Abstract
Although many functions and targets have been attributed to the histone and protein deacetylase SIRT1, a comprehensive analysis of SIRT1 binding proteins yielding a high-confidence interaction map has not been established. Using a comparative statistical analysis of binding partners, we have assembled a high-confidence SIRT1 interactome. Employing this method, we identified the deubiquitinating enzyme ubiquitin-specific protease 22 (USP22), a component of the deubiquitinating module (DUBm) of the SAGA transcriptional coactivating complex, as a SIRT1-interacting partner. We found that this interaction is highly specific, requires the ZnF-UBP domain of USP22, and is disrupted by the inactivating H363Y mutation within SIRT1. Moreover, we show that USP22 is acetylated on multiple lysine residues and that alteration of a single lysine (K129) within the ZnF-UBP domain is sufficient to alter interaction of the DUBm with the core SAGA complex. Furthermore, USP22-mediated recruitment of SIRT1 activity promotes the deacetylation of individual SAGA complex components. Our results indicate an important role of SIRT1-mediated deacetylation in regulating the formation of DUBm subcomplexes within the larger SAGA complex.
INTRODUCTION
The SIRT1 protein deacetylase belongs to a family of NAD+-dependent enzymes, often referred to as sirtuins (1–3). Its homology to the Saccharomyces cerevisiae silencing protein Sir2 led to the discovery that SIRT1, like Sir2, promotes the formation of heterochromatin by deacetylating histone substrates such as histones H1, H3, and H4 (4–6). In contrast to Sir2, SIRT1 is known to regulate lysine acetylation of a wide variety of nonhistone protein targets, including p53 (7, 8), FOXO (9–11), Ku70 (12), nuclear hormone receptors (COUP-TF [13], liver X receptor [LXR] [14], and AR [15]), AP-1 (16), c-Myc (17), NF-κB (18), PGC-1α (19), and others. By deacetylating such a broad spectrum of targets, SIRT1 controls a variety of physiological and pathological processes, ranging from metabolic homeostasis and aging to cancer and neurodegenerative diseases (20, 21).
Sirtuins are a well-studied class of deacetylases and are dependent on the cofactor NAD+, thereby linking their activity to the metabolic and redox states of the cell (22, 23). Indeed, SIRT1 has been shown to play roles in several key cellular and organismal metabolic pathways such as mitochondrial biogenesis (24–26), lipid metabolism (27), gluconeogenesis (19, 28), and insulin secretion (29). Additionally, SIRT1 activity is regulated through several alternative mechanisms. Transcriptional regulation of SIRT1 is altered by stress responses (30–32), metabolic homeostasis (33), as well as calorie restriction, a diet that promotes longevity and disease resistance in a variety of species (12, 34). It has also become increasingly apparent that a variety of posttranslation modifications occurs on SIRT1, including phosphorylation (35, 36), sumoylation (37), and ubiquitination (38), and that these modifications play roles in regulating its localization, substrate specificity, and enzymatic activity. Providing another level of control, it has recently been discovered that SIRT1 contains an autoregulatory domain within its C terminus (ESA) that is required for full deacetylase activity (39).
Protein lysine acetylation, like other posttranslational modifications, can impart regulatory control over various cellular processes through alterations in protein complex formation, DNA binding, localization, enzymatic activity, and stability. Because SIRT1 acts on both histone and nonhistone proteins, it is poised to regulate a variety of cellular processes. However, it remains unclear whether SIRT1 exhibits significant substrate selectivity in vivo (40, 41), and it is possible that substrate selectivity is accomplished through specific SIRT1-interacting proteins. Because the vast majority of previous studies investigating SIRT1 function have focused on non-chromatin-related substrates of SIRT1, the precise mechanisms by which SIRT1 regulates gene expression remain poorly understood.
Ubiquitin-specific protease 22 (USP22), the mammalian homolog of yeast Ubp8, has been shown to play a conserved role as the catalytic subunit within the deubiquitinating module (DUBm) of the SAGA (Spt-Ada-Gcn5-acetyltransferase) transcriptional coactivating complex (42–47). Beyond its SAGA-related functions, the study of USP22 has been relatively limited, although several reports have uncovered correlative links to human pathology. For instance, USP22 has been shown to be part of an 11-gene death-from-cancer signature that is predictive of treatment failure (48, 49), and its increased expression is associated with a poor prognosis in a variety of cancers (50, 51).
As a subunit of the SAGA complex, USP22 mediates transcriptional activation primarily by deubiquitination of histone H2B, although USP22 has also been shown to catalyze the removal of ubiquitin from H2A under some circumstances (45–47). In addition to histones, several novel substrates have been identified, including TRF1 (52) and FBP1 (53), further implicating USP22 in proliferation and oncogenesis. USP22 has also been shown to play a role in 3′-end processing of JAK-STAT-inducible genes through regulation of H2B ubiquitination (54).
Here we present evidence for a highly specific interaction between SIRT1 and USP22. This interaction occurs within the ZnF-UBP domain of USP22 and is disrupted by a mutation which renders SIRT1 catalytically inactive. In agreement with previous work, we determined that human USP22 absolutely requires formation of a DUBm complex for catalytic activity. We demonstrate that USP22 is an acetylated protein in vivo and that its acetylation status affects its binding to SAGA. Additionally, SIRT1 is recruited by its association with USP22 to the SAGA complex, where it deacetylates SAGA subunits and leads to a modulation of the DUBm-SAGA association.
MATERIALS AND METHODS
Antibodies.
For the production of the polyclonal anti-USP22 acetylated lysine 129 (Ac-K129; YZ1573), a rabbit was immunized with high-pressure liquid chromatography-purified acetylated cys-THY-conjugated peptide directed against USP22 (CMEIIA–Ac-K–EEQRKAW). The resulting serum was purified in a two-step fashion by affinity matrix positively against modified peptide and negatively against unmodified peptide. The polyclonal antibody against USP22 was a gift from Didier Devys (47). Antibodies for hemagglutinin (HA; H6908) and FLAG (F7425) were obtained from Sigma-Aldrich. Antibody for SIRT1 (1104-1) was procured from Epitomics. Antibody for GCN5 (20698) was obtained from Santa Cruz Biotechnology. Antibodies directed against TRRAP (A301-132A), ATXN7L3 (A302-800A), and USP10 (A300-900A) were obtained from Bethyl Labs. Antibodies for panacetyllysine (9441), monoclonal panacetyllysine (9681), total p53 (2524), and p53 Ac-K382 (2525) were procured from Cell Signaling Technology. Antibody against panacetyllysine (ICP0380) was obtained from ImmunChem. Histone H2A (39111), histone H2A.Z (39943), and histone macroH2A1.1 (39871) antibodies were from Active Motif. Antibody against monoubiquitinated H2B (MM029) was obtained from Medimabs. Antibody for beta-tubulin (05-661) was from EMD Millipore. Following normalization of protein by the Bradford assay (Bio-Rad), samples were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, blocked in 5% milk in Tris-buffered saline–Tween, and blotted with the indicated antibodies. Western blots using acetyl-specific antibodies were all performed using 5% bovine serum albumin as a replacement for powdered nonfat milk.
Plasmids and cell lines.
Open reading frame clones were obtained from the human ORFeome collection. SIRT1 was PCR cloned and recombined into pDONR223 using bacteriophage lambda (BP) recombinase from an available full-length cDNA. Sequence-validated open reading frames in pDONR223 were recombined into the Gateway destination vector MSCV-N-FLAG-HA-IRES-PURO (LTR-driven expression), pBB-N-GST, or pBB-N-HA using bacteriophage λ recombinase. Truncated protein expression vectors were produced by PCR cloning and recombined into pDONR223 using BP recombinase. Point mutants were generated by site-directed mutagenesis in pDONR223.1 vectors and then recombined into Gateway destination vectors. The GCN5-FLAG expression vector was a gift of Pere Puigserver.
HEK 293T, HCT116, and NCI-H460 cells were maintained in Dulbecco modified Eagle medium (DMEM; 11965-092; Life Technologies) plus 10% fetal bovine serum (FBS; Gemini Bio-Products) and penicillin-streptomycin (Life Technologies) at 37°C under a 5% CO2 atmosphere. Virus was produced by cotransfecting the protein of interest, GAG/POL, and VSVG plasmids into HEK 293T cells and was then used to infect HEK 293T cells for constitutive expression. Selection was accomplished by supplementing standard growth medium with 1 μg/ml puromycin (InvivoGen). Transient transfections were performed with either Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions or calcium phosphate.
For p53 acetylation assays, NCI-H460 cells were pretreated for 30 min with dimethyl sulfoxide (DMSO) or 10 μM EX527 (Tocris Bioscience) and subsequently treated with or without 0.5 μM doxorubicin (Sigma) for 6 h. Cells were subsequently washed with ice-cold phosphate-buffered saline (PBS) and lysed by scraping on ice in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, Roche Complete EDTA-free protease inhibitor cocktail, 1 mM phenylmethylsulfonyl fluoride [PMSF], 400 nM trichostatin A [TSA], 10 mM nicotinamide, 1 mM dithiothreitol [DTT]). The protein concentration was normalized by the bicinchoninic acid assay, samples were resolved by SDS-PAGE, and the indicated proteins were detected by immunoblotting.
siRNA.
Small interfering RNA (siRNA) oligonucleotides were purchased from Thermo Scientific (Dharmacon). Transfections were performed with Lipofectamine RNAiMAX (Life Technologies) per the manufacturer's instructions for reverse transfection. Briefly, in 12-well culture dishes, 1 μl of 20 μM siRNA oligonucleotide and 2 μl Lipofectamine RNAiMAX were added to 200 μl serum-free DMEM containing no antibiotics, and the mixture was incubated for 20 min at room temperature. Trypsinized cells diluted in DMEM plus 10% FBS containing no antibiotics were then added to DNA-lipid complexes and allowed to attach overnight. Fresh medium was changed at 24 h, and cells were harvested at 72 h. Oligonucleotide sequences are as follows: SIRT1 1, GUACAAACUUCUAGGAAUG; SIRT1 2, GUAGGCGGCUUGAUGGUAA; SIRT1 3, GCGAUUGGGUACCGAGAUA; SIRT1 4, GGAUAGGUCCAUAUACUUU; USP22 1, GGAGAAAGAUCACCUCGAA; USP22 2, CAAAGCAGCUCACUAUGAA; USP22 3, GGAAGAUCACCACGUAUGU; and USP22 4, CCUUUAGUCUCAAGAGCGA.
Recombinant protein purification.
