Abstract
Nairoviruses are responsible for numerous diseases that affect both humans and animal. Recent work has implicated the viral ovarian tumor domain (vOTU) as a possible nairovirus virulence factor due to its ability to edit ubiquitin (Ub) bound to cellular proteins and, at least in the case of Crimean-Congo hemorrhagic fever virus (CCHFV), to cleave the Ub-like protein interferon-stimulated gene 15 (ISG15), a protein involved in the regulation of host immunity. The prospective roles of vOTUs in immune evasion have generated several questions concerning whether vOTUs act through a preserved specificity for Ub- and ISG15-conjugated proteins and where that specificity may originate. To gain insight into the substrate specificity of vOTUs, enzymological studies were conducted on vOTUs from Dugbe, CCHFV, and Erve nairoviruses. These studies revealed that vOTUs originating from different nairoviruses display a significant divergence in their preference toward Ub and ISG15. In addition, a recently identified vOTU from turnip yellow mosaic tymovirus was evaluated to elucidate any possible similarities between vOTUs originating from different viral families. Although possessing a similar preference for certain polymeric Ub moieties, its activity toward Ub in general was significantly less then those of nairoviruses. Lastly, the X-ray crystallographic structure of the vOTU from the Dugbe nairovirus was obtained in complex with Ub to reveal structural commonalities of vOTUs originating from nairoviruses. The structure suggests that divergences between nairovirus vOTUs specificity originate at the primary structural level. Comparison of this structure to that originating from CCHFV identified key residues that infer the substrate specificity of vOTUs.
INTRODUCTION
Nairoviruses are negative-sense, single-stranded RNA [ssRNA(−)] viruses responsible for numerous diseases in both humans and animals. There are 34 known viruses belonging to the genus Nairovirus of family Bunyaviridae. Of these, the tick-born Crimean-Congo hemorrhagic fever virus (CCHFV), Dugbe virus (DUGV), and Nairobi sheep disease virus (NSDV) have been the most investigated. Although these viruses share considerable genomic similarity, their effects on both humans and animals vary from mild illness to loss of life. CCHFV, the most lethal to humans, is endemic across large swaths of sub-Saharan Africa, southeast Europe, and Asia. Fatality rates for CCHFV range from 5 to 33% depending on reported and confirmed case statistics. However, some outbreaks, to include the recent ones in India and the Sudan, suggest the rates could be up to 70 to 80% (1–5). CCHFV has also been reported to infect sheep, goats, cattle, horses, and donkeys in the wild; however, the disease does not manifest in a fatal form for these animals (6). Comparatively, infection of humans by NSDV and DUGV can cause a febrile illness but no associated fatalities (7, 8). NSDV is found in Eastern and Central Africa, and the Asian variant Ganjam nairovirus (GANV) found in India has been observed to infect sheep with a mortality of up to 90%, generating economic distress within infected areas (8–10). Alternatively, DUGV, which originates from sub-Saharan Africa, has not been found to be fatal for any known species. However, DUGV can cause mild febrile illness in species beyond humans, particularly cattle that it predominantly infects (11, 12). In addition to these human disease-causing nairoviruses, the Erve Virus (ERVEV) from northwest Europe has been implicated as a causative agent in human thunderclap headaches. ERVEV was isolated from the white-toothed shrew in 1982 and classified by indirect immunofluorescence assay (IFA) as belonging to the Nairovirus genus, ERVEV has only been recently sequenced limiting previous exploration of its proteome (13).
The genome of nairoviruses is partitioned into three RNA segments: small (S), medium (M), and large (L). Surprisingly, unlike other Bunyaviridae family members, the nairoviruses' L segment contains not only a RNA-dependent RNA polymerase but also a viral ovarian tumor domain protease homologue (vOTU) (14). As ssRNA(−) viruses, no viral protease is required for nairovirus genome replication (15). Instead, nairovirus vOTUs have been proposed to be one potential virulence factor (14–18). Unlike the previously identified Bunyaviridae family virulence factor, nonstructural protein NSs, which blocks transcription of alpha/beta interferon (IFN-α/β) and is absent in nairoviruses, vOTUs are suggested to impair innate immunity through deubiquitinating and deISGylating activity (19, 20).
As a deubiquitinating and deISGylating protease, nairovirus vOTUs fall into one of five protease superfamilies that facilitate a myriad of cellular processes such as proteasomal degradation, cell division, and regulation of the innate immune response through reversal of posttranslational modification by ubiquitin (Ub) (21). The multitude of outcomes is achieved by Ub's ability to form polymeric Ub (poly-Ub) chains through Ub's C-terminal glycine forming either a peptide bond via the N terminus (linear) or an isopeptide bond with one of the seven lysines (K6, -11, -27, -29, -33, -48, and -63) of a partnering Ub. Noncanonical forms of poly-Ub—linear and K6, -11, -27, -29, and -33—comprise over half of all poly-Ub-conjugated yeast proteins and have been implicated in immune system regulation and other key cellular functions (22). Specifically, K29-linked poly-Ub is known to facilitate trafficking proteins to the lysosome, whereas K11-linked poly-Ub has been implicated in regulating cell division (23). Although their functions have been less studied, K6, K27, and K33 appear to possibly act as modulators of immunologically relevant signaling pathways (24–27). Unlike the noncanonical linked poly-Ub, the effects of K48-linked and K63-linked poly-Ub modifications are better defined, with modification of host proteins by K48-linked poly-Ub leading to proteasomal degradation, whereas K63-linked poly-Ub modifications are implicated in the induction of the type I IFN (IFNI) response (28, 29). Upon entry into host cells, viruses need to outpace innate immunity before the adaptive immune system clears them from the organism. The type I IFN-α/β are critical orchestrators of innate immunity (30). Previously, a phosphorylation cascade was proposed to singularly instigate the production of type I IFNs and proinflammatory cytokines (NF-κB) and the type I IFN upregulation of several IFN-stimulated gene with antipathogenic properties through JAK1-STAT1/2 signaling (Janus-activated kinase 1–signal transducer and activator of transcription 1/2) (31). However, regulation of the IFNI response and activation of NF-κB has been shown to go beyond phosphorylation events and also relies on posttranslational modification by Ub and Ub-like IFN-simulated gene product 15 (ISG15) (32). In particular, JAK1, STAT1/2, TRAF, and RIG-I (retinoic acid-inducible gene 1), among other IFNI signaling and effector proteins, have been observed to undergo ubiquitination or ISGylation (33).
Recent studies have observed divergences in immunological response between human cells infected by DUGV and CCHFV underlining the differences in which the respective viruses influence cellular immunity (34, 35). Also, this phenomenon has been observed between GANV and NSDV. Specifically, Holzer et al. (36) recently narrowed IFN-α/β suppression to primarily the first 169 amino acids of the L-protein and suggested that the vOTU GANV may have less activity than that of CCHFV. However, the use of the recombinant overexpression system limited the ability to detect the extent of variability in deubiquitinating and deISGylating activity between the vOTUs originating from these viruses. This limitation has been observed in other studies, where unnaturally high expression of vOTUs, or when generously added in vitro, leading to a nearly 1:1 ratio of vOTU to Ub or ISG15 substrates, likely masks their specificity for poly-Ub and ISG15 conjugates, particularly during early viral replication (14, 37, 38). In addition, the increasing number of vOTUs being identified in ssRNA(−) and ssRNA(+) viral genomes, including the rice stripe tenuivirus (RSV), turnip yellow mosaic tymovirus (TYMV), and arteriviruses such as porcine reproductive and respiratory syndrome (PRRSV), have spurred speculation of whether their vOTUs' specificity and subsequent role is conserved (14, 38, 39).
To gain insight into the variability of deubiquitinating and deISGylating activity, the specificity among vOTUs from nairoviruses and how they may compare to non-nairovirus vOTUs, the enzymatic parameters and specificity of vOTUs from CCHFV, DUGV, ERVEV, and TYMV in regard to Ub, poly-Ub, and ISG15 moieties were determined. In addition, the ability for vOTUs to accommodate ISG15s from different species was examined. Finally, the X-ray crystal structure of the vOTU from DUGV bound with Ub was obtained to assess the potential origins of any detected variability among nairovirus vOTUs implicating primary structure as the main source of substrate specificity fluctuations. By comparing the structure of vOTUs bound to Ub from DUGV and CCHFV, key amino acid positions within nairovirus vOTUs that influenced activity were identified.
MATERIALS AND METHODS
Construction of vOTUs and Ub-Br3 expression vectors.
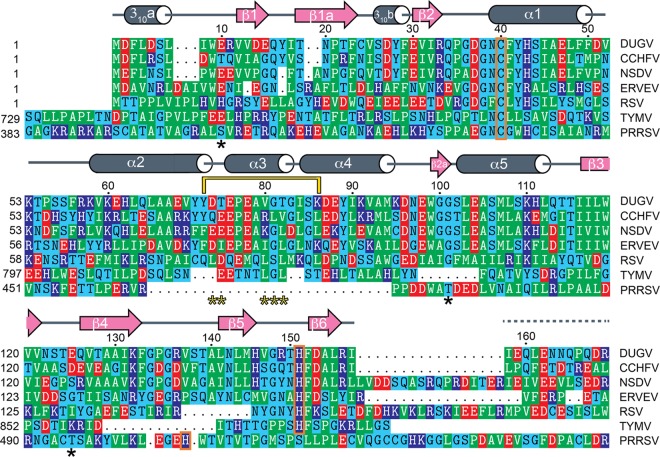
The production of the vOTU expression construct harboring the 169 amino acids from the L protein in CCHFV (CCHF vOTU; GenBank accession no. AAQ98866.2) is as previously described (17). The vOTUs from TYMV, ERVEV, and DUGV (TYM vOTU, ERVE vOTU, and DUG vOTU) were generated using the Escherichia coli BL21 codon optimized synthesis by GenScript, Inc. The DUG vOTU gene expression construct contained the first 169 amino acids from the L protein of DUGV (GenBank accession no. AAB18834.1). For the ERVEV vOTU expression construct, the first 171 amino acids from the L protein in ERVEV (GenBank AFH89032.1), which correspond to the first 169 amino acids of CCHF vOTU and DUG vOTU (Fig. 1) and includes the OTU domain as described by Dilcher et al. were utilized (13). Similarly to the CCHF vOTU construct, six histidines and a stop codon were added to ERVE vOTU and DUG vOTU constructs to generate a C-terminal histidine tag. For the TYM vOTU expression vector, the 151-amino-acid vOTU domain (residues 728 to 879) of TYMV (GenBank accession no. NP_663297.1) that included the vOTU domain as described in Chenon et al. with a six-histidine N terminus tag was synthesized (38). DUG vOTU, TYM vOTU, and ERVE vOTU were incorporated into pET11a plasmids using NdeI and BamHI restriction sites. Addgene plasmid 12448 harboring mouse ISG15 (mISG15) in pET15b as described by Kim et al. was obtained from Addgene (40). The vOTU constructs were introduced into E. coli BL21(DE3) competent cells by heat-shock transformation. The mISG15 construct was heat shock transformed into BL21(DE3)-CodonPlus competent cells. The resulting plasmids were then purified, restriction analyzed, and sequenced to verify the construct. Constructs of expression plasmids for truncated human Ub (hUb) to be C-terminally modified with 3-bromopropylamine were designed according to previously established studies (41, 42).
