Abstract
Oncolytic virus (OV) therapies of cancer are based on the use of replication-competent, tumor-selective viruses with limited toxicity. Newcastle disease virus (NDV), an avian paramyxovirus, is a promising OV and is inherently tumor selective and cytotoxic only to tumor cells. Replication is restricted in normal cells. Despite encouraging phase I/II clinical trials with NDV, further refinements for tumor-specific targeting are needed to enhance its therapeutic index. Systemically delivered NDV fails to reach solid tumors in therapeutic concentrations and also spreads poorly within the tumors due to barriers including complement, innate immunity, and the extracellular matrix. Overcoming these hurdles is paramount to realizing the exceptional oncolytic efficacy of NDV. We engineered the F protein of NDV and generated a recombinant NDV (rNDV) whose F protein is cleavable exclusively by prostate-specific antigen (PSA). The rNDV replicated efficiently and specifically in prostate cancer (CaP) cells and 3-dimensional prostaspheres but failed to replicate in the absence of PSA. Induction of intracellular PSA production by a synthetic androgen analog (R1881) enhanced fusogenicity in androgen-responsive CaP cells. Further, PSA-cleavable rNDV caused specific lysis of androgen-independent and androgen-responsive/nonresponsive CaP cells and prostaspheres, with a half-maximal effective concentration (EC50) ranging from a multiplicity of infection of 0.01 to 0.1. PSA-retargeted NDV efficiently lysed prostasphere tumor mimics, suggesting efficacy in vivo. Also, PSA-cleavable NDV failed to replicate in chicken embryos, indicating no pathogenicity for chickens. Prostate-specific antigen targeting is likely to enhance the therapeutic index of rNDV owing to tumor-restricted replication and enhanced fusogenicity.
INTRODUCTION
Prostate cancer (CaP) is the second leading cause of cancer-related deaths in the United States (1). Current treatment regimens for hormone-resistant CaP are palliative and marginally increase survival. Oncolytic virotherapy is a novel approach for treating CaP and became particularly attractive with the advent of techniques for genetic manipulation of RNA viruses (2, 3). Newcastle disease virus (NDV), an avian paramyxovirus, is an inherently tumor selective oncolytic virus (OV) for which compelling preclinical and phase I/II studies have been conducted with human subjects (4–6). Although naturally occurring strains of NDV used as oncolytic agents have shown some promising results and have been used in clinical trials, genetic modification of NDV affords the opportunity to improve its antitumor efficacy and therapeutic index.
Cleavage of the fusion (F) protein adjacent to its fusion peptide is required for the initiation of infection and is a major determinant of NDV virulence (7). We hypothesized that the F protein cleavage site of NDV could be made cleavable by prostate-specific antigen (PSA) so as to restrict its replication to PSA-producing CaP cells. PSA is a serine protease with chymotrypsin-like substrate specificity (8–11) that is found in high concentrations (mg/ml) in the seminal plasma (12). Increased serum PSA levels correlate with tumor volume (13) and most probably result from leakage of PSA from the prostatic ductal system into the prostatic stroma and subsequently into the bloodstream. The serum PSA test measures total PSA, which has been defined as consisting of all immunodetectable PSA and comprises mostly free PSA and PSA bound to the protein inhibitor α1-antichymotrypsin (ACT) (14). While it has been shown that explant tissues from human prostate tumors contained >80% active PSA (15), others have shown that the more prevalent form of PSA in tumor homogenates is in the ACT-bound state (16). The therapeutic index of NDV can be enhanced by specific activation through cancer cell type-specific proteases. To date, among oncolytic viruses, only Sendai virus and measles virus have been modified for cleavage by tumor-specific proteases such as matrix metalloproteases (MMP) or uroplasminogen activator (UPA) (17, 18).
The NDV F protein cleavage site possesses the consensus amino acid sequence 112(R/K)-R-Q-(R/K)-R//F117 (the double slash denotes the cleavage site) for virulent and mesogenic strains and the consensus sequence 112(G/E)-(K/R)-Q-(G/E)-R//L117 for strains of low virulence (19). To determine whether it would be possible to engineer recombinant NDV (rNDV) targeted to tumor-specific proteases, we modified the F protein cleavage site to target PSA and examined the ability of the mutant rNDV to replicate and kill CaP cells specifically. Denmeade et al. (20) reported HSSKLQ as a PSA-specific substrate that was not hydrolyzed by a variety of other proteases, including chymotrypsin and trypsin. Subsequently, these investigators reported that HP6SP5SP4KP3LP2QP1//LP1′ (where P1 to P6 stand for positions 1 to 6) was an efficient substrate for PSA (21). Further, molecular docking studies indicated that the amino acids at positions 1 to 3 (P1 to P3) are critical and that any amino acid with a smaller side chain at P4 would allow substrate specificity (21). We therefore mutated the fusion protein cleavage site sequence from 112(R/K)-R-Q-(R/K)-R//F117 to 112RHSSK//L117, 112RRKLQ//L117, or 112RRKLQ//F117 so as to retain phenylalanine or leucine at the P1′ position, because the presence of these amino acids at position 117 was reported to be essential for the induction of fusion in NDV (22). The 112RHSSK//L117 mutation was performed in order to understand whether a monobasic amino acid at the P1 position would result in F protein cleavage by proteases other than PSA. This mutated site was predicted not to be cleavable by PSA, since previous substrate docking studies claimed that glutamine at the P1 position was essential for a high degree of selectivity for PSA (21). We also constructed the F-Null protein by replacing the furin motif with the sequence 112GGPGGV117, changing the P1′ amino acid to valine, and by creating a neutral charge at the cleavage site with glycine to render it fusion defective (22). The peptide sequence of the NDV F-Null protein was therefore not expected to be cleaved by any known proteases, and it was used as a control protein in this study.
We show that only the 112RRKLQ//L117 sequence at the cleavage site of F was PSA specific and that rNDV with this PSA-specific site replicated efficiently in and was cytotoxic to CaP cells and 3-dimensional (3-D) prostaspheres, which are tumor mimics.
MATERIALS AND METHODS
Cells and reagents.
