Abstract
Nucleocytoplasmic shuttling of class IIa of histone deacetylases (HDACs) is a key mechanism that controls cell fate and animal development. We have identified the filamin B (FLNB) as a novel HDAC7-interacting protein that is required for temporal and spatial regulation of vascular endothelial growth factor (VEGF)-mediated HDAC7 cytoplasmic sequestration. This interaction occurs in the cytoplasm and requires monoubiquitination of an evolutionarily conserved lysine 1147 (K1147) in the immunoglobulin (Ig)-like repeat 10 (R10) of FLNB and the nuclear localization sequence of HDAC7. Inhibition of protein kinase C (PKC) blocks VEGF-induced ubiquitination of FLNB and its interaction with HDAC7. Small interfering RNA (siRNA) knockdown of FLNB or ubiquitin (Ub) in human primary endothelial cells blocks VEGF-mediated cytoplasmic accumulation of HDAC7, reduces VEGF-induced expression of the HDAC7 target genes Mmp-10 and Nur77, and inhibits VEGF-induced vascular permeability. Using dominant negative mutants and rescue experiments, we demonstrate the functional significance of FLNB K1147 to interfere with the ability of phorbol myristate acetate (PMA) to promote FLNB-mediated cytoplasmic accumulation of HDAC7. Taken together, our data show that VEGF and PKC promote degradation-independent protein ubiquitination of FLNB to control intracellular trafficking of HDAC7.
INTRODUCTION
Vascular endothelial growth factors (VEGFs) bind and activate their receptors to stimulate vessel formation in response to angiogenic stimuli such as hypoxia and inflammation. VEGF is also a potent vascular permeabilizing agent and dilator that promotes endothelial cell migration, proliferation, and vasculogenesis during embryonic development as well as angiogenesis under physiological and pathological conditions (1–4). VEGF-induced permeability is an ordered and coordinated process that includes VEGF receptor (VEGFR) activation, increased intracellular calcium, activation of downstream kinases such as Src, protein kinases C (PKCs), and mitogen-activated protein kinases (MAPKs), and subsequent altered gene expression. This leads to temporal disruption of endothelial cell-cell junctions, cytoskeleton remodeling, and increased vessel formation (2).
Class IIa histone deacetylases (HDACs), including HDAC4, -5, -7, and -9, are transcriptional corepressors implicated in animal development, cell differentiation, and glucose homeostasis (5–7). HDAC7 is the sole class IIa HDAC that is essential for life in rodents (8). Deletion of HDAC7 in mice resulted in embryonic lethality, primarily due to blood vessel disruption and dilation, indicating an important role of HDAC7 in vascular endothelial signaling. Of note, VEGF induces HDAC7 phosphorylation and cytoplasmic accumulation in human umbilical vein endothelial cells (HUVECs) (9, 10). Such activities involve VEGFR2 binding and activation/phosphorylation of downstream calcium-activated effectors PKC and protein kinase D (PKD). However, the detailed mechanisms underlying the VEGF-induced subcellular redistribution of HDAC7 remains poorly understood.
Filamins (FLNs) are actin-binding proteins that include three family members: FLNA, FLNB, and FLNC (11–13). While their roles in organizing actin filaments are well characterized, less is understood about their roles in cellular signaling. Among the family members, FLNA is the most intensively studied. In addition to its actin binding activity in the cytoskeleton, FLNA interacts with and modulates the activity of transcription factors such as androgen receptor (AR) (14), FOXC1 (15), and PEBP2β/CBFβ (16). Recent data indicate that FLNA interacts with prions and integrin β1 and thereby regulates cell spreading and migration (17, 18). In contrast, the role of FLNB in cell signaling and transcriptional regulation is less well understood.
In this study, we identified FLNB as a novel HDAC7-interacting protein. We show that FLNB mediates cytoplasmic retention of HDAC7 and regulates vascular permeability in response to VEGF. Interestingly, we have found that monoubiquitination of FLNB is required for its association with HDAC7 and that this monoubiquitination can be induced by VEGF, phorbol 12-myristate 13-acetate (PMA), and calcium/calmodulin-dependent protein kinase I (CaMK I). Our data support a model in which VEGF- and PKC-induced monoubiquitination of FLNB regulates intracellular HDAC7 trafficking and VEGF-mediated vascular endothelial permeability.
MATERIALS AND METHODS
Plasmid construction.
Expression plasmids for hemagglutinin (HA)-HDAC5 and -HDAC7, constitutively active CaMK I, green fluorescent protein (GFP)-HDAC7, glutathione S-transferase (GST)–HDAC7, HA-ubiquitin (Ub), FLAG-Ub, and the myocyte enhancer factor 2 (MEF2)-driven reporter construct have been previously described (19–21). HDAC7 constructs were generated from mouse cDNA. Full-length FLNB was a generous gift from Sandor S. Shapiro. Deletion, truncation, and point mutation expression plasmids of FLNB, HDAC7, and Ub were generated by PCR and site-directed mutagenesis according to the manufacturer's protocol (Invitrogen). All constructs were verified by DNA sequencing.
Reagents.
VEGF-A (catalog number 293-VE) was purchased from R&D Systems. Anti-GFP (B-2; sc-9996), anti-glyceraldehyde-3-phosphate dehydrogenase conjugated to horseradish peroxidase (anti-GAPDH-HRP) (FL-335; sc-25778 HRP), antihemagglutinin (anti-HA) (F7; sc-7392 and sc-805), anti-Ub (P4D1; sc-8017), and anti-lamin B (C-20; sc-6216) antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). PMA (P8139), Gö6983 (G1918), Gö6976 (G1171), anti-α-tubulin (T6074), anti-β-actin (A5441), anti-HDAC7 (H2662), and anti-FLAG (M2; F3165) antibodies were from Sigma. Anti-HA-HRP antibody (12013819001) was from Roche. Purified HDAC7 and pS178-specific HDAC7 antibodies were previously described (20). The purified antibody did not cross-react with HDAC4 or HDAC5. FLNB antibodies were generated using GST-FLNB (residues 743 to 1226) as an antigen and do not cross-react with FLNA. Anti-Ub antibodies used in the experiment shown in Fig. 3 for immunoprecipitation were generated using GST-Ub as an antigen and purified by GST and GST-Ub columns. The specificity of anti-FLNB and anti-Ub antibodies is shown in Fig. S10 in the supplemental material.
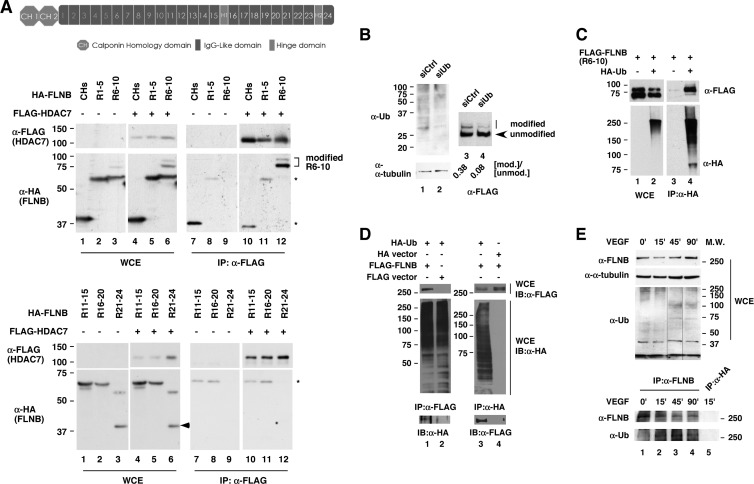
Fig 3.
HDAC7 interacts with ubiquitinated FLNB. (A) HDAC7 interacts with modified IgG-like repeats 6 to 10 (R6-10) of FLNB. A schematic representation of the FLNB domain structure is shown (top). The FLNB protein contains two calponin homology (CH) domains followed by 24 IgG-like repeats (R1 to R24) disconnected by two hinge domains (H1 and H2). HeLa cells were cotransfected with different HA-FLNB truncations and FLAG-HDAC7 or FLAG-vector expression plasmids, followed by coimmunoprecipitation using anti-FLAG antibodies. The precipitated samples were subjected to immunoblotting. Asterisks indicate calculated molecular masses (in kDa; shown at left) of HA-FLNB peptides. The arrowhead points to an unknown species on R21 to R24. (B) Knockdown of Ub inhibits FLNB(R6-10) modification. HeLa cells were transiently transfected with a nontargeting siRNA or a Ub siRNA, followed by transfection with FLAG-FLNB(R6-10). The cell extracts were subjected to Western blotting. The quantitative data indicate the ratio of modified to unmodified FLNB(R6-10) (indicated by the arrow). (C) FLNB(R6-10) is ubiquitinated. HeLa cells were cotransfected with FLAG-FLNB(R6-10), with HA-Ub, or HA-vector plasmids. Coimmunoprecipitation was carried out using anti-HA antibodies followed by Western blotting. (D) FLNB (full length) is ubiquitinated. HeLa cells were transiently transfected with FLAG-FLNB, FLAG-vector, HA-Ub, or HA-vector. Cells were harvested, and lysates were prepared. Whole-cell lysates were subjected to immunoprecipitation with anti-FLAG or anti-HA antibodies and Western blotting with anti-HA and anti-FLAG antibodies. (E) Induction of FLNB ubiquitination by VEGF in HMVECs. HMVECs were treated with 50 ng/ml of VEGFA and harvested at the times indicated (in minutes). Whole-cell lysates were prepared, and immunoprecipitation was performed with anti-FLNB, followed by immunoblotting with anti-Ub antibodies. The membrane was then stripped and reprobed with anti-FLNB antibodies. Anti-HA antibodies were used for immunoprecipitation of cell extracts prepared from 15-min treatments with VEGF. Anti-α-tubulin antibodies served as a loading control. M.W., molecular weights in thousands. The composite image shown in panel E was derived from a single immunoblot. WCE, whole-cell extract; IB, immunoblotting.
Cell culture.
HeLa cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (FBS), 50 units/ml penicillin G, and 50 μg of streptomycin sulfate at 37°C in 5% CO2. HUVECs and human microvascular endothelial cells (HMVECs) were purchased from Lonza and grown in endothelial cell basal medium (EBM-2) (Lonza) supplemented with FBS and growth factors (Lonza). For growth factor treatments, cells were grown in EBM-2 with 0.1% FBS for 16 to 24 h and then treated with growth factors for the periods indicated in the figure legends.