Fig 1.
Sequence alignment of several viral OTU proteases. The vOTUs are from Dugbe virus (DUGV; GenBank accession no. AAB18834.1), Crimean-Congo hemorrhagic fever virus (CCHFV; GenBank accession no. AAQ98866.2), Nairobi sheep disease virus (NSDV; GenBank accession no. ACH99799.1), Erve virus (ERVEV; GenBank accession no. AFH89032), rice stripe virus (RSV; GenBank accession no. ABC68333), turnip yellow mosaic virus (TYMV; GenBank accession no. NP_663297.1), and porcine reproductive and respiratory virus (PRRSV; GenBank accession no. Q9WJB2). The secondary structure of DUG vOTU according to Defined Secondary Structure of Proteins (DSSP) is shown as gray cylinders (helical regions), pink arrows (β-sheets), and gray lines (loops). Hashed gray line represents residues for which electron density was not defined in the crystal structure. Breaks denote regions were DUG vOTU does not have residues. Asterisks represent amino acid residues chosen for site-directed mutation. Orange brackets indicate residues that are part of vOTUs' catalytic triad. A yellow bracket outlines residues involved in a backbone shift between CCHFV and DUGV.
Expression and purification of vOTUs and Ub-Br3.
For vOTU enzymatic studies, E. coli strains harboring vOTUs from CCHFV, DUGV, ERVEV, and TYMV, as well as mISG15, were grown at 37°C in 6 liters of Luria-Bertani broth containing 100 μg of ampicillin/ml until the optical density at 600 nm reached 0.6. Expression of the vOTU or mISG15 gene was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 0.8 mM. The culture was further grown for 4 h at 37°C and then centrifuged at 6,000 × g for 10 min. The cells were collected and stored at −80°C until use. All vOTUs were purified according to the previously published protocol (17). Purification of mISG15 was performed according to an protocol identical to that used for the vOTUs with the exception that mISG15's buffer A contained 500 mM NaCl, 50 mM Tris (pH 7.5), and 1 mM Tris (2-carbozyethyl) phosphine hydrochloride(TCEP-HCl), and buffer B contained 200 mM NaCl and 50 mM HEPES (pH 8.0). All final protein concentrations were determined from the absorbance at 280 nm using an experimentally determined extinction coefficient (43).
Truncated hUb(1-75) was expressed according to previously established protocols with the exception that, after the IPTG induction, the culture was grown overnight at 18°C (41, 42). E. coli BL21(DE3)-CodonPlus pellets containing hUb(1-75) were lysed with buffer C (25 mM HEPES [pH 6.8], 50 mM sodium acetate, 75 mM NaCl) augmented with 0.16% Triton X-100. The solution was then sonicated on ice at 30% power with pulses of 5-s durations for 10 min. Insoluble debris was removed by centrifugation at 17,000 × g for 45 min. The clarified extract was filtered with a 0.80-μm-pore-size filter and then poured over a chitin column preequilibrated with buffer C. The column was washed with three column volumes of buffer C, followed by resuspension in two column volumes of buffer C supplemented with 100 mM sodium 2-mercaptoethanesulfonate (MESNA). The resuspension was rocked gently overnight at 4°C and eluted by filtration through a XK 26/40 GE column. The resulting hUb thioester was then derivatized with 3-bromopropylamine hydrobromide (Ub-Br3) according to previously described methods (41, 42). The purity of the Ub-Br3 was assessed by use of 10 to 20% Mini-Protean Tris-Tricine precast gels (Bio-Rad, CA).
Site-directed mutagenesis of the CCHF vOTU and DUG vOTU genes.
CCHF vOTU T10E, S101G, E128G, and E128T, mISG15 C76S, as well as DUG vOTU E10T, G101S, T128E, and α3-chimera (D74Q/T75E/V80R/G81L/T82V) protein mutants, were created by using QuikChange site-directed mutagenesis according to the manufacturer's protocol (Agilent Technologies, Inc.). The resulting mutant plasmids were introduced into E. coli XL1-Blue Supercompetent cells by heat shock transformation and were then propagated, purified for sequence verification, and transformed into E. coli BL21(DE3) cells for enzyme expression. The complete coding regions for all mutant enzymes were sequenced by GenScript, Inc., or Eton Bioscience, Inc.
Fluorescent vOTU deubiquitination and deISGylation assays.
All assays were performed in duplicate in buffer D (100 mM NaCl, 50 mM HEPES [pH 7.5], 0.01 mg of bovine serum albumin [BSA]/ml, 5 mM dithiothreitol [DTT]) using a Corning Costar half-volume black 96-well plate with a reaction volume of 50 μl. The rates of the reactions were observed using an Infinite M1000 series plate reader (Tecan, Inc.). Specifically, the increase in fluorescence (excitation λ, 360 nm; emission, 460 nm) of 7-amino-4-methylcourmarin (AMC) upon cleavage from hUb-AMC, human ISG15-AMC (hISG15-AMC) (Boston Biochem, MA), and ZRLRGG-AMC (Bachem) substrates was monitored for each of the vOTUs. The extinction coefficients for all three fluorescent substrates were determined by adding excess vOTU to various concentrations of each substrate, and the reactions were allowed to run until completion. The resulting maximum fluorescence values were plotted to determine the slope and subsequently each substrate's extinction coefficient.
To calculate the turnover rates for 1 μM hISG15-AMC, 1 μM hUb-AMC, and 50 μM ZRLRGG-AMC of DUG vOTU, ERVE vOTU, and TYM vOTU, as well as mutants originating from DUG vOTU and CCHF vOTU, various enzyme concentrations were used. This was to account for the varied activities of different vOTUs toward their hUb and hISG15 substrates. For DUG vOTU and associated mutants, 2 μM, 4 nM, and 4 μM concentrations of enzyme were used against hISG15-AMC, hUb-AMC, and ZRLRGG-AMC, respectively. The enzyme concentrations for CCHF vOTU and associated mutants were the same concentrations as those of DUG vOTU with the exception of 20 nM enzyme being utilized to observe the cleavage of hISG15-AMC. Similarly, 4 nM ERVE vOTU was used to cleave hUb-AMC and hISG15-AMC, with 1 μM enzyme used to cleave 50 μM ZRLRGG-AMC. For TYM vOTU, 2.2, 1.2, and 4 μM concentrations of enzyme were used to cleave hISG15-AMC, hUb-AMC, and ZRLRGG-AMC, respectively.
To determine the DUG vOTU's Vmax and Km values for hUb-AMC, DUG vOTU's concentration was maintained at 4 nM enzyme while the hUb-AMC concentration was varied from 0 to 75 μM. To determine DUG vOTU's Vmax and Km values for hISG15-AMC, DUG vOTU's concentration was maintained at 2 μM enzyme, while the hISG15-AMC concentration was varied from 0 to 32 μM. For calculation of the Vmax and Km values for TYM vOTU, an enzyme concentration of 1.2 μM was used while the hUb-AMC concentration was varied from 0 to 65 μM, and an enzyme concentration of 2.2 μM was used while the hISG15-AMC concentration was varied from 0 to 32 μM. ERVE vOTU's Vmax and Km values for hUb-AMC and hISG15-AMC were determined maintaining an enzyme concentration of 125 nM while the hUb-AMC concentration was varied from 0 to 51 μM, and an enzyme concentration of 4 nM was maintained while the hISG15-AMC concentration was varied from 0 to 5 μM. The initial rates were fitted to the Michaelis-Menten equation, υ = Vmax/[1 + (Km/[S])], using the Enzyme Kinetics module of SigmaPlot (v12.2; SPSS, Inc.). Vmax was translated into kcat using kcat = Vmax/[E].
The vOTUs from CCHFV, DUGV, ERVEV, and TYMV turnover rates for poly-Ub Förster resonance energy transfer (FRET) linkage substrates K11, K48, and K63 (Boston Biochem, MA) at 1 μM were determined by monitoring the increase in fluorescence (excitation λ, 544 nm; emission, 572 nm) resulting by the separation of a FRET TAMRA/QXL pair. The cleavage of three commercially available FRET TAMRA/QXL pair configurations per K48 and K63 poly-Ub linkage FRET substrates was assessed. Each di-Ub FRET substrate at 1 μM was evaluated against an enzyme concentration of 2 nM (CCHF and DUG vOTU), 125 nM (ERVE vOTU), or 500 nM (TYM vOTU). For the K11 di-Ub FRET substrate, enzyme concentrations of vOTUs from CCHFV (100 nM), DUGV (100 nM), ERVEV (2.6 μM), TYMV (20 nM) were used.
mISG15 competition assay.
The ability of 4 nM vOTU from CCHFV and ERVEV to cleave 2 μM hISG15-AMC in the presence of unlabeled mISG15 (120 μM) or hISG15 (120 μM) was observed by measuring the increased fluorescence of free AMC (excitation λ, 360 nm; emission, 460 nm) on an Infinite M1000 series plate reader (Tecan). Unlabeled mISG15 was obtained as described above, whereas hISG15 was acquired from a commercial source (Boston Biochem). All assays were performed in buffer E (200 mM NaCl, 50 mM HEPES [pH 8.0], 0.01 mg of BSA/ml, 5 mM DTT) using a Corning Costar half-volume black 96-well plate with a reaction volume of 50 μl.
Deubiquitinating gel shift assays.