The Vero, androgen receptor-negative (AR−) PC-3, and DU145 prostate cancer (CaP) cell lines were purchased from the ATCC and were maintained in Dulbecco modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals). BSR-T7/5 cells were obtained from Griffith D. Parks, Wake Forest University, and were maintained in DMEM supplemented with 10% FBS and G418 (Geneticin; Invitrogen) as described previously (23). Androgen receptor-positive (AR+), PSA-secreting C4-2b cells were obtained from Leland Chung, Emory University, and were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS. Another AR+, PSA-secreting cell line, WPE-int, was also obtained from the ATCC and was maintained in keratinocyte serum-free medium (K-SFM) supplemented with 0.05 mg/ml bovine pituitary extract (BPE) and 5 ng/ml epidermal growth factor (EGF) (K-SFM and supplements were from Invitrogen). AR+ and AR− CaP cells were supplemented with PSA (Sigma-Aldrich) at 100 ng/ml. A synthetic androgen analog, R1881 (Sigma-Aldrich), was used at a concentration of 1 nm/ml to induce intracellular PSA production only in androgen-responsive AR+ cells (Table 1). Cells were also maintained as nonadherent prostaspheres in serum-free medium with 1× glutamine, 10 ng/ml EGF, and 1× B27 supplement without vitamin A (Invitrogen). The full-length Beaudette C (BC) antigenome and the nucleoprotein (NP), phosphoprotein (P), and large polymerase protein (L) support plasmids have been described elsewhere (24). Anti-NDV chicken serum (Spafas) and horseradish peroxidase (HRP)- or fluorescein isothiocyanate (FITC)-conjugated goat anti-chicken antibodies (Kirkegaard & Perry Laboratories) were used at predetermined concentrations. The anti-NDV F antibody 39-D9 was a generous gift from Ronald Iorio, University of Massachusetts School of Medicine. Tosyl phenylalanyl chloromethyl ketone (TPCK) trypsin and tosyl lysine chloromethyl ketone (TLCK) chymotrypsin were obtained from Sigma-Aldrich.
Table 1.
Prostate cancer and non-prostate cancer cell lines used in this studya
| Cell line | Origin | AR status | Androgen responsive or nonresponsive for AR-mediated pathways | PSA production | Source or ATCC catalog no. |
|---|---|---|---|---|---|
| DU145 | Human prostate carcinoma | Negative | Nonresponsive | No | HTB-81 |
| PC3 | Human prostate carcinoma | Negative | Nonresponsive | No | CRL-1435 |
| C4-2B | Human prostate cancer cell line LnCaP | Positive | Responsive | Yes | Leland Chung, Emory University |
| WPE-int | Human prostate epithelium | Positive | Responsive | Yes | CRL-2888 |
| Vero | African green monkey kidney | Negative | Nonresponsive | No | CCL-81 |
| BSR-T7/5 | Baby hamster kidney | Negative | Nonresponsive | No | CCL-10 |
All cell lines are androgen independent for proliferation and survival.
Cell-based assays.
The mutant F genes and wild-type (wt) hemagglutinin (HN) gene were cloned into the NheI site of pCAGGS. Vero, BSR-T7/5, and PC-3 cells were seeded in 6-well plates and were cotransfected at 80% confluence with 1 μg of F and HN plasmids using Lipofectamine (Invitrogen). PSA, at a concentration of 100 ng/ml, was added at 12 h posttransfection to the wells transfected with F mutant plasmids. To determine the optimum PSA overlay concentration, PC-3 cells were cotransfected with mutant F and HN plasmids and were supplemented with exogenous PSA (1 to 400 ng/ml) at 12 h posttransfection. The optimum PSA overlay concentration was determined on the basis of the fusion index (25) and cell viability as determined by the trypan blue dye exclusion assay (26, 27). For determination of the fusion index, PC-3 cells were cotransfected with wt or mutant F genes and the wt HN gene. At 72 h posttransfection, the cells were fixed using methanol-acetone (1:1) and were washed with 1 mM EDTA solution in phosphate-buffered saline (PBS). After the PBS was decanted, cells were stained with hematoxylin-eosin stain (Fisher Scientific). The fusion areas in eight different fields were counted to determine the fusion index (mean number of nuclei per cell) as described previously (25, 28).
Generation and rescue of recombinant NDV.
The KLQL, HSSKL, KLQF, and F-Null cleavage site mutations were introduced into the full-length NDV BC genome by two-round PCR-directed mutagenesis. Primers were designed to introduce the desired mutations into the F cleavage site of a subclone carrying the SacII-to-NotI segment of the BC genome (29). The SacII-NotI fragment was then cloned into the full-length BC antigenome to generate full-length BC-HSSKL, BC-KLQL, BC-KLQF, and BC-F-Null clones (24). Further, the SacII-PmlI fragment bearing the enhanced green fluorescent protein (EGFP) gene was cloned into full-length BC-HSSKL, BC-KLQL, and BC-F-Null clones in order to produce infectious clones expressing EGFP. Transfection and rescue of recombinant NDVs were performed as described previously (29) with minor modifications. Briefly, cells were transfected with a full-length infectious clone of NDV along with NP, P, and L plasmids, with or without the wild-type BC F gene. BSR-T7/5 cells were overlaid with medium containing 100 ng/ml PSA 48 h before transfection. The medium was then removed, and the cells were washed three times with PBS and transfected with the plasmids. At 48 h posttransfection, PSA (100 ng/ml) was added again, and the medium was unchanged until the cells were harvested. BSR-T7/5 cells were overlaid with fresh Vero cells 4 days posttransfection, and the cells were examined for syncytia and fluorescence. At 7 days posttransfection, the entire-cell lysate was used for amplification of the virus in DU145 cells.
Immunofluorescence assay (IFA) and flow cytometry.
Vero cells were transfected with a wt or mutant F gene, with or without the HN gene. Following 48 h posttransfection, cells were fixed with methanol-acetone (1:1) for 30 min at room temperature. Live-cell staining was carried out for cells transfected with wt or mutant F proteins in order to detect cell surface fluorescence. Cells were washed with PBS and blocked in fluorescence-activated cell-sorting (FACS) buffer (PBS plus 1% bovine serum albumin and 0.02% sodium azide) for 30 min on ice. Blocking was followed by incubation with primary antibody 3-9D9 (diluted 1:500 in FACS buffer) for 1 h at 4°C. After three washes with FACS buffer, cells were incubated for 1 h at 4°C with an FITC-conjugated goat anti-chicken secondary antibody (Kirkegaard & Perry Laboratories) diluted 1:100 in FACS buffer. Following three more washes with FACS buffer, fixed cells were observed under a fluorescence microscope, while live cells were resuspended in FACS buffer and were subjected to flow cytometry.
Immunoblotting.
