Abstract
After entering a host cell, retroviruses such as simian immunodeficiency virus (SIV) uncoat, disassembling the viral capsid. Rates of uncoating that are too high and too low can be detrimental to the efficiency of infection. Rapid uncoating typically leads to blocks in reverse transcription, but the basis for replication defects associated with slow uncoating is less clear. Here we characterize the phenotypes of two SIVmac239 mutants with changes, A87E and A87D, in the helix 4/5 loop of the capsid protein. These mutant viruses exhibited normal capsid morphology but were significantly attenuated for infectivity. The infectivity of wild-type and mutant SIVmac239 was not decreased by aphidicolin-induced growth arrest of the target cells. In the cytosol of infected cells, the A87E and A87D capsids remained in particulate form longer than the wild-type SIVmac239 capsid, suggesting that the mutants uncoat more slowly than the wild-type capsid. Both mutants exhibited much higher levels of autointegrated DNA forms than wild-type SIVmac239. Thus, some changes in the helix 4/5 loop of the SIVmac239 capsid protein result in capsid hyperstability and an increase in autointegration.
INTRODUCTION
Lentiviruses differ from most other retroviruses in that they have evolved to infect mature individuals with competent immune systems (1). Indeed, lentiviruses rely upon the cells of the host immune system to provide functions necessary for their replication and dissemination. In so doing, they have evolved the ability to integrate their genome into cells that are terminally differentiated, accessing the nucleus through the nuclear pore (2). Only a fraction of viruses that gain entry to a cell are successful in navigating to the nuclear compartment and creating a provirus capable of supporting the late phase of retrovirus replication. This excess of defective events has complicated the characterization of early-stage infection.
Following entry into the host cell, lentiviruses must proceed through several steps on the way to generating a provirus. These early-phase processes include uncoating of the core, reverse transcription of the RNA genome, nuclear entry of the preintegration complex (PIC), and integration. Changes in lentivirus capsid proteins have been shown to accelerate or slow the uncoating process (3–7). Less expectedly, alterations in the capsid protein can affect subsequent steps in the early phase of lentivirus infection. An appreciation of the contribution of the capsid to multiple steps in the lentiviral life cycle has made capsid an attractive target of therapeutic intervention (6, 8–18).
Two host proteins that bind the human immunodeficiency virus type 1 (HIV-1) capsid and influence the uncoating process have been identified. TRIM5α is a protein that mediates the premature, deleterious disassembly of the capsid and therefore acts as a restriction factor (19). The prolyl cis-trans isomerase cyclophilin A (CypA) binds the HIV-1 capsid and modulates capsid stability in either a positive or negative manner, depending on the type of infected cell and the sequence of the helix 4/5 loop (CypA-binding loop) of the capsid protein (CA) (5, 16, 20–24). In T lymphocytes, CypA stabilizes the HIV-1 capsid and promotes infection (5). The CypA domain of the nuclear pore protein Nup358 has been reported to be important for proper nuclear targeting of HIV-1 preintegration complexes (5, 16, 20–24).
Simian immunodeficiency viruses (SIVs) infect feral African monkeys and apes and are the lentivirus ancestors of human immunodeficiency viruses (25–28). The capsids of monkey SIVs do not interact with CypA (29, 30). In permissive cells without a restricting TRIM5α protein, SIV infection can be studied in the absence of either of these host capsid-binding proteins. We took advantage of this characteristic to assess the function of the flexible helix 4/5 loop that extends above the capsid surface. In HIV-1, this loop is the site of CypA binding, near CA residues Gly89 and Pro90 (31). In contrast, the helix 4/5 loop of SIVmac239 does not contain a Gly-Pro motif and does not bind CypA. However, a structurally similar Ala-Pro motif occurs at capsid residues Ala87 and Pro88 in the SIVmac239 helix 4/5 loop. Interestingly, the helix 4/5 loop of the HIV-1 CA has an Ala-Pro pair (Ala92 and Pro93) C terminal to the Gly-Pro motif. Alteration of alanine 92 to glutamic acid, or the nearby glycine at position 94 to aspartic acid, renders HIV-1 CypA independent in certain cell types (24, 32). In other cell types, the infectivity of the A92E and G94D capsid mutants is inhibited by CypA binding. The replication phenotype of the A92E and G94D capsid mutants is predicted by the level of CypA in the target cell (5, 22, 24).
Because changes in the helix 4/5 loop of the HIV-1 CA can affect sensitivity to CypA and TRIM5α (15), the interpretation of the mechanisms that underlie the phenotypes of HIV-1 CA helix 4/5 mutants is complicated. Here we study the effects of acidic substitutions in the Ala-Pro motif in the helix 4/5 loop of the SIVmac239 CA. We characterize the phenotypes of the SIVmac239 A87E and A87D mutants, observing decreases in the early phase of infection and slower capsid uncoating. Although reverse transcription of these mutants was efficient, striking increases in the amounts of autointegrated viral DNA were seen for both mutants. Models that fit this heretofore unknown capsid function into current knowledge of early events in lentiviral infection are discussed.
MATERIALS AND METHODS
Cell lines and culture.
Adherent cell lines used in this study were grown in Dulbecco's modified Eagle's medium (DMEM) (high glucose) (catalog number 11965; Invitrogen) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. HEK293T/17 (ATCC CRL-11268) and HeLa (ATCC CCL-2) cells are of human origin. Cf2Th cells (ATCC CRL-1430) are of canine origin.
Site-directed mutagenesis.
All mutants were created by site-directed mutagenesis of either the full-length pSIVmac239ΔnefΔenvEGFP plasmid (see below) or the shuttle vector SGag3 (33), utilizing Pfu Ultra Hotstart 2× Master Mix (catalog number 600630-51; Stratagene), 125 ng of each mutagenic primer, and the following cycling protocol: 95°C for 1 min and 18 cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 12 min (full-length) or 5.5 min (SGag3), with a final 1-min extension step at 72°C. SGag3 sequences were then inserted into the full-length vector by DraIII/SbfI digestion and T4 ligation. All mutations were verified by sequencing. Mutagenic primers (Integrated DNA Technologies) were based on the forward wild-type (WT) sequence GCAGCACCCACAACCAGCTCCACAACAAGGACAACTTAGG.
Production of recombinant SIV variants expressing GFP.
The SIV-based pSIVmac239ΔnefΔenvEGFP plasmid was derived from pSIVΔnefEGFP (34) (a kind gift from Ronald Desrosiers, Harvard Medical School). Env sequences were deleted by first changing the ATG start codon to TCC to create a BspEI site. The 1.1-kb fragment between this and the native BspEI site at position 8013 (GenBank accession number M33262.1) was excised, leaving the tat and rev genes and the Rev-responsive element intact. This construct and derivative mutants were cotransfected into the desired cell type with a Rev-expressing construct and the vesicular stomatitis virus G glycoprotein (VSV-G)-expressing construct pHCMV-G, as described previously (15). HEK293T/17 cells were transfected by using Lipofectamine Plus or Lipofectamine 2000 according to the manufacturer's protocol. Recombinant viruses in the transfected-cell supernatants were filtered (0.45-μm filter), and titers were determined by a reverse transcriptase (RT) assay, as previously described (35). If necessary, viruses were concentrated by polyethylene glycol (PEG) precipitation using PEG-it (catalog number LV810A; SBI) and resuspended in an appropriate volume of 1× phosphate-buffered saline (PBS).
Infection assays.
One day before infection, HeLa and Cf2Th cells at a density of 1 × 105 to 3 × 105 cells/well were seeded into 24-well plates. On the day of infection, virus was added at the desired titer, as determined by RT activity, expressed in counts per minute (cpm). The medium was changed 2 to 4 h afterwards, and 48 h following infection, the percentage of green fluorescent protein (GFP)-positive cells and median fluorescence were determined by fluorescence-activated cell sorter (FACS) analysis.
Gag processing assay.
HEK293T/17 cells in 24-well plates were transfected with a Rev-expressing plasmid and the pSIVmac239ΔnefΔenvEGFP plasmids expressing the wild-type and mutant Gag proteins. At 16 h posttransfection, the medium was replaced by DMEM plus 10% dialyzed FBS, 1% penicillin-streptomycin, 1× l-glutamine, 1× nonessential amino acids, and 100-fold-diluted Express35S [35S]Met-Cys labeling mix (NEG072; PerkinElmer). At 48 h posttransfection, viruses were purified by filtering (0.45-μm filter) followed by pelleting through 20% sucrose for 1 h at 100,000 × g. Viral pellets were resuspended directly into loading buffer. Approximately 25,000 cpm was loaded into each well of a 12% Bis-Tris gel, which was run and exposed to film.
Electron microscopy and core morphology quantitation.
Envelope-free viruses were produced and purified by anion-exchange chromatography, as described previously (36). Purified virions were prepared for thin-section microscopy by the Harvard Medical School EM Facility. Fields containing multiple identifiable virions were selected for analysis. Virions were binned into one of three categories based upon the shape of their electron-dense core in the sectioning plane: conical cores, well-defined circular cores, and unidentifiable “other.”