Poly-Ub linked by the different isopeptide bonds (K6, K11, K27, K29, K33, K48, and K63), as well as the N-terminal peptide bond (linear), were purchased from Boston Biochem, MA. Dimeric-Ub substrates (10 μM) were incubated with each vOTU (4 nM CCHF vOTU, 4 nM DUG vOTU, 100 nM ERVE vOTU, and 6 μM TYM vOTU) in reaction buffer F (100 mM NaCl, 50 mM HEPES [pH 7.5], 2 mM DTT) at 37°C. The reactions were stopped at various times of > 1 h by mixing 9 μl of each reaction with 2× sodium dodecyl sulfate-Tricine sample buffer, followed by boiling at 95°C for 5 min. The results were visualized on 10 to 20% Mini-Protean Tris-Tricine precast gels (Bio-Rad). For vOTU cleavage of trimeric K48 and K63 linkages, 20 μM tri-Ub substrates were tested and analyzed in the same manner as the di-Ub.
DUG vOTU-Ub formation and crystallization.
DUG vOTU was combined with Ub-Br3 in equal molar ratios, incubated at 37°C for 2 h and left overnight at 4°C. Complex (DUG vOTU-Ub) formation was monitored using 10 to 20% Mini-Protean Tris-Tricine precast gels. DUG vOTU-Ub was dialyzed overnight against 1 liter of buffer G (100 mM NaCl, 50 mM Tris-HCl [pH 8.0]) and then purified using a GE Mono-Q column with a linear gradient of buffer G to buffer H (1 M NaCl, 50 mM Tris-HCl [pH 8.0]). Fractions were pooled according to the chromatogram, loaded onto an AP-1 (Waters) column packed with Superdex-75 resin preequilibrated with buffer I (150 mM NaCl, 5 mM HEPES [pH 7.4], 1 mM TCEP-HCl), and eluted at a flow rate of 1.0 ml/min. Resulting DUG vOTU-Ub was concentrated in a GE Vivaspin 6 10-kDa MWCO concentrator and filtered using a 0.22-μm-pore-size Costar spin filter.
The initial crystal conditions for DUG vOTU-Ub were determined from high-throughput screening of Qiagen Classics I and II screens in a 96-well sitting drop format using an Art Robbins Phoenix robot. Drops contained 0.4 μl of protein solution and 0.4 μl of precipitate with a 100-μl reservoir volume. Initial screening resulted in several hits; however, a solution containing 0.25 M LiSO4, 0.10 M Bis-Tris (pH 5.5), and 29% PEG 3350 produced the most viable crystals. These crystals were optimized using the Additive HT Screen from Hampton Research. Final DUG vOTU-Ub crystals were obtained through vapor diffusion using a 500-μl reservoir with 4-μl hanging drops mixed 1:1 with protein solution and 0.25 μl of 40% (vol/vol) 1,3-butanediol.
Data collection and X-ray structural determination of DUG vOTU-Ub.
An X-ray data set was collected using a crystal mounted onto a nylon loop flash frozen in liquid nitrogen. The frozen crystal was mounted under a stream of dry N2 at 100°K. A DUG vOTU-Ub data set with resolution to 2.85 Å was collected at the advanced light source (ALS) beam line 4.2.2 at 1.00 Å with a NOIR-1 MBC detector (NOIR-1). X-ray images were indexed, processed, integrated, and scaled using HKL2000 (44), and phases were determined and refined using Phaser (45). An initial phase solution was elucidated using a homology model based on the CCHF vOTU-Ub structure 3PRP for molecular replacement using Phaser (45). The structure was refined using iterative cycles of model building and refinement using COOT and REFMAC, respectively (45, 46). Water molecules were added to 2Fo-Fc density peaks of >1 σ using the Find Water COOT program function. The final model was checked for structural quality using the CCP4 suite programs Procheck and Sfcheck. The data refinement statistics are shown in Table 1. Structure factors and coordinates have been assigned PDB code 4HXD.
Table 1.
Crystallographic data for DUG vOTU-Ub complex
| Parametera | DUG vOTU-Ubb |
|---|---|
| Data collection | |
| Space group | C2 |
| Unit cell dimensions | |
| a, b, c (Å) | 113.5, 40.0, 114.2 |
| β (°) | 97.3 |
| Resolution (Å) | 50.0–2.85 |
| No. of reflections observed | 38,714 |
| No. of unique reflections | 11,887 |
| Rmerge (%) | 5.8 (14.1) |
| I/σI | 18.4 (6.1) |
| % Completeness | 95.8 (79.0) |
| Refinement | |
| Resolution range | 50–2.85 |
| No. of reflections in the working set | 11,301 |
| No. of reflections in the test set | 567 |
| Rwork (%) | 21.0 |
| Rfree (%) | 27.8 |
| RMSD | |
| Bond length (Å) | 0.01 |
| Bond angle (°) | 1.2 |
| No. of protein/water atoms | 3,706/46 |
| Avg B-factors (Å2) | |
| Total | 20.8 |
| Protein | 20.8 |
| Water | 12.9 |
| Ions | 41.6 |
Rmerge = ΣhΣI |II(h) − <I(h)>|/ΣhΣi> II(h), where II(h) is the ith measurement and <I(h)> is the weighted mean of all measurements of I(h). Rwork and Rfree = h(|F(h)obs| − |F(h)calc|)/h|F(h)obs| for reflections in the working and test sets, respectively. RMSD, root mean square deviation.
Where applicable, the last resolution shell is indicated in parentheses.
RESULTS
Comparison of nairovirus vOTUs specificity for Ub and ISG15.
To assess whether nairovirus vOTUs have a conserved specificity for both hUb and hISG15 conjugates, vOTUs from CCHFV, DUGV, and ERVEV were evaluated for their ability to remove 7-amino-4-methylcourmarin (AMC) from the C terminus of 1 μM hUb-AMC, hISG15-AMC, or a peptide that contains the last five highly conserved amino of hISG15 and hUb (ZRLRGG-AMC; Fig. 2). In addition, the recently identified vOTU from TYMV was evaluated along side the nairovirus vOTUs to determine whether the activity and specificity of negative single stranded nairoviruses for these substrates is potentially similar to a vOTU from a positive single-stranded RNA virus. Initially, the four vOTUs were assessed against the ZRLRGG-AMC conjugate to observe their affinity for the minimal recognition component these proteases use to identify hUb and hISG15 (Fig. 2a). The vOTUs of ERVEV and TYMV have a substantially more robust activity toward this substrate than those of CCHFV and DUGV (Fig. 2a). As observed in our previous study, CCHF vOTU cleaves the hUb-AMC 5-fold faster than its hISG15-AMC counterpart (17). Unexpectedly, the other three vOTUs have widely divergent activities from CCHF vOTU toward these two substrates (Fig. 2b and c). Specifically, CCHF vOTU cleaves hUb-AMC with a turnover rate twice that of DUG vOTU, which is the second most active vOTU toward this substrate. However, DUG vOTU possesses 200-fold less activity toward hISG15-AMC, suggesting that every vOTU may not be able to robustly cleave hISG15 conjugates. Intriguingly, the vOTU from ERVEV exhibits the reverse specificity, overwhelmingly preferring hISG15 to hUb. Not surprisingly, vOTU from the plant virus TYMV lacks any appreciable activity for hISG15, for which a plant homologue has yet to be identified. However, its 3-orders-of-magnitude-lower activity for hUb-AMC was interesting since there are only three amino acid differences, which are not located on the surface, between hUb and Ub originating from the yellow turnip, Brassica napobrassica. Overall, TYM vOTU has 6-fold higher activity for ZRLRGG-AMC versus hUb-AMC and 30-fold more than hISG15-AMC.
Fig 2.
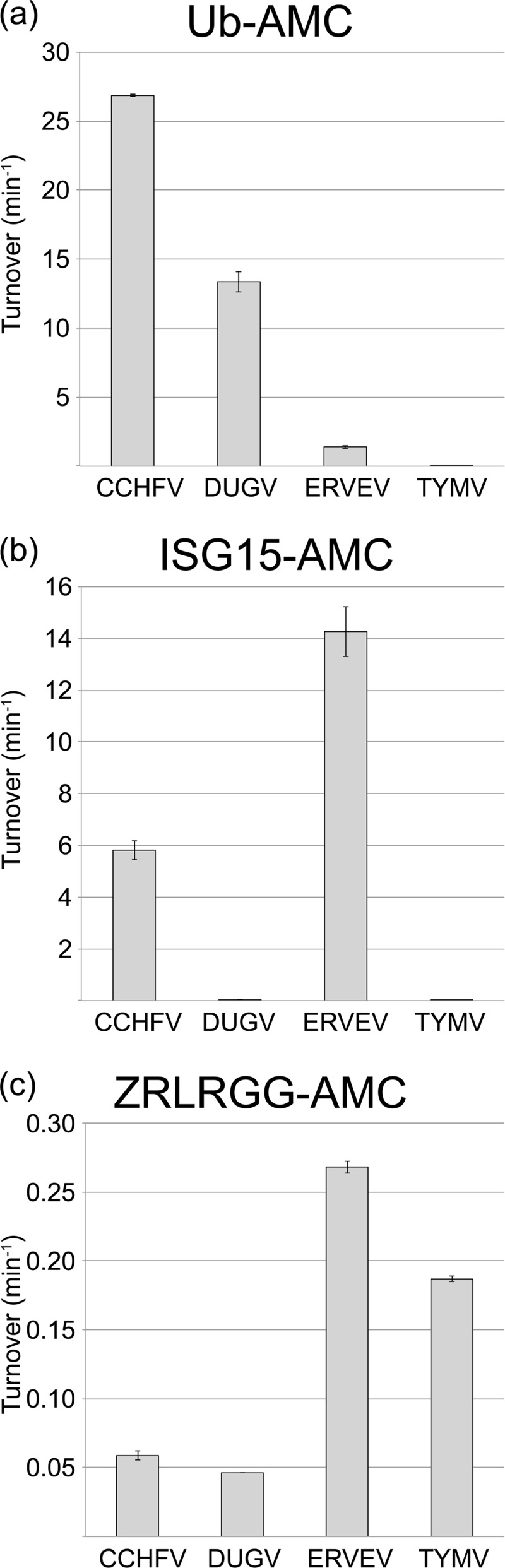
vOTU cleavage of peptide, hUb, and hISG15 AMC conjugates. The cleavage activities of vOTUs from CCHFV, DUGV, and TYMV for 50 μM ZRLRGG-AMC (a), 1 μM hUb-AMC (b), and 1 μM hISG15-AMC (c) were determined. Error bars represent standard deviations from the average.
