Abstract
The microtubule (MT) plus-end tracking protein (+TIP) CLASP mediates dynamic cellular behaviors and interacts with numerous cytoplasmic proteins. While the influence of some CLASP interactors on MT behavior is known, a comprehensive survey of the proteins in the CLASP interactome as MT regulators is missing. Ultimately, we are interested in understanding how CLASP collaborates with functionally linked proteins to regulate MT dynamics. Here, we utilize multiparametric analysis of time-lapse MT +TIP imaging data acquired in Drosophila melanogaster S2R+ cells to assess the effects on individual microtubule dynamics for RNA interference-mediated depletion of 48 gene products previously identified to be in vivo genetic CLASP interactors. While our analysis corroborates previously described functions of several known CLASP interactors, its multiparametric resolution reveals more detailed functional profiles (fingerprints) that allow us to precisely classify the roles that CLASP-interacting genes play in MT regulation. Using these data, we identify subnetworks of proteins with novel yet overlapping MT-regulatory roles and also uncover subtle distinctions between the functions of proteins previously thought to act via similar mechanisms.
INTRODUCTION
The orchestration of cytoskeletal dynamics is critical for a broad range of cellular behaviors, including mitosis, polarity, motility, morphogenesis, and cell-cell interaction (1–3). Microtubule (MT) polymer networks participate in numerous signaling pathways, often helping to assemble and/or deliver effector protein complexes and to define the spatial organization of cellular responses. Many classes of cytoskeletal binding proteins regulate the configuration of MT arrays and often interact with other protein networks. However, our understanding of how these extended effector networks function to control cytoskeletal dynamics is still limited. Large-scale screens for MT regulators have primarily relied on endpoint phenotypes that affect mitosis (4–6). The mitotic spindle is a unique apparatus whose gross architecture can be severely disturbed by accumulated effects of altered MT dynamics and thus offers a simple readout for such studies. However, these readouts report screening hits only on the basis of indirect MT phenotypes in a large complex system without pinpointing the actual role that they play in terms of bona fide MT regulation. Direct detection of altered MT dynamics has been much more challenging. For this reason, we adopted a quantitative live-imaging approach that allowed us to identify with single-MT resolution shifts in MT dynamics induced by RNA interference (RNAi)-mediated depletion of putative MT regulators.
CLASP (cytoplasmic linker protein [CLIP]-associated protein) is a well-conserved MT plus-end interacting protein (+TIP), which modulates dynamic instability and facilitates the interaction of MTs with other cellular structures, including the cell cortex (7, 8) and kinetochores (9–11). CLASP functions as an MT-stabilizing factor, promoting MT rescue both in cultured cells and in vivo (12–15). While CLASP has mostly been studied in the context of mitosis (16, 17), it is clear that CLASP plays major roles during interphase and important stages of cellular differentiation. For example, Drosophila melanogaster CLASP (also known as Chromosome bows [Chb], Multiple Asters [MAST], and Orbit) is required in the nervous system during key axon guidance decisions, where highly conserved cues such as Slit and Netrin regulate growth cone navigation (14). In this context, CLASP is necessary for axon guidance functions of the Abelson (Abl) nonreceptor tyrosine kinase (2), a key signaling component downstream of multiple cell surface receptors (18, 19). In addition to actin (20) and MTs (21), several CLASP binding partners have been identified, including the signaling proteins GSK3β, LL5β, and Abl (7, 14, 22) and cytoskeletal regulators such as the MT +TIPs EB1, CLIP, and XMAP215/TOG (Minispindles [Msps] in Drosophila) (21–23). While detailed studies for several of these have shed light on the mechanisms of CLASP protein complex function in MT regulation, they have also illustrated the wide variety of functions that CLASP plays during development and cellular behavior. Further exploration of how the CLASP interactome influences MT dynamics outside the mitotic spindle not only should provide a deeper understanding of CLASP as an integrator of upstream and downstream factors but also may identify novel MT-regulatory molecules.
We recently published a screen for CLASP interactors that combined analysis of ommatidial morphogenesis in the Drosophila retina with proteomics in Drosophila cell culture (23). This screen identified a series of potential partners for CLASP, including several conserved cytoskeletal regulators such as the MT +TIP and polymerase Msps (24, 25). However, for gene products not previously known to interact with MTs, we required an effective means to survey the CLASP interactome for novel genes required to support normal MT behavior in interphase cells. Thus, to define the MT-regulatory subnetwork of the CLASP interactome at sufficient resolution to distinguish different functional subclasses, we adopted an ex vivo live-cell imaging approach followed by multiparametric analysis of MT dynamics. After performing an expanded in vivo screen to define a more complete set of CLASP-interacting genes, we examined the MT-regulatory function of over 50 genes. Changes in MT dynamics were analyzed through systematic quantification of MT dynamics using plusTipTracker, a Matlab-based open-source software (26, 27) that allows fully automated and unbiased detection, classification, and analysis of changes in MT behavior. This permitted us to gain insight into the functional role of CLASP interactors in regulating MT dynamics. Our analysis confirmed functions for several proteins known to be involved in MT regulation and identified novel proteins not previously connected to MT dynamics. In addition, our data revealed subtle phenotypic differences between genes thought to act via similar mechanisms, suggesting that rather than functional redundancy between similar MT-associated proteins, they may function in distinct ways to regulate MT behavior.
MATERIALS AND METHODS
Genetic screen.
A total of 11,646 lines from the Exelixis transposon insertion subset (28, 29) were screened for genes that modify the gain-of-function phenotype that occurs upon GMR-Gal4-directed expression of CLASP using the GMR-Gal4 upstream activation sequence (UAS)-CLASP-GFP line. The GMR-Gal4 driver line expresses the yeast transcription factor GAL4 under the control of glass multiple reporter (GMR) promoter. Several studies have previously described the use of the transposon insertion strains for genome-wide genetic interaction screens (30–32). Transposon lines were crossed to the GMR-Gal4 UAS-CLASP-GFP line and evaluated for eye phenotypes to identify enhancers or suppressors of the UAS-CLASP phenotype. Two additional screens were used to identify CLASP interactors: one was an in vivo genetic screen utilizing the collection of Exelixis deletions (28, 33), and the other was a proteomic screen utilizing tandem affinity purification of the pMK33-CLASP-C-TAP vector expressed in Kc167 cells (23). Candidate CLASP interactors were categorized and selected for further analysis of microtubule dynamics by assigning molecular function on the basis of gene ontology (GO) terms (www.flybase.org; www.ncbi.nlm.nih.gov/gene).
Cell culture and transfection with dsRNA and GFP-tagged EB1 (EB1-GFP).
Drosophila S2R+ cells were grown and maintained in Schneider's Drosophila medium (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen) and penicillin-streptomycin. Cells were then treated with double-stranded RNAi as previously described (6). Briefly, primers for 300- to 600-bp fragments of target sequences were designed using PrimerQuest software (Integrated DNA Technologies, Coralville, IA) (see Table S1 in the supplemental material). Control double-stranded RNA (dsRNA) targeting green fluorescent protein (GFP) was designed. PCR products, flanked at their 5′ and 3′ ends by T7 sequences, were generated using Platinum Pfx DNA polymerase (Invitrogen), and dsRNA was produced by in vitro transcription using MEGAscript T7 kits (Ambion, Austin, TX). Drosophila S2R+ cells were plated, and cultured in six-well tissue culture plates overnight before transfection with 500 ng dsRNA using Effectene reagent according to the manufacturer's protocol (Qiagen, Valencia, CA), and incubated for 3 days. Several primer sets were designed for each candidate, and cells were transfected with dsRNA and tested for knockdown efficiency (see below). dsRNA resulting in 85 to 100% knockdown of candidate proteins or RNA after 3 days was used for all subsequent experiments. For live-cell time-lapse imaging, cells were transfected with 1 μg of pMT EB1-GFP (a generous gift of Steve Rogers, University of North Carolina, Chapel Hill, NC) 24 h before imaging, using the same protocol used to transfect dsRNA.
Western blotting and quantitative real-time PCR.
Western blotting and quantitative real-time PCR were used to evaluate knockdown of target protein and RNA, respectively, in S2R+ cells treated with both control and target-specific dsRNA. To measure protein levels, cells were lysed in lysis buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100) supplemented with cOmplete protease inhibitor (Roche, Indianapolis, IN). Electrophoresis and semidry membrane transfer onto nitrocellulose membrane were performed using a constant protein load. Membranes were blocked for 1 h (Tris-buffered saline–0.05% Tween 20 [TBST], 5% nonfat dry milk) and incubated in primary antibody solution at 4°C overnight (see Table S2A in the supplemental material for antibodies and concentrations). Tubulin (mouse monoclonal antibody DM1A; Sigma-Aldrich, St. Louis, MO) was used as a normalizing protein. Membranes were washed and incubated in the appropriate secondary antibody (horseradish peroxidase-conjugated goat anti-rabbit and goat anti-mouse secondary antibodies; Jackson ImmunoResearch, West Grove, PA) for 1 h at room temperature. Horseradish peroxidase was detected with enhanced chemiluminescence (GE Healthcare, Piscataway, NJ), and images were captured on autoradiographic film (GE Healthcare). The relative intensities of bands representing proteins of interest were quantified using NIH Image J software.
To evaluate changes in RNA levels, total RNA was harvested from S2R+ cells using an RNeasy kit (Qiagen, Valencia, CA) and reverse transcribed (iScript; Bio-Rad Laboratories, Hercules, CA). Quantitative reverse transcription-PCR (qRT-PCR) was carried out on a Life Technologies 7900HT system (Life Technologies, Grand Island, NY) using Power SYBR green PCR master mix (Applied Biosystems, Life Technologies) and primer sets designed using PrimerQuest software (Integrated DNA Technologies, Coralville, IA) (see Table S2B in the supplemental material). β-Actin or Rps38 was used as an internal control to normalize samples. qRT-PCR for each primer set was performed on both control and target dsRNA-treated cells for 27 cycles, and following amplification, melt curve analysis and ethidium bromide agarose gel electrophoresis were performed to evaluate the PCR products. Relative quantification of fold change in mRNA expression was calculated using the 2−ΔΔCT threshold cycle method.
Tubulin and GFP immunofluorescence.
One hour prior to fixation, cells treated with either control or target-specific dsRNA were resuspended and allowed to spread for 1.5 h on 35-mm no. 1.5 coverslip-bottomed MatTek dishes (MatTek Corp., Ashland, MA) that were treated with 0.5 mg/ml concanavalin A (ConA) and allowed to air dry (Sigma-Aldrich, St. Louis, MO). Cells were fixed with ice-cold methanol and rehydrated with phosphate-buffered saline (PBS) with 0.5% Triton X-100 (PBST). Samples were blocked for 1 h in PBS–4% normal goat serum–0.3% Triton X-100 and subsequently transferred into primary antibody solution overnight at 4°C. To investigate tubulin morphology, samples were incubated with a general antibody against tubulin (1:1,000 mouse monoclonal antibody DM1A; Sigma-Aldrich) (see Fig. 2). To evaluate tubulin stability, two samples were prepared per dsRNA and stained with antitubulin antibody (DM1A) and antibodies specific to either the tyrosinated or the detyrosinated form of tubulin (1:1,000-diluted rat monoclonal antitubulin tyrosinated YL1/2 antibody [Abcam]; 1:1,000-diluted rabbit polyclonal antitubulin detyrosinated AB3201 antibody [Millipore]). Cells were washed in PBS and incubated in secondary antibody solution for 1 h at room temperature (1:1,000-diluted anti-mouse Alexa Fluor 488-conjugated antibody and 1:1,000-diluted anti-rabbit Alexa Fluor 568-conjugated antibody; Invitrogen). Images were collected on a Nikon Ti inverted microscope equipped with a ×60 Plan Apo (numerical aperture, 1.4) objective lens. Ten multichannel images per sample were acquired using a Hamamatsu ORCA-ER-cooled charge-coupled device (CCD) camera controlled with NIS Elements software (Nikon Instruments, Inc., Melville, NY). For quantitative determination of microtubule stability, the analysis package of NIS Elements was used to threshold individual images and apply size restrictions to limit analysis only to whole cells. Total fluorescence intensity in both the red and green channels was measured for all individual cells in each image. Tubulin stability was calculated as the average ratio of red/green intensity, which corresponds to the amount of stable/dynamic tubulin.
