Abstract
Muscle atrophy occurs under various catabolic conditions, including insulin deficiency, insulin resistance, or increased levels of glucocorticoids. This results from reduced levels of insulin receptor substrate 1 (IRS-1), leading to decreased phosphatidylinositol 3-kinase activity and thereby activation of FoxO transcription factors. However, the precise mechanism of reduced IRS-1 under a catabolic condition is unknown. Here, we report that C1-Ten is a novel protein tyrosine phosphatase (PTPase) of IRS-1 that acts as a mediator to reduce IRS-1 under a catabolic condition, resulting in muscle atrophy. C1-Ten preferentially dephosphorylated Y612 of IRS-1, which accelerated IRS-1 degradation. These findings suggest a novel type of IRS-1 degradation mechanism which is dependent on C1-Ten and extends our understanding of the molecular mechanism of muscle atrophy under catabolic conditions. C1-Ten expression is increased by catabolic glucocorticoid and decreased by anabolic insulin. Reflecting these hormonal regulations, the muscle C1-Ten is upregulated in atrophy but downregulated in hypertrophy. This reveals a previously unidentified role of C1-Ten as a relevant PTPase contributing to skeletal muscle atrophy.
INTRODUCTION
Insulin receptor (IR) substrate 1 (IRS-1) is indispensable for propagating insulin action from the IR to final cellular effects, such as glucose uptake and protein synthesis (1, 2). Upon phosphorylation of tyrosine residues via an activated insulin receptor, IRS-1 acts as a docking site for Src homology 2 (SH2) domain-containing proteins such as phosphatidylinositol 3-kinase (PI3K), Grb2, and SHP-2 (3). Reduced responsiveness to insulin typically occurs at the level of IRS-1 (4), which, consequently, decreases the IRS-1-associated PI3K/Akt pathway caused by a reduction of IRS-1 tyrosine phosphorylation and the IRS-1 level itself (5–7).
Skeletal muscle accounts for ∼75% of whole-body glucose uptake and comprises ∼40% of body mass contributing to body movement. Both glucose uptake and muscle mass are under the control of IRS-1-associated PI3K activity. Therefore, defects in IRS-1-associated PI3K activity in skeletal muscle contribute to impaired glucose metabolism (8, 9) and loss of muscle mass, known as muscle atrophy (10, 11).
Skeletal muscle atrophy occurs in patients with various catabolic diseases, such as diabetes mellitus, sepsis, Cushing's syndrome, and cancer (10–12). These conditions activate the ubiquitin-proteasome pathway, which is associated with increased expression of muscle RING finger 1 (MuRF1) and muscle atrophy F-box (MAFbx, also called atrogin-1). These E3 ligases are high-fidelity markers of muscle atrophy not only in catabolic diseases but also upon denervation or disuse (13–17). The Forkhead box O (FoxO) class of transcription factors is necessary for inducing both MuRF1 and atrogin-1 (16). The transcriptional activity of FoxOs is inhibited in growing muscles by activation of the insulin-like growth factor 1 (IGF-1)/PI3K/Akt pathway, which promotes muscle hypertrophy (13, 18). During atrophy, the PI3K/Akt pathway is inhibited by the reduced anabolic activity of insulin and IGF-1 and by increased catabolic glucocorticoid, consequently activating FoxOs (12, 18–20). Both in vivo and in vitro data have demonstrated reduced levels of IRS-1 under atrophied conditions (20–22), although the mechanism of IRS-1 downregulation under catabolic conditions is unclear.
C1-Ten (also known as tensin2) is a member of the tensin family, which also includes tensin1, tensin3, and cten. All four tensins contain C-terminal SH2 and PTB domains. tensin1, C1-Ten, and tensin3 have a protein tyrosine phosphatase (PTPase) domain, similar to PTEN (23). However, tensin1 lacks a cysteine residue which is critical for any PTPase activity. In addition, C1-Ten possesses a C1 domain. Recent papers suggested that only C1-Ten, but not tensin3, has a PTEN-like activity in the cells (23, 24). However, there is no evidence that C1-Ten is a direct lipid phosphatase like PTEN is. Additionally, the direct protein substrate of C1-Ten remains unknown.
In this study, we demonstrated for the first time that C1-Ten is a relevant PTPase of IRS-1. We also suggest a novel regulatory mechanism for IRS-1 stability via IRS-1 Y612 dephosphorylation by C1-Ten which explains the reduction of IRS-1 under catabolic conditions. Furthermore, we found an inverse relationship between C1-Ten and IRS-1 levels in muscles undergoing atrophy. These results suggest the role of C1-Ten as a mediator of muscle atrophy under a catabolic condition through degradation of IRS-1 via its PTPase activity.
MATERIALS AND METHODS
Antibodies and reagents.
Anti-phospho-Akt (S473, T308), anti-phospho-extracellular signal-regulated kinase (anti-phospho-ERK; T202/Y204), anti-phospho-S6 kinase 1 (anti-phospho-S6K1; T389), anti-phospho-IRS (S636/S639), anti-phospho-FoxO1/3a (T24/T32), anti-FoxO1, anti-S6K1, and anti-ERK antibodies were purchased from Cell Signaling (Danvers, MA). Anti-IRS-1 and anti-phospho-IRS-1 (Y612) were obtained from Millipore (Darmstadt, Germany). Anti-Akt1/2, anti-myosin heavy chain 1/2/3 (anti-MYH1/2/3), anti-PTEN, anti-phospho-IRS (Y632, Y896), anti-py20, and anti-py99 were acquired from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-laminin-2 was purchased from Sigma-Aldrich (St. Louis, MO). Anti-myosin heavy chain (anti-MYH; MF20) was acquired from Developmental Studies Hybridoma Bank (Iowa City, IA). Anti-Tenc1 was obtained from Sigma and GeneTex Inc. (Irvine, CA). Anti-Tenc1 from GeneTex was used at a 1:100 dilution for studying C1-Ten in adult skeletal muscle because the anti-Tenc1 from Sigma detected the highly abundant muscle protein MYH in adult muscle. IGF-1 (Long–R3–IGF-1), insulin, and dexamethasone (Dex) were purchased from Sigma Chemical Co. (St. Louis, MO).
Cell culture.
HEK293 cells and L6 myoblasts were grown and maintained in high-glucose Dulbecco's modified Eagle's medium (DMEM) or low-glucose α-minimal essential medium (α-MEM) with 10% (vol/vol) fetal bovine serum (FBS; Lonza). As the ability of myoblast fusion into myotubes declines with passage, cells were used at low passages (≤6) for all experiments. The medium was switched to α-MEM with 2% (vol/vol) FBS for differentiation and was replaced every 2 days. Atrophy experiments were initiated on day 8, when myotubes were of constant size and myotube fusion ceased.
Animals.
All animal experiments were performed in our animal facility under POSTECH institutional guidelines. All animals, including 6-week-old male, obese, diabetic leptin receptor-deficient (db/db) mice and lean littermate control (+/+ or +/db) mice were maintained on a 12-h light–12-h dark cycle with regular unrestricted diets.
Plasmids.
The full-length coding region of human C1-Ten (Tenc1 isoform 2, KIAA1075) cDNA (a gift from Kazusa DNA Research Institute, Chiba, Japan) was subcloned into the EcoRI/XbaI site of pFlag-CMV2 (Sigma) using the following primers: 5′-TAGAATTCATGAAGTCCAGCGGCCCTGTGGAG-3′ and 5′-GCTCTAGATCATTTTCTCTGGCCCAGTAGAAC-3′. Flag–C1-Ten C231S was generated with a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) using the following primers: 5′-GTCGTACTATACTCCAAGGGAAACAAG-3′ and 5′-CTTGTTTCCCTTGGAGTATAGTACGAC-3′. The Flag–C1-Ten wild type (WT) was subcloned into the EcoRI/SacII site of pEYFP-C1 (Clontech, Palo Alto, CA) for the yellow fluorescent protein (YFP) constructs using the following primers: 5′-TAGAATTCTATGAAGTCCAGCGGCCCTGTGGAG-3′ and 5′-TCCCCGCGGGGATCATTTTCTCTGGCCCAGTAG-3′. The full-length coding region of human IRS-1 cDNA was kindly provided by Michael J. Quon (University of Maryland, Baltimore, MD). IRS-1 cDNA was subcloned into the EcoRI/XbaI site of pFlag-CMV2 (Sigma) using the following primers: 5′-TAGAATTCAATGGCGAGCCCTCCGGAGAGCGAT-3′ and 5′-GCTCTAGACTACTGACGGTCCTCTGGCTGCTT-3′. Flag–IRS-1 Y612F and Y632F were generated with a QuikChange site-directed mutagenesis kit (Stratagene) using the following primer pairs, respectively: primers 5′-ACGGATGATGGCTTCATGCCCATGTCCCCA-3′ and 5′-TGGGGACATGGGCATGAAGCCATCATCCGT-3′ and primers 5′-GGCAGTGGAGACTTTATGCCCATGAGCCCC-3′ and 5′-GGCAGTGGAGACTTTATGCCCATGAGCCCC-3′.
Purification of C1-Ten and in vitro PTPase assay.