Identification of components of cytoplasmic HDAC7 complex.
pBabe-FLAG-HDAC7 expression plasmid was stably transfected into HeLa cells. The resulting cells, HeLa-FLAG-HDAC7, were grown in large scale according to the provider's protocol (National Cell Culture Center). Nuclear and cytoplasmic extracts were prepared, and immunocomplexes were purified and resolved by SDS-PAGE. The distinct bands in the complexes were excised, and the proteins were identified by liquid chromatography tandem mass spectrometry (LC-MS/MS) at the Sloan-Kettering Institute Proteomics Facility.
Transient-transfection and luciferase assays.
Transient transfections and luciferase assays were performed in 48-well culture plates according to our previously published protocol (22). HeLa cells were transfected with an muscle creatine kinase (MCK) promoter-containing reporter construct and CMX–β-galactosidase (CMX–β-Gal) expression plasmids. Lipofectamine 2000 was used as the transfection reagent. The amount of DNA was kept constant by addition of empty pCMX vector. The cells were harvested 48 h after transfection, and luciferase and β-Gal activities were measured by using a luciferase assay system (Promega). Each reaction was performed in triplicate and repeated at least two times. Results shown indicate luciferase activity normalized to β-Gal levels.
siRNA transfection and qRT-PCR.
Small interfering RNA (siRNA) transfection and quantitative reverse transcription-PCR (qRT-PCR) were carried out according to our published protocol (23). Control (nontargeting siRNA 1; D-001810-01), FLNB siRNAs (J-020175-05 and J-020175-06), and ubiquitin (Ub) siRNAs (J-013382-05 and J-013382-08) were purchased from Dharmacon. For transfection, siRNA (200 pmol/well) and DharmaFECT 1 (Dharmacon) were used and transfected to HUVECs, HMVECs, or HeLa cells for 72 h. Cells were harvested and lysed with NETN buffer (containing 100 mM NaCl, 1 mM EDTA, 20 mM Tris-Cl [pH 8.0], 0.1% Nonidet P-40, and 10% glycerol in sterile water) and were subjected to Western blotting, probing with appropriate antibodies. Total RNA extract was prepared using PrepEase RNA spin kits (USB). cDNA was generated using Superscript II reverse transcription and oligo(dT) primers (Invitrogen). The cDNAs were quantified by real-time PCR by using an iCycler (Bio-Rad, Hercules, CA) with an iQ SYBR green Supermix kit (Bio-Rad) and appropriate primer sets. Subsequently, the melting curves were examined to ensure the homogeneity of the PCR products. The relative quantities of genes of interest were normalized to 18S rRNA.
Immunofluorescence microscopy.
Indirect immunofluorescence microscopy was conducted according to our published protocol (22). The cells were plated onto coverslips of 12-well plates and transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. For cotransfection, 200 pmol of siRNA was transfected into each well for 48 h, and then plasmids encoding the proteins indicated in the figures were cotransfected into HUVECs or HeLa cells. After 24 h, cells were washed with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde for 20 min at room temperature. Immunostaining of endogenous and transfected proteins was carried out using antibodies indicated below and anti-mouse or anti-rabbit IgG conjugated with Alexa Fluor 488 or 594 (Molecular Probes). DAPI (4′,6′-diamidino-2-phenylindole) was applied to the samples after the final wash to visualize cell nuclei. Images were visualized using a LEICA fluorescence microscope equipped with a camera. Prior to fixation, the cells were treated with VEGF (20 ng/ml) or PMA (50 ng/ml) for the time periods indicated in the figure legends depending on experimental requirements. For histograms of subcellular distributions, a total of more than 100 transfected cells for each set of experiments were scored unless specified otherwise.
Coimmunoprecipitation.
Transient transfection and coimmunoprecipitation were carried out according to our published protocol (22). Cells were transfected with 1 to 10 μg of the appropriate plasmids using Lipofectamine 2000 (Invitrogen) or transfected with targeting siRNAs or nontargeting siRNAs using DharmaFECT 1 (Dharmacon). Cells were harvested and lysed in NETN buffer at 4°C, scraped, sonicated, and centrifuged for 15 min at 2,500 × g. The supernatant was kept as the whole-cell extract. After samples were precleared by incubation with protein A/G-agarose (Pharmacia), immunoprecipitations were carried out using anti-FLAG (F2426) or anti-HA (E6779) antibody-conjugated agarose (Sigma) for 2 h at 4°C. Alternatively, antibodies were incubated with whole-cell extracts for 2 h, and then protein A/G-agarose was added into each sample for another 1 h at 4°C. Beads were washed with NETN buffer, and immunoprecipitates were run on SDS-PAGE gels, subjected to Western blotting, and probed with appropriate antibodies.
GST pulldown assays.
GST-FLNB, GST-ACTN4, and GST-HDAC7 fusion proteins and pulldown assays were prepared and performed according to our published protocol (24). For whole-cell lysate preparation, HeLa cells were transfected with an expression plasmid encoding HA-FLNB repeats 8 to 10 [HA-FLNB(R8-10)] or HA-HDAC7, harvested, and incubated with immobilized GST fusion proteins for 1 h at 4°C. After several washes with NETN buffer, retained fractions were subjected to Western blotting with anti-HA antibodies.
Fractionation of nuclear and cytoplasmic extracts.
Nuclear and cytoplasmic fractionation has been described previously (21). Briefly, cells were lysed in cytosolic lysis buffer for 1 (HUVECs) or 10 (HeLa cells) min on ice and centrifuged at 2,000 × g for 5 min at 4°C to yield cytoplasmic extracts. The pelleted nuclei were washed twice with cytosolic lysis buffer, resuspended in nuclear lysis buffer for 10 min on ice, and then centrifuged at 2,500 × g for 15 min at 4°C. The supernatants were kept as nuclear extracts and subjected to SDS-PAGE and Western blotting, and blots were probed with anti-FLNB, anti-HDAC7, anti-GAPDH, anti-α-tubulin, and anti-lamin B antibodies.
Endothelial cell permeability assays.
An in vitro vascular permeability assay kit was used according to the company's protocol (ECM 640; Chemicon). Briefly, HMVECs were transiently transfected with nontargeting siRNA or siRNA targeting FLNB for 72 h. The cells were washed with PBS, trypsinized, and counted by a hemocytometer. A total of 106 cells were seeded onto each insert that contained a semipermeable membrane coated with collagen. The cells were starved in 0.5% FBS in EBM-2 medium for 24 h after an endothelial cell monolayer was formed on the inserts. As a control for the formation of cell monolayer, a sample in which no cells were added was included for comparison. The cells were treated with VEGF for 4 h, and then cell basal medium containing fluorescein isothiocyanate (FITC)-dextran was added to the wells. The FITC-dextran in the solution was transferred to 96-well plates. Fluorescence was quantified using a fluorescence plate reader with 485-nm and 530-nm filters (25).
Identification of FLNB ubiquitination.
HeLa cells were transiently transfected with FLAG-FLNB(R6-10) or FLAG-vector with HA-CaMK I expression plasmids. Whole-cell lysates were prepared, followed by immunoprecipitation using anti-FLAG antibodies. The immunoprecipitated samples were resolved on SDS-PAGE gels and subjected to Coomassie blue staining or Western blotting. After staining, the bands were cut and sent for LC-MS/MS analysis at the Center for Proteomics and Bioinformatics at Case Western Reserve University.
RESULTS
Identification of FLNB as a novel cytoplasmic HDAC7-interacting protein.
In order to isolate cytoplasmic HDAC7 complexes, we generated a HeLa cell line, HeLa-FLAG-HDAC7, that stably expresses FLAG-HDAC7. Cytoplasmic extracts of HeLa-FLAG-HDAC7 cells were subjected to affinity chromatography using anti-FLAG-conjugated beads to purify FLAG-mHDAC7 complexes. After samples were washed, the bound fraction was eluted with FLAG peptides and resolved by SDS-PAGE. Several polypeptides were isolated from the gels and subjected to mass spectroscopy. One of these polypeptides was identified as FLNB, a putative novel HDAC7-interacting protein in the cytoplasmic complexes. In addition, polypeptides derived from several members of the 14-3-3 protein family, previously reported to bind HDAC7, were identified (see Fig. S1A and B in the supplemental material).