Plasmid-transfected cell lysates were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under nonreducing conditions. Following electrophoretic separation, the gels were transferred to nylon membranes using the iBlot kit (Invitrogen) according to the manufacturer's instructions. The membranes were then blocked in PBS containing 0.5% Tween 20 (PBST) and 1% nonfat dry milk for 2 to 3 h at room temperature or overnight at 4°C. Membranes were washed in PBST and were incubated overnight at 4°C with the primary antibody, anti-NDV chicken serum (purchased from Kirkegaard & Perry Laboratories), diluted in PBST with 0.5% nonfat dry milk (1:2,000). Membranes were further washed and were incubated with the secondary antibody, an IRDye 800CW-conjugated donkey anti-chicken antibody (Li-Cor), diluted in PBST with 0.5% nonfat dry milk (1:15,000), for 1 h at room temperature. Membranes were washed extensively, and the bound antibody was detected using the Odyssey CLX infrared system.
Virus titration.
Supernatants from virus-infected DU145 cells were clarified at 3, 000 × g and were used as the virus stock. Virus titers were obtained by calculating the 50% tissue culture infective dose (TCID50) using the method of Reed and Muench as described elsewhere (30).
Mean death time.
Ten 10-day-old specific-pathogen-free (SPF) chicken embryos were each inoculated with five different dilutions of the test viruses in order to calculate the mean death time as described previously (31).
Growth kinetics.
Cells and spheres were seeded in 6-well plates at 5 × 105 cells per well and were infected with recombinant BC-KLQL-GFP at a multiplicity of infection (MOI) of 0.01, 0.1, 1, or 10 for multicycle growth studies. After virus adsorption for 1 h at 37°C, cells were washed with PBS to remove any unabsorbed virions, and serum-free medium containing PSA (100 ng/ml) or R1881 (1 nm/ml) was added. At various time points after infection, 100 μl of supernatants was removed, and the TCID50 was determined by infecting fresh DU145 cells.
RT-PCR and sequencing.
Undiluted BC-KLQL-GFP virus was serially propagated 10 times in WPE-int cells. To analyze the stability of the introduced F mutation, the F-KLQL sequence was confirmed by performing reverse transcription-PCR (RT-PCR) on infectious supernatants using NDV genome-specific primers spanning the mutated region.
Cell viability.
Cells and spheres were plated as five replicates in 6-well plates at 5 × 105 cells per well and were infected with recombinant BC-KLQL-GFP at MOIs of 0.01, 0.1, 1.0, and 10.0. Cells and spheres were trypsinized at 24, 48, 72, 96, and 120 h postinfection and were checked for viability using the trypan blue dye exclusion assay (26, 27). The viability of PSA- and R1881-treated, uninfected control cells was set at 100% for different time points.
Statistical analysis.
One-way analysis of variance and the Student t tests were performed using JMP software (version 9; SAS). The half-maximal effective concentrations (EC50s) were calculated using the dose response-versus-inhibitor analysis with four parameters in GraphPad Prism (version 5; GraphPad Software).
RESULTS AND DISCUSSION
We reported previously that the recombinant Beaudette C (rBC) strain of Newcastle disease virus (NDV) specifically kills human tumor cells while sparing normal cells in an interferon-independent manner (24). We showed that NDV kills tumor cells by intrinsic and extrinsic pathways of apoptosis (24). We have also shown that rBC is safe and inherently oncolytic in a preclinical mouse model. We demonstrated that a single dose of interferon-resistant or -sensitive rBC with wt F effectively eradicated the tumor burden in human fibrosarcoma xenografts in nude mice (32). Recently, we reported the absence of retinoic acid-inducible gene I (RIG-I), a cytosolic RNA sensor, in cells sensitive to NDV and significantly higher levels of proinflammatory cytokines and chemokines in infected normal cells than in tumor cells (33). To make the NDV F protein cleavable by PSA, we constructed several F protein cleavage site mutants that are potentially cleavable by PSA and one mutant (F-Null) that is not cleavable by any known protease, and we tested their abilities to be transported to the cell surface and to induce fusion.
NDV fusion protein mutants are transported to the cell surface.
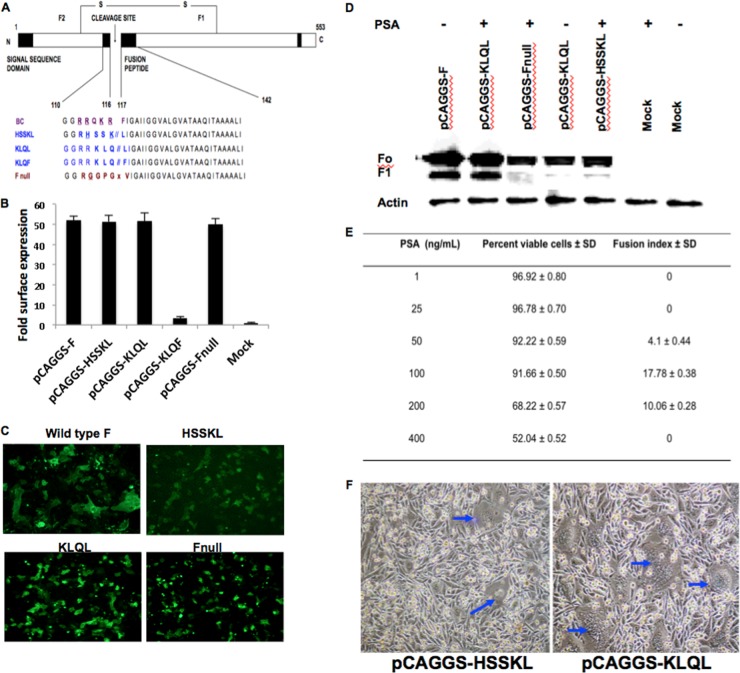
Proteins with the putative PSA-cleavable motif HSSKL, KLQL, or KLQF, or with the noncleavable F-Null mutant (Fig. 1A), were cloned into the expression vector pCAGGS. We screened these NDV fusion protein mutants in a plasmid system for their PSA specificities. Immunofluorescent staining was performed to analyze the cell surface expression of fusion protein mutants, and cell surface expression was quantified by flow cytometry. All fusion proteins, except the KLQF mutant, were transported to the cell surface. The cell surface expression of the HSSKL, KLQL, and F-Null mutants was within the same range as that of the wild-type (wt) F protein. The KLQF mutant, on the other hand, was undetectable (Fig. 1B). Coexpression of NDV wt F and hemagglutinin (HN) proteins in Vero cells resulted in multinucleated syncytia. The mutant proteins, however, were not fusogenic (Fig. 1C). This suggested that the mutant fusion proteins were functionally inactive and were not cleaved by ubiquitous proteases. The F proteins of virulent NDV strains with multibasic cleavage site amino acids are cleaved by ubiquitous subtilisin-like proteases, such as furin, PC6, and PACE 4, while nonpathogenic strains with monobasic cleavage sites are cleaved by trypsin-like enzymes found only in specific tissues (19, 34). The HSSKL mutant, with a monobasic cleavage site, was not cleavable by trypsin-like proteases, either (data not shown). Modification of the NDV F cleavage site to HSSKL or KLQL did not interfere with protein synthesis, maturation, or cell surface transport pathways. However, the presence of a phenylalanine after a glutamine at the cleavage site interfered with cell surface expression. Therefore, the KLQF mutant was excluded from further analysis.
