Abstract
The minichromosome maintenance protein homologs MCM8 and MCM9 have previously been implicated in DNA replication elongation and prereplication complex (pre-RC) formation, respectively. We found that MCM8 and MCM9 physically associate with each other and that MCM8 is required for the stability of MCM9 protein in mammalian cells. Depletion of MCM8 or MCM9 in human cancer cells or the loss of function MCM9 mutation in mouse embryo fibroblasts sensitizes cells to the DNA interstrand cross-linking (ICL) agent cisplatin. Consistent with a role in the repair of ICLs by homologous recombination (HR), knockdown of MCM8 or MCM9 significantly reduces HR repair efficiency. Chromatin immunoprecipitation analysis using human DR-GFP cells or Xenopus egg extract demonstrated that MCM8 and MCM9 proteins are rapidly recruited to DNA damage sites and promote RAD51 recruitment. Thus, these two metazoan-specific MCM homologs are new components of HR and may represent novel targets for treating cancer in combination with DNA cross-linking agents.
INTRODUCTION
Homologous recombination (HR) is critical for the repair of DNA damage induced by endogenous or exogenous agents. For example, ionizing radiation or the DNA-damaging agent doxorubicin induces double-stranded DNA breaks (DSBs) that are repaired by HR and nonhomologous end joining (NHEJ) (1). On the other hand, the repair of DNA interstrand cross-links (ICLs), induced by cisplatin, or by natural cellular metabolites such as lipid peroxides (2) usually occurs during the S phase of the cell cycle of proliferating cells (3, 4) and is dependent on translesion DNA synthesis (TLS), followed by HR (4–6). Several proteins, including the Fanconi anemia-related proteins (3, 7) and structure-specific endonucleases such as FAN1 (8–10), MUS81-EME1 (11), XPF-ERCC1 (12), and SLX1-SLX4 (13), as well as translesion DNA polymerases (4, 6) and HR-related proteins (5), are essential for the repair of ICLs.
HR consists of three steps: presynapsis, synapsis, and postsynapsis (1). During presynapsis, the MRN complex (MRE11, RAD50, and NBS1) interacts with CtIP (14, 15) and recognizes DNA breaks to make a short 3′ overhang structure. In yeast, the SGS1-DNA2 helicase/nuclease complex, as well as exonuclease 1 (EXO1), further resect the DNA ends to produce extended 3′ overhangs (16). Protein-protein interaction studies and in vitro assays suggest that the human BLM helicase may function as an ortholog of SGS1 (17), but it remains unclear whether BLM promotes the DNA resection step in vivo. After DNA resection, the RPA single-stranded DNA-binding protein is recruited to single-stranded DNA to stabilize the structure, and mediator proteins, including RAD51C, RAD52, and BRCA2, promote the formation of a RAD51 filament. RAD51, a key element in HR, binds single-stranded DNA at DNA breaks and facilitates the search for a homology donor (18). During the synapsis stage, strand invasion makes the D-loop, followed by RAD51 displacement, to promote DNA synthesis. Branch migration occurs during the postsynapsis stage, and the repair is completed via crossover or noncrossover pathways (1).
In addition to the helicases involved in DNA repair, the MCM2-MCM3-MCM4-MCM5-MCM6-MCM7 (MCM2-7) complex replicative helicase unwinds DNA prior to replication (19). Members of the MCM2-7 complex have an MCM helicase domain, which contains Walker A and Walker B motifs essential for ATP binding and hydrolysis, respectively. Two additional MCM family proteins, MCM8 and MCM9, have been reported, but their exact function remains controversial. MCM8 has significant homology with the human MCM7 (20) and was identified in a cancer-related gene screen using hepatitis B virus-based DNA tagging (21, 22). Human MCM8 has been implicated as a component of prereplication complexes (pre-RC) and was shown to interact with CDC6 protein (23), an ATPase important for the licensing step of DNA replication. Another study, however, did not find a role for Xenopus MCM8 in licensing but showed that it has helicase activity and plays a significant role during the elongation phase of DNA replication (24). In contrast to these studies, mutants of the Drosophila MCM8 homolog, REC, do not exhibit defects in S phase but have meiotic crossover defects, suggesting a role for MCM8 in meiosis but not in DNA replication (25). Knockdown of MCM8 in Drosophila S2 cells showed a 30% reduction in the number of replication forks but no effect on the cell cycle or viability, arguing that the protein was not as critical for DNA replication as the MCM2-7 complex (26).
The MCM9 protein, another helicase with significant homology with MCM8 (27, 28), is suggested to be another pre-RC component that interacts with CDT1 and is essential for MCM2-7 loading onto replication origins in Xenopus egg extract (29). Mice with homozygous deletions of Mcm9, however, are viable, ruling out an essential function for mammalian MCM9 in cell cycle progression or DNA replication (30). Interestingly, Mcm9-null mice exhibit deficits in the germ cell lineage, suggesting that MCM9 may play a role in meiosis. Therefore, although early studies suggest an essential role of these two helicases in DNA replication, the conclusions remain controversial.
Interestingly, Mcm8 is present only in Drosophila or higher eukaryotes, and Mcm9 is either absent (Drosophila) or present only in organisms with Mcm8 (31), suggesting that these two proteins may function together.
In this study, we demonstrate that human MCM8 and MCM9 proteins form a stable complex. Mammalian cells depleted of MCM8 and MCM9 or Mcm9-null mouse embryo fibroblast (MEF) cells are hypersensitive to the ICL-inducing agent cisplatin and exhibit defects in HR. In addition, MCM9 protein forms foci upon cisplatin treatment and MCM8 and MCM9 are recruited to DNA damage sites. Using immunoprecipitation, immunofluorescence, and chromatin immunoprecipitation (ChIP) assays in mammalian cells and in Xenopus egg extracts, we demonstrate that the MCM8-9 complex has a novel function in the recruitment of RAD51 to sites of DNA damage during HR.
MATERIALS AND METHODS
Cell culture and siRNA transfection.
U2OS, HeLa DR13-9, and 293T cells were grown in Dulbecco modified Eagle medium (DMEM; Cellgro) with 10% donor calf serum (Sigma-Aldrich) and penicillin/streptomycin (Cellgro). Mcm9-null MEF cells (30) were grown in DMEM with 10% fetal bovine serum (Gibco), 1 mM sodium pyruvate (Gibco), and penicillin-streptomycin and transformed using simian virus 40 T antigen. The small interfering RNA (siRNA) sequences are as follows: siMCM8-ORF, 5′-AGAAGACGCUGAGGAUAUA-3′; siMCM8-UTR, 5′-CAUAUCAGAUGUAGGCAUA-3′; siMCM9, 5′-GAUGACUUAGUGGAUAGUU-3′; siMCM6, 5′-GAAGAGGACGAGTCAGCAT-3′; siRAD51, 5′-GAGCUUGACAAACUACUUC-3′; and siBRCA2, 5′-GAAGAAUGCAGGUUUAAUA-3′. siRNAs were purchased from Invitrogen and transfected using Lipofectamine RNAi MAX (Invitrogen) according to the manufacturer's protocol.
Nuclear extract preparation.
293T cells were collected and resuspended in hypotonic lysis buffer (5 mM PIPES [pH 8], 85 mM KCl, 0.5% NP-40, and protease inhibitor), incubated for 10 min on ice, and homogenized by 10 strokes of the Dounce homogenizer. Cells were spun down for 10 min at 2,500 rpm at 4°C, and the supernatant (cytoplasmic extract) was collected. The pelleted nuclei were resuspended in NP-40 lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1% NP-40, 50 mM NaF, 1 mM Na3VO4, 5 mM EDTA, and protease inhibitor) and incubated on ice for 30 min. Nuclear extract was collected after the debris was cleared by centrifugation for 30 min at 13,000 rpm at 4°C.
Cell lysis and antibodies.
For immunoblotting, cells were lysed using radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate [SDS], 50 mM NaF, 1 mM Na3VO4, 1 mM EDTA, 1% deoxycholate, and protease inhibitor cocktail [Roche]). For coimmunoprecipitation assay, cells were lysed using modified cytoskeleton buffer as described previously (32). Anti-MCM8 (for Western blotting) and anti-MCM9 (for Western blotting and immunoprecipitation) antibodies were raised in rabbits against the N-terminal (MCM8) or C-terminal (MCM9) 100 amino acids of each protein. For MCM8 immunoprecipitation and ChIP experiments, anti-MCM8 antibody from Novus Biologicals (NB100-325) was used. Anti-β-actin, anti-α-tubulin, anti-RAD51 (Santa Cruz), anti-lamin A/C (Cell Signaling), anti-ORC2 (BD Pharmingen), and anti-RPA1, anti-BRCA2 (Calbiochem) were used for immunoblotting, immunofluorescence, and ChIP assays.