To explore the complexities in the affinities of vOTUs for hUb and hISG15, the Km and kcat values of the four vOTUs were ascertained for these substrates (Table 2). DUG vOTU shows a slightly higher kcat than that of CCHF vOTU for hUb-AMC but with a substantially increased Km. These results suggest that DUG vOTU can process hUb-AMC at rates beyond those of CCHF vOTU but appears to possess a weaker affinity for the substrate leading it to have a 6-fold reduction in hUb-AMC catalytic efficiency. For hISG15-AMC, DUG vOTU possesses 3 orders of magnitude lower kcat and double the Km for this substrate compared to CCHF vOTU. As a result, DUG vOTU's catalytic efficiency for hISG15 approaches that of the TYM vOTU, a vOTU that has likely not evolved to process this substrate. TYM vOTU's affinity for hUb-AMC does not appear to be particularly robust either, with saturating conditions of the hUb-AMC substrate beyond the maximal substrate concentrations achievable. In contrast to both the vOTUs from TYMV and DUGV, ERVE vOTU possesses a high level of ability to cleave hISG15 conjugates with a kcat within the same magnitude and a Km that was half that of CCHF vOTU. This translates into ERVE vOTU having the highest catalytic efficiency for hISG15 of any known to vOTU. Surprisingly, ERVE vOTU proves to be a poor deubiquitinating enzyme by nairovirus standards, to the point where saturating concentrations of hUb-AMC are beyond the maximal substrate concentrations currently achievable. Using the data available would suggest at least a 4-fold decrease in hUb-AMC kcat compared to CCHF vOTU, as well as a Km that is 2-fold that for DUG vOTU and an order of magnitude compared to CCHF vOTU.
Table 2.
Kinetic characterization of vOTUsa
| Virus | Ub-AMC |
ISG15-AMC |
||||
|---|---|---|---|---|---|---|
| kcat (min−1) | Km (M) | kcat/Km (min−1 M−1) | kcat (min−1) | Km (M) | kcat/Km (min−1 M−1) | |
| CCHFV | 208 ± 15 | (3.8 ± 0.8) × 10−6 | (5.5 ± 12) × 107 | 32.5 ± 3.0 | (2.2 ± 0.5) × 10−6 | (1.5 ± 3.6) × 107 |
| DUGV | 306 ± 13 | (35.7 ± 3.2) × 10−6 | (8.6 ± 0.86) × 106 | 0.244 ± 0.014 | (5.21 ± 0.9) × 10−6 | (4.7 ± 0.01) × 104 |
| ERVEV | 55.6 ± 3.1 | (64.9 ± 5.6) × 10−6 | (8.6 ± 0.088) × 105 | 18.5 ± 0.98 | (0.66 ± 0.14) × 10−6 | (2.8 ± 6.1) × 107 |
| TYMV | 1.79 ± 0.094 | (70.2 ± 6.0) × 10−6 | (2.6 ± 0.003) × 104 | 0.092 ± 0.007 | (13.5 ± 2.3) × 10−6 | (6.1 ± 0.001) × 103 |
Data for CCHFV were obtained from Capodagli et al. (17). Boldface values indicate that saturating conditions were not met.
Poly-Ub linkage specificity of nairovirus vOTUs.
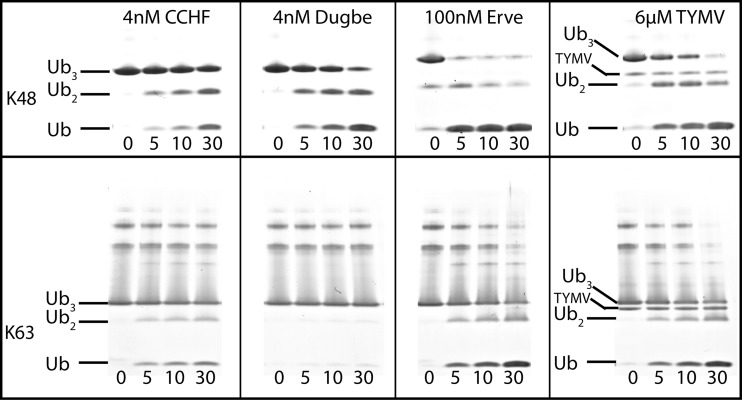
Since host cellular proteins are found to be bound to chains of ubiquitin (poly-Ub), the vOTUs of DUGV, CCHFV, and ERVEV were assessed for their ability to cleave the eight different linkage types of poly-Ub, K6, K11, K27, K33, K48, K63, and linear, side by side. In addition, TYMV vOTU was similarly evaluated to observe whether there was a potentially conserved preference among divergent viral families. With the vOTUs of DUGV and CCHFV exhibiting robust ability to separate hUb conjugates, only nanomolar quantities were necessary to observe poly-Ub cleavage within a 60-min time frame. Conversely, vOTUs from ERVEV and TYMV, which have limited activity toward hUb conjugates, require substantial quantities of protease. In the case of TYM vOTU, almost equal molar ratios were required to observe substantial cleavage (Fig. 3). Evaluation of the ability for the four vOTUs to sever the eight linkage forms between two hUb monomers unexpectedly revealed that all four vOTUs possess a relatively clear preference for K6 poly-Ub linkages. Also, between vOTUs of DUGV and CCHFV that were assessed at the same protease concentration, DUG vOTU appears to process K6 poly-Ub linkages at a faster rate. Interestingly, the four vOTUs have a mixed preference for their secondarily preferred poly-Ub linkage substrate. As was observed previously, CCHF vOTU appears to favor K63 poly-Ub linkage (17). However, the other three vOTUs seem to favor K48 poly-Ub linkages. In addition to severing K6, K48, and K63 poly-Ub linkages, all four vOTUs shared a robust preference for K11 poly-Ub linkages. The remaining K27, K29, K33, and linear poly-Ub linkages proved to be a relatively poor substrate for each of the four vOTUs to various degrees. CCHF vOTU appears to be the most selective of the four vOTUs assessed, only preferring the aforementioned K6, K63, K48, and K11 poly-Ub linkages (Fig. 3). Minor relative activities toward K33 and K27 poly-Ub linkages were observed for the remaining three vOTUs, with ERVE vOTU possibly having some extremely low relative preference toward K29 poly-Ub linkages.
Fig 3.
Gel shift assay of vOTU polyubiquitination linkage specificity. A 10 μM concentration of each di-Ub linkage was incubated with either 4 nM CCHFV, 4 nM DUGV, 100 nM ERVEV, or 6 μM TYMV at 37°C for an hour with samples taken at the indicated time points. The samples were heat inactivated at 95°C for 5 min and then run on a 10 to 20% Mini-Protean Tris-Tricine precast gels (Bio-Rad). The bands were visualized by staining with Coomassie blue.
Since the relative preference of vOTUs from CCHFV, DUGV, ERVEV, and TYMV appear to diverge for K63 and K48 poly-Ub linkages, a quantitative approach was used to understand the extent of the differences for these two substrates among these vOTUs. Specifically, the increase in fluorescence upon the vOTU-facilitated separation of a TAMRA/QXL FRET pair located between 1 μM two hUb molecules with either K48 or K63 linkages was examined. To minimize the possibility of FRET pair interference with vOTU-Ub interactions, separation of three different FRET pair configurations for K48 and K63 FRET substrates were observed using vOTUs from CCHFV, DUGV, ERVEV, and TYMV (data not shown). The rate for FRET pair configuration that is cleaved most efficiency by each vOTU for K48 and K63 FRET substrates were compared side by side (Fig. 4a to c). Although CCHF vOTU and DUG vOTU demonstrate comparable turnover rates for K48 poly-Ub linkages, CCHF vOTU severs K63 poly-Ub linkages at a rate of 7-fold that of DUG vOTU. Also, CCHF vOTU's turnover rate for the K63 FRET substrate is 3.5-fold elevated over that of hUb-AMC. DUG vOTU exhibited a similar phenomenon for its preferred K48 FRET substrate. This trend of K48 and K63 specificity was also present with the larger tri-Ub substrates (Fig. 5). Not surprisingly, TYM vOTU and ERVE vOTU cleaved these poly-Ub linkage substrates poorly, with the two vOTUs having 2 to 3 orders of magnitude less activity than vOTUs from CCHFV and DUGV. In addition to the use of K63 and K48 FRET substrates, a K11 FRET substrate was subsequently used to gauge the cleavage rates of the four vOTUs for K11 poly-Ub linkages. These results generally mirrored those observed in the separation of unlabeled poly-Ub linkages substrates and were all significantly below those observed for cleavage of hUb-AMC (Fig. 3 and 4c).
Fig 4.
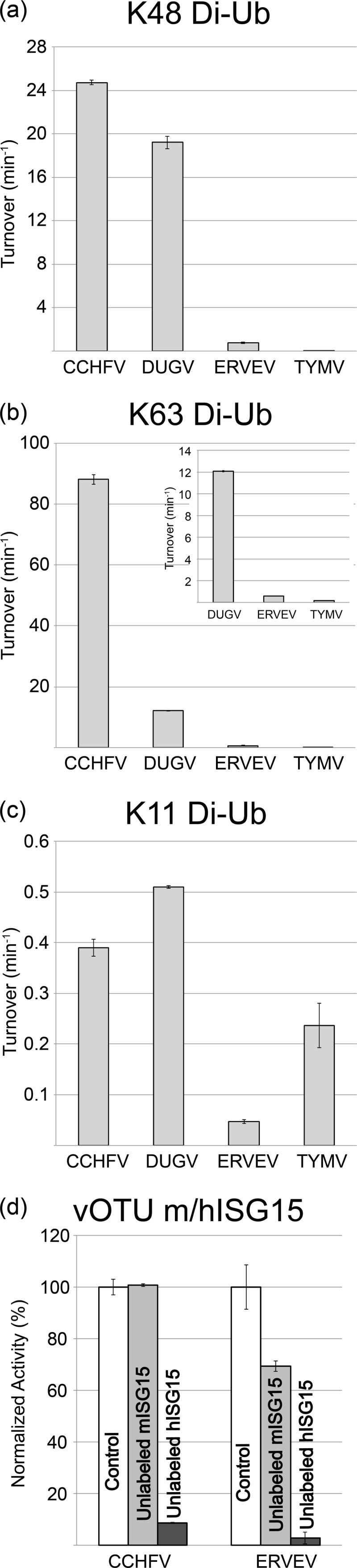
vOTU preference for FRET poly-Ub linkage substrates, as well as human and mouse ISG15. (a to c) vOTU cleavage activity for K48-linked (a), K63-linked (b), or K11-linked di-Ub (c). Turnover values were determined based on the increase in emission upon cleavage of 1 μM di-Ub in the presence of the vOTU from CCHFV, DUGV, and TYMV. (d) Relative rates of cleavage activity of 4 nM CCHF vOTU and ERVE vOTU for 1 μM hISG15-AMC (white) in the presence of 120 μM unlabeled ISG15 from either mouse (light gray) or human (dark gray) sources. Error bars represent standard deviations from the average.