Baculovirus to express glutathione S-transferase (GST)- or HA-tagged proteins was produced in Sf9 cells using a Bac-N-Blue expression system (Life Technologies) per the manufacturer's instruction. Infected Sf9 cells were grown to the necessary volumes in 15-cm2 culture dishes or glass spinner flasks at 27°C for 3 to 5 days. Expressing cells were collected by centrifugation, washed in ice-cold PBS, and lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% Nonidet P-40, Roche Complete EDTA-free protease inhibitor cocktail, 1 mM PMSF, 1 mM DTT [Roche]) for 1 h with gentle rocking at 4°C. Lysates were cleared by centrifugation (13,000 rpm, 10 min), and the resulting material was subjected to immunoprecipitation (IP) with immobilized glutathione-agarose (Pierce) or anti-HA-agarose (Sigma) overnight at 4°C with gentle inversion. Resin containing bound protein was washed extensively with ice-cold lysis buffer, followed by several washes with ice-cold PBS containing 1 mM DTT. Proteins were eluted with a molar excess of reduced glutathione (Sigma) or HA peptide (Sigma) and subsequently dialyzed overnight against PBS–β-mercaptoethanol. Proteins were tested for purity by SDS-PAGE, followed by Coomassie brilliant blue staining.
Protein purification.
For mass spectrometry purifications, cells from four 15-cm tissue culture dishes at ∼80% confluence (∼107 cells) were lysed in a total volume of 4 ml of lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, Roche Complete EDTA-free protease inhibitor cocktail) for 1 h with gentle rocking at 4°C. Lysates were cleared using centrifugation (13,000 rpm, 10 min), the supernatant was filtered through 0.45-μm-pore-size spin filters (Millipore) to further remove cell debris, and the resulting material was subjected to immunoprecipitation with 60 μl of immobilized anti-HA (Sigma) resin (50% slurry) overnight at 4°C with gentle inversion. Resin containing immune complexes was washed with 1 ml ice-cold lysis buffer 8 times, followed by three 1-ml PBS washes. Proteins were eluted with three 50-μl incubations with 250 μg/ml HA-peptide (Sigma) in PBS for 30 min each at 22°C, and elutions were pooled for a final volume of 150 μl. Proteins in each elution were precipitated with 20% trichloroacetic acid (TCA), and the resulting pellet was washed once with 10% TCA and 3 times with cold acetone.
Smaller-scale purifications used in non-mass spectrometry (non-MS) experiments were similar but generally used one 10-cm2 culture dish; however, 1 mM PMSF (Sigma) and 1 mM DTT were added to the lysis buffer. For experiments to examine acetylation of USP22 or SAGA components, two 15-cm2 culture dishes were typically treated overnight with TSA, nicotinamide (NAM), FK866, or EX527 (Tocris Bioscience) and lysed in lysis buffer, which additionally contained 1 mM PMSF, 1 mM DTT, 400 nM TSA (Sigma), and 10 mM nicotinamide (Sigma). Where indicated, FLAG-M2 resin (Sigma) was used in place of HA-agarose. Elutions from this resin were performed with 250 μg/ml 3× FLAG peptide (Sigma).
Gel filtration was performed on an Akta fast-performance liquid chromatograph (GE Healthcare Life Sciences) using a prepacked Superose 6, 10/300 GL column (GE Healthcare Life Sciences). The flow rate for the elution buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% Nonidet P-40) was 0.5 ml per minute, and 0.5-ml fractions were collected with an automated fraction collector. Fractions containing complexes of interest were pooled and immunoprecipitated with FLAG-M2 resin (Sigma), washed with lysis buffer, and eluted with 250 μg/ml 3× FLAG peptide (Sigma). Eluted complexes were then used in downstream assays, resolved by SDS-PAGE, and Western blotted.
Endogenous immunoprecipitations were performed by adding 1 μg of the indicated antibodies to 1 mg lysate and rocking gently at 4°C overnight. Immune complexes were incubated with anti-rabbit IgG–agarose (eBioscience) for 1 h at 4°C, washed 8 times with lysis buffer, and eluted with SDS-PAGE loading dye. Resulting proteins were separated by SDS-PAGE and immunoblotted. Endogenous immunoprecipitation of SIRT1 with GCN5 was performed by cross-linking cells with 1 mM Dithiobis(succinimidyl propionate) (DSP; Thermo Scientific) for 15 min at room temperature, followed by quenching with excess Tris, pH 7.4. Lysates were generated by scraping cells in RIPA buffer supplemented with 1 mM PMSF and Roche Complete EDTA-free protease inhibitor cocktail and rocking for 1 h at 4°C. Samples were cleared by centrifugation and passed through a 0.45-μm-pore-size spin filter (Millipore), followed by addition of anti-SIRT1 antibody (1104-1; Epitomics) or an equivalent amount of normal rabbit IgG (Millipore) and incubated overnight at 4°C. Immune complexes were incubated with anti-rabbit IgG agarose (eBioscience) for 1 h at 4°C; washed five times with RIPA buffer, three times with RIPA buffer with 500 mM NaCl, two times with standard IP lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40), and two times with ice-cold PBS; and then eluted with 10% ammonium hydroxide. Subsequent to drying in a Speed-Vac apparatus, samples were resuspended in SDS loading dye supplemented with 50 mM DTT, de-cross-linked, separated by SDS-PAGE, and immunoblotted.
Mass spectrometry.
Our MS analyses were performed as previously described (55). Briefly, TCA-precipitated proteins were resuspended in 30 μl 100 mM ammonium bicarbonate at pH 8.0 with 10% acetonitrile and 500 ng sequencing-grade trypsin (Promega) and incubated at 37°C for 4 h. Digested samples were then loaded onto stage tips and washed as described previously (56). Peptides were eluted with 50% acetonitrile, 5% formic acid, dried using a Speed-Vac apparatus, and resuspended in 10 μl of 5% acetonitrile, 5% formic acid. For each liquid chromatography (LC)-MS/MS run using an LTQ linear ion trap mass spectrometer (Thermo), 4 μl was loaded onto a C18 column (18 cm by 125 μm [inner diameter]), and peptides were eluted using a 50-min 8 to 26% acetonitrile gradient. Spectra were acquired in a data-dependent fashion using a Top-10 method. Each sample was loaded twice in succession, followed by a wash with 70% acetonitrile, 30% isopropanol. The resulting MS/MS spectra were recorded for each run and then searched against a target-decoy database of human tryptic peptides using the Sequest program (57). The resulting list of identifications for each was loaded into the Comparative Proteomics Analysis software suite (CompPASS) for further processing and analysis.
Data processing and CompPASS analysis.
Mass spectral data were processed using CompPASS, as previously described, with modifications (55, 58). Sequest summary files were processed into a high-threshold data set on the basis of a 2% protein false-positive rate as described earlier. These processed data sets were merged for each duplicate run and used to populate a stats table consisting of each data set for SIRT1 as well as 204 unrelated proteins (deubiquitinating enzymes [Dubs] and proteins from the Autophagy Interaction Network and several unique runs [https://harper.hms.harvard.edu/]). The normalized D scores (DN) were calculated from average assembled peptide spectral matches (APSMs) for each protein found in association with each bait. Proteins identified in each LC-MS/MS experiment with a DN score of ≥1 are considered to be high-confidence candidate interaction proteins (HCIPs).
Additional bioinformatic analysis.
Interactions used for generating protein networks were from the STRING database, found at http://string.embl.de/. Output files from CompPASS for network analysis are compatible with the Cytoscape software platform (http://www.cytoscape.org), and additional files containing both node and edge attributes were generated. Attribute files were used in Cytoscape to assign values for nodes and edges, as indicated. Gene ontology (GO) biological process terms were hand collated for HCIPs in the SIRT1 wild-type (WT) IPs. High-level tree biological processes that displayed an overlap for at least two HCIPs were assigned a colored annotation.
SAGA acetyllysine mapping.
For acetyllysine mapping purifications, cells from tissue culture dishes (60 by 15 cm2) treated overnight with 10 μM EX527 (Tocris Bioscience) were lysed in a total volume of 25 ml of lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, Roche Complete EDTA-free protease inhibitor cocktail) supplemented with 400 nM TSA, 20 mM NAM, 1 mM DTT, and 1 mM PMSF for 1 h with gentle rocking at 4°C. Lysates were cleared by centrifugation (16,000 × g, 15 min), the supernatant was filtered through 0.45-μm-pore-size spin filters (Millipore) to further remove cell debris, and the resulting material was subjected to immunoprecipitation with 300 μl of immobilized anti-HA (Sigma) resin (50% slurry) overnight at 4°C with gentle inversion. Resin containing FLAG-HA-USP22 immune complexes was washed 8 times with ice-cold lysis buffer, followed by three PBS washes and one wash with UltraPure distilled water (Invitrogen). Proteins were eluted with 300 μl of 10% ammonium hydroxide for 30 min at 4°C, spun through 0.45-μm-pore-size spin filters (Millipore) to separate eluted complexes from resin, and subsequently dried in a Speed-Vac. Lysates for quantitative mass spectrometry were generated as described above, except for those from plates (10 by 15 cm2) treated with DMSO or 10 μM EX527.
Digestion and isotopic labeling.
Samples were resuspended in reduction buffer (30 μl 8 M urea, 5 mM DTT, 100 mM Tris, pH 8.5) and incubated at 56°C for 30 min. After cooling to room temperature, iodoacetamide (15 mM) was added and the samples were incubated in the dark for 30 min. DTT was added to a final concentration of 10 mM and samples were incubated in the dark for 15 min to quench the alkylation reaction. After dilution with 100 mM Tris (pH 8.5) such that the final urea concentration was 3.3 M, 700 ng of endoproteinase Lys-C (Wako) was added and the samples were incubated at 37°C for 2 h. Samples were further diluted with 100 mM Tris (pH 8.2) to a final urea concentration of 1.6 M, 500 ng of trypsin (Promega) was added, and digestion was continued at 37°C overnight. After 12 h, trifluoroacetic acid (TFA) was added to a final concentration of 0.5%, the samples were centrifuged at 15,000 × g for 15 min, and the supernatant was collected. For isotopic labeling, samples were loaded onto C18 stage tips (59). C18-bound peptides were isotopically labeled via reductive dimethylation essentially as described previously (60). After washing and elution, peptides from control and drug-treated paired experiments were mixed prior to immunoprecipitation.
Peptide immunoprecipitation.
Immunoprecipitation was carried out as described previously (61). Settled agarose (50 μl) of conjugated antiacetyllysine antibody (ImmuneChem) was washed twice with acetyllysine IP buffer (50 mM MOPS [morpholinepropanesulfonic acid], pH 7.2, 10 mM sodium phosphate, 50 mM sodium chloride). After 10% of each labeled, mixed digest was set aside for quantification of protein levels prior to acetyllysine IP, the remainder of each mixed digest was diluted with 1 ml of acetyllysine IP buffer, added to the washed antiacetyllysine-agarose, and allowed to bind overnight at 4°C with gentle rocking. Samples were washed three times with acetyllysine wash buffer (50 mM MOPS, pH 7.2, 10 mM sodium phosphate, 150 mM sodium chloride) and twice with deionized water, before elution 3 times with 0.1% TFA. Prior to MS analysis, eluted peptides were desalted using C18 stage tips as described above.