Fate-of-capsid assays.
Cf2Th cells were seeded into 75-cm2 flasks to achieve 80 to 90% confluence at the time of infection. Flasks were prechilled on ice 10 min prior to the addition of 5 × 106 cpm of virus in a 4-ml volume. Flasks and virus were kept at 4°C for a half hour, and time zero was then marked by shifting them to a 37°C incubator. Virus was washed off the cells, and fresh medium was added at 4 h postinfection. At the appropriate time, flasks were chilled on ice and washed three times with ice-cold 1× PBS. Cells were released from the flask by pronase digestion (1 ml of 7 μg/ml pronase in 1× PBS for 5 min), transferred into 15-ml conical tubes, and washed three times with 10 ml cold PBS. The pellet was resuspended in 250 μl cold hypolysis solution (10 mM Tris [pH 8], 10 mM KCl, 1 mM EDTA), transferred into Eppendorf tubes, and kept on ice for 15 min. Cells were then mechanically disrupted for 1 min by using a pellet pestle (catalog number K49540; Kontes), and noncytosolic material was removed by centrifugation at 1,500 × g for 3 min. The supernatant was mixed with 250 μl fresh hypolysis buffer, and 400 μl of this mixture was layered on top of a 7-ml column of 50% sucrose–1× PBS in an Ultraclear SW41 tube (catalog number 344059; Beckman Coulter); the remaining lysate was preserved in 1× Laemmli buffer. The column was centrifuged for 2 h at 125,000 × g. The top 100 μl was collected, and the pellet was resuspended in 100 μl 1× PBS; concentrated Laemmli buffer was added to bring both samples to a 1× final concentration. Samples were run on 4 to 12% Bis-Tris gels and transferred onto nitrocellulose filters. The filters were probed with a 1:1,000 dilution of anti-SIVmac251 polyclonal serum (AIDS Reagent Program number 2773) and a 1:40,000 dilution of horseradish peroxidase (HRP)-conjugated goat anti-human IgG (catalog number A0293; Sigma) and developed with the SuperSignal West Femto chemiluminescence reagent (catalog number 34095; Thermo). Densitometry was performed by using ImageQuantTL, using a consistent 50-pixel rolling-ball background reduction.
Quantitative real-time PCR.
HeLa cells were seeded into 6-well plates to achieve 80% confluence at the time of infection. Viral suspensions were treated with DNase Turbo (catalog number AM2238; Ambion), according to the manufacturer's protocol, for 1 h at 37°C, and the titer was then determined by an RT assay. Approximately 100,000 cpm of virus, expected to result in a multiplicity of infection of ∼0.5, was added to each well at time zero. For negative controls, viruses were heat inactivated at 56°C for 1 h, and the infected cells were maintained in 100 μM zidovudine (AZT) prior to incubation with the virus and for the duration of the experiment. In some experiments, raltegravir was added to cells at a concentration of 1 μM for 1 h prior to addition of the virus and maintained at that concentration throughout the experiment (unless otherwise noted). Unbound virus was washed off the cells 2 h after virus-cell incubation was initiated. Cells were collected by trypsin digestion at 2, 6, 12, 24, and 48 h after virus-cell incubation. Total DNA was extracted by using the Wizard SV 96 Genomic system (catalog number A2371; Promega) and quantitated with a Nanodrop spectrophotometer. DNA was diluted to a final concentration of 10 ng/μl using double-distilled autoclaved water. Each quantitative real-time PCR (qPCR) mixture contained 100 ng of total DNA in a 50-μl reaction volume. The accumulation of product was assayed by a TaqMan probe on an Applied Biosystems 7300 instrument. The primers and probe for the analysis of the early stage of reverse transcription were early forward primer GTC AAC TCG GTA CTC AAT AAT AAG, early reverse primer GCG CCA ATC TGC TAG GGA T, and early probe 6-carboxyfluorescein (FAM)–CTG TTA GGA CCC TTT CTG CTT TGG GAA ACC GAA G–6-carboxytetramethylrhodamine (TAMRA). The primers and probe for the analysis of the late stage of reverse transcription were late forward primer TTG GGA AAC CGA AGC AGG, late reverse primer TCT CTC ACT CTC CTT CAA GTC CCT, and late probe FAM-AAA TCC CTA GAC GAT TGG CGC CTG AA-TAMRA. The primers and probes for the analysis of two-long terminal repeat (2-LTR) circles were 2-LTR forward primer GGA AAC CGA AGC AGG AAA AT, 2-LTR reverse primer CTG TGC CTC ATC TGA TAC, 2-LTR standard probe FAM-ATT GGC AGG ATT ACA CCT CAG GAC CAG-TAMRA, and 2-LTR junction probe FAM-ATT CCC TAG CAG ATA CTG GAA GGG AT-TAMRA. The primers and probe for the analysis of integrated viral DNA were primary (1°) Alu PCR forward primer ATG CCA CGT AAG CGA AAC TCT GTT CCC ATC TCT CCT AGC C, 1° Alu PCR reverse primer TCC CAG CTA CTC GGG AGG CTG AGG, secondary (2°) Alu PCR forward primer ATG CCA CGT AAG CGA AAC TC, 2° Alu PCR reverse primer ATT TTC CTG CTT CGG TTT CC, and Alu PCR probe FAM-CGC CTG GTC AAC TCG GTA CTC AAT AA-TAMRA.
Alu qPCR.
Using the samples collected for single-round qPCR, we optimized protocols for assaying integrated SIVmac239 proviruses (by Alu PCR) and autointegrated SIVmac239 genomes. For Alu PCR, we performed a primary amplification using 100 ng total DNA, 0.2 μM each primer, and Pfu UltraII Hotstart 2× Master Mix (catalog number 600850; Agilent) in a final volume of 20 μl. Cycling parameters for touchdown PCR (TD-PCR) were as follows: 95°C for 2 min; 15 cycles of 95°C for 30 s, 70°C to 55°C for 30 s, and 72°C for 1 min; and 11 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min. Primary PCR samples were diluted 1:5 into 100 μl water, and 10 μl was used in a standard 50-μl qPCR mixture. The number of integrants was determined by calibrating the signal versus a standard curve of integrated viruses of known copy number. The material for this standard curve was created by using HeLa cells transduced with recombinant SIV expressing GFP. The transduced cells were passaged for 4 weeks, the total DNA was collected as described above, and the proviral copy number was determined by utilizing the late qPCR primer set.
Characterization of PCR products amplified by the 2-LTR primer set.
Products from single-time-point qPCR amplifications with the 2-LTR primers, representing three independent infections, were taken directly from the postrun 96-well qPCR assay plate and pooled. Ten microliters of each resulting 150-μl sample was run on a 2% agarose gel and stained with ethidium bromide. Samples of interest were excised from the gel, extracted, and cloned directly into the TOPO-TA vector pCR4-TOPO (catalog number K457501; Invitrogen). After transforming Escherichia coli TOP10 cells, well-isolated ampicillin-resistant colonies were picked and grown in 200 μl Luria-Bertani (LB) medium with ampicillin overnight at room temperature in 96-well plates. These plates were then shipped to Beckman Coulter Genomics for single-pass sequencing using flanking T7 primers. All sequences were aligned and collated, comprising almost 500 individual clones. No sequences were discarded.
RESULTS
Effects of capsid changes on SIV production and infectivity.
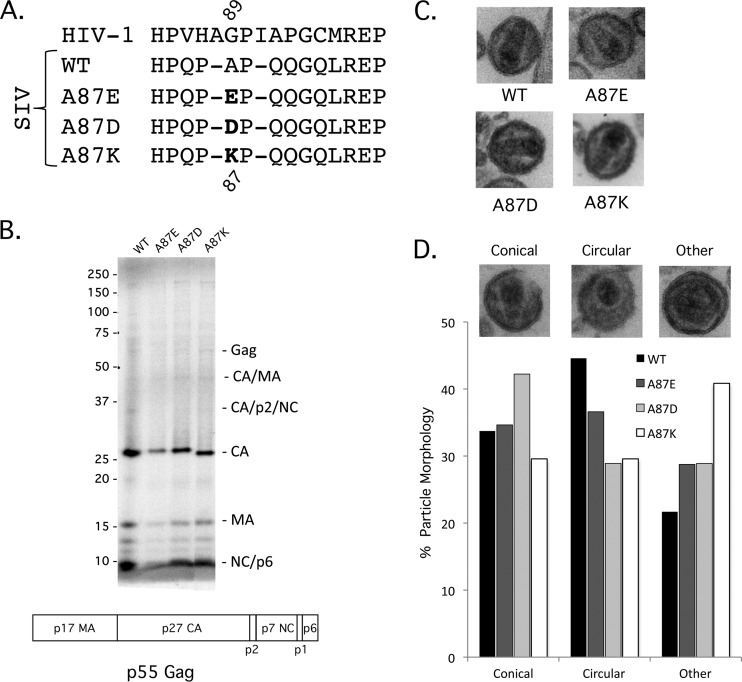
The primary structure of the helix 4/5 loop of the SIVmac239 CA differs from that of the HIV-1 CA but shares a proline-rich character and a (G/A)P motif at a similar position (Fig. 1A). We altered the alanine residue at position 87 in the SIVmac239 CA to an aspartic acid, glutamic acid, or lysine residue to investigate the function of the helix 4/5 loop in the SIV capsid. Recombinant enhanced GFP (EGFP)-expressing viruses pseudotyped with the VSV-G envelope protein were generated by transfection of HEK293T/17 cells. Viruses were collected, filtered, and concentrated (if necessary), and reverse transcriptase activity was measured. Equivalent amounts of reverse transcriptase activity associated with the wild-type and mutant viruses were detected in the medium of transfected cells (data not shown), suggesting that the assembly and release of the virus variants were comparable. SDS-PAGE analysis of metabolically labeled virion proteins revealed similar levels of processing of the Gag polyprotein (Fig. 1B). A87E, A87D, and A87K virions exhibited wild-type core morphology (Fig. 1C). The documented frequency of virions with ideal core morphology and virion size was only slightly reduced for the mutants compared with wild-type SIVmac239 (Fig. 1D).