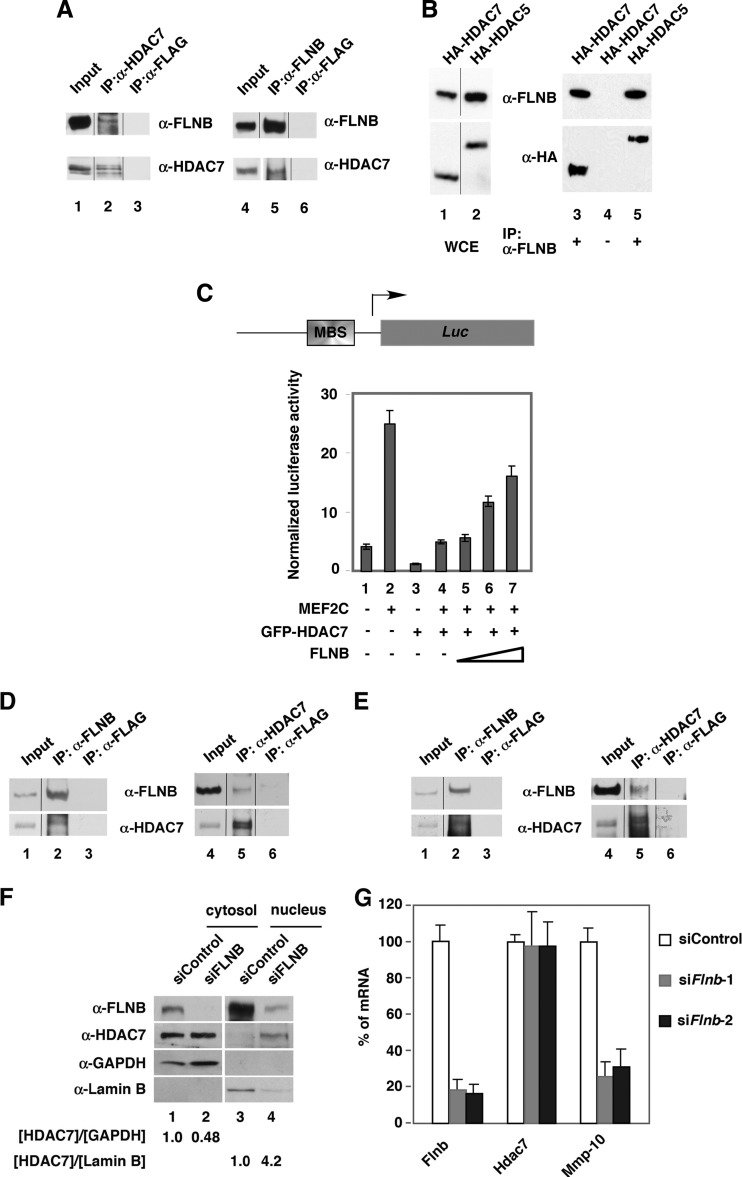
In order to verify the interaction between HDAC7 and FLNB, we carried out coimmunoprecipitation of endogenous and transfected proteins. We found that HDAC7 and FLNB were coprecipitated in the cytoplasmic fraction (Fig. 1A, lanes 2 and 5) and whole-cell extracts (see Fig. S1C in the supplemental material). In addition, endogenous FLNB interacted with ectopically expressed HDAC7 and HDAC5, another member of the class IIa HDAC family (Fig. 1B). As controls, FLNB did not coprecipitate with ectopically expressed SMRT or ACTN4 (see Fig. S1D). Because HDAC7 is known to repress myocyte enhancer factor 2 (MEF2)-mediated transcriptional activity (17, 26), we investigated the functional significance of the interaction between HDAC7 and FLNB by determining whether FLNB could affect transcription activity of an MEF2-driven reporter (Fig. 1C). The results showed that FLNB was capable of overcoming HDAC7-mediated repression activity on the reporter (Fig. 1C, lanes 4 to 7). The underlying mechanism for this effect was suggested by immunofluorescence microscopy, which showed that overexpression of FLNB promoted cytoplasmic accumulation of cotransfected HDAC7 in HeLa cells (see Fig. S1E). Importantly, knockdown of FLNB resulted in nuclear accumulation of HDAC7 (see Fig. S1F). We next asked whether the functional interaction between FLNB and HDAC7 that was observed in HeLa cells would also be detected in endothelial cells (ECs), which are noted for high levels of HDAC7 expression. Coimmunoprecipitation demonstrated that HDAC7 and FLNB interacted in HUVECs (Fig. 1D). Consistent with our complex purification data, HDAC7 and FLNB were coprecipitated in the cytosolic fraction (Fig. 1E). Furthermore, knockdown of FLNB led to nuclear accumulation of HDAC7 in HUVECs (Fig. 1F) and a decrease in the expression of Mmp-10, an HDAC7 target gene (Fig. 1G). These data suggest that FLNB counteracts HDAC7 repression activity on Mmp-10 by promoting cytoplasmic retention of HDAC7.
Fig 1.
FLNB interacts with HDAC7 in the cytoplasm of mammalian cells. (A) Anti-FLNB, anti-HDCA7, and anti-FLAG antibodies were used to carry out coimmunoprecipitations using cytoplasmic extracts prepared from HeLa cells, and immune pellets were subjected to Western blotting. Input lanes contain 2 and 10% of extract in order to detect FLNB and HDAC7 using anti-FLNB and anti-HDAC7 antibodies, respectively. (B) Expression plasmids encoding HA-HDAC5 or HA-HDAC7 were transfected into HeLa cells. Whole-cell extracts were prepared, followed by coimmunoprecipitation and Western blotting. (C) A schematic diagram of the MEF2-driven (MBS, MEF2 binding sequence) reporter construct is shown. Overexpression of FLNB overcomes HDAC7-mediated transcriptional repression (bottom panel). The MEF2 reporter construct was cotransfected with an internal control, CMX–β-Gal, and plasmids expressing MEF2C, FLNB, or HDAC7. (D) Association of FLNB and HDAC7 in HUVECs. Immunoprecipitation was carried out using anti-HDAC7, anti-FLNB, or anti-FLAG antibodies followed by Western blotting. (E) Cytoplasmic HDAC7 and FLNB interact. The cytoplasmic fraction was prepared and used for immunoprecipitation with anti-HDAC7 or anti-FLNB antibodies followed by Western blotting with the indicated antibodies. Input lanes contain 2 or 10% of extract in order to detect FLNB and HDAC7 using anti-FLNB and anti-HDAC7 antibodies, respectively. (F) Knockdown of FLNB leads to nuclear accumulation of HDAC7 in HUVECs. The cells were transfected with nontargeting siRNA (siControl) or FLNB siRNA (siFLNB). Nuclear and cytoplasmic fractions were prepared and subjected to Western blotting. (G) Knockdown of FLNB decreased the expression of the HDAC7 target gene, Mmp-10. HUVECs were transfected with nontargeting siRNA (siControl) or two siRNAs against FLNB (siFlnb-1 and siFlnb-2) for 72 h. Total RNA was extracted, and cDNA was generated. The levels of mRNA were quantified by real-time PCR and normalized to 18S rRNA levels. The spliced lanes shown in panels A, B, D, and E were merged from a single immunoblot. α, anti; IP, immunoprecipitation.
FLNB is critical for VEGF-induced cytoplasmic accumulation of HDAC7.
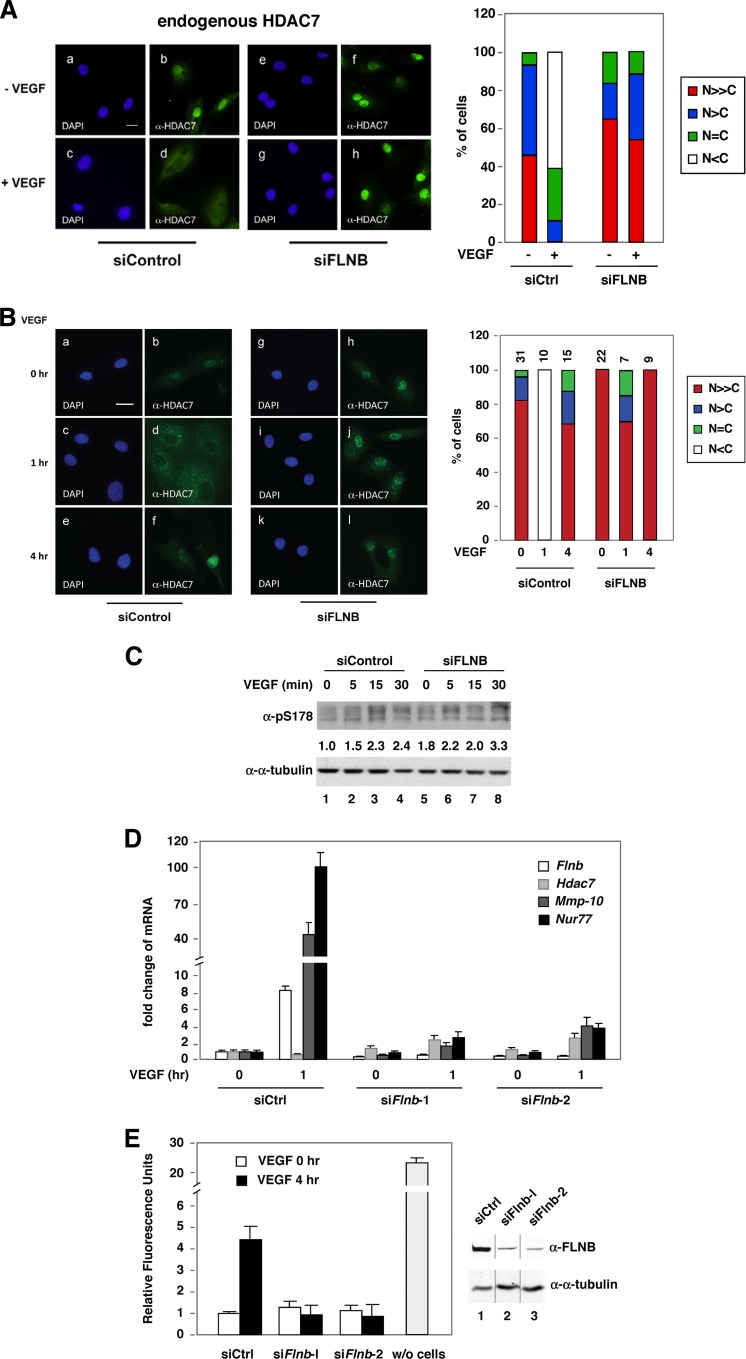
Evidence, including ours, has shown that transient accumulation of HDAC7 in the cytoplasm in response to VEGF exposure derepresses MEF target gene expression (9, 10). Because FLNB is known to bind F-actin filaments and localize predominantly in the cytoplasm, we sought to determine whether it played a role in HDAC7 redistribution in response to VEGF. Notably, knockdown of FLNB abolished the ability of VEGF to promote cytoplasmic accumulation of endogenous (Fig. 2A) and exogenous HDAC7 (see Fig. S2, top panel, in the supplemental material). However, knockdown of FLNB did not affect the subcellular localization of constitutively cytoplasmic mutant (deletion of the nuclear localization signal at amino acids 183 to 213[ΔNLS]) or a constitutive nuclear mutant (in which serines 178, 344, and 479 were replaced with alanines [S/A]) (20). In human microvascular endothelial cells (HMVECs), after 30 min of VEGF treatment, we observed cytoplasmic accumulation of endogenous HDAC7, and this effect peaked at 1 h (Fig. 2B). After 4 h of VEGF exposure, this effect was reversed, and HDAC7 was detected primarily in the nucleus. However, VEGF-induced HDAC7 phosphorylation was not blocked by FLNB knockdown (Fig. 2C). Furthermore, the ability of VEGF to induce HDAC7 cytoplasmic accumulation was significantly abolished, and VEGF-mediated induction of the HDAC7 repression targets, Mmp-10 and Nur77, was greatly reduced (Fig. 2D) in cells expressing siRNA targeting FLNB (siFLNB). Expression of Nur77 is critical for VEGF-mediated permeability (27), and thus our data suggest that FLNB is likely to be a critical modulator of VEGF-induced vascular permeability. Indeed, knockdown of FLNB significantly abolished VEGF-induced HMVEC permeability (Fig. 2E). Together, these data indicate that FLNB is required for VEGF-induced transient cytoplasmic accumulation of HDAC7, VEGF-induced upregulation of MEF2 and HDAC7 target genes, and VEGF-mediated increases in EC permeability.