Quantitative mass spectrometry.
Desalted samples were subjected to nano-LC-MS/MS using a 20-cm column composed of a 100-μm (inner diameter) fused-silica capillary that was flame pulled in-house to produce an approximately 5-μm tip and packed with Maccel C18 resin (3 μm; 200 Å; The Nest Group, Inc.). Data were collected on an LTQ Orbitrap Elite hybrid mass spectrometer (Thermo Fisher), operated in the data-dependent mode, essentially as described previously (62). MS/MS spectra were searched using the Sequest algorithm against a partially tryptic, sequence-reversed human proteome database (63). Peptide false detection rates (FDRs) were controlled for by filtering peptide spectral matches according to their cross correlation (XCorr), mass accuracy, tryptic state, charge state, and peptide length. Abundance ratios for isotope-labeled peptides were extracted with a software suite developed in-house. Quantitative measurements of peptide-level changes between control and drug-treated samples were normalized to the total protein levels quantified in a separate MS run using the sample reserved prior to antiacetyllysine IP.
Deubiquitination assays.
For ubiquitin vinylsulfone (UbVS) assays, recombinant proteins, cell lysates, or eluted protein complexes were diluted in Dub assay buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM DTT), and either DMSO or 5 μM ubiquitin vinylsulfone (U-202; Boston Biochem) was added for 1 h at 30°C, unless otherwise indicated. Reactions were stopped by the addition of SDS-PAGE loading dye and subsequent boiling for 5 min. Dubs were then resolved by SDS-PAGE and immunoblotted. For ubiquitin–7-amino-4-methylcoumarin (Ub-AMC) kinetic assays of recombinant USP22, equivalent amounts of recombinant USP2 catalytic core (USP2cc) or USP22 were diluted in Dub assay buffer and plated in opaque 96-well plates. Ub-AMC at a final concentration of 200 nM in Dub assay buffer was added to start the reaction. The resultant fluorescence at 342 nm/438 nm (excitation/emission) was detected on an EnSpire multimode plate reader (PerkinElmer). Recombinant proteins used were separated by SDS-PAGE and stained with Coomassie brilliant blue to normalize the concentrations and determine relative purity.
Intact mononucleosomes were generated by micrococcal nuclease treatment of chromatin fractions purified from HEK 293T cells. Purified Dubs or their catalytic mutant versions were tested for their ability to deubiquitinate equivalent amounts of these semipurified mononucleosomes at 30°C for 1 h. Reactions were stopped by the addition of SDS-PAGE loading dye and subsequent boiling for 5 min. Ubiquitinated histones were then resolved by SDS-PAGE and immunoblotted.
In vitro deacetylation.
Acetylated SAGA components were purified as described above by immunoprecipitating FLAG-HA-USP22 from cells treated for 24 h with 10 μM EX527 (Tocris Bioscience) with FLAG-M2 resin. Protein complexes were washed 8 times with ice-cold lysis buffer containing deacetylase inhibitors, washed 3 times with ice-cold deacetylation buffer (50 mM Tris-HCl, pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2), and eluted with 250 μg/ml 3× FLAG peptide (Sigma). Eluted complexes were used as the substrates for recombinant GST-SIRT1 produced in Sf9 cells with or without 1 mM NAD+ treated for 1 h at 37°C. Reactions were stopped by the addition of SDS-PAGE loading dye. Samples were boiled for 5 min, resolved by SDS-PAGE, and immunoblotted with the indicated antibodies.
RESULTS
Identification and characterization of the SIRT1 interactome.
To better understand the role of SIRT1 in the regulation of physiological and pathological processes, we employed a proteomic approach to identify novel interacting partners for SIRT1. Wild-type human SIRT1 was expressed as an N-terminal FLAG-HA fusion protein in HEK 293T cells. Proteins associated within anti-HA immune complexes were subsequently identified by mass spectrometry (LC-MS/MS). Identified SIRT1-interacting proteins were processed using a modified version of CompPASS to identify HCIPs (55). CompPASS utilizes an interacting protein database and three specificity metrics (DN score, an associated P value, and Z score) to identify HCIPs.
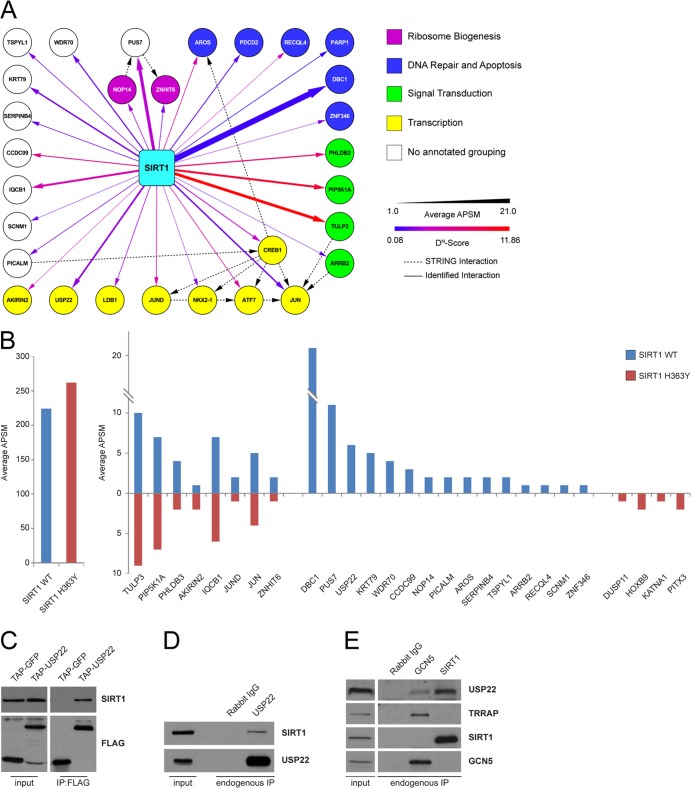
The resulting SIRT1 interaction network included several well-known binding partners such as Jun (16, 64–66), CREB (67), and AROS (68), thereby validating our experimental approach. Peptides for DBC1 (69, 70) and PARP1 (71) were also observed; however, these did not meet statistical criteria because they were identified as proteins interacting across a large number of baits. The screen also identified a wide array of novel SIRT1-interacting proteins involved in a variety of cellular functions, including transcription, DNA repair, apoptosis, signal transduction, and ribosome biogenesis (Fig. 1A).
Fig 1.
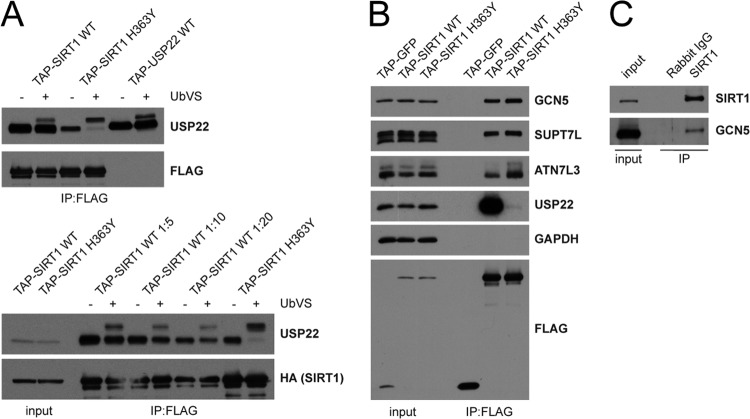
Identification and characterization of the SIRT1 interactome. (A) An interaction network for HCIPs (DN scores > 1) found in SIRT1 IP-MS/MS was created using networking tools in CompPASS. Protein matches for DBC1 and PARP1 did not meet DN-score cutoffs but were included in the network map. Maps were generated in Cytoscape with attribute files that reflect interactor abundance (thickness of the line) and DN score (color of the line). STRING database interactions are indicated as dashed lines. (B) Interactor abundance presented as average APSMs for bait proteins in SIRT1 WT and H363Y IP-MS/MS (left) and for selected interacting proteins (right). (C) FLAG-HA-USP22 (TAP) was immunoprecipitated with anti-FLAG-coupled resin and immunoprecipitates, and inputs were probed with antibodies against endogenous SIRT1. (D and E) Antibodies were used to immunoprecipitate endogenous USP22, GCN5, or SIRT1 from HEK 293T cells. Immunoblots were then probed with the indicated antibodies.
As a way to readily identify potential enzymatic substrates of SIRT1 within the interactome, we compared the wild-type SIRT1 interaction map to that of a catalytically inactive mutant of SIRT1 (H363Y). The H363Y mutation did not significantly alter many of the interactions observed with wild-type SIRT1, while a small subset of interactions was either ablated or enhanced by the H363Y mutation. Specifically, PUS7, CCDC99, USP22, and the known SIRT1 interactor DBC1 were present only within wild-type SIRT1 immune complexes (Fig. 1B). Given that USP22 and its homologs control important epigenetic functions from yeast to mammals through regulation of histone ubiquitination, together with the fact that USP22 immune complexes have also been shown to contain SIRT1 (55), we chose to focus on understanding the relevance of the interaction between SIRT1 and USP22.
Validation of the interaction between SIRT1 and USP22.
To confirm the interaction between SIRT1 and USP22, we immunoprecipitated FLAG-HA-USP22 or a tagged GFP control from HEK 293T cells and observed that endogenous SIRT1 interacted with overexpressed USP22 but not GFP (Fig. 1C). Consistent with our results with exogenous expression of fusion proteins, antibodies used to immunoprecipitate endogenous USP22 copurified endogenous SIRT1 (Fig. 1D). Moreover, when we examined endogenous immune complexes for either GCN5, a core component of the SAGA complex, or SIRT1, we determined that GCN5 interacted with TRRAP and USP22, whereas the interaction between SIRT1 and USP22 appeared to be exclusive of the larger SAGA complex (Fig. 1E).
Specificity and mapping of the SIRT1-USP22 interaction.
