Abstract
KMT2B (MLL2/WBP7) is a member of the MLL subfamily of H3K4-specific histone lysine methyltransferases (KMT2) and is vital for normal embryonic development in the mouse. To gain insight into the molecular mechanism underlying KMT2B function, we focused on MagohB, which is controlled by a CpG island promoter. We show that in cells lacking Mll2—the gene encoding KMT2B—the MagohB promoter resides in inaccessible chromatin and is methylated. To dissect the molecular events leading to the establishment of silencing, we performed kinetic studies in Mll2-conditional-knockout embryonic stem cells. KMT2B depletion was followed by the loss of the active chromatin marks and progressive loss of RNA polymerase II binding with a concomitant downregulation of MagohB expression. Once the active chromatin marks were lost, the MagohB promoter was rapidly methylated. We demonstrate that in the presence of KMT2B, neither transcription elongation nor RNA polymerase II binding is required to maintain H3K4 trimethylation at the MagohB promoter and protect it from DNA methylation. Reexpression of KMT2B was sufficient to reinstate an active MagohB promoter. Our study provides a paradigm for the idea that KMT2 proteins are crucial components for establishing and maintaining the transcriptionally active and unmethylated state of CpG island promoters.
INTRODUCTION
Histone H3 lysine 4 trimethylation (H3K4me3) has been associated with transcriptionally active promoters (1, 2). Mammals have at least 6 different H3K4-specific methyltransferases; in mice these are KMT2A (ALL1/CXXC7/MLL1), KMT2B (MLL2/WBP7), KMT2C (MLL3), KMT2D (MLL4), KMT2E (SET1A), and KMT2F (SET1B). Murine Mll2 is a paralogue of Mll1 and is the orthologue of human MLL4. Mll2 resides on mouse chromosome 7 (mm9 chr7: 31353874 to 31369215) and codes for the KMT2B methlytransferase. These enzymes have at least some nonredundant functions, as depletion of either KMT2A or KMT2B is embryonically lethal (3–5). KMT2A is the most studied of the H3K4 methyltransferases, as it is involved in various leukemogenic translocations (6, 7). These aberrant fusion proteins function by recruiting a constitutively active, transcriptional elongation-promoting complex to promoters (8–10).
In Saccharomyces cerevisiae, Set1 is the only enzyme responsible for H3K4me3 deposition and is recruited to the initiating form of RNA polymerase II (Pol II) (11), suggesting that the H3K4me3 mark is deposited after transcription has initiated. In mice, depletion of KMT2A results in transcriptional silencing, abnormal RNA polymerase II distribution, loss of histone acetylation, and CpG island promoter methylation on a subset of the Hox genes (12–14). These results suggest that KMT2A recruitment is crucial for the association of the basal transcription machinery and is involved in setting up a permissive chromatin landscape for transcription as well as preventing DNA methylation. However, direct evidence for this idea has so far been lacking.
In contrast to KMT2A, the KMT2B function is less characterized. In embryonic stem (ES) cells, only a small number of genes are significantly downregulated in response to KMT2B depletion, the most prominent of which is MagohB (15). MagohB is regulated by a CpG island promoter, and it is expressed in all tissues examined so far, including ES cells (15). MagohB expression is absolutely dependent on KMT2B in ES cells, with no apparent functional redundancy from other H3K4 methyltransferases. MagohB transcriptional silencing in the absence of KMT2B is characterized by loss of H3K4me3 and increased DNA methylation over the MagohB CpG island promoter (15). This phenomenon is similar to what has been observed on the Hox genes in cells lacking KMT2A (12–14). However, the order of events leading to the establishment of the silenced state is unknown in either case. In addition, the interdependency between RNA polymerase II binding and MLL2 association is unclear. We used ES cells carrying a conditional allele of Mll2 to address these questions and performed kinetic experiments aimed at dissecting the molecular events leading to MagohB silencing and DNA methylation. Here we present direct evidence that the presence of KMT2B is required for the stable maintenance of the basal transcription machinery and for maintaining low levels of DNA methylation. Our experiments demonstrate that DNA methylation is a rapid process that is secondary to gene silencing. Moreover, we demonstrate that reexpression of MLL2 is sufficient to reactivate the methylated MagohB promoter.
MATERIALS AND METHODS
Cell lines and cell culture.
Wild-type mouse ES cells (129P2 background; here termed E14) and Mll2−/−, Mll2F/F, and Mll2F/+ ES cells (5, 15, 16) were provided by A. F. Stewart and were grown in high-glucose Dulbecco modified Eagle medium supplemented with 15% serum (PAA Gold fetal bovine serum), 1,000 U/ml leukemia inhibitory factor (ESGRO; Millipore), 25 mM HEPES, 1 mM sodium pyruvate, 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 1× nonessential amino acids (Sigma), and 0.15 mM monothioglycerol at 37°C in 5% CO2 on tissue culture-grade plastics at a density of 104 cells/cm2. Cells were split every 2 to 3 days as necessary by brief trypsinization in an appropriate volume of 1× trypsin-EDTA at room temperature.
Genotyping.
The Mll2F/F and Mll2F/+ ES cells were tested for recombination of the Mll2 alleles twice by Southern blotting and after that by PCR and agarose gel electrophoresis every time that they were induced with 4-hydroxytamoxifen (OHT). OHT was added to the culture medium at a final concentration of 10−7 M. The genotyping PCR was performed using primers 145se (CGGAGGAAGAGAGCAGTGACG) and 147as (GGACAGGAGTCACATCTGCTAGG). The 145se/147as primer pair detects all Mll2 alleles.
Disruption of RNA polymerase II by α-amanitin and DRB.
Cells were grown as described above. 5,6-Dichlorobenzimidazole-1-β-d-ribofuranoside (DRB; Sigma) or α-amanitin (Sigma) was added to the culture medium to give a final concentration of 100 μM or 5 μg/ml, respectively. The cells were cultured for up to 24 h in DRB and up to 48 h in α-amanitin and then harvested using a method suited for the downstream analyses.
Transfections.
pCAGGS-FlpO-IRES-puro transfections were performed in 6-well plates using Lipofectamine LTX with Plus reagent (Invitrogen) according to the manufacturer's instructions. The transfected cells were grown in puromycin-containing medium for 24 h, harvested as a bulk population, and expanded for the time periods indicated in the relevant figures. Cells were cotransfected with 100-fold less of a green fluorescent protein (GFP)-expressing plasmid. Fluorescence-activated cell sorter analysis revealed that at least 95% of cells expressed GFP.
DNase I-hypersensitive site (DHS) mapping.
The method employed for DNase I treatment is an adaptation of the method described by Pfeifer and Riggs (17). Southern blot transfer and hybridization were performed as described by Cockerill (18).
In vivo footprinting.
Footprinting experiments were performed as described by Tagoh et al. (19), using Phusion polymerase (New England Biolabs) and the GC-specific buffer instead of the suggested Pfu polymerase. Micrococcal nuclease (MNase) digestions were performed using the same method used for DNase I. Primer sequences used in footprinting assays are available upon request.
Measurement of DNA methylation. (i) Pyrosequencing.
Genomic DNA was treated with sodium bisulfite using an EZ-DNA methylation kit from Zymo Research according to the manufacturer's instructions. Bisulfite-converted DNA was used as a PCR template to amplify the MagohB promoter using biotinylated primers specific for the converted DNA sequence (forward sequence, AGTAGAGAAGGTAGAAATTATTATTTATAG; reverse sequence, [biotin]-CTCTTAAAAATCTCTTACTTCCTCTTC). PCR products were purified using streptavidin-coated magnetic beads and prepared for sequencing using a third sequencing primer (SEQ [GGTAGAAATTATTATTTATAGATAT]).