Fig 1.
Gag precursor processing and core morphology of SIVmac239 capsid variants. (A) The central portion of the SIVmac239 helix 4/5 loop is shown, comparing the sequences of HIV-1 and wild-type (WT) SIVmac239 and the A87E, A87D, and A87K mutants. (B) The processing of the Gag precursor polyprotein was examined by metabolically labeling transfected HEK293T cells producing WT and mutant SIVmac239 virions. Secreted viruses were collected, purified over a 20% sucrose column, and run on a 12% Bis-Tris gel. Loading was normalized by the amount of 35S label. The domain organization of the SIVmac239 p55 Gag precursor polyprotein is illustrated beneath the gel. (C) To examine virion morphology, purified virions were pelleted, stained, and examined by electron microscopy. (D) Virion particles for each variant were identified and then binned according to their core morphology: conical, circular, or “other.” Examples of each are displayed above the category axis (wild type, n = 153; A87E, n = 166; A87D, n = 135; A87K, n = 213).
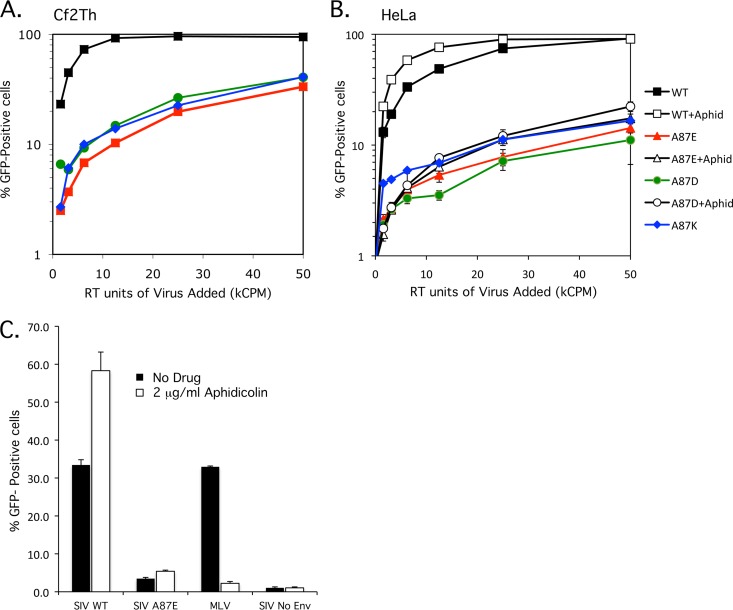
To examine the infectivity of the mutants, 2-fold dilutions of viral preparations normalized for RT activity were used to infect Cf2Th and HeLa cells. Although the human HeLa cells were infected less efficiently than the canine Cf2Th cells, the relative infectivities of the mutants, compared with that of wild-type SIVmac239, were similar in the two cell types. The A87E, A87D, and A87K mutants were 10 to 15% as infectious as the wild-type virus (Fig. 2A and B). The similarity of the results for human and canine cells suggests that the deficient phenotype is not due to increased susceptibility to species-specific restriction factors like TRIM5α.
Fig 2.
Infectivity of wild-type and mutant SIVmac239. (A and B) VSV-G-pseudotyped recombinant SIVmac239 viruses were produced in transfected HEK293T cells, and reverse transcriptase activity was measured. Increasing amounts of the WT, A87E, A87D, and A87K viruses, which express GFP, were used to infect Cf2Th (A) or HeLa (B) cells. The percentage of infected, GFP-positive cells was assayed at 48 h postinfection by FACS analysis. (B) Aphidicolin (2 μg/ml) was added to one set of the HeLa target cells (+aphid). The results shown are typical of those obtained in 3 independent experiments. (C) HeLa cells were infected with identical amounts (in RT units) of VSV-G-pseudotyped GFP-expressing recombinant WT and A87E SIV and, as a control, MLV. In some cases, 2 μg/ml aphidicolin was added to the HeLa cells. GFP-positive cells were measured by FACS analysis 48 h after infection. The means and standard deviations of the results obtained in 3 independent experiments are shown.
Multiple reports identified the capsid as a major determinant in allowing lentiviral preintegration complexes to transit through the nuclear pore (6, 8, 16, 24, 37). Some capsid alterations eliminate the ability of the lentivirus to infect growth-arrested cells and terminally differentiated macrophages. To assess whether the A87E or A87D CA mutant falls into this category, HeLa cells arrested in the G1/S phase of the cell cycle by the addition of 2 μg/ml aphidicolin were incubated with the wild-type and these mutant SIVs. The A87K mutant was not included due to results that indicated a block at or prior to initiation of reverse transcription (see below). The infection efficiency of the WT, A87E, and A87D viruses was insensitive to cell cycle arrest (Fig. 2B). In fact, the efficiency of infection increased slightly upon aphidicolin treatment, a phenomenon which was reported previously (38). Aphidicolin treatment reduced the infection of Moloney murine leukemia virus (MLV), a gammaretrovirus, by 95% (Fig. 2C).
Capsid stability in infected cells.
To examine the stability of the altered SIVmac239 capsids in infected cells, high-titer virus was added to Cf2Th cells, and fate-of-capsid assays were performed 16 h later, a time determined to be informative for this assay. At 16 h postinfection, A87E and A87D mutant viruses exhibited a significant increase in the amount of pelletable CA compared to wild-type SIVmac239; the A87K mutant had capsid stability indistinguishable from that of the wild-type virus (Fig. 3A). Two additional independent assays were performed, revealing a statistically significant increase in capsid stability at 16 h postinfection for the A87E and A87D mutants (Fig. 3B). To investigate the kinetics of the decrease in pelletable CA for the wild-type and mutant viruses, a fate-of-capsid assay was performed every 4 h after infection, up until 28 h postinfection (Fig. 3C). Whereas the amount of pelletable wild-type SIVmac239 CA is initially high, representing a pulse of entry, by 8 to 12 h postinfection, the majority of CA is found in soluble forms. Between 20 and 28 h after infection, the amount of pelletable wild-type CA decreases further and becomes undetectable. The amounts of pelletable CA in cells infected by the A87E and A87D mutants were larger than those seen in cells infected by the wild-type virus at all time points beyond 8 h postinfection. The persistence of signal in the pelletable CA fraction suggests that the A87E and A87D mutant capsids uncoat more slowly than the wild-type SIVmac239 capsid.
Fig 3.
Fate of the wild-type and mutant SIVmac239 capsids in infected cells. Cf2Th cells were incubated at 37°C with equivalent concentrations of VSV-G-pseudotyped viruses, based on RT units. At the indicated times, the infected cells were mechanically disrupted, and their cytosolic fractions were layered atop a 50% sucrose column, saving some to measure the “input.” After centrifugation, a “soluble” capsid sample was removed from near the top of the column, and the pelletable capsid was collected at the bottom of the tube following column removal. Input, soluble, and pelletable fractions were loaded onto a 4 to 12% Bis-Tris gel, Western blotted, and probed with anti-SIVmac251 polyclonal antibody. (A) The results of a single experiment are shown, with the fate of capsid measured at 16 h postinfection (p.i.). (B) Three independent infections were assayed for pelletable capsid at 16 h postinfection. The amounts of pelletable capsid were determined by densitometric analysis of Western blots, adjusted for input and normalized to the signal for WT SIVmac239, which was set at 100%. The A87E and A87D mutants had significantly more pelletable capsid than wild-type SIVmac239 (*, P < 0.01). (C) Time course of the fate of the capsid in infected cells. After the initiation of infection, the amounts of input and pelletable fractions containing CA were assayed every 4 h until 28 h postinfection. A parallel infection of recombinant WT SIVmac239 without a VSV-G glycoprotein (no Env) was used as a negative control. Data from one of two independent experiments, which generated similar results, are shown.
Synthesis of viral cDNA and proviruses.
