Abstract
DHX33 is a pivotal DEAH-box RNA helicase in the multistep process of RNA polymerase I-directed transcription of the ribosomal DNA locus. We explored the regulation of DHX33 expression by RasV12 and ARF to determine DHX33's role in sensing these opposing signals to regulate ribosome biogenesis. In wild-type primary fibroblasts, RasV12 infection induced a transient increase in DHX33 protein level, as well as an rRNA transcriptional rate that was eventually suppressed by a delayed activation of the ARF/p53 pathway. DHX33 expression was exclusively controlled at the level of translation. ARF caused a dramatic reduction in polysome-associated DHX33 mRNAs, while RasV12 led to a complete shift of existing DHX33 mRNAs to actively translating polysomes. The translation of DHX33 by RasV12 was sensitive to inhibitors of phosphatidylinositol 3-kinase, mTOR, and mitogen-activated protein and was pivotal for enhanced rRNA transcription and enhanced overall cellular protein translation. In addition, DHX33 knockdown abolished RasV12-induced rRNA transcription and protein translation and prevented both the in vitro and in vivo transforming properties of oncogenic RasV12. Our results directly implicate DHX33 as a crucial player in establishing rRNA synthesis rates in the face of RasV12 or ARF signals, adjusting ribosome biogenesis to match the appropriate growth or antigrowth signals.
INTRODUCTION
Cancers frequently harbor genetic mutations that activate oncogenes or inactivate tumor suppressors, leading to uncontrolled cell growth, evasion of apoptosis, and other enhanced cellular properties (1). To accommodate the rapid proliferation of cancer cells, several associated biological activities are also augmented in cancer cells (2). Recently, increasing evidence has shown that cancer cells often increase ribosome production to improve protein translation and cell growth (3–7). Ribosome biogenesis is frequently targeted by activated oncogenes and repressed by tumor suppressors (as reviewed in references 3 and 8). In fact, the link between nucleolar hypertrophy and tumorigenesis was recognized more than 100 years ago (8, 9). More recent data indicate that a marked increase in rRNA synthesis is a general attribute of many cancers (9, 10), which is consistent with the idea that changes in rRNA synthesis may be prerequisite alteration in the progression to cellular transformation. The rate of cancer cell proliferation in tumors is directly proportional to nucleolar size and RNA polymerase I (Pol I) activity, with overexpression of pre-rRNA correlating with poor prognosis in many cancers (10–13).
Ribosome biogenesis largely occurs in the nucleolus and is a highly coordinated biological process that includes rRNA synthesis, modification, processing, and assembly into ribosome subunits (10, 14–16). It is tightly controlled and directly linked to cell cycle events; defects in ribosome biogenesis often lead to apoptosis or cell cycle arrest (17–19). The initial step of ribosome biogenesis, ribosomal DNA (rDNA) transcription, is subject to numerous layers of regulation (20–22). Human rDNA contains >400 copies of the rRNA genes, organized in tandem arrays on five different human chromosomes. Initiation of rDNA transcription requires assembly of a specific multiprotein complex including Pol I and numerous associated proteins (3, 10). Two of these proteins are upstream binding factor (UBF) and the promoter selectivity factor, SL1/TIF-IB. Interaction of these two proteins at rDNA promoter leads to assembly of the preinitiation complex and subsequent transcriptional activation at the promoter (15, 23). Given its extreme importance in initiating ribosome biogenesis, rDNA transcription is greatly influenced by the Ras, Myc, and NPM oncogenes, as well as the ARF, p53, and PTEN tumor suppressors (14, 16, 24–29).
We previously identified the nucleolar DHX33 DEAH-box RNA helicase as an important mediator of RNA Pol I transcription through its interaction with UBF at rDNA loci following serum stimulation (30). In the present study, we explored the mechanism underlying DHX33 regulation. We now report that DHX33 is positioned at the crossroads of opposing Ras and ARF activities; oncogenic RasV12 stimulates but ARF represses translation of existing DHX33 mRNAs. In this manner we show that, DHX33 is used as an endpoint of contrasting signals to set ribosome biogenesis rates. Using xenograft models and established Ras mutant cancer cell lines, we demonstrate that DHX33 accumulation is pivotal for RasV12 to initiate tumor formation.
MATERIALS AND METHODS
Cell culture.
Wild-type mouse embryonic fibroblasts (MEFs), Arf−/− ear fibroblasts, Arf−/− MEFs, p53−/− MEFs, p53−/−; Mdm2−/− MEFs, and p53−/−; Mdm2−/−; Arf−/− MEFs were isolated from C57BL/6/Sv129 mixed mice. BxPC-3, Capan-2, Miapaca-2, and Panc-1 pancreatic cancer cell lines were kindly provided by Andrea Wang-Gillam (Washington University), BeaS2B cells were provided by Gregory Longmore (Washington University), and H441 and A549 lung cancer cells were provided by Steven Brody (Washington University). BxPC-3 and H441 cells were grown in RPMI 1640 containing 10% fetal bovine serum (FBS) with antibiotics and supplements (10 mmol of HEPES, 4.5 g of glucose, 2 mmol of l-glutamine, and 1 mmol of sodium pyruvate/liter). A549 cells were grown in F-12 medium supplemented with 10% FBS and 1% penicillin-streptomycin. All other cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS and 1% penicillin-streptomycin. All cell lines were incubated at 37°C with 5% CO2 in a humidified incubator. U0126 and LY294002 were purchased from Sigma. Rapamycin was purchased from LC laboratories.
Western blotting and antibodies.
Whole-cell lysates were prepared by incubation with 1× NP-40 buffer that included 0.5% NP-40 and 1% sodium dodecyl sulfate (SDS) supplemented with HALT protease and phosphatase inhibitors (Sigma). Lysates were cleared by centrifugation and protein concentration was tested by DC assay (Bio-Rad). Lysates were boiled with SDS sample buffer, separated by SDS-PAGE, and transferred to polyvinylidene difluoride membrane (Millipore). Membranes were blocked in 5% nonfat dry milk TBS-T buffer (10 mmol of Tris-HCl [pH 7.4]/liter, 150 mmol of NaCl/liter, 0.1% Tween 20) and incubated in primary antibodies diluted in blocking buffer at 4°C overnight. Blots were washed with TBS-T buffer and incubated with horseradish peroxidase-conjugated secondary antibodies (1:10,000; GE Healthcare) in blocking buffer at room temperature. Immune complexes were visualized with an enhanced chemiluminescence kit (GE Healthcare). Primary antibodies for immunodetection were sourced as follows: anti-ARF (rat; Santa Cruz), antitubulin (goat; Santa Cruz), anti-DHX33 (Novus), anti-S6 (Cell Signaling), anti-pS6 (Cell Signaling), anti-NF1 (Santa Cruz), anti-Ras (Santa Cruz), anti-p53 (Cell Signaling), anti-AKT (Cell Signaling), and anti-pS473-AKT (Cell Signaling).
Quantitative reverse transcription-PCR (qRT-PCR).
Primers were all designed by Primer Express 2.0 software and purchased from Integrated DNA technologies. Total RNA was extracted by NucleoSpin II (Clontech) RNA isolation kit and was reverse transcribed into cDNA by SuperScript III first-strand synthesis kit (Invitrogen). PCRs were performed on Bio-Rad C1000 thermal cycler and managed with Bio-Rad CFX96 software. For analysis of 47S rRNA transcript levels, SYBR green FastMix (Quanta Biosciences) was used and transcript quantification was performed by comparison with standard curves generated from dilution series of cDNA of human 47S rRNA (cloned in pCR2.1Topo). SYBR green mix from Bio-Rad was used for all other qRT-PCR analysis. Transcript quantification was calculated based on the value of ΔΔCT after normalization to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) values. Melting curve analysis confirmed that single products were amplified.
Focus assay.
Human cancer cell lines were infected by pLKO.1 lentivirus encoding shScrambled RNA or shRNA to knockdown DHX33, and cells were selected by puromycin for 2 days. Cells were then plated at a density of 104 per 100-mm dish and grown for 10 to 20 days. Colonies were washed with cold phosphate-buffered saline twice and fixed with 100% methanol for 10 min at room temperature. Colonies were then stained with Giemsa stain for 1 h at room temperature and washed with water before air-dried and photographed.
Soft agar assay.
A total of 104 cells were mixed in 4.0 ml of 0.3% agar–1× DMEM–10% FBS as the top agar and plated into 60-mm plates with 4.0 ml of 0.6% agar–1× DMEM–10% FBS as the base agar. Plates were incubated at 37°C and checked every 3 days, and the cells were fed with 2.0 ml of 0.3% agar–1× DMEM–10% FBS every week. The colonies were photographed and counted 2 to 3 weeks later.
Polysome profiles.
Cells (3 × 106) after transduction with the indicated virus for 96 h were treated with 10 μg of cycloheximide/ml prior to harvesting and counting. Cells were subjected to cytoplasmic ribosome fractionation as described previously using a sucrose density gradient system ranging from 7 to 47% (Teledyne ISCO). Fractions were collected, and RNA was extracted with TRIzol and converted into cDNA with superscript reverse transcriptase III (Invitrogen) before quantitative PCR analysis with the appropriate primers.