Fig 2.
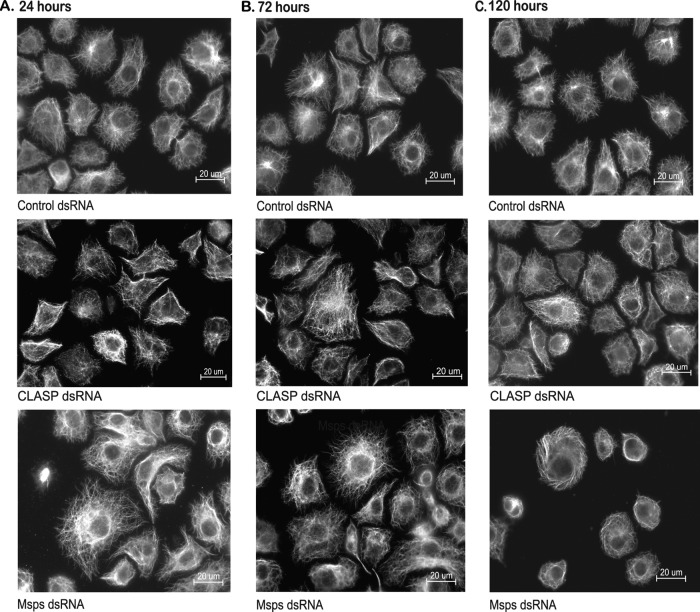
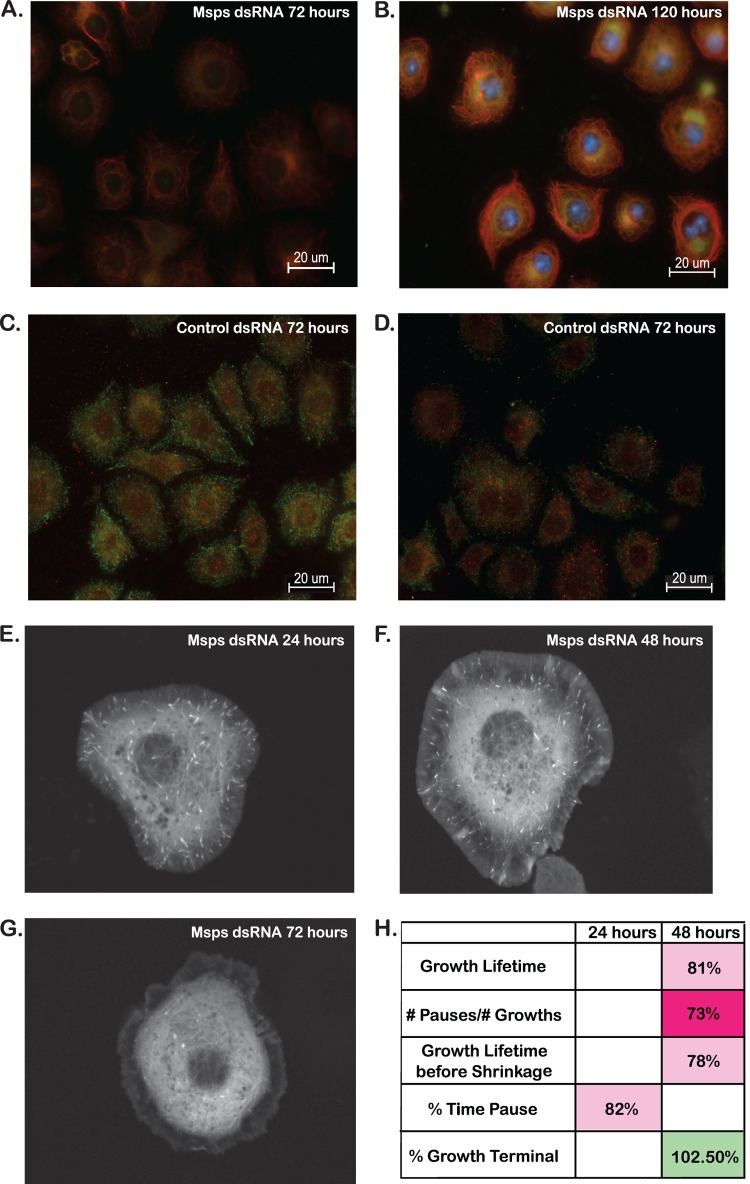
Ex vivo cell-based assay. (A to C) Immunofluorescence for antitubulin in S2R+ cells revealed changes in MT morphology upon prolonged knockdown of the MT +TIP Msps at late time points but not for the MT-associated protein CLASP. (A and B) No significant differences in CLASP or Msps knockdown cells were apparent compared to the control at 24 (A) or 48 h (B). (C) At 5 days after dsRNA treatment, CLASP knockdown cells showed no significant differences in MT morphology compared to the control. In contrast, Msps dsRNA-treated cells were less spread and MTs appeared to wrap around the periphery of the cell compared to the control.
In order to determine whether dsRNA treatment disrupts CLASP localization to the MT plus end, dsRNA-treated samples were transfected with 1 μg of pIZ EB1 CLASP-GFP (a generous gift of Steve Rogers, University of North Carolina, Chapel Hill, NC) 24 h before fixation, using the same protocol used to transfect dsRNA. Samples were fixed and blocked as described above before incubation with primary antibodies (1:1,000-diluted mouse monoclonal antibody DM1A [Sigma-Aldrich]; 1:500-diluted anti-rabbit Living Colors A.v. peptide antibody [Clontech, Mountain View, CA]). Cells were washed in PBS and incubated in secondary antibody solution for 1 h at room temperature (1:1,000-diluted anti-mouse Alexa Fluor 488-conjugated antibody and 1:1,000-diluted anti-rabbit Alexa Fluor 568-conjugated antibody; Invitrogen). Images were collected with a Yokogawa CSU-X1 spinning-disk confocal microscope with the Spectral Applied Research (Richmond Hill, ON, Canada) Borealis modification on a Nikon (Melville, NY) Ti-E inverted microscope using a ×60 Plan Apo (numerical aperture, 1.4) objective lens. The microscope was equipped with a Prior (Rockland, MA) Proscan II motorized stage. The Nikon Perfect Focus system was engaged to maintain a continuous plane of focus. EB1-GFP fluorescence was excited with 488-nm (for GFP) and 561-nm (for mCherry) 100-mW solid-state lasers from a Spectral Applied Research LMM-5 laser merge module and was selected and controlled with an acousto-optical tunable filter. Emission was collected with a Semrock (Rochester, NY) quad pass (405/491/561/642 nm) dichroic mirror and 525/50 nm (for GFP) and 620/60 nm (for mCherry) Chroma (Bellows Falls, VT) emission filters. Images were acquired using a Hamamatsu ORCA-ER-cooled CCD camera. Hardware was controlled with MetaMorph (version 7.7.9) software (Molecular Devices, Sunnyvale, CA.).
Live-cell imaging of MT dynamics.
For live-cell imaging, cells were again plated on coverslip-bottomed MatTek dishes (MatTek Corporation, Ashland, MA) coated with 0.5 mg/ml concanavalin A and allowed to spread for 1 h prior to imaging. Live-cell time-lapse data series were acquired as described above (for CLASP-GFP imaging) using a 488-nm laser to excite EB1-GFP. Time-lapse image series were acquired for a period of 1 min at a frame capture rate of every 750 ms using a 400-ms exposure. Three independent experiments were performed on different days for each treatment, and 6 to 8 cells per experiment were imaged for subsequent analysis.
EB1 detection and tracking.
All image analysis concerning the detection of individual EB1-GFP-positive comets, comet tracking, identification of growth tracks, extraction of MT dynamics, statistical analysis of MT dynamics, and graphical representation of the results was performed using an updated version of plusTipTracker (26, 27). Detection was performed using methods described previously (26, 27). Parameters for detection were set to the default parameters in the plusTipGetTracks graphical user interface of plusTipTracker (26): s1 = 1 (the sigma of the small Gaussian kernel of the difference of Gaussian [DoG] band-pass filter), s2 (the sigma of the large Gaussian kernel of the DoG band-pass filter)= 4, and K (the multiplication factor for the minimum threshold) = 3. Microtubule trajectories were reconstructed using the generic single-particle-tracking framework described previously (34). Optimization of tracking parameters was facilitated via the use of the plusTipParamSweepGUI tool included in the plusTipTracker package (26). Parameter sweeps were performed for several control movies on different days, and optimal tracking parameters based on these results were subsequently employed for all movies in the screening data set. Tracking accuracy was verified by visual inspection of select movies of EB1 comet dynamics with final track overlays. Frame-to-frame subtrack linking parameters were set as follows: minimum growth subtrack length, 3 frames (2.25 s); minimum search radius, 2 pixels (200 nm); maximum search radius, 8 pixels (800 nm). Note that unlike in previous studies (26), no posttracking linearity constraint on frame-to-frame subtrack linkages was employed in order to allow the tracking of both highly curved MTs and MTs undergoing abrupt side displacements due to local intracellular forces (i.e., all frame-to-frame subtrack linkages were maintained regardless of the angle between the two subsequent displacement vectors). Visual inspection of the EB1 movies revealed that these two dynamic behaviors were characteristic of the Drosophila S2R+ cell system, and removal of this constraint was necessary for appropriate tracking of MTs undergoing such events. The constraint was introduced in previous applications primarily to reduce the number of incorrect linkages between parallel arrays of densely packed growing MTs (26). MTs in Drosophila S2R+ cells did not generally exhibit such behavior, and therefore, this potential error was of less concern.
Note that in this tracking framework, MT pausing and shrinking are considered gaps in the MT growth trajectory, as the EB1 comet typically dissociates from the MT tip during these events. Forward gaps (Fgaps) correspond to MT pause events, and backward gaps (Bgaps) correspond to MT shrinkage events. Gap-tracking parameters were set as follows: the maximum gap length was set to 12 frames (9 s), establishing an upper temporal limit on acceptable subtrack pair linking. As described previously (26, 27), pairs of subtracks considered for linkage were likewise limited by certain spatial constraints; to be considered for gap closing, subtrack initiation must fall within a specified geometrical area relative to the site of subtrack termination. Here, these spatial constraints were set as follows: maximum forward angle for gap closing, 35 degrees; maximum backward angle for gap closing, 10 degrees; fluctuation radius, 2 pixels (200 nm); and maximum shrinkage factor, 1.5 times the velocity of MT growth.
Final classification of pause, shrinkage, and undetected growth events.
Accurate measurement of MT pause events via EB1 tracking requires separation of the total observed Fgap population into those resulting from true MT pause events, marked by significant EB1 comet disassembly and subsequent reassembly, and those arising from undetected EB1-decorated MT growth events that may arise if the decorated MT transiently leaves the focal plane. In the past, two methods have been applied in efforts to distinguish between these two populations: (i) unimodal thresholding of the total Fgap velocity distribution (35) and (ii) a local reclassification scheme where the velocity of each Fgap is compared to the average microtubule growth rate 2 to 3 frames before the comet disappearance (26). Here, the former unimodal thresholding method was employed, as there was a concern that the local scheme requires too many a priori assumptions regarding MT behavior. A consistent threshold for the upper bound of the true pause Fgap population was calculated via unimodal thresholding of the pooled control Fgap velocity distribution for a given day. Fgaps with velocities above this threshold were reclassified as high-velocity undetected growths, and the time and spatial information corresponding to these reclassified Fgaps was included as part of the flanking growth subtracks. The same threshold was applied for all dsRNA treatment conditions from the same day. Note that pooling of the control cell population was necessary because of the relatively small sample size of observed Fgaps in individual S2R+ cells (∼30 events per cell). Pause or rescue events of extremely short duration (micropause/rescue) were not separately detectable by plusTipTracker under our image-capture conditions due to the small number of EB1 molecules that assemble at MT plus ends during these transient events. Pooled data comprised ∼200 to 400 Fgap events, distribution numbers large enough to achieve an accurate threshold.