Fig 2.
The effect of FLNB knockdown on VEGF-mediated cytoplasmic accumulation of HDAC7 in endothelial cells. (A) Knockdown of FLNB blocks VEGF-mediated cytoplasmic accumulation of HDAC7 in HUVECs. HUVECs were transiently transfected with a control or FLNB siRNA (siControl or siFLNB, respectively), followed by treatment with or without VEGF (20 ng/ml) and immunofluorescence microscopy with anti-HDAC7 and DAPI staining. Subcellular distribution of endogenous HDAC7 in the absence or presence of PMA in HUVECs was scored (right panel). A total of more than 100 HUVECs for each condition were scored, and percentages of cells in different categories are shown. N≫C, predominately nuclear; N>C, mostly nuclear; N = C, equal intensity between nucleus and cytoplasm; N<C, primarily cytoplasmic. Scale bar, 20 μm. (B) Knockdown of FLNB inhibits the ability of VEGF to mediate the cytoplasmic accumulation of HDAC7 in HMVECs. HMVECs were transiently transfected with a control or FLNB siRNA followed by immunofluorescence microscopy. The numbers of transfected cells examined for each set of experiments were scored as described for panel A, and the percentages of cells in different categories are shown. Scale bar, 20 μm. (C) HMVECs were transiently transfected with a control or FLNB siRNA followed by VEGF treatment, and cells were harvested at indicated time points, followed by Western blotting with antibodies against HDAC7 phosphorylated at S178. (D) Knockdown of FLNB inhibits the ability of VEGF to mediate induction of HDAC7 target genes, Mmp-10 and Nur77, in HMVECs. The cells were transfected with nontargeted or two FLNB-targeted siRNAs and treated with VEGF for 1 h. Total RNA was extracted, and cDNA was generated and quantified by real-time PCR and normalized to the 18S rRNA level. Although VEGF induces Flnb mRNA levels in 1 h, it does not affect its protein abundance (see Fig. S6 in the supplemental material). (E) Fluorescence-based permeability assays were performed according to the manufacturer's protocol (36). HMVECs were transiently transfected with a control siRNA or two independent FLNB siRNAs for 72 h. A sample in which no cells were added (w/o cells) was used as a control for the formation of the cell monolayer. The immunoblot shows FLNB abundance after knockdown (right panel), and the composite image was derived from a single immunoblot.
HDAC7 interacts with ubiquitinated FLNB.
To further dissect the molecular basis of the interaction between FLNB and HDAC7, we mapped the interaction domains by expressing fragments of FLNB and performing coimmunoprecipitation with cotransfected FLAG-HDAC7 (Fig. 3A). Interestingly, we found that, in addition to the band corresponding to the calculated molecular size, several slower-migrating FLNB species in the whole-cell extracts of immunoglobulin (Ig)-like repeats 6 to 10 (R6-10) were also detected, suggesting that this fragment might be posttranslationally modified. However, HDAC7 coimmunoprecipitated only the slower-migrating species of FLNB(R6-10), suggesting that HDAC7 binds to modified FLNB. In accordance with this notion, bacterially expressed immobilized purified GST-FLNB did not pull down HDAC7 (see Fig. S3 in the supplemental material). To determine the nature of this modification, we carried out immunoprecipitations and mass spectrometry (MS). We identified several ubiquitin (Ub) peptides present in the modified FLNB bands from the FLAG-FLNB-transfected sample. To confirm these data, we determined whether knockdown of Ub alters the ratio of the intensity of modified to unmodified FLAG-FLNB(R6-10). Our data show that knockdown of Ub by siRNA decreased the ratio of modified/unmodified FLNB(R6-10) (Fig. 3B). We also show that upon cotransfection, FLAG-FLNB(R6-10) or FLAG-FLNB (full length) was coprecipitated with HA-Ub (Fig. 3C and D). Importantly, we observed that in HMVECs, VEGF induced endogenous FLNB ubiquitination (Fig. 3E), and FLNB ubiquitination disappeared after prolonged VEGF treatment (see Fig. S4 in the supplemental material). In summary, these data demonstrate that VEGF promotes FLNB ubiquitination and that HDAC7 interacts with ubiquitinated R6 to R10 of FLNB.
Monoubiquitination of K1147 is essential and sufficient for FLNB to interact with HDAC7.
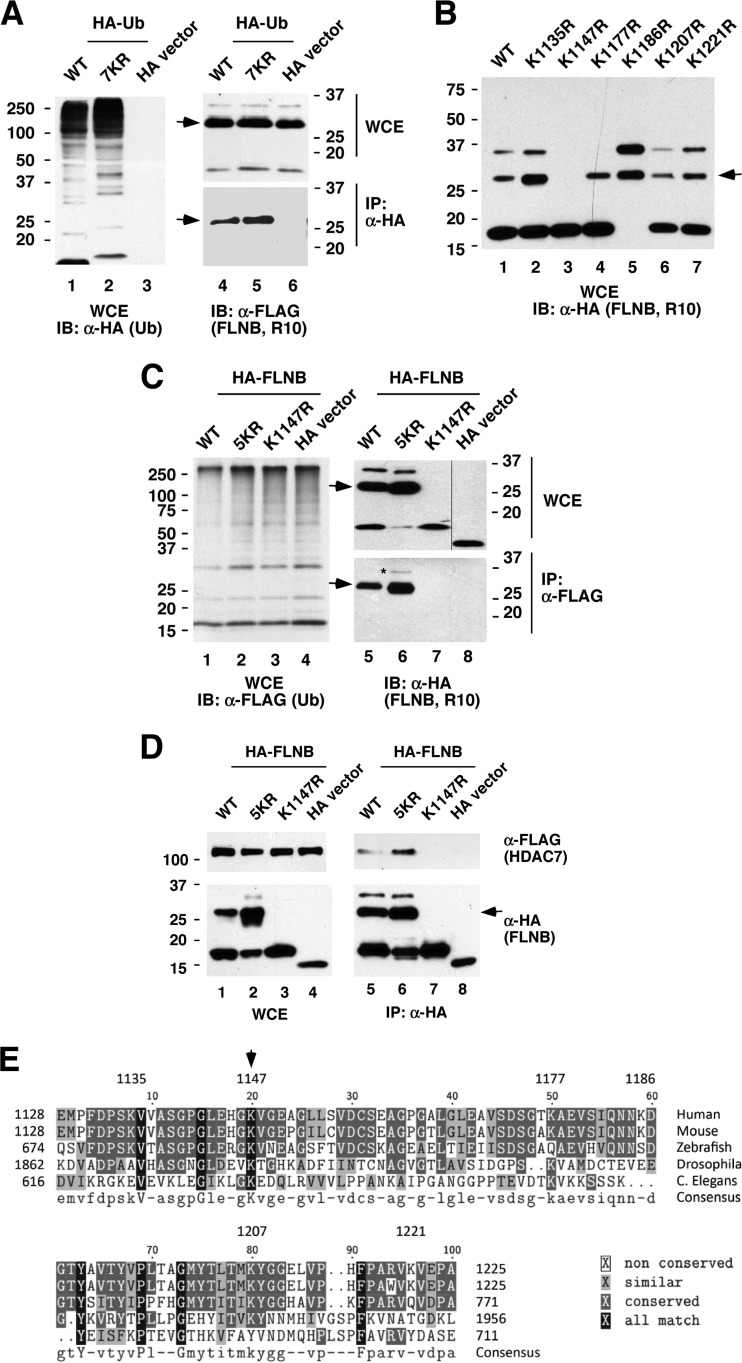
To identify the ubiquitinated lysine in FLNB(R6-10), we first determined which Ig-like repeat is ubiquitinated. We generated FLNB deletion and truncation constructs within R6 to R10 and found that R6 and R7 to R9 were not ubiquitinated, while R10 was ubiquitinated (see Fig. S5A and B in the supplemental material). Furthermore, a Ub mutant, 7KR, in which all (seven) possible polyubiquitinated lysines were replaced with arginines, was coimmunoprecipitated (conjugated) with FLNB(R10), indicating that FLNB(R10) is monoubiquitinated (Fig. 4A). We further generated amino acid substitutions in R10 by replacing individual lysines with arginines and found that the single mutation K1147R [FLNB(R10, K1147R)] abolished ubiquitination (second slowest migration species), while with K1177R we could not detect the slowest-migrating species (Fig. 4B). Interestingly, another mutant, K1186R, exhibited significantly more ubiquitination than the wild-type R10. We further confirmed that mutant K1147R lost ubiquitination, while an FLNB mutant, FLNB(R10, 5KR) in which only K1147 is intact, was ubiquitinated (Fig. 4C) and that coexpression of CaMK I enhanced ubiquitination of wild-type FLNB(R10) and FLNB(R10, 5KR) (see Fig. S5C in the supplemental material). Additionally, FLNB(R10, 5KR) is capable of supporting an association with HDAC7 (Fig. 4D), whereas FLNB(R10, K1147R), which is not ubiquitinated, is not capable of such an interaction. Alignment of the amino acid sequence of FLNB(R10) with R10 from other species indicates that K1147 is evolutionarily conserved, implying a potentially important function for this lysine (Fig. 4E). In summary, these data indicate that K1147 is ubiquitinated and that ubiquitination of K1147 is essential for FLNB to interact with HDAC7.
Fig 4.