To purify the full-length Flag–C1-Ten protein, three 10-cm dishes of HEK293 cells were transfected with 3 μg Flag–C1-Ten plasmids. After 48 h, the cells were lysed in 1 ml lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 20 mM sodium fluoride [NaF], 0.025% mercaptoethanol, 1% Triton X-100, 0.2% sodium deoxycholate, a protease inhibitor mixture). The cell lysates were sonicated and centrifuged at 14,000 rpm for 15 min at 4°C, and the supernatant was incubated with 40 μl of anti-Flag beads (Sigma) at 4°C for 4 h. After the incubation, the beads were washed three times with lysis buffer and once with wash buffer (50 mM Tris-HCl, 50 mM NaCl, 10% glycerol, 5 mM dithiothreitol [DTT]). After the wash buffer was discarded, the protein was eluted with 80 μl elution buffer (50 mM Tris-HCl, 50 mM NaCl, 10% glycerol, 5 mM DTT, 100 μg/ml Flag peptide) for 30 min, followed by a second elution with 40 μl elution buffer for 30 min. Approximately 8 ng/μl C1-Ten was obtained. The PTPase activity of C1-Ten was measured once, immediately following purification, as its activity quickly diminishes with time. A malachite green assay kit (17-125; Millipore, Milford, MA) was used to determine the PTPase activity of C1-Ten in vitro. A phosphate standard curve was generated for each experiment, and enzyme reactions were performed in a final volume of 50 μl reaction buffer (50 mM Tris-HCl [pH 7.4], 50 mM NaCl, 5 mM DTT, 2 mM MgCl2) in 96-well microtiter plates. The reaction was stopped by adding 120 μl malachite green detection solutions. Color was developed for 15 min prior to measuring the absorbance of each well at 620 nm. Negative-control wells contained enzyme only, and phosphate release was determined according to a standard curve. To prevent free phosphate contamination from cell lysates, HEK293 cells expressing the Flag-CMV2 vector were used and purification was performed in parallel with Flag–C1-Ten WT and Flag–C1-Ten C231S (CS) purification. The following equation was used to calculate PTPase activity: (pmol phosphate released from Flag–C1-Ten WT or Flag–C1-Ten CS − pmol phosphate released from Flag vector)/μg enzyme. Glutathione S-transferase–PTEN was used as a positive control.
Preparation of protein extracts.
Crushed, snap-frozen tissues, and harvested cells were lysed in buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1 mM Na3VO4, 20 mM NaF, 10 mM glycerophosphate, 1 mM PMSF, 10% glycerol, 1% Triton X-100, 0.2% sodium deoxycholate, and protease inhibitor cocktail.
Recombinant Ads.
Adenovirus (Ad)–C1-Ten WT and Ad–C1-Ten CS were generated through homologous recombination between the linearized pAd-Track-CMV vector carrying either Flag–C1-Ten WT or Flag–C1-Ten CS and the adenoviral backbone vector pAd-Easy, as described previously (25). Ad-green fluorescent protein (GFP) was used as a control for all experiments with L6 myotubes. Viruses were purified with an Adeno-X maxipurification kit (631532; Clontech) and titrated according to the manufacturer's instructions.
C1-Ten knockdown experiments.
AllStars negative-control small interfering RNA (siRNA) (con si) and C1-Ten siRNAs si1 (targeting the C1-Ten sequence TCAGTGGATTACAACATGACA) and si2 (targeting the C1-Ten sequence TCCAGTGGACACAGCACACTG) (Qiagen, Valencia, CA, and Bioneer, Daejeon, South Korea) were introduced into myotubes on day 6 of differentiation for hypertrophy experiments and on day 8 of culture for dexamethasone-induced atrophy experiments using a DeliverX Plus siRNA transfection kit (Panomics, Freemont, CA) according to the manufacturer's instructions.
In vivo transfection of adult skeletal muscle.
Adult male mice (weight, 30 g) were anesthetized, and the tibialis anterior muscle of shaved legs was injected with plasmids (25 μg in 30 μl sterile phosphate-buffered saline [PBS]) along the muscle's length. Then, 3 electric pulses were applied at 50 V/cm at 950-ms intervals with a square wave pulse electroporator (BEX Co., Ltd., Japan).
Lentivirus transduction.
293T cells were transfected with plasmids encoding control or C1-Ten short hairpin RNA (shRNA; sequence, GAGGAAATTCTGTGAGGACAA) in pLKO.1 vectors (Sigma-Aldrich), vesicular stomatitis virus glycoprotein, and delta 8.9 for 72 h. Media containing lentiviruses were harvested and used for transduction of target cells for 24 h. Stable cell lines were selected in the presence of 3 μg/ml puromycin (Sigma-Aldrich).
Quantitative real-time PCR (qRT-PCR) analysis.
Total RNA was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA), according to the manufacturer's protocol. Total RNA was reverse transcribed using an ImProm-II reverse transcription system (Promega, Madison, WI), according to the manufacturer's instructions. The MyiQ real-time PCR detection system (Bio-Rad, Hercules, CA) was used for detection and quantification. The PCR was performed using SYBR Premix Ex Taq II DNA polymerase (TaKaRa Bio, Shiga, Japan). The PCR was carried out in a final volume of 20 μl using 0.5 μM each primer (listed below), cDNA, and 10 μl of the supplied enzyme mixture containing the DNA double-strand-specific SYBR green I dye for detecting PCR products. PCR was performed with a 3-min preincubation at 95°C, followed by 40 cycles of 15 s at 95°C and 30 s at 60°C. PCR products were verified by melting curve analysis and agarose gel electrophoresis.
The sequences of the primers (intron-spanning primers) used were as follows: for GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′-CCATGACAACTTTGGCATTG-3′ and 5′-CCTGCTTCACCACCTTCTTG-3′; for C1-Ten, 5′-CTCAGTGGAGTTTGTTTTCTCCTC-3′ and 5′-GCTGATTGAAGTTTTCATAGGAGTC-3′; for p85α, 5′-GGCGATTACACTCTTACACTAAGGA-3′ and 5′-GAGTTGAAGGTTAATGGATCAGAGA-3′; for IRS-1, 5′-GTGAACCTCAGTCCCAACCATAAC-3′ and 5′-CCGGCACCCTTGAGTGTCT-3′; and for MuRF1, 5′-AGTGTCCATGTCTGGAGGTCGTTT-3′ and 5′-ACTGGAGCACTCCTGCTTGTAGAT-3′.
Statistical analysis.
Data are presented as means ± standard errors of the means (SEMs). Comparisons between two groups were made by unpaired two-tailed Student's t tests. P values of <0.05 were considered statistically significant.
RESULTS
C1-Ten is a candidate regulator of muscle function upregulated in catabolic muscle.
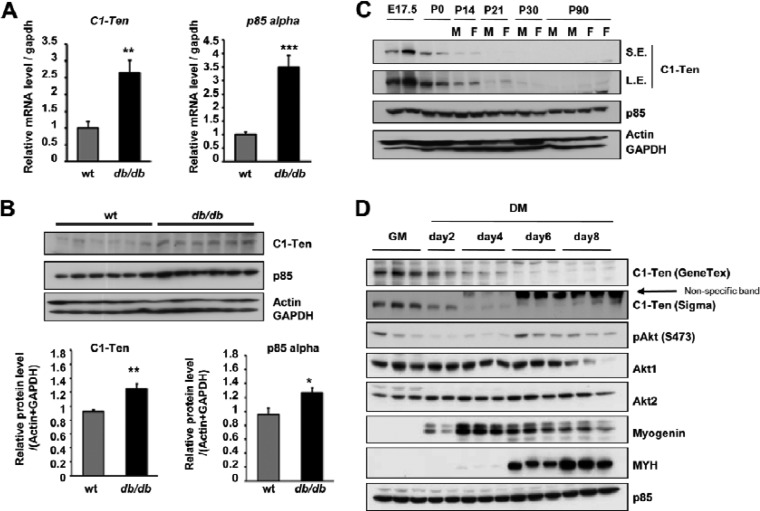
SH2 domain-containing proteins are crucial for propagating signals from the receptor to final cellular effects, and changes in the expression of many signaling molecules are frequently observed in various diseases, including diabetes. Thus, in an effort to identify novel proteins related to diabetes, we performed qRT-PCR analysis of SH2 domain-containing proteins in skeletal muscle from wild-type and db/db mice (see Fig. S1A in the supplemental material). Our screening identified C1-Ten as a potential protein that was significantly upregulated in diabetic skeletal muscle (Fig. 1A), and the p85α subunit of PI3K was used as a positive control (26, 27). To corroborate the observed increase in C1-Ten mRNA, we determined the C1-Ten protein level. Although commercially available C1-Ten antibodies did not work well for detecting the abundance of endogenous proteins corresponding to mRNA levels in tissues, a significant increase in C1-Ten protein levels was observed in diabetic skeletal muscle (Fig. 1B).
Fig 1.
Expression of C1-Ten in catabolic and anabolic muscle. (A) C1-Ten and PI3K-p85α mRNA levels in the gastrocnemius muscles of db/db mice were assessed by qRT-PCR and normalized to the gapdh level (n = 5); (B) C1-Ten and p85α protein levels in the gastrocnemius muscles of db/db mice (n = 6); (C and D) immunoblots showing C1-Ten expression during embryonic, postnatal development of skeletal muscle in mice (C) and L6 myogenesis (D). GM, growing medium; DM, differentiation medium; E number, embryonic day; P numbers, postnatal days; M, male; F, female. S.E., short exposure; L.E., long exposure; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To understand the functional meaning of the C1-Ten level in muscle physiology, we first monitored C1-Ten expression in postnatal development in vivo and myogenesis in vitro, during which anabolic pathway is activated (28, 29). Interestingly, C1-Ten expression was high during the embryonic stage 17.5 and on postnatal day 0 (P0) but decreasing on P21 (Fig. 1C), when significant myofiber hypertrophy begins (30). C1-Ten levels gradually decreased during myogenesis and were low on day 6 of differentiation (Fig. 1D), in which fused myotubes typically increase in mass (28, 29). Taken together, C1-Ten seems to be downregulated under the anabolic condition. Thus, we measured the C1-Ten level in the presence of catabolic glucocorticoid because glucocorticoid increase has been reported in diabetic mice (31). Incubating myotubes with synthetic glucocorticoid (i.e., Dex) caused the significant increase of C1-Ten and the induction of MuRF1 E3 ligase, a marker of muscle atrophy (see Fig. S1B in the supplemental material). Next, we examined whether the C1-Ten increase occurred under other catabolic conditions, like insulin deficiency. We injected rats with streptozotocin for 3 days, which induces acute type 1 diabetes. Under this insulin-deficient condition, we detected increased levels of C1-Ten and MuRF1 (see Fig. S1C in the supplemental material). In contrast to catabolic signals, anabolic insulin treatment resulted in decreased C1-Ten levels (see Fig. S1D in the supplemental material). Considering our findings that C1-Ten levels decreased in terminally differentiated myotubes and normal skeletal muscle but they were upregulated in catabolic muscle, we suggest that C1-Ten might be associated with muscle pathologies like muscle atrophy.