A simple thresholding scheme, set at zero gap velocity, was chosen to distinguish between observed gap events corresponding to shrinkage events and those corresponding to a pause event (i.e., in this study, all Bgap events were designated shrinkage events; see Fig. 4B). This was in contrast to previous studies that implemented a Bgap to pause the reclassification scheme as a means to distinguish backward gap events that likely arise due to measurement noise (i.e., the underlying MT end does not undergo a significant net loss during the gap time frame) from true shrinkage events where the underlying MT undergoes a measurable net polymer loss within the gap time frame. As the number of observed Bgaps before reclassification was only on the order of ∼20 events per S2R+ cell, this previous Bgap-to-pause reclassification scheme, when applied to the S2R+ EB1 comet data sets, resulted in a very high percentage of reclassifications (∼80%) that severely limited the statistical power available for measuring changes in shrinkage displacement, lifetime, and speed. Visual inspection of tracking showed that some Bgap events that could be classified as short shrinkage events via manual tracking were being reclassified as pauses, indicating that the previous Bgap-to-pause reclassification scheme might be overly conservative. Indeed, it was observed in multiple data sets that the combined Fgap and Bgap velocity distribution did not display a maximum value around zero, a fundamental assertion of the previous Bgap-to-pause reclassification scheme. Instead, the maximum of the gap velocity mode that most likely corresponds to the stabilized population of paused MTs appears to be consistently shifted to a small (subpixel) but systematically observed positive value of ∼2 μm/min (0.25 pixels per frame; see Fig. 4B). This shift likely arises due to latency between the reinitiation of net MT growth after a pause event and the accumulation of a detectable EB1-GFP comet. Therefore, simple inversion of the maximum velocity of the Fgap distribution for use as the threshold between Bgap shrinkage and pause events, as was done in previous studies, can result in an overestimation of stable pause events. This potential overestimation was less of a concern in previous systems analyzed (26, 27), given that the backward gap populations of the cellular systems under question were dominated by many shrinkage events with large displacements. In systems with very few shrinkage events with small displacements, the definition of this boundary becomes of more importance. A simple threshold at zero gap velocity was employed here to distinguish between shrinkage and pausing events in order to unambiguously monitor potential changes in these populations upon dsRNA treatment (Fig. 4B). While some of the pausing MT population may indeed still be convolved in the shrinkage population, we reasoned that maintaining constancy in the threshold between Fgap and Bgap when comparing experimental conditions is of more importance than attempting to calculate the absolute value of this boundary under each condition.
Fig 4.
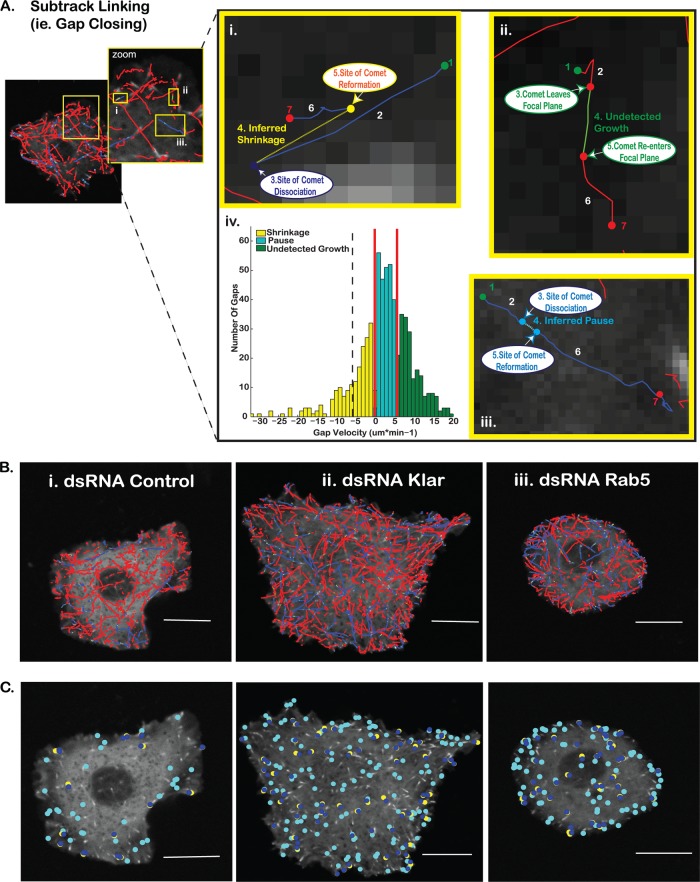
Classification of subtrack linkage as a shrinkage, pause, or out-of-focus growth event. (A) Compound tracks that represent dynamic MT trajectories exhibiting at least one nonterminal pause or shrinkage event (i.e., a pause or shrinkage event followed by robust EB1 comet-decorated regrowth) are generated by linking growth subtracks that meet spatial and temporal criteria consistent with a linear microtubule trajectory. Gaps in detected EB1 growth trajectories correspond to MT shrinkages (yellow) (i), undetected growth events (green) (ii), or MT pauses (cyan) (iii). Along each trajectory, initiation sites for each comet-decorated growth events are detected (i to iii, step 1). EB1 comet detections are linked between subsequent frames, creating the initial MT growth subtrack (i to iii, step 2). Comet disappearance marks the beginning of each gap (i to iii, step 3), and the beginning of the next growth subtrack is marked by the reappearance of EB1-GFP (i to iii, step 5). Gaps between subtracks likely belonging to the same MT are linked (i to iii, step 4) and subsequently classified on the basis of the magnitude and direction of the gap velocity (the distance between the comet disappearance and comet reappearance sites divided by time between these comet detections) relative to the growth direction of the originally detected microtubule growth subtrack. MT growth resumes (i to iii, step 6) until another dynamic event occurs or the MT trajectory is terminally lost from the field of view due to either terminal catastrophe or the permanent loss of the microtubule from the focal plane (i to iii, step 7). (iv) Gap classification was determined by thresholding the gap velocity distributions for all control cells of the same experimental day. Faster, positive gap velocities likely correspond to undetected growth events, while slower gap velocities are likely MT pause events. Gaps with negative velocities are classified as shrinkage events. Red lines indicate the upper and lower thresholds used to delineate the three gap populations in this study. The black dashed line indicates the velocity threshold for the backward gap to pause reclassification, assuming symmetry of the gap velocity distribution around 0. (B) Overlays of the MT tracking corresponding to control (i), Klar dsRNA-treated (ii), and Rab5 dsRNA-treated (iii) cells. Blue solid lines, linked growth subtracks (compound MT tracks); red solid lines, unlinked growth subtracks (trajectories with no dissociation/reassociation of EB1-GFP); dashed yellow lines, inferred shrinkage events (backward gaps); dashed cyan lines, inferred pauses (low-velocity forward gaps); green solid lines, higher-velocity forward gaps (reclassified as out-of-focus EB1-decorated MT growth events.) (C) Individual pause and shrinkage events in control (i), Klar dsRNA-treated (ii), and Rab5 dsRNA-treated (iii) cells. Cyan, pause initiation sites; dark blue, shrinkage initiation sites; yellow, sites of shrinkage rescue events. Bars = 10 μm.
Statistical analysis of MT dynamic parameters.
Significant differences in many MT dynamic parameters were observed among control cellular populations imaged on different experimental days, and therefore, pooling of data among multiple experiments was considered unjustified. Instead, we compared control and dsRNA-treated cellular distributions (n = ∼6 to 12 cells per condition) per experimental day. All parameters were calculated per cell sampled. Parameters derived from individual MT subtracks (e.g., subtrack lifetime, displacement, and average frame-to-frame velocity) were calculated as the median subtrack value per cell to minimize the effects of subtrack outliers, as subtrack distribution outliers potentially arise as a result of tracking errors. Growth tracks beginning in the first frame of the movie or ending in the last frame, as well as all flanking Bgaps and Fgaps, were not included in the final statistics. For many of the parameters, the distributions of the per cell parameter values passed tests for normality, making the use of a Student's t test appropriate for the comparison of the cellular distributions. However, as we could not confirm normality for all parameters, all statistics were performed using a one-tailed permutation t test of the means of the per cell values (1,000 repetitions). Note that P values obtained via this method typically corresponded well with those obtained using the Student t test, again confirming the normality of the data. For each dsRNA-treated population, consistent, statistically significant differences compared to the results for same-day controls across the majority of sampling days (>66%) were used as an indication that the parameter under question was likely being modulated due to the reduction of the protein. In total, analysis of 25 to 40 cells per dsRNA treatment across 3 to 5 sampling days was performed. A P value of 0.05 was used as a threshold for significance. Hits were defined in this manner, as the level of protein silencing for a given sample of cells may potentially be quite variable.
RESULTS
Selection of candidates.
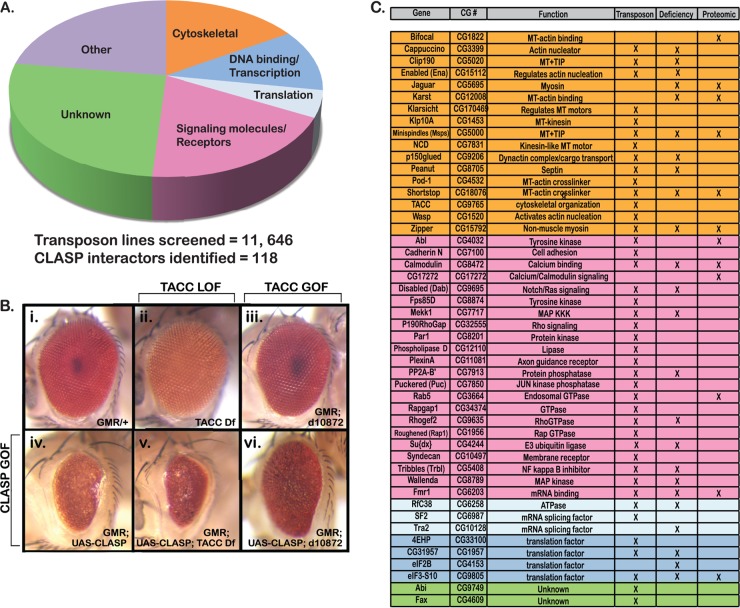
Forward genetic screening approaches in Drosophila using loss-of-function mutants have identified key components for a range of developmental processes (29, 30, 32, 36). In particular, screens for modifiers of sensitized mutant backgrounds offer a powerful strategy to define networks of functional interactions (37, 38). Using a CLASP overexpression phenotype in the Drosophila retina to produce a sensitized genetic background, we recently identified a novel set of CLASP-interacting genes (27). Further analysis of the CLASP-interacting MT +TIP Msps suggested that the approach was suitable to identify relevant functional partners for CLASP during nervous system development (23). To define the CLASP interactome more completely, we screened over 11,000 single-transposon-insertion strains in an isogenic background from the Exelixis collection covering nearly half of the Drosophila genome (30, 31). As previously described (23), CLASP overexpression was driven in the eye and transposon lines were crossed with GMR-GAL4 UAS-CLASP to identify enhancers and suppressors of the CLASP overexpression phenotype (39, 40). From the transposon strains screened, 118 genetic interactors of CLASP were identified, including 26 genes that we previously reported (23), thus significantly increasing the breadth of the CLASP interactome (Fig. 1A).
