is elicited by replicating viral DNA, which is sensed by the MRN complex. Taken together, the study provides, for the first time, a novel DNA replication model for autonomous parvovirus (Fig. 10).
Fig 10.
Proposed model for autonomous parvovirus DNA replication in the context of the intra-S-phase arrest. The proposed pathways utilized by autonomous parvovirus during viral DNA replication are described in detail in the Discussion. The question mark indicates steps not well understood.
In this model, MVC DNA replication triggers the intra-S-phase arrest through the MRN-ATM-SMC1 pathway. The replicating viral DNA mimics damaged DNA that is sensed by the MRN complex. The intra-S-phase arrest blocks cellular DNA synthesis and therefore prolongs S phase in infected cells, presumably through degradation or transcriptional regulation of DNA replication factors. In contrast, the MRN complex may coordinate DNA replication and repair factors through SMC1 activation to facilitate viral DNA synthesis. The feedback loop between viral DNA replication and the intra-S-phase arrest plays an essential role in modulation of the cellular environment by MVC to make it conducive to viral DNA replication.
One of the important findings of this study is that S phase is required but not sufficient for MVC DNA replication. It has been reported that MVM DNA replication is strictly dependent on cellular replication factors expressed in S phase (58, 59, 72). The basic replication machinery components, such as PCNA, RPA, pol α, pol δ, and cyclin A, all colocalized within the autonomous parvovirus-associated replication (APAR) bodies (59, 60). In vitro studies indicated that the cyclin A level directly affects MVM DNA replication efficiency (56) and that PCNA, RPA, and pol δ are essential for MVM DNA replication (73, 74); however, like many other DNA viruses, autonomous parvovirus infection blocks cellular DNA synthesis (43–45, 75, 76), which was thought to be due to competition for access to the cellular replication machinery by viral DNA replication (75, 76). Hence, cellular DNA replication is essential for autonomous parvovirus DNA. Here, we show that MVC DNA replicates poorly in both ATM inhibitor-treated and ATM-knockdown cells which have normal S-phase progression. Thus, we provide evidence that cellular DNA replication is not sufficient for MVC DNA replication. We conclude that, in addition to the requirement that infected cells be in S phase, which supplies DNA replication factors, the intra-S-phase arrest is necessary for autonomous parvovirus to compete with cellular DNA synthesis for viral DNA replication. We hypothesize that the intra-S-phase arrest facilitates the recruitment of DNA replication factors through a DNA repair pathway, since intra-S-phase arrest normally coordinates DNA repair following DDR induced by damaged cellular DNA (25, 77) and restarts of stalled DNA replication forks (28).
Inhibition of cellular DNA replication is a common strategy for DNA viruses to modulate the host cellular environment to make it conducive to viral DNA replication. Due to the limited genetic resource, parvoviruses neither encode their own polymerase nor drive infected cells into S phase through their viral components (75, 76). In comparison to parvoviruses, the inhibition processes of cellular DNA replication by other DNA viruses are often regulated by viral proteins that target the cellular DNA replication machinery. For instance, via viral protein pUL117, human cytomegalovirus (HCMV) blocks host DNA synthesis by delaying the accumulation of the mini-chromosome maintenance (MCM) complex proteins onto chromatin (41). Human papillomavirus (HPV) inhibits host DNA replication by viral early protein E4-mediated suppression of cellular replication origin licensing (42); however, polyomaviruses take advantage of DDR signaling to block cellular DNA synthesis. Simian virus 40 (SV40) infection uses the ATR-Δp53-p21 pathway to downregulate cyclin A-CDK2/1 activity, which forces the host cells to remain in S phase (78), whereas the polyomavirus RA strain has been shown to utilize ATM-SMC1 signaling to override cell cycle regulation and prolong S phase (79). As a result, viral infection-triggered intra-S-phase arrest slowed down cellular DNA synthesis; however, the intra-S-phase arrest induced by polyomaviruses is largely regulated by the viral large T antigen (16, 79–81). In contrast, none of the MVC-encoded proteins are involved in cell cycle regulation (3) (Fig. 9). Therefore, we have identified, for the first time, a viral DNA replication-dependent intra-S-phase arrest that is ATM mediated.
