Abstract
T follicular helper (Tfh) cells are a specialized subset of memory CD4+ T cells that are found exclusively within the germinal centers of secondary lymphoid tissues and are important for adaptive antibody responses and B cell memory. Tfh cells do not express CCR5, the primary entry coreceptor for both human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus (SIV), and therefore, we hypothesized that these cells would avoid infection. We studied lymph nodes and spleens from pigtail macaques infected with pathogenic strain SIVmac239 or SIVmac251, to investigate the susceptibility of Tfh cells to SIV infection. Pigtail macaque PD-1high CD127low memory CD4+ T cells have a phenotype comparable to that of human Tfh cells, expressing high levels of CXCR5, interleukin-21 (IL-21), Bcl-6, and inducible T cell costimulator (ICOS). As judged by either proviral DNA or cell-associated viral RNA measurements, macaque Tfh cells were infected with SIV at levels comparable to those in other CD4+ memory T cells. Infection of macaque Tfh cells was evident within weeks of inoculation, yet we confirmed that Tfh cells do not express CCR5 or either of the well-known alternative SIV coreceptors, CXCR6 and GPR15. Mutations in the SIV envelope gp120 region occurred in chronically infected macaques but were uniform across each T cell subset investigated, indicating that the viruses used the same coreceptors to enter different cell subsets. Early infection of Tfh cells represents an unexpected focus of viral infection. Infection of Tfh cells does not interrupt antibody production but may be a factor that limits the quality of antibody responses and has implications for assessing the size of the viral reservoir.
INTRODUCTION
T follicular helper (Tfh) cells are a subset of antigen-experienced CD4+ T cells with a unique ability to home to B cell follicles due to their expression of the chemokine receptor CXCR5, providing help to produce high-affinity, class-switched antibodies and B cell memory (1, 2). They therefore play a critical role in clearance of pathogens following infection, establishment of long-term humoral immunity, and efficacy of vaccines. In humans, Tfh cells in lymphoid tissue have a distinct cell surface membrane phenotype, including CXCR5, high levels of PD-1 (CD279), and low levels of the interleukin-7 receptor alpha (IL-7Rα) chain (CD127), associated with expression of the transcription factor Bcl-6 (reviewed in reference 2). Functionally, Tfh cells are characterized by high-level expression of interleukin-21 (IL-21) (1).
Primary human immunodeficiency virus (HIV) infection is diagnosed by increasing levels of HIV-specific antibodies, as measured by Western blotting, with IgM levels peaking at around 20 days after the onset of acute illness and disappearing around 60 days later (3), while IgG antibody levels continue to increase for months (3–6). This antibody response suggests that class-switching mechanisms mediated by HIV-specific Tfh cells are present and intact, while other HIV-specific CD4+ T cells, particularly Th1 cells that preferentially express CCR5, are relatively transient (7). However, only an extremely small proportion of the HIV-specific antibodies are neutralizing, and most of these are present at low titers (6). Broadly neutralizing anti-HIV-1 antibodies are characterized by the presence of surprisingly high levels of somatic hypermutation, which is believed to be the result of Tfh cell function in germinal centers (8–10). We hypothesized that in human subjects, CXCR5+ Tfh cells would be protected from HIV-1 infection due to their lack of CCR5 expression (2), thus allowing the full development of antibody responses to viral proteins.
Since Tfh cells are localized to secondary lymphoid organs, frequent sampling from patients during different phases of infection is not easily achieved. As an alternative, we have studied these cells isolated from spleen and lymph nodes of pigtail macaques infected with CCR5-dependent, pathogenic simian immunodeficiency virus (SIV) strain SIVmac239 or SIVmac251. We show that a subset of macaque lymphoid memory CD4+ T cells, which are PD-1high CD127low, have the characteristics of Tfh cells. Surprisingly, these cells are infected with SIV at a rate similar to those of other CD4+ memory T cell subsets, despite not expressing CCR5 or either of two alternative coreceptors for SIV, CXCR6/Bonzo and GPR15/BOB. Therefore, we compared the sequences of the SIV envelope gp120 region in Tfh cells with sequences isolated from other CD4+ T cell subsets and found that, as expected, mutations occurred during chronic infection, but these were consistent across different subsets, which indicates that the viruses use the same coreceptor for entry into Tfh cells. However, despite infection, Tfh cell numbers increase in relative terms during chronic infection. These findings impact the interpretation of data originating from primate models of SIV infection and our understanding of HIV-1 immunopathogenesis.
MATERIALS AND METHODS
Macaque tissues and SIV infections.
Spleen and lymph node (LN) samples were taken from 13 pigtail macaques (Macaca nemestrina) and are described in Table 1. Eight out of 13 animals were involved in a study of cytotoxic T lymphocyte (CTL) escape in which unvaccinated macaques were intramuscularly challenged with SIVmac239 (11, 12). Five out of 13 animals were involved in a vaccine study in which selected SIV CD8+ CTL epitopes were incorporated into an influenza virus vector and macaques were immunized with this vector and intravaginally challenged with SIVmac251. This approach provided no protective efficacy compared to unvaccinated controls (13) (data not shown). Spleen, mesenteric lymph nodes (MLNs), and inguinal lymph nodes (ILNs) were taken from macaques at the time of euthanasia. In 7 macaques, inguinal LNs were surgically removed at two time points (Table 1).
Table 1.
Pigtail macaques included in this studya
| Monkey ID | SIV strain | Vaccine received | Postinfection procedure | Plasma viral load (log10 copies/ml) | Peripheral CD4+ T cell count (% of baseline) | Peripheral CD4+ T cell count (cells/μl) |
|---|---|---|---|---|---|---|
| 5504 | SIVmac239 | None | ILN sample taken on day 28 | 5.4 | 85.5 | 1,266 |
| Euthanized on day 77 | 6.3 | 82.6 | 859 | |||
| 6176 | SIVmac239 | None | ILN sample taken on day 28 | 5.8 | 55.5 | 462 |
| Euthanized on day 77 | 7.0 | 52.8 | 316 | |||
| 7448 | SIVmac239 | None | ILN sample taken on day 28 | 5.7 | 99.8 | 1,844 |
| 7476 | SIVmac239 | None | ILN sample taken on day 28 | 5.3 | 87.7 | 1,193 |
| 3B14 | SIVmac239 | None | Euthanized on day 244 | 7.7 | 64.4 | 381 |
| 19341 | SIVmac251 | None | ILN sample taken on day 14 | 7.9 | 94.7 | 1,633 |
| ILN sample taken on day 182 | ND | ND | ND | |||
| 19530 | SIVmac251 | None | ILN sample taken on day −35 | NA | NA | ND |
| Euthanized on day 420 | 5.3 | 89.0 | 774 | |||
| C3754 | SIVmac251 | None | ILN sample taken on day 70 | 6.2 on day 56 | 62.8 | 590 |
| Euthanized on day 224 | 7.4 on Day 182 | 46.2 | 288 | |||
| C0933 | SIVmac251 | Flu-SIV | ILN sample taken on day 14 | 6.4 | 100.9 | 2,268 |
| ILN sample taken on day 469 | ND | 87.0 | 923 | |||
| 25377 | SIVmac251 | Flu-SIV | ILN sample taken on day 14 | 6.5 | 93.8 | 1,455 |
| Euthanized on day 182 | 7.6 | 31.0 | 242 | |||
| 45418 | SIVmac251 | Flu-SIV | ILN sample taken on day 70 | 4.0 on day 56 | 87.1 | 1,522 |
| B0517 | SIVmac251 | Flu-SIV | Euthanized on day 119 | 6.4 on day 105 | 46.2 | 318 |
| B0440 | SIVmac251 | Flu-SIV | Euthanized on day 273 | 6.9 on day 231 | 56.0 | 271 |
ND, not done; NA, not applicable.
Ethics statement.
Lymphoid tissue experiments on pigtail macaques were approved by the Animal Ethics Committee of the Australian Animal Health Laboratory, Geelong, Victoria, CSIRO Livestock Industries, a department of the Australian Government, and cared for in accordance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (14), issued by the National Health and Medical Research Council in conjunction with CSIRO Livestock Industries and the Australian Research Council. The Australian Animal Health Laboratory has Scientific Procedures Premises License (SPPL) number 113.
Single-cell isolation.
Single-cell suspensions were prepared from spleens and LNs within 12 h of resection. Tissues were minced and pushed through a 70-μm sieve for single-cell isolation. Excessive red cells from spleen single-cell preparation were removed by centrifugation with Ficoll-Paque. Single cells from spleens and LNs, along with peripheral blood mononuclear cells (PBMCs), were stained with antibodies for flow cytometry analysis in real time, and cells were also cryopreserved.
Flow cytometry.
Monoclonal antibodies to human or nonhuman primate (NHP) proteins used in this study were CD3-phycoerythrin (PE)-Cy7 (clone SP34-2), CD4-peridinin chlorophyll protein (PerCP)-Cy5.5 (clone L200), CD8-allophycocyanin (APC)-Cy7 (clone SK1), Bcl-6–PE (clone K112-91), ICOS (CD278)-PE (clone DX9), CD130-PE (clone AM64), and purified CCR5 (clone 3A9) from BD Biosciences (San Jose, CA); CD45RA-energy-coupled dye (ECD) (clone 2H4) and CD126-PE (clone M91) from Beckman Coulter (Hialeah, FL); CD127-eFluor450 (clone eBioRDR5) from eBioscience (San Diego, CA); PD-1 (PE or APC) (clone EH-12) and CD127-Brilliant Violet 421 (clone A019D5) from BioLegend (San Diego, CA); purified CXCR5 (clone 710D82.1) from the NIH Reagent Resource (Boston, MA); and purified CXCR6 (clone 56811) and GPR15 (clone 367902) from R&D Systems (Minneapolis, MN).
For detection of surface markers, macaque cells were stained with fluorochrome-conjugated antibodies, according to the manufacturers' directions, for 15 min at room temperature, washed once with PBA (phosphate-buffered saline [PBS] containing 0.5% bovine serum albumin [BSA] and 0.1% sodium azide), and resuspended in 0.5% PFA (PBS containing 0.5% paraformaldehyde) for fixation, as previously described for human immunophenotyping (15).