Thymidine incorporation assay.
siRNA oligonucleotides were transfected in U2OS cells in 10-cm plates, and after 6 h the cells were seeded in 12-well plates. At 10 min before harvest, the cells were pulsed with [methyl-3H]thymidine (2 μCi/ml). Cells were incubated with [2-14C]thymidine (0.02 μCi/ml) (Perkin-Elmer) for the preceding 24 h as a normalizing label, and the ratio of 3H to 14C incorporated into the cells gave a measure of the DNA synthesis at 24 h. The 10-min [methyl-3H]thymidine labeling was similarly done at 48 h posttransfection. Wild-type and Mcm9-null MEF cells were treated similar to U2OS cells. Cells were washed twice with ice-cold phosphate-buffered saline (PBS), incubated in stop solution (0.2 M sodium pyrophosphate, 10% [vol/vol] trichloroacetic acid [TCA]) for 20 min, washed twice with 95% ethanol, and subsequently solubilized with 1% SDS and 10 mM NaOH for 10 min at room temperature. Solubilized cells were absorbed in GF-A filters (24 mm; Whatman) and dried. The incorporated [methyl-3H]thymidine and [2-14C]thymidine were then measured using a liquid scintillation counter (Beckman).
Cell survival assay.
Cell survival after DNA damage was measured in the short term by MTT assay and in the long term by clonogenic assay. Indicated siRNAs were transfected in U2OS cells and after 24 h, various amount of cisplatin (Sigma-Aldrich) was added for 24 h. MEF cells were also treated with cisplatin or bleomycin (Selleckchem.com) for 24 h. Cell survival was subsequently measured by MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay according to the manufacturer's instruction (Promega, CellTiter96 nonradioactive cell proliferation assay). For the clonogenic assay, Mcm9 wild-type or null MEF cells were plated and treated with agents that cause DNA damage: cisplatin at the indicated concentration for 7 days, UV at the indicated J/m2, and doxorubicin (Sigma-Aldrich) at the indicated concentration for 1 h. At 7 days after initializing DNA damage, the colonies were stained with crystal violet and quantitated using Gene Tools software (Syngene).
In vivo homologous recombination assay.
HR assay was performed as described previously (33). Briefly, we transfected siRNA duplexes to HeLa DR13-9 cells, and 24 h later, transfected pCβA-SceI plasmid DNA. After another 48 h, the cells were collected, and green fluorescent protein (GFP)-expressing cells were counted using flow cytometry (BD FACSCalibur). Quantitation of GFP-positive cells was performed using FlowJo program (Tree Star, Inc.).
ChIP experiment at I-SceI cut site in cells.
We performed a ChIP assay on cellular chromatin as previously described (34), with some modifications. A total of 3 × 106 of HeLa DR13-9 cells were fixed with 1% formaldehyde for 10 min, followed by incubation with 0.125 M glycine for 5 min. The cells were lysed with SDS lysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA [pH 8.0], 0.1% SDS, and protease inhibitor cocktail [Roche]), incubated on ice for 20 min, and sonicated (30 s on and 30 s off, 6 times at an 11% amplitude using Sonic Dismembrator model 500 [Fisher Scientific]). Lysates were diluted in ChIP dilution buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Triton X-100, 0.1% SDS, and complete protease inhibitor cocktail [Roche]), and an equal volume of lysate was used for immunoprecipitation. Lysates were incubated with Dynabead-protein G (Invitrogen)-bound antibodies (anti-MCM8 [Novus Biologicals], anti-MCM9 raised against a C-terminal fragment of human MCM9 protein in rabbits, anti-RAD51 [Santa Cruz], and anti-BRCA2 [Calbiochem]) for 3 h. The beads were washed once with RIPA buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate) and once with 500 mM NaCl-RIPA buffer. Subsequently, the beads were washed once with LiCl wash buffer (10 mM Tris-HCl [pH 8.0], 0.25 M LiCl, 1 mM EDTA, 0.5% NP-40, 0.5% sodium deoxycholate) and twice with TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). The washed pellets were resuspended in ChIP elution buffer (10 mM Tris-HCl [pH 8.0], 300 mM NaCl, 5 mM EDTA, and 0.5% SDS), and incubated at 65°C for 6 h. DNA was recovered by regular ChIP protocol and analyzed by quantitative reverse transcription-PCR (qRT-PCR).
The primer sequences used for ChIP were as follows: F1, 5′-TACGGCAAGCTGACCCTGAA-3′; R1, 5′-CGTCCTCCTTGAAGTCGATG-3′; F2, 5′-GCCCATATATGGAGTTCCGC-3′; and R2, 5′-GGCCATTTACCGTCATTGAC-3′ (35). The F1-R1 primer-pair targets DNA right next to the I-SceI cut site, and the F2-R2 primer-pair targets DNA 2 kb upstream of the I-SceI cut site.
Immunofluorescence microscopy.
Cells were pre-extracted with 0.5% Triton X-100 for 1 min and fixed with 4% paraformaldehyde for 10 min. Antigens were detected with specific antibodies, and either Alexa Fluor 555–anti-rabbit (Invitrogen) or Alexa Fluor 488–anti-mouse (Invitrogen) IgG was used as a secondary antibody. Images were acquired using a Zeiss Axio observer A-1 equipped with a Zeiss LD Plan-Neofluar (×40/0.6Ph2CorrWD=3.3M27), an EC Plan-Apochromat (×63/1.4 oil), and a Zeiss AxioCam MRC. Acquired images were analyzed using Axiovision software.
Replication in Xenopus egg extracts.
Xenopus egg extracts (high-speed supernatant [HSS] and nucleoplasmic extract [NPE]) were prepared as described previously (36). Replication of pICL was performed as described previously (5) by first licensing DNA in HSS for 20 min, followed by the addition of NPE to initiate DNA replication. The reactions were supplemented with [α-32P]dATP for DNA labeling and an undamaged plasmid (pQuant) that serves as an internal standard for quantification. The preparation of pICL was described previously (4). Reaction products were purified as described previously (7), separated by 0.8% native agarose gel, and visualized using a phosphorimager. The replication efficiency was then calculated as described previously (5). For depletion of MCM9 from Xenopus egg extracts, protein A-Sepharose beads (GE Healthcare) were bound to anti-MCM9 serum at a 1:3 ratio. Extracts were incubated with antibody-bound beads at a 1:5 ratio for 20 min at 4°C for two rounds.
ChIP analyses in Xenopus egg extracts.
ChIP was performed as described previously (5). Briefly, reaction samples were cross-linked with formaldehyde, sonicated to yield DNA fragments roughly 300 to 500 bp in size, and immunoprecipitated with the indicated antibodies. Cross-links were then reversed and DNA was purified for analysis by quantitative real-time PCR with the following primers: ICL (5′-AGCCAGATTTTTCCTCCTCTC-3′ and 5′-CATGCATTGGTTCTGCACTT-3′), MID (5′-ACCCTGGGTTCTTTTCCAAC-3′ and 5′-CATTTCATCTGGAGCGTCCT-3′), FAR (5′-AACGCCAATAGGGACTTTCC-3′ and 5′-GGGCGTACTTGGCATATGAT-3′), and QNT (5′-TACAAATGTACGGCCAGCAA-3′ and 5′-GAGTATGAGGGAAGCGGTGA-3′).
RESULTS
MCM8 stabilizes MCM9, forming a stable nuclear complex.
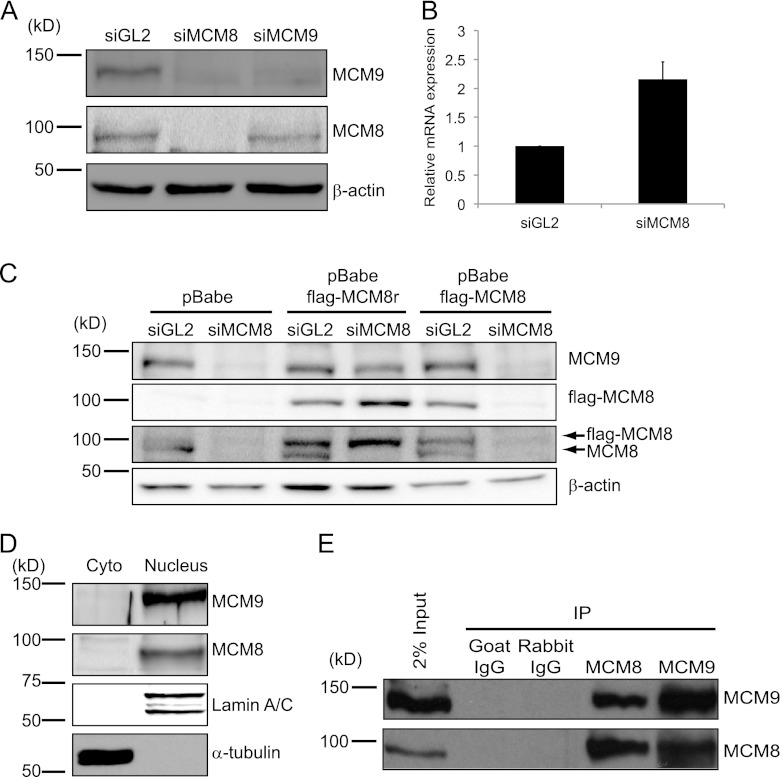
siRNA-mediated depletion of MCM8 from U2OS cells and several other human-derived cancer cells decreased MCM9 protein but not its mRNA (Fig. 1A and B and data not shown). On the other hand, siRNA-mediated depletion of MCM9 did not affect the steady-state level of MCM8 protein (Fig. 1A). To determine whether the reduction of MCM9 in MCM8-depleted cells was due to off-target activity of siMCM8, two U2OS cell lines were generated that stably express either wild-type (WT) Flag-tagged MCM8 or siRNA-resistant Flag-tagged MCM8 (Flag-MCM8r). siMCM8 decreased both endogenous MCM8 protein and ectopic Flag-MCM8 protein but did not decrease ectopic Flag-MCM8r protein (Fig. 1C). The Flag-MCM8r prevented the decrease of MCM9 protein after siMCM8 transfection, showing that the decrease of MCM9 was not due to an off-target activity of siMCM8. These results indicate that MCM8 is required for the stability of MCM9 protein and suggest that MCM9 coevolved with MCM8 to promote a related function.