(ii) HpaII digestion.
Five hundred micrograms of genomic DNA was digested to completion with HpaII. The digests were used as the template for quantitative PCRs (QPCRs) with primers flanking a single HpaII site on the MagohB promoter (MagohB HpaII forward primer, TTCTCTTGGGGGTCTCTTGCTTCC; MagohB HpaII reverse primer, CGCGTCACCAAGGGCGCGTT). Data were subsequently normalized against a PCR amplicon on the Oct4 promoter that does not contain an HpaII site.
(iii) Methylated DNA immunoprecipitation (MeDIP) and hydroxymethylated DNA immunoprecipitation (HmeDIP).
Five micrograms per immunoprecipitation (IP) of anti-methylated C (ab10805; Abcam) and anti-hydroxymethylated C (ab106918; Abcam) antibody each was incubated with 10 μl protein G Dynabeads (10004D; Life Technologies) for 2 h at 4°C with rotation. One hundred microliters purified genomic DNA (2 μg/IP) was added to the bead-antibody complexes, and the mixture was incubated overnight at 4°C with rotation. The beads were separated using a magnetic separator and washed once with each of the following: wash buffer 1 (20 mM Tris, pH 8.0, 2 mM EDTA, 1% Triton X-100, 0.1% SDS, 150 mM NaCl), wash buffer 2 (20 mM Tris, pH 8.0, 2 mM EDTA, 1% Triton X-100, 0.1% SDS, 500 mM NaCl), and TE-NaCl buffer (10 mM Tris, pH 8.0, 1 mM EDTA, 50 mM NaCl). Finally, the immunoprecipitated DNA was eluted in 0.1 M NaHCO3, 1% SDS, 200 mM NaCl containing 200 μg/ml proteinase K by a 4-h incubation at 65°C. The eluted DNA was purified using an Agencourt Ampure XP PCR cleanup kit and used as the template for a QPCR with primers specific for the MagohB promoter (forward primer, TACTCCGGTAGGAACGAAA; reverse primer, CTCCAATGCGAACCTTCAGT).
RNA and protein purification.
Total RNA and protein were purified using an RNeasy kit (Qiagen) according to the manufacturer's instructions.
cDNA synthesis and QPCR.
Moloney murine leukemia virus reverse transcriptase (Invitrogen) was used to reverse transcribe 1 μg total RNA according to the manufacturer's instructions. QPCR was performed on an ABI 7900HT or ABI 7500 real-time PCR instrument using ABI SYBR green master mix. Initial data analysis (threshold/baseline) was performed by ABI SDS software, and further analyses were performed in the Microsoft Excel program. Standard curve quantitation was performed for all QPCR experiments. The primer sequences used in mRNA expression measurements were as follows: for Gapdh (glyceraldehyde-3-phosphate dehydrogenase gene), forward primer ACCTGCCAAGTATGATGACATCA and reverse primer GGTCCTCAGTGTAGCCCAAGAT; for Oct4, forward primer AGGTGGAACCAACTCCCGAG and reverse primer GCTTCAGCAGCTTGGCAAC; and for MagohB, forward primer CGGGCATAAGGGCAAGTTT, primary RNA forward primer TGGGACTATGAAATTGTCTTTACC, and reverse primer AATTACTGTTGTTGGCATATGTAAGCTT.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (20) with modifications. ES cells were fixed in situ by addition of formaldehyde (final concentration, 1%) in the culture medium and incubation for 12 min. Nuclei were prepared from the fixed cells, and the contained chromatin was sheared by sonication in a Diagenode Bioruptor (high setting, 8 cycles of 30 min of pulse and 30 min of pause at 4°C). Immunoprecipitation was performed using antibodies against the RNA Pol II carboxy-terminal domain (CTD) (ab5408; Abcam), H3K4me3 (07-473; Millipore), and acetylated H3K9 (H3K9ac; ab4441; Abcam). The immunoprecipitated DNA was amplified by QPCR as described above. Primer sequences used in ChIP-QPCR were as follows: for the MagohB promoter, forward primer TACTCCGGTAGGAACGAAA and reverse primer CTCCAATGCGAACCTTCAGT; for the Oct4 promoter, forward primer TGGGCTGAAATACTGGGTTC and reverse primer TTGAATGTTCGTGTGCCAAT.
SDS-PAGE and Western blotting.
Whole-cell extracts from equal amounts of cells were separated on a Bio-Rad 4 to 20% gradient mini-Protean TGX precast gel. Proteins were transferred onto a nitrocellulose membrane by wet transfer. Membranes were blocked with 5% (wt/vol) bovine serum albumin in Tris-buffered saline–Tween. West-pico chemiluminescence (Thermo Fisher) was used to visualize the results according to the manufacturer's instructions. The primary antibodies used were ab5408 (Pol II CTD), ab5095 (serine-2-phosphorylated [phospho-S2] elongating form of Pol II), and ab86601 (Nup188) and are available from Abcam. The appropriate horseradish peroxidase-conjugated secondary antibodies were sourced from Jackson ImmunoResearch.
RESULTS
MagohB expression requires KMT2B.
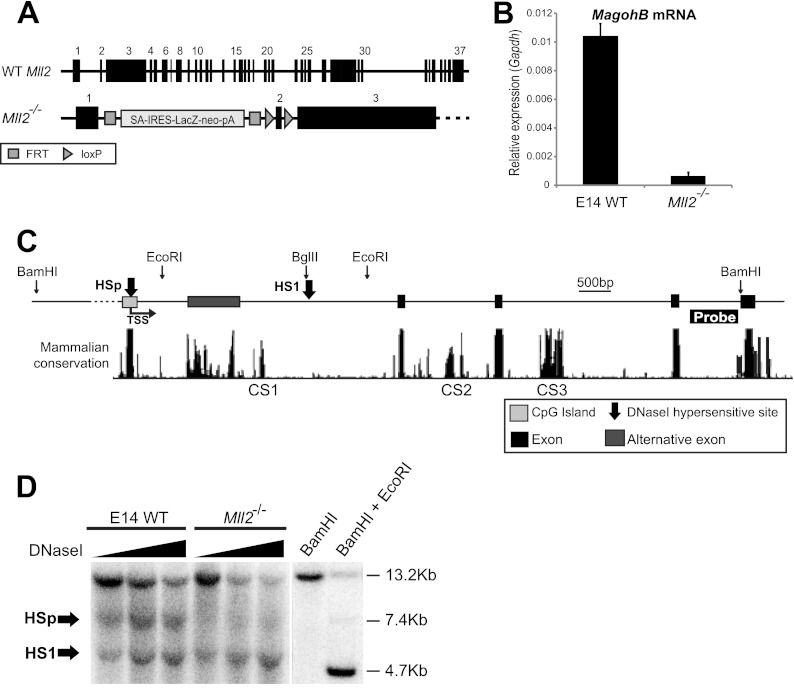
Mll2−/− ES cells were employed to investigate the mechanism of MagohB transcriptional regulation. These cells harbor a gene-trapping cassette in intron 1 of both alleles of Mll2 (Fig. 1A). The nascent Mll2 transcript is spliced to the cassette, thus preventing expression of wild-type Mll2. We measured MagohB steady-state mRNA levels by reverse transcription-QPCR in the wild-type ES cell line E14 and Mll2−/− ES cells. As reported previously (15), MagohB expression was abolished in Mll2−/− ES cells (Fig. 1B), suggesting that KMT2B is absolutely required for MagohB expression.
Fig 1.