[3H]uridine pulse-chase labeling.
Cells were first infected with the indicated virus. At 3 days postinfection, the cells were replated at a confluence of 60 to 70% per plate. On the following day, the cells were then pulsed with [3H]uridine at a concentration of 2.5 μCi/ml for 30 min and chased with unradiolabeled uridine at a concentration of 5 mM for the indicated time points. Approximately 2 × 106 cells were pelleted, and the total RNA was isolated after dissolving cells in RNAsolv (Omega Biotek, Norcross, GA). Formaldehyde RNA denaturing gel was run to separate different species of rRNA and then transferred to nylon Hybond+ membrane. After UV cross-linking and spraying with Enhancer (Perkin Elmer), the membrane was exposed to film and subjected to autoradiography.
[35S]methionine incorporation.
Cells were starved in cysteine-methionine-free medium for 4 h and then pulsed with [35S]methionine (50 μCi/ml)-containing medium for 30 min before being harvested. The cells were lysed, and supernatants were precipitated with trichloroacetic acid at a concentration of 10%. Protein pellets were subsequently dissolved by 1% SDS and analyzed for protein concentration. The samples were then analyzed for radioactivity by liquid scintillation counting. The data presented were normalized based on equal amount of protein in each sample.
Mouse xenografts.
Animals were handled according to protocols approved by the Washington University Animal Studies Committee. Nude mice were purchased from Jackson laboratories. Arf-null cells after transduction with the indicated virus were injected subcutaneously with 106 cells into the flanks of mice. Tumors were dissected after 2 weeks and photographed.
RESULTS
p19ARF induction during oncogenic stress lowers DHX33 protein levels.
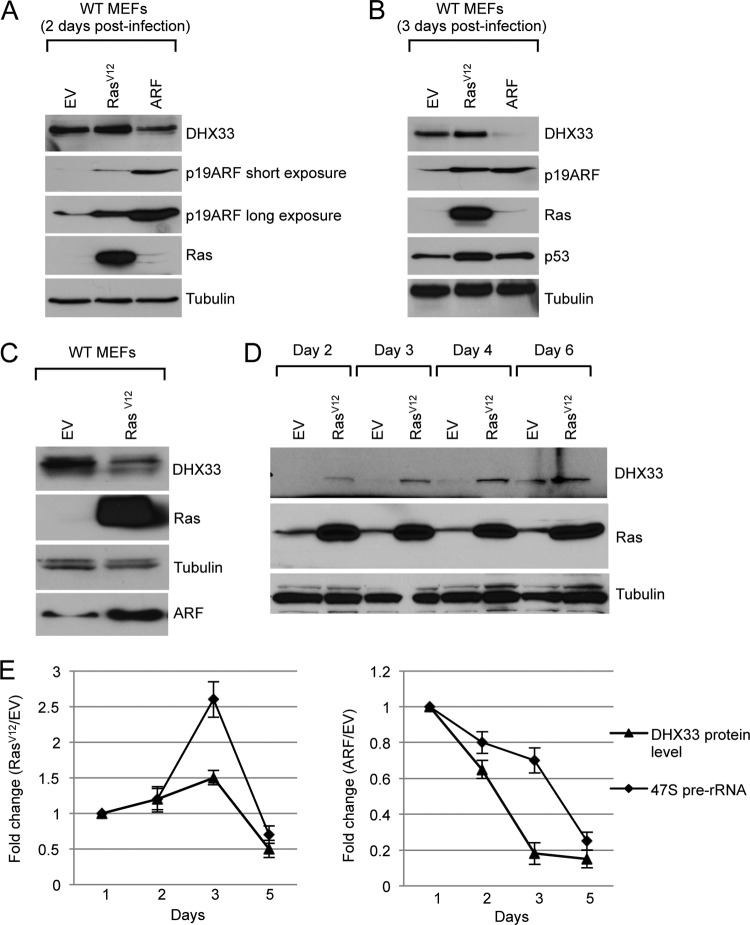
ARF is the principal tumor surveillance protein charge with preventing aberrant cell growth and proliferation during oncogenic insult (18, 31). In wild-type primary fibroblasts, oncogenic RasV12 induces ARF protein expression, resulting in subsequent p53 activation and cell cycle arrest. Numerous tumor suppressers and oncogenes are known to influence the levels and activities of key molecules involved in rDNA transcription, suggesting that rRNA transcription might be a focal point of opposing signaling moieties (3). Previously, we have shown that the DHX33 DEAH RNA box helicase is a novel regulator of rRNA transcription (30). To test whether p19ARF and RasV12 could affect DHX33 expression, wild-type (WT) MEFs were infected with either control, RasV12, or ARF-expressing retroviruses. DHX33 protein levels were analyzed at 2, 3, or 5 days postinfection. As shown, the ectopic expression of ARF resulted in a significant decrease in the DHX33 protein expression 2 days postinfection (Fig. 1A). Reduction of DHX33 by ARF continued through 3 days postinfection, when the DHX33 protein levels decreased 10-fold (Fig. 1B). In contrast, oncogenic stress by RasV12 infection resulted in slightly more DHX33 protein than control cells at 2 and 3 days postinfection (Fig. 1A and B). Noticeably, upregulation of ARF and p53 were only modest at these early time points following ectopic RasV12 expression. However, after 5 days of RasV12 infection, wild-type primary cells expressed significant levels of ARF with a resultant decrease in DHX33 protein (2-fold reduction) compared to control cells (Fig. 1C). These results indicate that two major pathways might regulate DHX33: ARF/p53 and oncogenic RasV12. The opposing activities from these two pathways caused DHX33 levels to be transiently increased and then significantly decreased. In accordance with this hypothesis, RasV12 infection of fibroblasts deficient in ARF (Arf-null MEFs) resulted in a robust increase in DHX33 levels from 2 days until 6 days postinfection (Fig. 1D). This is in contrast to a 2-fold reduction of DHX33 in WT MEFs in which endogenous ARF function is intact and induced during RasV12 infection. Thus, our results indicate that endogenous ARF is a key regulator of DHX33 expression during oncogenic stress.
Fig 1.
Regulation of DHX33 during oncogenic stress. (A) Wild-type MEFs were infected with retroviruses encoding RasV12, p19ARF, or empty vector, whole-cell lysates were prepared at 2 days postinfection and were subjected to Western blot analysis with the indicated antibodies. (B) The above-mentioned cells were harvested 3 days postinfection, and whole-cell extracts were subjected to Western blot analysis with the indicated antibodies. (C) Wild-type MEFs were infected with retroviruses encoding RasV12 or empty vector, whole-cell extracts were prepared 5 days postinfection and were subjected to Western blot analysis with the indicated antibodies. (D) Arf-null MEFs were infected with retroviruses encoding RasV12 or empty vector, whole-cell extracts were prepared from 2 days till 6 days postinfection and were subjected to Western blot analysis with the indicated antibodies. (E) Wild-type MEFs were infected with the above-mentioned retroviruses, and total RNA was extracted at 2, 3, or 5 days postinfection. Mouse 47S pre-rRNA levels were analyzed by real-time PCR and graphed in a time-dependent manner. Changes in DHX33 protein levels in the time course were also graphed after quantitation of DHX33 signals in panels A to C after normalization to the empty vector control. Error bars were taken from three independent experiments.
We next performed quantitative real-time PCR (qRT-PCR) to determine ribosome RNA transcriptional rates by analyzing 47S pre-rRNA transcript levels in both RasV12 and ARF-infected wild-type MEFs. As shown in Fig. 1E, ARF infection resulted in the downregulation of rRNA transcription in a time-dependent manner. At 5 days postinfection, 47S rRNA levels dropped to 30% of that in the control sample. Moreover, RasV12 infection first resulted in a transient increase in pre-rRNA synthesis at 2 and 3 days postinfection (up to 2.5-fold), but after 5 days postinfection, after endogenous ARF induction, the pre-rRNA levels dropped to 70% of empty vector control (Fig. 1E). This trend is in agreement with the increase in DHX33 followed by its decrease over time after RasV12 infection.
ARF regulation of DHX33 is dependent on Mdm2 and p53.