Fig 1.
MCM8 interacts with MCM9 and forms a stable nuclear complex. (A) Immunoblots of lysates of U2OS cells transfected with siRNA targeting the luciferase gene (siGL2, control) or the Mcm8 or Mcm9 genes (siMCM8 [siMCM8-ORF] and siMCM9, respectively). β-Actin protein serves as a loading control. (B) qRT-PCR of Mcm9 mRNA using total RNA from U2OS cells after siRNA transfection, normalized to GAPDH mRNA. Means + the standard deviations (SD) of triplicate measurements are shown. (C) Ectopic expression of siMCM8-resistant MCM8 protein (MCM8r) restores MCM9 levels in U2OS cells depleted of endogenous MCM8 protein. Wild-type and N-terminally Flag-tagged MCM8 (flag-MCM8) or siRNA resistant Flag-MCM8 protein (flag-MCM8r) were stably expressed in U2OS cells and transfected with siRNA targeting endogenous MCM8 or ectopic Flag-MCM8 but not ectopic MCM8r. Lysates were harvested 48 h after transfection and analyzed by SDS-PAGE and Western blotted with antibodies against the indicated proteins. The β-actin blot shows equal protein loading. (D) MCM8 and MCM9 are nuclear proteins. Immunoblots of cytoplasmic and nuclear proteins from 293T cells show that MCM8 and MCM9 are nuclear proteins. α-Tubulin and lamin A/C were used as controls of fractionation, respectively. Cyto, cytoplasmic proteins; Nucleus, nuclear proteins. (E) MCM8 associates with MCM9. Immunoprecipitates of endogenous MCM8 or MCM9 from 293T nuclear lysates coimmunoprecipitate MCM9 or MCM8, respectively.
Nuclear and chromatin fractionation of 293T cell extracts indicated that MCM8 and MCM9 proteins localized mainly to the nucleus (Fig. 1D). Furthermore, endogenous MCM8 was readily detectable in the anti-MCM9 immunoprecipitates of 293T nuclear extracts and vice versa (Fig. 1E). Addition of the DNA intercalating chemical ethidium bromide (EtBr) did not disrupt the interaction (see Fig. S1 in the supplemental material), indicating that MCM8 and MCM9 proteins form a stable nuclear complex without binding to DNA.
MCM8-9 is not essential for DNA replication.
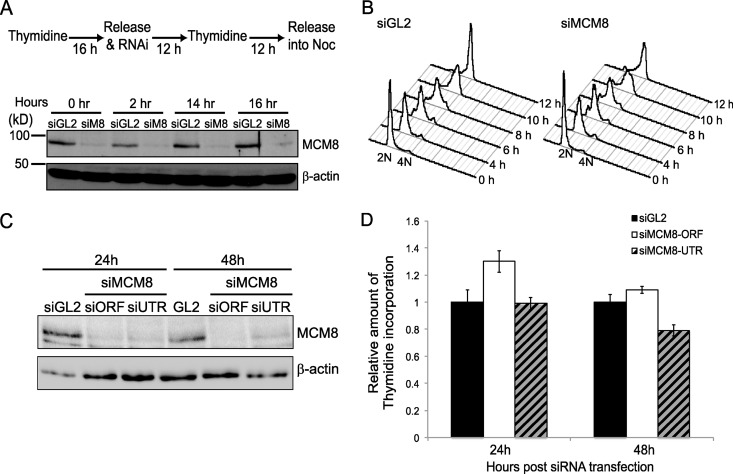
We next tested whether MCM8 or MCM9 is required for DNA replication. We investigated the requirement for MCM8 in S-phase progression by synchronizing cells at the G1/S transition using double thymidine block and monitoring their progression through the next S phase after release from the double thymidine block into nocodazole-containing medium (to arrest them in M phase) (Fig. 2A and B). MCM8 protein was effectively depleted by the time cells were released from the G1-S block (Fig. 2A). Despite this, there was no delay in S-phase progression in MCM8-depleted cells (Fig. 2B).
Fig 2.
MCM8-9 is not essential for DNA replication. (A) Schematic of the synchronization procedure used in U2OS cells. The extent of MCM8 protein knockdown by siRNA at each indicated time point following release from the double thymidine block was tested by SDS-PAGE and immunoblotting. β-Actin is shown as a loading control. siM8, siMCM8-UTR. (B) Propidium iodide fluorescence-activated cell sorting histograms are shown at the indicated hours after release from double thymidine block. (C) U2OS cells were harvested at the indicated time point after transfection of the indicated siRNAs. An immunoblot shows the amount of MCM8, and β-actin serves as a loading control. siORF, siMCM8-ORF; siUTR, siMCM8-UTR. (D) Thymidine incorporation assay in U2OS cells after knockdown by siMCM8-ORF or siMCM8-UTR. At the indicated times after siRNA transfection, cells were labeled with [methyl-3H]thymidine for 10 min. Cells were labeled with [2-14C]thymidine for the preceding 24 h to normalize for cell recovery. The histogram shows the ([methyl-3H]thymidine) uptake normalized to total DNA ([2-14C]thymidine) after transfection of siGL2 or two different siRNAs against MCM8. The results are expressed relative to that in control cells (siGL2). Means ± the SD of triplicates are shown. The result was confirmed by five independent experiments (data not shown) and shows that the depletion of MCM8 does not affect DNA synthesis at 24 and 48 h.
We also tested whether MCM8 depletion affected DNA replication by monitoring [methyl-3H]thymidine incorporation, a measure of DNA synthesis. MCM8 was depleted equally at 24 and 48 h using two different siRNA oligonucleotides (Fig. 2C), but DNA synthesis was comparable with control knockdown cells (siGL2) (Fig. 2D). This result suggests that the bulk of MCM8 is dispensable for DNA replication, S-phase progression, or cell proliferation. Because MCM8 knockdown also depletes MCM9 from cells, these results also rule out an essential function for the bulk of MCM9 in DNA replication. Knockdown of MCM9 alone or MCM9 and MCM8 together also did not inhibit DNA replication (Fig. 3D). Lack of an absolute requirement of the two proteins for DNA replication is consistent with the observation that homozygous deletion of Mcm9 gene still produces viable mice.
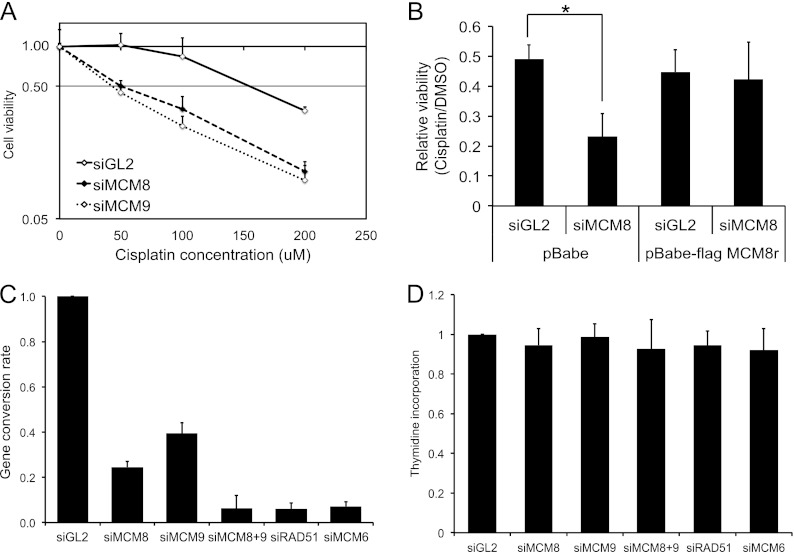
Fig 3.