Fig 1.
Characterization of fusion protein mutants by using cell-based assays. (A) Schematic of the NDV fusion protein and the PSA activation mutants. (B) Fold change in cell surface expression, measured quantitatively by flow cytometry. The percentages of fluorescent cells were determined by setting the gates using mock-transfected cells as a negative control and wild-type F as a positive control. (C) Immunofluorescent staining of Vero cells cotransfected with NDV F and HN plasmids, using an antibody against F protein (magnification, ×10). (D) Activation of mutant F proteins with an exogenous PSA overlay (100 ng/ml) (at 12, 24, and 48 h posttransfection) compared to the activation of wild-type F protein with no overlay. Lysates collected after 48 h posttransfection were used to detect the F1 peptide using a polyclonal anti-NDV chicken serum. (E) Determination of the optimal PSA overlay concentration. The exogenous PSA overlay concentration was calculated based on the percentage of cell viability and the fusion index in PC-3 cells. Cotransfection of expression plasmids (mutant pCAGGS-KLQL and wt pCAGGS-HN) in PC-3 cells, followed by an exogenous PSA overlay (1 ng/ml to 400 ng/ml), resulted in variable fusion indices and cell viabilities. Results are means ± standard deviations (SD) obtained from five independent experiments. (F) Syncytium formation in BSR-T7/5 cells 72 h posttransfection (blue arrows).
The KLQL mutant is activated by exogenous PSA overlay.
To further analyze the functional properties of mutant fusion proteins, the cell culture medium was supplemented with PSA (100 ng/ml) at 12, 24, and 48 h posttransfection. The addition of PSA resulted in efficient cleavage of the KLQL but not the HSSKL mutant (Fig. 1D). It was found that exogenous supplementation with PSA at a concentration of 100 ng/ml was well tolerated by the cells, and a mean fusion index of 17.78, similar to that of the wild-type F plasmid (data not shown), was observed for the KLQL mutant (Fig. 1E). The fusion index did not increase when PSA concentrations above 100 ng/ml were used. At higher doses, PSA was cytotoxic to cells, which could be due to the chymotrypsin-like protease activity of PSA on adherent cells. The HSSKL mutant F protein formed poor syncytia even after a PSA overlay, while the KLQL mutant was fusogenic (Fig. 1F). This result indicated that the KLQL mutant F protein was activated by PSA.
rNDV is replication restricted for PSA.
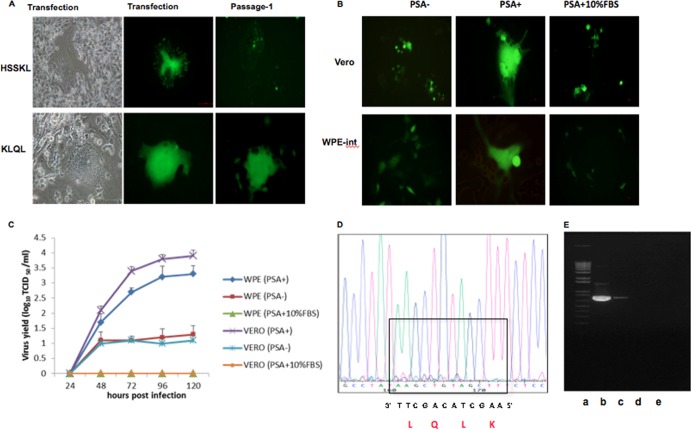
Having shown that the HSSKL and KLQL motifs are cleavable by PSA (Fig. 1D), we attempted to rescue NDV containing HSSKL or KLQL at the F protein cleavage site. Recombinant NDV (rNDV) was recovered using reverse genetics as described previously (29) with the addition of an exogenous overlay of PSA (100 ng/ml) for the 48-h period prior to transfection and after transfection. rNDV with the HSSKL motif was recoverable only by including a plasmid expressing wt NDV F. This rNDV failed to undergo multicycle replication in CaP cells (Fig. 2A) and therefore was excluded from further studies. The rNDV with the KLQL motif (BC-KLQL-GFP) was successfully recovered from BSR-T7/5 cells supplemented with exogenous PSA (Fig. 2A). BC-KLQL-GFP replicated only when PSA was added to the media of Vero and WPE-int cells (Fig. 2B).
Fig 2.
Specificity of recombinant NDV for PSA. (A) Virus rescue and sequential passage of the BC-HSSKL-GFP and BC-KLQL-GFP mutant viruses in BSR-T7/5 cells. Magnification, ×20. Note the absence of multicycle replication in the HSSKL virus. (B and C) Replication kinetics of the BC-KLQL-GFP virus (MOI, 1) in Vero and WPE-int cells with or without a PSA overlay or with an overlay of PSA supplemented with 10% FBS. (D) BC-KLQL-GFP was passaged serially in WPE-int cells. The sequence of the KLQL mutation in the virus was confirmed after 10 serial passages at an MOI of 1. (E) Reverse transcription-PCR gel confirming the absence of viral RNA in the allantoic fluid of BC-KLQL-GFP-infected specific-pathogen-free chicken embryos. Lanes: a, DNA marker; b, allantoic fluid from an embryo infected with Lasota-EGFP; c, cell culture supernatant infected with BC-KLQL-GFP; d and e, allantoic fluid from embryos infected with BC-KLQL-GFP.