Fig 5.
Poly-Ub linkage gel shift assay. A 20 μM concentration of each tri-Ub linkage was incubated with either 4 nM CCHF vOTU, 4 nM DUG vOTU, 100 nM ERVE vOTU, or 6 μM TYM vOTU at 37°C for an hour with samples taken at the indicated minute time points. The samples were heat inactivated at 95°C for 5 min and then run on a 10 to 20% Mini-Protean Tris-Tricine precast gels.
Species specificity of CCHF and ERVE vOTU for ISG15.
Unlike Ub, which is highly conserved in eukaryotes, ISG15 and its known homologues contain significant sequence variability, even among mammals. Previously, studies have suggested that vOTUs may prefer to cleave hISG15 or equivalent homologues specific to the species they infect (16). To investigate the effect of species diversity in ISG15s on vOTUs' ability to cleave these conjugates, unlabeled hISG15 and mISG15 were used as competitive inhibitors against vOTUs from CCHFV and ERVEV. Predictably, both vOTUs show >90% decreases in their abilities to cleave hISG15-AMC in the presence of unlabeled hISG15. However, the presence of unlabeled mISG15 has a divergent effect among the two vOTUs. For CCHF vOTU, mISG15 lacks appreciable ability to interfere with this vOTU's cleavage of hISG15-AMC, revealing that CCHF vOTU substantially prefers hISG15 to mISG15 (Fig. 4d). Conversely, ERVE vOTU is inhibited by ca. 30% at the same mISG15 concentration, suggesting that affinity for certain species' ISG15s, or homologues, may vary among nairovirus vOTUs.
X-ray crystallographic elucidation of DUG vOTU-Ub complex.
Previous structural elucidation of CCHF vOTU by others and our lab suggested that CCHF vOTU's ability to cleave hISG15, which was unexpected for an OTU, was primarily the result of the orientation in which CCHF vOTU bound to its hUb or hISG15 substrate (16, 17, 47). This suggested that vOTUs might, as a class, possess deISGylating activity. However, although the vOTUs from DUGV and ERVEV have high amino acid similarity to CCHF vOTU, 79 and 68%, respectively, a significant divergence in their preference between hUb and hISG15 conjugates was observed. To investigate the potential structural origins of these partialities, a covalent complex was formed between DUG vOTU and a bromolyated C-terminal hUb. Subsequently, the DUG vOTU-Ub complex was screened against several suites of commercially available precipitant screens and further optimized via an additive screen resulting in a set of crystal conditions. The initial condition contained PEG 3350, Bis-Tris (pH 5.5), and lithium sulfate and was optimized with 1,3-butanediol, yielding a 2.85 Å data set in a C2 space group (Table 1).
Structural comparison of DUG vOTU and CCHF vOTU complexes.
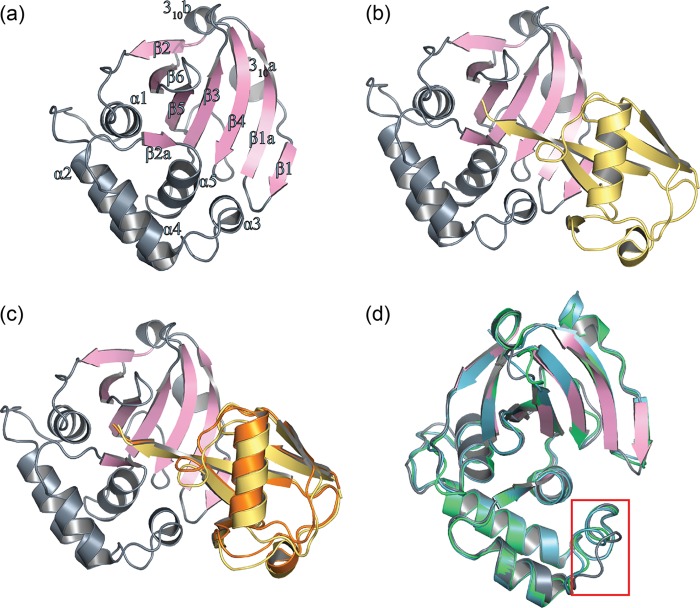
Comparison between the structures of DUG vOTU and CCHF vOTU in complex with hUb revealed that the two complexes adopted a comparable fold. Similar to CCHF vOTU, DUG vOTU is comprised of a core of seven β-sheets flanked by five α-helices and three 310-helices with an additional antiparallel sheet, β2a, formed between residues 100 to 102 of the protease and residues 73 to 75 of the hUb (Fig. 6a and b). In addition, DUG vOTU appears to accommodate hUb in the same manner as CCHF vOTU, suggesting that DUG vOTU employs no overtly tertiary structure rearrangement to account for its partiality between hUb and hISG15 (Fig. 6c).
Fig 6.
Diagram representation of DUG vOTU-Ub. (a) DUG vOTU monomer labeled and colored according to secondary structure: helices and loops are indicated in gray, and β-sheets are indicated in pink. (b) Complex of DUG vOTU rendered as in panel a, and hUb is rendered in yellow. (c) Overlay of DUG vOTU-Ub complex colored as in b, with hUb bound to CCHF vOTU (orange). (d) Overlay of DUG vOTU, CCHF vOTU bound to hUb (green, PDP code 3PRP), and CCHF vOTU bound to hISG15 (teal, PDB code 3PSE) DUG vOTU is as in panel a, with the α3 helix of each structure rendered as loops and boxed in red.
Despite their similarities, the overlay of DUG vOTU and CCHF vOTU bound to either hUb or hISG15 does highlight two variances between the vOTUs (Fig. 6d) (17, 47). One is the β3-4 loop, which encompasses DUG vOTU residues 121 to 126. This loop has a higher than average B-factor in both structures of vOTUs from DUGV and CCHFV and lacks any direct interaction with other parts of the vOTU, or its bound substrate, implying that the divergence observed is a result of inherent loop flexibility. This is not the case with the second dissimilar region that contains DUG vOTU residues 73 to 85, which comprises its α3 helix and surrounding residues. A clear divergence of this region in DUG vOTU can be observed compared to that of either comparable regions in CCHF vOTU-Ub or CCHF vOTU-ISG15 structures (Fig. 6d). Interestingly, this region forms a considerable interface with hUb in the DUG vOTU structure, as well as one between CCHF vOTU bound to hUb and hISG15 (Fig. 6c). Two driving forces readily appear to be responsible for the divergence between the two proteases' α3 helix. One is that DUG vOTU has a glycine at position 81, similar to many other vOTUs, instead of a leucine as in CCHF vOTU. The glycine results in breaking the α3 helix secondary structure, making DUG vOTU's α3 helix shorter than the one found in CCHF vOTU. The second appears to be the amino acid sequence of this region. CCHF vOTU possesses not only a leucine at position 81 but also a valine at position 82. The hydrophobic side chains of these two amino acids appear to insert into hydrophobic patches on hUb and internally on CCHF vOTU, respectively (Fig. 7a).
Fig 7.
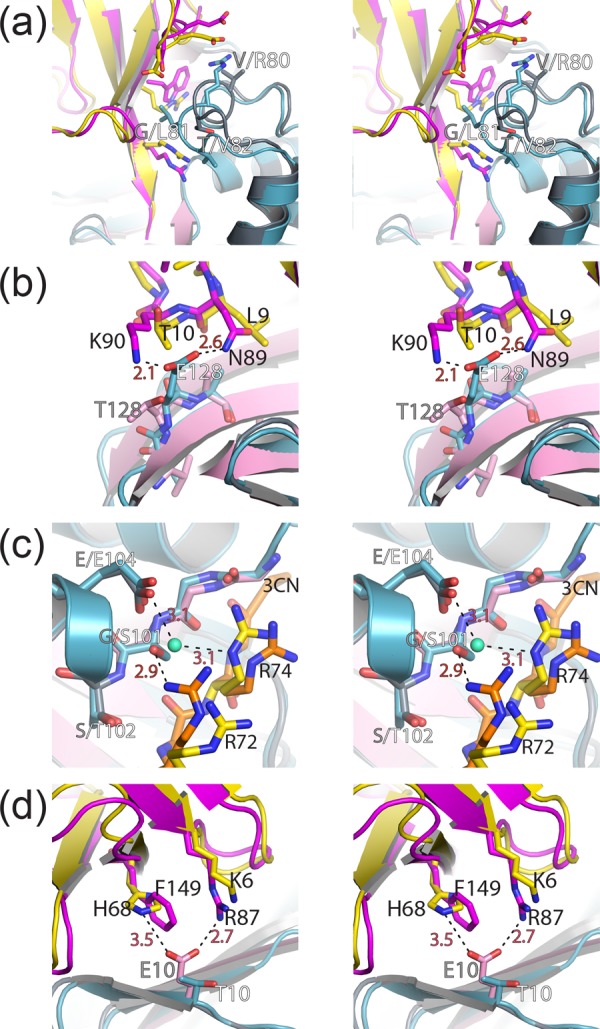
Locations of selective mutagenesis in vOTUs of CCHFV and DUGV. Wall-eyed stereo views of the interactions between DUG vOTU (gray/pink) and CCHF vOTU (teal) with hUb (yellow) and hISG15 (magenta) for the α3-chimera (a) and residues 128 (b), 100 to 102 (c), and 10 (d) are shown. Gray labels indicate CCHF vOTU residues, white labels indicate DUG vOTU residues, black labels indicate hUb and hISG15 residues, and red labels and dashed lines indicate distances. All of the distances are measured in angstroms.
Primary structural origins of Ub and ISG15 specificity between vOTUs of DUGV and CCHFV.