Fig 1.
Identification of CLASP interactors. The CLASP interactome was defined through in vivo genetic modifier screens using both the deletion and transposon insertion subsets of the Exelixis collection. (A) A total of 11,646 individual transposon insertions were screened to identify genes that modified the fully penetrant dominant rough eye phenotype observed upon CLASP misexpression. Overall, 118 CLASP interactors were identified. Interactors could be grouped into functional categories on the basis of gene ontology: cytoskeletal regulators, DNA binding and transcriptional regulators, translational regulators, signaling molecules and receptors, unknown function, and others. Other functions included ion transport, metabolism, and general protein binding. (B) TACC loss-of-function (LOF) (ii; TACC deficiency [Df]) and gain-of-function (GOF) (iii; d10872) mutants were used to evaluate genetic interactions with CLASP in the Drosophila eye. Loss of TACC expression enhanced the CLASP overexpression phenotype (iv and v), while overexpression of TACC suppressed the phenotype (iv and vi). (C) Forty-eight interactors were selected for further analysis in S2R+ cells on the basis of gene ontogeny and/or reported physical/functional interactions with cytoskeletal proteins (group colors correspond to the functional categories described for panel A).
Gene ontology (GO) analysis was performed to group the CLASP modifier loci according to molecular function, which included cytoskeletal regulation, signaling molecules/receptors, transcription/DNA binding, and translational regulation (Fig. 1A). Novel interactors identified in our current screen included a mutation in the transforming acidic coiled coil (TACC; Fig. 1B), a conserved protein known to associate with Msps (41–43). While TACC loss of function reproducibly enhanced the CLASP overexpression phenotype in the retina, overexpression of TACC was a potent suppressor (Fig. 1B). This confirmed a strong antagonistic genetic interaction highly reminiscent of the interaction between CLASP and the TACC-associated +TIP Msps (10). Although TACC has been proposed to influence MTs at the centrosome through recruitment of Msps (42), it has not itself been studied extensively as a regulator of interphase MT dynamics, highlighting the value of our current screen. In addition to TACC, multiple novel hits in our in vivo transposon screen were previously connected to some additional components in the CLASP interactome, such as the MT +TIP p150glued, the Abl substrate p190RhoGAP, or the guidance molecules plexin A and syndecan (Fig. 1C), suggesting that the screen recovered relevant functional candidates.
Single-cell analysis of MT morphology.
Our primary genetic screens used retinal development as a crude yet efficient assay for abnormal cellular morphogenesis but provided no direct information on the role(s) that CLASP interactors play in MT regulation. Therefore, we developed an ex vivo secondary screen to evaluate how CLASP interactors influence MT dynamics. Forty-eight candidates were chosen from the transposon screen (Fig. 1C) or a previous deficiency screen (23) on the basis of direct or indirect links to cytoskeletal protein networks and signaling pathways available in the literature. For the majority of these genes, MT dynamics had not been directly tested. Candidates were distributed into functional categories in proportions similar to those for the larger group of interactors (Fig. 1C). Within these broad functional categories, several candidates, including MT +TIPs Msps, Clip190, p150glued, and CLASP itself, are known MT regulators and thus served as positive controls.
For an ex vivo screening platform, we utilized a Drosophila S2R+ cell model system that provides the advantage of rapid and efficient disruption of protein function through RNAi to manipulate protein levels. Using primers specific to 300- to 600-bp regions of candidate genes of interest, dsRNA was generated through in vitro transcription (see Table S1 in the supplemental material) (6, 44). Following dsRNA treatment, Western blotting and/or PCR was used to evaluate the extent of knockdown (see Table S2 in the supplemental material for antibodies and primers). Within 72 h, expression levels were reduced 85% or more for all candidates (data not shown). To achieve maximal knockdown and dramatic phenotypes, S2R+ cells are typically evaluated 5 or more days after dsRNA treatment (5, 25, 45). However, such extended treatment raises the possibility of epiphenomena or compensation secondary to the primary impact of reducing target gene function. After treatment for 24 and 72 h, tubulin immunofluorescence showed that for the majority of dsRNAs tested, interphase MT morphology was indistinguishable compared to that for control cells (e.g., CLASP dsRNA and Msps dsRNA) (Fig. 2A and B). Although knockdown of a few candidates, including Msps (Fig. 2A to C, bottom row) and Klarsicht (Klar) (data not shown), resulted in detectable disruption of MT morphology, the fact that efficient knockdown of the well-established MT regulator CLASP did not show visible effects even after 120 h (Fig. 2C, top, middle row) made it evident that endpoint morphology was not a sufficiently sensitive assay. Therefore, it was essential to establish a direct method that would detect the effects of candidate gene knockdown at stages when no gross alteration in MT organization is observed.
In interphase cells, MTs exist as a dense network of filaments from which individual MTs can be resolved only in the cell periphery, making tracking of MT dynamics extremely challenging. Assuaging this challenge, robust observation of MT growth is enabled by fluorescently tagged MT +TIPs, such as EB1, which localize specifically to all assembling MT plus ends throughout the cell (46). Previous studies established that the dynamics of MTs observed in cells expressing low levels of GFP-tagged EB1 were comparable to the dynamics observed in cells injected with small amounts of fluorescently labeled tubulin (47–49). Therefore, the use of GFP-tagged EB1 as a marker for MT growth rates does not significantly alter the native MT behavior, even though EB1 participates in numerous protein complexes that localize to the MT plus end. EB1 is a particularly appropriate +TIP for observing MT dynamics, because it is one of the first proteins to bind to growing MTs and completely dissociates when the MT pauses or shrinks (50–53). Thus, by tracking the appearance and disappearance of EB1-GFP particles, it is possible to measure not only MT growth rates but also the duration of growth phases. To visualize the EB1-GFP association with growing MTs throughout the cell, S2R+ cells were plated on ConA to promote a high degree of adhesion and spreading (Fig. 3A and B) before live-cell time-lapse image series were captured. As overexpression of fluorescently tagged +TIPs may impact MT dynamics (54), cells were selected for imaging on the basis of a low and consistent median gray-scale value for each experimental day. For each dsRNA, 18 to 25 time-lapse movies were acquired at a 1.2-Hz frame rate using a spinning-disk confocal microscope (see Materials and Methods).
Fig 3.
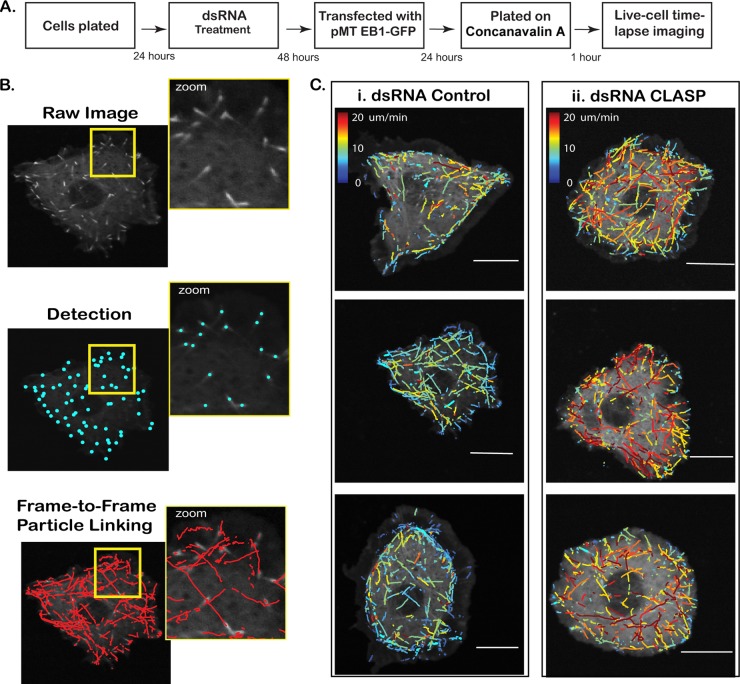
Analysis using plusTipTracker. (A) Work flow of ex vivo assay evaluating the effect of candidate CLASP-interacting proteins on MT dynamics. (B) (i) Cells expressing EB1-GFP selectively mark the plus end of growing MT forming an EB1 comet. (ii) Comet detection identifies EB1-GFP-positive MT ends. (iii) MT growth subtracks are reconstructed using a single-particle-tracking algorithm, followed by linkage of detected comets with a high probability for correspondence between consecutive frames. Velocity, lifetime, and displacement of each MT growth subtrack are extracted. (C) Growth subtracks color coded by average frame-to-frame velocity for a small population of control (i) and CLASP dsRNA-treated (ii) cells. CLASP knockdown results in higher average growth subtrack velocities compared to that for the control. Bars = 10 μm.
Recently, it was shown that automated tracking of EB1-GFP trajectories allows not only the direct measurement of growth rate and duration but also the indirect inference of the frequency, duration, and speed of pause and shrinkage events where the EB1 signal dissociates (26, 27). This approach, implemented in the open-source software plusTipTracker, combines robust single-particle tracking of fluorescent EB1 tags with a model that connects MT growth trajectories that have a high probability of belonging to the same MT but that were interrupted by a rescued pause or shrinkage event (26, 27). In this manner, the algorithm allows indirect inference of the frequency, duration, and speed of pause and shrinkage events at sites where the EB1 signal dissociates and subsequently reassociates (see Materials and Methods) (26, 27). This algorithmic framework provided an amenable platform to perform a robust and methodical exploration of how those proteins associated with the CLASP interactome may affect multiple facets of microtubule dynamics. Importantly, by design of the algorithm, neither terminal shrinkage events nor pauses during shrinkage events are detected. Unlike analysis of MT dynamics by tracking labeled MTs, where shifts in the frequency of rescue events are detected as changes in the duration of shrinkage events, plusTipTracker detects such shifts primarily as changes in the ratio between the occurrence of compound trajectories (linking several growth tracks interrupted by more frequent pause or shrinkage events) and single growth tracks. Thus, while our application of +TIP tracking to a screen for CLASP-interacting MT regulators offers great power in terms of automation and completeness in measuring the dynamics of MT populations in a growth state, it cannot deliver statistics on the dynamics of shrinkage and pausing on an absolute scale. Instead, our screen relied on the assumption that dsRNA treatments that affect MT shrinking and pausing cause shifts in the relative distribution of parameters associated with the probabilistic connecting of tracked growth events into compound trajectories.
Applied to S2R+ cells expressing small amounts of EB1-GFP, plusTipTracker detects and tracks every growing MT marked by an EB1 comet (Fig. 3B; see Movie S1 in the supplemental material). This permits direct comparison of growth speed (Fig. 3C) as well as other parameters derived from growth tracks (Table 1). Knockdown of CLASP, for example, resulted in a significant increase in growth speed (Fig. 3C; see Movie S2 in the supplemental material), supporting a role for CLASP in stabilizing the MT plus end and validating the use of plusTipTracker as an efficient means of extrapolating and quantifying variations in MT dynamic instability.
Table 1.