To confirm that virus replication was dependent on active PSA, we inactivated PSA by the addition of 10% FBS. Human and animal sera are enriched with protease inhibitors, including serpins (serine protease inhibitors), which bind and inactivate proteases, including trypsin and chymotrypsin (35, 36). It is well known that the proteolytic activity of PSA is inhibited in the bloodstream by the formation of complexes with serine protease inhibitors such as α1-antichymotrypsin and protein C inhibitor (37, 38). The multicycle replication of BC-KLQL-GFP was severely impaired with the addition of FBS (Fig. 2B and C), indicating PSA specificity. Exogenous TPCK trypsin (0.5 μg/ml) or TLCK chymotrypsin (1 μg/ml) overlay did not support virus replication as well (data not shown). This further confirmed the PSA specificity of the recombinant BC-KLQL-GFP virus.
The PSA cleavage motif in the PSA mutant virus was stable, and mutant rNDV did not replicate in chicken embryos.
The BC-KLQL-GFP virus was serially passaged in AR+ WPE-int cells, and the PSA-cleavable KLQL mutation was confirmed by sequencing after 10 passages. A representative sequence trace is shown in Fig. 2D to indicate the presence of the introduced KLQL mutation. The AR+ WPE-int cells were specifically used for stability studies because they are androgen responsive, and intracellular PSA production could be induced with R1881 (a synthetic androgen analog) (39). A PSA enzyme-linked immunosorbent assay (ELISA) was used to detect increased levels of unbound PSA in the supernatants of R1881-treated WPE-int cells (data not shown). The mean death time following BC-KLQL-GFP infection of 10-day-old specific-pathogen-free (SPF) chicken embryos was >168 h, and no embryo death could be recorded. There was no hemagglutinating activity in the allantoic fluid of infected chicken embryos (data not shown). No viral RNA was detected in the allantoic fluid of infected SPF embryos by RT-PCR, confirming the absence of virus replication (Fig. 2E). These results indicate that BC-KLQL-GFP does not replicate in SPF chicken embryos and is likely to be nonpathogenic to chickens.
PSA-targeted rNDV replicates in CaP cells and prostaspheres.
To determine whether the PSA-dependent mutant would replicate in an in vitro mimic of CaP, we performed growth kinetics in CaP cells and prostaspheres. CaP cells were cultured in serum-free medium to form spheroids (14). These prostaspheres represent 3-D clusters of tumor cells that develop into fairly large multicellular aggregates. Spheroids contain different subpopulations of cells that can be quiescent, hypoxic, and necrotic, closely mimicking a tumor (40–42). Spheres have been shown to be promising in vitro models for testing various anticancer compounds, since they represent more natural cellular dynamics and architecture than 2-dimensional (2-D) monolayer cultures (43–46).
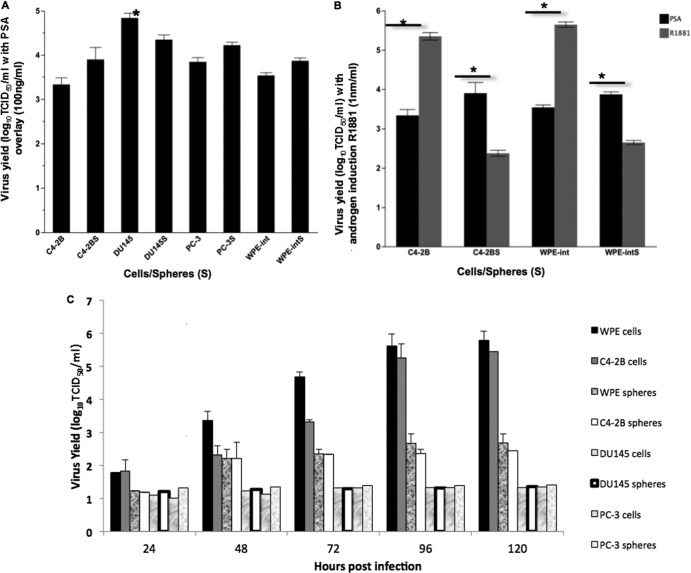
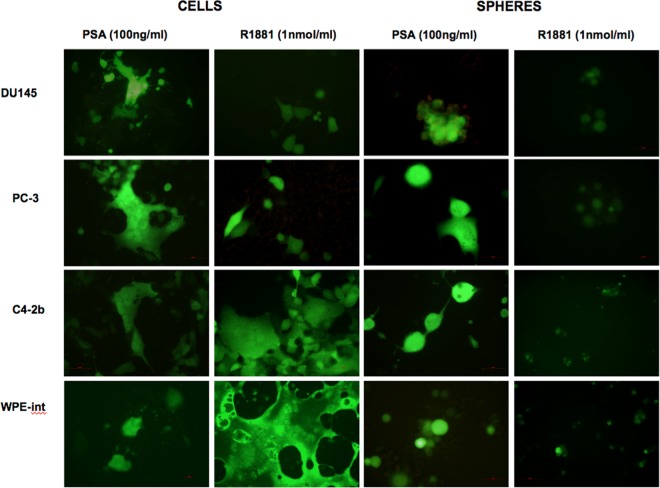
In the presence of exogenous PSA, the androgen-independent, AR− cell line DU145 was the most permissive cell type (Fig. 3A). As expected, virus yields in both AR+ and AR− cells and spheres were significantly higher at an MOI of 10 than at an MOI of 1 (data not shown). Interestingly, the induction of intracellular PSA by treatment with R1881 in androgen-responsive AR+ cells (WPE-int and C4-2B) resulted in viral titers significantly greater than those with a PSA overlay (Fig. 3B). Engaging the AR with the synthetic androgen analog R1881 induced intracellular PSA secretion (39) and thereby increased virus replication in AR+ WPE-int and C4-2B cells (MOI, 1) (Fig. 3C). We also confirmed PSA secretion by ELISA (data not shown). On the other hand, treatment of AR+ prostaspheres with R1881 led to disaggregation, low virus yields, and nonspecific cell death (Fig. 3C and 4). The remarkable fusogenicity of WPE-int and C4-2B androgen-responsive cells (Fig. 4) suggests the ability of the PSA-cleavable rNDV to spread efficiently. The fact that this mutant replicates efficiently in AR+ and AR− cells underscores its potential for oncolytic virotherapy of both hormone-sensitive and hormone-refractory CaP.
Fig 3.