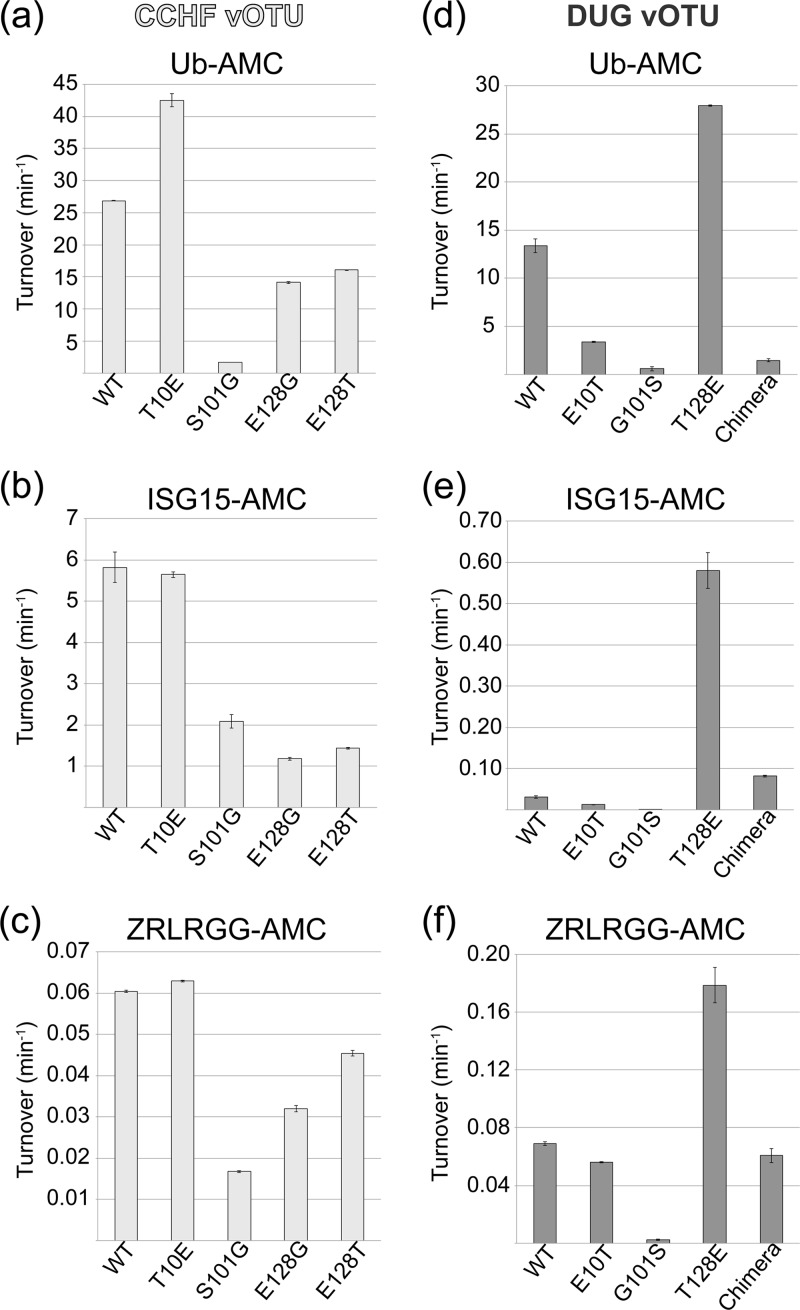
In order to examine the influence of the α3 helix on vOTUs' ability to cleave hISG15 conjugates, several positions within DUG vOTU—Asp74, Thr75, Val80, Gly81, and Thr82—were mutated to their corresponding residues in the CCHF vOTU. The resulting DUG vOTU α3 helix chimera has comparable activity to the wild-type DUG vOTU enzyme for the small peptide ZRLRGG-AMC, suggesting that the mutations did not impact catalytic activity. As expected, the DUG vOTU α3 helix chimera possesses a relatively significant increase of 150% activity for hISG15-AMC. Interestingly, however, the DUG vOTU α3-chimera's ability to cleave the hUb-AMC conjugate decreases significantly, suggesting other primary structural elements are necessary to confer robust deISGylating activity (Fig. 8d to f). One such region might include Glu128, which in CCHF vOTU forms hydrogen bonds with hISG15's Asn89 and Lys90 (Fig. 7b). However, these bonds are absent in DUG vOTU because the glutamate is replaced by a threonine. Introduction of the T128E mutation in DUG vOTU increases the activity toward hUb-AMC by 110%, hISG15-AMC by almost 2,000%, and ZRLRGG-AMC by 160%. Conversely, replacement of this glutamate in CCHF vOTU to a threonine resulted in a drop in activity of 40, 75, and 25% for hUb-AMC, hISG15-AMC, and ZRLRGG-AMC, respectively. In addition, a strain of CCHFV, UG3010, has been isolated from a patient in Uganda that contains a glycine polymorphism at position 128 (48). The introduction of this residue resulted in a mutant that followed the same pattern as the threonine mutation but with a greater effect. These results reveal the influence position 128 plays in determining specificity and activity within vOTUs (Fig. 8a to c).
Fig 8.
Effects of vOTU mutagenesis on vOTU activity toward peptide, hUb, and hISG15 AMC. The cleavage activity of mutants of CCHF vOTU (light gray) or DUG vOTU (dark gray) were determined at 1 μM against 1 μM hUb-AMC (a and d), 1 μM hISG15-AMC (b and e), or 50 μM ZRLRLGG-AMC (c and f). Error bars represent standard deviations from the average.
Beyond position 128 in the vOTU structures of CCHFV and DUGV, position 101 was identified as a potential key residue. The serine at this position in the structure of CCHF vOTU partially comprises β2a that forms an antiparallel β-sheet with hUb. In addition, Ser101 in CCHF vOTU also forms key H-bonds with the hUb arginine residues 72 and 74 side chains. However, this position is filled by a glycine in DUG vOTU, potentially impeding DUG vOTU's ability to interact with its hUb and hISG15 substrates (Fig. 7c). When CCHF vOTU's Ser101 was mutated to glycine, an expected drop in activity toward all assessed substrates of >60% was found. Unexpectedly, when Gly101 in DUG vOTU was mutated to serine, a reduction in activity by >90% was noticed for all substrates, while also resulting in a lower protein expression (data not shown). In vOTUs from DUGV, NSDV, and ERVEV, residue 101 is glycine and 102 is serine, whereas CCHF vOTU has a serine and threonine at these positions. Upon closer examination of CCHF vOTU's Thr102, it appears to be twisted in relation to the corresponding residue in DUGV via surrounding hydrophobic environment created by Ile118, Ile131, and Phe133. This twist results in fixing the orientation of Ser101 in manner that the side chain points toward CCHF vOTU's bound substrate.
Unlike structurally revealed differences in the vOTUs of CCHFV and DUGV that appear to limit DUG vOTU's ability to cleave either hUb or hISG15, DUG vOTU possesses at least one interaction that enhances deubiquitinating activity absent in CCHF vOTU. Specifically, DUG vOTU's Glu10 forms hydrogen bonds to His68 in hUb and is within hydrogen bonding range of Arg87 in hISG15 (Fig. 7d). Not surprisingly, alteration of Glu10 to threonine in DUG vOTU causes decreases of 75 and 60% in hUb-AMC and hISG15-AMC activity, while not affecting the overall catalytic activity of the enzyme. Also, when the T10E mutation was introduced into CCHF vOTU an increase of 50% activity toward hUb-AMC is observed.
DISCUSSION
Structural origins of nairovirus vOTU substrate specificity.
The broad divergence in specificity observed between nairovirus vOTUs appears to originate predominantly from their primary structure differences. This is based on the conserved β-sheet and α-helix tertiary structural scaffold of vOTUs from DUGV and CCHFV, their sequence similarity with other nairovirus vOTUs, and the conserved manner in which hUb is bound to these two vOTUs. In addition, the DUG vOTU-Ub structure implies that although the 30° twist observed between the nairovirus vOTUs and bound hUb and other OTU superfamily members is conserved, it does not necessarily imply that a nairovirus vOTU may process ISG15 conjugates, at least not human ones (17, 41, 49). Several key amino acids have been implicated in affecting CCHF vOTU substrate specificity. Specifically, mutation of Q16R and P77D were shown to ablate CCHF vOTU affinity for hUb and hISG15, respectively (16, 47). However, in spite of these residues being conserved in many nairoviruses, DUG vOTU still demonstrates solely deubiquitinating activity illustrating that these conserved sites do not solely indicate substrate preference. Through the CCHF vOTU-Ub and DUG vOTU-Ub structures, plus selective cross mutagenesis between vOTUs from CCHFV and DUGV, the affinity of nairovirus vOTUs for hUb or hISG15 appears to be largely synergistic. Specifically, the sequence compositions within the α3 helix along with additional favorable amino acids, such as Glu128, or other key positions, appear necessary to infer substantial activity toward hISG15 between vOTUs from DUGV and CCHFV. Also, residues such as Thr102 in CCHF vOTU, which do not directly interact with hUb or hISG15, seem necessary for the positive influence of serine at the preceding position. Inclusion of serine at a similar position in DUG vOTU, which lacks a threonine directly preceding it, highlights that, although affinity appears to be synergistic, single unfavorable amino acid changes within nairovirus vOTUs can have deleterious effects. An additional example of CCHF vOTU residue Glu128, which is a glycine in a CCHFV strain, UG3010, exemplifies this point by suggesting that even vOTUs that harbor polymorphisms between strains can have considerably divergent enzymatic behavior (Fig. 8).
Understandably, with only monomeric hUb bound to DUG vOTU-Ub and CCHF vOTU-Ub, these structures have limitations in directly revealing any specifics on what may drive nairovirus vOTUs to cleave only K6, K11, K48, and K63 poly-Ub linkages or accounting for these vOTUs' greater rates of cleavage of certain linkages over those observed for hUb-AMC, such as those observed for CCHF vOTU's and DUG vOTU's cleavage of K63 and K48 poly-Ub linkages, respectively. Although the precise origins surrounding poly-Ub specificity may require additional structural studies, the tertiary structural similarities between the vOTUs of DUGV and CCHFV suggests that these differences between nairovirus vOTUs are likely primarily structural in nature.
Nairovirus vOTUs and other vOTUs.