Parameters analyzed with plusTipTrackera
| Tracking parameter | Description |
|---|---|
| No. of growths | No. of growth subtracks (omitting those that begin in the first frame and end in the last frame) |
| Growth speed | Median speed (μm/min−1) of growth subtracks |
| Growth lifetime | Median lifetime (seconds) of growth tracks |
| Growth length | Median displacement (μm) of growth tracks |
| No. of pauses/no. of growths | Percent growth events ending in pause (after reclassification) |
| Pause lifetime | Median lifetime (seconds) of pauses |
| Growth velocity before pause | Mean velocity (μm/min−1) of all growth subtracks preceding a pause (after reclassification) |
| Growth lifetime before pause | Mean lifetime (seconds) of all growth subtracks preceding a pause (after reclassification) |
| No. of shrinkages/no. of growths | Percent growth subtracks ending in a shrinkage event |
| Shrinkage speed | Median speed (μm/min−1) of shrinkages |
| Shrinkage lifetime | Median lifetime (seconds) of shrinkages |
| Shrinkage length | Median displacement (μm) of shrinkages |
| Growth velocity before shrinkage | Mean velocity (μm/min−1) of all growth subtracks preceding a shrinkage event |
| Growth lifetime before shrinkage | Mean lifetime (seconds) of all growth subtracks preceding a shrinkage event |
| % time in pause | Sum of all pause lifetimes (after reclassification) divided by the sum of the total time spent in all states (growth, pause, shrinkage) |
| % time in shrinkage | Sum of all shrinkage lifetimes (after reclassification) divided by the sum of the total time spent in all states (growth, pause, shrinkage) |
| No. of compound tracks/no. of single tracks | No. of tracks with gaps over the no. of tracks without gaps |
| % growth terminal | Percent growth events not linked to gaps |
| Median nearest neighbor | Distance between a comet and its nearest neighbor (μm) |
Nineteen individual parameters were analyzed per dsRNA-treated cell, and mean values were compared to the values for control dsRNA-treated cells per experimental day. Fgaps (forward gaps) correspond to MT pause events, and Bgaps (backward gaps) correspond to MT shrinkage events.
Links between growth tracks are associated with one of three types of events: (i) rescued shrinkage events (Fig. 4Ai), (ii) undetected growth phases (Fig. 4Aii), and (iii) pauses during growth phases (Fig. 4Aiii). Undetected growth phases correspond to failure in comet detection, either because the comet temporarily moves out of the focal plane or because the low signal-to-noise-ratio (SNR) conditions bring comet signals close to the noise floor. The distinction of these events is accomplished on the basis of the inferred velocities within the linked segments, measured as the displacement between the site of comet disappearance and the site of comet reappearance divided by the number of frames between these events (Fig. 4Aiv), as previously described (26, 27). Compared to previous publications, we revised the classification scheme to better account for the relatively unstable EB1-GFP comet signal in S2R+ cells (see Materials and Methods). Of note, pause events during growth phases have apparently positive velocities due to the latency in comet detection after growth is reinitiated. In contrast, with the exception of rearward sliding, shrinkage events most frequently correspond to negative inferred gap velocities as the MT plus end undergoes negative net displacement (Fig. 4Aiv, yellow velocity population). Note that compared to previous studies, we revised the classification scheme thresholds slightly to better account for the relatively short shrinkage events observed in S2R+ cells (see Materials and Methods). After classification of these links, we then derived from them statistics of parameters that reflect the frequency and dynamics of pausing and shrinkage events (Table 1; Fig. 4B and C).
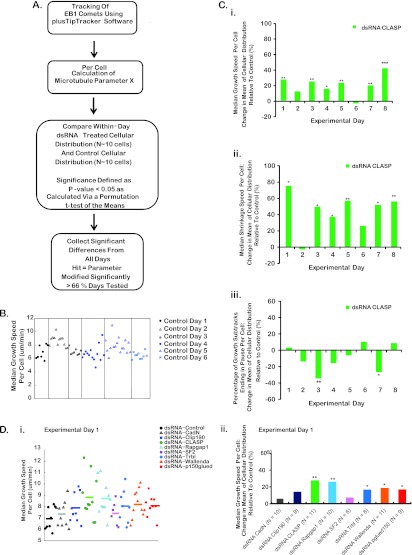
Overall, we calculated 19 individual parameters of MT dynamics as per cell values for each gene (Table 1). Cellular distributions corresponding to each parameter were compared for cells imaged on the same day (Fig. 5A and B). Similar to other ex vivo systems, we observed significant cell-to-cell and day-to-day heterogeneity in both control and experimental dsRNA-treated cells (Fig. 5C). To account for this variability, the median of the distribution for each gene was calculated and the average percent change relative to the median of the same-day control population was recorded (Fig. 5A, C, and D). Statistical significance was determined using a permutation t test (see Materials and Methods). Overall, candidate genes were scored as significant hits if their cellular distribution for a given parameter was statistically different from that for the same-day control population on the majority of experimental days tested (Fig. 5A and D).
Fig 5.
Criteria used to define screen hits and example results for select experimental days, quantified parameters, and dsRNA treatments. (A) Outline of the approach used to identify strong modulators of MT dynamics. (B and C) Scatter plots (B) and bar graphs (C) of median growth speed. Data were plotted for each cell imaged for a sample experimental day. (B) Median growth speed for control dsRNA-treated Drosophila S2R+ cells across six arbitrary experimental days. (C) Percent difference in the mean of the cellular distribution relative to the control for CLASP dsRNA-treated cells. Data are plotted for a subset of measured parameters of MT dynamic instability: median growth speed per cell (i), median shrinkage speed per cell (ii), and percentage of growth subtracks terminating in a pause event per cell (iii). Each bar represents the percent difference in the mean of the cellular distribution relative to the control for a given experimental day. All P values were calculated using a one-tailed permutation t test of the means. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D) (i) Scatter plot of median growth speeds per cell on a given experimental day showing the cell-to-cell variability of control and experimental dsRNA-treated cells. Different colors indicate the different dsRNA treatments. Lines indicate the mean of the cellular distribution for a given treatment. (ii) Percent difference in the mean of the cellular distribution of a dsRNA treatment relative to the control population for those cellular distributions plotted in panel B.
MT regulation by CLASP-interacting proteins.
Encouraged by the agreement between known CLASP functions and the increase in MT growth speed that we observed upon CLASP knockdown, we investigated the effects of CLASP knockdown on other, more indirect parameters of MT dynamics produced by plusTipTracker. CLASP promotes MT pause by binding to and stabilizing the MT plus end (8, 12, 13, 15, 21, 55). Consistent with this, we observed a trend toward decreased pausing upon CLASP knockdown, although the reduction fell below statistical significance (Fig. 5C), possibly due to the limited sensitivity of EB1 tags to indicate micropauses. CLASP knockdown significantly increased shrinkage speed. Additionally, we observed an increased length of shrinkage events and the percentage of time that compound trajectories spent in shrinkage (Fig. 6; see Movie S2 in the supplemental material), supporting a role for CLASP in promoting MT rescue.
Fig 6.
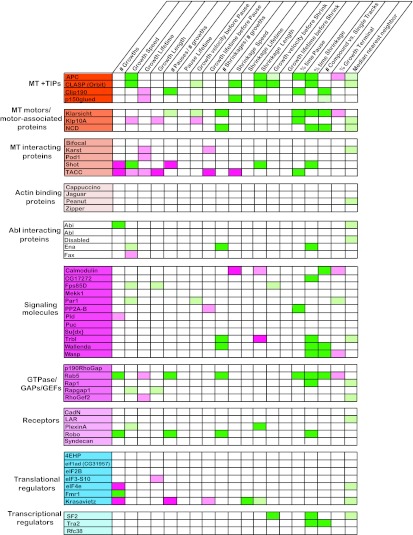
Quantitative analysis of MT dynamics using plusTipTracker. Quantitative analysis was performed on 19 different parameters of MT dynamics in S2R+ cells treated with 48 individual dsRNAs targeting CLASP-interacting proteins. All values were compared to those for control dsRNA-treated cells per experimental day, and the data shown represent the population averages across at least three separate experimental days. Column headings define the individual parameters measured, while row headings identify the dsRNA tested. Dark green, increases compared to control of 25% or greater; dark pink, decreases compared to control of 25% or greater; light green and pink, increases and decreases compared to control of 25% or less, respectively. All data shown represent changes compared to control that are statistically significant. Statistical significance is defined as a P value of <0.05.
CLASP colocalizes with other MT +TIPs at MT plus ends (46), including the potent MT polymerase Msps (24, 56). As described above, our results confirmed that chronic (5-day) knockdown of Msps produced significant morphological changes in MT (Fig. 2C). These changes in MT organization are due to alterations in MT stability, which was evaluated through immunofluorescence using antibodies directed against tyrosinated and detyrosinated tubulin; these alterations reflect the dynamic state of MT (Fig. 7A to D) (57). In contrast to MT morphology, visible changes in MT stability upon Msps knockdown were visible after 72 h (Fig. 7A and C), and by 5 days, in addition to a visible ring of bundled MT in the cell periphery, substantial tyrosinated tubulin was visible in the cell cortex, suggesting region-specific stabilization of MTs upon Msps knockdown (Fig. 7B and D). We were interested in whether these alterations in MT stability influenced EB1-GFP localization to the plus end. While EB1-GFP was visible at the plus end 24 and 48 h after Msps dsRNA treatment (Fig. 7E and F), by 72 h, EB1-GFP comets failed to localize to MT plus ends (Fig. 7G), likely due to increased MT stability and a lack of MT polymerization. As a result, we analyzed Msps dsRNA-treated cells after 24 and 48 h, when protein levels (as detected by Western blotting; data not shown) were decreased approximately 50% and 80%, respectively. By 48 h, Msps knockdown resulted in decreased growth lifetimes (Fig. 7H), consistent with previously published reports of Msps functioning as an MT polymerization factor (23, 24). However, we also observed a decrease in the frequency of pauses, which contrasts with previously published data (see Discussion).
Fig 7.
Analysis of MT dynamics in S2R+ cells treated with Msps dsRNA. (A to D) Antibodies against tyrosinated (green) and detyrosinated (red) tubulin were used to evaluate the dynamic state of MT in fixed S2R+ cells. (A) Cells treated with Msps dsRNA for 3 days showed significant increases in detyrosinated tubulin immunofluorescence (red) compared to that for the control (C). (B) By 5 days after dsRNA treatment, a visible ring of stable MT (marked by red detyrosinated tubulin fluorescence) was visible surrounding the entire cell. (C and D) Cells treated for 3 (C) and 5 (D) days with control dsRNA showed populations of dynamic tyrosinated MT at the cell periphery with more stable detyrosinated MT in the cell cortex. (E) Upon knockdown of the MT +TIP Msps for 24 h, EB1-GFP-positive comets are visible at the end of growing MT and are able to be tracked and analyzed. (F) By 48 h, the EB1-GFP comets remain visible and are able to be tracked, although they appear fainter and thinner than they did at 24 h. (G) By 72 h after dsRNA treatment, EB1-GFP comets are not visible in Msps dsRNA-treated cells. (H) Quantitative analysis was performed on 19 different parameters of MT dynamics in S2R+ cells treated with Msps dsRNA for 24 and 48 h. Parameters shown represent significant differences compared to the control. Statistical significance is defined as a P value of <0.05.
We also characterized the knockdown of adenomatous polyposis coli 1 (APC1) and Shortstop (Shot), both of which localize to the MT plus end. Several alleles of Shot were identified in our CLASP interactome screen, consistent with its known role in motor and central nervous system (CNS) axon guidance (58, 59). While APC1 was not identified in the CLASP interactome screen, it is a +TIP protein that, like CLASP and Shot, is reported to stabilize MTs and prevent catastrophe (60, 61). Thus, APC1 was included as a positive control to validate our analysis. APC1 knockdown showed a phenotype consistent with its reported role, resulting in increased growth speed and the percentage of time spent in shrinkage (Fig. 6). Knockdown of Shot, a known actin-MT cross-linker, altered MT dynamics and resulted in an increased growth speed and a decreased frequency of pause. Similar to APC1, these changes, coupled with an increase in shrinkage length and the percentage of time spent in shrinkage, support a role for Shot as an MT stabilizer. Interestingly, while genes grouped on the basis of GO terms (e.g., +TIPs) often showed similar functional profiles (fingerprints), subtle but mutually exclusive phenotypic variations emerged among members of the groups. For example, knockdown of two other EB1-associated +TIPs, Clip190 and p150glued, caused a significant reduction in growth lifetime (Fig. 6). However, knockdown of Clip190 primarily increased the probability of an MT growth trajectory ending in a pause, while p150glued knockdown increased the shrinkage speed (Fig. 6), suggesting that the precise MT-regulatory mechanisms of these molecules differ.