Upregulated C1-Ten induces atrophy of myotubes under the catabolic condition.
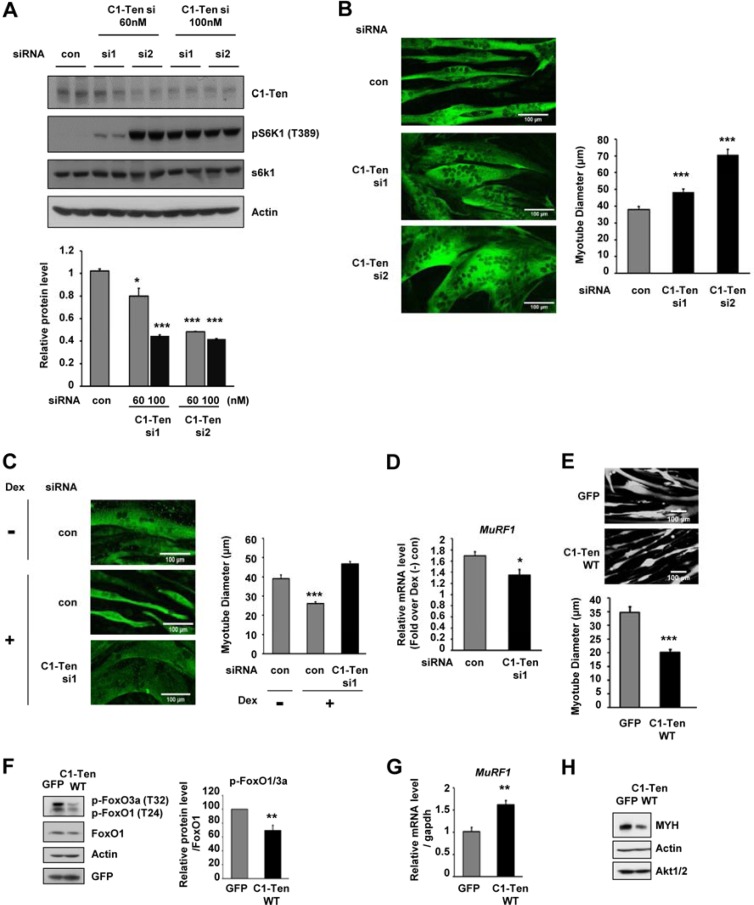
Based on the decreased levels of C1-Ten in anabolic muscle, we examined the effect of C1-Ten downregulation on muscle function. Two distinct nonoverlapping siRNAs against C1-Ten were tested. Both si1 and si2 reduced the basal C1-Ten level, but si2 was more potent (Fig. 2A; see Fig. S2A in the supplemental material). When the siRNA concentration was increased to 100 nM, the knockdown efficiency of si1 on the C1-Ten level was as effective as that of si2 (Fig. 2A). C1-Ten knockdown produced thicker myotubes in the order of knockdown efficiency (Fig. 2B). The mechanism of action seemed to involve the activation of anabolic pathways (i.e., Akt/S6K1) (Fig. 2A; see Fig. S2A in the supplemental material). Thus, we asked whether C1-Ten depletion is sufficient to reverse glucocorticoid-induced atrophy of myotubes. We chose si1 for the restoration experiments to limit the knockdown effects under the glucocorticoid-induced condition because si2 itself had a significant impact on myotube hypertrophy (Fig. 2B). This depletion of C1-Ten prevented glucocorticoid-induced atrophy (Fig. 2C) and blunted the induction of MuRF1 (Fig. 2D).
Fig 2.
Muscle size regulations by C1-Ten expression or depletion. (A) (Top) Effect of C1-Ten depletion on S6K1 signaling. si1 (TCAGTGGATTACAACATGACA) and si2 (TCCAGTGGACACAGCACACTG) were used to deplete C1-Ten. Knockdown was done for 24 h on day 6 of differentiation. (Bottom) Levels of C1-Ten protein relative to those of actin (n = 3). con, control. (B) C1-Ten knockdown was performed in myotubes after 6 days of culture. Cells were fixed in 4% paraformaldehyde in PBS for 20 min, washed twice with PBS, and permeabilized with 0.2% Triton X-100 for 5 min. After blocking with 3% bovine serum albumin, cells were stained for the myosin heavy chain with MF20 antibody. Images were obtained with a confocal microscope (LSM-510 Meta; Carl Zeiss, Jena, Germany). Myotube diameter was measured in three spots along a 50-μm length of a myotube. At 48 h after knockdown, the diameters of >50 myotubes were measured and their mean diameters were calculated using ImageJ software. (C) Effect of C1-Ten depletion on atrophy induced by glucocorticoid. C1-Ten knockdown was performed in myotubes after 8 days of culture. After 48 h, cells were stained with myosin heavy chain antibody. Images were obtained using a confocal microscope (LSM-510 Meta). The diameters of over 50 myotubes were measured. P values are compared to C1-Ten siRNA-treated (Dex-positive [Dex +]) cells. (D) The effect of C1-Ten knockdown on glucocorticoid-induced MuRF1 induction was determined by qRT-PCR. Data were normalized to gapdh expression and are presented as the fold induction relative to dexamethasone-untreated [Dex (−)] controls (n = 3). (E) The diameter was determined by infecting myotubes with GFP- or C1-Ten-expressing virus vectors. (Top) Images were taken 60 h after infection using a confocal microscope (LSM-510 Meta). Bars, 100 μm. (Bottom) More than 30 myotubes from independent experiments were measured. (F) (Left) Effect of C1-Ten on FoxO activation; (right) quantification of pFoxO1 and pFoxO3a over FoxO1 (n = 4). (G) Effect of C1-Ten on MuRF1 (n = 6) expression. (H) Effect of C1-Ten on MYH levels (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Next, we examined whether C1-Ten expression is sufficient to induce atrophy of myotubes. We used the adenovirus constructs expressing GFP (see Fig. S2B in the supplemental material), which allowed us to visualize the morphology of construct-expressing myotubes. Interestingly, C1-Ten overexpression for 60 h resulted in a 40% reduction in mean myotube diameter compared to that in cells expressing GFP (Fig. 2E). Mammalian cells contain three members of the FoxO family, FoxO1, FoxO3, and FoxO4 (32). Activation of FoxO1 and FoxO3 is sufficient to induce atrophy (16, 18). C1-Ten overexpression resulted in the activation of FoxO (reduction of FoxO1/3a phosphorylation) (Fig. 2F) and the subsequent MuRF1 induction downstream of FoxO activation (Fig. 2G). In line with this, myosin heavy chain (MYH), a MuRF1 E3 ligase target (33), was also reduced in C1-Ten-expressing cells (Fig. 2H), demonstrating that C1-Ten induces muscle atrophy by activating the catabolic FoxO pathway. These results suggest that increased C1-Ten induces muscle atrophy and that maintaining it at a low level may be required for normal muscle function.
C1-Ten activates the catabolic pathway at the level of IRS-1.
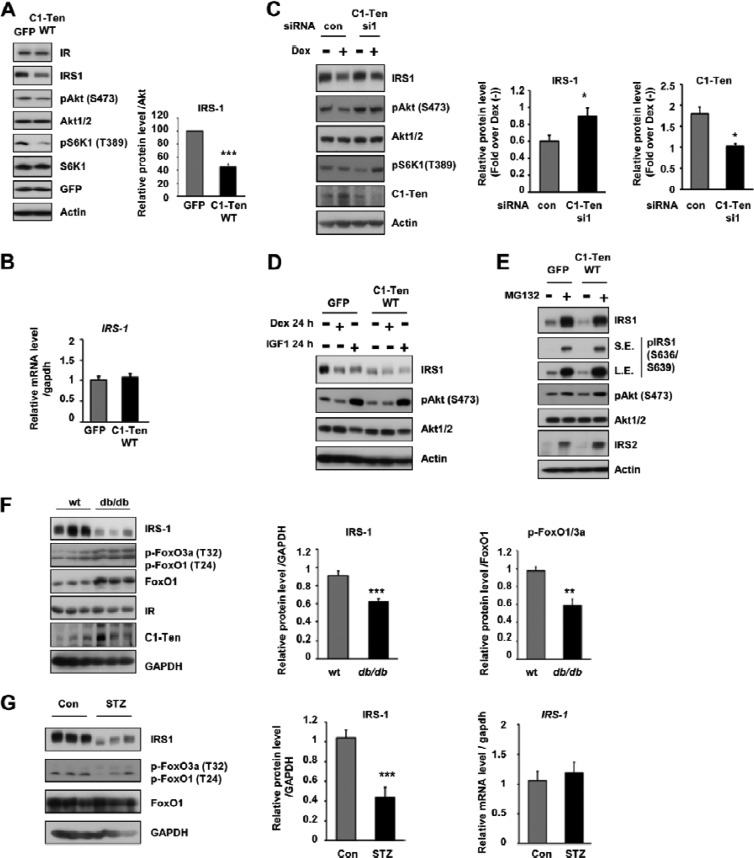
Next, we wanted to know which step of the atrophy pathway is modulated by C1-Ten. Reduced IRS-1 protein levels are observed in various types of muscle atrophy (16, 18, 20). Under our conditions, Dex treatment reduced the level of IRS-1 but not that of IR, p85, or PTEN (see Fig. S2C in the supplemental material). Given that Dex treatment caused IRS-1 reduction and C1-Ten knockdown prevented Dex-induced muscle atrophy and activated Akt/S6K1 pathways, we examined the effect of C1-Ten expression on IRS-1 and signaling downstream of IRS-1 (Akt/S6K1 phosphorylation). In C1-Ten-expressing myotubes, the IRS-1 protein level decreased significantly and a subsequent reduction in Akt/S6K1 phosphorylation was observed (Fig. 3A). However, no change in IRS-1 mRNA (Fig. 3B) or IR protein (Fig. 3A) levels was observed, as in Dex-treated myotubes.