Knockdown of MCM8 or MCM9 renders cells sensitive to cisplatin. (A) U2OS cells were transfected with the indicated siRNAs and treated for 24 h with the indicated doses of cisplatin 24 h posttransfection. Cell viability was measured by an MTT assay. The values represent means ± the SD from triplicates, and the result was confirmed by three independent experiments. (B) U2OS cells infected with empty vector (pBabe) or one expressing siRNA resistant Flag-MCM8 were transfected with siRNAs and treated with dimethyl sulfoxide (DMSO) or 200 μM cisplatin for 24 h. Cell viability was measured by an MTT assay. Values represent the means + the SD from triplicates. *, P = 0.008. (C) MCM8 and MCM9 are required for HR. An in vivo HR assay in HeLa DR13-9 after transfection of I-SceI expressing plasmid was performed. HR efficiency was calculated by subtracting the percentage of GFP-positive cells in the nontransfected control from that of the I-SceI-transfected experimental samples, followed by normalizing to the siGL2 treated cells. Means ± the SD from three independent experiments. (D) Thymidine incorporation assay in DR13-9 cells after knockdown by the indicated siRNAs. At 72 h after siRNA transfection, the cells were labeled with [methyl-3H]thymidine for 10 min. The cells were labeled with [2-14C]thymidine for the preceding 24 h to normalize for cell recovery. The histogram shows the ([methyl-3H]thymidine) uptake normalized to total DNA ([2-14C]thymidine) after transfection of the indicated siRNAs. The results are expressed relative to that in control cells (siGL2). Means + the SD of triplicates are shown. The result was confirmed by three independent experiments.
MCM8-9 complex is required for resistance to cisplatin and for HR.
Because Drosophila MCM8 has been shown to play a role in meiotic crossover (25), we hypothesized that the human MCM8-9 complex may be involved in homologous recombination (HR) repair. Because ICLs are mainly repaired in a process that involves HR, we treated control U2OS cells or cells depleted of MCM8-9 with cisplatin, which induces inter- and intrastrand cross-links (ICLs) and monitored the cells' viability by MTT assays. Cells depleted of MCM8 or MCM9 by siRNA transfection were significantly more sensitive to cisplatin than control (siGL2) cells (Fig. 3A). The enhanced cisplatin sensitivity of cells transfected with siMCM8 was lost in cells expressing MCM8r, which is resistant to siMCM8 (Fig. 3B). These results strengthened our hypothesis that MCM8-9 may be involved in HR repair.
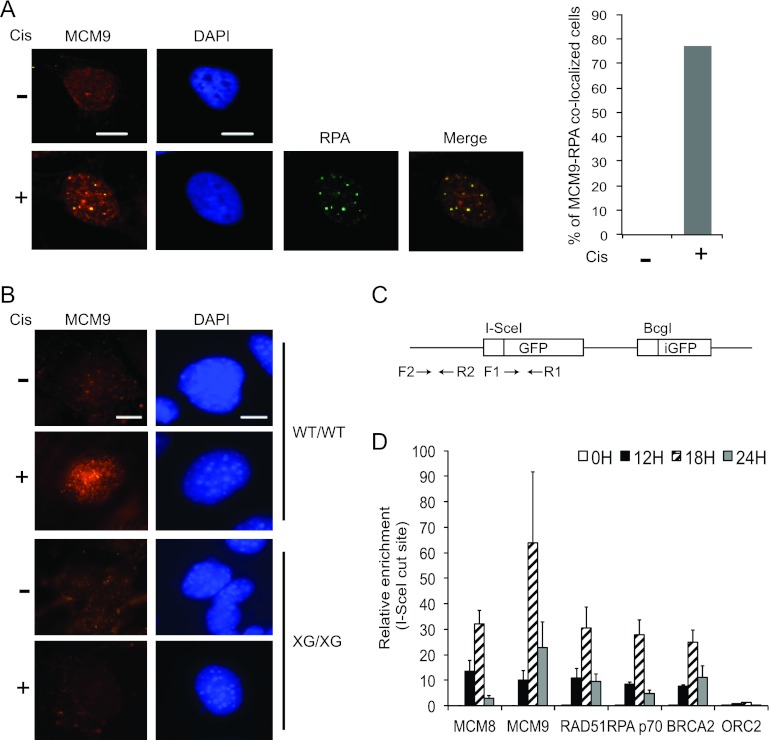
To test whether MCM8 and MCM9 are required for HR, we performed HR assays using HeLa DR13-9 cells, which have a pDR-GFP construct stably integrated in the genome (see Fig. 5C) (33). This cell line measures the homology-directed repair of a single DSB induced by I-SceI endonuclease to generate a functional GFP gene (37). In control (siGL2) cells, the expression of I-SceI generated 5 to 10% GFP-positive cells (data not shown). Consistent with its central role in HR, depletion of RAD51 inhibited HR to almost undetectable levels (Fig. 3C). Significantly, depletion of MCM8 or MCM9 by siRNA reduced the efficiency of HR repair to ca. 60 to 80% of control cells and codepletion of MCM8 and MCM9 almost completely shut down HR repair (Fig. 3C), without decreasing [methyl-3H]thymidine incorporation, a measure of DNA replication (Fig. 3D). siRNAs do not completely eliminate their targets, so it is not surprising that knockdown of MCM8 and MCM9 together has a greater effect on HR than either knockdown alone, even though the two proteins act at the same step on the pathway.
The role of MCM8-9 in HR is independent of any role as part of the replicative helicase.
Since MCM8-9 has been suggested to be part of the MCM2-7 replicative helicase, we wondered whether the requirement of MCM8-9 in HR was because they were necessary for optimal function of the replicative helicase. Indeed, depletion of MCM6 by siRNA inhibited the HR repair to levels comparable to that seen after depletion of RAD51 or MCM8-9 (Fig. 3C). The requirement of MCM6 for HR raised the possibility that MCM8-9 (like the MCM2-7 complex) is required for HR indirectly because it helps maintain the number or movement of replication forks that are necessary to sense DNA lesions and activate HR. Although knockdown of MCM8 and MCM9 did not decrease DNA replication (Fig. 3D), knockdown of MCM6 also, unfortunately, did not decrease [methyl-3H]thymidine incorporation, suggesting that siRNA experiments, being hypomorphic genetics, are unable to eliminate the role of a protein in DNA replication.
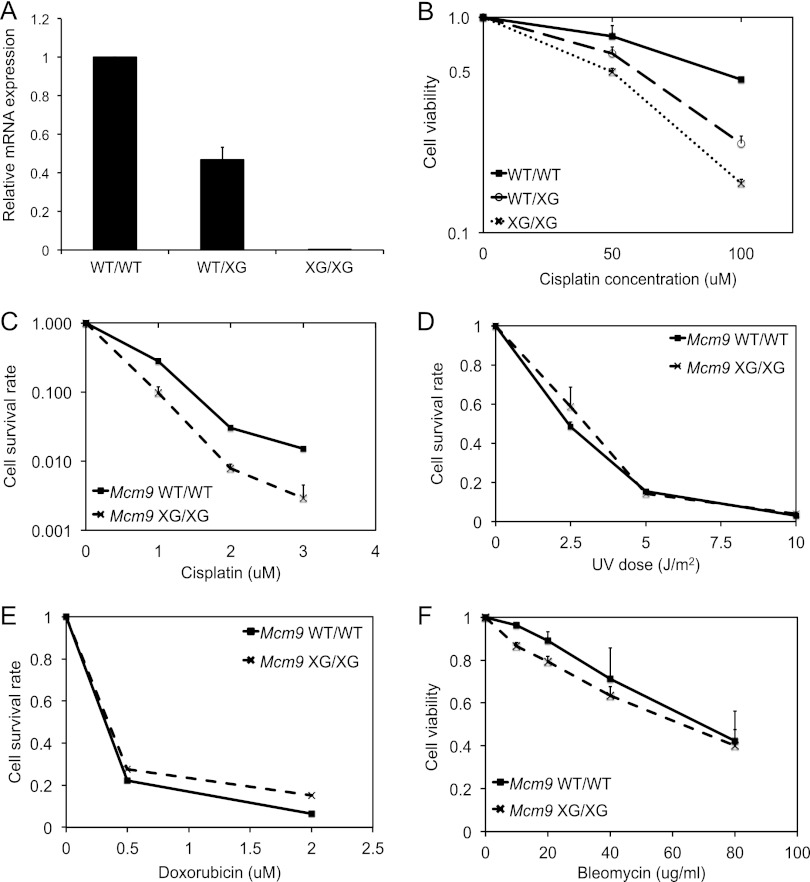
To determine whether MCM8-9 has a direct role in HR repair independent of a role as part of the replicative helicase complex, we turned to genetically engineered mice with null mutation of MCM9. Mcm6 knockout mice are nonviable early in embryogenesis, a finding consistent with the MCM2-7 complex being essential as the replicative helicase (38). In contrast, Mcm9-null mice are viable (30), which is consistent with MCM8-9 being not required for the function of the replicative helicase. We confirmed that MCM9 loss does not impair replication fork movement by examining the rate of DNA replication in mouse embryonic fibroblasts (MEFs). DNA synthesis rate in MEFs from Mcm9-null mice (XG743/XG743; hereafter, XG/XG) was unchanged compared to MEFs from Mcm9-wt mice (see Fig. S2A in the supplemental material). Mcm9-null MEFs (XG/XG) or heterozygous MEFs (WT/XG) were, however, hypersensitive to cisplatin both by the MTT assay for short-term viability (Fig. 4B) and by the colony formation assay for long-term viability (Fig. 4C). The sensitivity to cisplatin is correlated to the level of MCM9 mRNA expression because the heterozygous MEFs (WT/XG) had an intermediate level of expression between the WT and XG/XG MEFs (Fig. 4A) and an intermediate level of sensitivity to cisplatin (Fig. 4B). Thus, MCM9 (and, by extension, MCM8) are required for repair completely independent of any function as part of the replicative helicase.