We also analyzed TACC, a conserved Msps partner involved in mitotic spindle assembly and regulation of translation. The TACC homologs Xenopus Maskin and Caenorhabditis elegans TAC1 localize along spindle MTs, while Drosophila TACC (dTACC) localizes to both astral and spindle MTs (62). TACC function has primarily been implicated in MT regulation during mitosis through centrosomal recruitment of Msps. During mitosis, TACC is thought to promote MT stability and assembly via recruitment of Msps (42). In S2R+ cells, TACC knockdown produced a strong consistent interphase phenotype with a decreased growth speed, length, and lifetime as well as an increased amount of time spent in pause (Fig. 6), similar to the Msps phenotype seen at 48 h.
In addition to detection of novel MT-regulatory functions of CLASP interactors, we hoped to uncover new themes of functional connections among our candidates to gain more insight into possible subnetworks in the interactome. To this end, strikingly similar MT-regulatory fingerprints emerged from genes not previously characterized as regulators of dynamic instability, suggesting convergent pathways in which CLASP may also participate. For example, knockdown of Klar, which interacts with both MTs and dynein, produced an increased frequency of shrinkage events and an increased percentage of time that compound tracks spent in both pause and shrinkage (Fig. 6; see Movie S3 in the supplemental material), suggesting that Klar may stabilize MTs by inhibiting rescue and pause events. Knockdown of the small GTPase Rab5 produced a robust functional fingerprint nearly identical to that of knockdown of Klar, resulting in an increased number of rescued pauses and shrinkages as well as an increased percentage of time spent in shrinkage (Fig. 4C and D and 6). Finally, knockdown of the mitogen-activated protein (MAP) kinase kinase kinase (KKK) Wallenda that emerged as a hit in our CLASP interactome screen showed robust increases in the number of shrinkage events, the percentage of time in shrinkage, and the ratio of compound to single tracks (Fig. 6). Thus, knockdown of these three novel MT regulators produces strikingly similar phenotypes, raising the possibility of a common signaling partner or pathway (see Discussion).
Overall, nearly half of the tested CLASP-interacting proteins displayed significant and reproducible effects on multiple parameters of MT dynamics (Fig. 6). These included MT +TIPs, MT-associated protein, and signaling molecules (e.g., GTPase/GTPase-activating proteins [GAPs]/guanine nucleotide exchange factors [GEFs]). Among other proteins previously known to interact with MTs, our in vivo CLASP interaction screens identified several MT motors or motor-associated proteins that showed significant changes in MT behavior in our cell-based screen. To determine if any of the hits in our cell-based screen act upstream to enable the MT +TIP localization of CLASP, we costained cells for tubulin and CLASP after RNAi knockdown (Fig. 8); however, CLASP localized to plus ends in every case, consistent with the fact that no phenotypic fingerprints overlapped completely with the fingerprint of CLASP itself (Fig. 6). Somewhat surprisingly, we did not observe a phenotype upon knockdown of actin-associated proteins (e.g., Peanut), Abl signaling pathway components, or the majority of CLASP-interacting receptors (e.g., Plexin), raising the question of whether context-specific information (e.g., ligands) might be essential for the action of some candidates. Notable exceptions included the conserved axon guidance receptor Roundabout (Robo) and the Robo-associated Abl substrate Ena, a conserved actin regulator required for Robo receptor signaling (63, 64).
Fig 8.
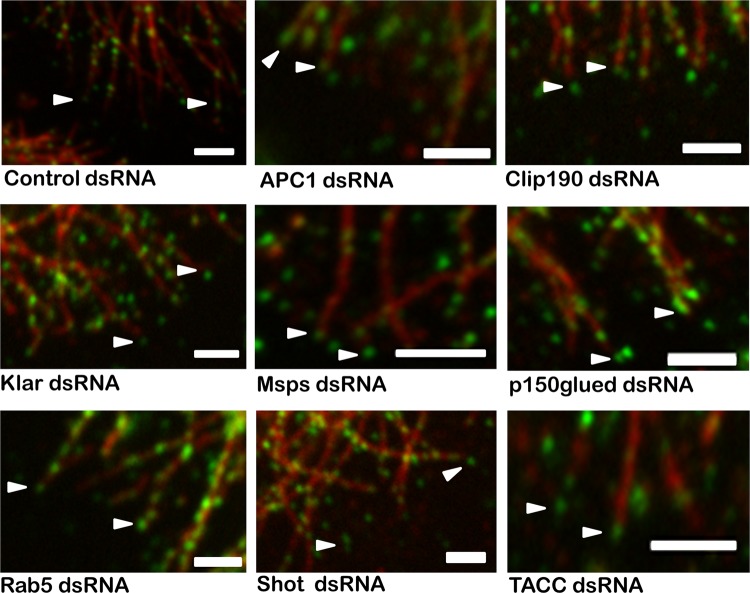
Localization of CLASP-GFP at the MT plus end. Immunofluorescence for antitubulin and CLASP-GFP in S2R+ cells was used to determine if any of the hits in our ex vivo screen influences the MT +TIP localization of CLASP. The 33 candidate interactors that showed significant MT phenotypes were evaluated for plus-end CLASP localization. Representative images of nine candidates, including the +TIPs and those showing the strongest MT phenotypes, show that CLASP localization to the MT plus end was unchanged for all candidates tested. Bars = 2 μm.
DISCUSSION
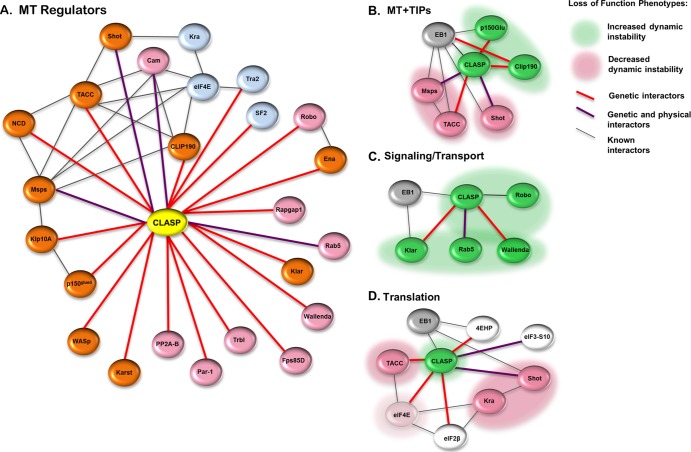
Studies of CLASP and several of its binding partners indicate that CLASP acts via parallel signaling pathways and multiple effectors to regulate cytoskeletal behavior (7, 11, 14, 23, 65). Our extensive genetic modifier screens in the fly retina provided a sensitive means to detect functional interaction with CLASP in vivo (27; this study), revealing an interactome of over 100 genes, but we needed a live-cell time-lapse approach to distinguish interactors that control MT dynamics from other functions downstream of or associated with CLASP. In addition to identifying novel regulators of MT dynamics, this approach offered sufficient resolution to detect significant phenotypic differences even among genes with similar functions. Our analysis of nearly 50 genes revealed MT-regulatory activity for a large subset of CLASP interactors in S2R+ cells (Fig. 9A). On the basis of overlapping functional fingerprints and GO term relationships, several clusters of proteins linked to CLASP and MT plus ends emerged from the CLASP network, including MT +TIPs themselves (CLIP190, p150glued, Shot, Msps, and TACC), proteins linked to MT-dependent transport (Klar, Rab5, and Wallenda), and proteins associated with translational regulation (Shot, Kra, TACC, and eukaryotic initiation factor 4E [eIF4E]) (Fig. 9B to D).
Fig 9.
Loss-of-function phenotypes within functional nodes of the CLASP interactome. (A) Of the 48 CLASP interactors investigated in this study, approximately 1/2 showed significant alterations in MT behavior upon knockdown. In addition to genetic and physical interactions with CLASP, several cytoskeleton-associated proteins are known to interact with each other, in addition to CLASP, supporting a role for CLASP as a coordinating node during MT regulation. Red lines, genetic interaction; purple lines, interaction supported by both genetic and proteomic data; black lines, previously identified interactions. Significant loss-of-function alterations in MT dynamics were apparent in several ontological networks, including MT +TIPs (B), signaling/transport components (C), and translational regulators (D). Green shading, increased dynamic instability upon knockdown; pink shading, decreased dynamic instability upon knockdown.
CLASP-interacting MT +TIPs.
The most immediate partners of CLASP are other +TIPs known to colocalize at MT plus ends and regulate aspects of MT dynamics and/or MT association with other cellular structures. The +TIP complex is highly multifunctional; not only are many +TIPs linked downstream to a diverse array of effector proteins, but also each effector protein often has multiple functions (46). Knockdown of two EB1-associated +TIPs that did emerge as genetic modifiers of CLASP, Clip190 and p150glued, exhibited changes in growth lifetime, consistent with previous data, suggesting that, like CLASP, these proteins suppress MT catastrophe (55, 66). However, the level of quantitative resolution afforded by our assay demonstrated that while p150glued primarily affects the rate of MT disassembly, Clip190 seems to inhibit MT pausing. This suggests that these CLASP interactors act in analogous but mechanistically distinct ways to regulate MTs.
An additional CLASP-interacting +TIP useful for assay validation was Msps, a well-known MT polymerase that promotes MT assembly and elongation during both mitosis and interphase (24, 67, 68). Overall, our data confirm observations made in previous studies of both vertebrate and invertebrate members of the XMAP215/TOG family and support a role for Msps in promoting MT polymerization (67, 68). In addition to enhancing MT assembly, it has also been suggested that XMAP215 family members also act as depolymerization factors in multiple species, including Xenopus spp. (69) and Saccharomyces cerevisiae (20). Previous studies in S2R+ cells put forth the hypothesis that Msps may regulate the transition between growth and shrinkage as an antipause factor, because depletion of Msps dramatically increases MT pausing and bundled MTs (30). Consistent with these results, we found increased relative MT stability in fixed S2R+ cells after 3 to 5 days of knockdown (Fig. 7A and B). However, under more acute Msps knockdown at 48 h, we found decreased MT pausing, along with decreased growth parameters predicted for a key MT polymerase (Fig. 7H). The contrast in pausing behavior at early and late time points raises the intriguing possibility that distinct Msps functions with different concentration thresholds exist. Recent evidence suggests that Msps may regulate several aspects of dynamic instability via interactions with the MT, both at the plus end and along the lattice, and that both interactions are required for normal dynamic instability (25). Thus, it is also possible that these multiple roles of Msps, as a polymerase, depolymerizing factor, and a stabilizing factor facilitating the transition between these phases, are regulated in part by Msps localization along the MT in a region-specific manner within the cell, not unlike the behavior of vertebrate CLASP in nonneuronal cells (e.g., see reference 7).
During mitosis, proper Msps localization and function require recruitment by the conserved MAP TACC (41, 42, 62). At the centrosome, the Msps-TACC complex localizes to both the plus and minus ends of MT to influence stability and assembly (41, 42). In the yeast Schizosaccharomyces pombe, TACC functions as an MT cross-linker and influences MT stability by promoting MT bundling (70). Our data are the first to report a role for TACC in modulating dynamic instability, and similar to the loss of Msps, TACC knockdown in S2R+ cells decreased parameters corresponding to MT growth (speed, lifetime, and length) (Fig. 6 and 9A). In contrast to mitosis, during interphase, TACC is not required for Msps localization to the plus end (24). Together, these observations raise the intriguing possibility that TACC regulation of interphase MT behavior is Msps independent, likely by promoting MT assembly and elongation at the plus end.
CLASP interactors linked to MT-dependent transport and signaling.