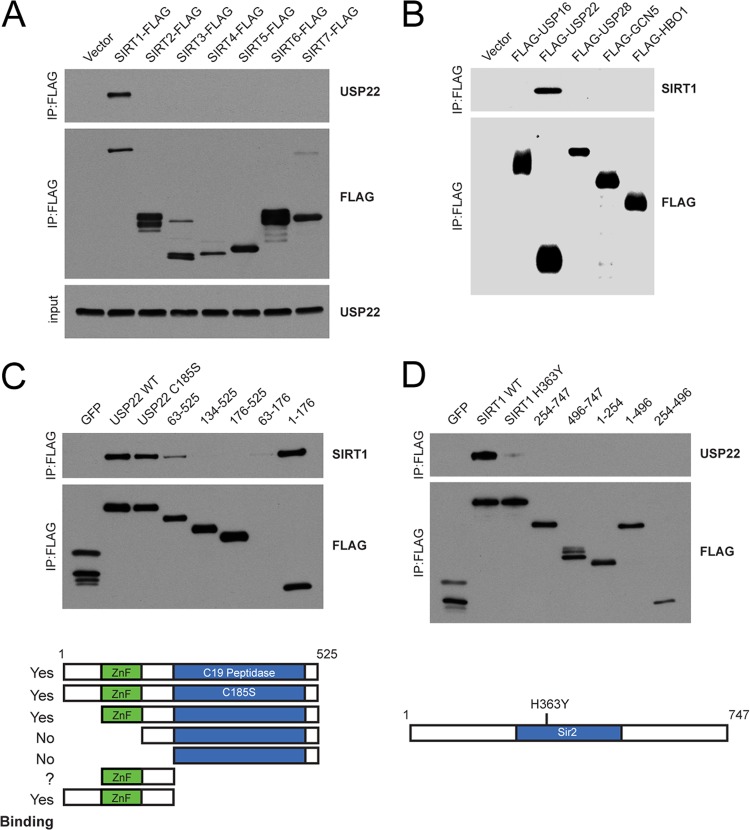
In order to test the specificity of the interaction, we expressed C-terminally FLAG-tagged fusions of each of the sirtuins, SIRT1 to SIRT7, in HEK 293T cells and found that of the seven sirtuins, only SIRT1 was capable of copurifying USP22 (Fig. 2A). Additionally, unrelated Dubs or histone acetyltransferases (HATs) were unable to pull down SIRT1 (Fig. 2B). Moreover, a reciprocal study of the Dub interaction landscape did not uncover SIRT1 as an interacting partner for 74 other deubiquitinating enzymes (55). These results demonstrate that the interaction between SIRT1 and USP22 is highly specific.
Fig 2.
Interaction of USP22 with SIRT1 is highly specific and requires the ZnF-UBP domain. (A) C-terminally FLAG-tagged SIRT1 to SIRT7 were overexpressed in HEK 293T cells, immunoprecipitated with FLAG resin, and immunoblotted for endogenous USP22. (B) Various N-terminal FLAG-tagged deubiquitinating enzymes and acetyltransferases were overexpressed, purified as described for panel A, and immunoblotted for endogenous SIRT1. (C and D) Truncations of FLAG-HA-USP22 (C) and FLAG-HA-SIRT1 (D) were generated and tested for their ability to interact with endogenous SIRT1 or USP22, respectively. Domain maps of USP22 (C) and SIRT1 (D) indicate truncations used in interaction studies.
USP22 consists of an N-terminal ZnF-UBP domain and a C-terminal C19 peptidase domain containing the catalytic cysteine required for enzymatic activity (46, 47). The ZnF-UBP domain of USP22 is unique since, unlike other ZnF-UBP domains, it cannot bind free ubiquitin due to differences in several key ubiquitin binding residues (72). It has been postulated that this domain might serve to facilitate protein-protein interactions. To gain insights into the domains within USP22 that mediate the interaction with SIRT1, we generated a series of USP22 truncations and a catalytic mutant in the C19 peptidase domain, C185S (Fig. 2C). Interestingly, SIRT1 binding occurred within the ZnF-UBP domain of USP22 and did not require the presence of the catalytic cysteine residue. In agreement with our initial screen results, the catalytically inactive SIRT1 (H363Y) displayed significantly reduced binding to USP22, suggesting that SIRT1 activity may be important for facilitating the interaction with USP22 (Fig. 2D). Reciprocal studies with SIRT1 did not reveal a single linear domain that mediated the interaction with USP22.
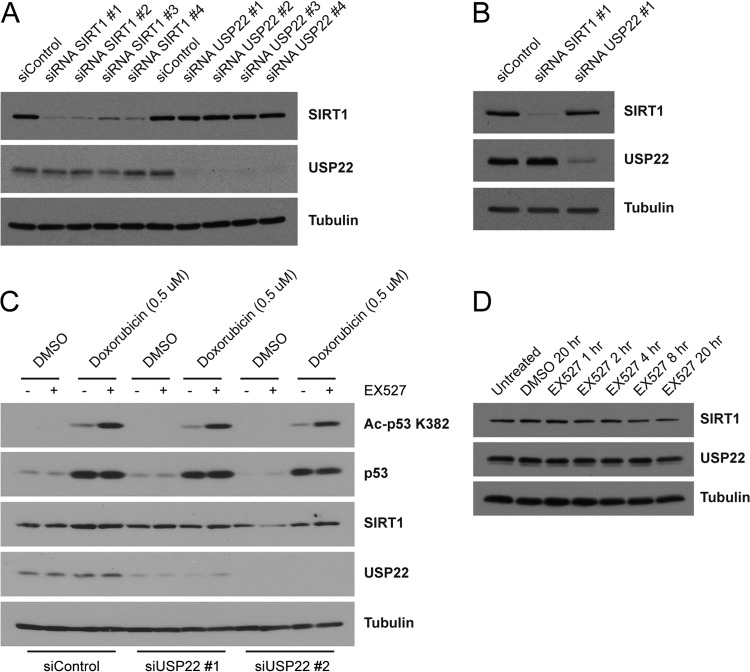
USP22 does not alter steady-state levels or deacetylase activity of SIRT1.
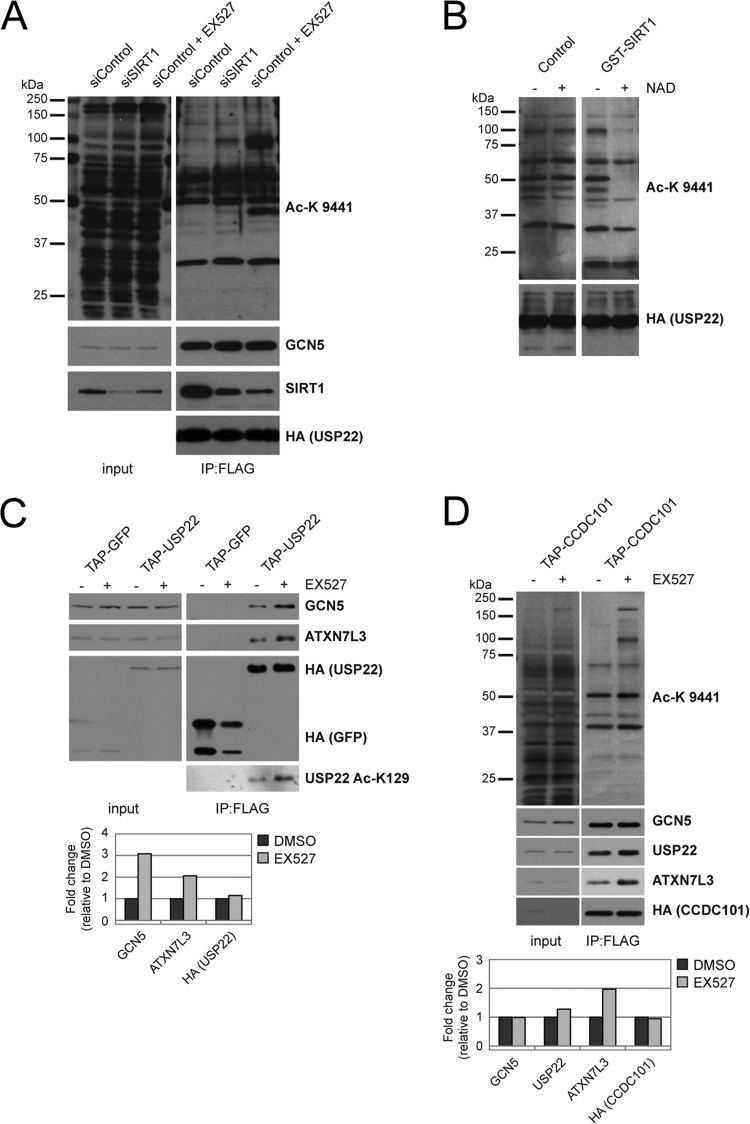
During preparation of the manuscript, a study was published indicating that USP22 deubiquitinates SIRT1, thereby increasing SIRT1 steady-state protein levels and suppressing apoptosis by decreasing p53-K382 acetylation (73). In our studies, we did not observe a difference in SIRT1 levels in either HEK 293T or HCT116 cells following siRNA-mediated knockdown of USP22 (Fig. 3A and B). Furthermore, USP22 knockdown in doxorubicin-treated cells had no effect on acetylated p53-K382 levels in either the presence or absence of the specific SIRT1 inhibitor EX527 (Fig. 3C). Additionally, although the binding of SIRT1 H363Y to USP22 is severely compromised compared to that of wild-type SIRT1, the stability of the H363Y mutant SIRT1 was identical to that of wild-type SIRT1. These results suggest that, in these cell types, both SIRT1 activity and SIRT1 stability are unaffected by the loss of USP22. A reciprocal examination using EX527 to inhibit SIRT1 activity found no evidence that SIRT1 activity modulates the steady-state stability of USP22 (Fig. 3D).
Fig 3.
USP22 does not alter steady-state levels or deacetylase activity of SIRT1. siRNA oligonucleotides directed against SIRT1, USP22, or the control were transfected into HEK 293T cells (A) or HCT116 cells (B) for 72 h. Lysates were immunoblotted with the indicated antibodies. (C) NCI-H460 cells were transfected as described for panel A with control or USP22 targeting siRNA oligonucleotides and then treated with or without doxorubicin (0.5 μM) in the presence or absence of the SIRT1 inhibitor EX527 (10 μM). (D) Lysates generated from HEK 293T cells treated for various times with or without EX527 (10 μM) were immunoblotted with the indicated antibodies.