Fig 4.
Mcm9-null MEF cells are sensitive to cisplatin but not UV, doxorubicin, or bleomycin. (A) qRT-PCR of Mcm9 mRNA using total RNA from MEF cells normalized to β-actin mRNA. Means ± the SD of triplicate measurements are shown. (B) MEFs with wild-type Mcm9 (WT/WT) or hetero- or homozygously mutated for Mcm9 (WT/XG or XG/XG, respectively) were treated with indicated doses of cisplatin for 24 h. Viability was measured by MTT assays. Values represent means ± the SD from triplicates. (C to E) Wild-type (WT/WT) or Mcm9-null (XG/XG) MEFs were plated and treated with the indicated doses of cisplatin (C), UV (D) or doxorubicin (E). Cisplatin was treated for 7 days, and doxorubicin was treated for 1 h and washed away. After 7 days, the cells were stained with crystal violet, and colonies were quantitated. Values represent means ± the SD from triplicates. (F) MEFs with wild-type Mcm9 (WT/WT) or homozygously truncated for Mcm9 (XG/XG) were treated with the indicated doses of bleomycin for 24 h. Viability was measured by MTT assays. Values represent means ± the SD from triplicates.
Mcm9-null MEF (XG/XG) were as resistant as wild-type MEFs (WT/WT) to UV-induced lesions, which do not require HR for repair (Fig. 4D). Although doxorubicin also produces DSBs by inhibiting DNA topoisomerase II, Mcm9 XG/XG cells were not hypersensitive to doxorubicin (Fig. 4E), most likely because DSBs induced by doxorubicin can be repaired by HR or NHEJ and so are not exclusively dependent on HR. Similarly, Mcm9 XG/XG cells were not hypersensitive to other DSB-inducing agents: bleomycin (Fig. 4F) or ionizing radiation (data not shown). Therefore, MCM8-9 is required specifically for the HR mode of DNA repair.
MCM8-9 is recruited to sites of homologous recombination repair.
To understand the function of MCM8-9 complex in HR repair, we first determined whether DNA damage affects the cellular distribution of MCM8 or MCM9 proteins. Upon cisplatin treatment, MCM9 protein formed multiple nuclear foci in both human cancer cells and MEFs (Fig. 5A and B) and colocalized with RPA1 foci (Fig. 5A). The absence of MCM9 foci in Mcm9-null MEFs (XG/XG) demonstrates the specificity of the anti-MCM9 immunofluorescence signal (Fig. 5B). Our anti-MCM8 antibodies, on the other hand, did not produce a specific signal, and thus we were not able to determine whether MCM8 was similarly redistributed after cisplatin treatment (data not shown).
Fig 5.
MCM8 and MCM9 localize to DNA breaks and are required for efficient HR. (A and B) MCM9 forms nuclear foci upon cisplatin treatment. U2OS cells or MEF cells (WT/WT and XG/XG) were treated with 20 μM of cisplatin for 16 h. Representative images are shown. Scale bar, 10 μm. The bar graph in panel A shows the percentage of cells that showed MCM9-RPA1 colocalization. (C) Schematic of substrate for HR assay (HeLa DR13-9) showing the primers used for the ChIP experiments relative to the I-SceI site. (D) ChIP results represent the relative enrichment of the indicated proteins at the DSB site (F1-R1) relative to distant site (F2-R2) and expressed as a ratio of immunoprecipitated DNA at different time points following the expression of I-SceI. Means ± the SD of triplicate measurements are shown, and the results were confirmed by three independent experiments.
Using I-SceI endonuclease to induce site-specific DSBs (Fig. 5C), we performed ChIP to test whether MCM8-9 proteins localized at the DSB site (Fig. 5D). Since RAD51 is a key player of HR, we first investigated the recruitment of RAD51 at DSBs. The I-SceI cut is extended by nucleases to generate a 3′ overhang structure that is expected to recruit more RAD51 to the F1-R1 site relative to the more distant F2-R2 site (Fig. 5C). RAD51 was recruited to the cut site within 12 h, with maximal accumulation at 18 h. RPA1 (p70) and BRCA2 also bind to the I-SceI cut site with the same kinetics as RAD51 protein. Consistent with the hypothesis that MCM8-9 plays a role in HR repair, both MCM8 and MCM9 proteins were recruited, with similar kinetics as RAD51, to the F1-R1 site. Another DNA-binding protein, ORC2, was not recruited to the I-SceI cut site. These results demonstrate that MCM8 and 9 accumulate at the DSB specifically at about the same time that RAD51 begins to be recruited.
MCM8-9 associates with RAD51 and promotes the recruitment of RAD51 to DNA break sites.
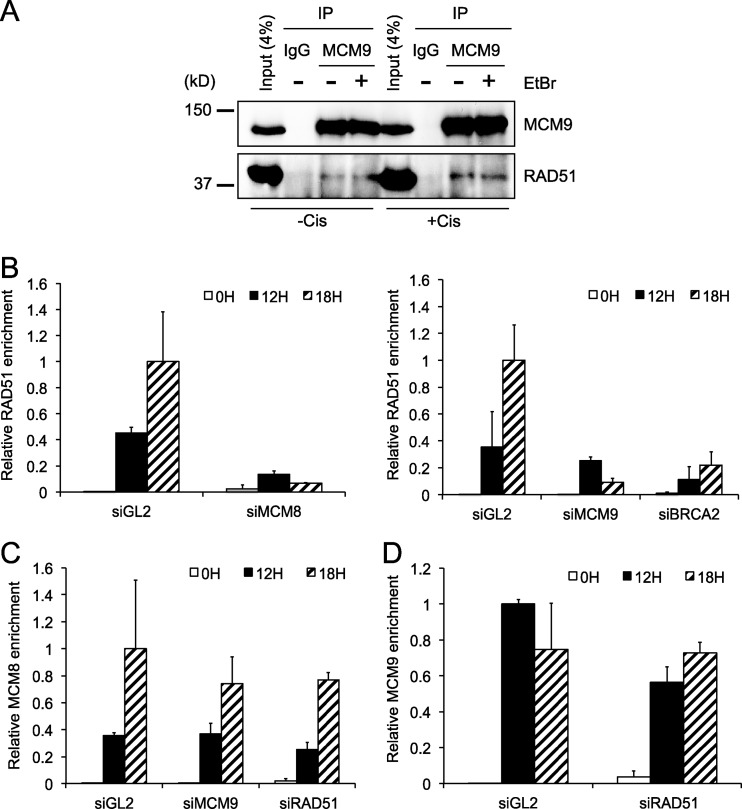
Since MCM8-9 and RAD51 are recruited at the DNA break site with similar kinetics, we next tested whether MCM8 or MCM9 interacts with RAD51. MCM8-9 immunoprecipitates from cells contain RAD51 and vice versa (Fig. 6A; also see Fig. S3 in the supplemental material). The interaction is not diminished by addition of EtBr, suggesting that the interaction is not due to the two DNA-binding proteins nonspecifically binding to DNA. Therefore, a small but significant fraction of endogenous MCM8-9 and RAD51 physically associate with each other.
Fig 6.
MCM8 and MCM9 associate with RAD51 and are required for RAD51 recruitment to damage sites. (A) MCM9 interacts with RAD51. Endogenous MCM9 protein is immunoprecipitated from U2OS cells and immunoblotted for RAD51 or MCM9 as indicated. EtBr was added to the immunoprecipitation (IP) reaction to exclude the possibility of DNA-dependent interaction. (B to D) ChIP results represent the relative enrichment of RAD51 (B), MCM8 (C), or MCM9 (D) at DSBs (F1-R1) relative to the distant site (F2-R2) at the indicated time points following the expression of I-SceI. We used either control cells or cells depleted of the indicated proteins by siRNA for 24 h. Values represent the means ± the SD of triplicate measurements.
To test whether MCM8-9 was required for RAD51 recruitment or vice versa, we analyzed the recruitment of RAD51, MCM8, or MCM9 to the I-SceI induced DSB in cells depleted of the individual proteins by siRNA. Figure 6B demonstrates that the depletion of MCM8 (which also decreases MCM9) or MCM9 alone reduced the recruitment of RAD51 to DSBs comparable to that seen upon BRCA2 depletion, without affecting the steady-state levels of RAD51 protein (see Fig. S4 in the supplemental material). On the other hand, the depletion of RAD51 did not affect the final amount of MCM8 (Fig. 6C) or MCM9 (Fig. 6D) recruited to the DSBs. Although required for RAD51 recruitment at DSBs, MCM9 was not required for MCM8 localization at DSBs (Fig. 6C). Because depletion of MCM8 destabilizes MCM9 (Fig. 1A), we could not test whether MCM8 alone was required for the recruitment of MCM9 to DSBs. Nevertheless, our results indicate that MCM8 and MCM9 are recruited to DNA DSBs independent of RAD51 but that MCM8-9 plays an important role in the recruitment of RAD51 at DSBs.