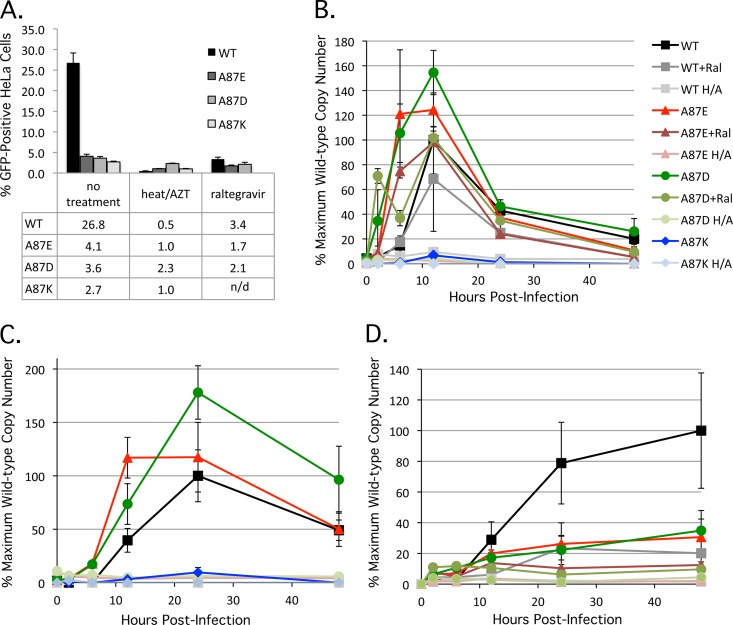
To determine whether the A87E, A87D, and A87K mutants efficiently reverse transcribe their genomes, we measured SIV cDNA production by real-time quantitative PCR (39, 40). HeLa cells were infected at a multiplicity of infection of approximately 0.5, and total DNA was collected from infected cells at 2, 6, 12, 24, and 48 h postinfection. As a negative control, viruses were heat inactivated at 56°C for 1 h, and 100 μM AZT was added to the target cells throughout the experiment. As an additional control, one subset of infected cells was treated with 1 μM (10× the reported 95% effective concentration [EC95]) raltegravir, an active-site inhibitor of retroviral integrases (41, 42). The infection levels achieved for the experimental and control assays are shown in Fig. 4A. Late RT products were detected in cells infected by the wild-type and A87E and A87D mutant viruses, indicating that reverse transcription of complete, late-stage genomes occurred; in contrast, the A87K mutant produced detectable but near-baseline levels of late-stage genomes (Fig. 4B). Similarly, the levels of the A87K early RT product, minus-strand strong-stop DNA, were well below those observed for wild-type SIVmac239 (data not shown). These data strongly suggest that the defect resulting from the introduction of a lysine at residue 87 of the SIVmac239 capsid manifests very soon after virus entry. A87K infection would therefore not be expected to result in normal levels of 2-LTR circles, and this was demonstrated by using a primer designed to amplify this product. Infections with wild-type SIVmac239 and the A87E and A87D mutants all yielded high levels of 2-LTR products, an observation which is discussed in more detail below (Fig. 4C). The number of proviruses integrated into the cells infected by the wild-type, A87E, and A87D viruses (Fig. 4D) corresponded to the observed number of GFP-positive cells in the infectivity assay (Fig. 4A). As expected, raltegravir did not eliminate reverse transcription but reduced both the infection efficiency and the number of proviruses.
Fig 4.
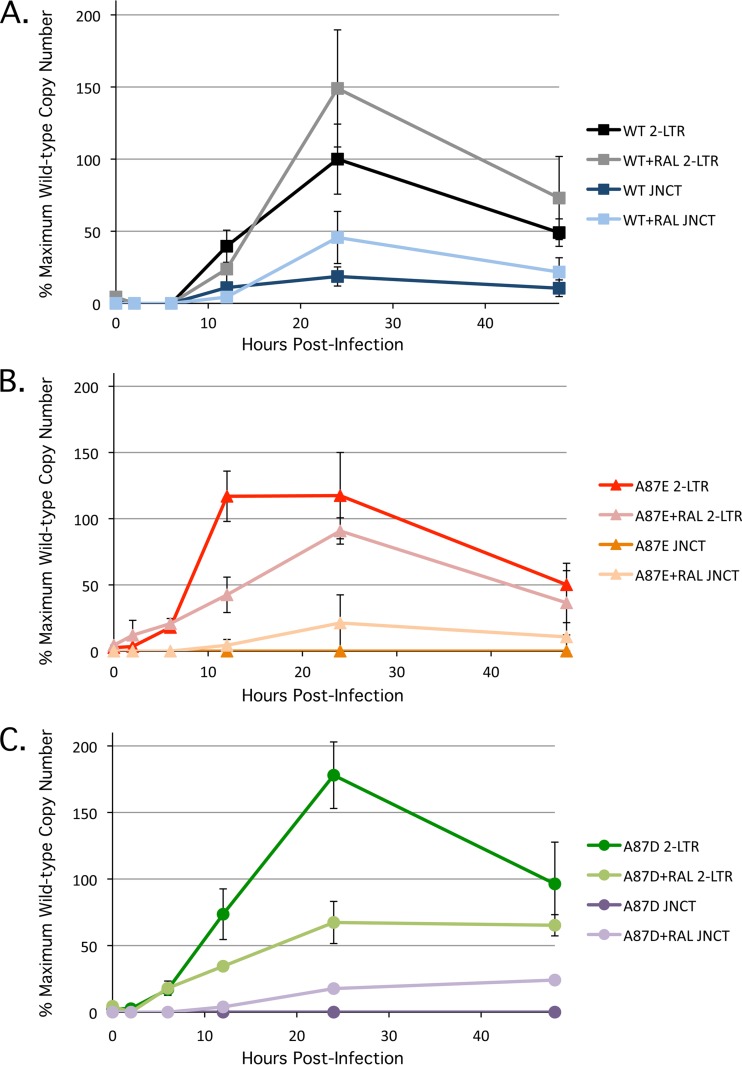
Reverse transcription and integration of SIVmac239 variants. (A) Three independent infections of HeLa cells with wild-type and mutant SIVmac239 at a multiplicity of infection of approximately 0.5 were performed, and the percentage of infected, GFP-positive cells was assayed at 48 h postinfection, as described in the Fig. 2 legend. In some experiments, the viruses were incubated at 56°C for 1 h, and the cells were treated with 100 mM AZT (“heat/AZT”). In another set of experiments, 1 μM raltegravir was added to the infected cells. In the experiment shown, the value obtained for the heat-inactivated, AZT-treated A87D mutant is atypically high due to stochastic autofluorescence of the HeLa cells after treatment with 100 μM AZT. n/d, not determined. (B) To assess the production of late reverse transcripts, total DNA was extracted from infected HeLa cells at 2, 6, 12, 24, and 48 h postinfection. Late reverse transcript copy numbers were determined by quantitative real-time PCR (H/A, heat inactivation of virus plus AZT treatment of target cells, as described above; Ral, raltegravir treatment [1 μM] of target cells, as described above). The values shown represent the percentages of the maximum wild-type SIVmac239 value (7,245 copies/100 ng DNA at 12 h postinfection). (C) The production of 2-LTR circles was assessed as described in Materials and Methods. Heat inactivation of viruses plus AZT treatment of target cells was carried out as described above. The values shown represent the percentages of the maximum wild-type SIVmac239 value (552 copies/100 ng DNA at 24 h postinfection). (D) The production of integrated proviruses was assessed as described in Materials and Methods. Heat inactivation of viruses plus AZT treatment of target cells and raltegravir treatment of target cells were carried out as described above. The values shown represent the percentages of the maximum wild-type SIVmac239 value (4,725 copies/100 ng DNA at 48 h postinfection).
Production of 2-LTR circles.
2-LTR circles are dead-end products of retroviral infection that result from the joining of viral cDNA ends by host ligases. During natural infection, circular viral cDNA forms are seen exclusively in the nucleus; the production of 2-LTR circles is therefore considered an indicator of successful nuclear import (43). Supporting the results obtained with aphidicolin, which suggested that the A87E and A87D mutants can transit across an intact nuclear membrane, abundant 2-LTR circles were observed for all three viruses (Fig. 5). The A87E and A87D mutants demonstrated earlier production and higher overall levels, respectively, of 2-LTR circles than wild-type SIVmac239. In cells infected with wild-type SIVmac239, the presence of 2-LTR circles with the canonical junction expected from end-to-end ligation was confirmed by the use of a specific probe (Fig. 5A). Despite the high overall levels of 2-LTR circles in the cells infected with the A87E and A87D mutants, 2-LTR circles with a canonical junction were not detected (Fig. 5B and C). This result indicated that a portion of the products being amplified by our 2-LTR qPCR primer set might be associated with processes other than end-to-end ligation of viral cDNA. We considered autointegrated viral DNA as a potential source of these amplified products.
Fig 5.