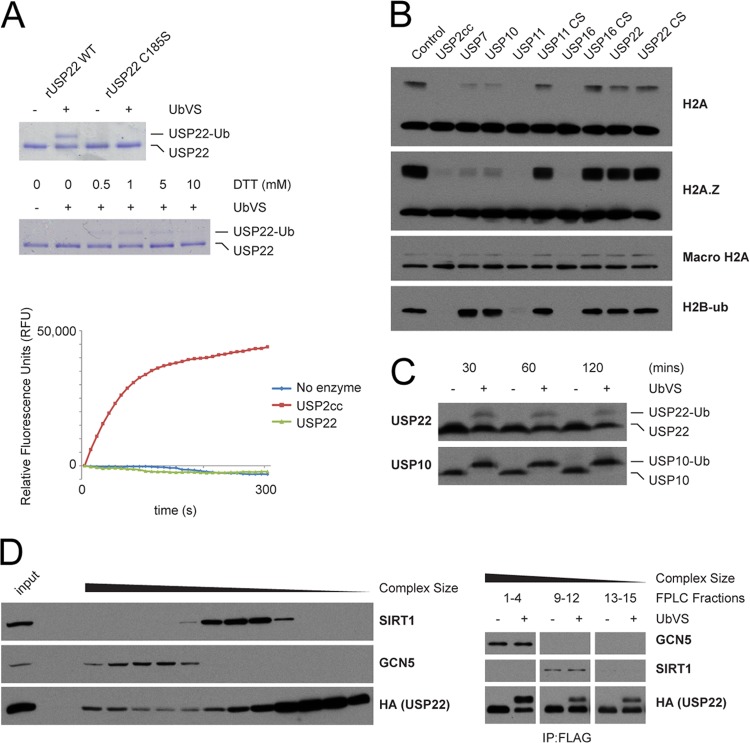
Mammalian USP22 requires complex formation for deubiquitinating activity.
Previous studies of DUBm in yeast and mammals have demonstrated that USP22 and its yeast homolog Ubp8 form heterotetrameric complexes that are absolutely required for catalytic activity (72, 74–76). In agreement with these studies, our initial characterization of recombinant USP22 produced in baculovirus-infected Sf9 insect cells showed that USP22 is relatively inactive in deubiquitination assays using ubiquitin-AMC or the suicide substrate ubiquitin vinylsulfone (UbVS) (Fig. 4A). When recombinant USP22 was tested for its ability to deubiquitinate purified intact nucleosomes, it was unable to remove ubiquitin conjugated to any of the tested histones (Fig. 4B). Endogenous cellular USP22 in whole-cell lysates was also primarily in the inactive state compared to USP10, an unrelated Dub (Fig. 4C). Extending the incubation times of the enzymatic reactions in cell lysates did not increase the mobility shift, suggesting that only a subset of USP22 is active in cells.
Fig 4.
Human USP22 requires complex formation for deubiquitinating activity. (A) (Top) Purified recombinant human USP22 or catalytically inactive USP22-C185S was incubated with either DMSO or 5 μM UbVS. A representative gel stained with Coomassie brilliant blue is shown. (Middle) Reactions were performed as described above with various concentrations of DTT. (Bottom) Kinetic deubiquitinating activity assay of recombinant USP22 or the catalytic core of USP2 (USP2cc) with the substrate Ub-AMC. Data are presented in relative fluorescence units (RFU). (B) Purified intact mononucleosomes were incubated with full-length or catalytic inactive (CS) recombinant human Dubs and immunoblotted with the indicated antibodies. (C) Equal amounts of whole-cell lysates from HEK 293T cells were treated with DMSO or 5 μM UbVS for the indicated times and immunoblotted with the indicated antibodies. (D) (Left) Whole-cell lysates from FLAG-HA-USP22-expressing HEK 293T cells were separated by gel filtration and immunoblotted; (right) fractions from gel filtration samples were pooled on the basis of size, purified by IP, and treated with DMSO or 5 μM UbVS. FPLC, fast-performance liquid chromatography.
To gain insights into whether complex formation and the activity of USP22 might be regulated by SIRT1, we fractionated USP22-containing complexes by molecular mass using gel filtration (Fig. 4D, left). Immunoprecipitation of FLAG-HA-USP22 from pooled Superose 6 fractions revealed USP22 to be present in three separate pools: monomeric, SIRT1-containing, and SAGA-containing pools. These immunoprecipitated pools were then subjected to Dub activity assays using UbVS to analyze USP22 enzymatic activity. The USP22 in high-molecular-mass fractions containing SAGA components was significantly more active than that in lower-molecular-mass pools containing either SIRT1-USP22 or monomeric USP22 (Fig. 4D, right). These data confirm the previously published reports (72, 74–76) demonstrating that USP22 deubiquitinating activity requires DUBm assembly.
To assess more directly the activity of USP22 associated with SIRT1, we treated immunoprecipitated SIRT1 with UbVS and blotted for the endogenous interacting USP22. Consistent with our fractionation results, the activity of the USP22 bound to SIRT1 was unaltered compared to that of the total material (Fig. 5A). Surprisingly, although the SIRT1 H363Y interaction with USP22 was drastically reduced, the remaining associated Dub was almost completely active. Considering that binding of the DUBm components ATXN7, ATXN7L3, and ENY2 is required to induce the conformational change necessary for USP22 enzymatic activity, we predicted that SIRT1 might be interacting with SAGA. Both SIRT1 WT and H363Y were capable of interacting with SAGA components when retrovirally expressed (Fig. 5B). Although overexpressed SIRT1 is capable of binding SAGA, our initial studies did not reveal an interaction between endogenous proteins (Fig. 1). Due to the potential catalytic and transient nature of this interaction, we postulated that we might capture the endogenous interaction by cross-linking with DSP. Using this technique, we observed an interaction between endogenous SIRT1 and GCN5 (Fig. 5C). These results suggest that SIRT1 directly interacts with SAGA components, most likely transiently.
Fig 5.
SIRT1 associates with the SAGA complex. (A) (Top) The indicated FLAG-HA-tagged proteins were isolated from HEK 293T cells, and Dub activity was analyzed by reactions with UbVS; (bottom) samples were prepared as described above, but wild-type SIRT1 samples were diluted as indicated in assay buffer before adding UbVS. (B) FLAG-HA-tagged proteins were isolated from HEK 293T cells and immunoblotted for the indicated proteins. (C) Control IgG or SIRT1-specific antibodies were used to IP endogenous SIRT1 from DSP-cross-linked HEK 293T cells. Immunoblots were probed with the indicated antibodies.
Acetylation of USP22 regulates protein-protein interaction.
Due to the observed differences in USP22 association between wild type and catalytically inactive SIRT1, we hypothesized that USP22 and, potentially, SAGA itself are substrates of SIRT1. Using several pan-acetyl-specific antibodies, we assessed the global acetylation of USP22 in the absence or presence of TSA and NAM, broad-spectrum histone deacetylase (HDAC) and sirtuin inhibitors, respectively (77, 78). An increase in USP22 acetylation in the presence of NAM and TSA was detected (Fig. 6A). High-resolution mass spectrometry was then used to identify several novel acetylation sites on USP22, in addition to the single site previously determined by large-scale human acetylome studies (61) (Fig. 6B).
Fig 6.
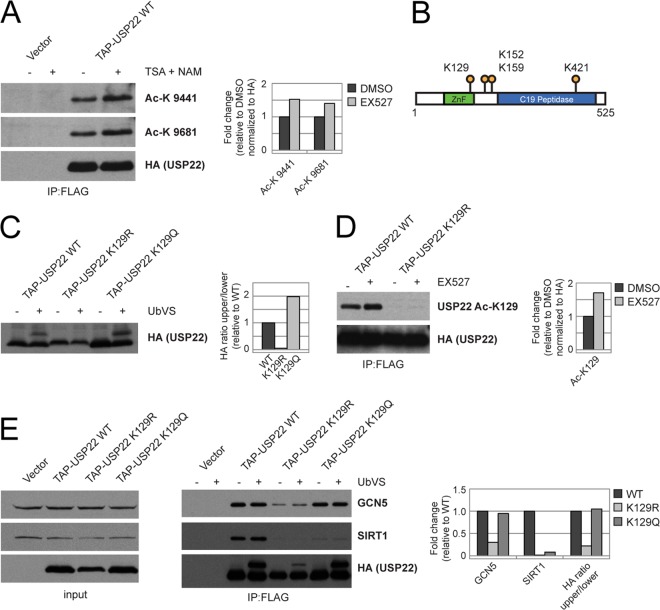
Acetylation of USP22 regulates protein-protein interaction. (A) FLAG-HA-USP22 was purified from HEK 293T cells treated with DMSO or a combination of TSA (400 nM) and nicotinamide (20 mM) and immunoblotted with panacetyllysine (Ac-K) antibodies 9441 and 9681. (B) Map of identified acetyllysine residues on USP22. (C) Cell lysates from HEK 293T cells stably expressing FLAG-HA-USP22 wild type, K129R, or K129Q were treated with 5 μM UbVS and then immunoblotted for HA. (D) FLAG-HA-USP22 WT or K129R was purified from HEK 293T cells treated with DMSO or EX527 (10 μM) and immunoblotted with the indicated antibodies. (E) The indicated FLAG-HA-USP22 mutants were purified from HEK 293T cells and subjected to DMSO or 5 μM UbVS and immunoblotted as indicated. Densitometry was performed using the ImageJ program (NIH).
The identification of lysine 129 (K129) as an acetylated residue was particularly relevant due to its presence within the ZnF-UBP domain, a domain that is necessary for the interaction with SIRT1 (Fig. 2). Mutation of K129 to arginine, to mimic the deacetylated state, resulted in a decrease in USP22 activity, as measured by the UbVS activity assay. However, upon mutation of K129 to glutamine, to mimic the acetylated state, we saw little, if any, change in USP22 activity (Fig. 6C). To monitor the acetylation status of this residue, we generated an acetyl-specific antibody that failed to detect this residue when mutated to an arginine. Treatment of FLAG-HA-USP22-expressing cells with EX527, a specific SIRT1 inhibitor, resulted in an increase in K129 acetylation, providing evidence that SIRT1 might alter acetylation of USP22 (Fig. 6D). These results suggested that K129 acetylation may be important for regulating the activity of USP22. As USP22 deubiquitinase activity is directly associated with SAGA complex formation, it was possible that K129 mutants would display altered binding to the SAGA complex. Consistent with this, mutation of K129 to arginine resulted in decreased binding of USP22 to GCN5 as well as decreased activity of USP22 (Fig. 6E). We cannot rule out the possibility that this lysine is critical to the structure of the UBP-ZnF. However, the fact that USP22 lysine 129 acetylation is regulated indicates that this modification may be important for modulating USP22 function.
SAGA components are acetylated in response to SIRT1 inhibition.
The observed SIRT1-mediated acetylation changes on USP22, taken together with our evidence that SIRT1 can associate with the SAGA complex, suggest a model in which USP22 recruits SIRT1 enzymatic activity to SAGA. Consistent with this possibility, we observed that several proteins that copurified with USP22 also displayed increased acetylation when cells were treated with the sirtuin inhibitor NAM or EX527 but not the HDAC inhibitor TSA (Fig. 7A). Interestingly, USP22-associated proteins isolated from cells treated with the nicotinamide phosphoribosyltransferase (NAMPT) inhibitor FK866 exhibited a pattern of acetylation similar to that for EX527, indicating that alterations in physiological levels of NAD+ can impact acetylation of these proteins, ostensibly by regulating SIRT1 activity (Fig. 7B).