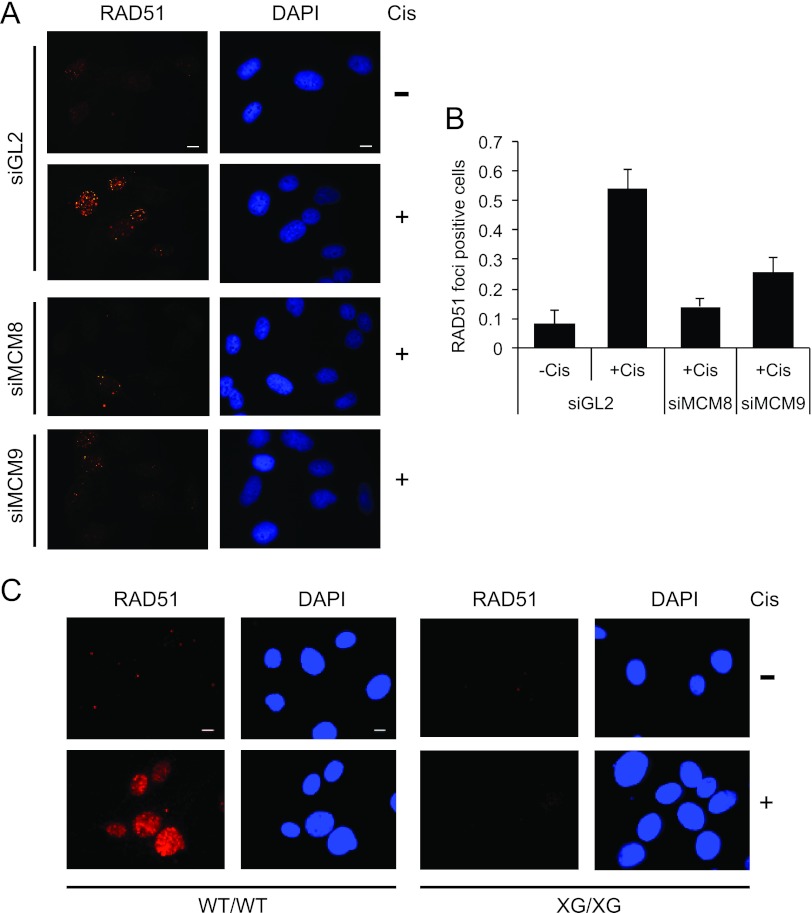
Cisplatin-induced DNA damage results in the recruitment of RAD51 to DNA damage sites, producing new RAD51 foci detectable by immunofluorescence. In control siGL2 transfected cells, RAD51 foci were formed upon cisplatin treatment (Fig. 7A and B). RAD51 focus formation was decreased upon depletion of MCM8 or MCM9 by siRNA (Fig. 7A and B) with MCM8 depletion having the greater effect. Consistent with these results, Mcm9-null MEF cells (XG/XG) showed a clear defect in RAD51 focus formation upon cisplatin treatment compared to wild-type MEF cells (WT/WT) (Fig. 7C). Foci were not formed even at early time points after cisplatin addition (see Fig. S5 in the supplemental material). RAD51 focus formation is also decreased in WT/XG heterozygous MEF cells (see Fig. S5 in the supplemental material) most likely due to an intermediate level of expression of MCM9 in these cells compared to the WT. This result can explain why heterozygous MEF cells showed intermediate hypersensitivity to cisplatin in Fig. 4B (also see Discussion). Together, these results suggest that MCM8-9 is required to recruit RAD51 to DSBs and to cisplatin-induced lesions.
Fig 7.
MCM8-9 is required for RAD51 focus formation upon ICLs. (A and B) MCM8 and MCM9 are required for RAD51 focus formation after cisplatin treatment. Cells were treated with DMSO (−Cis) or with 20 μM cisplatin (+Cis) for 16 h. Representative images are shown in panel A, and the histogram (B) shows the fraction of cells with RAD51 foci in control U2OS cells (siGL2) or U2OS cells depleted of MCM8 or MCM9 proteins. A total of 50 cells were counted per field over three randomly selected fields. The y axis is the fraction of RAD51-positive cells (>20 foci/cell), and bars represent the averages of all fields ± the SD. Scale bar, 10 μm. (C) Immunofluorescence of RAD51 in MEFs. Representative images of Mcm9 wild-type (WT/WT), and Mcm9-null (XG/XG) MEFs with RAD51 foci before or after treatment with 20 μM cisplatin for 16 h. Scale bar, 10 μm.
The Xenopus MCM8-9 complex is recruited to ICLs.
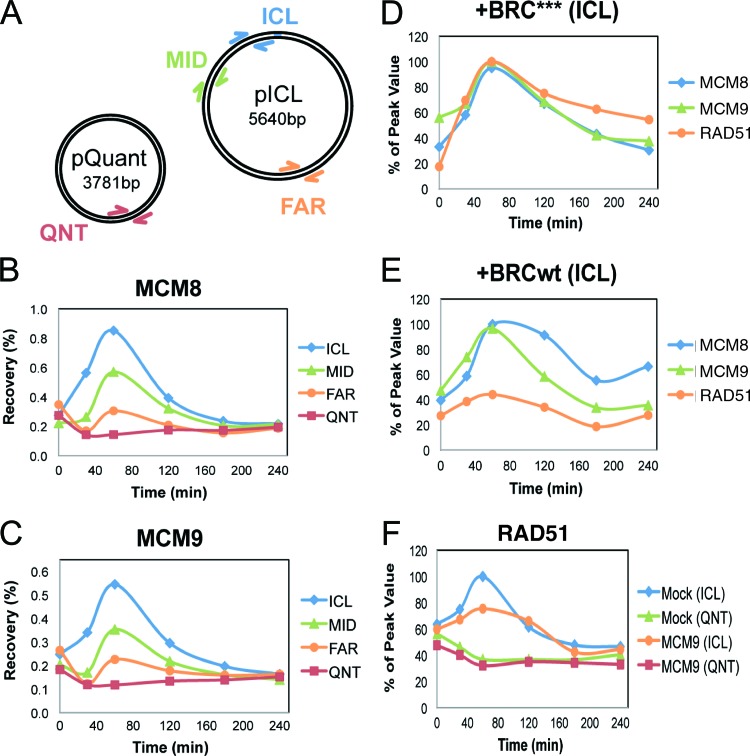
We previously developed a cell-free system based on Xenopus egg extracts that supports the replication-dependent repair of a plasmid that contains a single site-specific ICL (pICL) (4, 5). Repair is triggered when two replication forks converge on the cross-link Initially, fork stalling leads to an accumulation of RPA, which is quickly followed by loading of RAD51 onto the fork. Next, dual incisions on either side of the ICL create a DSB in one sister chromatid to unhook the cross-linked base. After repair of the other sister (with the unhooked ICL) via TLS, the broken sister is repaired by inter-sister HR. To examine the function of MCM8 and MCM9 in ICL repair, the binding of MCM8 and MCM9 to pICL or an undamaged control plasmid that also replicates (pQuant) was examined during replication-dependent repair in Xenopus egg extracts (Fig. 8A). The binding of MCM8 and MCM9 peaked 1 h after initiation of the repair reaction and was highly enriched near the ICL (Fig. 8B and C). While limited binding of MCM8 and MCM9 was also detected far from the cross-link, no replication-dependent binding was detected on pQuant (QNT), suggesting that in Xenopus egg extract, MCM8-9 is not significantly associated with the replication fork.
Fig 8.
MCM9 is required for efficient RAD51 binding to ICLs in Xenopus egg extracts. (A) Plasmid schematic showing the locations of different primer pairs used to analyze protein recovery during ChIP: ICL (25 to 132 bp away from the cross-link), MID (663 to 775 bp away), FAR (2,523 to 2,622 bp away), and QNT (undamaged control plasmid). (B and C) ChIP was used to measure the kinetics of MCM8 (B) or MCM9 (C) recruitment during replication of pICL and pQuant. A representative sample of three separate experiments is shown. (D and E) Inhibition of RAD51 loading does not interfere with MCM8 or MCM9 recruitment to the ICL. ICL localization of MCM8, MCM9, and RAD51 was measured by ChIP during replication in extracts supplemented with 14 μM BRC*** (D) or BRCwt peptide (E). (F) MCM9 depletion interferes with the accumulation of RAD51 at the ICL. pICL and pQuant were replicated in mock- or MCM9-depleted extracts, and recruitment of RAD51 was analyzed by ChIP.