The ATM-SMC1 pathway is intimately involved in slowing down the cellular DNA replication rate in response to DSBs (25); however, how phosphorylated SMC1 interferes with cellular DNA replication remains unclear. At least in the intra-S-phase arrest induced during MVC infection, RFC1, which is a key component of the RFC complex that loads PCNA to replicating DNA (82, 83), is a target for downregulation (Fig. 4). Notably, during the very early phase of infection, RFC1 colocalized within the viral replication centers and later disappeared from the centers when viral DNA was actively replicating. This led us to hypothesize that RFC1 is required for the conversion of viral ssDNA to the double-stranded replicative form (RF DNA) (Fig. 10, Step 1) upon virus infection. Nevertheless, the downregulation of RFC1 during the intra-S-phase arrest provides a candidate for linking SMC1 activation with downregulation of cellular DNA replication. The function of RFC1 in MVC DNA replication and in SMC1-mediated intra-S-phase arrest warrants further investigation.
Studies of virus infection-induced DDR have uncovered novel mechanisms underlying virus-host interaction (15, 18). Although early studies indicated that infection by most DNA viruses was able to create lesions on cellular DNA involving viral proteins (44, 80, 84–87), whether this is common and the major cause of the DDR signaling is not clear. MVC infection did not cause obvious damage to cellular DNA (Fig. 9); hence, the DNA damage signaling induced during MVC infection must come from viral DNA. We and others previously have shown that replication of autonomous parvovirus is required for triggering a DDR (4, 23, 63, 64). Here, we provide evidence, for the first time, that replicating viral genomes (or intermediates) mimic damaged DNA (likely DSBs), which, in the case of autonomous parvovirus, likely involves the unique hairpin structures, thereby recruiting the MRN complex and DDR proteins. However, due to the difficulty of isolating such intermediate DNA, we are not able to provide direct evidence to show that such DNA structures can directly induce DDR signaling. Nevertheless, in addition to the fact that the DNA damage sensor, the MRN complex, is directly associated with the replicating viral ssDNA, Nbs1 was phosphorylated in the viral replication centers (Fig. 9), strongly suggesting that a DNA repair pathway followed by the intra-S-phase arrest is involved in MVC DNA replication. Interestingly, accumulating evidence has shown that DNA repair factors localize in the replication compartments of many DNA viruses; for instance, the homologous recombinational repair (HRR) factors are recruited into the replication centers of Epstein-Barr virus (EBV), SV40, and HPV (80, 88, 89). It was suggested that HRR factors are recruited to repair DSBs on the viral genome in the viral replication compartments but not for viral DNA replication. It is understandable that the DSB-initiated repair pathways of homologous recombination and nonhomologous end joining (NHEJ) are involved in the replication of DNA viruses whose genome is dsDNA, since their replication often involves a step of circularization; however, DNA replication of autonomous parvoviruses, whose genome is ssDNA, follows a rolling-hairpin strategy of DNA replication which does not involve circularization of any replication intermediates (54). The fact that SMC1, a cohesion protein of chromosome DNA, plays a key role in MVC DNA replication may also suggest that it maintains proper alignment of the parvoviral minichromosome (90, 91) for terminal resolution of RF DNA (54), in addition to its role in the intra-S-phase arrest. How these DNA repair factors accumulated in the viral replication centers facilitate viral DNA replication, in particular during autonomous parvovirus infection, remains unknown and is a central question in parvovirus DNA replication.
In summary, MVC infection triggers a MRN-ATM-SMC1-mediated intra-S-phase arrest to create an S-phase environment and to recruit the cellular DNA replication machinery, and perhaps the DNA repair machinery, to facilitate MVC DNA replication. Such a strategy may represent a common feature of the DDR induced by other autonomous parvoviruses, which are dependent on S phase for replication in host cells.
ACKNOWLEDGMENTS
We thank David Pintel for suggestions during this study and critical reading of the manuscript. We are indebted to Aaron Chen for his work in initiating the BrdU incorporation assay in the lab.
