Abstract
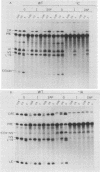
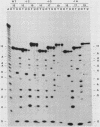
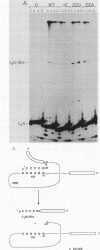
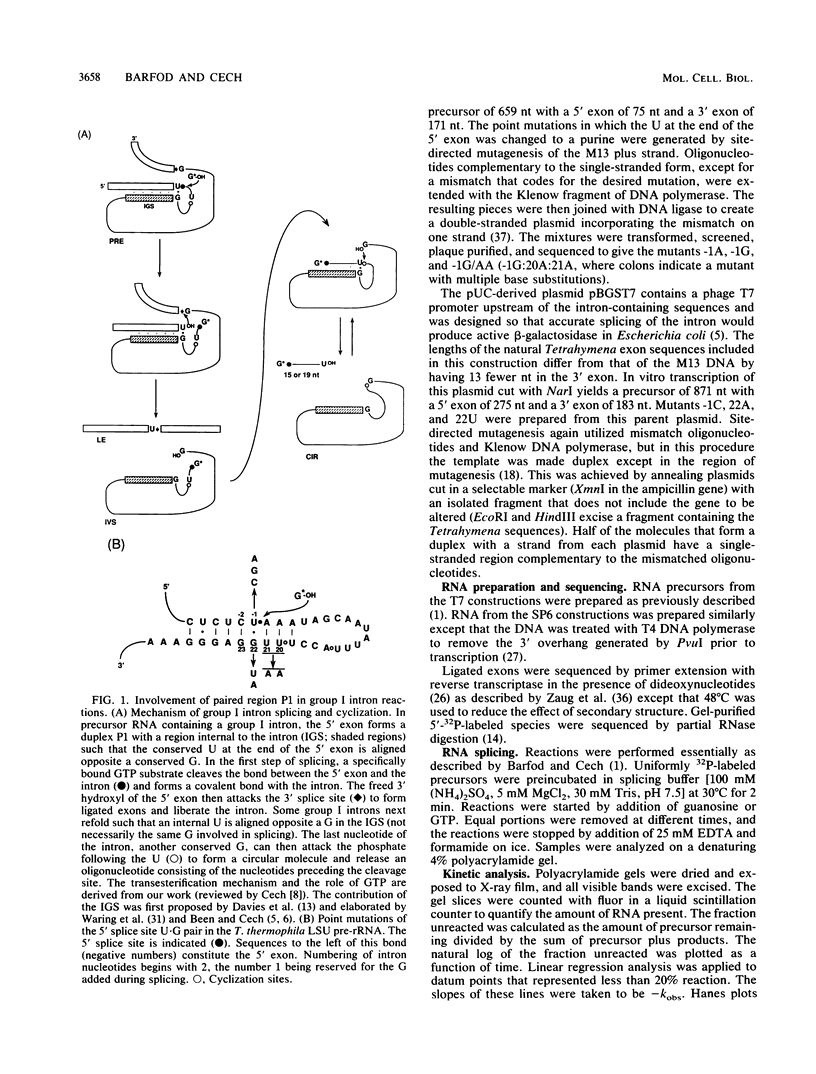
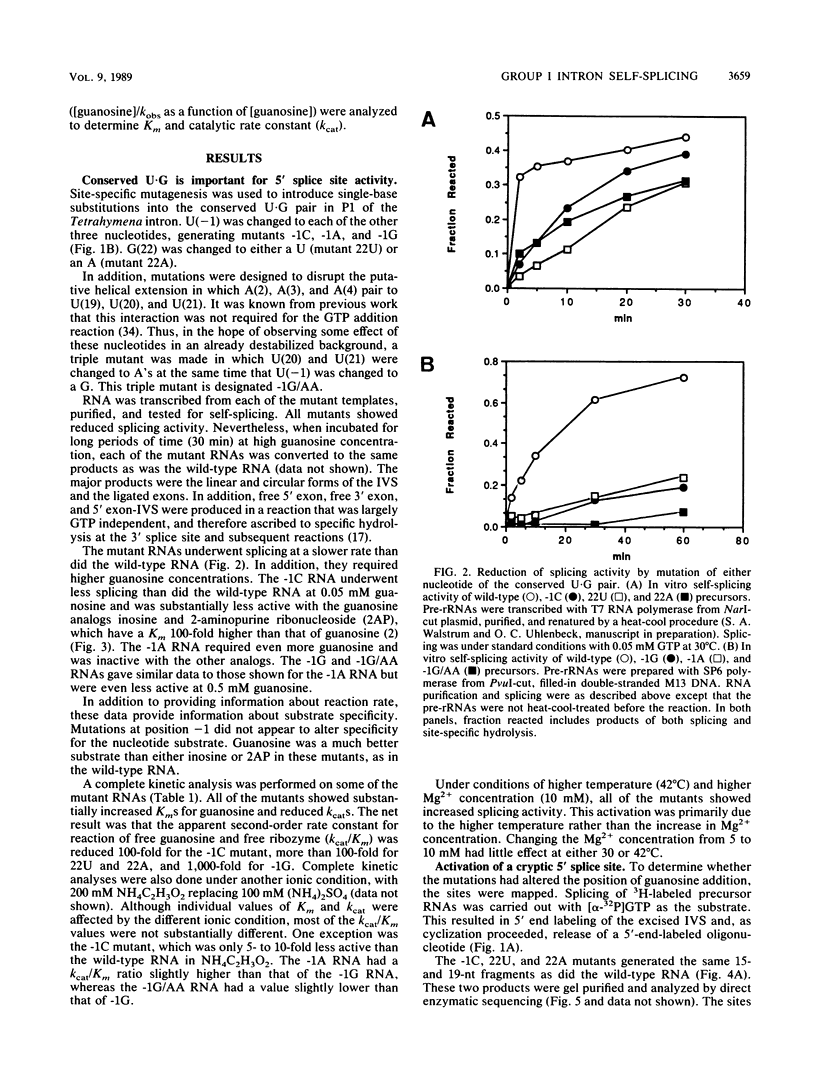
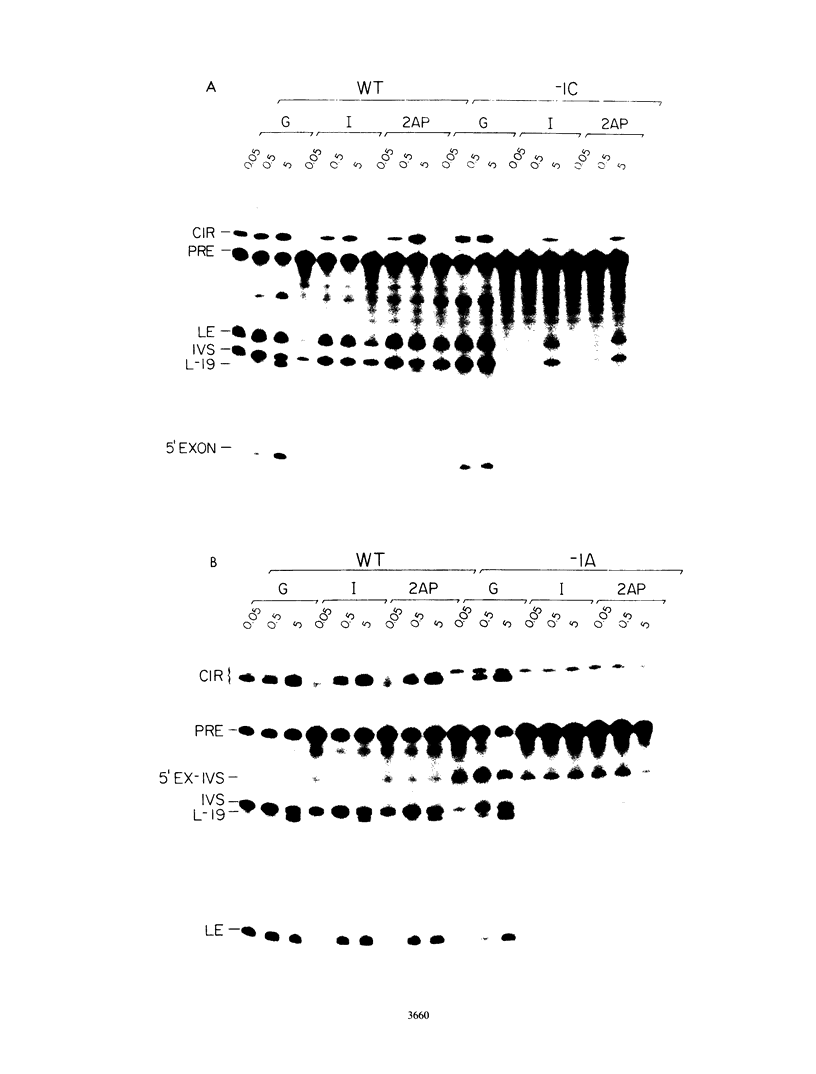
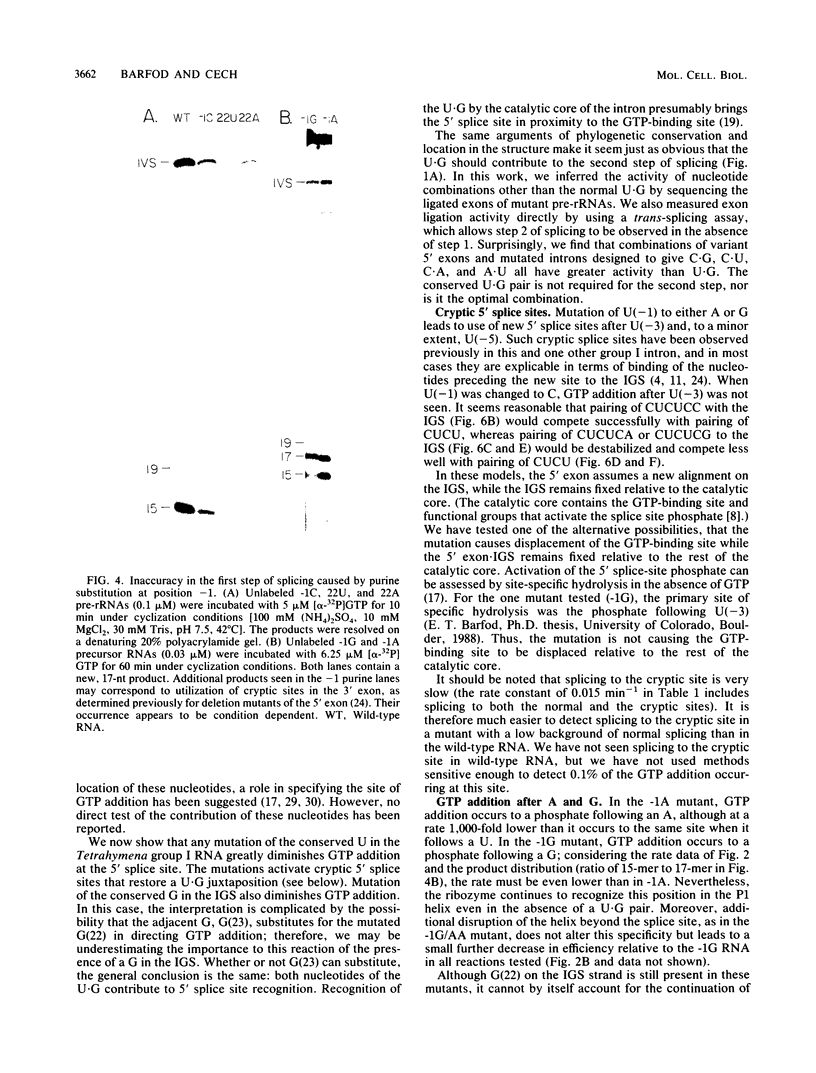
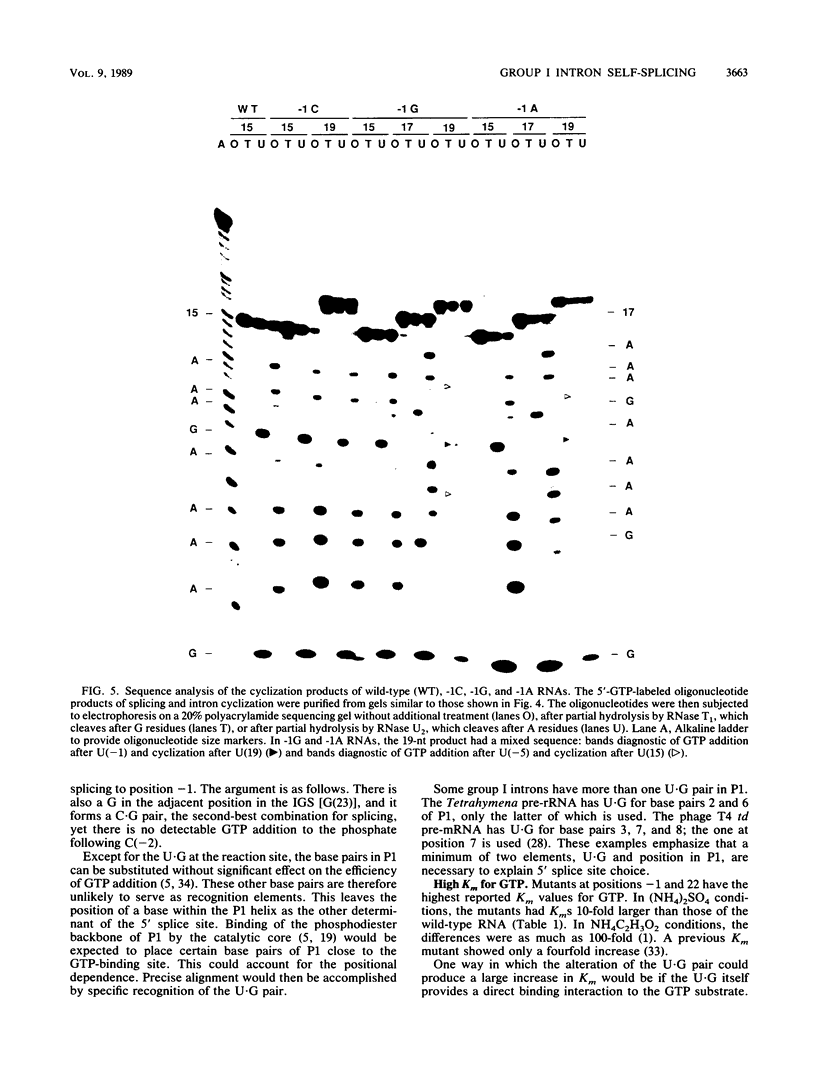
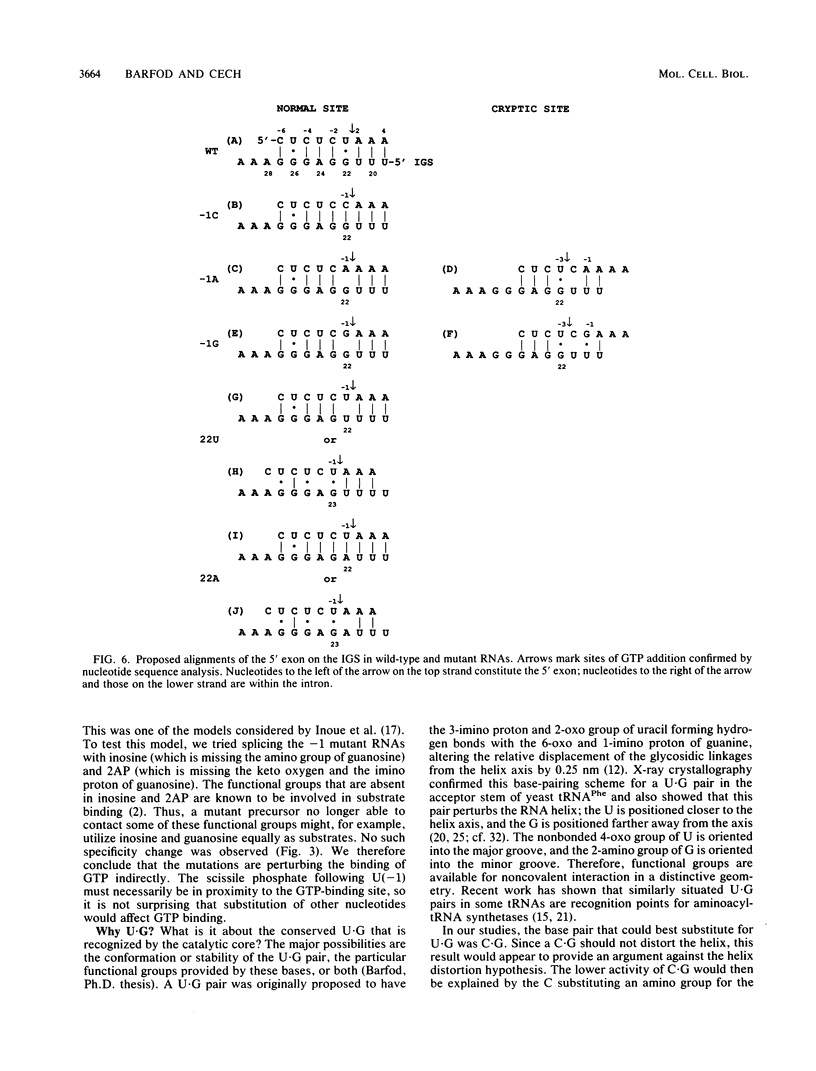
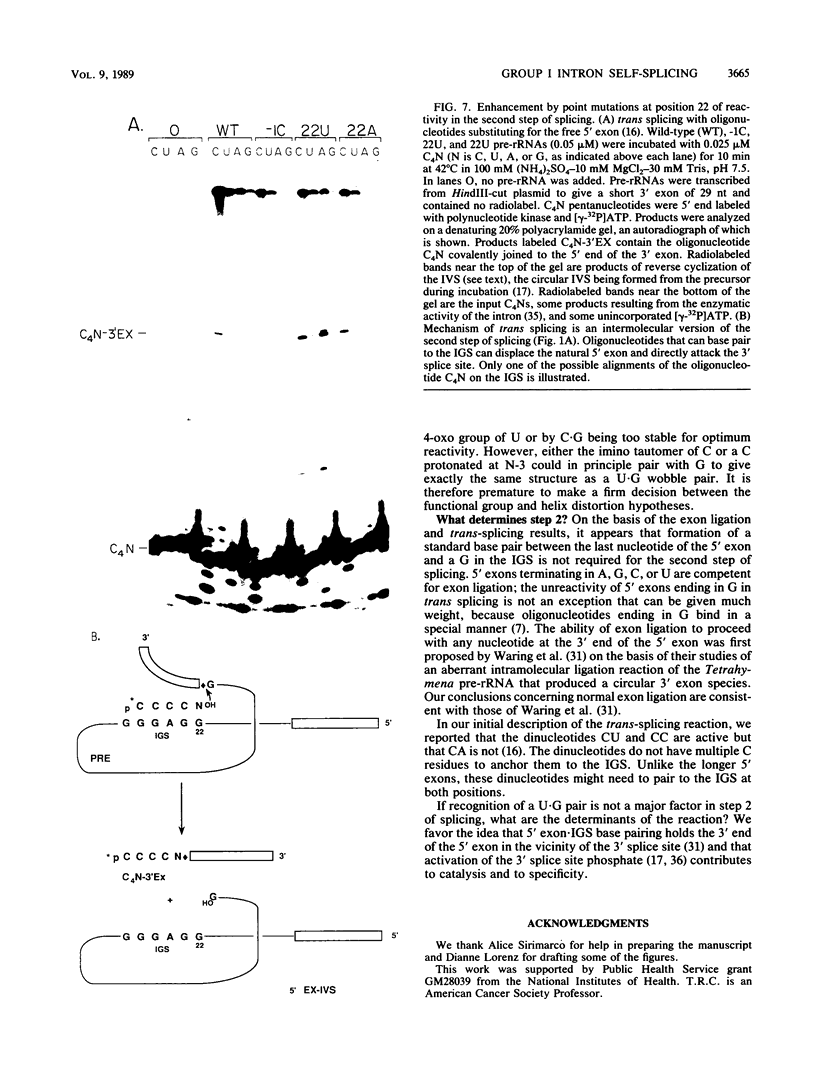
Group I self-splicing introns have a 5' splice site duplex (P1) that contains a single conserved base pair (U.G). The U is the last nucleotide of the 5' exon, and the G is part of the internal guide sequence within the intron. Using site-specific mutagenesis and analysis of the rate and accuracy of splicing of the Tetrahymena thermophila group I intron, we found that both the U and the G of the U.G pair are important for the first step of self-splicing (attack of GTP at the 5' splice site). Mutation of the U to a purine activated cryptic 5' splice sites in which a U.G pair was restored; this result emphasizes the preference for a U.G at the splice site. Nevertheless, some splicing persisted at the normal site after introduction of a purine, suggesting that position within the P1 helix is another determinant of 5' splice site