We next evaluated the interdependence of MCM8-9 and RAD51 binding to ICLs. To this end, Xenopus egg extracts were incubated with a BRCA2-derived peptide that blocks the formation of RAD51 filaments on DNA (BRCwt) or a mutant version of the peptide that does not interact with or inhibit RAD51 (BRC***) (5). Although BRCwt severely reduced RAD51 localization at the ICLs as expected (5, 39), it did not affect MCM8 or MCM9 binding, indicating that RAD51 is not required for MCM8 or MCM9 loading at the ICL (compare Fig. 8D and E). On the other hand, the depletion of MCM9 from the extract (see Fig. S6 in the supplemental material) reduced RAD51 binding to the ICL by 40% (after subtracting the background level of binding seen on pQuant [Fig. 8F]). Depletion of MCM9 from egg extract did not inhibit ICL repair, either due to residual MCM9 in the extract or redundant pathways that load sufficient amounts of RAD51 for ICL repair. Consistent with the lack of a significant role for MCM9 in mammalian DNA replication (Fig. 2), DNA replication in Xenopus egg extract was not decreased by immunodepletion of Xenopus MCM9 protein (see Fig. S2B and S6 in the supplemental material). Although MCM9 depletion significantly codepleted MCM8 from the extract (data not shown), we could not immunodeplete Xenopus MCM8 sufficiently to determine whether MCM8 itself was required for RAD51 recruitment in this system. Due to difficulties in preparing recombinant MCM8 and MCM9 proteins, we have not been able to rescue the RAD51 loading defect observed in MCM9-depleted egg extract. These results support the conclusions from experiments in mammalian cells that MCM8-9 is recruited to sites of DSBs, that loading occurs independently of RAD51, and that MCM8-9 helps to recruit RAD51 to DNA damage sites (Fig. 6B and 7).
DISCUSSION
MCM8 and MCM9 form a nuclear complex that is not essential for DNA replication.
Although MCM8 and MCM9 proteins contain a well-conserved MCM domain, they are only present in metazoans (31) and so are not as central to eukaryotic DNA replication as MCM2-7. We found that MCM8 interacts with MCM9, both in mammalian cells (Fig. 1E) and in Xenopus egg extract (data not shown) and that they form a stable nuclear complex. Consistent with a role in DNA damage, MCM9 protein forms nuclear foci in cells treated with the DNA-damaging and cross-linking agent cisplatin and both MCM8 and MCM9 proteins colocalize to sites of DSBs generated during the repair of these ICLs in the S phase of the cell cycle. The formation of this complex may not only confer stability on MCM9 but also generate catalytically active conformations of MCM8 and MCM9. Cells depleted of MCM9 alone, or MCM8, which coincidently destabilizes MCM9, exhibit similar phenotypes, including the inability to recruit RAD51 or promote HR, further demonstrating that the two helicases likely function as a heterocomplex.
An important question is whether MCM8-9 plays a role in normal DNA replication. As discussed in the introduction, the evidence implicating MCM8-9 in DNA replication is contradicted by evidence from other papers. In our experiments, despite a significant effect of MCM8 or MCM9 depletion in human cells on HR after 48 h, there was minimal effect of such depletion on DNA replication after 48 h. In addition, Mcm9-null mice (XG/XG) are viable, and the corresponding MEFs (XG/XG) do not show a decrease of thymidine incorporation (see Fig. S2A in the supplemental material) or cell proliferation (30). In contrast, mice with knockout of components of the replicative helicase MCM2-7 die early in embryogenesis (38). Likewise, the depletion of MCM9 from Xenopus egg extracts had no significant effect on DNA replication, and MCM8-9 is detected at background levels on a replicating plasmid without an ICL (see Fig. S2B in the supplemental material and pQuant in Fig. 8B and C). We postulate that MCM8-9 has a direct role in HR, while its role in DNA replication is less direct, if any.
MCM8 and MCM9: new components of the HR repair pathway.
HR repair is an important mechanism to maintain genome stability. HR is particularly important for the repair of ICLs and for the repair of DSBs. In the present study, we found that MCM9 is recruited to cisplatin-dependent DNA damage foci colocalized with RPA foci, and MCM8-9 is recruited to DSBs and ICLs to promote RAD51 recruitment. An important question that remains is what is the precise function of MCM8-9 in HR? Helicases have been reported to be important for HR. In contrast to DNA replication helicase, multiple helicases work redundantly in HR repair. RECQ5L and PARI act as antirecombinases (40, 41) to prevent nonspecific recombination events. Other helicases, such as SGS1 or BLM, act concurrently with nucleases to produce the 3′ overhang structure necessary for RAD51 filament formation (16, 42–44). Additional helicases (BLM, SRS2, RTEL1, etc.) are required for the events after D loop formation such as strand displacement (16). At the I-SceI cut site in mammalian cells, MCM8-9 depletion inhibits the recruitment of RAD51 and immunofluorescence microscopy showed the depletion of MCM8-9 prevents cisplatin-induced RAD51 focus formation. Based on this, we suggest that MCM8-9 may be involved in bringing RAD51 to the break site. This could be through protein-protein interactions between MCM8-9 and RAD51. Alternatively, MCM8-9, like SGS1 or BLM, may be involved in the excision step that generates single-stranded DNA at DSBs to produce the substrate for RAD51 loading.
RAD51-depleted cells or cells depleted of both MCM8 and MCM9 showed a >90% inhibition of HR repair, which is consistent with a role of MCM8-9 in RAD51 recruitment to DSBs. The smaller effect of Xenopus MCM8-9 depletion on RAD51 recruitment in Xenopus egg extracts (Fig. 8F) could be explained by the high concentration of RAD51 in the extracts or by the less-than-perfect depletion of the MCM8-9 proteins.
The cisplatin sensitivity and the decrease in RAD51 focus formation in the WT/XG heterozygous MEFs is most likely due to the lower level of MCM9 in these cells. It is worth mentioning, however, that we cannot demonstrate that the MCM9 protein is decreased in the heterozygotes because the anti-human MCM9 antibody cannot detect MCM9 protein on immunoblots, while the immunofluorescence experiments shown in Fig. 5B are insensitive to a 50% decrease of protein. Thus, there is a low possibility that the decrease in HR in the heterozygous MEFs is due to a dominant-negative action of the XG allele of Mcm9. We consider this unlikely, however, because siRNA knockdown of MCM8-9 in human cells and antibody depletion in Xenopus egg extracts also suppressed RAD51 recruitment to damage sites.
While this manuscript was under preparation, two other groups reported a function of MCM8-9 in HR. Lutzmann et al. reported that MCM8-9 knockout mice showed defects in gametogenesis and were sensitive to DSBs (45). Nishimura et al. reported a role for MCM8-9 in HR using chicken DT40 cells (46). Although these two reports suggest a role of MCM8-9 in HR, they propose different roles for MCM8-9 in this process. Lutzmann et al. suggest that Mcm8- or Mcm9-knockout MEF cells have a slow growth rate (45), and so at least part of the phenotype could stem from defective replication fork movement. We, however, did not see significant defect of DNA replication or cell proliferation in MCM9-depleted human cells, Mcm9-null MEFs or MCM9-depleted Xenopus egg extract (30). On the other hand, and consistent with our result, Nishimura et al. did not see any significant cell growth defect in chicken DT40 cells with Mcm8 or Mcm9 deletion (46). Thus, we favor the idea that MCM8-9 has a role in HR independent of any role as part of a replicative helicase.
Nishimura et al. suggest that MCM8-9 works downstream of RAD51, because Mcm9 deletion did not decrease RAD51 focus formation in chicken DT40 cells (46). In contrast, Lutzmann et al. show a decrease of RAD51 binding on chromatin after treating Mcm9-deleted MEFs with aphidicolin (45). We found that MCM8-9 is required for RAD51 recruitment at the DSB or ICL in three different organisms, lending support to the idea that MCM8-9 acts upstream from RAD51 recruitment.
In summary, the cisplatin sensitivity of MCM8-9-depleted cells, the localization of MCM8-9 to DSBs in cells, the localization to ICLs in Xenopus egg extracts, and the requirement of MCM8-9 for RAD51 recruitment and HR at I-SceI cut sites, along with RAD51 focus formation assay in human cancer cells and MEF cells together provide strong evidence for a proximal role for MCM8-9 in HR. Along with our previous report that Mcm9-deficient mice showed a high incidence of cancer compared to wild-type mice (30), we hypothesize that MCM8-9 has a tumor-suppressing function by recruiting RAD51 to DNA damage sites for efficient HR repair and is important for genome stability. Furthermore, the role of MCM8-9 in HR is consistent with genetics in Drosophila (25) and mice (30) that suggest a role of MCM8 or MCM9 in meiosis or in germ cell viability. It is intriguing that a nonsynonymous genetic change in the Mcm8 gene has been associated with early menopause, which is consistent with a role of the protein in germ cell biology (47, 48). In contrast, we found minimal evidence that MCM8-9 is critical for the licensing or elongation steps of DNA replication. Finally, our discovery that inactivation of MCM8-9 sensitizes cancer cells to ICL-producing agents, such as cisplatin, suggest that these two helicases may serve as new targets of chemotherapy in combination with ICL-producing anticancer agents.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Parvin for providing HeLa DR13-9 cells and members of the Dutta lab for helpful discussions.