This work was supported by PHS grants AI070723 and AI085236 from the National Institute of Allergy and Infectious Diseases (NIAID) and P30 GM103326 from the National Institute of General Medical Sciences (NIGMS).
Footnotes
Published ahead of print 30 January 2013
REFERENCES
- 1. Bern KI, Parrish CR. 2007. Parvoviruses, p 2437–2477 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA. (ed), Fields virology, fifth ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2. Young NS, Brown KE. 2004. Parvovirus B19. N. Engl. J. Med. 350:586–597 [DOI] [PubMed] [Google Scholar]
- 3. Chen AY, Luo Y, Cheng F, Sun Y, Qiu J. 2010. Bocavirus infection induces mitochondrion-mediated apoptosis and cell cycle arrest at G2/M phase. J. Virol. 84:5615–5626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Luo Y, Chen AY, Qiu J. 2011. Bocavirus infection induces a DNA damage response that facilitates viral DNA replication and mediates cell death. J. Virol. 85:133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sukhu L, Fasina O, Burger L, Rai A, Qiu J, Pintel DJ. 2013. Characterization of the nonstructural proteins of the bocavirus minute virus of canines. J. Virol. 87:1098–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sun Y, Chen AY, Cheng F, Guan W, Johnson FB, Qiu J. 2009. Molecular characterization of infectious clones of the minute virus of canines reveals unique features of bocaviruses. J. Virol. 83:3956–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Binn LN, Lazar EC, Eddy GA, Kajima M. 1970. Recovery and characterization of a minute virus of canines. Infect. Immun. 1:503–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U. S. A. 102:12891–12896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang Q, Deng X, Yan Z, Cheng F, Luo Y, Shen W, Lei-Butters DC, Chen AY, Li Y, Tang L, Soderlund-Venermo M, Engelhardt JF, Qiu J. 2012. Establishment of a reverse genetics system for studying human bocavirus in human airway epithelia. PLoS Pathog. 8:e1002899 doi:10.1371/journal.ppat.1002899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Allander T, Jartti T, Gupta S, Niesters HG, Lehtinen P, Osterback R, Vuorinen T, Waris M, Bjerkner A, Tiveljung-Lindell A, van den Hoogen BG, Hyypia T, Ruuskanen O. 2007. Human bocavirus and acute wheezing in children. Clin. Infect. Dis. 44:904–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Edner N, Castillo-Rodas P, Falk L, Hedman K, Soderlund-Venermo M, Allander T. 2012. Life-threatening respiratory tract disease with human bocavirus-1 infection in a four-year-old child. J. Clin. Microbiol. 50:531–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jartti T, Hedman K, Jartti L, Ruuskanen O, Allander T, Soderlund-Venermo M. 2012. Human bocavirus—the first 5 years. Rev. Med. Virol. 22:46–64 [DOI] [PubMed] [Google Scholar]
- 13. Meriluoto M, Hedman L, Tanner L, Simell V, Makinen M, Simell S, Mykkanen J, Korpelainen J, Ruuskanen O, Ilonen J, Knip M, Simell O, Hedman K, Soderlund-Venermo M. 2012. Association of human bocavirus 1 Infection with respiratory disease in childhood follow-up study, Finland. Emerg. Infect. Dis. 18:264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ursic T, Jevsnik M, Zigon N, Krivec U, Beden AB, Praprotnik M, Petrovec M. 2012. Human bocavirus and other respiratory viral infections in a 2-year cohort of hospitalized children. J. Med. Virol. 84:99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lilley CE, Schwartz RA, Weitzman MD. 2007. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 15:119–126 [DOI] [PubMed] [Google Scholar]
- 16. Nikitin PA, Lutig MA. 2011. At a crossroads: human DNA tumor viruses and the host DNA damage response. Future Virol. 6:813–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Turnell AS, Grand RJ. 2012. DNA viruses and the cellular DNA-damage response. J. Gen. Virol. 93:2076–2097 [DOI] [PubMed] [Google Scholar]