Virus yields in prostate cancer cells and spheres. (A) The virus yield in DU145 cells was significantly higher than those in other cells. Spheres are designated DU145S, PC-3S, WPE-intS, and C4-2BS. (B) PSA induction using the synthetic androgen analog R1881 significantly increased virus replication in AR+ cells (C4-2B and WPE-int) and decreased virus yield in AR+ spheres (C4-2BS and WPE-intS), *, P < 0.0001; n = 8. (C) Multistep growth kinetics of BC-KLQL-GFP, at an MOI of 1, after PSA induction with R1881 (1 nm/ml).
Fig 4.
Fusogenicity of PSA-cleavable rNDV in CaP cells and spheres. AR+ and AR− CaP cells and spheres were infected with recombinant BC-KLQL-GFP virus at an MOI of 1, with or without R1881 induction, and fusogenicity was examined 72 h postinfection. Note the enhanced fusogenicity in androgen-responsive C4-2B and WPE-int cells after R1881 induction. Spheres disintegrate after treatment with R1881.
The PSA-cleavable rNDV is cytotoxic to CaP cells and prostaspheres.
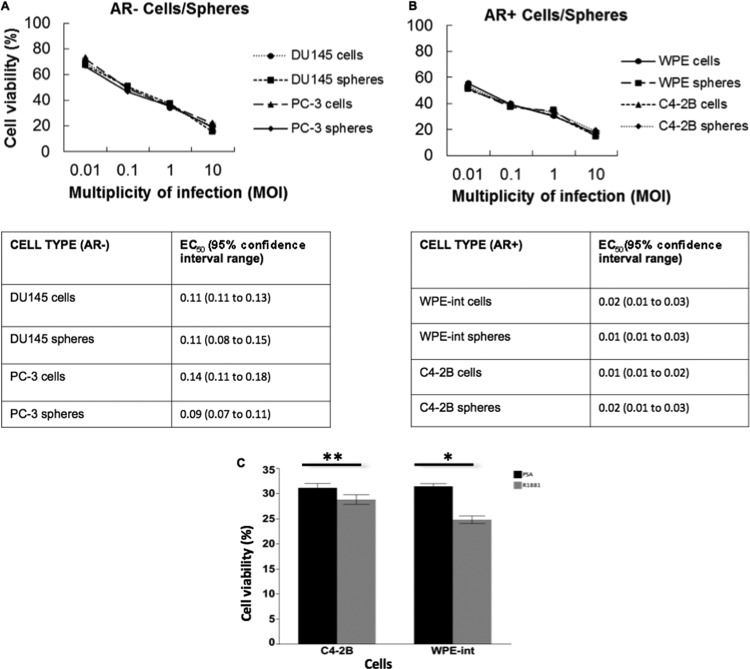
Cytotoxicity induced by rNDV was assessed by trypan blue staining of virus-infected CaP cells and spheres. The EC50 ranged from an MOI of 0.01 for AR+ cells/spheres (treated with R1881) to an MOI of 0.1 for AR− cells/spheres (supplemented with exogenous PSA) at 120 h postinfection (Fig. 5A and B). Cytotoxicity in AR+ WPE-int cells was significantly higher with R1881 treatment than with the exogenous PSA overlay, while C4-2B cells showed a less significant difference (Fig. 5C). Differential tumor selectivity may be due to differences in the cellular antiviral response to infection with NDV and in RIG-I expression (33). These results demonstrate that the PSA-cleavable rNDV will be activated only in the CaP tumor microenvironment, where free and active PSA is available, and will undergo selective multicycle replication and mediate cytotoxicity. We showed that intracellular PSA production induced by R1881 treatment enhanced virus spread, suggesting that intracellular F cleavage is important for enhanced fusion and oncolysis.
Fig 5.
Cytotoxicity of PSA-cleavable rNDV in CaP cells and spheres. Cell viability was assessed by the trypan blue dye exclusion assay at 120 h postinfection. The EC50 in AR+ and AR− cells/spheres was calculated by the sigmoidal dose response-versus-inhibitor analysis with four parameters, using GraphPad Prism, version 5 (GraphPad Software). (A) AR− cells and spheres with an exogenous PSA overlay (100 ng/ml). (B) AR+ cells (treated with R1881 at 1 ng/ml) and AR+ spheres (with PSA overlaid at 100 ng/ml). (C) AR+ cells (WPE-int and C4-2B) responded to PSA or R1881 supplementation. The viability of WPE-int cells treated with R1881 was significantly lower than that with the PSA overlay at 120 h postinfection. MOI, 1. *, P < 0.0001; **, P ≤ 0.0002; n = 6.
CaP is especially suited as a target for OVs because the prostate gland is easily accessible for inoculating viruses or for obtaining tissue samples. PSA, a serine protease, is abundant in the seminal plasma, and increased serum PSA levels are known to correlate with prostate tumor volumes (9–13). A major setback in using naturally occurring OVs for cancer therapy is off-targeting and failure to reach maximum tolerated doses. Since most of the proteolytically active PSA is present only in CaP (8–11), off-targeting will result in an abortive replication cycle with PSA-dependent rNDV. Although evidence exists to support PSA-targeted prodrugs (47–49), we have obtained “proof of principle” for the concept that a recombinant virus could be replication restricted for PSA. Tumor-specific targeting of rNDV can be combined with its inherent oncolytic and immunostimulatory properties, resulting in a therapeutic index enhanced by tumor-specific replication and efficient intratumoral spread. The inability of the PSA-cleavable mutant NDV to replicate in chicken embryos could be construed as a setback for virus stock production. However, our recent studies indicate that embryo-passaged NDV, but not human cell-grown NDV, was severely restricted by human complement (50). Human cell-grown NDV was protected against complement-mediated neutralization by the incorporation of host complement-regulatory proteins CD46 and CD55 on the viral envelope (50). Therefore, chicken embryos may not be suitable for clinical-grade virus production for human clinical trails. However, clinical-grade and complement-protected PSA-cleavable mutant virus stocks could be generated in WPE-int cells, and virus yield was higher after R1881 supplementation. During the course of our study, the mesogenic Beaudette C strain was classified as a “select agent” by the U.S. Department of Agriculture, with delayed regulatory approvals for performing preclinical tumor regression studies with rNDV in mouse models. Select agents are federally regulated agents that have potential use in biological warfare. Although we did not show efficacy in preclinical models, our virus replication and cytotoxicity results in a range of AR+ and AR− CaP cells and 3-D prostaspheres suggest that PSA-cleavable rNDV is a promising candidate for immediate preclinical trials in mouse models and phase I/II human clinical trials in hormone-sensitive and hormone-refractory CaP.