A two-step staining protocol was adopted for optimal detection of CXCR5, CCR5, CXCR6, and GPR15 (16). Macaque cells were stained with 10 μg/ml purified antibody against macaque CXCR5 or human CCR5, CXCR6, and GPR15 for 1 h on ice; washed once with PBA; and incubated with a 1:50 dilution of PE- or Alexa Fluor 647-conjugated AffiniPure F(ab′)2 fragment goat anti-mouse IgG(H+L) secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) for 45 min on ice. Cells were then washed twice with PBA, blocked with 10% mouse serum for 10 min at room temperature, and stained with antibodies to other surface proteins, as described above. A negative control was performed in parallel, except that the first antibody was not added.
For intracellular detection of the intranuclear transcription factor Bcl-6, the human FoxP3 buffer set (BD Biosciences) was used to permeabilize cells. Macaque cells were first stained with antibodies to surface proteins and then permeabilized according to the manufacturer's instructions. Cells were then stained with Bcl-6–PE antibody for 30 min at room temperature, washed once with PBA, and resuspended in 0.5% PFA.
Stained macaque cells were analyzed on a 3-laser BD LSR II instrument with FACSDiva 6.0 software (BD Biosciences), as previously described (15, 17).
Tfh cell sorting, nucleic acid extraction, and cDNA synthesis.
Cryopreserved single-cell preparations from macaque spleen or LN were thawed and stained with CD3-PE-Cy7, CD4-PerCP-Cy5.5, CD8-APC-Cy7, CD45RA-ECD, CD127-eFluor450, and PD-1–PE for 15 min at room temperature; washed once with PBS with 10% heat-inactivated fetal calf serum (FCS); and resuspended in PBS with 10% FCS. Tfh cells were defined as CD3+ CD4+ CD8− CD45RA− PD-1high CD127low. Tfh cells along with 3 other memory CD4 T cell subsets, PD-1med CD127low, PD-1low CD127low, and CD127+, as well as CD45RA+ naïve CD4 T cells in some cases, were sorted on a 3-laser FACSAria instrument using FACSDiva 5 software (BD Biosciences) and collected into polystyrene tubes coated with FCS.
Aliquots of sorted cells were washed twice with PBS, pelleted, and resuspended in appropriate lysis buffer for nucleic acid extraction. Genomic DNA (gDNA) was prepared from one aliquot by using the Promega Wizard SV genomic DNA purification system, and total RNA was extracted from a second aliquot by using TRIzol (Invitrogen) according to the manufacturer's directions. Purified total RNA was treated with 2 units of DNase I (Invitrogen) per 1 μg of RNA for 15 min at room temperature, followed by the addition of a 1/10 volume of 25 mM EDTA and heating at 60°C for 10 min.
qPCR assay for SIV proviral DNA and cell-associated RNA.
SIV proviral DNA and cell-associated RNA were measured by a quantitative PCR (qPCR) assay and a two-step reverse transcription (RT)-qPCR assay, respectively, with primers that target a highly conserved region of the SIV-gag p17 matrix (18). RT reactions were performed with iScript and an oligo(dT)20 primer according to the manufacturer's instructions (Bio-Rad, Hercules, CA). qPCRs were performed with SYBR green I Master (Roche, Basel, Switzerland) and 0.4 μM both sense and antisense primers for SIV-gag and the reference gene β-actin on a LightCycler 480 instrument (Roche). Samples were run in duplicate. A no-template control and, in the RT-qPCRs, a no-RT control were included in each run. Standards that consisted of 10-fold serial dilutions of plasmid DNA containing SIV-gag or β-actin or 10-fold serial dilutions of gDNA prepared from uninfected macaque PBMCs were also included in each run to validate amplification efficiencies and to determine the copy numbers of SIV-gag and β-actin in the samples.
Quantification of CXCR5, IL-21, IL-21Rα, CCR5, CXCR6, and GPR15 mRNAs.
Expression of mRNAs for CXCR5, IL-21, IL-21Rα, CCR5, CXCR6, and GPR15 was quantified by RT-qPCR and normalized to β-actin levels, as described above. Primers for CXCR5, IL-21 and IL-21Rα, CCR5 (19), CXCR6, and GPR15 were specifically designed for macaque sequences. Sequences of sense and antisense primers for CXCR5, IL-21, IL-21R, CCR5, CXCR6, and GPR15 are as follows: CXCR5 sense primer 5′-ATTCCTGCTGCCCATGCT-3′, CXCR5 antisense primer 5′-CCACCCTGACTGCCTTCTG-3′, IL-21 sense primer 5′-CATGCCCTTCATGTGATTCTT-3′, IL-21 antisense primer 5′-TTCTAGAGGACAGATGCTGATGA-3′, IL-21Rα sense primer 5′-GGTCATCTTTCAGACCCAGTC-3′, IL-21Rα antisense primer 5′-TCCAGAAGGCAGGAATGAAG-3′, CCR5 sense primer 5′-GTCCCCTTCTGGGCTCACTAT-3′, CCR5 antisense primer 5′-CCCTGTCAAGAGTTGACACATTGTA-3′, CXCR6 sense primer 5′-TGTCTGTGGCCTGGTGGGGA-3′, CXCR6 antisense primer 5′-GGCCCAGAAGGGCAGAGTGC-3′, GPR15 sense primer 5′-AACCCGGCAGCCGAAGACTG-3′, and GPR15 antisense primer 5′-GTAGGCAACCCCAGCAGGCA-3′ (GPR15 sense and antisense primers were used at 0.2 μM for qPCRs).
qPCR assay for spliced SIV-tat RNA.
Levels of spliced SIV-tat RNA were measured by RT-qPCR as previously described (20). The sense primer (5′-AGTGTTGCTACCATTGCCAGT-3′) binds in tat exon 1, and the antisense primer (5′-ATATGGGTTTGTTTGATGCAGA-3′) binds a sequence across the splice junction of tat exon 1 and tat exon 2 (letters in boldface type indicate the binding site in exon 2). For biological positive and negative controls of spliced SIV-tat qPCR assays, we infected CEMx174 cells with SIVmac251 and cultured the cells for 7 days until syncytia formed. Culture supernatant that contained high titers of free virus was separated from cells by spinning in a bench-top centrifuge at 13,000 rpm for 5 min. Viral RNA was extracted from the culture supernatant by using a Total RNA extraction kit (Promega, Madison, WI) according to the manufacturer's instructions. cDNA derived from viral RNA was prepared from the oligo(dT)20 primer, as described above, and used as a biological negative control. cDNA was also prepared from total RNA extracted from infected CEMx174 cells, as described above, and used as a biological positive control.
Determination of viral load.
SIV viral load was determined as previously described, with some modifications (21). EDTA-treated whole blood was centrifuged at 950 × g for 12 min at room temperature. Plasma was recovered from the top layer and stored at −80°C. RNA was isolated from 140 or 280 μl plasma by using a QIAamp viral RNA kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. SIV viral load was determined by qPCR by Superscript III RT-PCR (Invitrogen) using TaqMan Universal PCR Mastermix (Applied Biosystems Inc., Foster City, CA) according to the manufacturers' instructions. A 142-bp product in the SIV-gag gene was amplified by using primers 5′-AATTAGATAGATTTGGATTAGCAGAAAGC-3′ (forward) and 5′-CACCAGATGACGCAGACAGTATTAT-3′ (reverse) and detected by using the TaqMan probe 5′-CAACAGGCTCAGAAAA-3′, using a Mastercycler ep Realplex4 instrument (Eppendorf, Hamburg, Germany). The sample viral load was determined by using a standard curve made from in vitro-transcribed SIV RNA. To make the standards, a 670-nucleotide (nt) SIV-gag RNA product was in vitro transcribed by using a MEGAscript T7 kit (Ambion Inc., Austin, TX) and treated with Turbo DNase (Ambion), and RNA of the correct size was quantified by using an Agilent 2100 Bioanalyser (Agilent, Santa Clara, CA). Negative controls were included where the reverse transcriptase enzyme was omitted.
Bulk sequencing and clonal sequencing of the SIV envelope gene.
The SIV gp120 region was amplified from gDNA extracted from purified CD4+ T cell subsets by using Platinum Taq High Fidelity DNA polymerase (Life Technologies, Grand Island, NY). Sense primer gp120F (5′-CAGCTGCTTATCGCCATCTTGCT-3′) binds to a constant region upstream of the first variable region (V1), and antisense primer gp120R (5′-CGAGAAAACCCAAGAACCCTAGCAC-3′) binds to a constant region downstream of the last variable region (V5) of gp120, generating an amplicon of ∼1.6 kb. Following initial denaturation at 94°C for 2 min, 45 cycles of 94°C for 15 s, 62.5°C for 30 s, and 68°C for 1 min 45 s were performed. Amplicons were purified by using a PCR cleanup kit (Promega) before setting up bulk sequencing reactions or TA cloning into pCR4 (Life Technologies) for clonal sequencing.
For bulk sequencing, primers gp120F and gp120R plus the following additional four primers that bind to the constant regions within gp120 (see below) were used to sequence each sample, to ensure that all regions were covered by a high quality of sequencing signals: C4R (5′-TCTGCTCTAGTTCCATTAAAGCCAAACC-3′), C4F (5′-GGTTTGGCTTTAATGGAACTAGAGCAGA-3′), C3F (5′-ACAGGCTTGGAACAAGAGCAAATGA-3′), and C5R (5′-GATCTCCTCCTCCAGGAGCCGT-3′).