- 18. Weitzman MD, Lilley CE, Chaurushiya MS. 2010. Genomes in conflict: maintaining genome integrity during virus infection. Annu. Rev. Microbiol. 64:61–81:61–81 [DOI] [PubMed] [Google Scholar]
- 19. Ciccia A, Elledge SJ. 2010. The DNA damage response: making it safe to play with knives. Mol. Cell 40:179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petrini JH, Stracker TH. 2003. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 13:458–462 [DOI] [PubMed] [Google Scholar]
- 21. Durocher D, Jackson SP. 2001. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr. Opin. Cell Biol. 13:225–231 [DOI] [PubMed] [Google Scholar]
- 22. Yang J, Yu Y, Hamrick HE, Duerksen-Hughes PJ. 2003. ATM, ATR and DNA-PK: initiators of the cellular genotoxic stress responses. Carcinogenesis 24:1571–1580 [DOI] [PubMed] [Google Scholar]
- 23. Adeyemi RO, Landry S, Davis ME, Weitzman MD, Pintel DJ. 2010. Parvovirus minute virus of mice induces a DNA damage response that facilitates viral replication. PLoS Pathog. 6:e1001141 doi:10.1371/journal.ppat.1001141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ruiz Z, Mihaylov IS, Cotmore SF, Tattersall P. 2011. Recruitment of DNA replication and damage response proteins to viral replication centers during infection with NS2 mutants of minute virus of mice (MVM). Virology 410:375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bartek J, Lukas C, Lukas J. 2004. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell. Biol. 5:792–804 [DOI] [PubMed] [Google Scholar]
- 26. Gottifredi V, Prives C. 2005. The S phase checkpoint: when the crowd meets at the fork. Semin. Cell Dev. Biol. 16:355–368 [DOI] [PubMed] [Google Scholar]
- 27. Grallert B, Boye E. 2008. The multiple facets of the intra-S checkpoint. Cell Cycle 7:2315–2320 [DOI] [PubMed] [Google Scholar]
- 28. Hu J, Sun L, Shen F, Chen Y, Hua Y, Liu Y, Zhang M, Hu Y, Wang Q, Xu W, Sun F, Ji J, Murray JM, Carr AM, Kong D. 2012. The intra-s phase checkpoint targets dna2 to prevent stalled replication forks from reversing. Cell 149:1221–1232 [DOI] [PubMed] [Google Scholar]
- 29. Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, Hershko A, Pagano M, Draetta GF. 2003. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature 426:87–91 [DOI] [PubMed] [Google Scholar]
- 30. Falck J, Petrini JH, Williams BR, Lukas J, Bartek J. 2002. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat. Genet. 30:290–294 [DOI] [PubMed] [Google Scholar]
- 31. Karnani N, Dutta A. 2011. The effect of the intra-S-phase checkpoint on origins of replication in human cells. Genes Dev. 25:621–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mailand N, Falck J, Lukas C, Syljuasen RG, Welcker M, Bartek J, Lukas J. 2000. Rapid destruction of human Cdc25A in response to DNA damage. Science 288:1425–1429 [DOI] [PubMed] [Google Scholar]
- 33. Errico A, Costanzo V. 2012. Mechanisms of replication fork protection: a safeguard for genome stability. Crit. Rev. Biochem. Mol. Biol. 47:222–235 [DOI] [PubMed] [Google Scholar]
- 34. Strunnikov AV, Jessberger R. 1999. Structural maintenance of chromosomes (SMC) proteins: conserved molecular properties for multiple biological functions. Eur. J. Biochem. 263:6–13 [DOI] [PubMed] [Google Scholar]
- 35. Antoccia A, Sakamoto S, Matsuura S, Tauchi H, Komatsu K. 2008. NBS1 prevents chromatid-type aberrations through ATM-dependent interactions with SMC1. Radiat. Res. 170:345–352 [DOI] [PubMed] [Google Scholar]
- 36. Kim ST, Xu B, Kastan MB. 2002. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 16:560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. 2004. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 18:1423–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lehmann AR. 2005. The role of SMC proteins in the responses to DNA damage. DNA Repair (Amst.) 4:309–314 [DOI] [PubMed] [Google Scholar]