Ubiquitination of FLNB K1147 is sufficient for the interaction with HDAC7. (A) FLNB(R10) is monoubiquitinated. HA-tagged wild-type (WT) Ub or mutants (7KR) were cotransfected with FLAG-vector or FLAG-FLNB(R10). Whole-cell extracts were immunoprecipitated, followed by immunoblotting. As a control, HA-vector alone was included for parallel analyses (lane 3). Ub(7KR) harbors all seven lysine-to-arginine mutations and cannot be polyconjugated. The arrows mark the ubiquitinated FLNB(R10) (A to D). (B) K1147 is essential for FLNB(R10) modification. Wild-type or the indicated HA-FLNB(R10) mutant expression plasmids were transiently transfected into HeLa cells, and whole-cell extracts were subjected to Western blotting. (C) FLNB K1147 is ubiquitinated. Wild-type FLNB or the indicated HA-FLNB(R10) mutant was cotransfected with FLAG-Ub. Whole-cell extracts were prepared, followed by immunoprecipitation and immunoblotting. FLNB(R10, 5KR) is a mutant in which each lysine, except K1147, is replaced with arginine. FLNB (R10, K1147R) is a mutant in which lysine 1147 is replaced with arginine. (D) Ubiquitination of K1147 is sufficient to interact with HDAC7. The wild type or the indicated HA-FLNB(R10) mutant was cotransfected with or without FLAG-HDAC7. Immunoprecipitation with anti-HA antibodies was carried out, followed by immunoblotting. (E) K1147 is evolutionarily conserved. Alignment of FLNB(R10) sequence with IgG-like repeats from different species was performed using Biology WorkBench, version 3.2, software. The asterisk in panel C marks the slowest-migrating band, suggesting that this FLNB species is ubiquitinated. The spliced lanes shown in panel C were merged from a single immunoblot. C. Elegans, Caenorhabditis elegans.
Mapping of FLNB interacting domain on HDAC7.
In order to further elucidate the mechanisms underlying FLNB-mediated VEGF-induced cytoplasmic accumulation of HDAC7, we mapped the domain in HDAC7 that interacts with FLNB. We found that amino acids 2 to 254 of HDAC7 [HDAC7(2–254)] interacted with endogenous FLNB (see Fig. S6 in the supplemental material). However, amino acids 72 to 172 did not. We further demonstrated that HDAC7(2–254) interacts with ubiquitinated FLNB(R6-10) in HeLa cells (Fig. 5A) and that recombinant GST-HDAC7(157–206) was sufficient to interact with ubiquitinated R8 to R10 of FLNB (Fig. 5B). This region contains several functional domains, including a 14-3-3 binding site at serine 178 and a functional NLS within amino acids 183 to 213 (20, 28). Further mapping indicated that deletion of amino acids 190 to 201 in HDAC7 abolished its association with ubiquitinated FLNB(R6-10) (Fig. 5C, lane 5). Interestingly, when exogenously expressed, this deletion mutant was exclusively localized to the cytoplasm (Fig. 5D), implying that these amino acids are responsible for nuclear localization of HDAC7. However, the subcellular localization of another deletion construct, HDAC7(Δ223–237), exhibited a similar distribution to that of the wild-type protein, and the protein accumulated in the cytoplasm when CaMK I was cotransfected (see Fig. S7D). In summary, our data indicate that the FLNB(R6-10) interacting domain largely overlaps with the HDAC7 NLS.
Fig 5.
FLNB-interacting domain in HDAC7 largely overlaps with its nuclear localization sequence. (A) Amino acids 2 to 254 of HDAC7 interact with FLNB(R6-10). HeLa cells were cotransfected with FLAG-FLNB(R6-10) with HA-HDAC7(2–254) or HA-vector plasmids. Cells lysates were coimmunoprecipitated, followed by Western blotting. Note that HDAC7 interacts only with the slower ubiquitinated FLNB(R6-10). (B) GST-HDAC7(157–206) interacts with ubiquitinated HA-FLNB(R8-10). Immobilized GST-HDAC7 fusion proteins were incubated with extracts expressing ubiquitinated FLNB, followed by immunoblotting. (C) Amino acids 190 to 201 of HDAC7 are required for the interaction with FLNB in R6 to R10. Expression plasmids encoding HA-vector, HA-HDAC7, HA-HDAC7(Δ190–201), or HA-HDAC7(Δ223–237) were cotransfected with a FLAG-FLNB(R6-10) expression plasmid. Whole-cell extracts were incubated with anti-HA beads, and the immune pellets were subjected to Western blotting. The arrows indicate the ubiquitinated FLNB(R6-10). (D) HeLa cells were transiently transfected with HA-HDAC7, HA-HDAC7(Δ190–201), or HA-HDAC7(Δ223–237) expression plasmids, followed by immunostaining with anti-HA antibodies. The numbers of HA-HDAC7-transfected cells for each set of experiments were scored as described in the legend of Fig. 2A, and the percentages of cells in different categories are shown. IB, immunoblotting.
Knockdown of Ub blocks VEGF-mediated HDAC7 cytoplasmic accumulation, induction of MEF2 and HDAC7 target genes, and endothelial cell permeability.
In order to demonstrate the role of Ub in cytoplasmic retention of HDAC7, we determined the effect of Ub knockdown on the HDAC7-FLNB interaction and subcellular distribution of HDAC7 in response to VEGF. We found that knockdown of Ub abolished VEGF-induced HDAC7-FLNB interaction (Fig. 6A, compare lanes 6 and 8) and cytoplasmic accumulation of HDAC7 in HUVECs (Fig. 6B). Furthermore, depletion of Ub in HeLa cells significantly inhibited PMA-mediated (Fig. 6C) and CaMK I-mediated (see Fig. S8 in the supplemental material) cytoplasmic retention of HDAC7. From these results, we speculated that cytoplasmic redistribution of HDAC7 would be critical for VEGF-induced expression of the MEF2/HDAC7 target genes Mmp-10 and Nur77. Consistent with our observation, FLNB is essential for VEGF-mediated induction of Mmp-10 and Nur77 (Fig. 2); knockdown of Ub significantly compromised the ability of VEGF to induce expression of Mmp-10 and Nur77 (Fig. 6D). Similar to the observations in FLNB knockdown cells, knockdown of Ub potently inhibited VEGF-induced permeability in HMVECs (Fig. 6E). In conclusion, we demonstrate that Ub is required for VEGF-mediated HDAC7 cytoplasmic accumulation, HDAC7 and FLNB interaction, induction of MEF2 and HDAC7 target genes, and endothelial cell permeability.
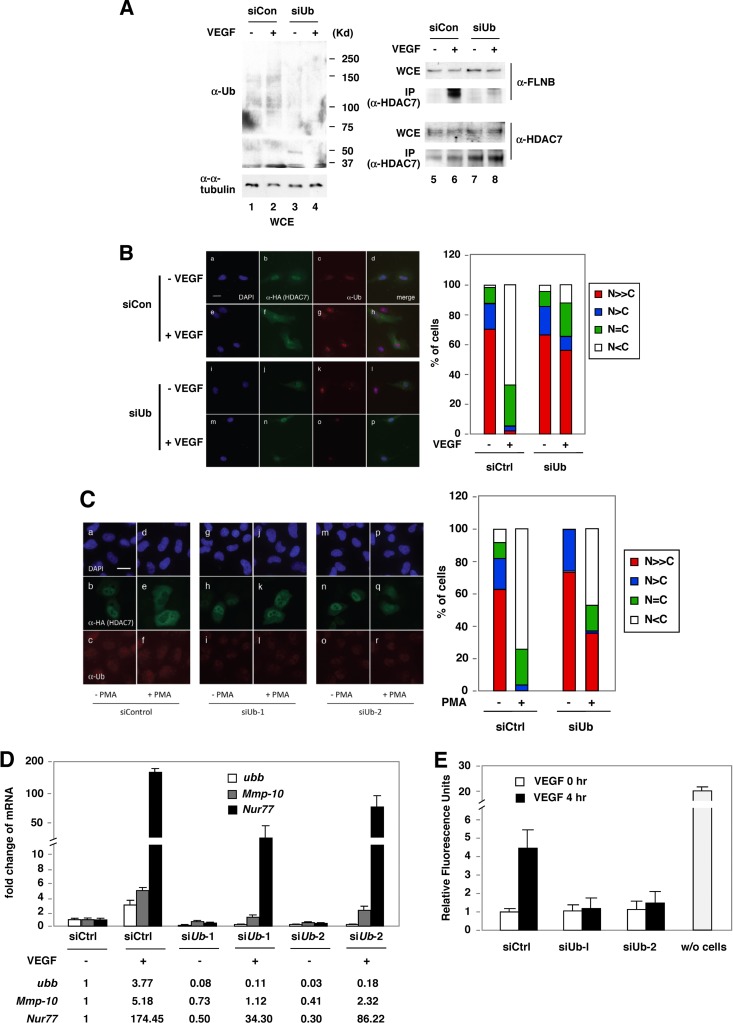
Fig 6.
Depletion of Ub blocks VEGF-induced endothelial cell permeability. (A) Depletion of Ub blocks VEGF-induced HDAC7-FLNB interaction in HMVECs. HMVECs were transiently transfected with control siRNA (siCon) or siRNA against Ub (siUb) for 48 h in regular EC medium. Cells were starved with 0.1% FBS in EBM-2 medium for another 24 h and then were treated with VEGF (50 ng/ml) for 20 min. Immunoprecipitation was performed with anti-HDAC7 antibodies, followed by Western blotting. (B) Knockdown of Ub abolishes VEGF-mediated cytoplasmic accumulation of HA-HDAC7 in HUVECs. HUVECs were transiently transfected with control siRNA or siRNA against Ub as described in panel A except that cells were treated with VEGF (20 ng/ml) for 2 h, followed by fluorescence microscopy. For quantification, the numbers of HA-HDAC7-transfected cells for each set of experiments were scored as described in the legend of Fig. 2A, and the percentages of cells in different categories are shown. Scale bar, 20 μm. (C) Knockdown of Ub blocks PMA-induced cytoplasmic sequestration of HA-HDAC7 in HeLa cells. Cells were transfected with control siRNA or siRNA against Ub and then transfected with HA-HDAC7 plasmids. Cells were treated with 10 ng/ml of PMA for 2 h, followed by fluorescence microscopy. The percentages of cells in different categories were scored as described in the legend of Fig. 2A. Scale bar, 20 μm. (D) Ub is critical for VEGF-mediated induction of MEF2/HDAC7 target genes, Mmp-10 and Nur77. The experiment was performed as described in the legend of Fig. 2C except that Ub siRNAs were used. ubb, Ub gene. (E) Permeability assays were performed as described in the legend of Fig. 2G except that Ub siRNAs were used. A sample in which no cells were added (w/o cells) was used as a control for the formation of the cell monolayer.