For clonal sequencing, gp120 amplicons were TA cloned into pCR4 and transformed into Escherichia coli Top10 cells (Life Technologies) by heat shock, according to manufacturer's instructions. On the next day, 20 colonies from each subset were used as templates for amplification using Platinum Taq High Fidelity DNA polymerase and universal primers M13-forward and M13-reverse. Following initial denaturation at 94°C for 2 min, 30 cycles of 94°C for 15 s, 52°C for 30 s, and 68°C for 1 min 45 s were performed. PCR products were mixed with 2 volumes of DNA binding buffer (4 M guanidine-HCl, 0.75 M potassium acetate [KAc; pH 4.6]), loaded onto a 96-well plate with Whatman filter paper (catalog no. 7700-2810; Whatman), washed with 400 μl of wash buffer (40% [vol/vol] 100 mM Tris, 20 mM EDTA, 0.4 M NaCl [pH 7.5], 60% [vol/vol] ethanol [EtOH]), and eluted with 50 μl of double-distilled water into a collection plate (catalog no. 7701-5250; Whatman). Purified PCR products were used to set up sequencing reactions with primers C4F, C4R, and C3F. Sequencing was performed by the Australian Genome Research Facility at Westmead Millennium Institute, Sydney, Australia.
Sequence alignment was performed by using high-quality sequences with MacVector with Assembler (version 12.5.0).
Anti-SIV antibody levels.
SIV antibody levels to HIV-2 p36 in macaque plasma were measured by an enzyme-linked immunosorbent assay (ELISA), using a GS HIV-2 EIA kit (Bio-Rad) according to the manufacturer's instructions. Western blotting was also performed to confirm the result, as previously described (22).
Statistics.
Statistical significance was analyzed by a Kruskal-Wallis test, Wilcoxon signed-rank test, or Mann-Whitney test with Prism 5.0 (GraphPad, La Jolla, CA). Unless otherwise indicated, data represent the median ± interquartile, with a P value of <0.05 considered statistically significant.
RESULTS
PD-1high CD127low memory CD4+ T cells in macaque lymphoid tissue have the phenotypic characteristics of Tfh cells.
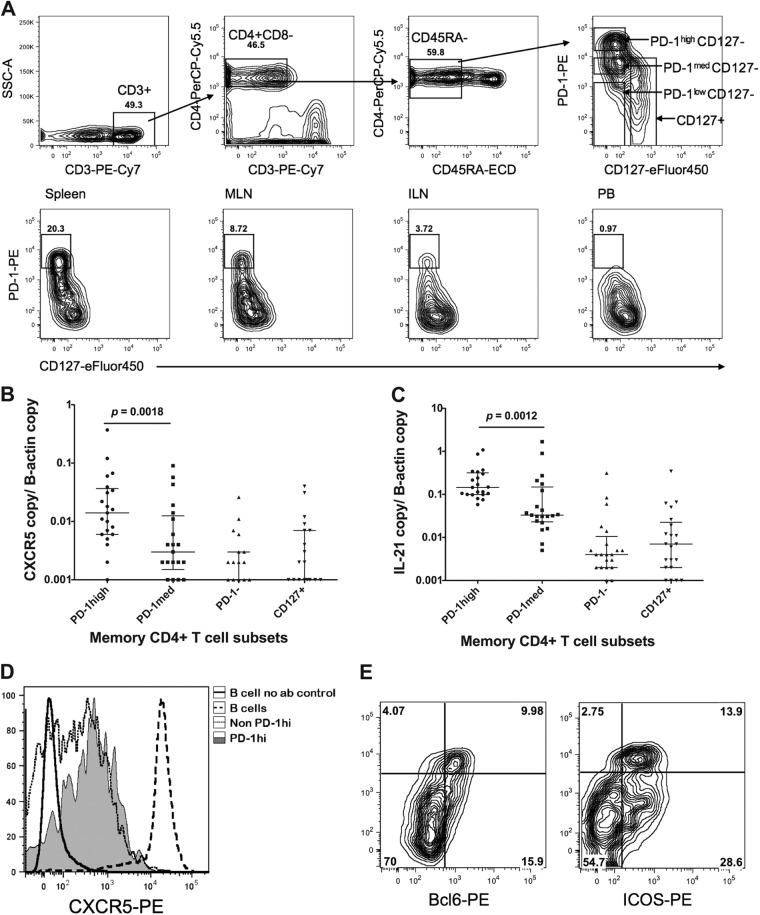
Although Tfh cells have been well characterized as CXCR5+ PD-1high ICOS+ Bcl-6+ memory CD4+ T cells in mice and humans (2, 23), the phenotype of macaque Tfh cells has not been extensively investigated. Despite a thorough screen, we found no antibody to human CXCR5 that cross-reacted with macaque CXCR5 available at the time when we commenced this study (data not shown). Therefore, we investigated other antibody combinations to define macaque Tfh cells based on the human cell surface phenotype and antibody cross-reactivity with macaques. PD-1 has been shown to be highly upregulated on Tfh cells in both mice and humans (2, 24) and to have a role in optimally sustaining germinal centers (25). We found that the anti-human PD-1 antibody clone EH12.2H7 cross-reacted with the macaque protein, and it clearly divided CD45RA-negative memory CD4+ T cells from macaque lymphoid tissues into three populations (Fig. 1A; see also Fig. S1 in the supplemental material for a more detailed gating strategy). The addition of an antibody to CD127, the α-chain of the IL-7 receptor, allowed us to objectively set the cutoff between PD-1high and PD-1med cells (see Fig. S2 in the supplemental material), as the cells that express the highest level of PD-1 do not express this long-term memory marker, consistent with the highly activated status of human Tfh cells (26) and as confirmed by recent microarray data for rhesus macaques (2, 27). As PD-1high CD127low memory CD4+ T cells were present in single-cell suspensions of mononuclear cells from spleen and lymph node but not in peripheral blood mononuclear cells (PBMCs) (Fig. 1A), we believed that this cell population was highly enriched for Tfh cells.
Fig 1.
Identification of Tfh cells in pigtail macaques. (A, top) Representative plots acquired from a BD FACSAria cell sorter showing the gating strategy for sorting of putative Tfh cells. Lymphocytes were first gated based on forward and side scatter (not shown) and then displayed on the first plot (SSC, side scatter). CD3+ T cells were subsequently gated as CD45RA− memory cells and then displayed as PD-1–PE versus CD127-eFluor 450 (top right). (Bottom) Representative PD-1-versus-CD127 plots, acquired by using a BD LSRII flow cytometer, for memory CD4+ T cells from macaque spleen, mesenteric lymph node, inguinal lymph node, and peripheral blood, from left to right. PD-1high CD127low cells were present at high levels in lymphoid tissues, but they were rare in peripheral blood. (B) Significantly higher copy numbers of CXCR5 mRNA, normalized to β-actin copy numbers, in purified PD-1high CD127low putative Tfh cells than in other subsets of lymphoid tissue memory CD4+ T cells, sorted using gating as depicted in panel A, top (n = 21). (C) Significantly higher copy numbers of IL-21 mRNA, normalized to β-actin copy numbers, in purified PD-1high CD127low putative Tfh cells than in other subsets of lymphoid tissue memory CD4+ T cells, sorted using gating as depicted in panel A, top (n = 21). (D) PD-1high CD127low putative Tfh cells are CXCR5+ by flow cytometry, with intermediate expression compared to B cells and a higher mean fluorescence intensity than other memory CD4+ T cells. The no-antibody control (no ab) shows negative staining of B cells. Results are representative of two independent experiments. (E) PD-1high CD127low putative Tfh cells are both Bcl-6+ (left) and ICOS+ (right) by flow cytometry. Data are representative of two experiments.
To confirm that these PD-1high CD127low memory CD4+ T cells in macaque lymphoid tissues had other typical characteristics of Tfh cells, we sorted memory CD4+ T cells (CD3+ CD4+ CD45RA−) into highly purified (purity, >97%) populations of PD-1high CD127low, PD-1med CD127low, PD-1low CD127low, and CD127+ cells and measured ex vivo mRNA levels of CXCR5 and IL-21 by reverse transcription–quantitative real-time PCR (RT-qPCR). PD-1high CD127low memory CD4+ T cells contained significantly higher levels of mRNA for CXCR5 and IL-21 than any of the other three purified memory CD4+ T cell subsets (Fig. 1B and C).
Subsequently, a monoclonal antibody specific for macaque CXCR5 (clone 710D82.1) was produced and made available by the NIH Nonhuman Primate Reagent Resource Program. Macaque PD-1high CD127low memory CD4+ T cells from spleen and lymph node demonstrated intermediate to high levels of CXCR5 surface expression, consistent with these cells being Tfh cells, while B cells from lymphoid tissues had the highest expression levels (Fig. 1D). However, although over 80% of PD-1high CD127low memory CD4+ T cells expressed CXCR5 at the protein level, some PD-1med CD127low memory CD4+ T cells were also stained with this CXCR5 antibody (Fig. 1D). Therefore, although CXCR5 staining confirmed our real-time PCR data and verified our flow cytometric strategy for isolation of Tfh cells, staining of CXCR5 with this particular antibody may not be adequate for definitive identification or purification of Tfh cells in macaque lymphoid tissues. This is consistent with recently reported data (27).
Finally, intracellular staining with monoclonal antibody to Bcl-6 showed that the expression of this transcription factor, characteristically expressed in Tfh cells, was restricted to PD-1high CD127low memory CD4+ T cells from macaque lymphoid tissues (Fig. 1E). Similarly, PD-1high CD127low memory CD4+ T cells also expressed the important Tfh-costimulatory molecule ICOS (Fig. 1E).
Taken together, these data confirm that PD-1high CD127low memory CD4+ T cells from macaque spleen and lymph nodes had phenotypic characteristics typical of Tfh cells. Therefore, we used this combination in sorting experiments in order to investigate the susceptibility of macaque Tfh cells (PD-1high CD127low memory CD4+ T cells) to SIV infection.
Macaque Tfh cells contain SIV proviral DNA and cell-associated viral RNA.