- 39. Wakeman TP, Kim WJ, Callens S, Chiu A, Brown KD, Xu B. 2004. The ATM-SMC1 pathway is essential for activation of the chromium[VI]-induced S-phase checkpoint. Mutat. Res. 554:241–251 [DOI] [PubMed] [Google Scholar]
- 40. Hermanns J, Schulze A, Jansen D, Kleinschmidt JA, Schmidt R, HHzur 1997. Infection of primary cells by adeno-associated virus type 2 results in a modulation of cell cycle-regulating proteins. J. Virol. 71:6020–6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Qian Z, Leung-Pineda V, Xuan B, Piwnica-Worms H, Yu D. 2010. Human cytomegalovirus protein pUL117 targets the mini-chromosome maintenance complex and suppresses cellular DNA synthesis. PLoS Pathog. 6:e1000814 doi:10.1371/journal.ppat.1000814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roberts S, Kingsbury SR, Stoeber K, Knight GL, Gallimore PH, Williams GH. 2008. Identification of an arginine-rich motif in human papillomavirus type 1 E1^E4 protein necessary for E4-mediated inhibition of cellular DNA synthesis in vitro and in cells. J. Virol. 82:9056–9064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Op De Beeck A, Anouja F, Mousset S, Rommelaere J, Caillet-Fauquet P. 1995. The nonstructural proteins of the autonomous parvovirus minute virus of mice interfere with the cell cycle, inducing accumulation in G2. Cell Growth Differ. 6:781–787 [PubMed] [Google Scholar]
- 44. Op De Beeck A, Caillet-Fauquet P. 1997. The NS1 protein of the autonomous parvovirus minute virus of mice blocks cellular DNA replication: a consequence of lesions to the chromatin? J. Virol. 71:5323–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Op De Beeck A, Sobczak-Thepot J, Sirma H, Bourgain F, Brechot C, Caillet-Fauquet P. 2001. NS1- and minute virus of mice-induced cell cycle arrest: involvement of p53 and p21cip1. J. Virol. 75:11071–11078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guan W, Wong S, Zhi N, Qiu J. 2009. The genome of human parvovirus B19 virus can replicate in nonpermissive cells with the help of adenovirus genes and produces infectious virus. J. Virol. 83:9541–9553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guan W, Cheng F, Yoto Y, Kleiboeker S, Wong S, Zhi N, Pintel DJ, Qiu J. 2008. Block to the production of full-length B19 virus transcripts by internal polyadenylation is overcome by replication of the viral genome. J. Virol. 82:9951–9963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hirt B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365–369 [DOI] [PubMed] [Google Scholar]
- 49. Ueda J, Saito H, Watanabe H, Evers BM. 2005. Novel and quantitative DNA dot-blotting method for assessment of in vivo proliferation. Am. J. Physiol. Gastrointest. Liver Physiol. 288:G842–G847 [DOI] [PubMed] [Google Scholar]
- 50. Hang H, Fox MH. 2004. Analysis of the mammalian cell cycle by flow cytometry. Methods Mol. Biol. 241:23–35 [DOI] [PubMed] [Google Scholar]
- 51. Gratzner HG. 1982. Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: a new reagent for detection of DNA replication. Science 218:474–475 [DOI] [PubMed] [Google Scholar]
- 52. Lachmann S, Rommeleare J, Nuesch JP. 2003. Novel PKCeta is required to activate replicative functions of the major nonstructural protein NS1 of minute virus of mice. J. Virol. 77:8048–8060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oleksiewicz MB, Wolfinbarger JB, Bloom ME. 1998. A comparison between permissive and restricted infections with Aleutian mink disease parvovirus (ADV): characterization of the viral protein composition at nuclear sites of virus replication. Virus Res. 56:41–51 [DOI] [PubMed] [Google Scholar]
- 54. Cotmore SF, Tattersall P. 2005. A rolling-haipin strategy: basic mechanisms of DNA replication in the parvoviruses, p 171–181 In Kerr J, Cotmore SF, Bloom ME, Linden RM, Parrish CR. (ed), Parvoviruses. Hoddler Arond, London, United Kingdom [Google Scholar]