ACKNOWLEDGMENTS
This work was supported by the Department of Defense Congressionally Directed Medical Research Program Concept Award to S.E. (award PC060855).
Footnotes
Published ahead of print 23 January 2013
REFERENCES
- 1. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. 2009. Cancer statistics, 2009. CA Cancer J. Clin. 59:225–249 [DOI] [PubMed] [Google Scholar]
- 2. Cattaneo R, Miest T, Shashkova EV, Barry MA. 2008. Reprogrammed viruses as cancer therapeutics: targeted, armed and shielded. Nat. Rev. Microbiol. 6:529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guo ZS, Thorne SH, Bartlett DL. 2008. Oncolytic virotherapy: molecular targets in tumor-selective replication and carrier cell-mediated delivery of oncolytic viruses. Biochim. Biophys. Acta 1785:217–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Freeman AI, Zakay-Rones Z, Gomori JM, Linetsky E, Rasooly L, Greenbaum E, Rozenman-Yair S, Panet A, Libson E, Irving CS, Galun E, Siegal T. 2006. Phase I/II trial of intravenous NDV-HUJ oncolytic virus in recurrent glioblastoma multiforme. Mol. Ther. 13:221–228 [DOI] [PubMed] [Google Scholar]
- 5. Lam HY, Yeap SK, Rasoli M, Omar AR, Yusoff K, Suraini AA, Alitheen NB. 2011. Safety and clinical usage of Newcastle disease virus in cancer therapy. J. Biomed. Biotechnol. 2011:718710 doi:10.1155/2011/718710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lorence RM, Roberts MS, O'Neil JD, Groene WS, Miller JA, Mueller SN, Bamat MK. 2007. Phase 1 clinical experience using intravenous administration of PV701, an oncolytic Newcastle disease virus. Curr. Cancer Drug Targets 7:157–167 [DOI] [PubMed] [Google Scholar]
- 7. Sergel-Germano T, McQuain C, Morrison T. 1994. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J. Virol. 68:7654–7658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Akiyama K, Nakamura T, Iwanaga S, Hara M. 1987. The chymotrypsin-like activity of human prostate-specific antigen, gamma-seminoprotein. FEBS Lett. 225:168–172 [DOI] [PubMed] [Google Scholar]
- 9. Christensson A, Laurell CB, Lilja H. 1990. Enzymatic activity of prostate-specific antigen and its reactions with extracellular serine proteinase inhibitors. Eur. J. Biochem. 194:755–763 [DOI] [PubMed] [Google Scholar]
- 10. Lilja H, Christensson A, Dahlen U, Matikainen MT, Nilsson O, Pettersson K, Lovgren T. 1991. Prostate-specific antigen in serum occurs predominantly in complex with α1-antichymotrypsin. Clin. Chem. 37:1618–1625 [PubMed] [Google Scholar]
- 11. Otto A, Bar J, Birkenmeier G. 1998. Prostate-specific antigen forms complexes with human α2-macroglobulin and binds to the α2-macroglobulin receptor/LDL receptor-related protein. J. Urol. 159:297–303 [DOI] [PubMed] [Google Scholar]
- 12. Lilja H, Abrahamsson PA, Lundwall A. 1989. Semenogelin, the predominant protein in human semen. Primary structure and identification of closely related proteins in the male accessory sex glands and on the spermatozoa. J. Biol. Chem. 264:1894–1900 [PubMed] [Google Scholar]
- 13. Weir EG, Partin AW, Epstein JI. 2000. Correlation of serum prostate specific antigen and quantitative immunohistochemistry. J. Urol. 163:1739–1742 [PubMed] [Google Scholar]
- 14. McCormack RT, Rittenhouse HG, Finlay JA, Sokoloff RL, Wang TJ, Wolfert RL, Lilja H, Oesterling JE. 1995. Molecular forms of prostate-specific antigen and the human kallikrein gene family: a new era. Urology 45:729–744 [DOI] [PubMed] [Google Scholar]
- 15. Denmeade SR, Sokoll LJ, Chan DW, Khan SR, Isaacs JT. 2001. Concentration of enzymatically active prostate-specific antigen (PSA) in the extracellular fluid of primary human prostate cancers and human prostate cancer xenograft models. Prostate 48:1–6 [DOI] [PubMed] [Google Scholar]
- 16. Vukmirovic-Popovic S, Escott NG, Duivenvoorden WC. 2008. Presence and enzymatic activity of prostate-specific antigen in archival prostate cancer samples. Oncol. Rep. 20:897–903 [PubMed] [Google Scholar]
- 17. Kinoh H, Inoue M. 2008. New cancer therapy using genetically-engineered oncolytic Sendai virus vector. Front. Biosci. 13:2327–2334 [DOI] [PubMed] [Google Scholar]
- 18. Springfeld C, von Messling V, Frenzke M, Ungerechts G, Buchholz CJ, Cattaneo R. 2006. Oncolytic efficacy and enhanced safety of measles virus activated by tumor-secreted matrix metalloproteinases. Cancer Res. 66:7694–7700 [DOI] [PubMed] [Google Scholar]
- 19. Collins MS, Bashiruddin JB, Alexander DJ. 1993. Deduced amino acid sequences at the fusion protein cleavage site of Newcastle disease viruses showing variation in antigenicity and pathogenicity. Arch. Virol. 128:363–370 [DOI] [PubMed] [Google Scholar]
- 20. Denmeade SR, Lou W, Lovgren J, Malm J, Lilja H, Isaacs JT. 1997. Specific and efficient peptide substrates for assaying the proteolytic activity of prostate-specific antigen. Cancer Res. 57:4924–4930 [PMC free article] [PubMed] [Google Scholar]
- 21. Singh P, LeBeau AM, Lilja H, Denmeade SR, Isaacs JT. 2009. Molecular insights into substrate specificity of prostate specific antigen through structural modeling. Proteins 77:984–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morrison T, McQuain C, Sergel T, McGinnes L, Reitter J. 1993. The role of the amino terminus of F1 of the Newcastle disease virus fusion protein in cleavage and fusion. Virology 193:997–1000 [DOI] [PubMed] [Google Scholar]
- 23. Buchholz UJ, Finke S, Conzelmann KK. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Elankumaran S, Rockemann D, Samal SK. 2006. Newcastle disease virus exerts oncolysis by both intrinsic and extrinsic caspase-dependent pathways of cell death. J. Virol. 80:7522–7534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kohn A. 1965. Polykaryocytosis induced by Newcastle disease virus in monolayers of animal cells. Virology 26:228–245 [DOI] [PubMed] [Google Scholar]