Fig 3.
C1-Ten-induced muscle atrophy via IRS-1 reduction. (A) Effect of C1-Ten on Akt/S6K1 signaling and IRS-1 levels. (Left) Myotubes at 8 days of differentiation were infected with Ad-GFP constructs for 48 h and subjected to immunoblotting; (right) levels of IRS-1 protein relative to those of Akt (n = 5). (B) The effect of C1-Ten on IRS-1 mRNA was determined by qRT-PCR (n = 3). (C) (Left) Effect of C1-Ten knockdown on glucocorticoid-induced IRS-1 reduction in myotubes. At 4 h posttransfection, myotubes were treated with 100 nM dexamethasone for 16 h. (Middle and right) protein levels presented as fold induction relative to dexamethasone-untreated [Dex (−)] sets (n = 5). (D) To determine the effect of dexamethasone or IGF-1 on IRS-1 and Akt, myotubes were infected with GFP- or C1-Ten-expressing virus vectors for 24 h and treated with 100 nM dexamethasone or 10 ng/ml IGF-1 for an additional 24 h. (E) The effects of MG132 on C1-Ten-induced IRS-1 reduction were determined by treating L6 myotubes with 20 μM MG132 for 2 h. (F) IRS-1 protein levels and FoxO activation in gastrocnemius muscles of db/db mice. IRS-1 and pFoxO1/3a protein levels were quantified relative to those of GAPDH (n = 6) and FoxO1 (n = 3), respectively. (G) Protein level (left and middle) and mRNA level (right) of IRS-1 in gastrocnemius muscles of streptozotocin (STZ)-treated rats. Streptozotocin (100 mg/kg of body weight) was dissolved in 0.1 M citrate buffer (pH 4.5) and injected into male rats (∼180 g) by the tail vein. Three days later, the rats were anesthetized and muscle tissue was harvested (n = 5). P values are compared to the control pair of fed rats. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Because Dex caused the downregulation of IRS-1 and Akt/S6K1 pathways and C1-Ten expression reduced IRS-1 levels and inhibited the activation of Akt/S6K1, we tested whether the prevention of Dex-induced atrophy by C1-Ten knockdown occurs via restoration of the IRS-1 level. Depletion of Dex-induced C1-Ten by si1 partially preserved the IRS-1 level and prevented the subsequent decrease in Akt activation (Fig. 3C), suggesting that C1-Ten induces muscle atrophy at the level of IRS-1 reduction. Interestingly, si2 treatment, which was potent at reducing the basal C1-Ten level, slightly reduced the basal IRS-1 level (see Fig. S2A in the supplemental material). However, IRS-1 reduction by Dex or si2 resulted in the opposite effects on signaling downstream of IRS-1 and muscle size (Fig. 3D and 2C versus Fig. S2A in the supplemental material and Fig. 2A and B). Thus, we compared the effect of IGF-1 or Dex on the IRS-1 level in myotubes. This is based upon the finding that C1-Ten knockdown by si2 exerted effects on Akt activation and hypertrophy of myotubes similar to the effect of IGF-1 treatment (Fig. 2A and 3D; see Fig. S2A in the supplemental material) and C1-Ten overexpression emulated the effects of Dex treatment (Fig. 2E). Incubation of myotubes with anabolic IGF-1 activated Akt signaling, but Dex treatment inhibited Akt activation (Fig. 3D). In spite of this opposite action on Akt, both hormones reduced the IRS-1 level (Fig. 3D), as in C1-Ten si2-expressing or C1-Ten-overexpressing myotubes. Prolonged activation of Akt and S6K1 by IGF-1 is known to induce IRS-1 reduction via negative feedback (34, 35), and a recent paper showed that IGF-1 accelerated IRS-1 degradation in C2C12 myotubes via a proteasome-dependent pathway (36). Thus, IRS-1 reduction by C1-Ten knockdown seemed to be caused by an S6K1-dependent negative-feedback loop to restrain the anabolic effects of IGF-1, as previously reported (36).
Next, we determined how C1-Ten overexpression affects the IRS-1 level. Treatment with MG132, a proteasome inhibitor, blocked the C1-Ten-induced reduction of IRS-1, indicating the involvement of proteasomal degradation (Fig. 3E). C1-Ten-mediated IRS-1 reduction was independent of IRS-1 serine phosphorylation, demonstrated by IRS-1 serine phosphorylation (Fig. 3E) and treatment with inhibitors of PI3K and the mammalian target of rapamycin (mTOR) (see Fig. S2E in the supplemental material) in C1-Ten-expressing L6 myotubes. In C2C12 myotubes, glucocorticoid was reported to reduce IRS-1 protein stability, which is independent of S6K1 (37), as seen in our data. Taken together, these results indicate that the C1-Ten-mediated IRS-1 reduction is mechanistically different from canonical insulin/IGF-1-induced IRS-1 degradation.
As we observed increased C1-Ten levels in skeletal muscle of mice with acute type 1 or chronic type 2 diabetes (Fig. 1A and B; see Fig. S1C in the supplemental material) and muscle atrophy has been reported to be accelerated in these mice (10), we asked whether the in vivo levels of IRS-1 decrease. There was a significant reduction in the IRS-1 protein level (Fig. 3F). Next, we determined whether FoxO activation occurs under our conditions. Although phosphorylated FoxO1/3a levels were increased, total FoxO1 protein levels were increased to an even greater extent, resulting in a reduction of the pFoxO1 and pFoxO3a/total FoxO1 ratio (Fig. 3F). Because dephosphorylated FoxOs can enter the nucleus and turn on their target genes, FoxOs are activated in the skeletal muscle of db/db mice. However, not only enhanced levels of cortisol but also hyperinsulinemia are evident in these mice (38), and both of these can contribute to decreased IRS-1 levels in db/db mice. In insulin-deficient acute diabetes-induced muscle atrophy, we observed a marked decrease in IRS-1 protein levels (Fig. 3G). This led us to speculate that there must be a distinct IRS-1 degradation mechanism possibly mediated by C1-Ten under a catabolic condition. Taken together, the reduction in IRS-1 in muscle atrophy in diabetes (i.e., acute type 1 or chronic type 2) can be due in part to the reduced stability of IRS-1 as a result of increased C1-Ten levels under catabolic conditions.
C1-Ten reduces IRS-1 stability via its PTPase activity.
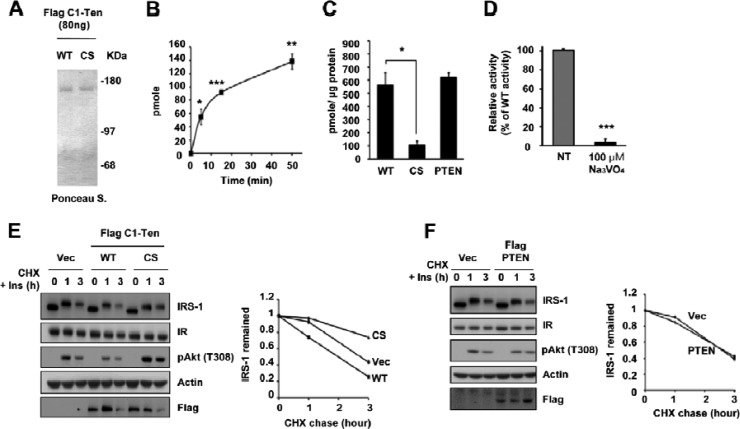
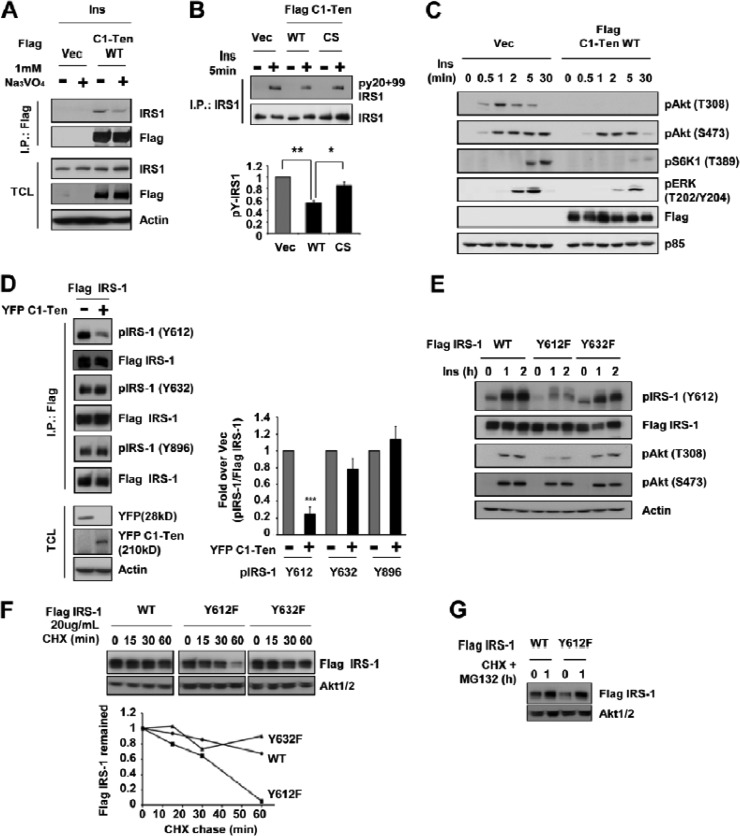
A report has shown partial preservation of the reduction in IRS-1 levels by maintaining tyrosine phosphorylation via an unidentified mechanism (34). Incubating C1-Ten-expressing cells with vanadate, a PTPase inhibitor, restored IRS-1 levels (see Fig. S2F in the supplemental material), supporting the role of PTPase activity in IRS-1 stability. Therefore, we asked whether C1-Ten is a PTPase responsible for the C1-Ten-induced reduction in IRS-1. Even if C1-Ten has the PTPase active-site signature motif, there is no direct evidence for C1-Ten PTPase activity in vitro (23). Therefore, we purified the full-length C1-Ten protein from HEK293 cells using a Flag immunoprecipitation/Flag peptide elution system (Fig. 4A). This protein, together with phosphotyrosine-containing peptides, demonstrated the time-dependent PTPase activity of C1-Ten (Fig. 4B). C1-Ten WT had activity comparable to that of PTEN when the same amount of protein was used, but a C1-Ten CS mutant in which catalytic residue cysteine 231 was replaced by serine showed insignificant levels of PTPase activity (Fig. 4C). Also, C1-Ten PTPase activity was abrogated by a general PTPase inhibitor, vanadate, which suggests that C1-Ten is a relevant PTPase (Fig. 4D).