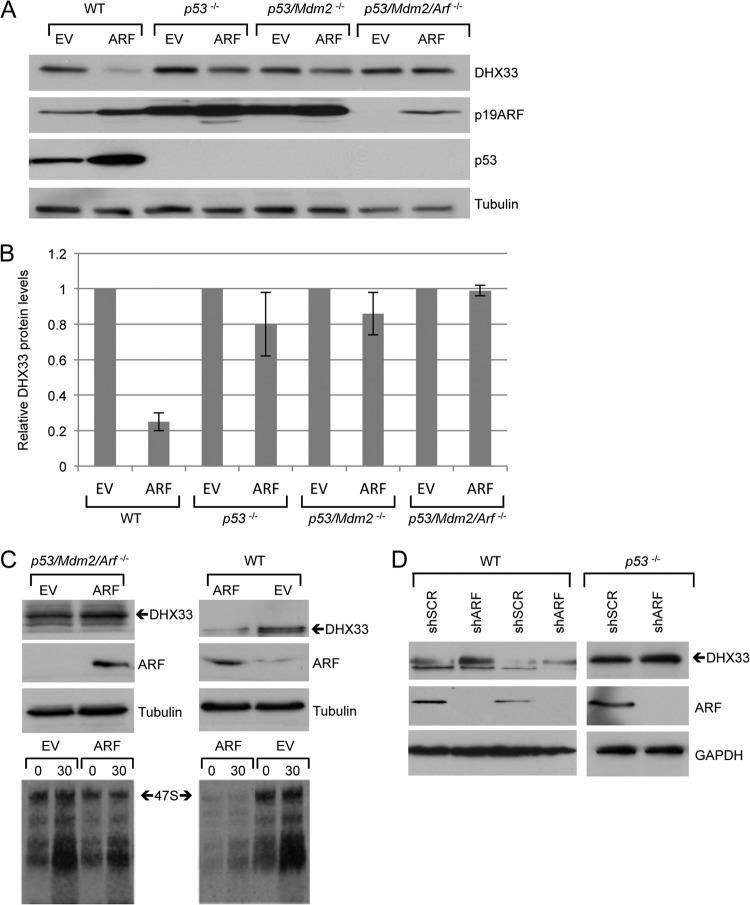
The ARF tumor suppresser has p53-dependent and -independent functions (32). Wild-type MEFs maintain an intact p53 pathway downstream of ARF, suggesting that regulation of DHX33 by ARF in WT MEFs could be p53 dependent. To study whether this regulation occurs in a p53-dependent manner, we infected p53−/− MEFs, p53−/−; Mdm2−/− (DKO) MEFs, and p53−/−; Mdm2−/−; Arf−/− (TKO) MEFs with ARF-expressing retroviruses. As shown in Fig. 2, ARF overexpression in WT MEFs resulted in a significant reduction in DHX33 levels. However, the reduction of DHX33 was far less significant in p53-null MEFs and DKO MEFs. In TKO MEFs, we observed no reduction in DHX33 protein levels, indicating that reduction of DHX33 by ARF requires p53 (Fig. 2B). In addition, we found that the infection of ARF in wild-type MEFs resulted in a much greater inhibition of ribosome RNA synthesis than in TKO MEFs (Fig. 2C). Our results indicate that ARF inhibits ribosome biogenesis not only in a p53-independent manner but also in a p53-dependent manner. Knockdown of endogenous ARF only mildly enhanced DHX33 protein expression in p53−/− MEFs (Fig. 2D), suggesting that p53 is required for DHX33 induction following loss of ARF.
Fig 2.
Reduction of DHX33 by ARF infection is dependent on p53 and Mdm2. (A) Wild-type MEFs, p53-null MEFs, p53−/−; Mdm2−/− MEFs, and p53−/−; Mdm2−/−; Arf−/− MEFs were infected with retroviruses encoding either pBABE empty vector or pBABE-HA-ARF. Whole-cell lysates were prepared 5 days postinfection and were subjected to Western blot analysis by the indicated antibodies. (B) Quantitation of the DHX33 protein levels after normalization to empty vector in each group, error bars were taken from three independent experiments. (C) MEFs were infected by retrovirus encoding pBABE empty vector or pBABE-HA-ARF. At 4 days postinfection, cells were harvested and analyzed by Western blotting with the indicated antibodies. At 4 days postinfection, cells were also pulsed by [3H]uridine and chased at the indicated time points. Total RNA were analyzed for 47S pre-rRNA levels. (D) WT MEFs or p53-null MEFs were infected with lentivirus encoding either shSCR or shARF. At 4 days postinfection, the cells were harvested and subjected to Western blot analysis with the indicated antibodies.
ARF reduces DHX33 protein levels through a translational control mechanism.
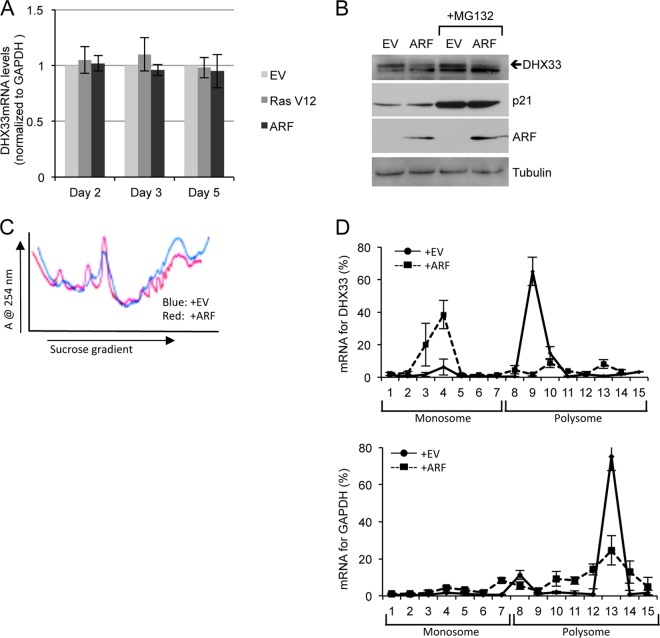
To dissect the mechanism of DHX33 reduction by ARF, we first analyzed DHX33 mRNA levels. qRT-PCR was performed on total RNAs isolated from ARF- and RasV12-infected cells at 2, 3, and 5 days postinfection. Both GAPDH mRNA and actin mRNAs were used as internal controls. We observed no significant change in DHX33 mRNA expression at each time point after ARF or RasV12 infection of WT MEFs compared to the empty vector control (Fig. 3A). These results indicate that reduction of DHX33 by ARF does not occur by transcriptional regulation. ARF has been previously shown to influence the stability of several proteins (33, 34). To determine whether DHX33 protein reduction was due to accelerated protein degradation upon ARF induction, cells were treated with MG132, a 26S proteasome inhibitor, for 6 h. As shown in Fig. 3B, we found that DHX33 was not stabilized in the presence of MG132. As a positive control, p21CIP1 was stabilized to a significant degree with MG132 treatment, demonstrating that MG132 is functioning as expected to inhibit 26S proteasome. These results imply that reduction of DHX33 in the presence of ARF is not due to accelerated protein degradation.
Fig 3.
Induction of ARF inhibits DHX33 translation. (A) Wild-type MEFs were infected with retroviruses encoding empty vector, p19ARF, or RasV12, and the total RNA was extracted from each sample at 2, 3, or 5 days postinfection. DHX33 mRNA levels were analyzed by qPCR with GAPDH as an internal control. Bars represent the standard deviation taken from three separate experiments. (B) Wild-type MEFs were infected with retroviruses encoding empty vector (EV) or p19ARF. At 3 days postinfection, the cells were treated with 50 μM MG132 for 6 h, and total cell lysates were prepared and subjected to Western blot analysis with the indicated antibodies. p21 protein stabilization was used as a positive control to monitor MG132 function. (C) A total of 1.5 × 106 wild-type cells infected with retroviruses encoding empty vector or p19ARF at 3 days postinfection were subjected to cytosolic polysome fractionation. The absorbance was monitored at 254 nm, and resultant ribosome profiles are shown for each sample. (D) The above-mentioned fractions from monoribosomes or polysomes were subjected to total RNA isolation and qPCR analysis to detect DHX33 mRNA levels. GAPDH mRNA levels were used as a control. The data presented are the percentages of mRNA from each fraction calculated from a standard curve generated by a series of diluted DHX33 plasmid. Error bars were taken from two independent experiments.
To determine whether DHX33 reduction was due to translational repression of existing DHX33 mRNAs, we chose to analyze polysome-associated DHX33 mRNAs. We performed a polysome fractionation by sucrose gradient after lysis of WT MEFs that were either transduced with vector control or ARF overexpressing retroviruses (Fig. 3C). We analyzed the mRNA distribution of DHX33 in monosome and polysome fraction by qRT-PCR. As shown in Fig. 3D, we found that in ARF-infected WT MEFs, a large portion of DHX33 mRNAs (up to 60% of total mRNA) had moved into the monoribosome fractions. Conversely, empty vector-infected WT MEFs exhibited a majority of their DHX33 mRNAs associated with polysomes (70%). These data clearly indicate that ARF induction causes a translational repression of DHX33 in the cytoplasm.
DHX33 protein reduction decreases protein translation, while DHX33 overexpression enhances protein translation.