- 55. Moran R, Darzynkiewicz Z, Staiano-Coico L, Melamed MR. 1985. Detection of 5-bromodeoxyuridine (BrdUrd) incorporation by monoclonal antibodies: role of the DNA denaturation step. J. Histochem. Cytochem. 33:821–827 [DOI] [PubMed] [Google Scholar]
- 56. Bashir T, Horlein R, Rommelaere J, Willwand K. 2000. Cyclin A activates the DNA polymerase delta-dependent elongation machinery in vitro: A parvovirus DNA replication model. Proc. Natl. Acad. Sci. U. S. A. 97:5522–5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cotmore SF, Tattersall P. 1987. The autonomously replicating parvoviruses of vertebrates. Adv. Virus Res. 33:91–174 [DOI] [PubMed] [Google Scholar]
- 58. Wolter S, Richards R, Armentrout RW. 1980. Cell cycle-dependent replication of the DNA of minute virus of mice, a parvovirus. Biochim. Biophys. Acta 607:420–431 [DOI] [PubMed] [Google Scholar]
- 59. Bashir T, Rommelaere J, Cziepluch C. 2001. In vivo accumulation of cyclin A and cellular replication factors in autonomous parvovirus minute virus of mice-associated replication bodies. J. Virol. 75:4394–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cziepluch C, Lampel S, Grewenig A, Grund C, Lichter P, Rommelaere J. 2000. H-1 parvovirus-associated replication bodies: a distinct virus-induced nuclear structure. J. Virol. 74:4807–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. 2004. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 64:9152–9159 [DOI] [PubMed] [Google Scholar]
- 62. Delia D, Fontanella E, Ferrario C, Chessa L, Mizutani S. 2003. DNA damage-induced cell-cycle phase regulation of p53 and p21waf1 in normal and ATM-defective cells. Oncogene 22:7866–7869 [DOI] [PubMed] [Google Scholar]
- 63. Lou S, Luo Y, Cheng F, Huang Q, Shen W, Kleiboeker S, Tisdale JF, Liu Z, Qiu J. 2012. Human parvovirus B19 DNA replication induces a DNA damage response that is dispensable for cell cycle arrest at G2/M phase. J. Virol. 86:10748–10758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Luo Y, Lou S, Deng X, Liu Z, Li Y, Kleiboeker S, Qiu J. 2011. Parvovirus B19 infection of human primary erythroid progenitor cells triggers ATR-Chk1 signaling, which promotes B19 virus replication. J. Virol. 85:8046–8055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Olive PL. 2002. The comet assay. An overview of techniques. Methods Mol. Biol. 203:179–194 [DOI] [PubMed] [Google Scholar]
- 66. Ostling O, Johanson KJ. 1984. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem. Biophys. Res. Commun. 123:291–298 [DOI] [PubMed] [Google Scholar]
- 67. Shaposhnikov SA, Salenko VB, Brunborg G, Nygren J, Collins AR. 2008. Single-cell gel electrophoresis (the comet assay): loops or fragments? Electrophoresis 29:3005–3012 [DOI] [PubMed] [Google Scholar]
- 68. Singh NP, McCoy MT, Tice RR, Schneider EL. 1988. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 175:184–191 [DOI] [PubMed] [Google Scholar]
- 69. Chen AY, Cheng F, Lou S, Luo Y, Liu Z, Delwart E, Pintel D, Qiu J. 2010. Characterization of the gene expression profile of human bocavirus. Virology 403:145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Paull TT, Lee JH. 2005. The Mre11/Rad50/Nbs1 complex and its role as a DNA double-strand break sensor for ATM. Cell Cycle 4:737–740 [DOI] [PubMed] [Google Scholar]