choice. When the U was changed to a C, the accuracy of splicing was not affected, but the Km for GTP was increased by a factor of 15 and the catalytic rate constant was decreased by a factor of 7. Substitution of U.A, U.U, G.G, or A.G for the conserved U.G decreased the rate of splicing by an even greater amount. In contrast, mutation of the conserved G enhanced the second step of splicing, as evidenced by a trans-splicing assay. Furthermore, a free 5' exon ending in A or C instead of the conserved U underwent efficient ligation. Thus, unlike the remainder of the P1 helix, which functions in both the first and second steps of self-splicing, the conserved U.G appears to be important only for the first step.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barfod E. T., Cech T. R. Deletion of nonconserved helices near the 3' end of the rRNA intron of Tetrahymena thermophila alters self-splicing but not core catalytic activity. Genes Dev. 1988 Jun;2(6):652–663. doi: 10.1101/gad.2.6.652. [DOI] [PubMed] [Google Scholar]
- Bass B. L., Cech T. R. Specific interaction between the self-splicing RNA of Tetrahymena and its guanosine substrate: implications for biological catalysis by RNA. 1984 Apr 26-May 2Nature. 308(5962):820–826. doi: 10.1038/308820a0. [DOI] [PubMed] [Google Scholar]
- Been M. D., Barfod E. T., Burke J. M., Price J. V., Tanner N. K., Zaug A. J., Cech T. R. Structures involved in Tetrahymena rRNA self-splicing and RNA enzyme activity. Cold Spring Harb Symp Quant Biol. 1987;52:147–157. doi: 10.1101/sqb.1987.052.01.019. [DOI] [PubMed] [Google Scholar]
- Been M. D., Cech T. R. One binding site determines sequence specificity of Tetrahymena pre-rRNA self-splicing, trans-splicing, and RNA enzyme activity. Cell. 1986 Oct 24;47(2):207–216. doi: 10.1016/0092-8674(86)90443-5. [DOI] [PubMed] [Google Scholar]
- Been M. D., Cech T. R. RNA as an RNA polymerase: net elongation of an RNA primer catalyzed by the Tetrahymena ribozyme. Science. 1988 Mar 18;239(4846):1412–1416. doi: 10.1126/science.2450400. [DOI] [PubMed] [Google Scholar]
- Been M. D., Cech T. R. Selection of circularization sites in a group I IVS RNA requires multiple alignments of an internal template-like sequence. Cell. 1987 Sep 11;50(6):951–961. doi: 10.1016/0092-8674(87)90522-8. [DOI] [PubMed] [Google Scholar]
- Been M. D., Cech T. R. Sites of circularization of the Tetrahymena rRNA IVS are determined by sequence and influenced by position and secondary structure. Nucleic Acids Res. 1985 Dec 