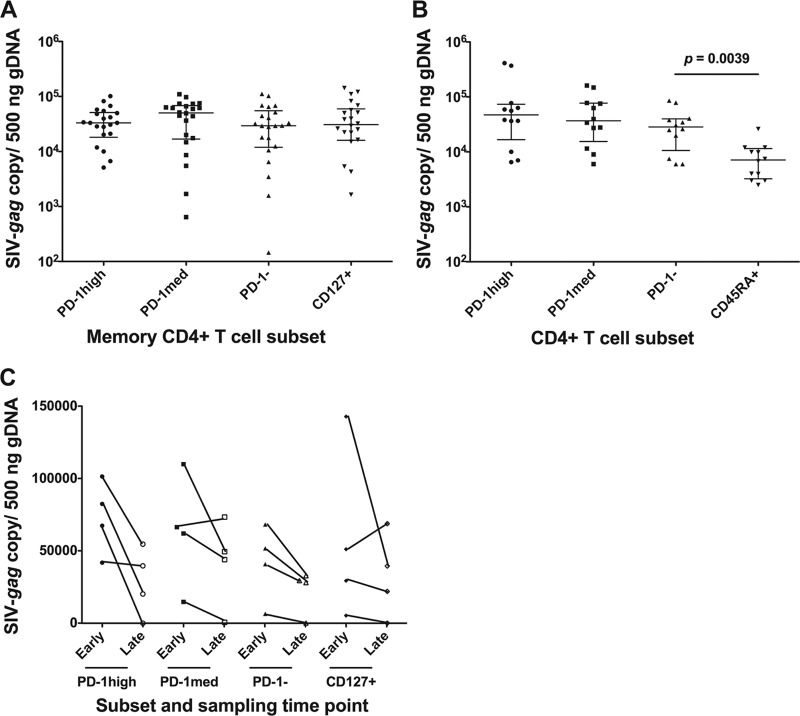
We sorted PD-1high CD127low, representing Tfh cells, as well as PD-1med CD127low, PD-1low CD127low, and CD127+ memory CD4+ T cell populations from single-cell suspensions derived from macaque lymphoid tissues. Genomic DNA (gDNA) was extracted, and total SIV-gag was measured by qPCR. SIV-gag DNA was present in PD-1high CD127low Tfh cells from spleen and/or lymph node tissue from all 13 macaques, and the copy number normalized to the β-actin copy number was comparable to that found in the other sorted memory CD4+ T cell subsets (Fig. 2A). As controls, CD45RA+ CD4+ T cells, which are predominantly naïve cells in lymphoid tissues, with a median of over 97% being CD27+ (n = 8) (a representative plot is shown in Fig. S1B in the supplemental material), from 12 samples were sorted, and SIV-gag DNA was measured. As expected, the tissue naïve CD4+ T cell subset contained SIV-gag DNA at significantly lower levels than in the Tfh, PD-1med, or PD-1low memory CD4+ T cell subset (median difference, 5.9-, 6.4-, and 3.4-fold less, respectively; P < 0.0039) (Fig. 2B). As an additional control, CD4− CD8+ T cells from three samples were purified by sorting, and SIV-gag DNA was below the level of detection in these purified cells (data not shown).
Fig 2.
SIV-gag proviral DNA copy numbers normalized to gDNA input. (A) SIV-gag proviral DNA levels in the four memory CD4+ T cell subsets, sorted as described in the legend of Fig. 1A. Twenty-two samples from nine infected macaques were sorted. (B) SIV-gag proviral DNA levels in three memory CD4+ T cell subsets, PD-1high CD127low, PD-1med CD127low, and PD-1low CD127low, and in naïve CD4+ T cells were obtained from 12 samples from 6 infected macaques. Naïve CD4+ T cells had significantly lower SIV-gag proviral DNA levels than the other three memory CD4+ T cell subsets. (C) Longitudinal data from ILN samples from 4 macaques taken at an earlier time point (14 to 28 days) and then again at a later time point (77 to 469 days).
We also found that there was a trend toward higher infection levels at an earlier phase (14 to 28 days postinfection [p.i.]) than at a later phase (77 to 469 days postinfection) when we compared sequential inguinal lymph node samples from 4 macaques (Fig. 2C); however, there were insufficient samples for formal statistical analysis. Importantly, this high level of infection seen in lymph node Tfh cells within 2 weeks to 1 month of infection indicates that the infection of this subset is established early and at rates comparable with those of other memory CD4+ T cell subsets.
Since SIV DNA in Tfh cells may have been transcriptionally inactive, we then measured cell-associated viral RNA in macaque Tfh cells by RT-qPCR for SIV-gag and spliced tat. Macaque Tfh cells contained low but detectable levels of both SIV-gag RNA (Fig. 3A) and spliced-tat RNA (Fig. 3B). The expression levels of both viral genes in PD-1high CD127low memory CD4+ T cells was not significantly different from those in other memory CD4+ T cell subsets, suggesting that they were as likely as other memory cell populations not only to be infected with SIV but also to transcribe viral genes.
Fig 3.
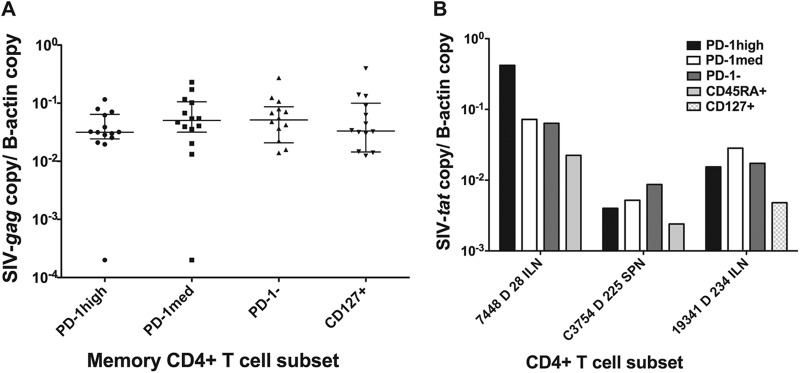
(A) SIV-gag cell-associated RNA copy numbers normalized to β-actin copy numbers. There was no significant difference (n = 14) among the 4 memory CD4+ T cell subsets. (B) Spliced-tat RNA copy numbers normalized to β-actin copy numbers from three macaque samples. Spliced-tat levels in cells from another three macaque samples were above the limit of detection but below the limit of quantification and are not shown.
Percentages of Tfh cells in macaque lymph nodes increase in chronic SIV infection.
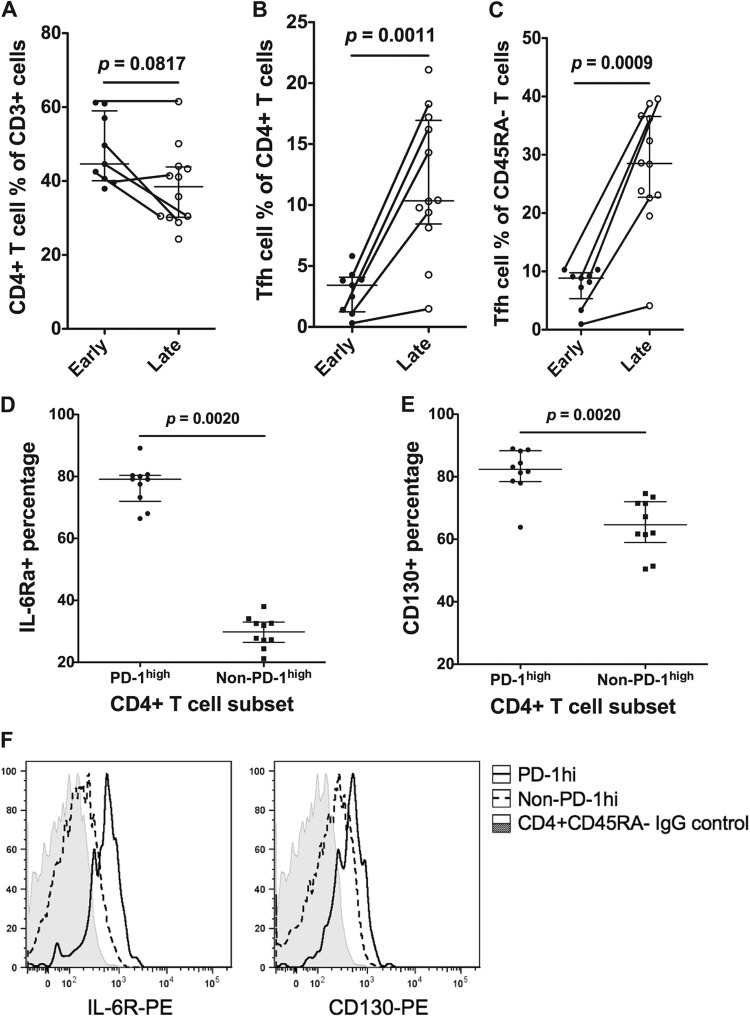
Since Tfh cells are infected with SIV early after inoculation, we studied whether percentages of these cells decreased during the course of progressive SIV infection in inguinal lymph node (ILN) biopsy specimens taken either early in infection (≤28 days p.i.) or later in infection (≥77 days p.i.). In five individuals, sequential ILN biopsy specimens were obtained both early and late. The results show that, as expected, there was a slightly lower percentage of CD3+ T cells that were CD4+, although this was not statistically significant (Fig. 4A). In contrast, the percentage of CD4+ T cells that exhibited the PD-1high CD127low Tfh phenotype increased significantly, from a median of 4% to 10% (P = 0.0011) (Fig. 4B). The large increase was observed not only cross-sectionally but also in 4 out of 5 pigtail macaques that had serial ILN biopsy specimens (Fig. 4B). The increase in the percentage of Tfh cells was even more dramatic relative to other memory CD45RA− CD4+ T cells, increasing from a median of 9% to 28% memory CD4+ T cells (P = 0.0009) (Fig. 4C).
Fig 4.