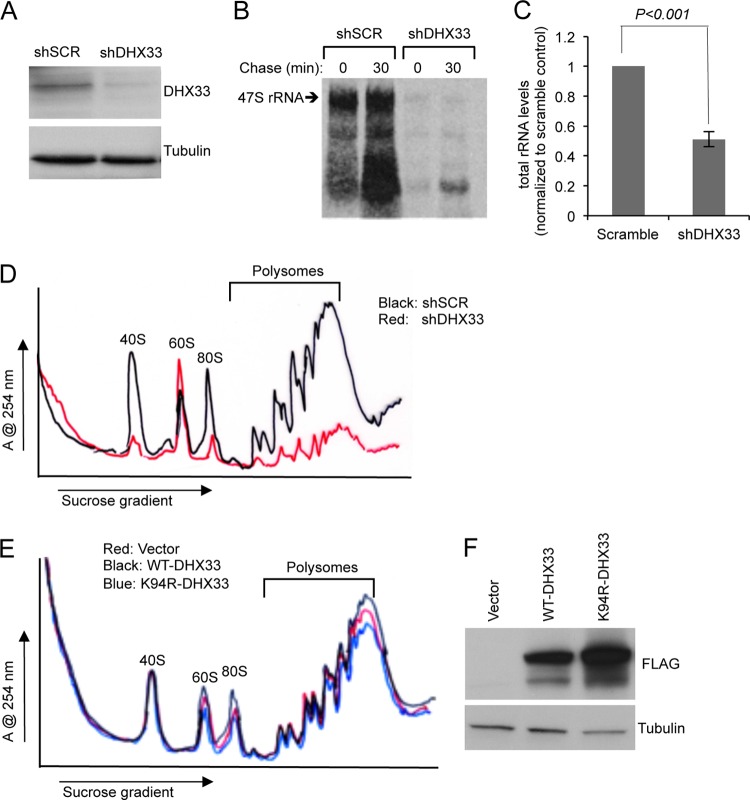
Our previous data has shown that DHX33 is an important regulator of rRNA transcription; DHX33 knockdown reduced rRNA production, while DHX33 overexpression enhanced rRNA synthesis (30). In TKO MEFs, we manipulated DHX33 levels by utilizing lentivirus infection to knockdown (Fig. 4A) or overexpress (Fig. 4E) DHX33 protein. As shown in Fig. 4B, knockdown of DHX33 nearly abolished all rRNA production. Since rRNA is the key component for ribosome assembly, we hypothesized that DHX33 knockdown should result in less available ribosomes and thus decrease overall protein translation. mRNAs undergoing active translation are bound to multiple ribosomes, forming polysomes. The level of polysomes is widely regarded as an indicator of overall protein translational activity. Therefore, we performed cytosolic ribosome fractionation using sucrose gradients to monitor polysome levels. Strikingly, we noted a significant reduction of polysomes in TKO MEFs infected with DHX33 knockdown lentiviruses (Fig. 4D). The cytosolic 40S and 80S ribosomes were also decreased dramatically. Interestingly, the 60S ribosome peak was enhanced, indicating a different dynamic regulation of 40S and 60S, even though all of the rRNA species were decreased significantly (Fig. 4C).
Fig 4.
DHX33 protein knockdown or overexpression influences ribosome biogenesis and protein translation. (A) Arf/p53/Mdm2−/− MEFs were infected by lentivirus encoding shRNA-DHX33 or shScrambled (shSCR). At 3 days after infection, the cell lysates were subjected to Western blot analysis with anti-DHX33 and tubulin antibodies. (B) Infected cells from above were pulsed with [3H]uridine and chased for the indicated time points to monitor newly synthesized rRNA. Equal numbers of cells were pelleted for total RNA extraction. RNA was separated and transferred onto nylon membranes for autoradiography. (C) Equal numbers of the above-mentioned cells were subjected to total RNA isolation and then isolated by formaldehyde RNA denaturing gel. 28S and 18S rRNA were visualized by ethidium bromide staining and quantified. Bars were taken from three different experiments. (D) Equal numbers of Arf/p53/Mdm2−/− MEFs infected by the indicated short-hairpin lentiviruses were subjected to cytosolic ribosome profile analysis at 4 days postinfection. (E and F) Arf/p53/Mdm2−/− MEFs were infected with lentiviruses encoding empty vector, DHX33 (wild type) or mutant DHX33 (K94R). At 4 days postinfection, infected cells were subjected to cytosolic polysome profile analysis (E) and Western blot analysis with the indicated antibodies (F).
Previously, we have found that wild-type DHX33 overexpression enhanced rRNA synthesis, while helicase-defective mutant of DHX33 (K94R) inhibited it (30). To determine the effect of DHX33 overexpression on cell growth, we transduced wild-type DHX33 and helicase-dead K94R mutant of DHX33 in TKO MEFs by lentivirus infection. Wild-type DHX33 only slightly enhanced 80S formation and polysome formation, whereas the K94R DHX33 mutant resulted in decreased levels of polysomes (Fig. 4E). This suggests that DHX33 is important for translation, but its overexpression might not be sufficient to significantly enhance protein synthesis. Western blot analysis showed levels of overexpressed wild-type and K94R mutant DHX33 (Fig. 4F).
RasV12 stimulates DHX33 mRNA translation.
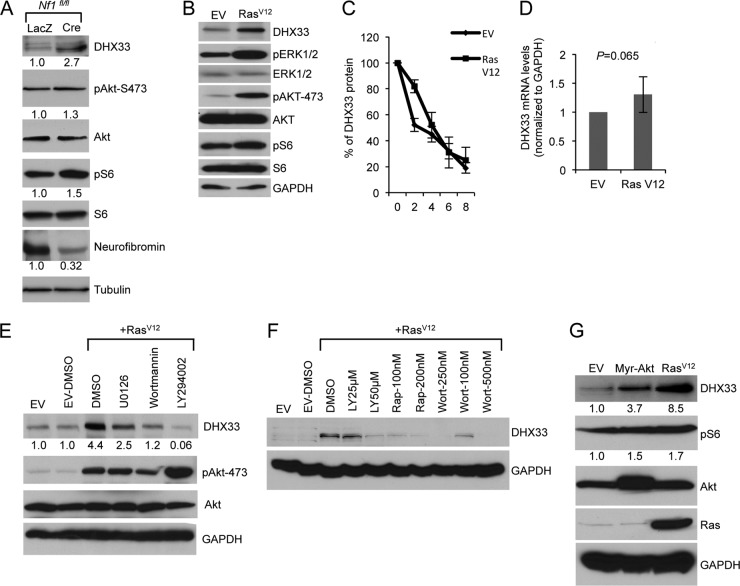
We previously showed that RasV12 expression caused a significant increase in DHX33 protein expression in Arf-null MEFs, implying that Ras hyperactivation regulates DHX33 levels in the absence of Arf. The neurofibromin (Nf1 gene) tumor suppressor protein is an upstream regulator of Ras signaling; loss of Nf1 results in irreversible activation of Ras and results in subsequent heightened growth and proliferation in vitro and in vivo tumor formation (35–37). We isolated MEFs from Nf1fl/fl mice and used adenovirus to overexpress Cre recombinase, resulting in the successful deletion of Nf1 alleles (Fig. 5A). Compared to the control Lac Z adenovirus-infected Nf1fl/fl MEFs, we found that DHX33 was upregulated 2.7-fold following Nf1 loss (Fig. 5A). As a confirmation for the activation of Ras signaling, we also detected increased levels of phospho-S6 and phospho-AKT (Fig. 5A).
Fig 5.
Ras activity induces DHX33 protein expression. (A) Nf1fl/fl MEFs were infected with adenoviruses encoding either LacZ or Cre recombinase at a multiplicity of infection of 200. At 2 days postinfection, the cells were then serum starved for 72 h. Equal amount of cell lysates were subjected to Western blot analysis with the indicated antibodies. (B) Arf-null ear fibroblasts from 2-month-old mice were infected with retroviruses encoding either pBABE empty vector (EV) or pBABE-RasV12. At 3 days postinfection, infected cells were subjected to Western blot analysis with the indicated antibodies. (C) The above-mentioned cells were treated with cycloheximide at a concentration of 80 μg/ml for the indicated times. Protein extracts from the cells pelleted from the indicated time points was subjected to Western blot analysis. Signals of DHX33 protein was graphed after normalization to GAPDH control. Bars represent errors from two independent experiments. (D) Total RNA was isolated from the above-mentioned cells and changes of DHX33 mRNA levels were analyzed by qPCR with GAPDH as a control. P is derived from five separate experiments. (E) Arf-null ear fibroblasts were infected with empty vector or RasV12. At 3 days postinfection, the cells were treated with U0126 (20 μM), wortmannin (100 nM), or LY294002 (50 μM) for 24 h. Cell lysates were subjected to Western blot analysis with the indicated antibodies. (F) Arf-null cells infected with empty vector or RasV12 were treated with rapamycin, wortmannin, or LY294002 as indicated for 24 h. Cell lysates were prepared and analyzed for DHX33 protein levels with GAPDH as a loading control. (G) Arf-null MEFs were infected with retroviruses encoding myristoylated Akt (Myr-Akt), RasV12, or empty vector. Cell lysates were prepared at 4 days postinfection after puromycin selection and analyzed by Western blotting for DHX33, pAkt-473, Akt, and GAPDH protein levels. The fold change is indicated below identified blots.