Production of 2-LTR circles in infected cells. Recombinant viruses were used to infect HeLa cells, and at the indicated times, the levels of 2-LTR circles were measured by real-time PCR. Both a standard probe (2-LTR), which detects all 2-LTR circles, and a probe (JNCT) specific for canonical 2-LTR junctions were used. Some infected cells were treated with 1 μM raltegravir (Ral). The results for infections by wild-type (WT) SIVmac239 (A) and the A87E (B) and A87D (C) mutants are shown. The values shown represent the percentages of the maximum wild-type SIVmac239 value (823 copies/100 ng DNA at 24 h postinfection after raltegravir treatment). The results shown represent the averages and standard deviations derived from three independent infections.
Autointegration.
Like 2-LTR and 1-LTR circles, autointegrated forms of viral cDNA are end products of a defective pathway for retroviral infection. Unlike 1- and 2-LTR circles, which are created by the action of host enzymes, autointegrated genomes result from the viral integrase-catalyzed attack of the viral DNA genome on itself (43). Host proteins within the preintegration complex of retroviruses serve to stimulate efficient intermolecular integration and suppress autointegration (44). The hallmarks of autointegration are the creation of 1- and 2-LTR circles with heterogeneity resulting from differences in the orientation of the intasome attack and the creation of junctions joining processed 5′ and 3′ LTRs to viral DNA (45). One can distinguish the 2-LTR circles that result from autointegration from “true” 2-LTR circles formed by end-to-end ligation by PCR amplification across the 2-LTR junction and gel analysis of the products. Although primer sets may vary, the analysis remains the same: true 2-LTR circles will produce uniform products, whereas autointegrated 2-LTR circles will produce smears due to the random sites of integration (43, 46). For this purpose, the primer set that we used in our 2-LTR qPCRs would be expected to generate both types of products. PCR products from each time point of the wild-type and mutant SIVmac239 infections were run on 2% agarose gels. The 210-bp product expected for a true 2-LTR circle junction was clearly present in the DNA from wild-type SIVmac239-infected cells at 12 to 48 h following virus-cell incubation (Fig. 6). In contrast, under identical amplification conditions, total DNA extracted from the A87E- and A87D-infected cells at 12 to 48 h postinfection yielded a heterogeneous population of products that migrated as a smear. As the heterogeneous products amplified during A87E and A87D infection hybridized with the TaqMan probe to yield a signal in the qPCR assay, these products likely represent 2-LTR circles with various junction lengths. If these represent autointegration products, their levels should decrease when the activity of the viral integrase is inhibited. Raltegravir is a strand transfer inhibitor of retroviral integrases that also modestly affects 3′-end processing (47). Raltegravir potently reduces proviral formation but also increases the amount of episomal 2-LTR circles; the latter effect probably results from an increased availability of linear unintegrated viral DNA, which is the substrate for the host ligases that produce true 2-LTR circles (39, 48). Addition of raltegravir at 10 times the inhibitory concentration (EC95) reduced the number of cells infected by the wild-type and mutant viruses (Fig. 4A). The number of integrated genomes was reduced proportionately (Fig. 4C). Raltegravir treatment boosted the number of true 2-LTR circles in wild-type SIVmac239-infected cells (Fig. 5A and 6). Interestingly, raltegravir treatment of A87E- and A87D-infected cells resulted in patterns of 2-LTR circle formation that resembled those of the untreated wild-type-virus-infected cells. Notably, although the levels of total 2-LTR circles in A87E- and A87D-infected cells were reduced by raltegravir treatment, the levels of 2-LTR circles with canonical junctions were increased by raltegravir treatment (Fig. 5B and C). Strikingly, raltegravir treatment of A87E- and A87D-infected cells resulted in the appearance of ∼210-bp bands typical of the junctional PCR product expected for true 2-LTR circles; these discrete bands replaced the smears associated with the heterogeneous 2-LTR circle junctions in the untreated cells infected by these mutants (Fig. 6B and C).
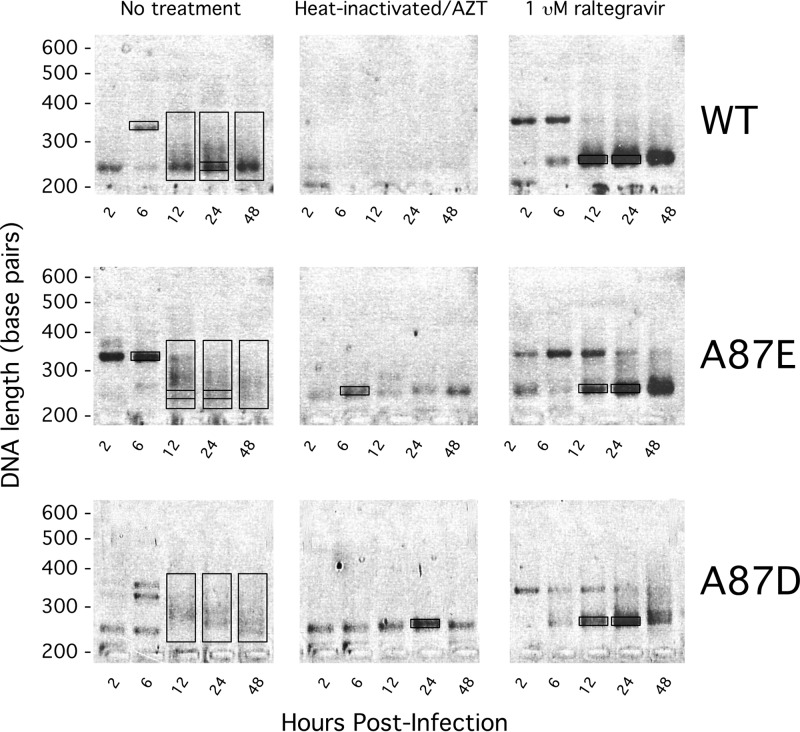
Fig 6.
Size analysis of 2-LTR products. HeLa cells were infected by recombinant wild-type (WT) and A87E and A87D mutant SIVmac239. At the indicated times after infection, 2-LTR circles were amplified by qPCR using the standard 2-LTR primer set. The products of the qPCRs were run on a 2% agarose gel and stained with ethidium bromide. For some of the experiments, the viruses were heated at 56°C for 1 h, and the cells were treated with 100 mM AZT. In another set of experiments, cells were treated with 1 μM raltegravir. The boxes indicate the material excised and sequenced and are denoted (from left to right and top to bottom) as follows: WT 6-h background, WT 12-h smear, WT 24-h smear and ∼210 bp (within smear), WT 48-h smear, WT plus raltegravir at 12 h and 24 h and ∼210 bp, A87E 6-h background, A87E 12-h smear and ∼210 bp, A87E 24-h smear and ∼210 bp, A87E 48-h smear, A87E 6-h heat-inactivated background, A87E plus raltegravir at 12 h and 24 h and ∼210 bp, A87D 12-h smear, A87D 24-h smear, A87D 48-h smear, A87D 24-h heat-inactivated background, and A87D plus raltegravir at 12 h and 24 h and ∼210 bp.
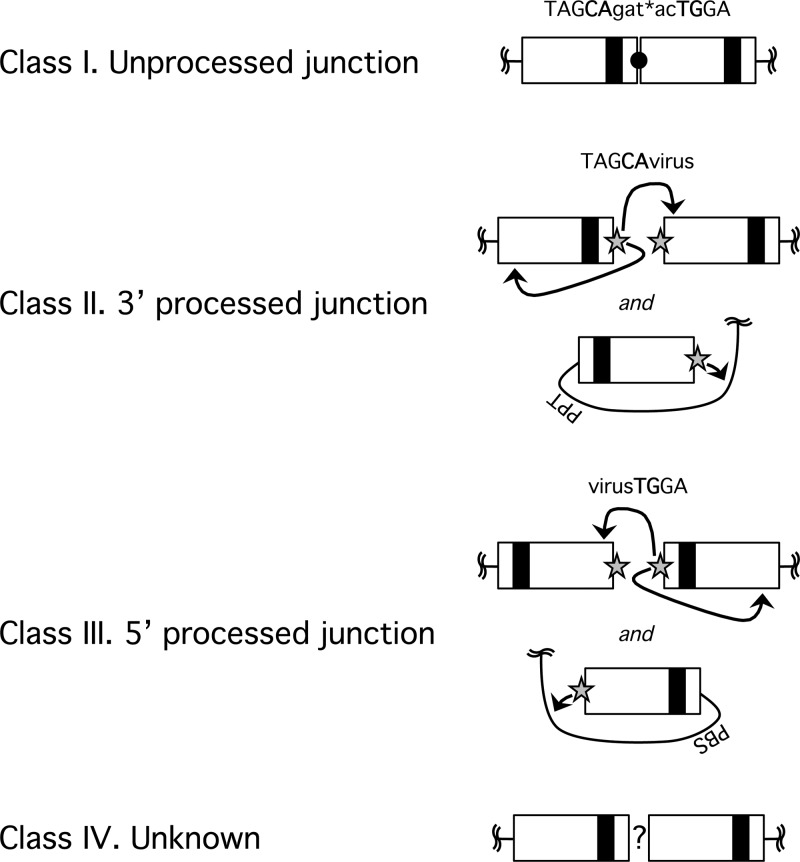
The 2-LTR junctions created by host processes result mostly from fusion of unprocessed or improperly processed viral cDNA (49). The junctions created by autointegration, on the other hand, should look identical to those created by bona fide proviral formation; i.e., a processed “CA” dinucleotide should be joined to a random viral sequence. To support the hypothesized origin of the different 2-LTR forms detected in the experiment shown in Fig. 6, we excised and sequenced representative amplicons from the agarose gel. Almost 500 independent sequences were aligned and analyzed, discarding none, allowing us to define four classes of 2-LTR junctions (Fig. 7). The class I junction represents the canonical 2-LTR circle junction that results from end-to-end joining of unprocessed LTR ends; thus, class I junctions are associated with “true” 2-LTR circles. Class II and class III junctions result from autointegration events in which the 3′- and 5′-end CA dinucleotides, respectively, of the LTR are involved in the nucleophilic attack on the viral target DNA. Class IV junctions were created by various poorly defined processes. The majority of these class IV genomes were fusions of deleted 3′ and 5′ ends. Sequences that apparently retain some undigested tRNALys primer for reverse transcription in the junction were grouped into class IV as well.