CD4+ T cell and Tfh cell frequencies during the early stage (≤28 days postinfection) and late stage (≥77 days postinfection) in ILNs. Lines connecting two data points indicate sequential ILN biopsy specimens from the same macaque. (A) The CD4+ T cell proportion as a percentage of CD3+ T cells decreased in ILNs from the early stage (n = 9) to the late stage (n = 12) but was not statistically significant. (B) The Tfh cell proportion as a percentage of CD4+ T cells significantly increased in ILNs from the early stage (n = 9) to the late stage (n = 12). (C) The Tfh cell proportion as a percentage of memory (CD45RA−) T cells significantly increased in ILNs from the early stage (n = 9) to the late stage (n = 12). (D) In memory CD4+ T cells, the PD-1high subset expressed significantly higher levels of IL-6Rα than the non-PD-1high subset (n = 10). (E) In memory CD4+ T cells, the PD-1high subset expressed significantly higher levels of CD130 than the non-PD-1high subset (n = 10). (F) Representative staining of CD126 (left) and CD130 (right) on macaque memory CD4+ T cells. The IgG staining control is shown as shaded histograms, PD-1high CD127low cells are shown as solid lines, and the remaining memory CD4+ T cells are shown as dashed lines. Results are representative of 10 experiments.
IL-6 and IL-21 have been reported to play important roles in Tfh cell development (28). We found that a substantial proportion of macaque Tfh cells highly expressed the two components of the receptor for IL-6, CD126 and CD130 (Fig. 4D to F) but not receptors for IL-2, IL-4, or IL-10 (data not shown). Furthermore, Tfh cells did not preferentially express the receptor for IL-21 at either the protein or mRNA level (data not shown).
Macaque Tfh cells do not express known SIV coreceptors.
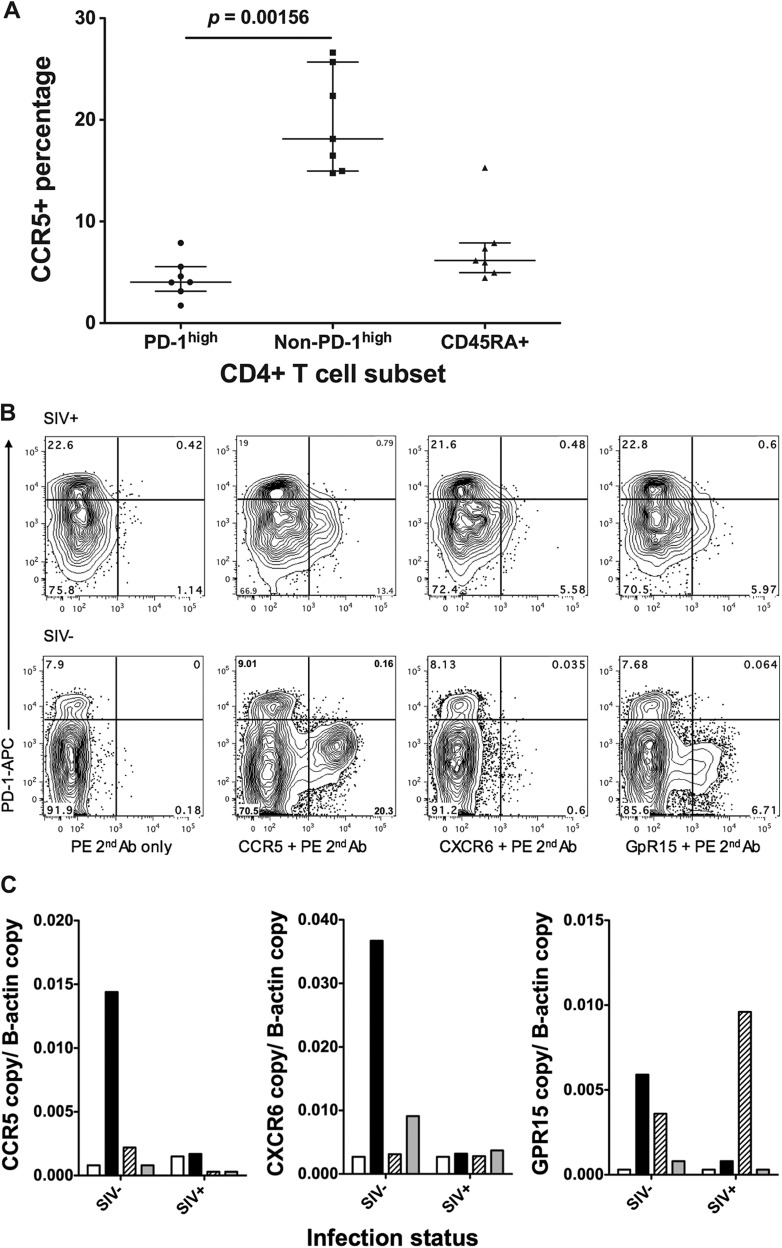
Given that the rates of SIV infection were similar in each of the CD4+ memory T cell subsets purified from macaque lymphoid tissue, we asked whether the PD-1high CD127low cells expressed any of the coreceptors for SIV, particularly CCR5 or CXCR6/Bonzo and GPR15/BOB, which have been reported to be alternative coreceptors for certain SIVmac strains (reviewed in reference 29).
We stained macaque spleen and lymph node samples with antibodies against CCR5, CXCR6, and GPR15 but could not detect substantial expression on Tfh cells despite demonstrating clear expression of the coreceptors on non-Tfh memory CD4+ T cells in both uninfected and infected macaques (Fig. 5A and B). Furthermore, RT-qPCR confirmed low expression levels of these genes in Tfh cells (Fig. 5C), but we also observed downregulation of CCR5 expression in other memory CD4+ T cell subsets during chronic infection (Fig. 5B). This confirms that macaque Tfh cells lack expression of CCR5, as previously reported for murine Tfh cells (2), and is consistent with our observations of human lymph nodes (data not shown).
Fig 5.
Expression levels of CCR5, CXCR6, and GPR15 determined by flow cytometry and RT-qPCR. (A) CCR5 protein levels on CD4+ T cell subsets on macaque tissues (n = 7), measured by flow cytometry. (B) Representative plots for CCR5 (second panel), CXCR6 (third panel), and GPR15 (far right) staining on memory CD4+ T cells from SIV+ (top) and SIV-negative (bottom) macaque tissues. Plots on the left show negative controls, in which the first antibody was not added. (C) mRNA levels of CCR5 (left), CXCR6 (middle), and GPR15 (right) on CD4+ T cell subsets measured by RT-qPCR (white bars, PD-1high; black bars, PD-1med; patterned bars, CD127+; gray bars, CD45RA+).
Within each macaque, SIV envelope gp120 sequences from Tfh cells were highly similar to the sequences from other CD4+ T cell subsets.
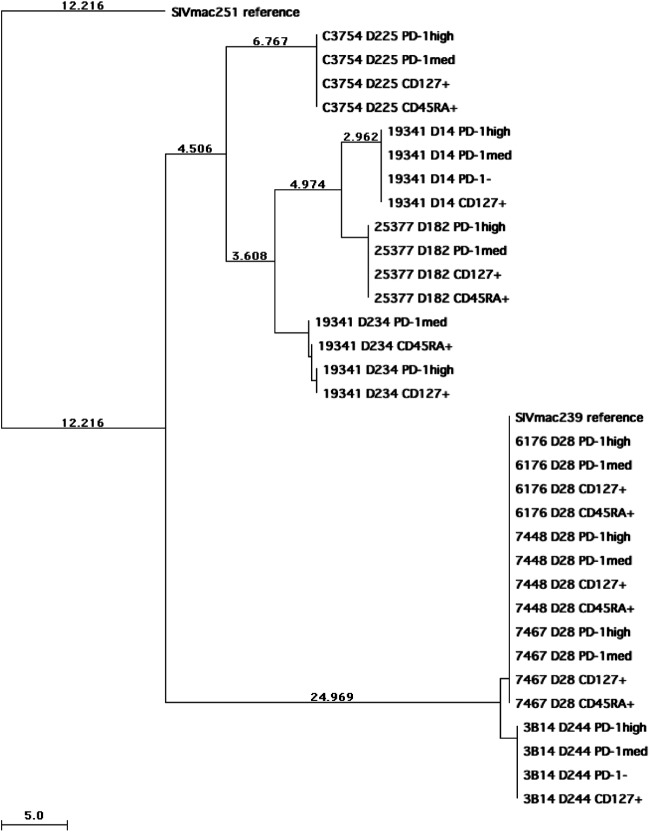
To investigate whether the virus may have entered macaque Tfh cells via a coreceptor substantially different from that used in other memory CD4+ T cell subsets, probably CCR5, we first bulk sequenced the PCR products of the SIV envelope gp120 region, amplified from proviral DNA isolated from purified Tfh cells, from eight pigtail macaques acutely or chronically infected with SIVmac239 or SIVmac251. We also sequenced proviral DNA from other memory CD4+ T cell subsets or naïve CD4+ T cells from the same animals. Consistent with previous reports, mutations were observed during chronic infection, compared to the reference SIVmac239 (GenBank accession no. M33262) or SIVmac251 (GenBank accession no. M19499) sequence. Although mutations occurred with different patterns within gp120 in each chronically infected macaque, these mutations were highly similar among each T cell subset isolated from the same macaque, as determined by bulk sequencing (Fig. 6).
Fig 6.
Neighbor-joining tree of gp120 sequences from proviral DNA isolated from CD4+ T cell subsets (PD-1high CD127low, PD-1med CD127low, PD-1− CD127low, and CD127+ memory CD4+ T cells as well as CD45RA+ naïve CD4+ T cells) of macaques infected with SIVmac239 or SIVmac251. SIVmac239 and SIVmac251 reference sequences were downloaded from GenBank, and SIVmac251 was assigned to be the root of the tree. The sample name shows the macaque identification, days postinoculation, and subset name in order. Branches are scaled in absolute numbers of differences according to the scales at the bottom of the trees.