Fig 4.
Negative regulation of IRS-1 stability via C1-Ten PTPase activity. (A) Ponceau S staining revealed the quantity, integrity, and purity of the full-length C1-Ten protein. (B) Time-dependent PTPase activity of C1-Ten. The malachite green assay was performed with 200 ng of C1-Ten and 400 μM EEEEpYEEEE peptide (n = 3). (C) Cysteine-based PTPase activity of C1-Ten. C1-Ten CS and PTEN were used as the negative and positive controls, respectively. Error bars indicate the SEMs of four independent experiments, each performed in triplicate. (D) Vanadate-mediated inhibition of C1-Ten PTPase activity. Error bars indicate the SEMs of three independent experiments, each performed in duplicate. (E) CHX chase. HEK293 cells were transfected with vector (Vec), Flag–C1-Ten WT, or Flag–C1-Ten CS. After 48 h, CHX (20 μg/ml) and 10 nM insulin (Ins) were added and chased for the times indicated, followed by immunoblot analysis (left) and graphical analysis (right). (F) HEK293 cells were transfected with vector or Flag-PTEN. After 48 h, CHX and insulin were added and chased, followed by immunoblot analysis (left) and graphical analysis (right). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In contrast to C1-Ten WT, C1-Ten CS activated signaling downstream of IRS-1 (i.e., Akt/S6K1) in myotubes (see Fig. S2G in the supplemental material). There was also a reduction of IRS-1 in C1-Ten CS-expressing myotubes, as in IGF-1-treated or C1-Ten si2 expressing cells (Fig. 3D; see Fig. S2A and G in the supplemental material). As expected, treatment with rapamycin or LY294002 blocked C1-Ten CS-induced IRS-1 degradation, indicating that this degradation is dependent on mTOR and PI3K activity (see Fig. S2H in the supplemental material).
Due to the large accumulation of IRS-1 after just 2 h of MG132 treatment, we hypothesized that the turnover rate of IRS-1 in myotubes is fast. This might be because L6 cells produce approximately 10 ng/ml IGF-2 (39). Therefore, we determined the effects of C1-Ten on IRS-1 degradation after long-term treatment of HEK293 cells with IGF-1 in order to trace C1-Ten WT- or C1-Ten CS-mediated IRS-1 reduction. Following 12 h of IGF-1 treatment, IRS-1 protein was degraded in vector- or C1-Ten WT-expressing cells, but C1-Ten CS blocked this degradation; however, it no longer blocked IRS-1 degradation after 48 h of IGF-1 treatment (see Fig. S2I in the supplemental material). Instead, there was doublet of IRS-1 in C1-Ten CS-expressing cells because of the mobility shift induced by hyperphosphorylation, which was most evident under 10 nM IGF-1 treatment (see Fig. S2I in the supplemental material). Different from myotubes, preservation of tyrosine phosphorylation by C1-Ten CS blocked IRS-1 degradation to some extent, which in turn led to prolonged activation of Akt and mTOR, causing IRS-1 degradation, as seen in 3T3-L1 adipocytes (34). This result in HEK293 cells helps our understanding of the results seen in C1-Ten CS-expressing, C1-Ten si2-expressing, or IGF-treated myotubes. The differences in the role of tyrosine phosphorylation in IRS-1 stability between myotubes and other cell types might be caused by the presence of cell type- or tissue-specific E3 ligases targeting IRS-1 for degradation (36).
To clarify the role of C1-Ten PTPase activity on IRS-1 stability, we monitored IRS-1 protein decay in the presence of the protein synthesis inhibitor cycloheximide (CHX) for a subsequent 3-h period in HEK293 cells. The rate of IRS-1 decay was accelerated by C1-Ten WT (Fig. 4E) but not by PTEN (Fig. 4F), supporting the specificity of C1-Ten-induced IRS-1 reduction. IRS-1 decayed more slowly in C1-Ten CS-expressing cells than in C1-Ten WT-expressing cells (Fig. 4E), suggesting a role for tyrosine phosphorylation on IRS-1 stability. Reflecting these IRS-1 level changes, Akt phosphorylation was decreased by C1-Ten WT but was sustained by C1-Ten CS (Fig. 4E). Taken together, we propose that C1-Ten regulates IRS-1 stability via its PTPase activity, which is the distinctive mechanism to reduced IRS-1 stability via anabolic pathways.
C1-Ten dephosphorylates Y612 of IRS-1, inducing IRS-1 degradation.
Next, we checked the possibility that IRS-1 is a substrate of C1-Ten. In general, the PTPase CS mutant traps its substrate and forms a more stable complex with its substrate than the WT PTPase. Immunoprecipitation experiments with IRS-1 after insulin treatment demonstrated that C1-Ten CS had a more stable interaction with IRS-1 than C1-Ten WT (see Fig. S3A in the supplemental material). To further validate that IRS-1 is a direct substrate of C1-Ten, immunoprecipitation was performed in the absence or presence of vanadate, a phosphotyrosine mimetic. If the interaction between the C1-Ten PTPase domain and IRS-1 is direct, excess vanadate will compete with IRS-1 for binding to the C1-Ten PTPase domain. In the presence of vanadate, less IRS-1 coimmunoprecipitated with C1-Ten (Fig. 5A), suggesting a direct interaction between the two proteins. Reflecting the PTPase-substrate relationship of C1-Ten and IRS-1, C1-Ten WT caused a 40% reduction in total IRS-1 tyrosine phosphorylation, which was restored by C1-Ten CS (Fig. 5B), without affecting IR tyrosine phosphorylation (see Fig. S3B in the supplemental material). C1-Ten prevented Akt and ERK activation upon insulin stimulation (Fig. 5C) and IGF-1 stimulation (see Fig. S3C in the supplemental material), but it did not affect epidermal growth factor-mediated Akt and ERK activation (see Fig. S3D in the supplemental material), suggesting that C1-Ten specifically affects IRS-1-mediated signaling.
Fig 5.
Accelerated degradation of Y612-dephosphorylated IRS-1. (A) IRS-1 as a substrate of C1-Ten. After HEK293 cells were treated with insulin for 5 min, immunoprecipitation (I.P.) was performed with anti-Flag antibody in the absence or presence of Na3VO4 and then subjected to immunoblotting with anti-IRS-1 antibody. TCL, total cell lysate. (B) By using HEK293 cells, immunoprecipitation was performed with anti-IRS-1 antibody and the immunoprecipitates were analyzed by immunoblotting with phosphotyrosine antibodies (py20 + py99). (Top) Representative immunoblot; (bottom) quantitation of pY-IRS-1 normalized to the amount for the insulin-stimulated vector (Vec) controls (n = 3). (C) Effect of C1-Ten on the signaling downstream of IRS-1 in HEK293 cells treated with insulin. HEK293 cells were transfected with vector or Flag–C1-Ten WT for 24 h. The cells were then serum starved for 18 h prior to treatment with 50 nM insulin. (D) (Left) HEK293 cells were cotransfected with Flag–IRS-1 and YFP vector or YFP–C1-Ten WT. After 48 h, immunoprecipitation was performed with anti-Flag antibody, and the immunoprecipitates were subjected to immunoblotting with pY612, pY632, or pY896 antibodies of IRS-1. (Right)The amount of pIRS-1 was normalized to that of immunoprecipitated Flag–IRS-1 and represented as the fold change over the result for the YFP vector (n = 3). (E) Effect of IRS-1 YF mutants on insulin signaling. HEK293 cells were transfected with Flag–IRS-1 WT, Y612F, or Y632F. (F) Regulation of IRS-1 stability by IRS-1 Y612. CHX chase was used. Serum-starved HEK293 cells were incubated with CHX for 1 h, followed by immunoblotting (top). The remaining amount of IRS-1 was normalized to the amount of Akt1/2 (bottom). (G) CHX and MG132 were added to IRS-1 WT- or Y612F-expressing cells for 1 h. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
IRS-1 contains 16 potential tyrosine sites (3); thus, we asked whether C1-Ten could affect specific IRS-1 tyrosine residues. We observed that C1-Ten significantly reduced the phosphorylation of IRS-1 Y612 (human)/Y608 (mouse and rat) but not that of IRS-1 Y632 or Y896 (Fig. 5D). C1-Ten-expressing myotubes also had reduced IRS-1 Y608 (human Y612) phosphorylation compared to GFP-expressing control cells after accumulating IRS-1 with MG132 (see Fig. S3E in the supplemental material). In contrast, C1-Ten shRNA-expressing myoblasts (see Fig. S3F in the supplemental material) increased IRS-1 Y608 phosphorylation (see Fig. S3G in the supplemental material). On Akt phosphorylation, IRS-1 Y612F showed effects (Fig. 5E) similar to those that it showed in C1-Ten-expressing HEK293 cells (Fig. 5C; see Fig. S3C in the supplemental material). Thus, these results indicate that C1-Ten reduces IRS-1 tyrosine phosphorylation and preferentially acts on IRS-1 Y612.