9;13(23):8389–8408. doi: 10.1093/nar/13.23.8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R. Conserved sequences and structures of group I introns: building an active site for RNA catalysis--a review. Gene. 1988 Dec 20;73(2):259–271. doi: 10.1016/0378-1119(88)90492-1. [DOI] [PubMed] [Google Scholar]
- Cech T. R. The chemistry of self-splicing RNA and RNA enzymes. Science. 1987 Jun 19;236(4808):1532–1539. doi: 10.1126/science.2438771. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Zaug A. J., Grabowski P. J. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell. 1981 Dec;27(3 Pt 2):487–496. doi: 10.1016/0092-8674(81)90390-1. [DOI] [PubMed] [Google Scholar]
- Chandry P. S., Belfort M. Activation of a cryptic 5' splice site in the upstream exon of the phage T4 td transcript: exon context, missplicing, and mRNA deletion in a fidelity mutant. Genes Dev. 1987 Nov;1(9):1028–1037. doi: 10.1101/gad.1.9.1028. [DOI] [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Davies R. W., Waring R. B., Ray J. A., Brown T. A., Scazzocchio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982 Dec 23;300(5894):719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988 May 12;333(6169):140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- Inoue T., Sullivan F. X., Cech T. R. Intermolecular exon ligation of the rRNA precursor of Tetrahymena: oligonucleotides can function as 5' exons. Cell. 1985 Dec;43(2 Pt 1):431–437. doi: 10.1016/0092-8674(85)90173-4. [DOI] [PubMed] [Google Scholar]
- Inoue T., Sullivan F. X., Cech T. R. New reactions of the ribosomal RNA precursor of Tetrahymena and the mechanism of self-splicing. J Mol Biol. 1986 May 5;189(1):143–165. doi: 10.1016/0022-2836(86)90387-6. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Cech T. R. Three-dimensional model of the active site of the self-splicing rRNA precursor of Tetrahymena. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8788–8792. doi: 10.1073/pnas.84.24.8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J. E., Jack A., Robertus J. D., Brown R. S., Rhodes D., Clark B. F., Klug A. Structure of yeast phenylalanine transfer RNA at 2.5 A resolution. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4414–4418. doi: 10.1073/pnas.72.11.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3' acceptor end. Science. 1988 May 6;240(4853):793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- Michel F., Jacquier A., Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982 Oct;64(10):867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- Price J. V., Cech T. R. Coupling of Tetrahymena ribosomal RNA splicing to beta-galactosidase expression in Escherichia coli. Science. 