This study was supported by National Institutes of Health grants R01 CA60499 and R01 GM84465 to A.D., HL098316 to J.C.W., and R00CA140774 to T.A. D.T.L. was supported by American Cancer Society postdoctoral fellowship PF-10-146-01-DMC. A.G. was supported by Sir Henry Wellcome Fellowship 082716/Z/07/Z.
Footnotes
Published ahead of print 11 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01503-12.
REFERENCES
- 1. Heyer WD, Ehmsen KT, Liu J. 2010. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44:113–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scharer OD. 2005. DNA interstrand crosslinks: natural and drug-induced DNA adducts that induce unique cellular responses. Chembiochem 6:27–32 [DOI] [PubMed] [Google Scholar]
- 3. Deans AJ, West SC. 2011. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer 11:467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. 2008. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134:969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Long DT, Raschle M, Joukov V, Walter JC. 2011. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 333:84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moldovan GL, Madhavan MV, Mirchandani KD, McCaffrey RM, Vinciguerra P, D'Andrea AD. 2010. DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol. Cell. Biol. 30:1088–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. 2009. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326:1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kratz K, Schopf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavo E, Sartori AA, Hengartner MO, Jiricny J. 2010. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell 142:77–88 [DOI] [PubMed] [Google Scholar]
- 9. Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiacovo MP, Elledge SJ. 2010. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol. Cell 39:36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. MacKay C, Declais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, Lilley DM, Rouse J. 2010. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell 142:65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. 2006. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 25:4921–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ciccia A, McDonald N, West SC. 2008. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu. Rev. Biochem. 77:259–287 [DOI] [PubMed] [Google Scholar]
- 13. Svendsen JM, Smogorzewska A, Sowa ME, O'Connell BC, Gygi SP, Elledge SJ, Harper JW. 2009. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138:63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. 2007. Human CtIP promotes DNA end resection. Nature 450:509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, Tainer JA. 2009. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell 139:87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mimitou EP, Symington LS. 2009. Nucleases and helicases take center stage in homologous recombination. Trends Biochem. Sci. 34:264–272 [DOI] [PubMed] [Google Scholar]
- 17. Imamura O, Campbell JL. 2003. The human Bloom syndrome gene suppresses the DNA replication and repair defects of yeast dna2 mutants. Proc. Natl. Acad. Sci. U. S. A. 100:8193–8198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. San Filippo J, Sung P, Klein H. 2008. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77:229–257 [DOI] [PubMed] [Google Scholar]
- 19. Bochman ML, Schwacha A. 2009. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol. Mol. Biol. Rev. 73:652–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson EM, Kinoshita Y, Daniel DC. 2003. A new member of the MCM protein family encoded by the human MCM8 gene, located contrapodal to GCD10 at chromosome band 20p12.3-13. Nucleic Acids Res. 31:2915–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gozuacik D, Chami M, Lagorce D, Faivre J, Murakami Y, Poch O, Biermann E, Knippers R, Brechot C, Paterlini-Brechot P. 2003. Identification and functional characterization of a new member of the human MCM protein family: hMcm8. Nucleic Acids Res. 31:570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gozuacik D, Murakami Y, Saigo K, Chami M, Mugnier C, Lagorce D, Okanoue T, Urashima T, Brechot C, Paterlini-Brechot P. 2001. Identification of human cancer-related genes by naturally occurring hepatitis B virus DNA tagging. Oncogene 20:6233–6240 [DOI] [PubMed] [Google Scholar]
- 23. Volkening M, Hoffmann I. 2005. Involvement of human MCM8 in prereplication complex assembly by recruiting hcdc6 to chromatin. Mol. Cell. Biol. 25:1560–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maiorano D, Cuvier O, Danis E, Mechali M. 2005. MCM8 is an MCM2-7-related protein that functions as a DNA helicase during replication elongation and not initiation. Cell 120:315–328 [DOI] [PubMed] [Google Scholar]
- 25. Blanton HL, Radford SJ, McMahan S, Kearney HM, Ibrahim JG, Sekelsky J. 2005. REC, Drosophila MCM8, drives formation of meiotic crossovers. PLoS Genet. 1:e40 doi:10.1371/journal.pgen.0010040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Crevel G, Hashimoto R, Vass S, Sherkow J, Yamaguchi M, Heck MM, Cotterill S. 2007. Differential requirements for MCM proteins in DNA replication in Drosophila S2 cells. PLoS One 2:e833 doi:10.1371/journal.pone.0000833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yoshida K. 2005. Identification of a novel cell-cycle-induced MCM family protein MCM9. Biochem. Biophys. Res. Commun. 331:669–674 [DOI] [PubMed] [Google Scholar]
- 28. Lutzmann M, Maiorano D, Mechali M. 2005. Identification of full genes and proteins of MCM9, a novel, vertebrate-specific member of the MCM2-8 protein family. Gene 362:51–56 [DOI] [PubMed] [Google Scholar]
- 29. Lutzmann M, Mechali M. 2008. MCM9 binds Cdt1 and is required for the assembly of prereplication complexes. Mol. Cell 31:190–200 [DOI] [PubMed] [Google Scholar]
- 30. Hartford SA, Luo Y, Southard TL, Min IM, Lis JT, Schimenti JC. 2011. Minichromosome maintenance helicase paralog MCM9 is dispensible for DNA replication but functions in germ-line stem cells and tumor suppression. Proc. Natl. Acad. Sci. U. S. A. 108:17702–17707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu Y, Richards TA, Aves SJ. 2009. Ancient diversification of eukaryotic MCM DNA replication proteins. BMC Evol. Biol. 9:60 doi:10.1186/1471-2148-9-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee KY, Bang SW, Yoon SW, Lee SH, Yoon JB, Hwang DS. 2012. Phosphorylation of ORC2 protein dissociates origin recognition complex from chromatin and replication origins. J. Biol. Chem. 287:11891–11898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ransburgh DJ, Chiba N, Ishioka C, Toland AE, Parvin JD. 2010. Identification of breast tumor mutations in BRCA1 that abolish its function in homologous DNA recombination. Cancer Res. 70:988–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Negishi M, Saraya A, Mochizuki S, Helin K, Koseki H, Iwama A. 2010. A novel zinc finger protein Zfp277 mediates transcriptional repression of the Ink4a/arf locus through polycomb repressive complex 1. PLoS One 5:e12373 doi:10.1371/journal.pone.0012373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. 2006. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 8:91–99 [DOI] [PubMed] [Google Scholar]
- 36. Walter J, Sun L, Newport J. 1998. Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell 1:519–529 [DOI] [PubMed] [Google Scholar]
- 37. Pierce AJ, Johnson RD, Thompson LH, Jasin M. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13:2633–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chuang CH, Wallace MD, Abratte C, Southard T, Schimenti JC. 2010. Incremental genetic perturbations to MCM2-7 expression and subcellular distribution reveal exquisite sensitivity of mice to DNA replication stress. PLoS Genet. 6:e1110 doi:10.1371/journal.pgen.1001110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, Venkitaraman AR, Kowalczykowski SC. 2009. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell 136:1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schwendener S, Raynard S, Paliwal S, Cheng A, Kanagaraj R, Shevelev I, Stark JM, Sung P, Janscak P. 2010. Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J. Biol. Chem. 285:15739–15745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moldovan GL, Dejsuphong D, Petalcorin MI, Hofmann K, Takeda S, Boulton SJ, D'Andrea AD. 2012. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell 45:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gravel S, Chapman JR, Magill C, Jackson SP. 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22:2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. 2008. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. U. S. A. 105:16906–16911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Symington LS, Gautier J. 2011. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45:247–271 [DOI] [PubMed] [Google Scholar]
- 45. Lutzmann M, Grey C, Traver S, Ganier O, Maya-Mendoza A, Ranisavljevic N, Bernex F, Nishiyama A, Montel N, Gavois E, Forichon L, de Massy B, Mechali M. 2012. MCM8- and MCM9-deficient mice reveal gametogenesis defects and genome instability due to impaired homologous recombination. Mol. Cell 47:523–534 [DOI] [PubMed] [Google Scholar]
- 46. Nishimura K, Ishiai M, Horikawa K, Fukagawa T, Takata M, Takisawa H, Kanemaki MT. 2012. Mcm8 and Mcm9 form a complex that functions in homologous recombination repair induced by DNA interstrand crosslinks. Mol. Cell 47:511–522 [DOI] [PubMed] [Google Scholar]
- 47. Murray A, Bennett CE, Perry JR, Weedon MN, Jacobs PA, Morris DH, Orr N, Schoemaker MJ, Jones M, Ashworth A, Swerdlow AJ. 2011. Common genetic variants are significant risk factors for early menopause: results from the Breakthrough Generations Study. Hum. Mol. Genet. 20:186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. He C, Kraft P, Chen C, Buring JE, Pare G, Hankinson SE, Chanock SJ, Ridker PM, Hunter DJ, Chasman DI. 2009. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat. Genet. 41:724–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.