To determine whether IRS-1 Y612 dephosphorylation affects IRS-1 stability, we monitored the rate of IRS-1 WT, Y612F, or Y632F protein decay. IRS-1 Y612F decayed at a very high rate during a subsequent 1-h period of CHX chasing compared to the rate of decay of IRS-1 WT or IRS-1 Y632F (Fig. 5F). However, incubation of IRS-1 Y612F-expressing cells with MG132 restored the stability of IRS-1 Y612F to a level similar to that of IRS-1 WT (Fig. 5G). Taken together, these results suggest that C1-Ten preferentially dephosphorylates IRS-1 Y612, which induces proteasome-mediated IRS-1 degradation.
C1-Ten causes atrophy of adult muscle fibers in a PTPase-dependent manner.
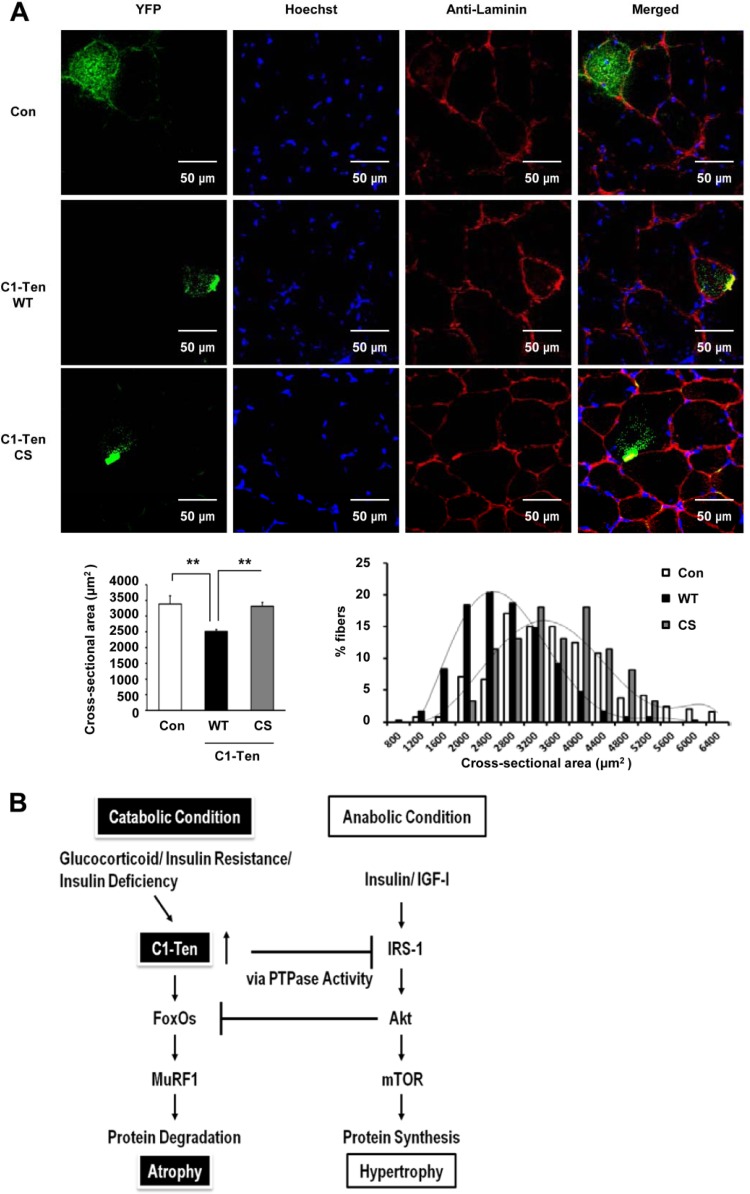
Because we observed that C1-Ten caused the atrophy of myotubes through IRS-1 reduction via its PTPase activity (Fig. 2C and E, 3A, and 4E) and C1-Ten levels increased in atrophied muscle in vivo (Fig. 1A and B; see Fig. S1C in the supplemental material), we asked whether C1-Ten affects muscle size in vivo in its PTPase-dependent way. We transfected YFP-tagged C1-Ten constructs (see Fig. S4A in the supplemental material) into the tibialis anterior muscles of mice via electroporation. As in the cellular localization, C1-Ten formed cytoplasmic puncta in adult skeletal muscle (40). We found that C1-Ten transfection into muscles for 12 days resulted in a 25% reduction in mean cross-sectional area (CSA) and a leftward shift in the distribution of myofiber CSA (Fig. 6A). This was dependent on its PTPase activity, as demonstrated by transfection with the C1-Ten CS mutant. Taken together, these results propose that C1-Ten PTPase can negatively regulate muscle function.
Fig 6.
C1-Ten PTPase-dependent regulation on adult muscle size. (A) Adult tibialis anterior muscles were transfected with YFP-, YFP–C1-Ten WT-, or YFP–C1-Ten CS mutant (C231S phosphatase-inactive mutant)-expressing vectors, and mice were killed after 12 days. Muscle cryosections (10 μm thick) were fixed with 4% paraformaldehyde at room temperature. After being rinsed with PBS, the cryosections were permeabilized with 0.2% Triton X-100 for 5 min and blocked with 3% bovine serum albumin. Cryosections were stained for Hoechst and laminin. Images were obtained with a confocal microscope (Olympus, Tokyo, Japan, or Leica sp5, Germany) and merged with the YFP signal. (Bottom left) Effect of C1-Ten expression on mean cross-sectional area (CSA). More than >200 fibers expressing each YFP construct were measured (n = 5). (Bottom right) Distribution of the CSA of fibers expressing YFP (control [Con]), YFP–C1-Ten WT (WT), or YFP–C1-Ten CS (CS). **, P < 0.01. (B) Schematic depicting skeletal muscle atrophy induced by C1-Ten under pathological conditions. The balance between anabolic and catabolic pathways is crucial for maintaining skeletal muscle mass. Anabolic pathways involving insulin and the IGF-1/IRS-1/Akt pathway not only promote protein synthesis (hypertrophy) but also suppress catabolic pathways, represented by FoxOs/MuRF1, leading to protein degradation (atrophy). Under pathological catabolic conditions, such as increased levels of glucocorticoids, insulin resistance, or insulin deficiency, C1-Ten is upregulated and induces IRS-1 degradation through its PTPase activity, subsequently downregulating the anabolic pathway and leading to atrophy.
DISCUSSION
In this study, we newly identified C1-Ten to be a PTPase of IRS-1, contributing to the degradation of IRS-1 and muscle atrophy under catabolic conditions.
IRS-1 is a key mediator of the anabolic action of insulin/IGF-1 and regulates the balance between the anabolic and catabolic pathways (1–4). Thus, a reduced level of IRS-1 has been regarded as one of the key contributors for the development of catabolic diseases, including diabetes (7, 11). Despite its importance, the IRS-1 degradation mechanism under catabolic status has not been fully understood. The prevalent explanation for IRS-1 degradation is the PI3K- and mTOR-dependent pathway after long-term insulin/IGF-1 stimulation (34, 35), which is one of the explanations for IRS-1 reduction under conditions of hyperinsulinemia (38). However, recent reports question the negative role of IRS-1 serine phosphorylation in insulin signaling because of conflicting data from animal studies and cell-based experiments (41, 42). Moreover, we and others have observed IRS-1 decreases under catabolic states, such as glucocorticoid excess and insulin deficiency, despite downregulation of the PI3K/mTOR pathway (20–22, 37). This suggests the possibility that there is another mechanism for IRS-1 degradation.
In this study, we showed for the first time that a novel IRS-1 degradation mechanism which is dependent on C1-Ten-mediated tyrosine dephosphorylation of IRS-1 exists. We suggest that reduced tyrosine phosphorylation of IRS-1 contributes to IRS-1 degradation via a PI3K- and mTOR-independent pathway. More specifically, we demonstrated that IRS-1 Y612, one of the most important sites for PI3K activity, induced IRS-1 degradation upon dephosphorylation by C1-Ten. However, it remains unclear how Y612 of IRS-1, upon dephosphorylation, targets IRS-1 for degradation. IRS-1 can be phosphorylated on tyrosine in both low-density microsomes (LDM) and cytosol, but C1-Ten is localized mainly in the cytosol, where the proteasome machinery exists and degradation occurs (40, 43, 44). Therefore, we assume that C1-Ten dephosphorylates and degrades cytosolic IRS-1. However, the dynamics of C1-Ten localization must be investigated to clarify this issue. Even if C1-Ten preferentially reduced Y612 phosphorylation of IRS-1 and the IRS-1 Y612F mutant showed similar impacts on insulin signaling as C1-Ten WT did, there remain other possibilities for the reduced IRS-1 stability conferred by C1-Ten. It might be meaningful to determine the changes in interacting partners or other phosphorylation sites of IRS-1 upon C1-Ten overexpression. Another remaining issue is related to our finding that the IRS-1 Y612F mutant itself exerted inhibitory effects on insulin signaling and reduced its stability under the basal state. This finding suggests that IRS-1 Y612 might be the most critical residue contributing to catabolic disease. Even if IRS-1 Y612 accounts for about 60% of PI3K activity (45), it needs to be determined whether this residue plays a key role in the progression of catabolic diseases.