Next, we performed clonal sequencing on proviral DNA from purified CD4+ T cell subsets from a macaque acutely infected with SIVmac239 (28 days p.i.). We sequenced a total of 79 clones of gp120 PCR products, comprising 16 to 23 clones from each of the four memory CD4+ T cell subsets. Some clones (20/79) were 100% identical to the reference sequence at the nucleic acid level, and 34/79 sequences had no predicted amino acid changes. The majority of the clones contained only 1 to 5 nucleic acid mutations, with 2 clones having 11 mutations, at this very early, day 28, time point (Table 2). These mutations were randomly distributed across the gp120 sequence and across the subpopulation of T cells without a particular pattern emerging, nor were any of these mutations overrepresented among sequences derived from a particular memory CD4+ T cell subset. Because each subset represented a very small minority population found within the sequences for each T cell population, these mutations were not readily detected by bulk sequencing. It was reported previously that an amino acid substitution at position 324 in the SIVmac239 envelope plays an important role in coreceptor choice, cell tropism, and CD4-independent cell fusion (30); however, no mutation at this position was found in any of the clones that we sequenced. Furthermore, there was no clustering of particular gp120 proviral DNA sequences on the phylogenetic tree within specific CD4+ T cell subsets (see Fig. S3 in the supplemental material).
Table 2.
Summary of gp120 clonal sequencing results
| T cell subset | No. of gp120 clones analyzed | No. of clones in which DNA sequence had 100% identity to reference | No. of clones in which protein sequence had 100% identity to reference | DNA mutation rate (per 1,000 bp) |
|---|---|---|---|---|
| PD-1high | 23 | 5 | 9 | 0.74 |
| PD-1med | 23 | 6 | 11 | 1.31 |
| CD127+ | 17 | 3 | 6 | 1 |
| CD45RA+ | 16 | 6 | 8 | 0.98 |
Both bulk sequencing and clonal sequencing results confirmed that gp120 sequences from Tfh cells are highly similar and distributed among the Tfh cells in a manner similar to those from other CD4+ T cell subsets. These results strongly suggest that the evolutionary pressures exerted within each of these populations are similar and that there has not been purifying selection driven by differential coreceptor usage among these populations.
Effect of SIV infection of Tfh cells on SIV-specific antibody production.
We longitudinally measured the level of anti-SIV antibodies in the pigtail macaques included in this study. SIV antibodies were detected in plasma samples from all 13 macaques from time points later than 2 to 3 weeks after infection (Fig. 7). Five of the 13 macaques were euthanized before the first 6 months postinfection, and in 4 out of 5 of these macaques, SIV antibody levels were increasing at the time of euthanasia (Fig. 7A). The other 8 macaques were euthanized at between 6 months and 1 year after infection. Half of these macaques had continuously increasing antibody levels until euthanasia (Fig. 7B), while the other half had peak antibody levels between days 35 and 97, which then decreased with a variable time course (Fig. 7C). These results show that these macaques were capable of making an SIV-specific antibody response, despite evidence of infection of Tfh cells with SIV.
Fig 7.
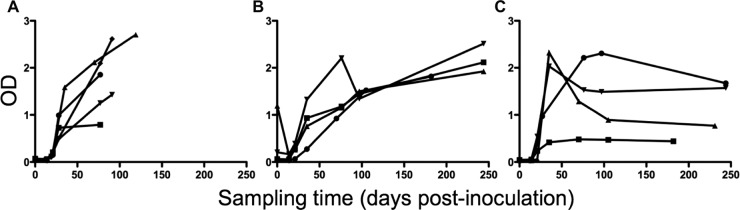
Anti-SIV antibody production in 13 SIV-infected macaques. (A) Five macaques were euthanized within 6 months postinfection. Antibody levels reached maximum at the time of euthanasia. (B) Four macaques, monitored for longer than 6 months postinfection, had antibody levels that were still increasing at the time of euthanasia. (C) Four macaques, monitored for longer than 6 months postinfection, had antibody levels that reached a maximum between days 35 and 97 postinfection and then decreased by the time of euthanasia. OD, optical density.
DISCUSSION
In order to study the effect of SIV infection on macaque Tfh cells, we have demonstrated that PD-1high CD127low memory CD4+ T cells in lymphoid tissue can be characterized as CXCR5+, IL-21+, ICOS+, Bcl-6+, and IL-6R+ cells, analogous to Tfh cells in mice and humans (2, 31). The phenotype of macaque Tfh cells has not been extensively studied. In particular, the expression of the transcription factor Bcl-6 is an important regulator of Tfh differentiation (2) and was clearly upregulated in pigtail macaque PD-1high CD127low memory CD4+ T cells. This definition of Tfh cells is consistent with that reported very recently by Petrovas and colleagues (27).
Subsequently, we showed that highly purified pigtail macaque PD-1high CD127low memory CD4+ Tfh cells were infected with SIV DNA and SIV RNA. The presence of spliced SIV mRNA indicates that these cells can be productively infected. Rapid progression of SIV infection occurs in 10 to 30% of rhesus macaques infected with SIVmac251 or SIVsm strains. These macaques produce little or no SIV-specific antibody response (32, 33), consistent with a complete loss of CD4+ T cell helper activity, and are characterized by high numbers of productively infected cells (33), including in germinal centers after primary infection (32), and significantly higher levels of apoptosis in the T cell areas of lymph nodes (34). Importantly, in the 13 macaques included in the current study, there were no such antibody-negative rapid progressors, suggesting that in our cohort, infection of Tfh cells was associated with typical progression and not related exclusively to rapid progression.
Our results confirm and extend a recent report that impairment of antibody responses in SIV-infected rhesus macaques was associated with viral infection of PD-1+ lymph node cells (35). However, that study did not distinguish cells on the basis of the extent of PD-1 expression, which we found was important in defining Tfh cells. As a result, the cells studied were described as ICOS+ and CTLA-4+, both of which can be expressed by both T regulatory cells (36) and activated Th1 cells in acute HIV-1 infection (7, 17). Our results confirm, and extend, a more recent report of infection of Tfh cells in rhesus macaques at rates similar to those of other memory subsets on the basis of viral DNA copies per cell (27).
Our results also demonstrate that in typical progressive SIV infection, while there is infection of Tfh cells, the function of these cells appears overall to be intact, since there is continuing development of anti-SIV antibody responses, and in fact, the proportion of Tfh cells in lymph nodes increases longitudinally. Two other very recent studies reported a progressive increase in numbers of PD-1+ Tfh cells in chronic SIV infection (27, 37), and similar increases have been noted for humans chronically infected with HIV-1 (38). In a previous study by Hong et al. (37), occasional CD4+ cells in germinal centers were also SIV Gag+ by immunohistochemistry, and in the study by Petrovas et al., similar rates of SIV DNA were detected in each sorted memory cell subset. Our results show similar rates of SIV DNA infection and furthermore show that this provirus is transcriptionally active, with SIV RNA being present in highly purified PD-1high CD127low memory CD4+ T cells. These observations are consistent with previous observations that there is an increased development of germinal centers in pathogenic primate models of primate SIV infection compared to nonpathogenic models (39).
A very recent report showed that pathogenic SIV infection of rhesus macaques and HIV infection of humans are associated with productive infection of Tfh cells, together with very large deposits of virions on follicular dendritic cells, in germinal centers, at much higher levels than those found in sooty mangabeys, a natural host for SIV (40). This intriguing finding suggests that germinal center reservoirs may indeed be an important defining pathogenic feature of SIV/HIV infection. Germinal centers in HIV-1 infection represent the largest reservoir of HIV-1 in the body within infected individuals (reviewed in reference 41).
The mechanisms for Tfh cell accumulation during chronic SIV/HIV infection are yet to be fully resolved. Recent studies of lymphocytic choriomeningitis virus (LCMV) infection in mice (42), SIV infection in rhesus macaques (27, 37), and HIV infection in humans (38) have shown that chronic viral persistence leads to an increase in the number of Tfh cells. In the LCMV-infected murine model, viral persistence and prolonged T cell receptor (TCR) stimulation were shown to progressively redirect CD4+ T cell development away from a Th1 response during acute infection and toward a Tfh-dominated response during the chronic phase (42). In this model, a post-acute-phase IL-6 response was associated with a resurgence in antibody-mediated clearance of viral infection (43). It is therefore intriguing that both IL-6 mRNA levels in lymph nodes and IL-6 protein levels in plasma are dramatically upregulated following SIVmac239 (44) and SIVmac251 (45) infections, respectively. Another molecular mechanism that could contribute to the skewing of effector CD4+ T cells toward Tfh cells was recently described (46). In this model, excess Bcl-6 represses its direct target Prdm1 when levels of IL-2 are limiting, allowing the switch-on of Tfh-like genes, including CXCR5, BTLA, and IL-6Rα. Taken together with the fact that IL-2-producing T cells are lost during chronic HIV/SIV infection and our finding of the presence of IL-6R on macaque Tfh cells, chronic exposure to IL-6 and the limited IL-2 production from CD4+ T cells which characterize SIV and HIV infections may both contribute to the relative accumulation of Tfh cells during SIV infection. In addition, a lack of the peripheral trafficking marker S1PR1 and expansion of germinal center B cells during chronic SIV/HIV infection may cause Tfh cells to accumulate in tissues and escape cell death through interactions with PD-L1, despite their low rates of expression of Ki-67 in vivo but high spontaneous caspase-3 activity in vitro (27, 38).
How SIV infects PD-1high CD127low Tfh cells remains an open question. We found that PD-1high CD127low Tfh cells did not express significant levels of any of the best-described coreceptors for SIV, namely, CCR5, CXCR6/Bonzo, or GPR15/BOB (29). Human Tfh cells have been shown to express CXCR4 (23), and we did find expression of CXCR4 on PD-1high CD127low Tfh cells, as expected (data not shown). CXCR4 acts as the major alternate coreceptor for HIV-1, but it is generally found that SIV strains and in particular SIVmac239 and SIVmac251 rarely use CXCR4 for entry (29). Furthermore, CXCR5 usage has not been found for HIV-1 isolates from PBMCs (47) or for SIVmac251 (48), although it was reported for 2 strains of HIV-2 (48). Despite rare usage of either CXCR5 or CXCR4 by SIV, macaque Tfh cells have never been specifically studied before. Therefore, further studies are required to clarify the coreceptor usage governing entry of SIV into Tfh cells.