1985 May 10;228(4700):719–722. doi: 10.1126/science.2986286. [DOI] [PubMed] [Google Scholar]
- Price J. V., Engberg J., Cech T. R. 5' exon requirement for self-splicing of the Tetrahymena thermophila pre-ribosomal RNA and identification of a cryptic 5' splice site in the 3' exon. J Mol Biol. 1987 Jul 5;196(1):49–60. doi: 10.1016/0022-2836(87)90510-9. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenborn E. T., Mierendorf R. C., Jr A novel transcription property of SP6 and T7 RNA polymerases: dependence on template structure. Nucleic Acids Res. 1985 Sep 11;13(17):6223–6236. doi: 10.1093/nar/13.17.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shub D. A., Gott J. M., Xu M. Q., Lang B. F., Michel F., Tomaschewski J., Pedersen-Lane J., Belfort M. Structural conservation among three homologous introns of bacteriophage T4 and the group I introns of eukaryotes. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1151–1155. doi: 10.1073/pnas.85.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan F. X., Cech T. R. Reversibility of cyclization of the Tetrahymena rRNA intervening sequence: implication for the mechanism of splice site choice. Cell. 1985 Sep;42(2):639–648. doi: 10.1016/0092-8674(85)90121-7. [DOI] [PubMed] [Google Scholar]
- Waring R. B., Davies R. W. Assessment of a model for intron RNA secondary structure relevant to RNA self-splicing--a review. Gene. 1984 Jun;28(3):277–291. doi: 10.1016/0378-1119(84)90145-8. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Williamson C. L., Tierney W. M., Kerker B. J., Burke J. M. Site-directed mutagenesis of core sequence elements 9R', 9L, 9R, and 2 in self-splicing Tetrahymena pre-rRNA. J Biol Chem. 1987 Oct 25;262(30):14672–14682. [PubMed] [Google Scholar]
- Zaug A. J., Been M. D., Cech T. R. The Tetrahymena ribozyme acts like an RNA restriction endonuclease. Nature. 1986 Dec 4;324(6096):429–433. doi: 10.1038/324429a0. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Cech T. R. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986 Jan 31;231(4737):470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Kent J. R., Cech T. R. A labile phosphodiester bond at the ligation junction in a circular intervening sequence RNA. Science. 1984 May 11;224(4649):574–578. doi: 10.1126/science.6200938. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]