We have uncovered a novel role for C1-Ten as a catabolic modulator of skeletal muscle. Here, we showed that C1-Ten led to the inhibition of an anabolic pathway (i.e., Akt/S6K1) and activation of catabolic pathways (i.e., FoxO and MuRF1) via IRS-1 reduction. This seemed to cause a marked reduction in the diameter of myotubes and, to a lesser extent, the CSA of adult myofibers. C1-Ten-induced atrophy of myotubes and inhibition of signaling downstream of IRS-1 in myotubes were fully restored by exogenous insulin/IGF-1 incubation (Fig. 3D). In healthy animals, insulin and IGF-1 are always present, and this might compensate for the C1-Ten overexpression effects in myofibers. Therefore, we do not think that C1-Ten itself causes insulin resistance, despite its ability to reduce IRS-1. This is also the case with IRS-1+/− mice, in which a 50% reduction in the IRS-1 protein occurs. Even in disease states, there is no case of total deficiency of IRS-1. Thus, studies using IRS-1+/− mice demonstrate that the IRS-1 reduction itself is not sufficient to cause the development of overt insulin resistance and diabetes. However, it is enough if there are other minor defects in insulin signaling (46, 47). Thus, upregulation of C1-Ten in combination with the catabolic condition, such as an increase in glucocorticoids or insulin deficiency, exacerbates catabolic states through IRS-1 reduction.
A recent study showed that inhibiting glucocorticoid hydroxysteroid dehydrogenase type 1 (11 β-HSD1), producing active cortisol, ameliorates the metabolic dysfunction in patients with type 2 diabetes who failed metformin monotherapy (48). Our data illustrate the hormonal regulation of C1-Ten expression by catabolic (i.e., glucocorticoid) and anabolic (i.e., insulin) signals. These findings suggest that C1-Ten reflects changes in hormonal homeostasis and acts as an atrophy mediator under catabolic conditions. Considering this result, the development of a specific C1-Ten inhibitor may be effective therapeutically to prevent muscle atrophy in patients with catabolic diseases. Future studies that genetically manipulate C1-Ten expression in animal models will provide insights into the role of C1-Ten not only in skeletal muscle but also in other tissues.
Our study contributes to data already published regarding the physiological function of C1-Ten. Some studies suggest that C1-Ten is a positive regulator of cell migration (49) and proliferation by Akt activation (50), but other studies suggest that C1-Ten is a negative regulator of cell migration and proliferation through inhibition of Akt (24). On the basis of our findings, the true effect of C1-Ten on Akt activation may depend on the contribution of IRS-1 and the basal state of cells, such as whether the IGF autocrine pathway is activated. In fact, we showed that C1-Ten expression in HEK293 cells inhibited insulin signaling via IRS-1 without altering IRS-1 expression under a basal state. However, long-term stimulation of C1-Ten-expressing HEK293 cells with insulin and CHX showed a significant impact on the IRS-1 level. C1-Ten expression in L6 myotubes induced degradation of IRS-1 under the basal condition, possibly because L6 cells produce approximately 10 ng/ml IGF-2 (39). C1-Ten activates Akt upon thrombopoietin stimulation in megakaryocytic cells (50), which do not express IRS-1 (51) but, rather, express IRS-2. However, C1-Ten may not regulate IRS-2, as C1-Ten caused proteasomal degradation of IRS-1 but increased IRS-2 mRNA in myotubes (see Fig. S2B in the supplemental material), and short-term insulin stimulation fully restored C1-Ten-dependent Akt inhibition in myotubes, possibly due to an elevation in IRS-2 (Fig. 3D and E). This is consistent with data for IRS-1−/− mouse embryonic fibroblasts and for skeletal muscle of IRS-1+/− mice (47, 52). A recent study using muscle-specific IRS-1- or IRS-2-knockout mice suggested that IRS-1 has a more important role than IRS-2 in skeletal muscle with respect to muscle mass and Akt signaling (53). However, both IRS-1 and IRS-2 are required for FoxO inhibition. Because IRS genes were deleted in both skeletal muscle and cardiac muscle, these mice showed sudden death at between 2 and 3 weeks of age because of cardiomyopathy. Therefore, the generation of inducible conditional IRS-1-knockout mice will be required for detailed understanding of the IRS-1 function in the progression of muscle atrophy.
Our study has identified a novel type of IRS-1 degradation mechanism that explains the reduction in IRS-1 under catabolic conditions and suggests that C1-Ten is a relevant IRS-1 PTPase and a new therapeutic target for the treatment of muscle atrophy.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the 21st Frontier Functional Proteomics Project (FPR08B1-300) and the Global Research Network Program (KPF-2008-005-C00036) of the South Korean Ministry of Education, Science, and Technology. This study was also supported by the World Class University program through the National Research Foundation of South Korea, funded by the Ministry of Education, Science and Technology (R31-2008-000-10105-0) and the Global Frontier Project (NRF-MI-AXA002-2010-0029764).
Footnotes
Published ahead of print 11 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01447-12.
REFERENCES
- 1. Sun XJ, Rothenberg P, Kahn CR, Backer JM, Araki E, Wilden PA, Cahill DA, Goldstein BJ, White MF. 1991. Structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature 352:73–77 [DOI] [PubMed] [Google Scholar]
- 2. Sun XJ, Wang L-M, Zhang Y, Yenush L, Myers MG, Jr, Glasheen E, Lane WS, Pierce JH, White MF. 1995. Role of IRS-2 in insulin and cytokine signalling. Nature 377:173–177 [DOI] [PubMed] [Google Scholar]
- 3. Myers MG, Jr, White MF. 1996. Insulin signal transduction and the IRS proteins. Annu. Rev. Pharmacol. Toxicol. 36:615–658 [DOI] [PubMed] [Google Scholar]
- 4. Saltiel AR. 2001. New perspectives into the molecular pathogenesis and treatment of type 2 diabetes. Cell 104:517–529 [DOI] [PubMed] [Google Scholar]
- 5. Goodyear LJ, Giorgino F, Sherman LA, Carey J, Smith RJ, Dohm GL. 1995. Insulin receptor phosphorylation, insulin receptor substrate-1 phosphorylation, and phosphatidylinositol 3-kinase activity are decreased in intact skeletal muscle strips from obese subjects. J. Clin. Invest. 95:2195–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bjornholm M, Kawano Y, Lehitihet M, Zierath JR. 1997. Insulin receptor substrate-1 phosphorylation and phosphatidylinositol 3-kinase activity in skeletal muscle from NIDDM subjects after in vivo insulin stimulation. Diabetes 46:524–527 [DOI] [PubMed] [Google Scholar]
- 7. Carvalho E, Jansson P-A, Axelsen M, Eriksson JW, Huang X, Groop L, Rondinone C, Sjöström L, Smith U. 1999. Low cellular IRS 1 gene and protein expression predict insulin resistance and NIDDM. FASEB J. 13:2173–2178 [DOI] [PubMed] [Google Scholar]
- 8. Zierath JR, Krook A, Wallberg-Henriksson H. 2000. Insulin action and insulin resistance in human skeletal muscle. Diabetologia 43:821–835 [DOI] [PubMed] [Google Scholar]
- 9. Leng Y, Karlsson HK, Zierath JR. 2004. Insulin signaling defects in type 2 diabetes. Rev. Endocr. Metab. Disord. 5:111–117 [DOI] [PubMed] [Google Scholar]
- 10. Wang X, Hu Z, Hu J, Du J, Mitch WE. 2006. Insulin resistance accelerates muscle protein degradation: activation of the ubiquitin-proteasome pathway by defects in muscle cell signaling. Endocrinology 147:4160–4168 [DOI] [PubMed] [Google Scholar]
- 11. Bailey JL, Zheng B, Hu Z, Price SR, Mitch WE. 2006. Chronic kidney disease causes defects in signaling through the insulin receptor substrate/phosphatidylinositol 3-kinase/Akt pathway: implications for muscle atrophy. J. Am. Soc. Nephrol. 17:1388–1394 [DOI] [PubMed] [Google Scholar]
- 12. Wing SS, Goldberg AL. 1993. Glucocorticoids activate the ATP-ubiquitin-dependent proteolytic system in skeletal muscle during fasting. Am. J. Physiol. 264:E668–E676 [DOI] [PubMed] [Google Scholar]
- 13. Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. 2001. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 3:1014–1019 [DOI] [PubMed] [Google Scholar]
- 14. Glass DJ. 2003. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat. Cell Biol. 5:87–90 [DOI] [PubMed] [Google Scholar]