PMA and CaMK I promote FLNB ubiquitination.
PMA, which activates the VEGFR2 downstream kinase PKC in HUVECs (9), was capable of inducing FLNB ubiquitination in a dose- and time-dependent manner in HeLa cells (Fig. 7A). Furthermore, PMA promoted HDAC7 cytoplasmic accumulation in HUVECs, and this activity was abolished when FLNB was knocked down (Fig. 7B). A similar result was obtained in HeLa cells (see Fig. S9 in the supplemental material). We further determined whether PKC activity is essential for VEGF-induced FLNB ubiquitination and its association with HDAC7. Indeed, pretreatment with PKC inhibitors, Gö6983 or Gö6976, blocked the ability of VEGF to induce FLNB ubiquitination and its association with HDAC7 (Fig. 7C). Since CaMK I promotes HDAC7 cytoplasmic retention (20, 28), we speculated that CaMK I might enhance the association between FLNB and HDAC7. Indeed, a constitutively active form of CaMK I significantly induced the association between FLNB and HDAC7 (see Fig. S7A in the supplemental material) and promoted FLNB(R6-10) ubiquitination (see Fig. S7B). Notably, knockdown of FLNB greatly compromised the ability of CaMK I to promote cytoplasmic sequestration of HDAC7 (compare frames f and o in Fig. S7C in the supplemental material). Because ubiquitinated FLNB(R6-10) binds the HDAC7 NLS (Fig. 5C), we hypothesized that overexpression of FLNB(R10) may affect the ability of PMA to promote HDAC7 cytoplasmic accumulation. To test this, HeLa cells were transiently cotransfected with FLAG-HDAC7 and HA-FLNB(R10), treated with or without PMA, and followed by immunofluorescence microscopy using anti-HA and anti-FLAG antibodies. Without PMA treatment, FLAG-HDAC7 predominantly localizes in the nucleus, regardless of the presence of wild-type or mutant HA-FLNB(R10) (Fig. 7D, left panel). In PMA treated cells with FLAG-HDAC7 singly transfected or cotransfected with HA-vector, FLAG-HDAC7 was localized in the cytoplasm (Fig. 7D, right panel, frame c). By itself, FLNB(R10) is distributed throughout the cell including the nucleus. However, coexpression of FLNB(R10) inhibited PMA-mediated HA-HDAC7 cytoplasmic accumulation (Fig. 7D, frame g). In contrast, cotransfection of HA-FLNB(R10, K1147R) was unable to block PMA-mediated HDAC7 cytoplasmic accumulation (frame j), while HA-FLNB(R10, 5KR) efficiently inhibited the effect of PMA on HDAC7 localization (frame n). Quantification of these data is also shown.
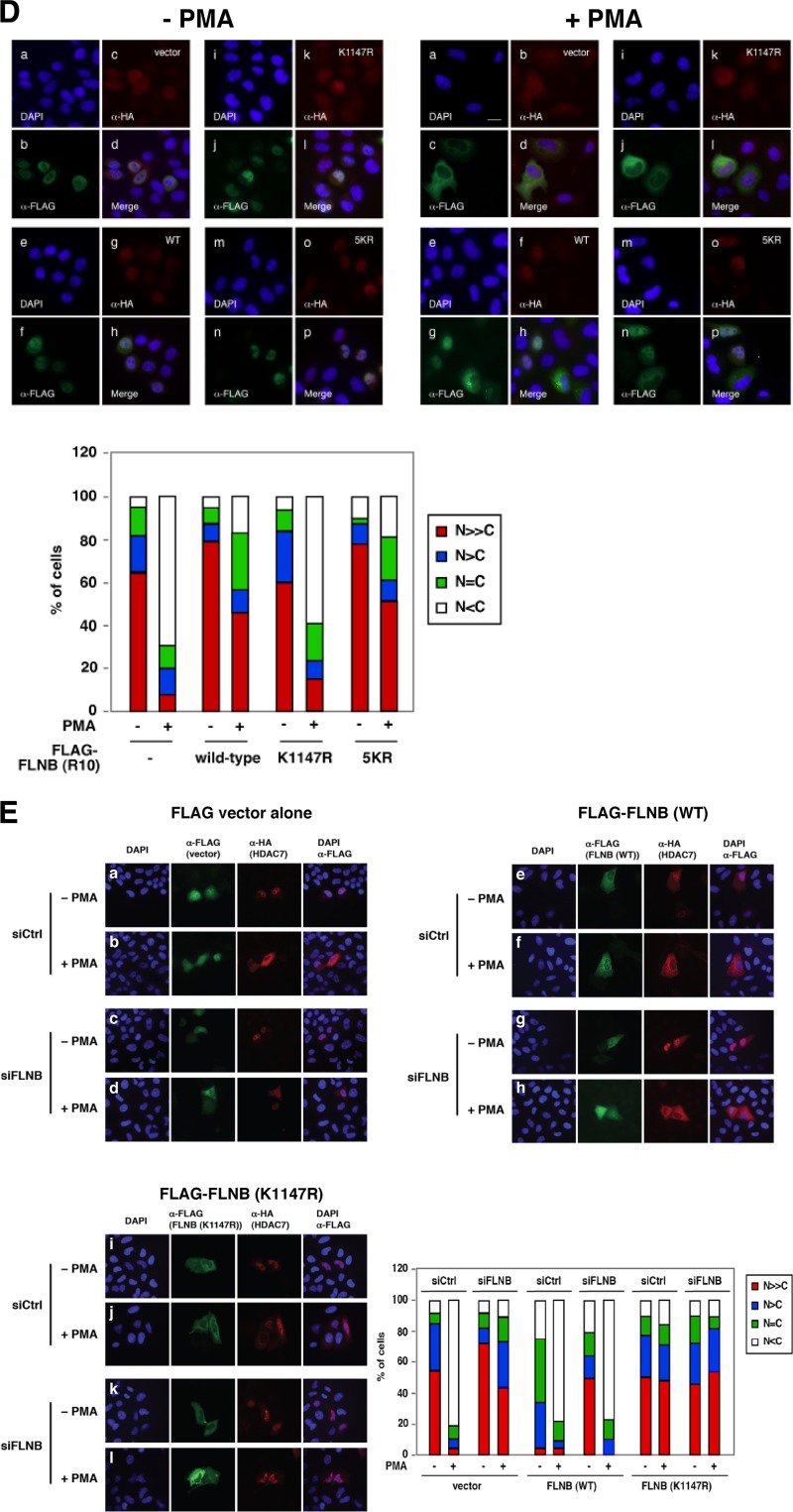
Fig 7.
The effects of PMA on FLNB ubiquitination and its association with HDAC7. (A) PMA enhances the ubiquitination of FLNB(R6-10) in a dose- and time-dependent manner. An expression plasmid encoding HA-FLNB(R6-10) was transfected into HeLa cells. The cells were treated with 10 ng/ml or 100 ng/ml PMA for the indicated times. Whole-cell extracts were prepared, followed by Western blotting. (B) FLNB is required for PMA-induced cytoplasmic accumulation of HDAC7. HUVECs were transiently transfected with a control or FLNB siRNA for 48 h, and then cells were transfected with an HA-HDAC7 expression construct for another 20 h, followed by treatment with 10 ng/ml PMA for 4 h. The data were analyzed by fluorescence microscopy (left panel). Statistical analysis of the subcellular distribution of HA-HDAC7 is shown (right panel). HUVECs transfected with both HA-HDAC7 and a nontargeting siRNA or siFLNB-1 were scored for the subcellular distribution of HA-HDAC7 with or without FLNB knockdown in the presence of PMA. A total of more than 100 HA-HDAC7-transfected cells for each set of experiments were scored as described in the legend of Fig. 2A, and the percentages of cells in different categories are shown. Scale bar, 20 μm. (C) Inhibition of PKC activity blocks VEGF-induced FLNB ubiquitination and its interaction with HDAC7 in HUVECs. HUVECs were serum starved by incubation in EBM-2 medium containing 0.1% FBS for 16 h. Prior to treatment with VEGF (50 ng/ml) for 20 min, the cells were pretreated with vehicle or PKC inhibitors (Gö6983, 0.5 μM; Gö6976, 5 μM) for 40 min. Immunoprecipitation was performed with anti-Ub antibodies, followed by Western blotting with the indicated antibodies. (D) HeLa cells were transiently cotransfected with FLAG-HDAC7 and HA-vector (−) (frames a to d), HA-FLNB(R10) (frames e to h), HA-FLNB(R10, K1147R) (frames i to l), or HA-FLNB(R10, 5KR) (frames m to p) in the absence (−) or presence (+) of PMA. Quantification of subcellular distribution of FLAG-HDAC7 is shown. HeLa cells transfected with both FLAG-HDAC7 and HA-FLNB were scored as described in the legend of Fig. 2A for subcellular distribution of FLAG-HDAC7 with or without PMA (50 ng/ml) treatment for 4 h. Scale bar, 20 μm. (E) HeLa cells were transiently transfected with siControl or siFLNB for 48 h, followed by cotransfection of HA-HDAC7 with FLAG-FLNB (wild-type) (rows e to h), FLAG-FLNB(K1147R) (rows i to l), or empty vector (FLAG-vector) (rows a to d) in the absence or presence of PMA (50 ng/ml) for 4 h. Immunofluorescence microscopy was performed with anti-HA (HDAC7; red), anti-FLAG (green), and DAPI staining. Quantification of subcellular distribution of HA-HDAC7 is shown (bottom right panel). Over 100 cells transfected with both HA-HDAC7 and FLAG-FLNBs were scored for the subcellular distribution of HA-HDAC7 using the system described in the legend of Fig. 2A. The spliced lanes shown in panel C were merged from a single immunoblot.