KMT2B is required for MagohB expression and maintenance of an open chromatin structure on the MagohB promoter. (A) Schematic depiction of the Mll2 alleles in wild-type (WT Mll2) and Mll2−/− cells. The Mll2− allele contains a STOP cassette composed of a splice acceptor (SA), an internal ribosome entry sequence (IRES), the β-galactosidase gene (lacZ), a neomycin resistance gene (neo), two polyadenylation signals (pA), and a transcriptional terminator sequence (not shown). Black boxes, Mll2 exons; E14, wild-type (WT) ES cell line; FRT, FLP recombination target. (B) MagohB mRNA expression in wild-type and knockout cells. MagohB expression is normalized against Gapdh mRNA expression. Bars represent the means plus SDs of at least 3 measurements. (C) Schematic of the structure of the MagohB locus. CS1, CS2, and CS3, conserved intronic regions. Arrows indicate the positions of the two DNase I-hypersensitive sites detected within an extended genomic region. (D) Southern blot visualizing the DHSs located within the BamHI fragment using the probe abutting the downstream BamHI site, as indicated in panel C. Arrows, the two DNase I-hypersensitive sites detected. The last two lanes serve as a DNase I-nondigested control (BamHI) and as size markers (BamHI plus EcoRI/BglII [BamHI + EcoRI]).
In order to dissect the molecular mechanism by which KMT2B activates or maintains transcription from the MagohB promoter, we first had to identify potential cis regulatory elements on the Magoh2B locus. Three conserved intronic regions which could be regulating MagohB expression were identified (through the UCSC genome browser's mammalian conservation plots) and were termed conserved sequence 1 (CS1), CS2, and CS3 (Fig. 1C). We then employed DNase I-hypersensitive site (DHS) mapping to examine the entire MagohB gene and the immediate 5′ flanking region. Hybridization with probes on either side of the restriction fragment revealed the presence of two DHSs, termed HSp and HS1 (Fig. 1D; results from hybridization with only one probe are shown). HSp forms over the MagohB CpG island promoter and is present only in E14 wild-type cells. Loss of HSp in the Mll2−/− cells suggests that KMT2B is involved in maintenance of the open chromatin conformation of the MagohB promoter. HS1 forms over a simple repeat element (Fig. 1D and data not shown), possibly because the DNA sequence of the repeat element is unfavorable for nucleosome assembly. In summary, from our data, it is highly likely that the main element driving expression of MagohB is its CpG island promoter.
MagohB is transcriptionally silenced 4 days after de novo Mll2 deletion.
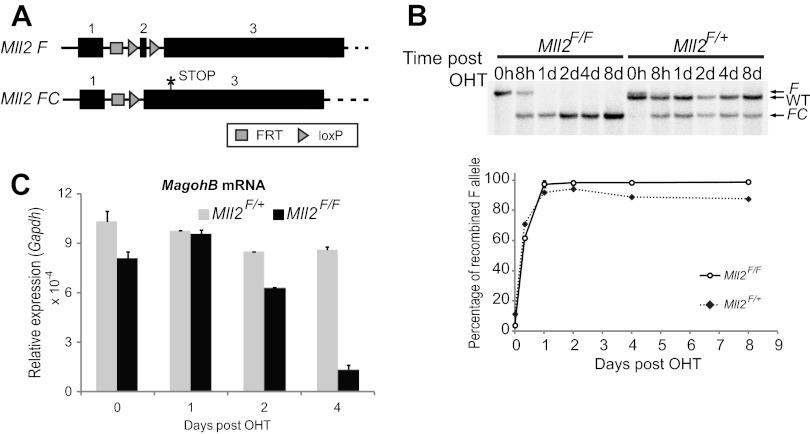
To establish the order of events that lead to MagohB silencing after KMT2B depletion, we employed ligand-dependent conditional mutagenesis (21). The Mll2F/+ and Mll2F/F ES cells ubiquitously express Cre-ERT2 from the Rosa26 locus and have one or two conditional Mll2 alleles, respectively (described in more detail in references 5 and 15). Cre-ERT2 activation by 4-hydroxytamoxifen (OHT) results in deletion of Mll2 exon 2, causing a frameshift and introducing a stop codon within exon 3 (Fig. 2A). We measured the efficiency and kinetics of Cre-mediated recombination by Southern blotting (Fig. 2B) and found that recombination was complete 24 h after OHT induction. These results confirm previous studies using the same ES cell system (15), showing similar recombination kinetics. The study described above had also demonstrated an approximately 50% reduction of KMT2B protein 12 h after Cre activation. Importantly, no outgrowth of nonrecombined cells was observed at up to 8 days postinduction, as detected by the absence of the Mll2F allele in either cell line.
Fig 2.
Cre-mediated deletion of Mll2 results in MagohB silencing 4 days after OHT treatment. (A) Cre-mediated deletion of Mll2 exon 2 results in a frameshift and generation of a STOP codon in exon 3. F, conditional allele; FC, null allele. (B) (Top) Mll2 Southern blot distinguishing the different Mll2 alleles before and after OHT treatment. The image shown is a representative of 2 experiments. WT, wild-type allele; d, days. (Bottom) Quantitation of the Southern blot. The intensity quantification was performed using the Quantity One software suite (Bio-Rad). (C) MagohB mRNA expression after Mll2 deletion. MagohB expression is normalized against Gapdh mRNA expression. Bars represent the means plus SDs of 4 measurements.
We measured MagohB steady-state mRNA levels over a 4-day period after OHT induction (Fig. 2C). Although Mll2 deletion was complete within 1 day after addition of OHT, the first transcriptional effects on MagohB were observed at 2 days postinduction. This discrepancy can be attributed to both the stability of the MagohB mRNA and the half-life of KMT2B. Subsequently, a nearly complete depletion of MagohB mRNA was observed 4 days after OHT induction. No significant downregulation of MagohB was observed in Mll2F/+ cells, indicating that one allele of Mll2 is sufficient to maintain MagohB expression.
Chromatin structure of the MagohB promoter is perturbed after KMT2B depletion.
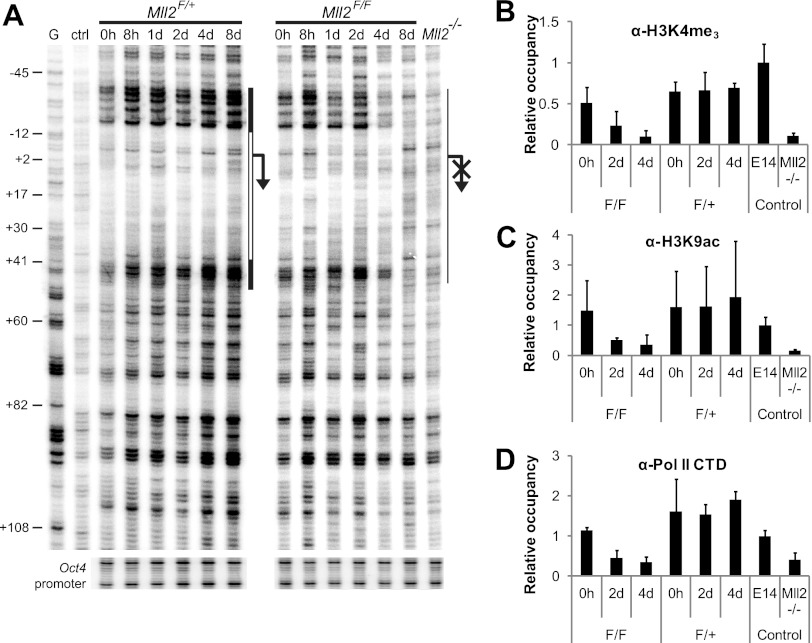
To gain insight into the mechanistic details of the different steps of MagohB silencing, we performed DNase I footprinting experiments over a time course of 8 days (Fig. 3A). We observed a characteristic protection from DNase I digestion around the transcription start site (TSS) flanked by hypersensitive regions, suggesting the presence of a protein assembly, most likely the RNA polymerase II preinitiation complex. This pattern was maintained in the Mll2F/+ cells throughout this time course, consistent with the aforementioned maintenance of MagohB mRNA levels in those cells. In contrast, the DNase I digestion pattern observed in Mll2F/F cells over the MagohB TSS was altered 4 days after OHT induction, coinciding with the observed MagohB mRNA depletion. The digestion pattern observed 8 days after OHT induction very closely resembles the pattern observed in Mll2−/− cells. This experiment demonstrates that the maintenance of DNase I-hypersensitive chromatin over the TSS requires the presence of KMT2B.