Fig 7.
Four classes of LTR junctions amplified by the standard 2-LTR primer set. Products excised from the gel shown in Fig. 6 were ligated into a TOPO-TA cloning vector and transformed into E. coli DH5α, and individual colonies were sent for sequencing. The sequenced LTR junctions could be grouped into four classes. Class I junctions are unprocessed and ligated LTRs, as expected for a true 2-LTR circle. Class II and class III junctions apparently result from autointegration. Class II junctions have a processed 3′ LTR that joins a random point within the viral genome. Class III junctions have a processed 5′ LTR joined to a viral sequence. Class IV junctions are created by an unknown process and most often consist of junctions in which both 5′ and 3′ LTR ends have undergone deletions.
The types of 2-LTR junctions associated with the PCR-amplified DNA bands in the gel shown in Fig. 6 are summarized in Table 1. The majority of species in the bands of ∼210 bp exhibited class I junctions for both wild-type and mutant virus infections, with or without raltegravir treatment. These results support the conclusion that the major form of 2-LTR circles in untreated and raltegravir-treated cells infected by wild-type SIVmac239 results from end-to-end ligation of viral cDNA.
Table 1.
Sequence-based classification of 2-LTR junctionsa
| Parameter | Value for virus at time after infection (h) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT SIVmac239 |
A87E |
A87D |
|||||||||||||
| 12 | 24 | 24 | 48 | 12 + 24 | 12 | 24 | 12 | 24 | 48 | 12 + 24 | 12 | 24 | 48 | 12 + 24 | |
| Raltegravir (1 μM) treatment | No | No | No | No | Yes | No | No | No | No | No | Yes | No | No | No | Yes |
| Fragment size (bp) | Smear (230–330) | ∼210 | Smear (230–330) | Smear (230–330) | ∼210 | ∼210 | ∼210 | Smear (230–330) | Smear (230–330) | Smear (230–330) | ∼210 | Smear (230–330) | Smear (230–330) | Smear (230–330) | ∼210 |
| No. of sequenced products of class/total no. of sequenced products | |||||||||||||||
| Class I, unprocessed | 12/40 | 7/10 | 17/43 | 9/34 | 6/7 | 8/17 | 3/4 | 1/97 | 0/45 | 3/34 | 7/8 | 5/53 | 0/43 | 4/30 | 21/23 |
| Class II, 3′ processed | 9/40 | 1/10 | 19/43 | 14/34 | 0/7 | 8/17 | 1/4 | 59/97 | 31/45 | 26/34 | 1/8 | 18/53 | 29/43 | 18/30 | 0/23 |
| Class III, 5′ processed | 7/40 | 0/10 | 5/43 | 3/34 | 0/7 | 1/17 | 0/4 | 19/97 | 11/45 | 2/34 | 0/8 | 22/53 | 13/43 | 7/30 | 0/23 |
| Class IV, unknown | 12/40 | 2/10 | 2/43 | 8/34 | 1/7 | 0/17 | 0/4 | 21/97 | 3/45 | 3/34 | 0/8 | 8/53 | 1/43 | 1/30 | 2/23 |
The indicated PCR-amplified products from the experiment shown in Fig. 6 were excised from the gel, cloned, and sequenced. The number of sequenced products in each class divided by the total number of sequenced products is shown.
The 2-LTR circles associated with the heterogeneous PCR-amplified bands in the smears contained mostly class II and class III junctions (Table 1), supporting an autointegration origin. Some of these 2-LTR circles could be detected at 12 to 24 h following infection of cells by wild-type SIVmac239, as expected, representing 47% of the total sequences collected at 24 h postinfection (46, 50). These autointegrated forms, however, represented the vast majority of 2-LTR circles observed in cells infected by the A87E and A87D mutant viruses (88% and 98%, respectively, at 24 h postinfection). Comparing wild-type and mutant viruses, the observed differences in the distribution of 2-LTR circles among the classes at 24 h postinfection were highly significant (P = 5 × 10−6 for wild type versus A87E and P = 3 × 10−6 for wild type versus A87D, determined by a Fisher exact-probability test). Raltegravir treatment of the cells infected by these mutants eliminated autointegrants and restored the production of true 2-LTR circles with class I junctions. These results support a model in which autointegration occurs more efficiently during the course of A87E and A87D virus infection than during wild-type SIVmac239 infection.
In the experiment shown in Fig. 6, AZT treatment of cells incubated with heat-inactivated viruses resulted in a reduced level of 2-LTR circles. However, in AZT-treated cells incubated with the heat-inactivated A87E and A87D viruses, some bands of uncertain origin were PCR amplified. We sequenced these bands and a few other bands of uncertain identity that were PCR amplified from 2-LTR circles in the virus-infected cells. The bands in the AZT-treated cells incubated with heat-inactivated A87E and A87D viruses migrated near the ∼210-bp band expected for class I junctions; however, these bands exhibited mostly inexplicably amplified human chromosomal DNA and some class IV junctions. The ∼350-bp bands seen at some early time points following infection by both wild-type and mutant viruses appeared to result from either RT copy choice errors or packaging of improperly spliced genomes.
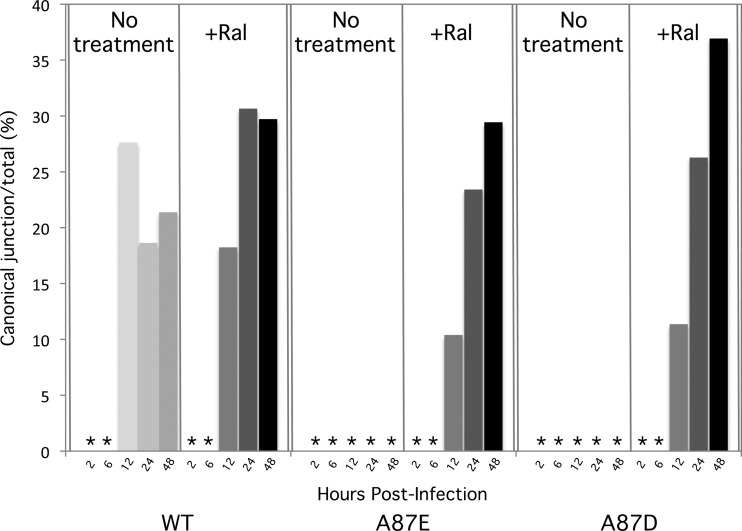
Reliable quantitation of the efficiency of retroviral autointegration is challenging because of the heterogeneity of potential products. However, we could readily measure the efficiency with which true 2-LTR circle junctions were formed during the course of wild-type and mutant SIVmac239 infection. The same primer set used to generate the data shown in Fig. 6 was used to amplify the 2-LTR junction fragments, and a TaqMan probe was designed to hybridize across the canonical 2-LTR junctions. At 12 to 48 h following infection of cells by wild-type SIVmac239, 17 to 27% of the total 2-LTR signal was captured by the canonical probe (Fig. 8). This percentage increased slightly at 24 and 48 h postinfection as a result of raltegravir treatment. No canonical 2-LTR circle junctions were detected in untreated cells infected by the A87E and A87D viruses. Only after raltegravir treatment were canonical 2-LTR circle junctions detected in cells infected by these mutant viruses. These results confirm that an integrase-dependent process during infection of cells by the A87E and A87D viruses results in a dramatic decrease in bona fide 2-LTR circles relative to wild-type SIVmac239 infection. The results are consistent with a greater proportion of the A87E and A87D viral cDNAs being channeled into autointegration events than wild-type SIVmac239 cDNA.
Fig 8.
Frequency of production of canonical 2-LTR junctions. For each time point following infection of HeLa cells by wild-type (WT) and mutant SIVmac239, the total number of 2-LTR circles was measured by real-time qPCR with a standard 2-LTR probe (see the Fig. 5 legend). In parallel, the number of 2-LTR circles with canonical 2-LTR circle junctions was measured with a specific junctional probe. The number of canonical 2-LTR junctions is shown as a percentage of the total 2-LTR circle junctions observed, with or without raltegravir treatment of the target cells. The asterisks indicate values less than 1%.