Several CLASP-interacting motor proteins or motor-associated proteins also altered MT behavior in S2R+ cells. Interactions between CLASP and dynein are important in MT regulation in both yeast and Drosophila cells, although the functional relationship between these two molecules depends on the mitotic state of the cell (11, 71). Thus, it is possible that the influence of MT motors on dynamic instability relies, at least in part, on additional MT-localized motor-associated proteins. For example, Klarsicht (Klar) interacts with both MTs and dynein and is required for proper nuclear positioning in Drosophila photoreceptor cells in a manner identical to that of p150glued (72). Klar is also required for bidirectional lipid transport along MTs, suggesting that it may interact with both dynein and kinesins (73). Our data now for the first time implicate Klar upstream of MT dynamics and suggest that Klar stabilizes MTs as a negative regulator of rescue and pause. Interestingly, Klar's robust functional fingerprint was nearly identical to that of the small GTPase Rab5, best known for its role in early endosome formation. Rab5 is implicated in chromosome alignment during mitosis, interacts with both dynein and MTs (74), and promotes actin remodeling during receptor tyrosine kinase-mediated circular ruffling, acting as a signaling GTPase (75). The functional profiles of both Rab5 and Klar are consistent with an observed change in the ratio of compound to single tracks (Fig. 6 and 9B), indicating that knockdown of these proteins increases the probability for rescue. Hence, we hypothesize that both endogenous Rab5 and Klar stabilize MTs as inhibitors of rescue and pause events and may function in a shared signaling pathway.
In addition to the overlap in Rab5 and Klar phenotypes, other lines of evidence connect and implicate these CLASP interactors in a MT rescue mechanism. For example, Rab5 regulates the activity of c-Jun N-terminal kinase (JNK) during neuronal migration (76). JNK is involved in MT dynamics during axonal transport via interactions with kinesin-1 to promote MT growth, elongation, and rescue (77). An additional upstream regulator of JNK, Wallenda, regulates the linkage between kinesin and its cargo during axonal transport, likely via JNK activation (78). In S2R+ cells, Wallenda knockdown suggests that endogenous Wallenda also acts to inhibit MT rescue. Therefore, the overlapping fingerprints of MT-regulatory activity observed for Klar, Rab5, and Wallenda may parallel a theme of linkage between JNK, MT motors, and transport. The finding that Klar and Rab5 fingerprints also overlap significantly with the fingerprint of Robo, a receptor clearly linked to CLASP function (58), raises the intriguing possibility that MT rescue is inhibited by basal or constitutive activation of Robo receptors by a low level of Slit ligand.
Translational regulatory interactors in the CLASP network.
One conspicuous set of novel CLASP interactors from our genetic screens was translational regulators (Fig. 1 and 9C). Rapid and local protein synthesis is required for the directional response to axon guidance cues (79–81), and both translation initiation and elongation factors physically bind to the cytoskeleton as an organizational scaffold (82, 83). Thus, we added several candidates to our screen to test the link between translational control and MT regulation. For example, the translation initiation factor eIF4E interacts with TACC/Maskin to regulate translation during mitosis (84) and with the Robo- and Shot-interacting gene Krasavietz (Kra) during axon guidance (58). In Drosophila embryos, Kra, eIF2β, and Shortstop (Shot) form a ternary complex that interacts with actin for proper chemorepulsion at the CNS midline (58, 83). The EB1-associated actin-MT cross-linker Shot (85, 86) was one of our strongest and most consistent hits, displaying a functional profile similar to that of other known MT stabilizers, including CLASP. Kra knockdown produced a significant phenotype in S2R+ cells with a reduced frequency of pause events while at the same time increasing shrinkage lifetime. Thus, Kra appears to stabilize MTs by promoting MT pause at the expense of shrinkage, which closely matches the Shot phenotype (Fig. 6 and 9C). However, knockdown of eIF4E showed a relatively subtle effect in our assay. This suggests that Kra and TACC regulate MT dynamics in S2R+ cells via principally nonprotein synthesis-dependent mechanisms, raising the possibility that the translational effectors linked to CLASP mediate downstream control of other outputs for the MT +TIP complex, such as actin regulation or signaling.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Jacqui Rho and Robert Obar for technical assistance. We are grateful to Jennifer Waters and the Nikon Imaging Center at Harvard Medical School for technical support for microscopy performed in this study. We thank the ICCB-Longwood Screening Facility at Harvard Medical School for assistance with quantitative real-time PCR to evaluate dsRNA efficiency.
Work in the D.V.V. lab was supported by grants from the NINDS (R01 NS035909) and NIMH (SBIR 5R44GM077774). Work in the G.D. lab was supported by a grant from NIGMS (U01 GM67230). J.B.L. was supported by an NCI fellowship (5T2CA009361-30). L.A.L. was supported by an NIH NRSA fellowship (5F32NS063512).
Footnotes
Published ahead of print 4 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01442-12.
REFERENCES
- 1. Heng YW, Koh CG. 2010. Actin cytoskeleton dynamics and the cell division cycle. Int. J. Biochem. Cell Biol. 42:1622–1633 [DOI] [PubMed] [Google Scholar]
- 2. Kodama A, Lechler T, Fuchs E. 2004. Coordinating cytoskeletal tracks to polarize cellular movements. J. Cell Biol. 167:203–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. 2003. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 5:599–609 [DOI] [PubMed] [Google Scholar]
- 4. Goshima G, Vale RD. 2003. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162:1003–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316:417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rogers SL, Rogers GC. 2008. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat. Protoc. 3:606–611 [DOI] [PubMed] [Google Scholar]
- 7. Lansbergen G, Grigoriev I, Mimori-Kiyosue Y, Ohtsuka T, Higa S, Kitajima I, Demmers J, Galjart N, Houtsmuller AB, Grosveld F, Akhmanova A. 2006. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev. Cell 11:21–32 [DOI] [PubMed] [Google Scholar]
- 8. Mimori-Kiyosue Y, Grigoriev I, Lansbergen G, Sasaki H, Matsui C, Severin F, Galjart N, Grosveld F, Vorobjev I, Tsukita S, Akhmanova A. 2005. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168:141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hannak E, Heald R. 2006. Xorbit/CLASP links dynamic microtubules to chromosomes in the Xenopus meiotic spindle. J. Cell Biol. 172:19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maiato H, Sampaio P, Lemos CL, Findlay J, Carmena M, Earnshaw WC, Sunkel CE. 2002. MAST/Orbit has a role in microtubule-kinetochore attachment and is essential for chromosome alignment and maintenance of spindle bipolarity. J. Cell Biol. 157:749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reis R, Feijao T, Gouveia S, Pereira AJ, Matos I, Sampaio P, Maiato H, Sunkel CE. 2009. Dynein and mast/orbit/CLASP have antagonistic roles in regulating kinetochore-microtubule plus-end dynamics. J. Cell Sci. 122:2543–2553 [DOI] [PubMed] [Google Scholar]
- 12. Al-Bassam J, Kim H, Brouhard G, van Oijen A, Harrison SC, Chang F. 2010. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev. Cell 19:245–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bratman SV, Chang F. 2007. Stabilization of overlapping microtubules by fission yeast CLASP. Dev. Cell 13:812–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee H, Engel U, Rusch J, Scherrer S, Sheard K, Van Vactor D. 2004. The microtubule plus end tracking protein Orbit/MAST/CLASP acts downstream of the tyrosine kinase Abl in mediating axon guidance. Neuron 42:913–926 [DOI] [PubMed] [Google Scholar]
- 15. Sousa A, Reis R, Sampaio P, Sunkel CE. 2007. The Drosophila CLASP homologue, Mast/Orbit regulates the dynamic behaviour of interphase microtubules by promoting the pause state. Cell Motil. Cytoskeleton 64:605–620 [DOI] [PubMed] [Google Scholar]
- 16. Inoue YH, do Carmo Avides M, Shiraki M, Deak P, Yamaguchi M, Nishimoto Y, Matsukage A, Glover DM. 2000. Orbit, a novel microtubule-associated protein essential for mitosis in Drosophila melanogaster. J. Cell Biol. 149:153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Inoue YH, Savoian MS, Suzuki T, Mathe E, Yamamoto MT, Glover DM. 2004. Mutations in orbit/mast reveal that the central spindle is comprised of two microtubule populations, those that initiate cleavage and those that propagate furrow ingression. J. Cell Biol. 166:49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hernandez SE, Krishnaswami M, Miller AL, Koleske AJ. 2004. How do Abl family kinases regulate cell shape and movement? Trends Cell Biol. 14:36–44 [DOI] [PubMed] [Google Scholar]
- 19. Moresco EM, Koleske AJ. 2003. Regulation of neuronal morphogenesis and synaptic function by Abl family kinases. Curr. Opin. Neurobiol. 13:535–544 [DOI] [PubMed] [Google Scholar]
- 20. van Breugel M, Drechsel D, Hyman A. 2003. Stu2p, the budding yeast member of the conserved Dis1/XMAP215 family of microtubule-associated proteins is a plus end-binding microtubule destabilizer. J. Cell Biol. 161:359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, Galjart N. 2001. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell 104:923–935 [DOI] [PubMed] [Google Scholar]
- 22. Watanabe T, Noritake J, Kakeno M, Matsui T, Harada T, Wang S, Itoh N, Sato K, Matsuzawa K, Iwamatsu A, Galjart N, Kaibuchi K. 2009. Phosphorylation of CLASP2 by GSK-3beta regulates its interaction with IQGAP1, EB1 and microtubules. J. Cell Sci. 122:2969–2979 [DOI] [PubMed] [Google Scholar]
- 23. Lowery LA, Lee H, Lu C, Murphy R, Obar RA, Zhai B, Schedl M, Van Vactor D, Zhan Y. 2010. Parallel genetic and proteomic screens identify Msps as a CLASP-Abl pathway interactor in Drosophila. Genetics 185:1311–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brittle AL, Ohkura H. 2005. Mini spindles, the XMAP215 homologue, suppresses pausing of interphase microtubules in Drosophila. EMBO J. 24:1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Currie JD, Stewman S, Schimizzi G, Slep KC, Ma A, Rogers SL. 2011. The microtubule lattice and plus-end association of Drosophila Mini spindles is spatially regulated to fine-tune microtubule dynamics. Mol. Biol. Cell 22:4343–4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Applegate KT, Besson S, Matov A, Bagonis MH, Jaqaman K, Danuser G. 2011. plusTipTracker: quantitative image analysis software for the measurement of microtubule dynamics. J. Struct. Biol. 176:168–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matov A, Applegate K, Kumar P, Thoma C, Krek W, Danuser G, Wittmann T. 2010. Analysis of microtubule dynamic instability using a plus-end growth marker. Nat. Methods 7:761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Artavanis-Tsakonas S. 2004. Accessing the Exelixis collection. Nat. Genet. 36:207. [DOI] [PubMed] [Google Scholar]
- 29. Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D, Killpack K, Laufer A, Mazzotta J, Smith RD, Stevens LM, Stuber C, Tan LR, Ventura R, Woo A, Zakrajsek I, Zhao L, Chen F, Swimmer C, Kopczynski C, Duyk G, Winberg ML, Margolis J. 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36:283–287 [DOI] [PubMed] [Google Scholar]