- 15. Li Y-P, Chen Y, John J, Moylan J, Jin B, Mann DL, Reid MB. 2005. TNF-α acts via p38 MAPK to stimulate expression of the ubiquitin ligase atrogin1/MAFbx in skeletal muscle. FASEB J. 19:362–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. 2004. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117:399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wray CJ, Mammen JMV, Hershko DD, Hasselgren P-O. 2003. Sepsis upregulates the gene expression of multiple ubiquitin ligases in skeletal muscle. Int. J. Biochem. Cell Biol. 35:698–705 [DOI] [PubMed] [Google Scholar]
- 18. Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. 2004. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell 14:395–403 [DOI] [PubMed] [Google Scholar]
- 19. Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857–868 [DOI] [PubMed] [Google Scholar]
- 20. Zheng B, Ohkawa S, Li H, Roberts-Wilson TK, Price SR. 2010. FOXO3a mediates signaling crosstalk that coordinates ubiquitin and atrogin-1/MAFbx expression during glucocorticoid-induced skeletal muscle atrophy. FASEB J. 24:2660–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Giorgino F, Almahfouz A, Goodyear LJ, Smith RJ. 1993. Glucocorticoid regulation of insulin receptor and substrate IRS-1 tyrosine phosphorylation in rat skeletal muscle in vivo. J. Clin. Invest. 91:2020–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saad MJA, Folli F, Kahn CR. 1993. Modulation of insulin receptor, insulin receptor substrate-1 and phosphatidylinositol 3-kinase in liver and muscle of dexamethasone-treated rats. J. Clin. Invest. 92:2065–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hafizi S, Ibraimi F, Dahlbäck B. 2005. C1-TEN is a negative regulator of the Akt/PKB signal transduction pathway and inhibits cell survival, proliferation, and migration. FASEB J. 19:971–973 [DOI] [PubMed] [Google Scholar]
- 24. Hafizi S, Gustafsson A, Oslakovic C, Idevall-Hagren O, Tengholm A, Sperandio O, Villoutreix BO, Dahlbäck B. 2010. Tensin2 reduces intracellular phosphatidylinositol 3,4,5-trisphosphate levels at the plasma membrane. Biochem. Biophys. Res. Commun. 399:396–401 [DOI] [PubMed] [Google Scholar]
- 25. He T-C, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. U. S. A. 95:2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bandyopadhyay GK, Yu JG, Ofrecio J, Olefsky JM. 2005. Increased p85/55/50 expression and decreased phosphotidylinositol 3-kinase activity in insulin-resistant human skeletal muscle. Diabetes 54:2351–2359 [DOI] [PubMed] [Google Scholar]
- 27. Tsuchida H, Björnholm M, Fernström M, Galuska D, Johansson P, Wallberg-Henriksson H, Zierath J, Lake S, Krook A. 2002. Gene expression of the p85a regulatory subunit of phosphatidylinositol 3-kinase in skeletal muscle from type 2 diabetic subjects. Pflügers Arch. 445:25–31 [DOI] [PubMed] [Google Scholar]
- 28. Florini JR, Magri KA, Ewton DZ, James PL, Grindstaff K, Rotwein PS. 1991. “Spontaneous” differentiation of skeletal myoblasts is dependent upon autocrine secretion of insulin-like growth factor-II. J. Biol. Chem. 266:15917–15923 [PubMed] [Google Scholar]
- 29. Rosen KM, Wentworth BM, Rosenthal N, Villa-Komaroff L. 1993. Specific, temporally regulated expression of the insulin-like growth factor II gene during muscle cell differentiation. Endocrinology 133:474–481 [DOI] [PubMed] [Google Scholar]
- 30. White R, Bierinx A-S, Gnocchi V, Zammit P. 2010. Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev. Biol. 10:21 doi:10.1186/1471-213X-10-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Livingstone DEW, Grassick SL, Currie GL, Walker BR, Andrew R. 2009. Dysregulation of glucocorticoid metabolism in murine obesity: comparable effects of leptin resistance and deficiency. J. Endocrinol. 201:211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tran H, Brunet A, Griffith EC, Greenberg ME. 2003. The many forks in FOXO's road. Sci. Signal. 2003:re5 doi:10.1126/stke.2003.172.re5 [DOI] [PubMed] [Google Scholar]
- 33. Clarke BA, Drujan D, Willis MS, Murphy LO, Corpina RA, Burova E, Rakhilin SV, Stitt TN, Patterson C, Latres E, Glass DJ. 2007. The E3 ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 6:376–385 [DOI] [PubMed] [Google Scholar]
- 34. Pederson TM, Kramer DL, Rondinone CM. 2001. Serine/threonine phosphorylation of IRS-1 triggers its degradation. Diabetes 50:24–31 [DOI] [PubMed] [Google Scholar]
- 35. Sun XJ, Goldberg JL, Qiao LY, Mitchell JJ. 1999. Insulin-induced insulin receptor substrate-1 degradation is mediated by the proteasome degradation pathway. Diabetes 48:1359–1364 [DOI] [PubMed] [Google Scholar]
- 36. Shi J, Luo L, Eash J, Ibebunjo C, Glass DJ. 2011. The SCF-Fbxo40 complex induces IRS1 ubiquitination in skeletal muscle, limiting IGF1 signaling. Dev. Cell 21:835–847 [DOI] [PubMed] [Google Scholar]
- 37. Kuo T, Lew MJ, Mayba O, Harris CA, Speed TP, Wang J-C. 2012. Genome-wide analysis of glucocorticoid receptor-binding sites in myotubes identifies gene networks modulating insulin signaling. Proc. Natl. Acad. Sci. U. S. A. 109:11160–11165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Orland MJ, Permutt MA. 1987. Quantitative analysis of pancreatic proinsulin mRNA in genetically diabetic (db/db) mice. Diabetes 36:341–347 [DOI] [PubMed] [Google Scholar]
- 39. Giorgino F, Smith RJ. 1995. Dexamethasone enhances insulin-like growth factor-I effects on skeletal muscle cell proliferation. Role of specific intracellular signaling pathways. J. Clin. Invest. 96:1473–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yam JWP, Ko FCF, Chan C-Y, Jin D-Y, Ng IO-L. 2006. Interaction of deleted in liver cancer 1 with tensin2 in caveolae and implications in tumor suppression. Cancer Res. 66:8367–8372 [DOI] [PubMed] [Google Scholar]
- 41. Lee DH, Shi J, Jeoung NH, Kim MS, Zabolotny JM, Lee SW, White MF, Wei L, Kim Y-B. 2009. Targeted disruption of ROCK1 causes insulin resistance in vivo. J. Biol. Chem. 284:11776–11780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Copps KD, Hancer NJ, Opare-Ado L, Qiu W, Walsh C, White MF. 2010. Irs1 serine 307 promotes insulin sensitivity in mice. Cell Metab. 11:84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hiratani K, Haruta T, Tani A, Kawahara J, Usui I, Kobayashi M. 2005. Roles of mTOR and JNK in serine phosphorylation, translocation, and degradation of IRS-1. Biochem. Biophys. Res. Commun. 335:836–842 [DOI] [PubMed] [Google Scholar]
- 44. Inoue G, Cheatham B, Emkey R, Kahn CR. 1998. Dynamics of insulin signaling in 3T3-L1 adipocytes. J. Biol. Chem. 273:11548–11555 [DOI] [PubMed] [Google Scholar]
- 45. Esposito DL, Li Y, Cama A, Quon MJ. 2001. Tyr612 and Tyr632 in human insulin receptor substrate-1 are important for full activation of insulin-stimulated phosphatidylinositol 3-kinase activity and translocation of GLUT4 in adipose cells. Endocrinology 142:2833–2840 [DOI] [PubMed] [Google Scholar]
- 46. Brüning JC, Winnay J, Bonner-Weir S, Taylor SI, Accili D, Kahn CR. 1997. Development of a novel polygenic model of NIDDM in mice heterozygous for IR and IRS-1 null alleles. Cell 88:561–572 [DOI] [PubMed] [Google Scholar]
- 47. Shirakami A, Toyonaga T, Tsuruzoe K, Shirotani T, Matsumoto K, Yoshizato K, Kawashima J, Hirashima Y, Miyamura N, Kahn CR, Araki E. 2002. Heterozygous knockout of the IRS-1 gene in mice enhances obesity-linked insulin resistance: a possible model for the development of type 2 diabetes. J. Endocrinol. 174:309–319 [DOI] [PubMed] [Google Scholar]
- 48. Hollis G, Huber R. 2011. 11β-Hydroxysteroid dehydrogenase type 1 inhibition in type 2 diabetes mellitus. Diabetes Obes. Metab. 13:1–6 [DOI] [PubMed] [Google Scholar]
- 49. Chen H, Duncan IC, Bozorgchami H, Lo SH. 2002. Tensin1 and a previously undocumented family member, tensin2, positively regulate cell migration. Proc. Natl. Acad. Sci. U. S. A. 99:733–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jung AS, Kaushansky A, Macbeath G, Kaushansky K. 2011. Tensin2 is a novel mediator of thrombopoietin (TPO)-induced cellular proliferation by promoting Akt signaling. Cell Cycle 10:1838–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Miyakawa Y, Rojnuckarin P, Habib T, Kaushansky K. 2001. Thrombopoietin induces phosphoinositol 3-kinase activation through SHP2, Gab, and insulin receptor substrate proteins in BAF3 cells and primary murine megakaryocytes. J. Biol. Chem. 276:2494–2502 [DOI] [PubMed] [Google Scholar]
- 52. Guo S, Dunn SL, White MF. 2006. The reciprocal stability of FOXO1 and IRS2 creates a regulatory circuit that controls insulin signaling. Mol. Endocrinol. 20:3389–3399 [DOI] [PubMed] [Google Scholar]
- 53. Long YC, Cheng Z, Copps KD, White MF. 2011. Insulin receptor substrates Irs1 and Irs2 coordinate skeletal muscle growth and metabolism via the Akt and AMPK pathways. Mol. Cell. Biol. 31:430–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.