We further determined whether an ectopically expressed FLNB was capable of rescuing the ability of PMA to promote cytoplasmic sequestration of HDAC7 in cells in which endogenous FLNB had been knocked down by siRNA. HeLa cells were first transiently transfected with a control siRNA or an FLNB-targeting siRNA, followed by cotransfection of an HA-HDAC7 expression plasmid with FLAG-tagged wild-type (WT) FLNB [FLAG-FLNB(WT)] or a mutant, FLNB(K1147R). Cells were treated with or without PMA, followed by immunofluorescence microscopy. In control siRNA knockdown cells, ectopic expression of wild-type FLNB resulted in a modest increase in cytoplasmic distribution of HDAC7 compared to cells transfected with vector alone, even in the absence of PMA (Fig. 7E, rows a and e). This observation was consistent with the data shown in Fig. S1E in the supplemental material. Treatment with PMA led to a further increase in the cytoplasmic distribution of HDAC7. In contrast, cells expressing FLNB(K1147R) displayed a largely nuclear distribution of the mutant compared to FLNB(WT)-transfected cells in the absence of PMA (Fig. 7E, rows e and i). Moreover, PMA failed to promote cytoplasmic sequestration of cotransfected HDAC7 in FLNB(K1147R)-transfected cells, indicating that FLNB(K1147R) behaved in a dominant negative manner (Fig. 7E, row j).
In FLNB knockdown cells transfected with empty vector, HDAC7 was primarily localized in the nucleus, regardless of PMA treatment (Fig. 7E, row c). Ectopic overexpression of FLNB resulted in a significant increase in the cytoplasmic localization of HDAC7 in FLNB knockdown cells, even in the absence of PMA. PMA further induced cytoplasmic sequestration of HDAC7. In contrast, expression of FLNB(K1147R) in FLNB knockdown cells had no effect on the HDAC7 subcellular distribution in the absence of PMA and failed to rescue the ability of PMA to induce cytoplasmic sequestration of HDAC7. These results demonstrate that the K1147 residue in FLNB is critical for PMA-induced cytoplasmic accumulation of HDAC7. Taking these results together, we conclude that PKC and CaMK I promote FLNB ubiquitination and its interaction with HDAC7 and that FLNB is essential for PKC-mediated cytoplasmic accumulation of HDAC7.
DISCUSSION
Temporal and spatial regulation of nucleocytoplasmic shuttling of class IIa HDACs is a key mechanism to control cell differentiation, animal development, and glucose homeostasis (29). HDAC7 plays a pivotal role in vasculogenesis and embryonic development, in part by repressing the expression of MEF2 target genes such as Mmp-10 and Nur77. Consistent with this role, knockdown or knockout of HDAC7 leads to increases in Mmp-10 expression and disruption of endothelial cell-cell adhesion and subsequent disruption of vascular integrity in vitro and in vivo (8). VEGF controls EC permeability and angiogenesis partly through temporal induction of MEF2 and HDAC7 target genes. Recent evidence in endothelial cells supports a model in which VEGF engages VEGFR2, leading to an activation cascade of downstream kinases that involves PKCs and PKD and increased phosphorylation of HDAC7. In an attempt to identify cytoplasmic factors that regulate the subcellular distribution of HDAC7, we report the isolation of FLNB as a pivotal player essential for VEGF-mediated cytoplasmic sequestration of HDAC7, VEGF-induced expression of MEF2 and HDAC7 target genes, and VEGF-mediated permeability. In addition, we found that monoubiquitination of FLNB is required for HDAC7 binding and cytoplasmic retention and that this is common to both ECs and HeLa cells. Thus, these data suggest a universal mechanism governing FLNB-mediated cytoplasmic accumulation of HDAC7.
HDAC7 binds monoubiquitinated FLNB.
HDAC7 was originally shown to mediate repression of the transcriptional corepressor SMRT (30). In vitro deacetylation assays also demonstrated that class IIa HDACs possess potent deacetylase activity (31). Recently, we along with other labs have demonstrated that class IIa HDACs, including HDAC4 and HDAC7, are able to potentiate sumoylation of a variety of proteins including PML, LXR, MEF2, and HIC1 (19, 25, 32–34). In this study, we show for the first time that HDAC7 binds FLNB in a ubiquitination-dependent manner. This observation uncovers a novel mechanism for regulating the nucleocytoplasmic shuttling of HDAC7. Because class IIa HDACs do not harbor a recognizable Ub binding motif (17), it is likely that this interaction is through an unconventional Ub binding motif. Alternatively, monoubiquitination of FLNB may alter a local conformation, thereby creating or unmasking an HDAC7 binding surface.
The role of FLNB monoubiquitination in VEGF-mediated cytoplasmic retention of HDAC7.
The PKC activator, PMA, promotes FLNB ubiquitination, while inhibition of PKCs blocks VEGF-mediated FLNB ubiquitination, supporting the idea that PKCs are essential mediators that promote FLNB ubiquitination. As VEGF, PKCs, and class II HDACs play important roles in animal development and hormonal homeostasis, we note that K1147 in FLNB(R10) is evolutionarily conserved from fly to human. From these observations we speculate that such ubiquitination-dependent regulation is likely well preserved throughout evolution.
In this study, we have demonstrated that VEGF-induced FLNB ubiquitination is transient, that its kinetics correlate with VEGF-mediated cytoplasmic accumulation of HDAC7, and that ubiquitinated FLNB binds the HDAC7 NLS. Consistent with this observation, siRNA knockdown of Ub blocks VEGF-mediated FLNB-HDAC7 interaction. Furthermore, our mutational analyses have demonstrated that K1147 in FLNB(R10) is essential and sufficient to support monoubiquitination and the interaction between FLNB and HDAC7. In order to demonstrate the functional significance of K1147, the only ubiquitinated residue in R10, we employed a dominant negative strategy. We found that overexpression of a FLAG-FLNB(R10) fragment significantly blocked PMA-mediated cytoplasmic accumulation of cotransfected HA-HDAC7 (Fig. 7D). This observation suggests that this fragment functions as a dominant negative mutant and that other domains of FLNB are required for PMA-mediated cytoplasmic sequestration of HDAC7. Furthermore, overexpression of a ubiquitination-competent mutant, FLNB(R10, 5KR), resulted in a dominant negative phenotype similar to that for the wild-type R10 fragment. However, the ubiquitination-deficient mutant, FLNB(R10, K1147R), lost the ability to block PMA-mediated cytoplasmic sequestration of HDAC7. Based on the evidence with the rescue experiments (Fig. 7E), we conclude that PMA-induced ubiquitination of K1147 is essential for PMA-mediated HDAC7 cytoplasmic sequestration (Fig. 8).
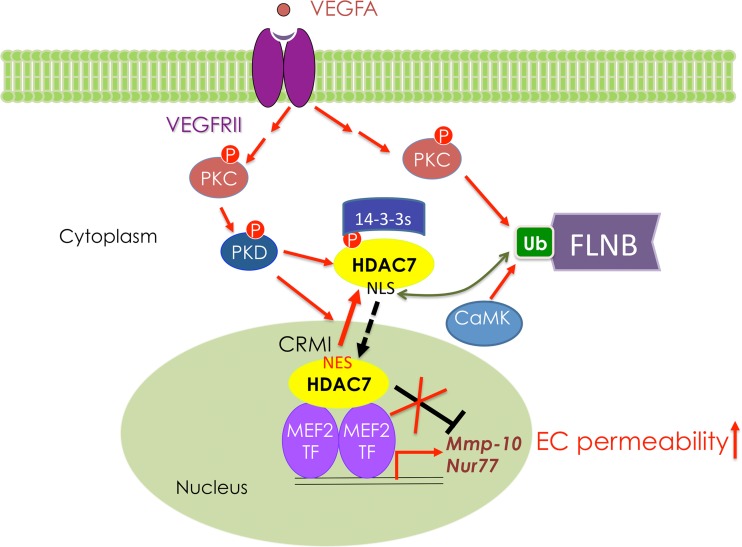
Fig 8.
A model in which VEGF promotes HDAC7 phosphorylation and FLNB ubiquitination to promote HDAC7 cytoplasmic sequestration. Subcellular distribution of HDAC7 is controlled by an N-terminal nuclear localization sequence (NLS) and a C-terminal nuclear export sequence (NES). CRM1 recognizes NES and promotes HDAC7 nuclear export, and importin binds the NLS and promotes HDAC7 nuclear localization (20). VEGF-A binds and induces activation of VEGFR2 downstream kinases including PKC and PKD. Activated PKD phosphorylates HDAC7 to generate 14-3-3 binding sites (9, 10), whereas activated PKC promotes ubiquitination of FLNB, which binds and masks the HDAC7 NLS from importin binding, thus blocking HDAC7 from nuclear entry.
The molecular details underlying VEGF-induced monoubiquitination.
In the absence of the proteasome inhibitor MG132, we were able to detect ubiquitination of endogenous FLNB. Similarly, when FLNB(R6-10) was cotransfected with exogenous Ub, only two distinct species of ubiquitinated FLNB were detected in the absence of MG132. These observations support the hypothesis that VEGF- and PKC-mediated FLNB monoubiquitination is not a mark for proteasome-mediated degradation. Rather, the observation that ubiquitination of FLNB is essential for HDAC7 interaction indicates that FLNB ubiquitination is a signaling event. Interestingly, our data show that CaMK I and PMA are capable of inducing FLNB ubiquitination, suggesting that phosphorylation of an unknown substrate promotes FLNB monoubiquitination. As monoubiquitination of FLNB is critical for HDAC7 interaction, the molecular mechanism underlying VEGF-, CaMK I-, and PKC-induced FLNB modification is under investigation.
When FLNB(R10, K1147R) is expressed, both slowly migrating modified FLNB(R10) species are lost (Fig. 4B), indicating that K1147 is required for ubiquitination or an additional unknown modification. Intriguingly, we found that FLNB(R10, K1177R) loses the slowest-migrating FLNB species, whereas K1186R exhibits more ubiquitination than the wild-type fragment. Furthermore, FLNB(R10, 5KR) exhibits much higher levels of ubiquitination than the wild-type fragment (Fig. 4C and D). These observations suggest a complex regulation of ubiquitination among these lysine residues that warrants further investigation in order to elucidate the mechanism underlying regulation of K1147 ubiquitination.
Several groups including ours have established that 14-3-3 proteins bind phosphorylated class II HDACs in vivo and in vitro and that this interaction is critical for cytoplasmic sequestration of class II HDACs (29, 35). Indeed, our survey of HDAC7 cytoplasmic complex components included several 14-3-3 family members. Unlike 14-3-3 binding, our in vitro pulldown assays indicated that the association of HDAC7 and FLNB does not require HDAC7 phosphorylation. Taken together, these observations indicate that 14-3-3 and FLNB interact independently with HDAC7 and that both interactions are essential for VEGF-mediated cytoplasmic sequestration of HDAC7.