In order to dissect the mechanism of DHX33 induction by Ras signaling, we first confirmed the activation of several conserved signaling events downstream of Ras. As shown in Fig. 5B, RasV12 expression in Arf-null cells induced activation of the mitogen-activated protein kinase (MAPK) pathway as indicated by increased phospho-ERK1/2 and activation of phosphatidylinositol 3-kinase (PI3K)/AKT pathway as indicated by phospho-AKT-S473, as well as activation of mTOR pathway by increased phospho-S6. To check whether upregulation of DHX33 in this setting was due to protein stability or mRNA level changes, we performed protein half-life assays. As shown in Fig. 5C, there was no significant change in DHX33 stability in empty vector or RasV12-transduced cells. Next, we analyzed mRNA levels of DHX33 by RT-PCR and found no significant change in DHX33 mRNA in RasV12-infected cells (Fig. 5D). To dissect the mechanism of DHX33 induction by Ras, we treated RasV12-infected Arf-null cells with PI3K/AKT or MAPK pathway inhibitors. Upregulation of DHX33 was completely abolished by the PI3K pathway inhibitors wortmannin and LY294002 but only partially by MEK inhibitor U0126 (Fig. 5E), demonstrating that Ras/PI3K is the main signaling pathway that regulates DHX33 protein induction. To determine whether DHX33 upregulation was controlled by mTOR activation, we treated cells with rapamycin. As shown in Fig. 5F, rapamycin inhibited the induction of DHX33 in a dose-dependent manner to a similar extent as wortmannin and LY294002, indicating that the Ras/PI3K/mTOR pathway is primarily responsible for upregulating DHX33 translation. To further confirm these results, we infected Arf-null MEFs with a constitutively active myristoylated Akt (Myr-Akt) retrovirus and found that activation of Akt alone was able to induce DHX33 protein levels but not to the levels seen in RasV12-infected cells (Fig. 5G).
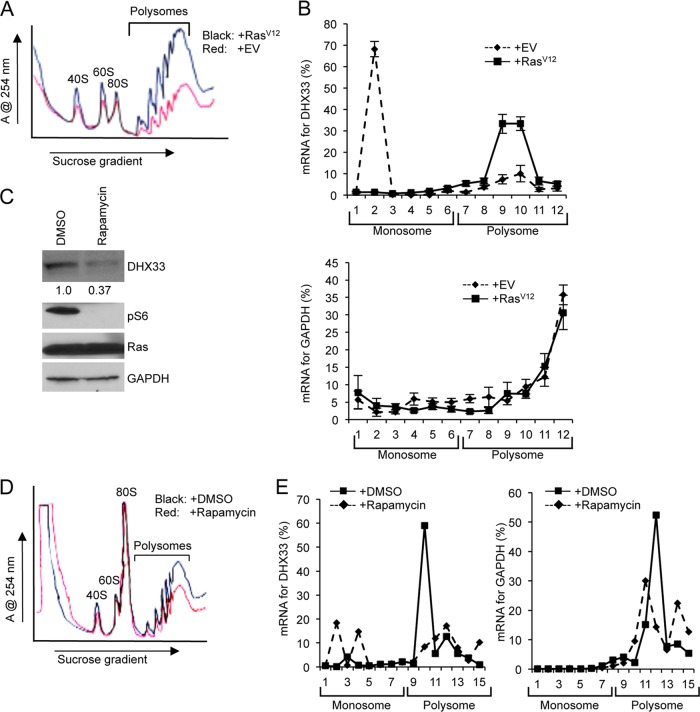
We also analyzed DHX33 mRNA distribution on polysomes. As expected, RasV12 infection significantly enhanced production of cytosolic ribosomes and polysome formation (Fig. 6A). Approximately 70% of DHX33 mRNA was not associated with polysomes in Arf-null cells (Fig. 6B). In contrast, a majority (75%) of DHX33 mRNAs associated with polysomes in Arf-null cells that were infected with RasV12 retroviruses (Fig. 6B). As a control, GAPDH mRNA distribution was also analysis and showed no significant difference between empty vector and RasV12 infection (Fig. 6B). This significant difference shows that DHX33 mRNAs are selectively translated upon RasV12 infection in the absence of Arf. To confirm that the Ras/PI3K/mTOR pathway indeed translationally regulates DHX33, we further treated the cells with rapamycin and analyzed DHX33 mRNA distribution on polysomes. As shown in Fig. 6C and D, rapamycin treatment resulted in a reduction of DHX33 protein levels and global protein translational repression. A significant proportion of DHX33 mRNA was shifted from polysomes to monoribosomes following rapamycin treatment (Fig. 6E).
Fig 6.
DHX33 protein induction is under translational control. (A) A total of 3 × 106 Arf-null cells infected with retroviruses encoding empty vector or RasV12 at 4 days postinfection were subjected to cytosolic ribosome profiling. (B) The resultant fractionations from above were analyzed by RT-PCR for DHX33 mRNA distribution on ribosomes. GAPDH was used as a negative control. Bar data were taken from three independent experiments. (C) RasV12-infected Arf-null cells at 4 days postinfection were treated with rapamycin at 100 nM for 24 h. Whole-cell extracts were then subjected to total protein analysis by Western blotting with the indicated antibodies. The fold change is indicated underneath the blots. (D) A total of 3 × 106 of RasV12-infected Arf-null cells after rapamycin treatment (100 nM) were subjected to cytosolic ribosome profiling. (E) The resultant fractions from above were analyzed by RT-PCR for DHX33 mRNA distribution on ribosomes. GAPDH was used as a negative control. The data represents a typical result from three independent experiments.
DHX33 upregulation is required for enhanced rRNA transcription during Ras activation.
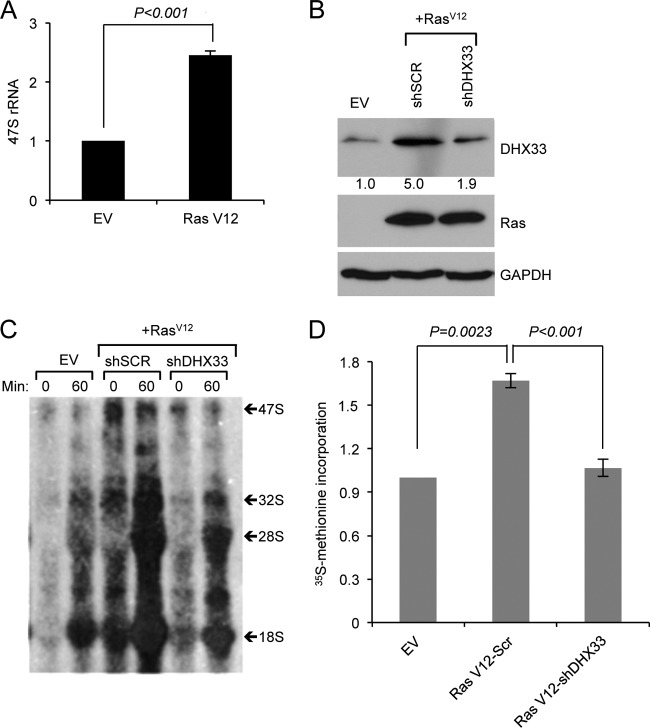
We have previously reported that DHX33 is an important factor in rRNA transcription (30). We hypothesized that elevated levels of DHX33 during Ras activation are important for Ras to promote rRNA synthesis. To test this hypothesis, we first detected pre-rRNA transcript levels by qRT-PCR in both empty vector and RasV12-infected Arf-null cells and saw a 2- to 3-fold increase in 47S rRNA levels (Fig. 7A). To test whether DHX33 was required for this observed increase in pre-rRNA levels, Arf-null fibroblasts were first infected with RasV12 retroviruses, followed by a second infection with lentiviruses expressing knockdown shRNAs for DHX33 (Fig. 7B). We performed pulse-chase labeling with [3H]uridine to detect ongoing rRNA synthesis. We found that reduction in DHX33 resulted in significantly lower 47S rRNA transcript levels that mirrored those seen in uninfected Arf-null cells (Fig. 7C). We also measured global protein synthesis by [35S]methionine incorporation into newly synthesized proteins for RasV12 transformed Arf-nulls cells after DHX33 knockdown and found that DHX33 knockdown caused a significant reduction in protein synthesis (Fig. 7D).
Fig 7.
DHX33 protein induction plays a crucial role in RasV12-enhanced rRNA transcription. (A) Arf-null ear fibroblasts from 2-month-old mice were infected with either empty vector or RasV12-encoding retroviruses. Total RNA was extracted and analyzed by RT-PCR for 47S pre-rRNA levels. Error bars indicate standard deviation from three independent experiments. (B, C, and D) Arf-null ear fibroblasts were infected with retroviruses encoding empty vector or RasV12. At 2 days postinfection, the cells were then infected with lentiviruses encoding shScrambled (SCR) or shRNA-DHX33 for 3 days. Cells were then subjected to Western blot analysis for DHX33 protein levels (B). Equal numbers of cells were pulsed with [3H]uridine and chased at the indicated time points, the total RNA was isolated and separated for rRNA synthesis analysis, and a representative result from three independent experiments is shown (C). The cells were then pulse-labeled with [35S]methionine incorporation and 35S-labeled proteins were measured. Error bars represent the standard deviation from three independent experiments (D). **, P < 0.001.