Fig 3.
KMT2B is required to maintain activating histone marks and a stable RNA polymerase II complex on the MagohB promoter. (A) DNase I in vivo footprinting of the MagohB promoter in OHT-treated Mll2F/F and Mll2F/+ cells. White bar, region protected from DNase I; black bars, hypersensitive regions; G, naked genomic DNA subjected to a Maxam-Gilbert G reaction prior to ligation-mediated PCR; ctrl, in vitro DNase I-digested DNA; arrow, TSS. Time after OHT treatment is indicated above the relevant lanes. Mll2−/− ES cells were included to demonstrate the DNase I digestion pattern in the inactive state. Annotation on the left (in base pairs) is relative to the TSS. Primers on the Oct4 promoter were used to verify equal DNase I digestion of all samples. The image shows a representative of 2 experiments. (B to D) ChIP-QPCR assays using antibodies against H3K4me3 (B), H3K9ac (C), and the RNA polymerase II CTD (D). Data for histone modifications were corrected for nucleosome density, as determined by a histone H3 ChIP assay. Data for RNA polymerase II are corrected for the signal obtained with input material. Data shown are internally normalized against the signal obtained with primers for the Oct4 promoter and presented in relation to the occupancy measured in E14 wild-type ES cells. Bars represent the means plus SDs of at least 4 measurements.
To correlate the reduction in nuclease accessibility with changes in histone modification patterns, we employed ChIP-QPCR studies. These assays revealed that loss of KMT2B and downregulation of gene expression are associated with loss of the active H3K4me3 and H3K9ac marks (Fig. 3B and C), which was first observed 2 days after OHT induction in Mll2F/F cells. The levels of both histone marks decreased to near background levels at 4 days postinduction, concomitantly with MagohB mRNA depletion. RNA polymerase II levels closely correlated with the levels of these two histone marks (Fig. 3D). Loss of only one Mll2 allele (Mll2F/+ cells) had no effect on the chromatin environment of the MagohB promoter. These results support the view that KMT2B and/or the H3K4me3 mark is required for RNA polymerase II association with the MagohB promoter.
Chromatin architecture at MagohB promoter.
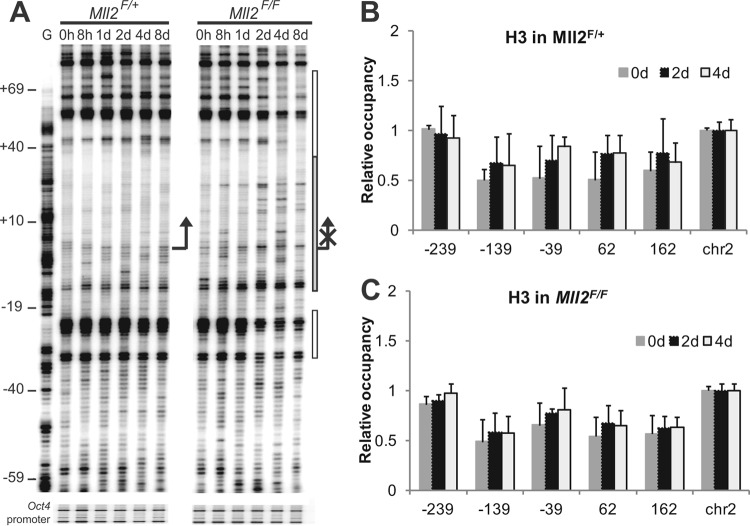
DHSs and active promoters have previously been reported to be devoid of nucleosomes (22 –26). MNase preferentially cuts in nucleosomal linker regions in chromatin (27, 28), which can be visualized as strong double-strand cuts in high-resolution mapping assays (19). In addition, large nonhistone DNA-protein complexes can also be indicated by flanking MNase cuts (29). We therefore employed MNase footprinting to examine the chromatin architecture of the MagohB promoter after deletion of Mll2. Identical MNase digestion patterns were observed in untreated Mll2F/F and Mll2F/+ cells with strong MNase cuts flanking the TSS and the active RNA polymerase II complex. This digestion pattern, especially the strong flanking sites, remained unchanged in Mll2F/+ cells up to 8 days after OHT induction. In Mll2F/F cells, however, it was altered 2 days after OHT induction (Fig. 4A). Changes in MNase accessibility were also observed further downstream (data not shown). These findings confirmed the DNase I data demonstrating that upon loss of KMT2B, nuclease accessibility on the MagohB promoter and surrounding region was altered. To clarify whether these changes were due to nucleosome repositioning or eviction of the basal transcription machinery and/or other protein factors, we performed ChIP experiments using an antibody raised against histone H3 (Fig. 4B and C). These experiments demonstrated that nucleosomes on the MagohB CpG island promoter were only loosely positioned, if at all, consistent with previously reported findings with CpG island promoters (30). Most importantly, we did not detect major alterations in the nucleosome occupancy pattern at the MagohB promoter up to 8 days after OHT treatment in Mll2F/+ (Fig. 4B) or Mll2F/F (Fig. 4C) ES cells.
Fig 4.
Nucleosome remodelling over the MagohB promoter 2 days after Mll2 deletion. (A) MNase in vivo footprinting visualizing the vicinity of the MagohB transcription start site in OHT-treated Mll2F/F cells. Gray bar covering the TSS, moderately increased MNase sensitivity; white bars, protected regions when comparing digestion patterns observed at the 8-day time point to those at the 0-h time point; G, G reaction; arrow, TSS. The time after OHT induction is indicated at the top of each lane. Annotation on the left (in base pairs) is relative to the TSS. Primers on the Oct4 promoter were used to verify equal MNase digestion of all samples. The image shows a representative of 2 experiments. (B and C) Nucleosome density over the MagohB promoter 0, 2, and 4 days after OHT treatment of Mll2F/+ (B) and Mll2F/F (C) cells. Nucleosome density was measured by histone H3 ChIP followed by QPCR with primers centered on the positions indicated on the x axis. chr2, chromosome 2, which represents a transcriptionally inactive control region. Bars represent the means plus SDs of at least 4 measurements.
DNA methylation occurs soon after transcriptional silencing.