We have previously shown that VEGF promotes cytoplasmic accumulation of wild-type HDAC7 but not in a mutant in which PKD-phosphorylated serines are mutated (9). In this study, we further demonstrate that inhibition of PKCs significantly compromises the ability of VEGF to induce FLNB ubiquitination and its association with HDAC7 (Fig. 7C). Thus, our data support a model in which activation by VEGF or PMA of PKC, a downstream effector of membrane-bound receptors including VEGFR2, promotes HDAC7 phosphorylation and FLNB ubiquitination and that both events are required for VEGF-mediated cytoplasmic accumulation of HDAC7 (Fig. 8).
Supplementary Material
ACKNOWLEDGMENTS
We thank David Samols and Edward Stavnezer for their comments on the manuscript.
This project is supported by RO1 HL093269 and DK078965 to H.-Y.K., P30 CA08748 to the Sloan-Kettering Proteomics Facility, and P30 EY11373 and funds from Case Western Reserve University and the Cleveland Foundation to M.M.
Footnotes
Published ahead of print 11 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01146-12.
REFERENCES
- 1. Bates DO. 2010. Vascular endothelial growth factors and vascular permeability. Cardiovasc. Res. 87:262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dejana E. 2004. Endothelial cell-cell junctions: happy together. Nat. Rev. 5:261–270 [DOI] [PubMed] [Google Scholar]
- 3. Gavard J, Gutkind JS. 2006. VEGF controls endothelial-cell permeability by promoting the beta-arrestin-dependent endocytosis of VE-cadherin. Nat. Cell Biol. 8:1223–1234 [DOI] [PubMed] [Google Scholar]
- 4. Wu H, Chen X, Xiong J, Li Y, Li H, Ding X, Liu S, Chen S, Gao S, Zhu B. 2011. Histone methyltransferase G9a contributes to H3K27 methylation in vivo. Cell Res. 21:365–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haberland M, Montgomery RL, Olson EN. 2009. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 10:32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kasler HG, Young BD, Mottet D, Lim HW, Collins AM, Olson EN, Verdin E. 2011. Histone deacetylase 7 regulates cell survival and TCR signaling in CD4/CD8 double-positive thymocytes. J. Immunol. 186:4782–4793 [DOI] [PubMed] [Google Scholar]
- 7. Yan W, Si Y, Slaymaker S, Li J, Zheng H, Young DL, Aslanian A, Saunders L, Verdin E, Charo IF. 2010. Zmynd15 encodes a histone deacetylase-dependent transcriptional repressor essential for spermiogenesis and male fertility. J. Biol. Chem. 285:31418–31426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chang S, Young BD, Li S, Qi X, Richardson JA, Olson EN. 2006. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell 126:321–334 [DOI] [PubMed] [Google Scholar]
- 9. Ha CH, Jhun BS, Kao HY, Jin ZG. 2008. VEGF stimulates HDAC7 phosphorylation and cytoplasmic accumulation modulating matrix metalloproteinase expression and angiogenesis. Arterioscler. Thromb. Vasc. Biol. 28:1782–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang S, Li X, Parra M, Verdin E, Bassel-Duby R, Olson EN. 2008. Control of endothelial cell proliferation and migration by VEGF signaling to histone deacetylase 7. Proc. Natl. Acad. Sci. U. S. A. 105:7738–7743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Christodoulides N, Feng S, Resendiz JC, Berndt MC, Kroll MH. 2001. Glycoprotein Ib/IX/V binding to the membrane skeleton maintains shear-induced platelet aggregation. Thromb. Res. 102:133–142 [DOI] [PubMed] [Google Scholar]
- 12. Popowicz GM, Schleicher M, Noegel AA, Holak TA. 2006. Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem. Sci. 31:411–419 [DOI] [PubMed] [Google Scholar]
- 13. Robertson SP. 2005. Filamin A: phenotypic diversity. Curr. Opin. Genet. Dev. 15:301–307 [DOI] [PubMed] [Google Scholar]
- 14. Loy CJ, Sim KS, Yong EL. 2003. Filamin-A fragment localizes to the nucleus to regulate androgen receptor and coactivator functions. Proc. Natl. Acad. Sci. U. S. A. 100:4562–4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berry FB, O'Neill MA, Coca-Prados M, Walter MA. 2005. FOXC1 transcriptional regulatory activity is impaired by PBX1 in a filamin A-mediated manner. Mol. Cell. Biol. 25:1415–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yoshida N, Ogata T, Tanabe K, Li S, Nakazato M, Kohu K, Takafuta T, Shapiro S, Ohta Y, Satake M, Watanabe T. 2005. Filamin A-bound PEBP2β/CBFβ is retained in the cytoplasm and prevented from functioning as a partner of the Runx1 transcription factor. Mol. Cell. Biol. 25:1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gao C, Liu Y, Lam M, Kao HY. 2010. Histone deacetylase 7 (HDAC7) regulates myocyte migration and differentiation. Biochim. Biophys. Acta 1803:1186–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu Y, Han D, Han Y, Yan Z, Xie B, Li J, Qiao N, Hu H, Khaitovich P, Gao Y, Han JD. 2011. Ab initio identification of transcription start sites in the Rhesus macaque genome by histone modification and RNA-Seq. Nucleic Acids Res. 39:1408–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao C, Ho CC, Reineke E, Lam M, Cheng X, Stanya KJ, Liu Y, Chakraborty S, Shih HM, Kao HY. 2008. Histone deacetylase 7 promotes PML sumoylation and is essential for PML nuclear body formation. Mol. Cell. Biol. 28:5658–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao C, Li X, Lam M, Liu Y, Chakraborty S, Kao HY. 2006. CRM1 mediates nuclear export of HDAC7 independently of HDAC7 phosphorylation and association with 14-3-3s. FEBS Lett. 580:5096–5104 [DOI] [PubMed] [Google Scholar]
- 21. Li X, Song S, Liu Y, Ko SH, Kao HY. 2004. Phosphorylation of the histone deacetylase 7 modulates its stability and association with 14-3-3 proteins. J. Biol. Chem. 279:34201–34208 [DOI] [PubMed] [Google Scholar]
- 22. Gao C, Cheng X, Lam M, Liu Y, Liu Q, Chang KS, Kao HY. 2008. Signal-dependent regulation of transcription by histone deacetylase 7 involves recruitment to promyelocytic leukemia protein nuclear bodies. Mol. Biol. Cell 19:3020–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheng X, Kao HY. 2009. G protein pathway suppressor 2 (GPS2) is a transcriptional corepressor important for estrogen receptor alpha-mediated transcriptional regulation. J. Biol. Chem. 284:36395–36404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reineke EL, Lam M, Liu Q, Liu Y, Stanya KJ, Chang KS, Means AR, Kao HY. 2008. Degradation of the tumor suppressor PML by Pin1 contributes to the cancer phenotype of breast cancer MDA-MB-231 cells. Mol. Cell. Biol. 28:997–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rivera-Del Valle N, Gao S, Miller CP, Fulbright J, Gonzales C, Sirisawad M, Steggerda S, Wheler J, Balasubramanian S, Chandra J. 2010. PCI-24781, a novel hydroxamic acid HDAC inhibitor, exerts cytotoxicity and histone alterations via caspase-8 and FADD in leukemia cells. Int. J. Cell Biol. 2010:207420 doi:10.1155/2010/207420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dressel U, Bailey PJ, Wang SC, Downes M, Evans RM, Muscat GE. 2001. A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J. Biol. Chem. 276:17007–17013 [DOI] [PubMed] [Google Scholar]
- 27. Gao DJ, Xu M, Zhang YQ, Du YQ, Gao J, Gong YF, Man XH, Wu HY, Jin J, Xu GM, Li ZS. 2010. Upregulated histone deacetylase 1 expression in pancreatic ductal adenocarcinoma and specific siRNA inhibits the growth of cancer cells. Pancreas 39:994–1001 [DOI] [PubMed] [Google Scholar]
- 28. Kao HY, Verdel A, Tsai CC, Simon C, Juguilon H, Khochbin S. 2001. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 276:47496–47507 [DOI] [PubMed] [Google Scholar]
- 29. Martin M, Kettmann R, Dequiedt F. 2009. Class IIa histone deacetylases: conducting development and differentiation. Int. J. Dev. Biol. 53:291–301 [DOI] [PubMed] [Google Scholar]
- 30. Kao HY, Downes M, Ordentlich P, Evans RM. 2000. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 14:55–66 [PMC free article] [PubMed] [Google Scholar]
- 31. de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. 2003. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370:737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang CC, Gao S, Sung LY, Corry GN, Ma Y, Nagy ZP, Tian XC, Rasmussen TP. 2010. Rapid elimination of the histone variant MacroH2A from somatic cell heterochromatin after nuclear transfer. Cell. Reprogram. 12:43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gao YS, Hubbert CC, Yao TP. 2010. The microtubule-associated histone deacetylase 6 (HDAC6) regulates epidermal growth factor receptor (EGFR) endocytic trafficking and degradation. J. Biol. Chem. 285:11219–11226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang J, Zhang M, Zhang Y, Kou Z, Han Z, Chen DY, Sun QY, Gao S. 2010. The histone demethylase JMJD2C is stage-specifically expressed in preimplantation mouse embryos and is required for embryonic development. Biol. Reprod. 82:105–111 [DOI] [PubMed] [Google Scholar]
- 35. Parra M, Verdin E. 2010. Regulatory signal transduction pathways for class IIa histone deacetylases. Curr. Opin. Pharmacol. 10:454–460 [DOI] [PubMed] [Google Scholar]
- 36. Byfield FJ, Wen Q, Leszczynska K, Kulakowska A, Namiot Z, Janmey PA, Bucki R. Cathelicidin LL-37 peptide regulates endothelial cell stiffness and endothelial barrier permeability. Am. J. Physiol. Cell Physiol. 300:C105–C112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen L, Petrelli R, Gao G, Wilson DJ, McLean GT, Jayaram HN, Sham YY, Pankiewicz KW. 2010. Dual inhibitors of inosine monophosphate dehydrogenase and histone deacetylase based on a cinnamic hydroxamic acid core structure. Bioorg. Med. Chem. 18:5950–5964 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.