DHX33 is required in RasV12-initiated tumor formation.
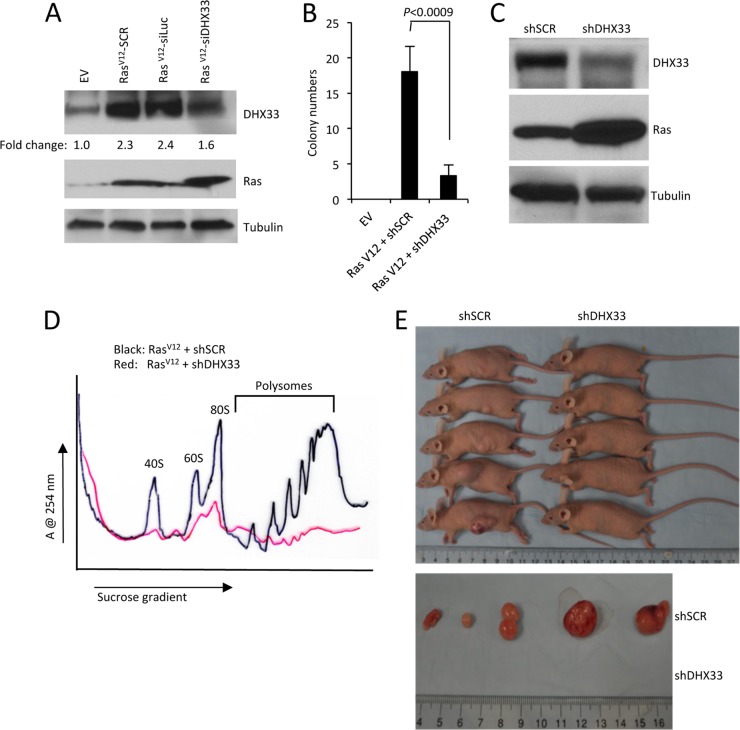
Given that we have shown a requirement for DHX33 in RasV12-initiated 47S rRNA transcription, we next sought to determine the contribution of DHX33 to RasV12-driven cellular transformation. Arf-null MEFs infected with RasV12 retroviruses were subjected to a second infection with shSCR or shDHX33 lentiviruses. DHX33 protein knockdown efficiency was analyzed by Western blotting (Fig. 8A). Importantly, DHX33 knockdown did not reduce DHX33 levels below those seen in control cells (Fig. 8A, lanes 1 and 4). After DHX33 knockdown, cells were plated in soft agar and grown for 2 weeks and resultant transformed cell colonies were counted. We observed a significant decrease in soft agar colonies in RasV12+shDHX33-infected cells, underscoring the importance of heightened DHX33 expression in RasV12-mediated cellular transformation (Fig. 8B). We next determined whether DHX33 knockdown influenced RasV12-initiated mRNA translation. Again, Arf-null MEFs were infected with RasV12 retroviruses and subjected to a second infection with lentiviruses encoding shRNAs for DHX33. Western blot analysis confirmed successful overexpression of RasV12 and knockdown of DHX33 (Fig. 8C). We measured significant decreases in cytosolic ribosome subunits and actively translating polysomes in the RasV12+shDHX33 cells (Fig. 8D), indicating that elevated DHX33 expression is required for enhanced ribosome production and mRNA translation following ectopic RasV12 expression. To assess the impact of DHX33 knockdown on RasV12-initiated tumor formation, we injected 106-infected cells into the flanks of immunocompromised mice. At 2 weeks postinjection, we detected significant tumor cell growth of the cells infected with RasV12+shSCR, while cells infected with RasV12+shDHX33 did not exhibit any measurable tumor formation (Fig. 8E). This striking difference indicates that DHX33 is a crucial target of oncogenic RasV12 and is required to enhance RasV12-mediated cell growth and tumor formation.
Fig 8.
DHX33 induction is required for RasV12-initiated tumor formation. (A) Arf-null ear fibroblasts from 2-month-old mice were infected with retroviruses encoding pBABE-empty vector (EV) or pBABE-RasV12. Cells were then infected with lentiviruses encoding shScrambled (shSCR), shLuciferase (shLuc), or shDHX33. Whole-cell lysates were extracted and analyzed by Western blotting with Ras, DHX33, and tubulin antibodies. (B) A total of 5 × 103 infected cells were plated onto soft agar 60-mm plates in triplicate to measure anchorage-independent cell growth after 14 days. Quantitation of the colony numbers is presented from three representative fields under ×4 magnification. Error bars represent the standard deviation calculated from three different fields of colonies on triplicate plates. (C) Arf-null NIH 3T3 cells were infected with retroviruses encoding RasV12, followed by infection with lentiviruses encoding shScrambled (shSCR) or shDHX33. Whole-cell lysates were subjected to Western blot analysis with Ras, DHX33, and tubulin antibodies. (D) A total of 3 × 106 infected NIH 3T3 cells were subjected to cytosolic ribosome profiles. (E) The upper panel shows NIH 3T3 cells infected with retroviruses encoding RasV12 that were then infected with shSCR or shDHX33 lentiviruses. A total of 106 infected NIH 3T3 cells were injected into the flanks of nude mice. Tumor formation was visualized and photographed after 14 days. For the lower panel, mice were sacrificed at day 14 postinjection, and tumors were excised and photographed.
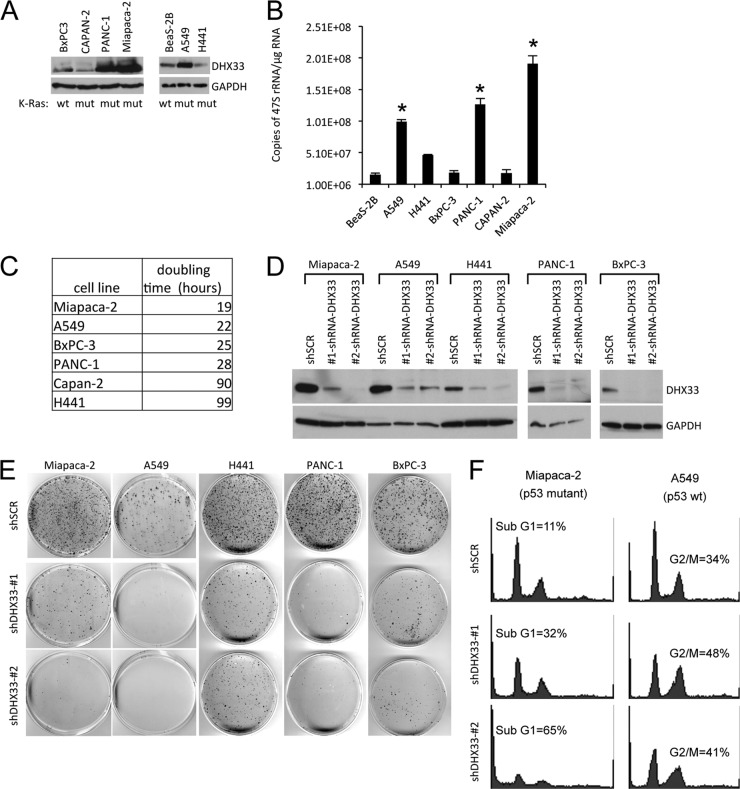
Correlation between DHX33 protein levels, 47S rRNA levels, and cell proliferation in K-Ras mutated human cancer cell lines.
Ras gene mutation has been frequently observed in human cancers (23). To determine whether endogenous DHX33 is upregulated in human cancers harboring mutant Ras alleles, we performed Western blot analysis for endogenous DHX33 protein levels on a panel of human cancer cell lines. As shown in Fig. 9A, we found elevated DHX33 protein levels in three of five K-Ras mutant cancer cell lines using wild-type K-Ras cell lines as a comparison. DHX33 protein levels were significantly upregulated in Miapaca-2, PANC-1, and A549 cells compared to wild-type K-Ras human cancer cell line, BxPC-3 or normal immortalized human lung epithelial cell line, BeaS-2B (Fig. 9A). Due to the pivotal role of DHX33 in rRNA transcription, we also measured 47S rRNA levels by qRT-PCR in a panel of K-Ras mutated cancer cell lines. We discovered that 47S rRNA transcript levels correlated with DHX33 protein levels (Fig. 9B). For example, 47S rRNA transcript levels were the highest in Miapaca-2 cells, where DHX33 protein level was also the highest. Although in Capan-2, where DHX33 levels were the lowest, 47S rRNA level were also the lowest (Fig. 9B). Moreover, we also noticed that cell proliferation rates were tightly correlated with DHX33 protein levels and 47S rRNA levels in these K-Ras mutated human cancer cell lines (Fig. 9C).