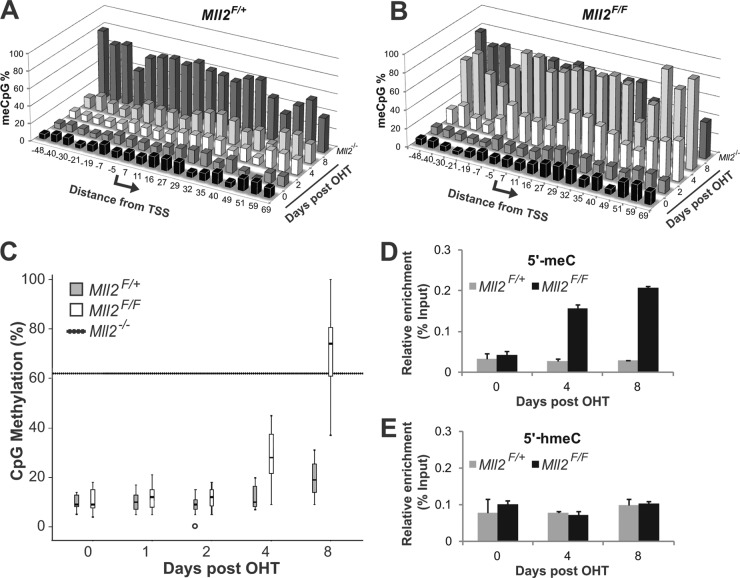
To directly investigate whether DNA methylation is a silencing initiating or maintaining event, we measured DNA methylation levels on the MagohB CpG island promoter region by bisulfite conversion and pyrosequencing over an 8-day time course. A total of 19 CpG sites distributed between bp −48 and +69 from the transcription start site were examined in this assay. The results showed no significant difference in DNA methylation in Mll2F/+ cells over the duration of this time course (Fig. 5A). In contrast, increased DNA methylation at the MagohB promoter was evident in Mll2F/F cells 4 days after OHT induction (Fig. 5B). At this time point, the H3K4me3 and H3K9ac histone marks along with RNA polymerase II had already been removed and transcription had ceased. We therefore conclude that DNA methylation cannot be responsible for initiating MagohB silencing in those cells. As no clear bias toward methylation (or protection from methylation) of specific CpG dinucleotides was observed, the three-dimensional graphs presented in Fig. 4A and B were summarized in a simpler box plot format (Fig. 5C). This representation revealed a significant (at 95% confidence) overall increase in CpG methylation on the MagohB promoter in Mll2F/F cells 4 days after OHT treatment. At the 8-day time point, CpG methylation on the MagohB promoter in Mll2F/F cells equalled the levels measured in Mll2−/− cells. As this method cannot distinguish between cytosine methylation and hydroxymethylation, we performed MeDIP and HmeDIP assays coupled with QPCR. We measured an increase in methylcytosine levels on the MagohB promoter exclusively in Mll2F/F cells at 4 and 8 days after OHT treatment (Fig. 5D), but hydroxymethylcytosine levels in both Mll2F/+ and Mll2F/F ES cells were low (Fig. 5E). Importantly, the levels of this modification did not change after KMT2B depletion, verifying that what we detected by bisulfite conversion and pyrosequencing is indeed DNA methylation.
Fig 5.
Rapid DNA methylation of the MagohB CpG island after KMT2B depletion. Cytosine modification levels on a part of the MagohB CpG island promoter in Mll2F/+ (A) and Mll2F/F (B) cells were measured by bisulfite conversion and subsequent pyrosequencing. The cells were induced with OHT at 0 days and harvested at the time points indicated on the z axis. DNA methylation levels measured in Mll2−/− cells are inserted as a separate point on the z axis for comparison. Each point on the x axis represents an individual CpG dinucleotide. (C) Data presented in panels A and B summarized in a box plot format. Analysis was performed using SPSS. Hatched line, median DNA methylation level in Mll2−/− cells; black bars, median CpG methylation over the region examined (bp −48 to +69); boxes, 95% confidence interval; whiskers, 99% confidence interval; open circle, outlier. (D) MeDIP-QPCR with primers specific for the MagohB promoter in Mll2F/+ and Mll2F/F ES cells 0, 4, and 8 days after OHT treatment. 5′-meC, 5′ methylated C. Bars represent the means plus SDs of 3 experiments. (E) HmeDIP-QPCR with primers specific for the MagohB promoter in Mll2F/+ and Mll2F/F ES cells 0, 4, and 8 days after OHT treatment. 5′-hmeC, 5′ hydroxymethylated C. Bars represent means + SDs of 3 experiments.
Transcription and RNA polymerase II are dispensable for the maintenance of the H3K4me3 mark and for protection from DNA methylation.
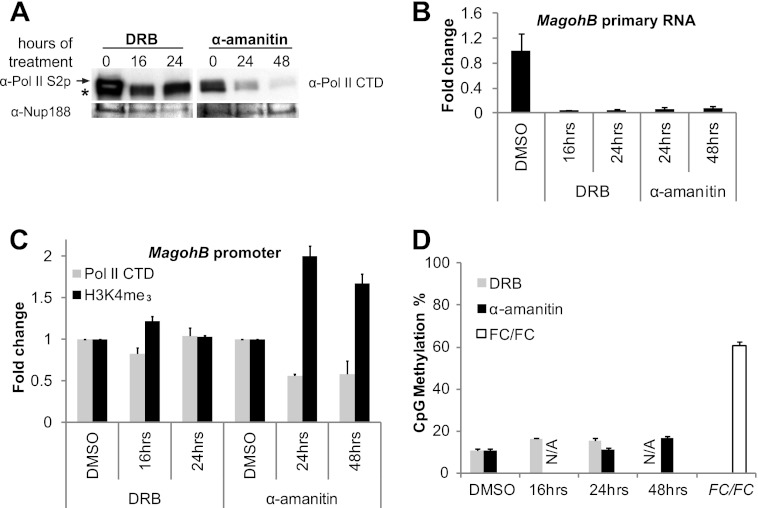
A recent study reports that the Drosophila homologue of KMT2B—Trx—is found to rapidly reassociate with nascent DNA after the passage of the replication fork, while H3K4-methylated histones are detected only much later (31). Additionally, a previous report has shown that KMT2A remains bound on highly condensed mitotic chromosomes and marks genes that will be rapidly and highly induced upon entry of the cell into G1 phase (32). During cell division, transcription ceases and RNA polymerase II is evicted from DNA. KTM2 proteins in higher eukaryotes therefore appear to be able to bind independently of RNA polymerase II association. To gain insight into the role of KMT2B in the absence of transcription and/or RNA polymerase II, Mll2F/F cells were treated with DRB or α-amanitin. DRB blocks transcriptional elongation by inhibiting p-TEFb enzymatic activity (33, 34). α-Amanitin binds the active site of RNA polymerase II, and this complex is subsequently targeted for degradation (35). Western blotting using whole-cell extracts demonstrated that the serine-2-phosphorylated elongating form of RNA polymerase II was depleted after 16 h of DRB treatment, while a 24- or 48-h α-amanitin treatment greatly reduced total RNA polymerase II levels (Fig. 6A). As expected, MagohB primary transcripts (Fig. 6B) and steady-state mRNA (data not shown) were barely detectable following treatment with either of the two drugs. Gapdh expression levels were found to be just above detection limits (data not shown), demonstrating that this effect was not confined to MagohB. Importantly, 18S rRNA could be readily detected, as RNA polymerase I is reported to be unaffected by either of these drugs (data not shown).
Fig 6.
The H3K4me3 mark and protection from DNA methylation persist on the MagohB promoter, despite a block in transcription and a decrease in RNA polymerase II association. (A) Western blot analysis of DRB- and α-amanitin-treated Mll2F/F cells, using whole-cell protein extracts. Arrow, band for phosphoserine 2 RNA polymerase II; asterisk, a nonspecific band. Dimethyl sulfoxide (DMSO)-treated samples were used as a control to exclude drug carrier effects. An antibody against nucleoporin Nup188 was used to verify equal sample loading. The image shown is a representative of 2 individual experiments. (B) Reverse transcription-QPCR detecting primary MagohB RNA in DRB- and α-amanitin-treated cells. Data presented here are normalized to the level of 18S rRNA, which is transcribed by RNA polymerase I. Bars represent the means plus SDs of 4 measurements. (C) ChIP with antibodies against H3K4me3 and RNA polymerase II CTD. Data for histone modifications are corrected for nucleosome density, as determined by a histone H3 IP assay. Data for RNA polymerase II are corrected for input. The data presented are plotted as the fold change over the results for dimethyl sulfoxide-treated cells. Bars represent the means plus SDs of 4 measurements. (D) DNA methylation levels were measured by QPCR with primers spanning a single HpaII site after HpaII digestion. Methylation levels of the same CpG dinucleotide 8 days after OHT treatment of Mll2F/F cells (FC/FC) are shown for comparison. Data presented here were normalized over an amplicon on the Oct4 promoter that does not contain an HpaII site. N/A, not assayed. Dimethyl sulfoxide-treated cells were used to exclude any drug carrier effects. Bars represent the means plus SDs of 4 measurements.