Alternatively, Tfh cells may be derived from activated CD4+ T cells in paracortical areas of lymphoid tissue and pass through an intermediate differentiation step (1, 2), where they may be susceptible to infection with SIV, prior to fully differentiating and migrating into B cell follicles. Our SIV envelope sequencing data seem to favor this hypothesis. Interestingly, preliminary data suggest that a proportion of PD-1med CD127low cells can upregulate surface PD-1 to a level comparable to that in Tfh cells upon in vitro TCR stimulation (data not shown), which suggests that Tfh cells may arise from these PD-1med CD127low cells, which highly express CCR5. Whether or not Tfh precursors are infected and continue to differentiate into PD-1high CD127low Tfh cells is a very important issue to be addressed. Further studies are still required to clarify the exact steps of the cellular pathway of Tfh cell differentiation (1, 2), in particular whether or not Tfh cells differentiate directly from activated naïve CD4+ T cells or via other lineages (1, 2).
It remains important to investigate possible infection of Tfh cells in human lymphoid tissue during HIV-1 infection, especially as similar increases in Tfh cell numbers in chronic HIV infection were recently noted (38), and a very recent paper reported that Tfh cells from human lymph node biopsy specimens from HIV-positive (HIV+) subjects contain HIV-1 DNA (49). Furthermore, purified Tfh cells from lymph nodes from HIV-uninfected subjects were readily infected in vitro (49) by a CXCR4-using strain of HIV-1, but coreceptor expression or usage was not studied. How Tfh cells may be infected in vivo during HIV-1 infection remains unknown. These results confirm and extend previous reports of productively infected CD4+ T cells in germinal centers (50) and infection of germinal center CD4+ cells with proviral HIV-1 DNA (51), but in those reports, these cells were not characterized as Tfh cells.
The recent association of vaccine protection with induced antibody responses (52) and findings that multiple mutations in Ig variable regions are required for broadly neutralizing antibodies (8–10) imply an extremely important role for Tfh cells and germinal center reactions in vaccine protection. Such neutralizing antibody responses are relatively rare and can take months to years to develop (6). Follicular hyperplasia and increased numbers of germinal centers are characteristic of early chronic HIV-1 infection (53) but are not found in very early primary HIV-1 infection, suggesting that the earliest detectable antibodies, appearing within days after the onset of symptoms (3, 4, 6), may come from extrafollicular B cell responses (54) and that germinal center reactions require additional time before they are initiated. Following primary HIV-1 infection, HIV-specific antibody responses continue to evolve for months, as judged by longitudinal diagnostic Western blots (4, 5), and also, very early intervention with antiretroviral therapy during primary HIV-1 infection can result in decreased titers of anti-HIV-1 antibodies (55), consistent with the protracted nature of the typical anti-HIV-1 humoral response. Correlating Tfh cell functionality with affinity maturation of anti-HIV-1 antibodies and somatic mutation of Ig genes in individual HIV-specific B cell clones will be a future research goal of the field.
Despite infection of human Tfh cells with HIV-1, antibody responses to HIV-1 envelope quasispecies continue to develop de novo as the virus evolves to escape earlier antibody responses (56, 57), possibly contributing to the equilibrium of steady-state viral loads (58). Also, whether infection of Tfh cells contributes to follicular abnormalities (59), B cell hyperactivity, and hypergammaglobulinemia (60), all of which are characteristic of chronic HIV-1 infection, is unclear. Most important is the issue of whether direct infection of Tfh cells is a major contributor to the general paucity of high-titer anti-HIV-1-neutralizing antibodies in natural infection. An improved understanding of the impact of HIV-1 infection on cell-cell interactions and germinal center dynamics in lymphoid tissue is likely to inform these unresolved questions.
Supplementary Material
ACKNOWLEDGMENTS
Anti-macaque CXCR5 monoclonal antibody was generously provided by the NIH Nonhuman Primate Reagent Resource Program.
This work was supported by Australian National Health and Medical Research (NHMRC) program grant no. 510488. The Kirby Institute is funded by the Australian Government Department of Health and Ageing and is affiliated with the Faculty of Medicine, The University of New South Wales.
Footnotes
Published ahead of print 16 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02497-12.
REFERENCES
- 1. King C. 2009. New insights into the differentiation and function of T follicular helper cells. Nat. Rev. Immunol. 9:757–766 [DOI] [PubMed] [Google Scholar]
- 2. Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663 [DOI] [PubMed] [Google Scholar]
- 3. Cooper DA, Imrie AA, Penny R. 1987. Antibody response to human immunodeficiency virus after primary infection. J. Infect. Dis. 155:1113–1118 [DOI] [PubMed] [Google Scholar]
- 4. Tindall B, Cooper DA. 1991. Primary HIV infection: host responses and intervention strategies. AIDS 5:1–14 [PubMed] [Google Scholar]
- 5. Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, Heldebrant C, Smith R, Conrad A, Kleinman SH, Busch MP. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 17:1871–1879 [DOI] [PubMed] [Google Scholar]
- 6. McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. 2010. The immune response during acute HIV-1 infection: clues for vaccine development. Nat. Rev. Immunol. 10:11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zaunders JJ, Munier ML, Kaufmann DE, Ip S, Grey P, Smith D, Ramacciotti T, Quan D, Finlayson R, Kaldor J, Rosenberg ES, Walker BD, Cooper DA, Kelleher AD. 2005. Early proliferation of CCR5+ CD38+++ antigen-specific CD4+ Th1 effector cells during primary HIV-1 infection. Blood 106:1660–1667 [DOI] [PubMed] [Google Scholar]
- 8. Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636–640 [DOI] [PubMed] [Google Scholar]
- 9. Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu Z, Qin HR, Chen W, Zhao Q, Shen X, Schutte R, Wang Y, Ofek G, Streaker E, Prabakaran P, Fouda GG, Liao HX, Owens J, Louder M, Yang Y, Klaric KA, Moody MA, Mascola JR, Scott JK, Kwong PD, Montefiori D, Haynes BF, Tomaras GD, Dimitrov DS. 2011. Cross-reactive HIV-1-neutralizing human monoclonal antibodies identified from a patient with 2F5-like antibodies. J. Virol. 85:11401–11408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Loh L, Reece JC, Fernandez CS, Alcantara S, Center R, Howard J, Purcell DF, Balamurali M, Petravic J, Davenport MP, Kent SJ. 2009. Complexity of the inoculum determines the rate of reversion of SIV Gag CD8 T cell mutant virus and outcome of infection. PLoS Pathog. 5:e1000378 doi:10.1371/journal.ppat.1000378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peut V, Kent SJ. 2009. Substantial envelope-specific CD8 T-cell immunity fails to control SIV disease. Virology 384:21–27 [DOI] [PubMed] [Google Scholar]
- 13. Sexton A, De Rose R, Reece JC, Alcantara S, Loh L, Moffat JM, Laurie K, Hurt A, Doherty PC, Turner SJ, Kent SJ, Stambas J. 2009. Evaluation of recombinant influenza virus-simian immunodeficiency virus vaccines in macaques. J. Virol. 83:7619–7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. National Health and Medical Research Council 2004. Australian code of practice for the care and use of animals for scientific purposes, 7th ed National Health and Medical Research Council, Canberra, Australia [Google Scholar]
- 15. Zaunders JJ, Dyer WB, Munier ML, Ip S, Liu J, Amyes E, Rawlinson W, De Rose R, Kent SJ, Sullivan JS, Cooper DA, Kelleher AD. 2006. CD127+ CCR5+ CD38+++ CD4+ Th1 effector cells are an early component of the primary immune response to vaccinia virus and precede development of interleukin-2+ memory CD4+ T cells. J. Virol. 80:10151–10161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reynes J, Portales P, Segondy M, Baillat V, Andre P, Reant B, Avinens O, Couderc G, Benkirane M, Clot J, Eliaou JF, Corbeau P. 2000. CD4+ T cell surface CCR5 density as a determining factor of virus load in persons infected with human immunodeficiency virus type 1. J. Infect. Dis. 181:927–932 [DOI] [PubMed] [Google Scholar]
- 17. Zaunders JJ, Ip S, Munier ML, Kaufmann DE, Suzuki K, Brereton C, Sasson SC, Seddiki N, Koelsch K, Landay A, Grey P, Finlayson R, Kaldor J, Rosenberg ES, Walker BD, Fazekas de St Groth B, Cooper DA, Kelleher AD. 2006. Infection of CD127+ (interleukin-7 receptor+) CD4+ cells and overexpression of CTLA-4 are linked to loss of antigen-specific CD4 T cells during primary human immunodeficiency virus type 1 infection. J. Virol. 80:10162–10172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shehu-Xhilaga M, Kent S, Batten J, Ellis S, Van der Meulen J, O'Bryan M, Cameron PU, Lewin SR, Hedger MP. 2007. The testis and epididymis are productively infected by SIV and SHIV in juvenile macaques during the post-acute stage of infection. Retrovirology 4:7 doi:10.1186/1742-4690-4-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, Roederer M. 2005. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature 434:1093–1097 [DOI] [PubMed] [Google Scholar]
- 20. Sirois M, Robitaille L, Allary R, Shah M, Woelk CH, Estaquier J, Corbeil J. 2011. TRAF6 and IRF7 control HIV replication in macrophages. PLoS One 6:e28125 doi:10.1371/journal.pone.0028125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leutenegger CM, Higgins J, Matthews TB, Tarantal AF, Luciw PA, Pedersen NC, North TW. 2001. Real-time TaqMan PCR as a specific and more sensitive alternative to the branched-chain DNA assay for quantitation of simian immunodeficiency virus RNA. AIDS Res. Hum. Retroviruses 17:243–251 [DOI] [PubMed] [Google Scholar]