Fig 7.
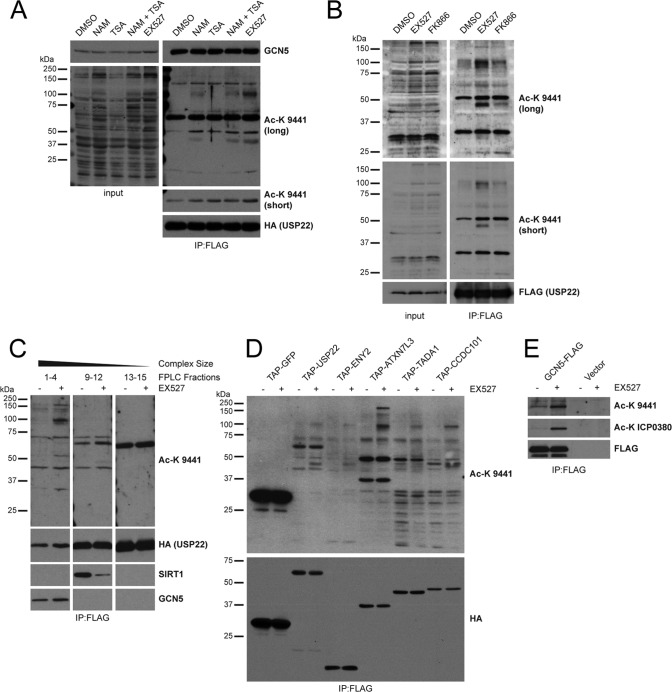
SAGA components are acetylated in response to SIRT1 inhibition. (A) FLAG-HA-USP22 purified from HEK 293T cells treated with DMSO, TSA (400 nM), NAM (20 mM), or EX527 (10 μM) was immunoblotted with the indicated antibodies. The lower Ac-K 9441 blot is a shorter exposure displaying only the main band found between 50 and 75 kDa. (B) FLAG-HA-USP22 was purified as described for panel A from HEK 293T cells treated with DMSO, EX527 (10 μM), or FK866 (10 nM). (C) Whole-cell lysates from HEK 293T cells stably expressing FLAG-HA-USP22 treated with DMSO or EX527 (10 μM) were separated by gel filtration, pooled by size, and further purified by FLAG IP. Eluted complex pools were immunoblotted with the indicated antibodies. (D) Several FLAG-HA-tagged SAGA components or a FLAG-HA-GFP control were purified from HEK 293T cells treated with DMSO or EX527 (10 μM) and immunoblotted for the indicated proteins. (E) GCN5-FLAG was purified from HEK 293T cells treated with DMSO or EX527 (10 μM) and immunoblotted with the indicated antibodies.
Subsequently, we set out to determine whether the acetylated proteins associated with USP22 were components of the SAGA complex using a variety of complementary approaches. First, USP22-containing complexes were isolated by gel filtration from FLAG-HA-USP22-expressing cells treated with DMSO or EX527. Acetylated proteins were bound to USP22 in higher-molecular-mass fractions which also contained SAGA complexes (Fig. 7C). Additionally, when we purified other components of either the DUBm (ENY2 and ATXN7L3) or the core SAGA complex (TADA1 and CCDC101) treated with and without EX527, we observed a similar acetylation pattern (Fig. 7D). Finally, acetylation of immunoprecipitated FLAG-GCN5 was also increased by SIRT1 inhibition (Fig. 7E). Taken together, these observations indicate that SAGA subunits become hyperacetylated in response to SIRT1 inhibition.
Quantitative proteomics reveals a SIRT1-regulated SAGA acetylome.
To directly measure the acetylation of SAGA subunits in response to SIRT1 inhibition, we utilized quantitative mass spectrometry techniques. We developed a work flow to provide in-depth quantitative analysis of SAGA acetylation (Fig. 8A). Using this technique, we discovered a wide array of novel acetylation sites on a number of SAGA subunits, including and greatly expanding on those that we previously observed for USP22 (Fig. 8B). Moreover, in agreement with our previous results, when we compared SIRT1 inhibitor-treated samples to controls, we found that many, but not all, SAGA complex acetylation sites were significantly increased in a SIRT1-dependent manner (Fig. 8C and D). These findings demonstrate that inhibition of SIRT1 results in increased acetylation of USP22-associated proteins and that these proteins are indeed SAGA complex components.
Fig 8.
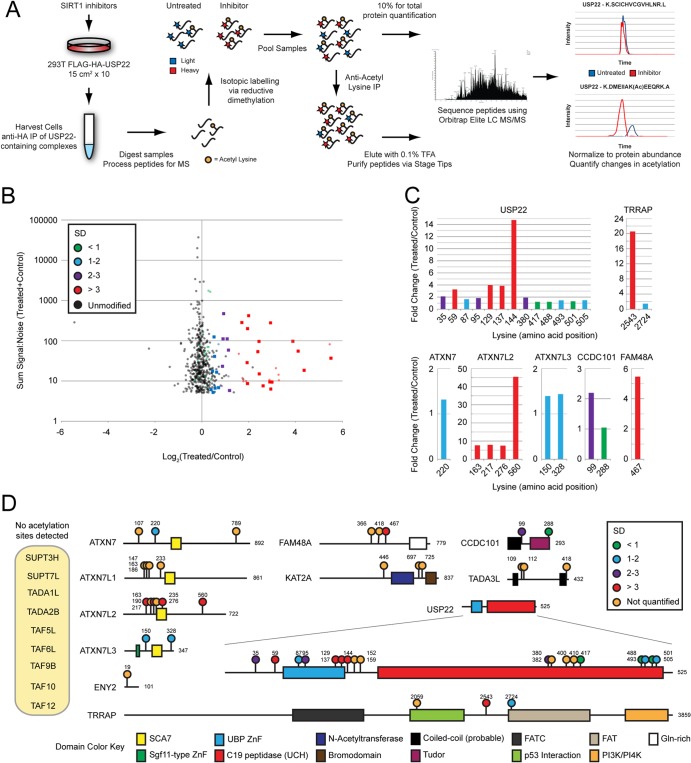
Quantitative mass spectrometry reveals a SIRT1-regulated SAGA acetylome. (A) Outline of the work flow used to analyze acetylation of USP22-associated proteins in response to SIRT1 inhibition. (B) Dot plot of individual peptides representing the sum of the signal to noise for a given peptide plotted against its fold change versus the results for the control. Both unmodified (black) and acetylated (colored) peptides were recovered following antiacetyllysine IP. Quantitative measurements of peptide-level changes following acetyllysine IP were normalized to pre-IP total protein measurements. Changes observed in the subset of unmodified peptides were assumed to represent a distribution of measurements unaffected by SIRT1 inhibition. The standard deviation (SD) of this subset was used to approximate the significance of changes in levels of acetylated peptides. Acetylated peptides representing SAGA components with changes greater than 1 standard deviation are indicated by colored squares. The dot color represents the standard deviation with respect to the unmodified peptide distribution. (C) Quantification of individual acetylation sites is displayed as the fold change versus the results for the control. Bar colors represent standard deviations, as described for panel B. (D) Domain map of SAGA components displaying acetylation sites discovered. Acetylation site colors represent standard deviations, as described for panel B. Orange acetylation sites were identified but not quantified. UCH, ubiquitin carboxyl-terminal hydrolase; FATC, FRAP, ATM, TRRAP C terminal; FAT, FRAP, ATM, TRRAP; PI3K, phosphatidylinositol 3-kinase; PI4K, phosphatidylinositol 4-kinase.
SIRT1 deacetylates SAGA subunits in vivo and in vitro.
To confirm the results observed with SIRT1 small-molecule inhibitors, siRNA oligonucleotides were used to deplete SIRT1 from HEK 293T cells expressing FLAG-HA-USP22. Removal of SIRT1 activity by siRNA induced a similar pattern of acetylation on USP22-associated proteins as EX527 (Fig. 9A). These results provided evidence that SIRT1 is proficient at deacetylating SAGA in vivo. To assess the ability of SIRT1 to directly deacetylate SAGA complex components in vitro, we purified acetylated USP22-associated proteins from HEK 293T cells treated with EX527. Acetylated complexes were then used as the substrates in deacetylation reaction mixtures containing recombinant wild-type SIRT1. SIRT1 catalyzed the removal of acetyl marks from several of the USP22-associated proteins in an NAD+-dependent fashion, thus providing further evidence that SIRT1 deacetylates SAGA components in vitro and in vivo (Fig. 9B).
Fig 9.
DUBm complex formation and SAGA interaction are regulated by SIRT1-mediated deacetylation. (A) FLAG-HA-USP22 was purified from HEK 293T cells transfected with siRNA against SIRT1 or a control siRNA treated with and without EX527 (10 μM) and immunoblotted with the indicated antibodies. (B) Eluted FLAG-HA-USP22 complexes from EX527 (10 μM)-treated cell lysates (performed as described for panel A) were incubated with or without recombinant human GST-tagged SIRT1 in the presence or absence of NAD+. Samples were immunoblotted with the indicated antibodies. (C) FLAG-HA-GFP or FLAG-HA-USP22 was purified from HEK 293T cells treated with DMSO or EX527 (10 μM). Samples were immunoblotted with the indicated antibodies. Densitometry on TAP-USP22 IP lanes was performed using the ImageJ program (NIH). (D) Samples were prepared as described for panel C from HEK 293T cells expressing FLAG-HA-tagged CCDC101. Densitometry on TAP-CCDC101 IP lanes was performed as described for panel C.
DUBm complex formation and SAGA interaction are regulated by SIRT1-mediated deacetylation.
Together, our results indicate that deacetylation of USP22 may disrupt DUBm interaction with SAGA. To test this, we immunoprecipitated FLAG-HA-USP22 from cells treated with DMSO or EX527 and examined if and to what extent the DUBm-SAGA interaction was altered. In cells treated with EX527, the increase in acetylation of USP22-K129 was associated with a modest increase in the interaction between USP22 and both ATXN7L3 and GCN5, indicating that inhibition of SIRT1 promoted DUBm/SAGA complex formation (Fig. 9C). Furthermore, upon isolation of a different core component of the SAGA complex, CCDC101, we observed a similar increase in acetylation of various SAGA components along with greater binding of USP22 and ATXN7L3, although GCN5 binding was unaltered (Fig. 9D). This indicates that the pool of DUBm-bound SAGA is affected by SIRT1-mediated deacetylation, while the GCN5 interaction with CCDC101 appears to be unaltered. Taken together, our results demonstrate that a highly specific interaction of USP22 with SIRT1 facilitates recruitment of SIRT1 deacetylase activity to the DUBm/SAGA complex, an event that regulates SAGA acetylation and complex formation.