We next examined the chromatin state at the MagohB promoter by ChIP-QPCR. We did not observe any difference in RNA polymerase II occupancy or H3K4me3 levels in DRB-treated cells (Fig. 6C). We measured an approximately 50% reduction in RNA polymerase II occupancy over the MagohB promoter in cells treated with α-amanitin, accompanied by an approximately 2-fold increase in H3K4me3 levels. The increase in H3K4me3 could be explained by the loss of H3K4-specific demethylases as a result of RNA polymerase II inhibition. These findings support the notion that H3K4me3 deposition and maintenance on the MagohB promoter do not require RNA polymerase II recruitment and transcription.
To test whether the reduction in elongation or RNA polymerase II occupancy increased DNA methylation at the MagohB CpG island promoter, we examined DNA methylation levels after DRB or α-amanitin treatment by HpaII digestion and QPCR. We did not observe any increase in methylation of this particular CpG dinucleotide following either treatment. This result suggests that in the presence of KMT2B, protection from DNA methylation does not depend on productive transcriptional elongation and is not affected by a severe depletion of RNA polymerase II binding. The presence of KMT2B and/or the H3K4me3 mark is sufficient to protect the MagohB promoter from the action of DNA methyltransferases.
Reactivation of endogenous Mll2 alleles is sufficient to revert MagohB to the active state.
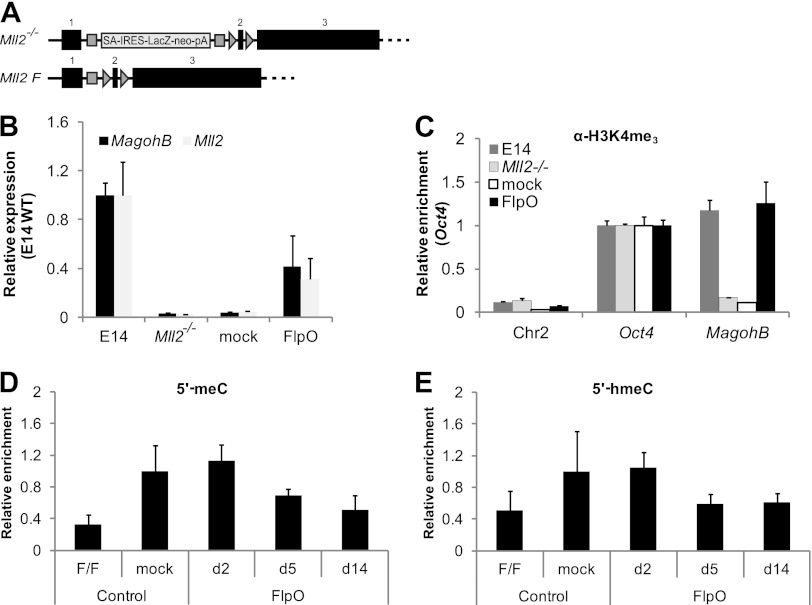
In order to examine whether DNA methylation permanently locks the MagohB promoter in the inactive state, we reactivated the endogenous Mll2 alleles in Mll2−/− cells by transient expression of the FlpO recombinase. Expression of FlpO results in removal of the stop cassette and expression of wild-type KMT2B from the endogenous locus (Fig. 7A). Mll2−/− cells were transfected with a pCAGGS-FlpO-puro plasmid and enriched by culturing in 1 μg/ml puromycin-containing medium for 24 h. Successful deletion of the stop cassette and reactivation of Mll2 were verified by genotyping PCR (data not shown) and Mll2 steady-state mRNA expression measurement (Fig. 7B). Upon reintroduction of KMT2B (14 days posttransfection), MagohB mRNA could be readily detected (Fig. 7B), suggesting that KMT2B action can overcome the repressive barrier posed by DNA methylation. MagohB reactivation was accompanied by the reestablishment of the H3K4me3 mark on the MagohB promoter (Fig. 7C), as shown by ChIP-QPCR assays performed at the same time point. Both cytosine methylation (Fig. 7D) and 5′ hydroxymethylation (Fig. 7E) were detected and progressively decreased on the MagohB promoter. Our results suggest that reactivation of the endogenous Mll2 alleles and physiological levels of KMT2B expression are sufficient to reestablish the normal H3K4me3 pattern and to induce DNA demethylation, most likely via a 5′ hydroxymethylcytosine intermediate.
Fig 7.
Reactivation of the endogenous Mll2 alleles in Mll2−/− ES cells is sufficient to reactivate MagohB. (A) The stop cassette was excised by FlpO-mediated recombination, allowing transcription of full-length Mll2. Black boxes, Mll2 exons. (B) MagohB and Mll2 expression relative to expression levels in E14 wild-type ES cells. pCAGGS-FlpO-puro-transfected Mll2−/− cells were harvested at 14 days posttransfection. Mock, pMAX-GFP-transfected Mll2−/− ES cells. Bars represent the means plus SDs of at least 4 measurements. (C) H3K4me3 ChIP followed by QPCR with primers specific for the loci indicated on the x axis. FlpO-transfected cells were cultured for 14 days posttransfection. Bars represent the means plus SDs of at least 4 measurements. (D) MeDIP-QPCR with primers specific for the MagohB promoter in Mll2F/F and mock- and FlpO-transfected Mll2−/− ES cells, as indicated. Data are presented in relation to the enrichment measured in mock-transfected Mll2−/− cells. FlpO-transfected cells were harvested at the time points indicated on the x axis. Bars represent the means plus SDs of 3 experiments. (E) HmeDIP-QPCR with primers specific for the MagohB promoter in Mll2F/F and mock- and FlpO-transfected Mll2−/− ES cells, as indicated. Data are presented in relation to the enrichment measured in mock-transfected Mll2−/− cells. FlpO-transfected cells were harvested at the time points indicated on the x axis. Bars represent the means plus SDs of 3 experiments.
DISCUSSION
KMT2B is required to establish and maintain an active chromatin environment and recruit RNA polymerase II to the MagohB promoter.
The inducible Mll2-knockout system allowed us to dissect the order of events leading to MagohB silencing and demonstrate that it occurs in a sequential fashion. After KMT2B depletion, the levels of the H3K4me3 mark decrease (herein and reference 15) with a concomitant decrease in both H3K9 acetylation and RNA polymerase II association with the MagohB promoter, resulting in transcriptional silencing. Additionally, the MagohB promoter lacks DNase I hypersensitivity in the absence of KMT2B, as shown by our DHS mapping experiments in Mll2−/− cells.