- 30. Chang HC, Dimlich DN, Yokokura T, Mukherjee A, Kankel MW, Sen A, Sridhar V, Fulga TA, Hart AC, Van Vactor D, Artavanis-Tsakonas S. 2008. Modeling spinal muscular atrophy in Drosophila. PLoS One 3:e3209 doi:10.1371/journal.pone.0003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kankel MW, Hurlbut GD, Upadhyay G, Yajnik V, Yedvobnick B, Artavanis-Tsakonas S. 2007. Investigating the genetic circuitry of mastermind in Drosophila, a notch signal effector. Genetics 177:2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shalaby NA, Parks AL, Morreale EJ, Osswalt MC, Pfau KM, Pierce EL, Muskavitch MA. 2009. A screen for modifiers of notch signaling uncovers Amun, a protein with a critical role in sensory organ development. Genetics 182:1061–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36:288–292 [DOI] [PubMed] [Google Scholar]
- 34. Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. 2008. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5:695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rosin PL. 2001. Unimodal thresholding. Pattern Recognition 34:2083–2096 [Google Scholar]
- 36. Valakh V, Naylor SA, Berns DS, DiAntonio A. 2012. A large-scale RNAi screen identifies functional classes of genes shaping synaptic development and maintenance. Dev. Biol. 366:163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bach EA, Vincent S, Zeidler MP, Perrimon N. 2003. A sensitized genetic screen to identify novel regulators and components of the Drosophila janus kinase/signal transducer and activator of transcription pathway. Genetics 165:1149–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith RK, Carroll PM, Allard JD, Simon MA. 2002. MASK, a large ankyrin repeat and KH domain-containing protein involved in Drosophila receptor tyrosine kinase signaling. Development 129:71–82 [DOI] [PubMed] [Google Scholar]
- 39. Karim FD, Chang HC, Therrien M, Wassarman DA, Laverty T, Rubin GM. 1996. A screen for genes that function downstream of Ras1 during Drosophila eye development. Genetics 143:315–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Therrien M, Morrison DK, Wong AM, Rubin GM. 2000. A genetic screen for modifiers of a kinase suppressor of Ras-dependent rough eye phenotype in Drosophila. Genetics 156:1231–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gergely F, Karlsson C, Still I, Cowell J, Kilmartin J, Raff JW. 2000. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc. Natl. Acad. Sci. U. S. A. 97:14352–14357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee MJ, Gergely F, Jeffers K, Peak-Chew SY, Raff JW. 2001. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat. Cell Biol. 3:643–649 [DOI] [PubMed] [Google Scholar]
- 43. Peset I, Seiler J, Sardon T, Bejarano LA, Rybina S, Vernos I. 2005. Function and regulation of Maskin, a TACC family protein, in microtubule growth during mitosis. J. Cell Biol. 170:1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Worby CA, Simonson-Leff N, Dixon JE. 2001. RNA interference of gene expression (RNAi) in cultured Drosophila cells. Sci. STKE 2001:pl1 doi:10.1126/stke.2001.95.pl1. [DOI] [PubMed] [Google Scholar]
- 45. Dean SO, Spudich JA. 2006. Rho kinase's role in myosin recruitment to the equatorial cortex of mitotic Drosophila S2 cells is for myosin regulatory light chain phosphorylation. PLoS One 1:e131 doi:10.1371/journal.pone.0000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Akhmanova A, Steinmetz MO. 2010. Microtubule +TIPs at a glance. J. Cell Sci. 123:3415–3419 [DOI] [PubMed] [Google Scholar]
- 47. Komarova YA, Vorobjev IA, Borisy GG. 2002. Life cycle of MTs: persistent growth in the cell interior, asymmetric transition frequencies and effects of the cell boundary. J. Cell Sci. 115:3527–3539 [DOI] [PubMed] [Google Scholar]
- 48. Piehl M, Cassimeris L. 2003. Organization and dynamics of growing microtubule plus ends during early mitosis. Mol. Biol. Cell 14:916–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stepanova T, Slemmer J, Hoogenraad CC, Lansbergen G, Dortland B, De Zeeuw CI, Grosveld F, van Cappellen G, Akhmanova A, Galjart N. 2003. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J. Neurosci. 23:2655–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dixit R, Barnett B, Lazarus JE, Tokito M, Goldman YE, Holzbaur EL. 2009. Microtubule plus-end tracking by CLIP-170 requires EB1. Proc. Natl. Acad. Sci. U. S. A. 106:492–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Green RA, Wollman R, Kaplan KB. 2005. APC and EB1 function together in mitosis to regulate spindle dynamics and chromosome alignment. Mol. Biol. Cell 16:4609–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Komarova Y, Lansbergen G, Galjart N, Grosveld F, Borisy GG, Akhmanova A. 2005. EB1 and EB3 control CLIP dissociation from the ends of growing microtubules. Mol. Biol. Cell 16:5334–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rogers SL, Wiedemann U, Hacker U, Turck C, Vale RD. 2004. Drosophila RhoGEF2 associates with microtubule plus ends in an EB1-dependent manner. Curr. Biol. 14:1827–1833 [DOI] [PubMed] [Google Scholar]
- 54. Skube SB, Chaverri JM, Goodson HV. Effect of GFP tags on the localization of EB1 and EB1 fragments in vivo. Cytoskeleton (Hoboken, N.J.) 67:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Komarova YA, Akhmanova AS, Kojima S, Galjart N, Borisy GG. 2002. Cytoplasmic linker proteins promote microtubule rescue in vivo. J. Cell Biol. 159:589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. 2008. XMAP215 is a processive microtubule polymerase. Cell 132:79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kreis TE. 1987. Microtubules containing detyrosinated tubulin are less dynamic. EMBO J. 6:2597–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee S, Nahm M, Lee M, Kwon M, Kim E, Zadeh AD, Cao H, Kim HJ, Lee ZH, Oh SB, Yim J, Kolodziej PA. 2007. The F-actin-microtubule crosslinker Shot is a platform for Krasavietz-mediated translational regulation of midline axon repulsion. Development 134:1767–1777 [DOI] [PubMed] [Google Scholar]
- 59. Van Vactor DV, Sink H, Fambrough D, Tsoo R, Goodman CS. 1993. Genes that control neuromuscular specificity in Drosophila. Cell 73:1137–1153 [DOI] [PubMed] [Google Scholar]
- 60. Mimori-Kiyosue Y, Shiina N, Tsukita S. 2000. Adenomatous polyposis coli (APC) protein moves along microtubules and concentrates at their growing ends in epithelial cells. J. Cell Biol. 148:505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zumbrunn J, Kinoshita K, Hyman AA, Nathke IS. 2001. Binding of the adenomatous polyposis coli protein to microtubules increases microtubule stability and is regulated by GSK3 beta phosphorylation. Curr. Biol. 11:44–49 [DOI] [PubMed] [Google Scholar]
- 62. Peset I, Vernos I. 2008. The TACC proteins: TACC-ling microtubule dynamics and centrosome function. Trends Cell Biol. 18:379–388 [DOI] [PubMed] [Google Scholar]
- 63. Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. 2003. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu. Rev. Cell Dev. Biol. 19:541–564 [DOI] [PubMed] [Google Scholar]
- 64. Yu TW, Hao JC, Lim W, Tessier-Lavigne M, Bargmann CI. 2002. Shared receptors in axon guidance: SAX-3/Robo signals via UNC-34/Enabled and a Netrin-independent UNC-40/DCC function. Nat. Neurosci. 5:1147–1154 [DOI] [PubMed] [Google Scholar]
- 65. Kumar P, Lyle KS, Gierke S, Matov A, Danuser G, Wittmann T. 2009. GSK3beta phosphorylation modulates CLASP-microtubule association and lamella microtubule attachment. J. Cell Biol. 184:895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Manna T, Honnappa S, Steinmetz MO, Wilson L. 2008. Suppression of microtubule dynamic instability by the +TIP protein EB1 and its modulation by the CAP-Gly domain of p150glued. Biochemistry 47:779–786 [DOI] [PubMed] [Google Scholar]
- 67. Gard DL, Kirschner MW. 1987. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J. Cell Biol. 105:2203–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vasquez RJ, Gard DL, Cassimeris L. 1994. XMAP from Xenopus eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J. Cell Biol. 127:985–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shirasu-Hiza M, Coughlin P, Mitchison T. 2003. Identification of XMAP215 as a microtubule-destabilizing factor in Xenopus egg extract by biochemical purification. J. Cell Biol. 161:349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Thadani R, Ling YC, Oliferenko S. 2009. The fission yeast TACC protein Mia1p stabilizes microtubule arrays by length-independent crosslinking. Curr. Biol. 19:1861–1868 [DOI] [PubMed] [Google Scholar]
- 71. Grallert A, Beuter C, Craven RA, Bagley S, Wilks D, Fleig U, Hagan IM. 2006. S. pombe CLASP needs dynein, not EB1 or CLIP170, to induce microtubule instability and slows polymerization rates at cell tips in a dynein-dependent manner. Genes Dev. 20:2421–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Patterson K, Molofsky AB, Robinson C, Acosta S, Cater C, Fischer JA. 2004. The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol. Biol. Cell 15:600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Guo Y, Jangi S, Welte MA. 2005. Organelle-specific control of intracellular transport: distinctly targeted isoforms of the regulator Klar. Mol. Biol. Cell 16:1406–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Capalbo L, D'Avino PP, Archambault V, Glover DM. 2011. Rab5 GTPase controls chromosome alignment through Lamin disassembly and relocation of the NuMA-like protein Mud to the poles during mitosis. Proc. Natl. Acad. Sci. U. S. A. 108:17343–17348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lanzetti L, Palamidessi A, Areces L, Scita G, Di Fiore PP. 2004. Rab5 is a signalling GTPase involved in actin remodelling by receptor tyrosine kinases. Nature 429:309–314 [DOI] [PubMed] [Google Scholar]
- 76. Kawauchi T, Sekine K, Shikanai M, Chihama K, Tomita K, Kubo K, Nakajima K, Nabeshima Y, Hoshino M. 2010. Rab GTPases-dependent endocytic pathways regulate neuronal migration and maturation through N-cadherin trafficking. Neuron 67:588–602 [DOI] [PubMed] [Google Scholar]
- 77. Daire V, Giustiniani J, Leroy-Gori I, Quesnoit M, Drevensek S, Dimitrov A, Perez F, Pous C. 2009. Kinesin-1 regulates microtubule dynamics via a c-Jun N-terminal kinase-dependent mechanism. J. Biol. Chem. 284:31992–32001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Horiuchi D, Collins CA, Bhat P, Barkus RV, Diantonio A, Saxton WM. 2007. Control of a kinesin-cargo linkage mechanism by JNK pathway kinases. Curr. Biol. 17:1313–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jung H, Holt CE. 2011. Local translation of mRNAs in neural development. Wiley Interdiscip. Rev. RNA 2:153–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tcherkezian J, Brittis PA, Thomas F, Roux PP, Flanagan JG. 2010. Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. Cell 141:632–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Van Horck FP, Holt CE. 2008. A cytoskeletal platform for local translation in axons. Sci. Signal. 1:pe11 doi:10.1126/stke.18pe11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Howe JG, Hershey JW. 1984. Translational initiation factor and ribosome association with the cytoskeletal framework fraction from HeLa cells. Cell 37:85–93 [DOI] [PubMed] [Google Scholar]
- 83. Kim S, Coulombe PA. 2010. Emerging role for the cytoskeleton as an organizer and regulator of translation. Nat. Rev. Mol. Cell Biol. 11:75–81 [DOI] [PubMed] [Google Scholar]
- 84. Barnard DC, Cao Q, Richter JD. 2005. Differential phosphorylation controls Maskin association with eukaryotic translation initiation factor 4E and localization on the mitotic apparatus. Mol. Cell. Biol. 25:7605–7615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Applewhite DA, Grode KD, Keller D, Zadeh AD, Slep KC, Rogers SL. 2010. The spectraplakin Short stop is an actin-microtubule cross-linker that contributes to organization of the microtubule network. Mol. Biol. Cell 21:1714–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Subramanian A, Prokop A, Yamamoto M, Sugimura K, Uemura T, Betschinger J, Knoblich JA, Volk T. 2003. Shortstop recruits EB1/APC1 and promotes microtubule assembly at the muscle-tendon junction. Curr. Biol. 13:1086–1095 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.