The increasing inclusion of new vOTU members originating across the viral genomes other than nairoviruses, including those of arteriviruses, tymoviruses, and tenuiviruses, has spurred speculation on whether there is possible conservation of functionality among these proteases (14, 38, 50). Supporting this assertion has been the sequence similarity beyond the first 20 to 30 equivalent amino acids of nairoviruses and vOTUs originating from these other viruses (Fig. 1). However, there are substantial differences between the four groups of viruses currently known to utilize vOTUs, including the necessity for vOTUs to cleave viral polypeptides in ssRNA(+) vOTU-containing viruses, the location of viral replication complexes within the host cells, and whether the host is plant or animal. Intriguingly from a protease prospective, the vOTU from the ssRNA(+) tymovirus TYMV has a very low deubiquitinating activity—almost 3 orders of magnitude less than vOTUs from DUGV and CCHFV for hUb and poly-Ubs—suggesting that deubiquitinating activity might be secondary to polypeptide cleavage. However, TYM vOTU does prefer the same four, K6, K11, K48, and K63 poly-Ub linkages to the others, as do nairovirus vOTUs. Unfortunately, vOTUs from arteriviruses, such as PRRSV, have not been evaluated against all eight poly-Ub types of linkages, nor have their respective rates toward Ub and ISG15 been established, but they have been observed to cleave both K63 and K48 poly-Ub linkages (50). This, with previously established sequence homology, could suggest arteriviruses may also have activity toward K6 and K11 poly-Ubs.
Potential influence of nairovirus vOTUs specificity on pathogenesis.
A wide array of disease outcomes has been observed for nairovirus infections of humans and animals ranging from transient viremia and migraines to mortality (34, 36, 51). Previous studies have proposed that vOTUs play a significant role in the ability of nairoviruses to evade host immune response through their deubiquitinating and deISGylating activities, prompting them to be considered potential virulence factors (14, 16, 17, 36, 47). This classification suggests that a significant divergence in vOTU activity, or specificity, possibly exists between vOTUs from different nairoviruses and potentially within species. The evaluation of deubiquitinating and deISGylating activities of vOTUs from CCHFV, DUGV, and ERVEV appears to support this assertion. Despite a reasonably high sequence similarity between three evaluated nairovirus vOTUs, orders of magnitude differences in proteolytic activity between them have been observed for different hUb moieties, as well for hISG15 conjugates. Distilling these preferences into catalytic efficiencies readily reflects that particular nairovirus vOTUs do not always possess robust dual human deubiquitinating and deISGylating functionality with a preference for ubiquitinated substrates as the previously studied vOTU from CCHFV may have proposed (16, 17, 47) (Table 2). Instead, the preference of nairovirus vOTUs potentially could be either for human ubiquitinated substrates, ISGylated ones, or both.
Curiously, among the nairovirus vOTUs evaluated, only the one from the often-human fatal CCHFV has coexisting robust deubiquitinating and deISGylating activities related to hUb and hISG15. The vOTUs from DUGV and ERVEV, which have currently not been observed to be fatal in humans, only possess substantial activity toward one or the other. For ERVE vOTU, the virtually complete amino acid conservation among Ub within animals suggests that ERVE vOTU predominantly acts as a deISGylase during infection, making it the first known viral protease that is incapable of effectively cleaving both monomeric and polymeric Ub conjugates while still retaining strong activity for hISG15. Although the ERVE vOTU shares deISGylating functionality with CCHF vOTU, the two vOTUs respond differently when mISG15 is used as a competitive inhibitor. In contrast to Ub, among higher animals there is relatively low sequence homology for ISG15. Interestingly, this diversity has previously been linked to influenza virus B's ability of NS1 to specifically bind ISG15 of human or nonhuman primate origin, explaining the susceptibility of ISG15-deficient mice to influenza virus B (52, 53). Previously, CCHFV has been observed to infect several mammals but not lead to any serious signs of disease outside of humans (6). In addition, mice unable to respond to IFNI developed a disease mimicking several aspects of the clinical course of CCHF in humans, including mortality (54). The significantly reduced affinity of CCHF vOTU for mISG15 appears to further support that ISG15 diversity could be a zoonotic barrier for viruses. Also, the ability of mISG15 to inhibit the vOTU from ERVEV, but not CCHFV, suggests that ERVE vOTU has a higher affinity for mISG15 and is possibly less selective for particular species ISG15. Alternatively, mISG15 may share more homology with currently unknown sequence composition of the white tooth shrew, which ERVE is commonly found to infect (55). Furthermore, correlation between nairovirus vOTUs and host tropism does offer the remote possibility that DUG vOTU may also be a deISGylase against a currently unknown host. One possibility could be cattle that it has been known to predominantly infect. Alternatively, DUG vOTU might also possess significant cleavage activity for mISG15, as Bakshi et al. have reported (37).
This unique ability of CCHF vOTU to cleave hISG15 conjugates at relatively high rates originally had differentiated CCHF vOTUs from other members of the OTU superfamily and likened CCHF vOTU to a virulence factor of severe acute respiratory coronavirus, papain-like protease (PLpro), which also has strong deubiquitinating and deISGylating activities (14, 17, 56). However, the PLpro and CCHF vOTU similarities go beyond their ability to cleave both hUb and hISG15. CCHF vOTU appears to be unique among the three evaluated nairovirus vOTUs for its ability to robustly cleave K63 poly-Ub linkages, while retaining the comparable ability to cleave K48 poly-Ub, such as is the case for PLpro (57). This is in contrast to other known viral deubiquitinases, such as herpesvirus M48, or vOTUs from other nairoviruses, including DUG vOTU (58). Also, the robust specificity for K63 poly-Ub that CCHF vOTU possesses may be of importance pathogenically since Bogunovic et al. recently suggested that ISGylation and ubiquitination might be redundant in humans for viral infections (59). With certain host immunologically related proteins having been observed to be modified with either ISG15 or K63 poly-Ub, the removal of both by a viral protease may ensure immunosuppression. Comparison of CCHF vOTU's poly-Ub preferences and those of other vOTUs also illustrates that CCHF vOTU is also the most specific to the four poly-Ub linkages—K6, K11, K48, and K63—that currently evaluated vOTUs prevailingly cleave (Fig. 3). This also likely downplays the need for nairoviruses to cleave these other forms of poly-Ub moieties for successful viral evasion. Overall, the substrate preferences of the nairovirus vOTUs evaluated here suggest that a virus containing a vOTU capable of not only deubiquitinase and species-specific deISGylase activities but also robust deubiquitinase activity toward K63 poly-Ub linkages might have the best chance for viral immune evasion and pathogenesis. Naturally, the full impact of diversity in specificity among vOTUs on viral pathogenesis can ideally be addressed when a reverse genetics system for nairoviruses is available.
ACKNOWLEDGMENTS
We thank Keith Wilkinson for his gift of Ub expression plasmids.
The Advanced Light Source is supported by the Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under contract DE-AC02-05CH11231. This study was supported with funding provided by National Institute of Health grants 1R03AI092249-01 and 1R03MH097507-01A1, as well as by the University of Denver's Partnership in Scholarship Program.
Footnotes
Published ahead of print 23 January 2013
REFERENCES
- 1. Karti SS, Odabasi Z, Korten V, Yilmaz M, Sonmez M, Caylan R, Akdogan E, Eren N, Koksal I, Ovali E, Erickson BR, Vincent MJ, Nichol ST, Comer JA, Rollin PE, Ksiazek TG. 2004. Crimean-Congo hemorrhagic fever in Turkey. Emerg. Infect. Dis. 10:1379–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mishra AC, Mehta M, Mourya DT, Gandhi S. 2011. Crimean-Congo haemorrhagic fever in India. Lancet 378:372. [DOI] [PubMed] [Google Scholar]
- 3. Ozkaya E, Dincer E, Carhan A, Uyar Y, Ertek M, Whitehouse CA, Ozkul A. 2010. Molecular epidemiology of Crimean-Congo hemorrhagic fever virus in Turkey: occurrence of local topotype. Virus Res. 149:64–70 [DOI] [PubMed] [Google Scholar]
- 4. World Health Organization 2001. Crimean-Congo haemorrhagic fever: fact sheet. WHO Media Centre, World Health Organization, Geneva, Switzerland [Google Scholar]
- 5. World Health Organization 2003. Crimean-Congo haemorrhagic fever (CCHF) in Mauritania: update. World Health Organization, Geneva, Switzerland [Google Scholar]
- 6. Nalca A, Whitehouse CA, Ergonul O, Whitehouse CA. (ed). 2007. Crimean-Congo hemorrhagic fever. Springer, Dordrecht, Netherlands [Google Scholar]
- 7. Burt FJ, Spencer DC, Leman PA, Patterson B, Swanepoel R. 1996. Investigation of tick-borne viruses as pathogens of humans in South Africa and evidence of Dugbe virus infection in a patient with prolonged thrombocytopenia. Epidemiol. Infect. 116:353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dandawate CN, Work TH, Webb JK, Shah KV. 1969. Isolation of Ganjam virus from a human case of febrile illness: a report of a laboratory infection and serological survey of human sera from three different states of India. Indian J. Med. Res. 57:975–982 [PubMed] [Google Scholar]
- 9. Marczinke BI, Nichol ST. 2002. Nairobi sheep disease virus, an important tick-borne pathogen of sheep and goats in Africa, is also present in Asia. Virology 303:146–151 [DOI] [PubMed] [Google Scholar]
- 10. Sudeep AB, Jadi RS, Mishra AC. 2009. Ganjam virus. Indian J. Med. Res. 130:514–519 [PubMed] [Google Scholar]
- 11. Sang R, Onyango C, Gachoya J, Mabinda E, Konongoi S, Ofula V, Dunster L, Okoth F, Coldren R, Tesh R, da Rossa AT, Finkbeiner S, Wang D, Crabtree M, Miller B. 2006. Tickborne arbovirus surveillance in market livestock, Nairobi, Kenya. Emerg. Infect. Dis. 12:1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Steele GM, Nuttall PA. 1989. Difference in vector competence of two species of sympatric ticks, Amblyomma variegatum and Rhipicephalus appendiculatus, for Dugbe virus (Nairovirus, Bunyaviridae). Virus Res. 14:73–84 [DOI] [PubMed] [Google Scholar]