- 26. Altman SA, Randers L, Rao G. 1993. Comparison of trypan blue dye exclusion and fluorometric assays for mammalian cell viability determinations. Biotechnol. Prog. 9:671–674 [DOI] [PubMed] [Google Scholar]
- 27. Tennant JR. 1964. Evaluation of the trypan blue technique for determination of cell viability. Transplantation 2:685–694 [DOI] [PubMed] [Google Scholar]
- 28. Huang Z, Panda A, Elankumaran S, Govindarajan D, Rockemann DD, Samal SK. 2004. The hemagglutinin-neuraminidase protein of Newcastle disease virus determines tropism and virulence. J. Virol. 78:4176–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krishnamurthy S, Huang Z, Samal SK. 2000. Recovery of a virulent strain of Newcastle disease virus from cloned cDNA: expression of a foreign gene results in growth retardation and attenuation. Virology 278:168–182 [DOI] [PubMed] [Google Scholar]
- 30. Flint SJ, Enquist LW, Racaniello VR, Skalka AM. 2009. Principles of virology: molecular biology, pathogenesis, and control of animal viruses, 3rd ed, p 32–37 ASM Press, Washington, DC. [Google Scholar]
- 31. Alexander DJ. 1988. Newcastle disease diagnosis. In Alexander DJ. (ed), Newcastle disease. Kluwer Academic Publishers, Boston, MA [Google Scholar]
- 32. Elankumaran S, Chavan V, Qiao D, Shobana R, Moorkanat G, Biswas M, Samal SK. 2010. Type I interferon-sensitive recombinant Newcastle disease virus for oncolytic virotherapy. J. Virol. 84:3835–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Biswas M, Kumar SR, Allen A, Yong W, Nimmanapalli R, Samal SK, Elankumaran S. 2012. Cell-type-specific innate immune response to oncolytic Newcastle disease virus. Viral Immunol. 25:268–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Leeuw OS, Koch G, Hartog L, Ravenshorst N, Peeters BP. 2005. Virulence of Newcastle disease virus is determined by the cleavage site of the fusion protein and by both the stem region and globular head of the haemagglutinin-neuraminidase protein. J. Gen. Virol. 86:1759–1769 [DOI] [PubMed] [Google Scholar]
- 35. Goettig P, Magdolen V, Brandstetter H. 2010. Natural and synthetic inhibitors of kallikrein-related peptidases (KLKs). Biochimie 92:1546–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Neri A, Bohoslawec O, Anderson TD, Tokes ZA. 1991. Differential release of active proteinase inhibitors by two rat mammary adenocarcinoma variants possessing different metastatic potentials. Cancer Res. 51:1318–1325 [PubMed] [Google Scholar]
- 37. Christensson A, Lilja H. 1994. Complex formation between protein C inhibitor and prostate-specific antigen in vitro and in human semen. Eur. J. Biochem. 220:45–53 [DOI] [PubMed] [Google Scholar]
- 38. Huber PR, Mattarelli G, Strittmatter B, van Steenbrugge GJ, Schmid HP, Maurer A. 1995. In vivo and in vitro complex formation of prostate specific antigen with α1-anti-chymotrypsin. Prostate 27:166–175 [DOI] [PubMed] [Google Scholar]
- 39. Bauer JA, Thompson TA, Church DR, Ariazi EA, Wilding G. 2003. Growth inhibition and differentiation in human prostate carcinoma cells induced by the vitamin D analog 1α,24-dihydroxyvitamin D2. Prostate 55:159–167 [DOI] [PubMed] [Google Scholar]
- 40. Freyer JP, Sutherland RM. 1980. Selective dissociation and characterization of cells from different regions of multicell tumor spheroids. Cancer Res. 40:3956–3965 [PubMed] [Google Scholar]
- 41. Kostarelos K, Emfietzoglou D, Papakostas A, Yang WH, Ballangrud A, Sgouros G. 2004. Binding and interstitial penetration of liposomes within avascular tumor spheroids. Int. J. Cancer 112:713–721 [DOI] [PubMed] [Google Scholar]
- 42. Sutherland RM. 1988. Cell and environment interactions in tumor microregions: the multicell spheroid model. Science 240:177–184 [DOI] [PubMed] [Google Scholar]
- 43. Han J, Chang H, Giricz O, Lee GY, Baehner FL, Gray JW, Bissell MJ, Kenny PA, Parvin B. 2010. Molecular predictors of 3D morphogenesis by breast cancer cell lines in 3D culture. PLoS Comput. Biol. 6:e1000684 doi:10.1371/journal.pcbi.1000684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Härmä V, Virtanen J, Mäkelä R, Happonen A, Mpindi JP, Knuuttila M, Kohonen P, Lötjönen J, Kallioniemi O, Nees M. 2010. A comprehensive panel of three-dimensional models for studies of prostate cancer growth, invasion and drug responses. PLoS One 5:e10431 doi:10.1371/journal.pone.0010431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, Gray JW, Bissell MJ. 2007. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 1:84–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee GY, Kenny PA, Lee EH, Bissell MJ. 2007. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods 4:359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chandran SS, Nan A, Rosen DM, Ghandehari H, Denmeade SR. 2007. A prostate-specific antigen activated N-(2-hydroxypropyl) methacrylamide copolymer prodrug as dual-targeted therapy for prostate cancer. Mol. Cancer Ther. 6:2928–2937 [DOI] [PubMed] [Google Scholar]
- 48. Denmeade SR, Nagy A, Gao J, Lilja H, Schally AV, Isaacs JT. 1998. Enzymatic activation of a doxorubicin-peptide prodrug by prostate-specific antigen. Cancer Res. 58:2537–2540 [PubMed] [Google Scholar]
- 49. Khan SR, Denmeade SR. 2000. In vivo activity of a PSA-activated doxorubicin prodrug against PSA-producing human prostate cancer xenografts. Prostate 45:80–83 [DOI] [PubMed] [Google Scholar]
- 50. Biswas M, Johnson JB, Kumar SR, Parks GD, Elankumaran S. 2012. Incorporation of host complement regulatory proteins into Newcastle disease virus enhances complement evasion. J. Virol. 86:12708–12716 [DOI] [PMC free article] [PubMed] [Google Scholar]