- 22. Kent SJ, De Rose R, Mokhonov VV, Mokhonova EI, Fernandez CS, Alcantara S, Rollman E, Mason RD, Loh L, Peut V, Reece JC, Wang XJ, Wilson KM, Suhrbier A, Khromykh A. 2008. Evaluation of recombinant Kunjin replicon SIV vaccines for protective efficacy in macaques. Virology 374:528–534 [DOI] [PubMed] [Google Scholar]
- 23. Yu D, Batten M, Mackay CR, King C. 2009. Lineage specification and heterogeneity of T follicular helper cells. Curr. Opin. Immunol. 21:619–625 [DOI] [PubMed] [Google Scholar]
- 24. Haynes NM, Allen CD, Lesley R, Ansel KM, Killeen N, Cyster JG. 2007. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J. Immunol. 179:5099–5108 [DOI] [PubMed] [Google Scholar]
- 25. Good-Jacobson KL, Szumilas CG, Chen L, Sharpe AH, Tomayko MM, Shlomchik MJ. 2010. PD-1 regulates germinal center B cell survival and the formation and affinity of long-lived plasma cells. Nat. Immunol. 11:535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lim HW, Kim CH. 2007. Loss of IL-7 receptor alpha on CD4+ T cells defines terminally differentiated B cell-helping effector T cells in a B cell-rich lymphoid tissue. J. Immunol. 179:7448–7456 [DOI] [PubMed] [Google Scholar]
- 27. Petrovas C, Yamamoto T, Gerner MY, Boswell KL, Wloka K, Smith EC, Ambrozak DR, Sandler NG, Timmer KJ, Sun X, Pan L, Poholek A, Rao SS, Brenchley JM, Alam SM, Tomaras GD, Roederer M, Douek DC, Seder RA, Germain RN, Haddad EK, Koup RA. 2012. CD4 T follicular helper cell dynamics during SIV infection. J. Clin. Invest. 122:3281–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. 2009. Bcl6 mediates the development of T follicular helper cells. Science 325:1001–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peden KWC, Farber JM. 2000. Coreceptors for human immunodeficiency virus and simian immunodeficiency virus. Adv. Pharmacol. 48:409–478 [DOI] [PubMed] [Google Scholar]
- 30. Pohlmann S, Davis C, Meister S, Leslie GJ, Otto C, Reeves JD, Puffer BA, Papkalla A, Krumbiegel M, Marzi A, Lorenz S, Munch J, Doms RW, Kirchhoff F. 2004. Amino acid 324 in the simian immunodeficiency virus SIVmac V3 loop can confer CD4 independence and modulate the interaction with CCR5 and alternative coreceptors. J. Virol. 78:3223–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma CS, Suryani S, Avery DT, Chan A, Nanan R, Santner-Nanan B, Deenick EK, Tangye SG. 2009. Early commitment of naive human CD4(+) T cells to the T follicular helper (TFH) cell lineage is induced by IL-12. Immunol. Cell Biol. 87:590–600 [DOI] [PubMed] [Google Scholar]
- 32. Chakrabarti L, Cumont MC, Montagnier L, Hurtrel B. 1994. Variable course of primary simian immunodeficiency virus infection in lymph nodes: relation to disease progression. J. Virol. 68:6634–6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hirsch VM, Santra S, Goldstein S, Plishka R, Buckler-White A, Seth A, Ourmanov I, Brown CR, Engle R, Montefiori D, Glowczwskie J, Kunstman K, Wolinsky S, Letvin NL. 2004. Immune failure in the absence of profound CD4+ T-lymphocyte depletion in simian immunodeficiency virus-infected rapid progressor macaques. J. Virol. 78:275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Monceaux V, Estaquier J, Fevrier M, Cumont MC, Riviere Y, Aubertin AM, Ameisen JC, Hurtrel B. 2003. Extensive apoptosis in lymphoid organs during primary SIV infection predicts rapid progression towards AIDS. AIDS 17:1585–1596 [DOI] [PubMed] [Google Scholar]
- 35. Klatt NR, Vinton CL, Lynch RM, Canary LA, Ho J, Darrah PA, Estes JD, Seder RA, Moir SL, Brenchley JM. 2011. SIV infection of rhesus macaques results in dysfunctional T- and B-cell responses to neo and recall Leishmania major vaccination. Blood 118:5803–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Herman AE, Freeman GJ, Mathis D, Benoist C. 2004. CD4+CD25+ T regulatory cells dependent on ICOS promote regulation of effector cells in the prediabetic lesion. J. Exp. Med. 199:1479–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hong JJ, Amancha PK, Rogers K, Ansari AA, Villinger F. 2012. Spatial alterations between CD4+ T follicular helper, B, and CD8+ T cells during simian immunodeficiency virus infection: T/B cell homeostasis, activation, and potential mechanism for viral escape. J. Immunol. 188:3247–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lindqvist M, van Lunzen J, Soghoian DZ, Kuhl BD, Ranasinghe S, Kranias G, Flanders MD, Cutler S, Yudanin N, Muller MI, Davis I, Farber D, Hartjen P, Haag F, Alter G, Wiesch JS, Streeck H. 2012. Expansion of HIV-specific T follicular helper cells in chronic HIV infection. J. Clin. Invest. 122:3271–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cumont MC, Diop O, Vaslin B, Elbim C, Viollet L, Monceaux V, Lay S, Silvestri G, Le Grand R, Muller-Trutwin M, Hurtrel B, Estaquier J. 2008. Early divergence in lymphoid tissue apoptosis between pathogenic and nonpathogenic simian immunodeficiency virus infections of nonhuman primates. J. Virol. 82:1175–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brenchley JM, Vinton C, Tabb B, Hao XP, Connick E, Paiardini M, Lifson JD, Silvestri G, Estes JD. 2012. Differential infection patterns of CD4+ T cells and lymphoid tissue viral burden distinguish progressive and nonprogressive lentiviral infections. Blood 120:4172–4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Haase AT. 1999. Population biology of HIV-1 infection: viral and CD4+ T cell demographics and dynamics in lymphatic tissues. Annu. Rev. Immunol. 17:625–656 [DOI] [PubMed] [Google Scholar]
- 42. Fahey LM, Wilson EB, Elsaesser H, Fistonich CD, McGavern DB, Brooks DG. 2011. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. J. Exp. Med. 208:987–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Harker JA, Lewis GM, Mack L, Zuniga EI. 2011. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science 334:825–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zou W, Lackner AA, Simon M, Durand-Gasselin I, Galanaud P, Desrosiers RC, Emilie D. 1997. Early cytokine and chemokine gene expression in lymph nodes of macaques infected with simian immunodeficiency virus is predictive of disease outcome and vaccine efficacy. J. Virol. 71:1227–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Campillo-Gimenez L, Cumont MC, Fay M, Kared H, Monceaux V, Diop O, Muller-Trutwin M, Hurtrel B, Levy Y, Zaunders J, Dy M, Leite-de-Moraes MC, Elbim C, Estaquier J. 2010. AIDS progression is associated with the emergence of IL-17-producing cells early after simian immunodeficiency virus infection. J. Immunol. 184:984–992 [DOI] [PubMed] [Google Scholar]
- 46. Oestreich KJ, Mohn SE, Weinmann AS. 2012. Molecular mechanisms that control the expression and activity of Bcl-6 in T(H)1 cells to regulate flexibility with a T(FH)-like gene profile. Nat. Immunol. 13:405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shimizu N, Tanaka A, Oue A, Mori T, Ohtsuki T, Apichartpiyakul C, Uchiumi H, Nojima Y, Hoshino H. 2009. Broad usage spectrum of G protein-coupled receptors as coreceptors by primary isolates of HIV. AIDS 23:761–769 [DOI] [PubMed] [Google Scholar]
- 48. Kanbe K, Shimizu N, Soda Y, Takagishi K, Hoshino H. 1999. A CXC chemokine receptor, CXCR5/BLR1, is a novel and specific coreceptor for human immunodeficiency virus type 2. Virology 265:264–273 [DOI] [PubMed] [Google Scholar]
- 49. Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. 2013. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J. Exp. Med. 210:143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hufert FT, van Lunzen J, Janossy G, Bertram S, Schmitz J, Haller O, Racz P, von Laer D. 1997. Germinal centre CD4+ T cells are an important site of HIV replication in vivo. AIDS 11:849–857 [DOI] [PubMed] [Google Scholar]
- 51. Gratton S, Cheynier R, Dumaurier MJ, Oksenhendler E, Wain-Hobson S. 2000. Highly restricted spread of HIV-1 and multiply infected cells within splenic germinal centers. Proc. Natl. Acad. Sci. U. S. A. 97:14566–14571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220 [DOI] [PubMed] [Google Scholar]
- 53. Racz P, Tenner-Racz K, van Vloten F, Schmidt H, Dietrich M, Gluckman JC, Letvin NL, Janossy G. 1990. Lymphatic tissue changes in AIDS and other retrovirus infections: tools and insights. Lymphology 23:85–91 [PubMed] [Google Scholar]
- 54. McHeyzer-Williams M, Okitsu S, Wang N, McHeyzer-Williams L. 2012. Molecular programming of B cell memory. Nat. Rev. Immunol. 12:24–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zaunders JJ, Cunningham PH, Kelleher AD, Kaufmann GR, Jaramillo AB, Wright R, Smith D, Grey P, Vizzard J, Carr A, Cooper DA. 1999. Potent antiretroviral therapy of primary human immunodeficiency virus type 1 (HIV-1) infection: partial normalization of T lymphocyte subsets and limited reduction of HIV-1 DNA despite clearance of plasma viremia. J. Infect. Dis. 180:320–329 [DOI] [PubMed] [Google Scholar]
- 56. Richman DD, Wrin T, Little SJ, Petropoulos CJ. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 58. Igarashi T, Brown C, Azadegan A, Haigwood N, Dimitrov D, Martin MA, Shibata R. 1999. Human immunodeficiency virus type 1 neutralizing antibodies accelerate clearance of cell-free virions from blood plasma. Nat. Med. 5:211–216 [DOI] [PubMed] [Google Scholar]
- 59. Schacker TW, Nguyen PL, Martinez E, Reilly C, Gatell JM, Horban A, Bakowska E, Berzins B, van Leeuwen R, Wolinsky S, Haase AT, Murphy RL. 2002. Persistent abnormalities in lymphoid tissues of human immunodeficiency virus-infected patients successfully treated with highly active antiretroviral therapy. J. Infect. Dis. 186:1092–1097 [DOI] [PubMed] [Google Scholar]
- 60. Moir S, Fauci AS. 2009. B cells in HIV infection and disease. Nat. Rev. Immunol. 9:235–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.