DISCUSSION
SIRT1 plays a critical role in the regulation of a wide array of physiological and pathological processes, including diabetes, cancer, inflammatory diseases, and neurological disorders (20, 21). Using biochemical and proteomic approaches, we sought to gain further insight into the functions of SIRT1 by identifying its interaction landscape. In the present study, we describe a specific interaction between the deubiquitinating enzyme USP22 and the NAD+-dependent deacetylase SIRT1 and present a comprehensive interaction network for SIRT1 through the use of the CompPASS method (55). We further characterize the USP22-SIRT1 complex as requiring the ZnF-UBP domain and confirm that mammalian USP22 enzymatic activity minimally requires its interaction with the other components of the DUB module (75, 76, 79).
Unlike the interaction with DUBm components, the interaction of USP22 with SIRT1 did not stimulate USP22 activity, and contrary to previous reports (73), we did not see a noticeable effect of the loss of USP22 activity on either the steady-state levels or deacetylase activity of SIRT1. We hypothesize that overexpression will not increase active USP22 since other DUBm components mandatory for USP22 deubiquitinating activity are limiting for complex formation. However, differences in cell lines, knockdown efficiency, or other factors may contribute to our inability to observe these changes in SIRT1 upon loss of USP22 function. Nonetheless, both modes of regulation could potentially coexist to further fine-tune the control of transcription in response to a variety of cellular stresses such as DNA damage, endoplasmic reticulum (ER) stress, or nutrient deprivation.
In this study, we show that USP22 is an acetylated protein and that interaction with both SIRT1 and the DUBm/SAGA complex is modulated by the acetylation of a single residue found within the ZnF-UBP domain of USP22. The observation that SIRT1 H363Y binds almost exclusively to active USP22 in the context of SAGA raised the possibility that SIRT1 deacetylase activity is recruited to SAGA via USP22. Indeed, inhibition of SIRT1, but not other HDAC family members, led to hyperacetylation of several subunits of the DUBm/SAGA complex. This increase in SAGA component acetylation parallels the ability of DUBm to form a functional complex with the main SAGA unit. These data are highly suggestive that acetylation is a positive regulator of USP22 activity and that SIRT1 may act to counteract this.
Yet, it is still unclear to what extent SIRT1 deacetylation impacts DUBm-SAGA function with respect to deubiquitination-acetylation cycles at actively transcribed genes. Recent studies suggest that, in yeast, several SAGA subunits (Sgf73p, Ada3p, and Spt7p) are acetylated by Gcn5p itself in response to a rise in intracellular acetyl coenzyme A (acetyl-CoA) during entry into growth (80). Our data suggest that in addition to acetyl-CoA abundance, the acetylation of the human SAGA complex may also be regulated in response to metabolic fluctuations by physiological changes in NAD+ through SIRT1. Moreover, the previous study also maintained that recruitment of SAGA to various oxidative-phase-growth genes might be facilitated by acetylation, indicating that only a subset of SAGA coactivator targets may be regulated by SIRT1. This is substantiated by the fact that not all SAGA complexes appear to contain DUB module components and that some potentially interchangeable orthologs of ATXN7, such as ATXN7L2, are seemingly deacetylated to different extents by SIRT1. Additionally, our data also agree with previous studies suggesting that DUBm binding is not essential for SAGA formation and, likewise, SIRT1 does not alter formation of the core SAGA complex per se (81).
Our work highlights the importance of acetylation as a modification that can regulate transcriptional processes by altering not only histone N-terminal tails but also a variety of coactivators, corepressors, and bona fide transcription factors (61). Our analysis of SAGA complex acetylation demonstrates that protein acetylation is indeed widespread among transcriptional regulating proteins and indicates that the size of the acetylome may be underestimated (61). Many known targets of SIRT1 are chromatin-modifying enzymes or transcription factors, indicating that SIRT1 may act to modulate these more direct writers of the histone code in response to various cellular and environmental stresses (82). Acetylation has previously been shown to act as a regulatory switch that can alter further coordinated modifications such as ubiquitination, sumoylation, and phosphorylation, thereby adjusting protein stability, localization, and signaling (83–88). We found that acetylation, in the context of the DUBm/SAGA complex, alters protein interactions. It will be interesting to further test whether it is the acetylation itself, possibly by regulating the surface charge, which affects this characteristic directly or, rather, if this effect is due to secondary modifications.
It has recently been shown that CCDC101 (Sgf29) recognizes H3K4me2/3 and, additionally, that SAGA and another similar complex called ATAC regulate distinct sets of genes (89–91). Although data suggesting that under some conditions DUBm is important for coactivation exist, a critical question remains whether the global subset of genes where DUBm components bind within the genome are actively regulated by DUBm components. Some of these data may be provided by examining the overlap between existing chromatin IP sequencing data sets for SAGA subunits (89, 91) with transcriptional profiles observed upon alteration of histone H2B ubiquitination (92, 93). Furthermore, we speculate that at some loci, there might be overlapping binding sites for SAGA, DUBm, and SIRT1. Such a mechanism would serve to differentiate the various pools of SAGA coactivator complexes and may underlie their potential roles in transcriptional activation/repression and epigenetic inheritance.
In summary, our study identifies a specific interaction between SIRT1 and USP22 that is dependent on acetylation of the ZnF-UBP domain of USP22. This results in the recruitment of deacetylase activity to regulate the acetylation of the SAGA coactivator complex and the formation of deubiquitinating-competent DUBm/SAGA complexes. This study provides a framework for understanding the expanding role of protein lysine acetylation in fine-tuning transcription and chromatin condensation in response to nutrient homeostasis and the fluctuation of metabolic intermediates.
ACKNOWLEDGMENTS
The work was supported by NIH/NIA grants AG19719 and AG027916-04S2, the Ellison Medical Foundation, and the Glenn Foundation for Medical Research to D.A.S., by NIH grant GM054137 to J.W.H., and by NIH grant GM67945 to S.P.G. E.J.B was supported by a postdoctoral fellowship from the Damon Runyon Cancer Research Foundation.
We thank Didier Devys for the USP22 antibody, Weiwei Dang and Shelley Berger for reagents and advice, and Mathew Sowa and John Lydeard for technical assistance.
Footnotes
Published ahead of print 4 February 2013
REFERENCES
- 1. Imai S, Armstrong CM, Kaeberlein M, Guarente L. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795–800 [DOI] [PubMed] [Google Scholar]
- 2. Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. U. S. A. 97:6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. 1999. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99:735–745 [DOI] [PubMed] [Google Scholar]
- 4. Frye RA. 1999. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 260:273–279 [DOI] [PubMed] [Google Scholar]
- 5. Guarente L. 2000. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 14:1021–1026 [PubMed] [Google Scholar]
- 6. Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. 2004. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell 16:93–105 [DOI] [PubMed] [Google Scholar]
- 7. Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. 2001. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107:137–148 [DOI] [PubMed] [Google Scholar]
- 8. Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149–159 [DOI] [PubMed] [Google Scholar]
- 9. Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. 2004. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303:2011–2015 [DOI] [PubMed] [Google Scholar]
- 10. Huang H, Tindall DJ. 2007. Dynamic FoxO transcription factors. J. Cell Sci. 120:2479–2487 [DOI] [PubMed] [Google Scholar]
- 11. Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. 2004. Mammalian SIRT1 represses forkhead transcription factors. Cell 116:551–563 [DOI] [PubMed] [Google Scholar]
- 12. Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. 2004. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305:390–392 [DOI] [PubMed] [Google Scholar]
- 13. Senawong T, Peterson VJ, Avram D, Shepherd DM, Frye RA, Minucci S, Leid M. 2003. Involvement of the histone deacetylase SIRT1 in chicken ovalbumin upstream promoter transcription factor (COUP-TF)-interacting protein 2-mediated transcriptional repression. J. Biol. Chem. 278:43041–43050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. 2007. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol. Cell 28:91–106 [DOI] [PubMed] [Google Scholar]
- 15. Fu M, Liu M, Sauve AA, Jiao X, Zhang X, Wu X, Powell MJ, Yang T, Gu W, Avantaggiati ML, Pattabiraman N, Pestell TG, Wang F, Quong AA, Wang C, Pestell RG. 2006. Hormonal control of androgen receptor function through SIRT1. Mol. Cell. Biol. 26:8122–8135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang R, Chen HZ, Liu JJ, Jia YY, Zhang ZQ, Yang RF, Zhang Y, Xu J, Wei YS, Liu DP, Liang CC. 2010. SIRT1 suppresses activator protein-1 transcriptional activity and cyclooxygenase-2 expression in macrophages. J. Biol. Chem. 285:7097–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yuan J, Minter-Dykhouse K, Lou Z. 2009. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J. Cell Biol. 185:203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. 2004. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23:2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. 2005. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434:113–118 [DOI] [PubMed] [Google Scholar]
- 20. Donmez G, Guarente L. 2010. Aging and disease: connections to sirtuins. Aging Cell 9:285–290 [DOI] [PubMed] [Google Scholar]
- 21. Haigis MC, Sinclair DA. 2010. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5:253–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Imai S. 2011. Dissecting systemic control of metabolism and aging in the NAD world: the importance of SIRT1 and NAMPT-mediated NAD biosynthesis. FEBS Lett. 585:1657–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu J, Auwerx J. 2010. Protein deacetylation by SIRT1: an emerging key post-translational modification in metabolic regulation. Pharm. Res. 62:35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. 2006. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell 127:1109–1122 [DOI] [PubMed] [Google Scholar]
- 25. Nemoto S, Fergusson MM, Finkel T. 2005. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha}. J. Biol. Chem. 280:16456–16460 [DOI] [PubMed] [Google Scholar]
- 26. Price NL, Gomes AP, Ling AJ, Duarte FV, Martin-Montalvo A, North BJ, Agarwal B, Ye L, Ramadori G, Teodoro JS, Hubbard BP, Varela AT, Davis JG, Varamini B, Hafner A, Moaddel R, Rolo AP, Coppari R, Palmeira CM, de Cabo R, Baur JA, Sinclair DA. 2012. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 15:675–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. 2004. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 429:771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, III, Olefsky J, Guarente L, Montminy M. 2008. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature 456:269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S. 2005. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2:105–117 [DOI] [PubMed] [Google Scholar]