- 71. Stracker TH, Petrini JH. 2011. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 12:90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Deleu L, Pujol A, Faisst S, Rommelaere J. 1999. Activation of promoter P4 of the autonomous parvovirus minute virus of mice at early S phase is required for productive infection. J. Virol. 73:3877–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Christensen J, Cotmore SF, Tattersall P. 1997. A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polypeptide to initiate parvovirus DNA replication. J. Virol. 71:1405–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Christensen J, Tattersall P. 2002. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J. Virol. 76:6518–6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hardt N, Dinsart C, Spadari S, Pedrali-Noy G, Rommelaere J. 1983. Interrelation between viral and cellular DNA synthesis in mouse cells infected with the parvovirus minute virus of mice. J. Gen. Virol. 64:1991–1998 [DOI] [PubMed] [Google Scholar]
- 76. Oleksiewicz MB, Alexandersen S. 1997. S-phase-dependent cell cycle disturbances caused by Aleutian mink disease parvovirus. J. Virol. 71:1386–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Branzei D, Foiani M. 2008. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 9:297–308 [DOI] [PubMed] [Google Scholar]
- 78. Rohaly G, Korf K, Dehde S, Dornreiter I. 2010. Simian virus 40 activates ATR-Δp53 signaling to override cell cycle and DNA replication control. J. Virol. 84:10727–10747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dahl J, You J, Benjamin TL. 2005. Induction and utilization of an ATM signaling pathway by polyomavirus. J. Virol. 79:13007–13017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Boichuk S, Hu L, Hein J, Gjoerup OV. 2010. Multiple DNA damage signaling and repair pathways deregulated by simian virus 40 large T antigen. J. Virol. 84:8007–8020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shi Y, Dodson GE, Shaikh S, Rundell K, Tibbetts RS. 2005. Ataxia-telangiectasia-mutated (ATM) is a T-antigen kinase that controls SV40 viral replication in vivo. J. Biol. Chem. 280:40195–40200 [DOI] [PubMed] [Google Scholar]
- 82. Bowman GD, O'Donnell M, Kuriyan J. 2004. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 429:724–730 [DOI] [PubMed] [Google Scholar]
- 83. Zhang G, Gibbs E, Kelman Z, O'Donnell M, Hurwitz J. 1999. Studies on the interactions between human replication factor C and human proliferating cell nuclear antigen. Proc. Natl. Acad. Sci. U. S. A. 96:1869–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chenet-Monte C, Mohammad F, Celluzzi CM, Schaffer PA, Farber FE. 1986. Herpes simplex virus gene products involved in the induction of chromosomal aberrations. Virus Res. 6:245–260 [DOI] [PubMed] [Google Scholar]
- 85. Fortunato EA, Spector DH. 2003. Viral induction of site-specific chromosome damage. Rev. Med. Virol. 13:21–37 [DOI] [PubMed] [Google Scholar]
- 86. Nichols WW, Bradt CI, Toji LH, Godley M, Segawa M. 1978. Induction of sister chromatid exchanges by transformation with simian virus 40. Cancer Res. 38:960–964 [PubMed] [Google Scholar]
- 87. zur Hausen H. 1967. Induction of specific chromosomal aberrations by adenovirus type 12 in human embryonic kidney cells. J. Virol. 1:1174–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gillespie KA, Mehta KP, Laimins LA, Moody CA. 2012. Human papillomaviruses recruit cellular DNA repair and homologous recombination factors to viral replication centers. J. Virol. 86:9520–9526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kudoh A, Iwahori S, Sato Y, Nakayama S, Isomura H, Murata T, Tsurumi T. 2009. Homologous recombinational repair factors are recruited and loaded onto the viral DNA genome in Epstein-Barr virus replication compartments. J. Virol. 83:6641–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ben-Asher E, Bratosin S, Aloni Y. 1982. Intracellular DNA of the parvovirus minute virus of mice is organized in a minichromosome structure. J. Virol. 41:1044–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Doerig C, McMaster G, Sogo J, Bruggmann H, Beard P. 1986. Nucleoprotein complexes of minute virus of mice have a distinct structure different from that of chromatin. J. Virol. 58:817–824 [DOI] [PMC free article] [PubMed] [Google Scholar]