- 13. Dilcher M, Koch A, Hasib L, Dobler G, Hufert F, Weidmann M. 2012. Genetic characterization of Erve virus, a European nairovirus distantly related to Crimean-Congo hemorrhagic fever virus. Virus Genes 45:426–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frias-Staheli N, Giannakopoulos NV, Kikkert M, Taylor SL, Bridgen A, Paragas J, Richt JA, Rowland RR, Schmaljohn CS, Lenschow DJ, Snijder EJ, Garcia-Sastre A, Virgin HW. 2007. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2:404–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bergeron E, Albarino CG, Khristova ML, Nichol ST. 2010. Crimean-Congo hemorrhagic fever virus-encoded ovarian tumor protease activity is dispensable for virus RNA polymerase function. J. Virol. 84:216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Akutsu M, Ye Y, Virdee S, Chin JW, Komander D. 2010. Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains. Proc. Natl. Acad. Sci. U. S. A. 108:2228–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Capodagli GC, McKercher MA, Baker EA, Masters EM, Brunzelle JS, Pegan SD. 2011. Structural analysis of a viral ovarian tumor domain protease from the Crimean-Congo hemorrhagic fever virus in complex with covalently bonded ubiquitin. J. Virol. 85:3621–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edelmann MJ, Kessler BM. 2008. Ubiquitin and ubiquitin-like specific proteases targeted by infectious pathogens: emerging patterns and molecular principles. Biochim. Biophys. Acta 1782:809–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bridgen A, Weber F, Fazakerley JK, Elliott RM. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 98:664–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Giorgi C, Accardi L, Nicoletti L, Gro MC, Takehara K, Hilditch C, Morikawa S, Bishop DH. 1991. Sequences and coding strategies of the S RNAs of Toscana and Rift Valley fever viruses compared to those of Punta Toro, Sicilian sandfly fever, and Uukuniemi viruses. Virology 180:738–753 [DOI] [PubMed] [Google Scholar]
- 21. Komander D, Rape M. 2012. The ubiquitin code. Annu. Rev. Biochem. 81:203–229 [DOI] [PubMed] [Google Scholar]
- 22. Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. 2009. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137:133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M, Kelley RF, Dixit VM. 2010. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol. Cell 39:477–484 [DOI] [PubMed] [Google Scholar]
- 24. Arimoto K, Funami K, Saeki Y, Tanaka K, Okawa K, Takeuchi O, Akira S, Murakami Y, Shimotohno K. 2010. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proc. Natl. Acad. Sci. U. S. A. 107:15856–15861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang H, Jeon MS, Liao L, Yang C, Elly C, Yates JR, III, Liu YC. 2010. K33-linked polyubiquitination of T cell receptor-zeta regulates proteolysis-independent T cell signaling. Immunity 33:60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shang F, Deng G, Liu Q, Guo W, Haas AL, Crosas B, Finley D, Taylor A. 2005. Lys6-modified ubiquitin inhibits ubiquitin-dependent protein degradation. J. Biol. Chem. 280:20365–20374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zucchelli S, Codrich M, Marcuzzi F, Pinto M, Vilotti S, Biagioli M, Ferrer I, Gustincich S. 2010. TRAF6 promotes atypical ubiquitination of mutant DJ-1 and alpha-synuclein and is localized to Lewy bodies in sporadic Parkinson's disease brains. Hum. Mol. Genet. 19:3759–3770 [DOI] [PubMed] [Google Scholar]
- 28. Herrmann J, Lerman LO, Lerman A. 2007. Ubiquitin and ubiquitin-like proteins in protein regulation. Circ. Res. 100:1276–1291 [DOI] [PubMed] [Google Scholar]
- 29. Pickart CM, Cohen RE. 2004. Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell. Biol. 5:177–187 [DOI] [PubMed] [Google Scholar]
- 30. Versteeg GA, García-Sastre A. 2010. Viral tricks to grid-lock the type I interferon system. Curr. Opin. Microbiol. 13:508–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weber F, Mirazimi A. 2008. Interferon and cytokine responses to Crimean Congo hemorrhagic fever virus; an emerging and neglected viral zoonosis. Cytokine Growth Factor Rev. 19:395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao C, Denison C, Huibregtse JM, Gygi S, Krug RM. 2005. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 102:10200–10205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ha BH, Kim EE. 2008. Structures of proteases for ubiquitin and ubiquitin-like modifiers. BMB Rep. 41:435–443 [DOI] [PubMed] [Google Scholar]
- 34. Peyrefitte CN, Perret M, Garcia S, Rodrigues R, Bagnaud A, Lacote S, Crance JM, Vernet G, Garin D, Bouloy M, Paranhos-Baccala G. 2010. Differential activation profiles of Crimean-Congo hemorrhagic fever virus- and Dugbe virus-infected antigen-presenting cells. J. Gen. Virol. 91:189–198 [DOI] [PubMed] [Google Scholar]
- 35. Rodrigues R, Paranhos-Baccala G, Vernet G, Peyrefitte CN. 2012. Crimean-Congo hemorrhagic fever virus-infected hepatocytes induce ER-stress and apoptosis crosstalk. PLoS One 7:e29712 doi:10.1371/journal.pone.0029712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holzer B, Bakshi S, Bridgen A, Baron MD. 2011. Inhibition of interferon induction and action by the nairovirus Nairobi sheep disease virus/Ganjam virus. PLoS One 6:e28594 doi:c17 10.1371/journal.pone.0028594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bakshi S, Holzer B, Bridgen A, McMullan G, Quinn D, Baron MD. 2012. The Dugbe virus ovarian tumour (OTU) domain interferes with ubiquitin/ISG15-regulated innate immune cell signalling. J. Gen. Virol. 94:298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chenon M, Camborde L, Cheminant S, Jupin I. 2011. A viral deubiquitylating enzyme targets viral RNA-dependent RNA polymerase and affects viral infectivity. EMBO J. 31:741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang HM, Yang J, Sun HR, Xin X, Wang HD, Chen JP, Adams MJ. 2007. Genomic analysis of rice stripe virus Zhejiang isolate shows the presence of an OTU-like domain in the RNA1 protein and a novel sequence motif conserved within the intergenic regions of ambisense segments of tenuiviruses. Arch. Virol. 152:1917–1923 [DOI] [PubMed] [Google Scholar]
- 40. Kim KI, Zhang DE. 2005. UBP43, an ISG15-specific deconjugating enzyme: expression, purification, and enzymatic assays. Methods Enzymol. 398:491–499 [DOI] [PubMed] [Google Scholar]
- 41. Messick TE, Russell NS, Iwata AJ, Sarachan KL, Shiekhattar R, Shanks JR, Reyes-Turcu FE, Wilkinson KD, Marmorstein R. 2008. Structural basis for ubiquitin recognition by the Otu1 ovarian tumor domain protein. J. Biol. Chem. 283:11038–11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wilkinson KD, Gan-Erdene T, Kolli N. 2005. Derivitization of the C terminus of ubiquitin and ubiquitin-like proteins using intein chemistry: methods and uses. Methods Enzymol. 399:37–51 [DOI] [PubMed] [Google Scholar]
- 43. Gill SC, von Hippel PH. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319–326 [DOI] [PubMed] [Google Scholar]
- 44. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. A 276:307–326 [DOI] [PubMed] [Google Scholar]
- 45. Bailey S. 1994. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 46. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 47. James TW, Frias-Staheli N, Bacik JP, Levingston-Macleod JM, Khajehpour M, Garcia-Sastre A, Mark BL. 2011. Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease. Proc. Natl. Acad. Sci. U. S. A. 108:2222–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Deyde VM, Khristova ML, Rollin PE, Ksiazek TG, Nichol ST. 2006. Crimean-Congo hemorrhagic fever virus genomics and global diversity. J. Virol. 80:8834–8842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang OW, Ma X, Yin J, Flinders J, Maurer T, Kayagaki N, Phung Q, Bosanac I, Arnott D, Dixit VM, Hymowitz SG, Starovasnik MA, Cochran AG. 2012. Phosphorylation-dependent activity of the deubiquitinase DUBA. Nat. Struct. Mol. Biol. 19:171–175 [DOI] [PubMed] [Google Scholar]
- 50. van Kasteren PB, Beugeling C, Ninaber DK, Frias-Staheli N, van Boheemen S, Garcia-Sastre A, Snijder EJ, Kikkert M. 2012. Arterivirus and nairovirus ovarian tumor domain-containing deubiquitinases target activated RIG-I to control innate immune signaling. J. Virol. 86:773–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Treib J, Dobler G, Haass A, von Blohn W, Strittmatter M, Pindur G, Froesner G, Schimrigk K. 1998. Thunderclap headache caused by Erve virus? Neurology 50:509–511 [DOI] [PubMed] [Google Scholar]
- 52. Sridharan H, Zhao C, Krug RM. 2010. Species specificity of the NS1 protein of influenza B virus: it binds only human and non-human primate ubiquitin-like ISG15 proteins. J. Biol. Chem. 258:7852–7856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Versteeg GA, Hale BG, van Boheemen S, Wolff T, Lenschow DJ, García-Sastre A. 2010. Species-specific antagonism of host ISGylation by the influenza B virus NS1 protein. J. Virol. 84:5423–5430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bente D, Alimonti J, Shieh W, Camus G, Stroher U, Zaki S, Jones S. 2010. Pathogenesis and immune response of Crimean-Congo hemorrhagic fever virus in a STAT-1 knockout mouse model. J. Virol. 84:11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Woessner R, Grauer MT, Langenbach J, Dobler G, Kroeger J, Mielke HG, Mueller P, Haass A, Treib J. 2000. The Erve virus: possible mode of transmission and reservoir. Infection 28:164–166 [DOI] [PubMed] [Google Scholar]
- 56. Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 79:15189–15198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ratia K. 2008. Structure, function, and inhibition of the papain-like protease from SARS coronavirus. Ph.D. thesis University of Illinois, Chicago, IL [Google Scholar]
- 58. Schlieker C, Weihofen WA, Frijns E, Kattenhorn LM, Gaudet R, Ploegh HL. 2007. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol. Cell 25:677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bogunovic D, Byun M, Durfee LA, Abhyankar A, Sanal O, Mansouri D, Salem S, Radovanovic I, Grant AV, Adimi P, Mansouri N, Okada S, Bryant VL, Kong XF, Kreins A, Velez MM, Boisson B, Khalilzadeh S, Ozcelik U, Darazam IA, Schoggins JW, Rice CM, Al-Muhsen S, Behr M, Vogt G, Puel A, Bustamante J, Gros P, Huibregtse JM, Abel L, Boisson-Dupuis S, Casanova JL. 2012. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science 337:1684–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]