- 30. Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, Furneaux H, Gorospe M. 2007. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell 25:543–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. 2005. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell 123:437–448 [DOI] [PubMed] [Google Scholar]
- 32. Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, Nemoto S, Finkel T, Gu W, Cress WD, Chen J. 2006. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat. Cell Biol. 8:1025–1031 [DOI] [PubMed] [Google Scholar]
- 33. Nemoto S, Fergusson MM, Finkel T. 2004. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science 306:2105–2108 [DOI] [PubMed] [Google Scholar]
- 34. Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, Carruba MO. 2005. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science 310:314–317 [DOI] [PubMed] [Google Scholar]
- 35. Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R, Bordone L. 2009. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS One 4:e8414 doi:10.1371/journal.pone.0008414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sasaki T, Maier B, Koclega KD, Chruszcz M, Gluba W, Stukenberg PT, Minor W, Scrable H. 2008. Phosphorylation regulates SIRT1 function. PLoS One 3:e4020 doi:10.1371/journal.pone.0004020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang Y, Fu W, Chen J, Olashaw N, Zhang X, Nicosia SV, Bhalla K, Bai W. 2007. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat. Cell Biol. 9:1253–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gao Z, Zhang J, Kheterpal I, Kennedy N, Davis RJ, Ye J. 2011. Sirtuin 1 (SIRT1) protein degradation in response to persistent c-Jun N-terminal kinase 1 (JNK1) activation contributes to hepatic steatosis in obesity. J. Biol. Chem. 286:22227–22234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kang H, Suh JY, Jung YS, Jung JW, Kim MK, Chung JH. 2011. Peptide switch is essential for Sirt1 deacetylase activity. Mol. Cell 44:203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blander G, Olejnik J, Krzymanska-Olejnik E, McDonagh T, Haigis M, Yaffe MB, Guarente L. 2005. SIRT1 shows no substrate specificity in vitro. J. Biol. Chem. 280:9780–9785 [DOI] [PubMed] [Google Scholar]
- 41. Garske AL, Denu JM. 2006. SIRT1 top 40 hits: use of one-bead, one-compound acetyl-peptide libraries and quantum dots to probe deacetylase specificity. Biochemistry 45:94–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17:2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, Emili A, Hughes TR, Greenblatt JF, Berger SL. 2005. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol. Cell. Biol. 25:1162–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pijnappel WW, Timmers HT. 2008. Dubbing SAGA unveils new epigenetic crosstalk. Mol. Cell 29:152–154 [DOI] [PubMed] [Google Scholar]
- 45. Zhang XY, Pfeiffer HK, Thorne AW, McMahon SB. 2008. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle 7:1522–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. 2008. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell 29:102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, Georgieva SG, Schule R, Takeyama K, Kato S, Tora L, Devys D. 2008. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell 29:92–101 [DOI] [PubMed] [Google Scholar]
- 48. Glinsky GV. 2006. Genomic models of metastatic cancer: functional analysis of death-from-cancer signature genes reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype with altered cell cycle control and activated polycomb group (PcG) protein chromatin silencing pathway. Cell Cycle 5:1208–1216 [DOI] [PubMed] [Google Scholar]
- 49. Glinsky GV, Berezovska O, Glinskii AB. 2005. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Invest. 115:1503–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu YL, Yang YM, Xu H, Dong XS. 2011. Aberrant expression of USP22 is associated with liver metastasis and poor prognosis of colorectal cancer. J. Surg. Oncol. 103:283–289 [DOI] [PubMed] [Google Scholar]
- 51. Zhang Y, Yao L, Zhang X, Ji H, Wang L, Sun S, Pang D. 2011. Elevated expression of USP22 in correlation with poor prognosis in patients with invasive breast cancer. J. Cancer Res. Clin. Oncol. 137:1245–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, Chang S, Dent SY. 2009. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol. Cell 35:352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Atanassov BS, Dent SY. 2011. USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep. 12:924–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chipumuro E, Henriksen MA. 2012. The ubiquitin hydrolase USP22 contributes to 3′-end processing of JAK-STAT-inducible genes. FASEB J. 26:842–854 [DOI] [PubMed] [Google Scholar]
- 55. Sowa ME, Bennett EJ, Gygi SP, Harper JW. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2:1896–1906 [DOI] [PubMed] [Google Scholar]
- 57. Elias JE, Gygi SP. 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4:207–214 [DOI] [PubMed] [Google Scholar]
- 58. Behrends C, Sowa ME, Gygi SP, Harper JW. 2010. Network organization of the human autophagy system. Nature 466:68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rappsilber J, Ishihama Y, Mann M. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75:663–670 [DOI] [PubMed] [Google Scholar]
- 60. Broberg A. 2007. High-performance liquid chromatography/electrospray ionization ion-trap mass spectrometry for analysis of oligosaccharides derivatized by reductive amination and N,N-dimethylation. Carbohydr. Res. 342:1462–1469 [DOI] [PubMed] [Google Scholar]
- 61. Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834–840 [DOI] [PubMed] [Google Scholar]
- 62. Haas W, Faherty BK, Gerber SA, Elias JE, Beausoleil SA, Bakalarski CE, Li X, Villen J, Gygi SP. 2006. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol. Cell. Proteomics 5:1326–1337 [DOI] [PubMed] [Google Scholar]
- 63. Eng JK, McCormack AL, Yates JR. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976–989 [DOI] [PubMed] [Google Scholar]
- 64. Dey S, Bakthavatchalu V, Tseng MT, Wu P, Florence RL, Grulke EA, Yokel RA, Dhar SK, Yang HS, Chen Y, St Clair DK. 2008. Interactions between SIRT1 and AP-1 reveal a mechanistic insight into the growth promoting properties of alumina (Al2O3) nanoparticles in mouse skin epithelial cells. Carcinogenesis 29:1920–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gao Z, Ye J. 2008. Inhibition of transcriptional activity of c-JUN by SIRT1. Biochem. Biophys. Res. Commun. 376:793–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang J, Lee SM, Shannon S, Gao B, Chen W, Chen A, Divekar R, McBurney MW, Braley-Mullen H, Zaghouani H, Fang D. 2009. The type III histone deacetylase Sirt1 is essential for maintenance of T cell tolerance in mice. J. Clin. Invest. 119:3048–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Qiang L, Lin HV, Kim-Muller JY, Welch CL, Gu W, Accili D. 2011. Proatherogenic abnormalities of lipid metabolism in SirT1 transgenic mice are mediated through Creb deacetylation. Cell Metab. 14:758–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kim EJ, Kho JH, Kang MR, Um SJ. 2007. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol. Cell 28:277–290 [DOI] [PubMed] [Google Scholar]
- 69. Kim JE, Chen J, Lou Z. 2008. DBC1 is a negative regulator of SIRT1. Nature 451:583–586 [DOI] [PubMed] [Google Scholar]
- 70. Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. 2008. Negative regulation of the deacetylase SIRT1 by DBC1. Nature 451:587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rajamohan SB, Pillai VB, Gupta M, Sundaresan NR, Birukov KG, Samant S, Hottiger MO, Gupta MP. 2009. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol. Cell. Biol. 29:4116–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bonnet J, Romier C, Tora L, Devys D. 2008. Zinc-finger UBPs: regulators of deubiquitylation. Trends Biochem. Sci. 33:369–375 [DOI] [PubMed] [Google Scholar]
- 73. Lin Z, Yang H, Kong Q, Li J, Lee SM, Gao B, Dong H, Wei J, Song J, Zhang DD, Fang D. 2012. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol. Cell 46:484–494 [DOI] [PubMed] [Google Scholar]
- 74. Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. 2008. Yeast ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat. Cell Biol. 10:707–715 [DOI] [PubMed] [Google Scholar]
- 75. Kohler A, Zimmerman E, Schneider M, Hurt E, Zheng N. 2010. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141:606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C. 2010. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328:1025–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277:45099–45107 [DOI] [PubMed] [Google Scholar]
- 78. Yoshida M, Kijima M, Akita M, Beppu T. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265:17174–17179 [PubMed] [Google Scholar]
- 79. Lang G, Bonnet J, Umlauf D, Karmodiya K, Koffler J, Stierle M, Devys D, Tora L. 2011. The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Mol. Cell. Biol. 31:3734–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cai L, Sutter BM, Li B, Tu BP. 2011. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 42:426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Koutelou E, Hirsch CL, Dent SY. 2010. Multiple faces of the SAGA complex. Curr. Opin. Cell Biol. 22:374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature 403:41–45 [DOI] [PubMed] [Google Scholar]
- 83. Ahn SH, Diaz RL, Grunstein M, Allis CD. 2006. Histone H2B deacetylation at lysine 11 is required for yeast apoptosis induced by phosphorylation of H2B at serine 10. Mol. Cell 24:211–220 [DOI] [PubMed] [Google Scholar]
- 84. Choi E, Choe H, Min J, Choi JY, Kim J, Lee H. 2009. BubR1 acetylation at prometaphase is required for modulating APC/C activity and timing of mitosis. EMBO J. 28:2077–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. de la Vega L, Grishina I, Moreno R, Kruger M, Braun T, Schmitz ML. 2012. A redox-regulated SUMO/acetylation switch of HIPK2 controls the survival threshold to oxidative stress. Mol. Cell 46:472–483 [DOI] [PubMed] [Google Scholar]
- 86. Palermo R, Checquolo S, Giovenco A, Grazioli P, Kumar V, Campese AF, Giorgi A, Napolitano M, Canettieri G, Ferrara G, Schinina ME, Maroder M, Frati L, Gulino A, Vacca A, Screpanti I. 2012. Acetylation controls Notch3 stability and function in T-cell leukemia. Oncogene 31:3807–3817 [DOI] [PubMed] [Google Scholar]
- 87. Stankovic-Valentin N, Deltour S, Seeler J, Pinte S, Vergoten G, Guerardel C, Dejean A, Leprince D. 2007. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psiKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol. Cell. Biol. 27:2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ullmann R, Chien CD, Avantaggiati ML, Muller S. 2012. An acetylation switch regulates SUMO-dependent protein interaction networks. Mol. Cell 46:759–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Krebs AR, Karmodiya K, Lindahl-Allen M, Struhl K, Tora L. 2011. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol. Cell 44:410–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nagy Z, Riss A, Fujiyama S, Krebs A, Orpinell M, Jansen P, Cohen A, Stunnenberg HG, Kato S, Tora L. 2010. The metazoan ATAC and SAGA coactivator HAT complexes regulate different sets of inducible target genes. Cell. Mol. Life Sci. 67:611–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M. 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142:967–980 [DOI] [PubMed] [Google Scholar]
- 92. Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. 2008. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell Biol. 10:483–488 [DOI] [PubMed] [Google Scholar]
- 93. Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, Hublarova P, Moyal L, Gana-Weisz M, Shiloh Y, Yarden Y, Johnsen SA, Vojtesek B, Berger SL, Oren M. 2008. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 22:2664–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]