Fig 9.
DHX33 is overexpressed in Ras-mutated cancer cell lines and is required for their efficient growth and proliferative properties. (A) A panel of K-Ras mutated or wild-type cancer cell lines (mutation status is shown at the bottom) were screened for total DHX33 protein expression, p14ARF status is also shown at the bottom. (B) 47S rRNA was measured by RT-PCR and normalized to total RNA levels. Error bars represent the standard deviation from three separate experiments. *, P < 0.001. (C) A total of 5 × 104 cells were plated onto six-well culture plates. Cell numbers were counted daily and graphed. The doubling time was calculated based on growth curves and is shown in the table. (D) The indicated cell lines were infected with lentiviruses encoding shScrambled (shSCR) or shDHX33. Whole-cell lysates were extracted 4 days postinfection and subjected to Western blot analysis with antibodies recognizing DHX33 and tubulin. (E) shSCR or shDHX33-infected cells (104) from indicated cancer cell lines were plated onto 100-mm culture dishes. The cells were fixed 10 or 20 days later with 100% methanol and incubated with Giemsa stain for 1 h. Stained colonies were air dried and photographed. (F) shSCR or shDHX33-infected cells (104) from Miapaca-2 and A549 cancer cells were subjected to cell cycle analysis by flow cytometry after propidium iodide staining.
To study the importance of DHX33 protein upregulation in K-Ras mutated human cancer cells, we utilized two unique shRNAs to knock down endogenous human DHX33 protein levels and measured cell growth over time. The knockdown efficiency of DHX33 for all five different cancer cell lines is shown in Fig. 9D. All cells exhibited some dependency on DHX33 for sustained proliferation (Fig. 9E). However, the negative impact of DHX33 on long-term proliferation was the most dramatic in the highly proliferative Miapaca-2 and A549 cells. The p53 mutational status might influence the different outcomes we observed for DHX33 knockdown. DHX33 knockdown in Miapaca-2 (mutant p53) resulted in significant cell death, while in p53 wild-type A549 cells, DHX33 knockdown resulted in a G2/M arrest (Fig. 9F). Taken together, our results show that elevated DHX33 protein expression in mutant K-Ras cancer cell lines is pivotal in enhancing rRNA transcription and proliferation.
DISCUSSION
Ras is one of the most frequently mutated oncogenes in human cancers. Three members of the Ras family, sharing 85% primary sequence identity, have been found to be activated in human cancers: H-Ras, N-Ras, and K-Ras (25). Up to 30% of human lung cancers harbor K-Ras mutations and, in pancreatic cancers, the K-Ras mutation rate is >90% (25). Ras signaling is a complex network whose downstream components play multiple roles in cell growth and cell proliferation. In its active, GTP-bound state, Ras is able to activate two major oncogenic signaling cascades: Raf/MEK/ERK and PI3K/AKT pathway (28). Aside from its role in promoting cell proliferation and cell survival, cell invasiveness and enhanced production of angiogenic factors, Ras activation also causes a significant elevation in the production of rRNA and increases in mRNA translation. Ras enhanced ribosome RNA synthesis are due to a variety of contributions from several Ras downstream effectors such as ERK (38–40), cyclin D1 (41, 42), and mTOR (43, 44), all of which can promote RNA Pol I transcription through different mechanisms. However, these enhancements and gains were observed in cells lacking an intact ARF/p53 pathway. The canonical roles of the ARF tumor suppressor reside in its ability to sense activated Ras alleles and prevent downstream cellular processes normally augmented by oncogenic Ras. Thus, it seems that proteins central to these processes must be under the control of both ARF and Ras regulators. Identifying these key players was our focus.
In this report, we identified a new downstream target of Ras, the DHX33 DEAH-box RNA helicase. DHX33 plays as an important role in promoting rRNA synthesis and ribosome biogenesis (30). In cells that maintain an intact Arf locus, oncogenic RasV12 overexpression resulted in a significant reduction in DHX33 protein expression without any lowering of DHX33 mRNA. The timing of DHX33 downregulation coincided with the classical induction of ARF expression by oncogenic RasV12 alleles. This negative regulation was not observed in cells lacking p53, arguing that the attenuation of DHX33 protein expression relied on the canonical ARF/p53 tumor suppressor pathway. Our results support the notion that other than cell cycle regulation, a p53-dependent role of ARF might also reside in inhibiting ribosome biogenesis. The ARF tumor suppressor has been found to inhibit rRNA synthesis (14, 24) through its ability to prevent UBF phosphorylation (24) and by translocating TTF-I, a RNA polymerase I termination factor, from the nucleolus into the nucleus (31). Nonetheless, in the absence of Arf, RasV12 was quite capable of dramatically increasing DHX33 protein expression, squarely placing DHX33 in the nexus of ARF and Ras regulation. In contrast to ARF, RasV12 considerably shifted existing DHX33 mRNAs onto translating polysomes. Thus, we have identified a new route through which ARF inhibits rRNA synthesis.
We have provided evidence that elevated expression of DHX33 is critical for RasV12-induced cellular transformation. Importantly, our experiments utilized shRNAs that target and reduce DHX33 expression back to just above baseline. As such, we are not entirely removing DHX33 from these cells. Reduction of DHX33 in RasV12-expressing Arf-null cells resulted in a return of 47S rRNA and mRNA translation back to levels normally seen in Arf-null cells. These cells no longer grow in soft agar and do not form tumors in immunocompromised mice. Much of the focus on ARF tumor biology has been on its ability to respond to oncogenic signals, such as those emanating from RasV12, to induce a potent p53-dependent cell cycle arrest. Even more recently, a significant amount of interest has also shifted to ARF's ability to directly inhibit ribosome biogenesis independent of p53. Our new findings herein imply that ARF and Ras are in a constant struggle for downstream target activation/inactivation. Normal cells harboring activated Ras alleles in the face of wild-type ARF are unable to gain access to critical downstream targets, such as DHX33, to fully activate critical processes required for tumorigenesis. ARF effectively eliminates these proteins by removing their mRNAs from actively translating polyribosomes. When Arf is lost, Ras gains access to these targets, and active translation of them ensues. How ARF might selectively repress mRNA translation remains to be investigated, but our findings with DHX33 are reminiscent of ARF's regulation of VEGFA translation (45).
Enhanced ribosome biogenesis is tightly correlated with enhanced cell proliferation in human cancers. Targeting RNA Pol I transcription has been regarded as a potential treatment for cancer patients (3). Several drugs, including actinomycin D, cisplatin, 5-fluorouracil, and camptothecin have been shown to inhibit RNA Pol I transcription (46–48). However, this field is still at the early stage and will require more selective targets in order to develop efficient therapeutic drugs that preferentially inhibit tumor growth while sparing normal cells. Recently, selective drugs that target rRNA synthesis have been developed, shedding a more positive light onto our ability to target rRNA synthesis as a way of treating cancers. One of the latest compounds, CX-3543, is a small molecule nucleolus-targeting agent that selectively disrupts nucleolin/rDNA G-quadruplex complexes in the nucleolus (49). CX-3543 inhibited Pol I transcription and induced apoptosis in cancer cells and is currently in phase II clinical trials. Another compound, CX-5461, selectively inhibits Pol I-driven relative to Pol II-driven transcription, DNA replication, and mRNA translation (50). CX-5461 inhibits the initiation stage of rRNA synthesis and induces both senescence and autophagy through a p53-independent process in solid tumor cell lines. Although more work needs to be done in order to develop efficient and more specific drugs to target RNA Pol I transcription as a way for cancer treatment, the validity of the approach itself has proven fruitful.
It seems uncertain whether other consensus targets of both ARF and oncogenic RasV12 exist. However, given the pleiotropic effects of ARF and Ras, identification of other common proteins seems likely. In fact, given the large number of RNA helicases in the DEAD/DEAH-box family, DHX33 may signal the first of many dually regulated helicases. Given our findings that the helicase activity of DHX33 was required for the RasV12-driven phenotype, generating novel compounds that inhibit its helicase activity seems a viable approach to prevent RasV12-induced transformation, especially in clinically relevant settings where Ras mutants, and thus aberrant DHX33 expression, drive the disease.
ACKNOWLEDGMENTS
We thank the members of the Weber laboratory for their advice and technical assistance. Lian-Fai Yee provided Arf-null ear fibroblasts. The Genome Institute and Children's Discovery Institute at Washington University provided lentiviral RNAi library constructs.
A.J.S. was supported by Komen for the Cure. This study was supported by National Institutes of Health grant CA120436 and by an Era of Hope Scholar Award in Breast Cancer Research (BC007304) to J.D.W.