DISCUSSION
During the early phase of retroviral infection, the capsid can influence many processes that are essential for the successful formation of the provirus. Changes in retroviral capsids can influence uncoating, reverse transcription, nuclear entry, chromatin targeting and binding, and integration (3, 4, 6, 7, 13, 14, 16, 18, 37). Here we describe two SIVmac239 capsid mutants, A87E and A87D, that uncoat less efficiently than wild-type SIVmac239 yet are capable of reverse transcription and nuclear entry. These mutants are blocked prior to integration and demonstrate an increase in autointegration. Thus, the SIV capsid may play a direct or indirect role in mitigating detrimental autointegration.
Several of the early steps in retrovirus infection are interdependent, working to ensure that essential functions occur at optimal times and locations. The relationship between uncoating and reverse transcription has been investigated most extensively. Accelerated uncoating resulting from capsid changes, restriction by TRIM5α, or destabilizing drugs has been associated with failure to initiate and/or complete reverse transcription (3, 4, 7, 9, 11, 17, 19). Conversely, blocking reverse transcription was reported to inhibit functional uncoating (51). Both the A87E and A87D SIV CA mutants exhibited significantly more pelletable capsid in the cytosol of infected cells than wild-type SIV, at multiple time points after infection. The basis for the apparent decrease in the rate of capsid uncoating is unknown but could result from increased stability of the capsid or from altered interactions with as-yet-unknown host factors. Notably, a decreased rate of uncoating resulted specifically from the introduction of an acidic residue at position 87 of CA. Viruses with a positively charged lysine at this position, although similarly reduced in infectivity, exhibited wild-type capsid stability. Whatever the basis for the slower uncoating of the A87E and A87D mutants, reverse transcription proceeded even more efficiently for both mutants than for wild-type SIV. Therefore, decreases in SIV uncoating do not necessarily lead to decreases in reverse transcription. Moreover, the phenotype of the A87K mutant demonstrates that efficient reverse transcription does not automatically follow timely uncoating and that CA changes can disrupt reverse transcription without apparently affecting capsid stability.
Of interest, some alterations in retroviral capsids have been reported to result in blocks to infection after reverse transcription has been completed (3, 37). For example, some HIV-1 CA mutants fail to negotiate passage through the nuclear pore complex and do not access the nuclear chromatin (6). Both SIV and HIV-1 utilize karyophilic host proteins to shuttle their preintegration complexes into the nucleus of the target cell (3, 4, 7, 9, 11, 17, 19). The A87E and A87D SIV CA mutants can form true 2-LTR circles in the presence of an integrase inhibitor, raltegravir. As the canonical 2-LTR circle junction is created by host nuclear ligases, this observation suggests that both mutants can access the nucleus, at least in the presence of raltegravir. Moreover, the infectivity of the mutants was not affected by aphidicolin-induced cell cycle arrest, consistent with the known ability of lentiviruses to enter the nucleus by means of the nuclear pore (2).
The reduced infectivity of the A87E and A87D mutants relative to wild-type SIVmac239 was accompanied by a proportionate decrease in the number of proviruses. Although apparently able to access the nucleus, the mutants do not effectively integrate their viral DNA into the host DNA. The 2-LTR circles formed during infection by the A87E and A87D viruses exhibited junctional heterogeneity. Analysis of these junctions revealed properly 3′-processed LTR ends ligated to different viral sequences, indicative of autointegration. Autointegration is a consequence of the retroviral life-style; like 1- and 2-LTR circles, autointegrated genomes are readily detectable end products even during efficient wild-type infections. Nevertheless, the number of autointegrated genomes created during a wild-type SIVmac239 infection is mitigated, presumably through either defense of the double-stranded-DNA (dsDNA) genome, enhancement of the desired intermolecular integration, or a combination of both. Both HIV-1 and murine leukemia virus (MLV) preintegration complexes (PICs) isolated from infected cells efficiently perform intermolecular integrations and avoid intramolecular autointegration (43, 52). In vitro autointegration of intact HIV-1 PICs can be stimulated upon integrase activation in the absence of an intermolecular target (43). In contrast, MLV autointegration is favored only upon salt stripping of proteins from the PIC; protection is reconstituted by adding back host cytoplasmic extracts. The protein BAF, for barrier-to-autointegration factor, was identified as the host factor that stimulated intermolecular MLV integration (52). BAF acts as a dimer to divalently bind dsDNA and induce its compaction. Although BAF has been found to be a component of HIV-1 PICs, it appears to play a minimal role in HIV-1 infection (53–55).
The two aberrant characteristics of the A87E and A87D phenotype, capsid hyperstability and autointegration vulnerability, may be linked. In one model, reverse transcription produces the viral cDNA of the A87E and A87D mutant viruses, but the lack of dissolution or restructuring of the capsid core prevents the active intasome from encountering an intermolecular target. This model presumes that in a wild-type infection, autointegration is prevented by the efficient transit of the PIC into the nucleus, where it acquires new host factors and encounters a target-rich environment. In another model, the low rate of uncoating reduces the recruitment of host factors that suppress autointegration. In a third model, capsid uncoating leads directly to conformational changes in the PIC that suppress autointegration. These models are not mutually exclusive.
Although our results do not rule out other contributing mechanisms, the high level of autointegration observed for the A87E and A87D mutants provides a natural explanation of the replication defects observed for these mutants. At least for the easily assessed 2-LTR circles, autointegrated products predominate at the expense of true 2-LTR circles for these mutants; together with the observation that raltegravir treatment of cells infected by these mutant viruses allows efficient formation of true 2-LTR circles, the results suggest that in this case, autointegration is largely complete before entry of the PIC into the nucleus. This model is consistent with previously reported observations that suggested that integrases become active as soon as a substrate is available and that 3′-end processing of viral cDNA occurs in the cytoplasm of infected cells (50). A corollary of this model is that although a significant fraction of the A87E and A87D viral genomes are rendered nonfunctional by cytoplasmic autointegration events, viral cDNAs that somehow escape this fate can form PICs that are functional with respect to nuclear pore transit and subsequent integration. In the absence of a specific autointegration inhibitor, it is difficult to demonstrate formally that preventing autointegration can rescue A87E and A87D virus infectivity, as mutant provirus formation remains dependent upon integrase activity.
Future work seeking to understand the early events in the lentiviral life cycle, especially studies using 2-LTR circle formation as a surrogate for nuclear entry, should include an assessment of autointegrated genomes.
ACKNOWLEDGMENTS
We thank Yvette McLaughlin and Elizabeth Carpelan for manuscript preparation.
We thank the National Institutes of Health (award AI063987 and Center for AIDS Research award AI06354), the International AIDS Vaccine Initiative, and the late William F. McCarty-Cooper.