In summary, from our data it is likely that KMT2B forms a platform that coordinates the assembly of the basal transcription machinery via multiple interactions. The transactivation domain of KMT2A has been shown to interact directly with CBP/p300-containing histone acetyltransferase (HAT) complexes (36). HAT complexes are known to act as large multiprotein scaffolds that can mediate interactions between transcription factors and the basal transcription machinery (37). KMT2B may be involved in stabilizing the recruitment of CBP/p300-containing complexes, which can in turn recruit the transcriptional apparatus. The preinitiation complex may be further stabilized by TFIID binding to H3K4-trimethylated nucleosomes (38, 39) flanking the MagohB promoter. This model predicts that loss of KMT2B would result in loss of histone acetylation and RNA polymerase II, which is precisely what we observed.
KMT2B action is not dependent on transcription or the presence of RNA polymerase II.
Our experiments using DRB and α-amanitin demonstrate that in the presence of KMT2B, RNA polymerase II recruitment and transcriptional elongation are largely dispensable for the maintenance of H3K4 trimethylation. This result is consistent with the hypothesis that MLL proteins (KMT2A to KMT2D) operate differently from yeast Set1 and have a causal role in transcriptional activation. Previous studies have hinted to such a mechanism, as depletion of KMT2A resulted in changes in RNA polymerase II distribution on KMT2A target genes (12, 13). Moreover, it has been demonstrated that CpG islands recruit H3K4 methyltransferases regardless of their transcriptional activity (40). Our experiments provide direct evidence that neither active transcription nor the presence of the RNA polymerase II preinitiation complex is required to maintain the H3K4me3 mark over a CpG island promoter, further suggesting that MLL (KMT2A to KMT2D) proteins act upstream of RNA polymerase II recruitment.
KMT2B confers protection from DNA methylation.
How CpG islands are protected from DNA methylation has been a long-standing subject of research. Our data clearly demonstrate that loss of the H3K4me3 mark results in the rapid onset of DNA methylation at multiple CpG dinucleotides over the MagohB CpG island promoter. Moreover, transcriptional elongation and RNA polymerase II did not appear to play a role in MagohB methylation protection, as illustrated by our experiments using DRB and α-amanitin. In turn, we show that reexpression of KMT2B is sufficient to induce demethylation of the methylated MagohB promoter, the reestablishment of its normal H3K4me3 pattern, and the activation of transcription (Fig. 7D and E). This indicates that once all activators are present, the RNA polymerase II complex efficiently reassociates and overrides the silent state. It has previously been demonstrated that H3K4me3 may confer protection from DNA methylation (41), as the DNA methyltransferase common subunit—DNMT3L—can interact only with unmethylated H3K4. As such, KMT2 catalytic activity would preclude DNA methyltransferase recruitment, thus conferring protection from DNA methylation to KMT2 target promoters. In addition, physically retaining KMT2B on chromatin during replication may further protect from DNA methyltransferase action. The detailed dissection of these processes is a current subject of further experimentation outside the scope of this study.
KMT2B in CpG island promoter regulation.
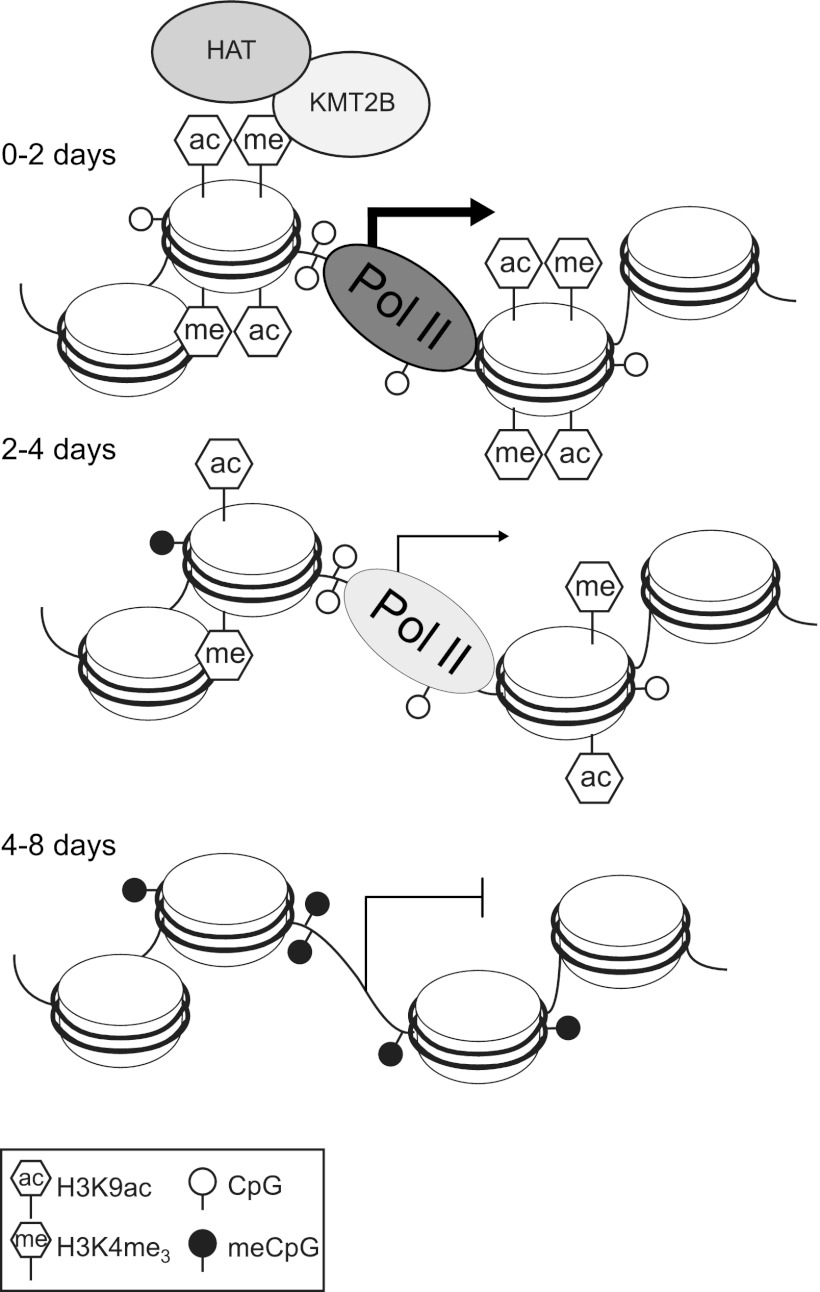
Based on our results, we propose a model for the role of KMT2B in MagohB transcriptional regulation (Fig. 8). After KMT2B depletion, the H3K4me3 mark is either actively removed or diluted over consecutive cell divisions, resulting in loss of the H3K9ac mark. RNA polymerase II can no longer associate with the MagohB promoter, as its recruitment may be facilitated or stabilized by direct interactions of TFIID with the H3K4me3 mark (38, 39) and interactions with CBP/p300-containing HAT complexes (36, 37). As the preinitiation complex is removed, transcription ceases. After the H3K4me3 mark and/or KMT2B has been removed from the MagohB promoter, DNA methyltransferases are able to target and de novo methylate the MagohB CpG island. We believe that such a scenario is avoided at most genes by the high level of redundancy within the KMT2 family of histone methyltransferases. However, the specific developmental defects of individual KMT2 family member knockouts indicate that windows in development exist where the dynamic activation of genes becomes dependent on a specific family member. Currently, the transcription factors recruiting KMT2A/B-containing complexes are not known. The MagohB promoter binds Sp1 and Sp3 in vitro, but no binding could be detected in ChIP experiments, and deletion of either factor individually did not influence MagohB mRNA levels (data not shown). Whatever recruits KMT2A/B complexes, our model helps to explain why once a transcriptional network has been destabilized in the context of tumorigenesis, CpG island promoters become vulnerable to gene silencing.
Fig 8.