Footnotes
Published ahead of print 11 February 2013
REFERENCES
- 1. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 144:646–674 [DOI] [PubMed] [Google Scholar]
- 2. Luo J, Solimini NL, Elledge SJ. 2009. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136:823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 50:131–156 [DOI] [PubMed] [Google Scholar]
- 4. Maggi LB, Jr, Weber JD. 2005. Nucleolar adaptation in human cancer. Cancer Invest. 23:599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Montanaro L, Trere D, Derenzini M. 2008. Nucleolus, ribosomes, and cancer. Am. J. Pathol. 173:301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. White RJ. 2008. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 24:622–629 [DOI] [PubMed] [Google Scholar]
- 7. White RJ. Transcription by RNA polymerase III: more complex than we thought. Nat. Rev. Genet. 12:459–463 [DOI] [PubMed] [Google Scholar]
- 8. Pianese G. 1896. Beitrag zur Histologie und Aetiologie der Carcinoma Histologische und experimentelle Untersuchungen. Beitr. Pathol. Anat. Allgem. Pathol. 142:1–193 [Google Scholar]
- 9. Williamson D, Lu YJ, Fang C, Pritchard-Jones K, Shipley J. 2006. Nascent pre-rRNA overexpression correlates with an adverse prognosis in alveolar rhabdomyosarcoma. Genes Chromosomes Cancer 45:839–845 [DOI] [PubMed] [Google Scholar]
- 10. Grummt I. 2003. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 17:1691–1702 [DOI] [PubMed] [Google Scholar]
- 11. Derenzini M, Trere D, Pession A, Govoni M, Sirri V, Chieco P. 2000. Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J. Pathol. 191:181–186 [DOI] [PubMed] [Google Scholar]
- 12. Derenzini M, Trere D, Pession A, Montanaro L, Sirri V, Ochs RL. 1998. Nucleolar function and size in cancer cells. Am. J. Pathol. 152:1291–1297 [PMC free article] [PubMed] [Google Scholar]
- 13. Trere D, Ceccarelli C, Montanaro L, Tosti E, Derenzini M. 2004. Nucleolar size and activity are related to pRb and p53 status in human breast cancer. J. Histochem Cytochem. 52:1601–1607 [DOI] [PubMed] [Google Scholar]
- 14. Ayrault O, Andrique L, Fauvin D, Eymin B, Gazzeri S, Seite P. 2006. Human tumor suppressor p14ARF negatively regulates rRNA transcription and inhibits UBF1 transcription factor phosphorylation. Oncogene 25:7577–7586 [DOI] [PubMed] [Google Scholar]
- 15. Learned RM, Learned TK, Haltiner MM, Tjian RT. 1986. Human rRNA transcription is modulated by the coordinate binding of two factors to an upstream control element. Cell 45:847–857 [DOI] [PubMed] [Google Scholar]
- 16. Zhai W, Comai L. 2000. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell. Biol. 20:5930–5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rubbi CP, Milner J. 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 22:6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sherr CJ. 1998. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 12:2984–2991 [DOI] [PubMed] [Google Scholar]
- 19. Yuan X, Zhou Y, Casanova E, Chai M, Kiss E, Grone HJ, Schutz G, Grummt I. 2005. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol. Cell 19:77–87 [DOI] [PubMed] [Google Scholar]
- 20. Chedin S, Laferte A, Hoang T, Lafontaine DL, Riva M, Carles C. 2007. Is ribosome synthesis controlled by pol I transcription? Cell Cycle 6:11–15 [DOI] [PubMed] [Google Scholar]
- 21. Laferte A, Favry E, Sentenac A, Riva M, Carles C, Chedin S. 2006. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 20:2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Palmero I, Pantoja C, Serrano M. 1998. p19ARF links the tumour suppressor p53 to Ras. Nature 395:125–126 [DOI] [PubMed] [Google Scholar]
- 23. Fernandez-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer 2:344–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ayrault O, Andrique L, Larsen CJ, Seite P. 2004. Human Arf tumor suppressor specifically interacts with chromatin containing the promoter of rRNA genes. Oncogene 23:8097–8104 [DOI] [PubMed] [Google Scholar]
- 25. Downward J. 2003. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3:11–22 [DOI] [PubMed] [Google Scholar]
- 26. Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. 2005. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 7:311–318 [DOI] [PubMed] [Google Scholar]
- 27. Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA. 2005. Myc-dependent regulation of rRNA synthesis during Drosophila development. Nat. Cell Biol. 7:295–302 [DOI] [PubMed] [Google Scholar]
- 28. Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. 2000. Understanding Ras: ‘it ain't over ‘til it's over’. Trends Cell Biol. 10:147–154 [DOI] [PubMed] [Google Scholar]
- 29. Zhang C, Comai L, Johnson DL. 2005. PTEN represses RNA Polymerase I transcription by disrupting the SL1 complex. Mol. Cell. Biol. 25:6899–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Y, Forys JT, Miceli AP, Gwinn AS, Weber JD. 2011. Identification of DHX33 as a mediator of rRNA synthesis and cell growth. Mol. Cell. Biol. 31:4676–4691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lessard F, Morin F, Ivanchuk S, Langlois F, Stefanovsky V, Rutka J, Moss T. The ARF tumor suppressor controls ribosome biogenesis by regulating the RNA polymerase I transcription factor TTF-I. Mol. Cell 38:539–550 [DOI] [PubMed] [Google Scholar]
- 32. Weber JD, Jeffers JR, Rehg JE, Randle DH, Lozano G, Roussel MF, Sherr CJ, Zambetti GP. 2000. p53-independent functions of the p19ARF tumor suppressor. Genes Dev. 14:2358–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, Zhang Y. 2003. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell 12:1151–1164 [DOI] [PubMed] [Google Scholar]
- 34. Zhang Y, Xiong Y, Yarbrough WG. 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92:725–734 [DOI] [PubMed] [Google Scholar]
- 35. Basu TN, Gutmann DH, Fletcher JA, Glover TW, Collins FS, Downward J. 1992. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature 356:713–715 [DOI] [PubMed] [Google Scholar]
- 36. Dasgupta B, Yi Y, Chen DY, Weber JD, Gutmann DH. 2005. Proteomic analysis reveals hyperactivation of the mammalian target of rapamycin pathway in neurofibromatosis 1-associated human and mouse brain tumors. Cancer research. 65:2755–2760 [DOI] [PubMed] [Google Scholar]
- 37. Johannessen CM, Reczek EE, James MF, Brems H, Legius E, Cichowski K. 2005. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc. Natl. Acad. Sci. U. S. A. 102:8573–8578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stefanovsky V, Langlois F, Gagnon-Kugler T, Rothblum LI, Moss T. 2006. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell 21:629–639 [DOI] [PubMed] [Google Scholar]
- 39. Stefanovsky VY, Pelletier G, Hannan R, Gagnon-Kugler T, Rothblum LI, Moss T. 2001. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell 8:1063–1073 [DOI] [PubMed] [Google Scholar]
- 40. Zhao J, Yuan X, Frodin M, Grummt I. 2003. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell 11:405–413 [DOI] [PubMed] [Google Scholar]
- 41. Voit R, Grummt I. 2001. Phosphorylation of UBF at serine 388 is required for interaction with RNA polymerase I and activation of rDNA transcription. Proc. Natl. Acad. Sci. U. S. A. 98:13631–13636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Voit R, Hoffmann M, Grummt I. 1999. Phosphorylation by G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 18:1891–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, Hannan RD. 2003. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol. Cell. Biol. 23:8862–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mayer C, Zhao J, Yuan X, Grummt I. 2004. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 18:423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kawagishi H, Nakamura H, Maruyama M, Mizutani S, Sugimoto K, Takagi M, Sugimoto M. ARF suppresses tumor angiogenesis through translational control of VEGFA mRNA. Cancer Res. 70:4749–4758 [DOI] [PubMed] [Google Scholar]
- 46. Ghoshal K, Jacob ST. 1997. An alternative molecular mechanism of action of 5-fluorouracil, a potent anticancer drug. Biochem. Pharmacol. 53:1569–1575 [DOI] [PubMed] [Google Scholar]
- 47. Jordan P, Carmo-Fonseca M. 1998. Cisplatin inhibits synthesis of rRNA in vivo. Nucleic Acids Res. 26:2831–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pondarre C, Strumberg D, Fujimori A, Torres-Leon R, Pommier Y. 1997. In vivo sequencing of camptothecin-induced topoisomerase I cleavage sites in human colon carcinoma cells. Nucleic Acids Res. 25:4111–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Drygin D, Siddiqui-Jain A, O'Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, Lim JK, Von Hoff D, Anderes K, Rice WG. 2009. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 69:7653–7661 [DOI] [PubMed] [Google Scholar]
- 50. Drygin D, Lin A, Bliesath J, Ho CB, O'Brien SE, Proffitt C, Omori M, Haddach M, Schwaebe MK, Siddiqui-Jain A, Streiner N, Quin JE, Sanij E, Bywater MJ, Hannan RD, Ryckman D, Anderes K, Rice WG. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits rRNA synthesis and solid tumor growth. Cancer Res. 71:1418–1430 [DOI] [PubMed] [Google Scholar]