Footnotes
Published ahead of print 23 January 2013
REFERENCES
- 1. Coffin JM, Hughes SH, Varmus H. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY: [PubMed] [Google Scholar]
- 2. Bukrinsky MI, Sharova N, Dempsey MP, Stanwick TL, Bukrinskaya AG, Haggerty S, Stevenson M. 1992. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc. Natl. Acad. Sci. U. S. A. 89:6580–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Forshey BM, von Schwedler U, Sundquist WI, Aiken C. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 76:5667–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leschonsky B, Ludwig C, Bieler K, Wagner R. 2007. Capsid stability and replication of human immunodeficiency virus type 1 are influenced critically by charge and size of Gag residue 183. J. Gen. Virol. 88:207–216 [DOI] [PubMed] [Google Scholar]
- 5. Li Y, Kar AK, Sodroski J. 2009. Target cell type-dependent modulation of human immunodeficiency virus type 1 capsid disassembly by cyclophilin A. J. Virol. 83:10951–10962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yamashita M, Perez O, Hope TJ, Emerman M. 2007. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 3:1502–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang R, Shi J, Byeon IJ, Ahn J, Sheehan JH, Meiler J, Gronenborn AM, Aiken C. 2012. Second-site suppressors of HIV-1 capsid mutations: restoration of intracellular activities without correction of intrinsic capsid stability defects. Retrovirology 9:30 doi:10.1186/1742-4690-9-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ambrose Z, Lee K, Ndjomou J, Xu H, Oztop I, Matous J, Takemura T, Unutmaz D, Engelman A, Hughes SH, KewalRamani VN. 2012. Human immunodeficiency virus type 1 capsid mutation N74D alters cyclophilin A dependence and impairs macrophage infection. J. Virol. 86:4708–4714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blair WS, Pickford C, Irving SL, Brown DG, Anderson M, Bazin R, Cao J, Ciaramella G, Isaacson J, Jackson L, Hunt R, Kjerrstrom A, Nieman JA, Patick AK, Perros M, Scott AD, Whitby K, Wu H, Butler SL. 2010. HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS Pathog. 6:e1001220 doi:10.1371/journal.ppat.1001220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Iaco A, Luban J. 2011. Inhibition of HIV-1 infection by TNPO3 depletion is determined by capsid and detectable after viral cDNA enters the nucleus. Retrovirology 8:98 doi:10.1186/1742-4690-8-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kortagere S, Madani N, Mankowski MK, Schon A, Zentner I, Swaminathan G, Princiotto A, Anthony K, Oza A, Sierra LJ, Passic SR, Wang X, Jones DM, Stavale E, Krebs FC, Martin-Garcia J, Freire E, Ptak RG, Sodroski J, Cocklin S, Smith AB., III 2012. Inhibiting early-stage events in HIV-1 replication by small-molecule targeting of the HIV-1 capsid. J. Virol. 86:8472–8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krishnan L, Matreyek KA, Oztop I, Lee K, Tipper CH, Li X, Dar MJ, Kewalramani VN, Engelman A. 2010. The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. J. Virol. 84:397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee K, Ambrose Z, Martin TD, Oztop I, Mulky A, Julias JG, Vandegraaff N, Baumann JG, Wang R, Yuen W, Takemura T, Shelton K, Taniuchi I, Li Y, Sodroski J, Littman DR, Coffin JM, Hughes SH, Unutmaz D, Engelman A, KewalRamani VN. 2010. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7:221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matreyek KA, Engelman A. 2011. The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid. J. Virol. 85:7818–7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Owens CM, Song B, Perron MJ, Yang PC, Stremlau M, Sodroski J. 2004. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J. Virol. 78:5423–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, Hue S, Fletcher AJ, Lee K, KewalRamani VN, Noursadeghi M, Jenner RG, James LC, Bushman FD, Towers GJ. 2011. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 7:e1002439 doi:10.1371/journal.ppat.1002439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi J, Zhou J, Shah VB, Aiken C, Whitby K. 2011. Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J. Virol. 85:542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vozzolo L, Loh B, Gane PJ, Tribak M, Zhou L, Anderson I, Nyakatura E, Jenner RG, Selwood D, Fassati A. 2010. Gyrase B inhibitor impairs HIV-1 replication by targeting Hsp90 and the capsid protein. J. Biol. Chem. 285:39314–39328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. U. S. A. 103:5514–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hatziioannou T, Perez-Caballero D, Cowan S, Bieniasz PD. 2005. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J. Virol. 79:176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qi M, Yang R, Aiken C. 2008. Cyclophilin A-dependent restriction of human immunodeficiency virus type 1 capsid mutants for infection of nondividing cells. J. Virol. 82:12001–12008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sokolskaja E, Sayah DM, Luban J. 2004. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J. Virol. 78:12800–12808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Towers GJ, Hatziioannou T, Cowan S, Goff SP, Luban J, Bieniasz PD. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138–1143 [DOI] [PubMed] [Google Scholar]
- 24. Ylinen LM, Schaller T, Price A, Fletcher AJ, Noursadeghi M, James LC, Towers GJ. 2009. Cyclophilin A levels dictate infection efficiency of human immunodeficiency virus type 1 capsid escape mutants A92E and G94D. J. Virol. 83:2044–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436–441 [DOI] [PubMed] [Google Scholar]
- 26. Gao F, Yue L, White AT, Pappas PG, Barchue J, Hanson AP, Greene BM, Sharp PM, Shaw GM, Hahn BH. 1992. Human infection by genetically diverse SIVSM-related HIV-2 in West Africa. Nature 358:495–499 [DOI] [PubMed] [Google Scholar]
- 27. Marx PA, Li Y, Lerche NW, Sutjipto S, Gettie A, Yee JA, Brotman BH, Prince AM, Hanson A, Webster RG, Desrosiers RC. 1991. Isolation of a simian immunodeficiency virus related to human immunodeficiency virus type 2 from a West African pet sooty mangabey. J. Virol. 65:4480–4485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peeters M, Honore C, Huet T, Bedjabaga L, Ossari S, Bussi P, Cooper RW, Delaporte E. 1989. Isolation and partial characterization of an HIV-related virus occurring naturally in chimpanzees in Gabon. AIDS 3:625–630 [DOI] [PubMed] [Google Scholar]
- 29. Braaten D, Luban J. 2001. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 20:1300–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh CT, Sodroski J, Gottlinger HG. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363–365 [DOI] [PubMed] [Google Scholar]
- 31. Vajdos FF, Yoo S, Houseweart M, Sundquist WI, Hill CP. 1997. Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein. Protein Sci. 6:2297–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aberham C, Weber S, Phares W. 1996. Spontaneous mutations in the human immunodeficiency virus type 1 gag gene that affect viral replication in the presence of cyclosporins. J. Virol. 70:3536–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Owens CM, Yang PC, Gottlinger H, Sodroski J. 2003. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J. Virol. 77:726–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alexander L, Veazey RS, Czajak S, DeMaria M, Rosenzweig M, Lackner AA, Desrosiers RC, Sasseville VG. 1999. Recombinant simian immunodeficiency virus expressing green fluorescent protein identifies infected cells in rhesus monkeys. AIDS Res. Hum. Retroviruses 15:11–21 [DOI] [PubMed] [Google Scholar]
- 35. Rho HM, Poiesz B, Ruscetti FW, Gallo RC. 1981. Characterization of the reverse transcriptase from a new retrovirus (HTLV) produced by a human cutaneous T-cell lymphoma cell line. Virology 112:355–360 [DOI] [PubMed] [Google Scholar]
- 36. Kutner RH, Zhang XY, Reiser J. 2009. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat. Protoc. 4:495–505 [DOI] [PubMed] [Google Scholar]
- 37. Dismuke DJ, Aiken C. 2006. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J. Virol. 80:3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Groschel B, Bushman F. 2005. Cell cycle arrest in G2/M promotes early steps of infection by human immunodeficiency virus. J. Virol. 79:5695–5704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Butler SL, Hansen MS, Bushman FD. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631–634 [DOI] [PubMed] [Google Scholar]
- 40. Munk C, Brandt SM, Lucero G, Landau NR. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. U. S. A. 99:13843–13848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Koh Y, Haim H, Engelman A. 2011. Identification and characterization of persistent intracellular human immunodeficiency virus type 1 integrase strand transfer inhibitor activity. Antimicrob. Agents Chemother. 55:42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Koh Y, Matreyek KA, Engelman A. 2011. Differential sensitivities of retroviruses to integrase strand transfer inhibitors. J. Virol. 85:3677–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Farnet CM, Haseltine WA. 1991. Circularization of human immunodeficiency virus type 1 DNA in vitro. J. Virol. 65:6942–6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Van Maele B, Busschots K, Vandekerckhove L, Christ F, Debyser Z. 2006. Cellular co-factors of HIV-1 integration. Trends Biochem. Sci. 31:98–105 [DOI] [PubMed] [Google Scholar]
- 45. Oh J, Chang KW, Wierzchoslawski R, Alvord WG, Hughes SH. 2008. Rous sarcoma virus (RSV) integration in vivo: a CA dinucleotide is not required in U3, and RSV linear DNA does not autointegrate. J. Virol. 82:503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yan N, Cherepanov P, Daigle JE, Engelman A, Lieberman J. 2009. The SET complex acts as a barrier to autointegration of HIV-1. PLoS Pathog. 5:e1000327 doi:10.1371/journal.ppat.1000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marinello J, Marchand C, Mott BT, Bain A, Thomas CJ, Pommier Y. 2008. Comparison of raltegravir and elvitegravir on HIV-1 integrase catalytic reactions and on a series of drug-resistant integrase mutants. Biochemistry 47:9345–9354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Svarovskaia ES, Barr R, Zhang X, Pais GC, Marchand C, Pommier Y, Burke TR, Jr, Pathak VK. 2004. Azido-containing diketo acid derivatives inhibit human immunodeficiency virus type 1 integrase in vivo and influence the frequency of deletions at two-long-terminal-repeat-circle junctions. J. Virol. 78:3210–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Randolph CA, Champoux JJ. 1993. The majority of simian immunodeficiency virus/mne circle junctions result from ligation of unintegrated viral DNA ends that are aberrant for integration. Virology 194:851–854 [DOI] [PubMed] [Google Scholar]
- 50. Miller MD, Farnet CM, Bushman FD. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hulme AE, Perez O, Hope TJ. 2011. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc. Natl. Acad. Sci. U. S. A. 108:9975–9980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee MS, Craigie R. 1998. A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl. Acad. Sci. U. S. A. 95:1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen H, Engelman A. 1998. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc. Natl. Acad. Sci. U. S. A. 95:15270–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang Y, Cai M, Clore GM, Craigie R. 2011. No interaction of barrier-to-autointegration factor (BAF) with HIV-1 MA, cone-rod homeobox (Crx) or MAN1-C in absence of DNA. PLoS One 6:e25123 doi:10.1371/journal.pone.0025123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shun MC, Daigle JE, Vandegraaff N, Engelman A. 2007. Wild-type levels of human immunodeficiency virus type 1 infectivity in the absence of cellular emerin protein. J. Virol. 81:166–172 [DOI] [PMC free article] [PubMed] [Google Scholar]