Model for MagohB silencing following KMT2B depletion. KMT2B is normally recruited to the MagohB promoter by transcription factors, by interaction of the PHD fingers with H3K4me3, or through CXXC domain binding to nonmethylated CpG dinucleotides. After Mll2 deletion, H3K4me3 and H3K9ac are gradually depleted from the MagohB promoter and RNA polymerase II is lost, leading to transcriptional silencing. This is finally followed by DNA methylation and loss of the DNase I-hypersensitive site.
ACKNOWLEDGMENTS
This work was funded by a studentship grant from European Community (FP6) Integrated Project EuTRACC, grant no. LSHG-CT-2007-037445 to C.B. and A.F.S., a grant from the City of Hope Medical Center to A.D.R. and C.B., and a Leukemia & Lymphoma Research grant to C.B.
V.L. performed experiments, M.H., H.H., A.F.S., and C.B. designed experiments, H.H. and A.F.S. provided essential reagents, and V.L., A.D.R., A.F.S., and C.B. wrote the paper.
Footnotes
Published ahead of print 28 January 2013
REFERENCES
- 1. Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res. 21: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ruthenburg AJ, Allis CD, Wysocka J. 2007. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 25: 15–30 [DOI] [PubMed] [Google Scholar]
- 3. Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ. 2004. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr. Biol. 14: 2063–2069 [DOI] [PubMed] [Google Scholar]
- 4. Yagi H, Deguchi K, Aono A, Tani Y, Kishimoto T, Komori T. 1998. Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood 92: 108–117 [PubMed] [Google Scholar]
- 5. Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, Tufteland KR, Aasland R, Anastassiadis K, Ang SL, Stewart AF. 2006. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development 133: 1423–1432 [DOI] [PubMed] [Google Scholar]
- 6. Collins EC, Rabbitts TH. 2002. The promiscuous MLL gene links chromosomal translocations to cellular differentiation and tumour tropism. Trends Mol. Med. 8: 436–442 [DOI] [PubMed] [Google Scholar]
- 7. Daser A, Rabbitts TH. 2005. The versatile mixed lineage leukaemia gene MLL and its many associations in leukaemogenesis. Semin. Cancer Biol. 15: 175–188 [DOI] [PubMed] [Google Scholar]
- 8. Slany RK. 2005. When epigenetics kills: MLL fusion proteins in leukemia. Hematol. Oncol. 23: 1–9 [DOI] [PubMed] [Google Scholar]
- 9. Slany RK. 2009. The molecular biology of mixed lineage leukemia. Haematologica 94: 984–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smith E, Lin C, Shilatifard A. 2011. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 25: 661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ng HH, Robert F, Young RA, Struhl K. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- 12. Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. 2005. MLL associates specifically with a subset of transcriptionally active target genes. Proc. Natl. Acad. Sci. U. S. A. 102: 14765–14770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, Ge K, Krumlauf R, Shilatifard A. 2009. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol. Cell. Biol. 29: 6074–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Terranova R, Agherbi H, Boned A, Meresse S, Djabali M. 2006. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc. Natl. Acad. Sci. U. S. A. 103: 6629–6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Glaser S, Lubitz S, Loveland KL, Ohbo K, Robb L, Schwenk F, Seibler J, Roellig D, Kranz A, Anastassiadis K, Stewart AF. 2009. The histone 3 lysine 4 methyltransferase, Mll2, is only required briefly in development and spermatogenesis. Epigenetics Chromatin 2: 5 doi:10.1186/1756-8935-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lubitz S, Glaser S, Schaft J, Stewart AF, Anastassiadis K. 2007. Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Mol. Biol. Cell 18: 2356–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pfeifer GP, Riggs AD. 1991. Chromatin differences between active and inactive X chromosomes revealed by genomic footprinting of permeabilized cells using DNase I and ligation-mediated PCR. Genes Dev. 5: 1102–1113 [DOI] [PubMed] [Google Scholar]
- 18. Cockerill PN. 2000. Identification of DNaseI hypersensitive sites within nuclei. Methods Mol. Biol. 130: 29–46 [DOI] [PubMed] [Google Scholar]
- 19. Tagoh H, Cockerill PN, Bonifer C. 2006. In vivo genomic footprinting using LM-PCR methods. Methods Mol. Biol. 325: 285–314 [DOI] [PubMed] [Google Scholar]
- 20. Forsberg EC, Downs KM, Christensen HM, Im H, Nuzzi PA, Bresnick EH. 2000. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. U. S. A. 97: 14494–14499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Logie C, Stewart AF. 1995. Ligand-regulated site-specific recombination. Proc. Natl. Acad. Sci. U. S. A. 92: 5940–5944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Workman JL, Roeder RG. 1987. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell 51: 613–622 [DOI] [PubMed] [Google Scholar]
- 23. Almer A, Rudolph H, Hinnen A, Horz W. 1986. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 5: 2689–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fletcher TM, Xiao N, Mautino G, Baumann CT, Wolford R, Warren BS, Hager GL. 2002. ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol. Cell. Biol. 22: 3255–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reik A, Schutz G, Stewart AF. 1991. Glucocorticoids are required for establishment and maintenance of an alteration in chromatin structure: induction leads to a reversible disruption of nucleosomes over an enhancer. EMBO J. 10: 2569–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Richard-Foy H, Hager GL. 1987. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 6: 2321–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Axel R. 1975. Cleavage of DNA in nuclei and chromatin with staphylococcal nuclease. Biochemistry 14: 2921–2925 [DOI] [PubMed] [Google Scholar]
- 28. Clark RJ, Felsenfeld G. 1971. Structure of chromatin. Nat. New Biol. 229: 101–106 [DOI] [PubMed] [Google Scholar]
- 29. Lefevre P, Witham J, Lacroix CE, Cockerill PN, Bonifer C. 2008. The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol. Cell 32: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fenouil R, Cauchy P, Koch F, Descostes N, Cabeza JZ, Innocenti C, Ferrier P, Spicuglia S, Gut M, Gut I, Andrau JC. 2012. CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters. Genome Res. 22: 2399–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Petruk S, Sedkov Y, Johnston DM, Hodgson JW, Black KL, Kovermann SK, Beck S, Canaani E, Brock HW, Mazo A. 2012. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell 150: 922–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blobel GA, Kadauke S, Wang E, Lau AW, Zuber J, Chou MM, Vakoc CR. 2009. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol. Cell 36: 970–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fraser NW, Sehgal PB, Darnell JE. 1978. DRB-induced premature termination of late adenovirus transcription. Nature 272: 590–593 [DOI] [PubMed] [Google Scholar]
- 34. Marshall NF, Peng J, Xie Z, Price DH. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271: 27176–27183 [DOI] [PubMed] [Google Scholar]
- 35. Nguyen VT, Giannoni F, Dubois MF, Seo SJ, Vigneron M, Kedinger C, Bensaude O. 1996. In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res. 24: 2924–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cosgrove MS, Patel A. 2010. Mixed lineage leukemia: a structure-function perspective of the MLL1 protein. FEBS J. 277: 1832–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chan HM, La Thangue NB. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114: 2363–2373 [DOI] [PubMed] [Google Scholar]
- 38. van Ingen H, van Schaik FM, Wienk H, Ballering J, Rehmann H, Dechesne AC, Kruijzer JA, Liskamp RM, Timmers HT, Boelens R. 2008. Structural insight into the recognition of the H3K4me3 mark by the TFIID subunit TAF3. Structure 16: 1245–1256 [DOI] [PubMed] [Google Scholar]
- 39. Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131: 58–69 [DOI] [PubMed] [Google Scholar]
- 40. Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, Turner DJ, Illingworth R, Bird A. 2010. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464: 1082–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]