Abstract
Immunoglobulin (Ig) class switch recombination (CSR) is initiated by activation-induced cytidine deaminase (AID) that catalyzes numerous DNA cytosine deaminations within switch regions. The resulting uracils are processed by uracil base excision and/or mismatch repair enzymes that ultimately generate switch region DNA double-strand breaks (DSBs). Uracil glycosylase 2 (UNG2) is required for CSR, most likely by removing uracils to generate abasic sites. Although it is presumed that the apurinic/apyrimidinic endonuclease 1 (APE1) generates DNA strand incisions (a prerequisite for CSR) at these abasic sites, a direct test of the requirement for APE1 in CSR has been difficult because of the embryonic lethality of APE1 ablation in mice. Here, we report the successful deletion of the APE1 gene in a mouse B cell line (CH12F3) capable of robust CSR in vitro. In contrast to the general assumption that APE1 is essential for cellular viability, deletion of APE1 in CH12F3 cells has no apparent effect on cell viability or growth. Moreover, CSR in APE1-null CH12F3 cells is drastically reduced, providing direct evidence for an essential role for APE1 in switch region cleavage and CSR. Finally, deletion of AP endonuclease 2 (APE2) has no effect on CSR in either APE1-proficient or -deficient cells.
INTRODUCTION
In antigen-stimulated B cells, somatic hypermutation (SHM) diversifies the variable regions of immunoglobulin (Ig) heavy (H) and light (L) chain genes, whereas class switch recombination (CSR) provides a mechanism to diversify the constant region of the Ig heavy chain (1, 2). SHM introduces point mutations into the Ig variable (V) regions, which is the mechanistic basis for Ig affinity maturation. CSR introduces DNA double-strand breaks (DSBs) into donor and acceptor switch (S) regions; subsequently a “cut-and-paste” mechanism results in intrachromosomal deletion. Thus, CSR juxtaposes a previously distal constant region to the V region, allowing a “switch” of the class (or isotype) of the expressed immunoglobulin without altering its antigen specificity (1, 2).
Both SHM and CSR require activation-induced cytidine deaminase (AID) (3, 4), which deaminates DNA cytosines to uracils at transcribed V and S regions (1, 2, 5). Uracils in DNA can be detected and repaired by either the uracil glycosylase (UNG2)-dependent base excision repair (BER) pathway or by the MSH2-dependent mismatch repair (MMR) pathway. Indeed, deletion of either UNG2 or MSH2 reduces CSR efficiency (6, 7) and deletion of both completely abolishes CSR (8). However, in contrast to the normal high-fidelity repair that restores DNA to its original state, processing of AID-generated uracils leads to mutations in the V region during SHM and DNA double-strand breaks in switch regions during CSR. It is therefore unclear whether the BER and MMR factors that are involved in SHM and CSR have been usurped to function differently (without their characteristic high fidelity) in germinal center B cells or whether perhaps only some of the components in either pathway are utilized.
A noticeable difference between SHM and CSR with regard to uracil processing is that SHM, but not CSR, can occur without uracil removal or DNA strand incision. DNA replication over uracil is sufficient to introduce C-to-T mutation, albeit not the entire spectrum of mutations is seen in the normal SHM process (7, 9). CSR, on the other hand, requires DNA cleavage of not one, but both DNA strands. It is generally conjectured that DNA incisions during CSR are mediated by APE1, which nicks the DNA at abasic sites after uracil removal by UNG2. It was hypothesized that closely positioned single-strand breaks across the two DNA strands can spontaneously form DSBs (1, 2, 10). Mammals have several uracil glycosylases (11), but only one enzyme that has substantial AP endonuclease activity (APE1) (12). APE1 cleaves the phosphodiester bond at the 5′ end of the abasic site to yield a 3′-OH and a 5′-deoxyribophosphate on either end of the single-strand break. Unlike UNG, the role of APE1 in CSR has been difficult to test by traditional genetic approaches because APE1 is essential for mouse embryonic development (13, 14) and was reported to be essential for viability of several human cell lines (15). Recently, a minor AP endonuclease (APE2) was identified based on homology search (16). APE2 is a very weak AP endonuclease in vitro, and its role in BER or CSR is uncertain (17).
Two earlier studies that attempted to address the role of APE1 in CSR yielded conflicting results (18, 19). In one study, a small decrease of CSR efficiency was reported in APE1-haplodeficient mice (18). In the other study, small interfering RNA was used to transiently deplete APE1 in a CSR-competent mouse B cell line (CH12F3); with this approach, CSR was not affected (19). Both studies also examined CSR in APE2-deficient mice, again with contradictory conclusions (18–20).
In this study, we attempted somatic gene targeting of APE1 and APE2 in the CH12F3 cell line. To our surprise, complete deletion of the APE1 gene is fully compatible with CH12F3 viability and growth. Here, we have measured CSR in cells completely lacking Ape1. Whereas ablation of APE1 drastically reduces (∼5-fold) CSR efficiency in CH12F3 cells, deletion of APE2 has no effect on CSR (in either APE1-proficient or -deficient cells). These data provide strong and direct evidence for an essential role for APE1 in CSR and further strengthen the DNA deaminase model for AID function in CSR.
MATERIALS AND METHODS
Reagents.
APE1 antibody (10203-1-AP) was purchased from Proteintech Group, Inc. (Chicago, IL). APE2 antibody was kindly provided by Janet Stavnezer (University of Massachusetts Medical School, Worcester, MA). Antibody against mouse β-actin (sc-47778) was purchased from Santa Cruz Biotech (Santa Cruz, CA).
Cell culture and CSR assay.
CH12F3 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum and 50 μM β-mercaptoethanol. For the CSR assay, healthy CH12F3 cells were seeded at 5 × 104 cells/ml in the presence of 1 μg/ml anti-CD40 antibody (eBioscience 16-0402-86), 5 ng/ml of interleukin-4 (IL-4) (R&D Systems 404-ML), and 0.5 ng/ml transforming growth factor β1 (TGF-β1) (R&D Systems 240-B), and grown for 72 h. Cells were stained with a fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgA antibody (BD Biosciences 559354) and analyzed on an LSR II flow cytometer (BD Biosciences). CSR efficiency is determined as the percentage of IgA-positive cells.
APE1 gene targeting.
A 10.8-kb BglII fragment containing the APE1 gene was isolated from bacterial artificial chromosome (BAC) RP23-243M12 (GenBank accession no. AC136376) and cloned into the targeting vector backbone. The 1.7-kb BclI-BsrGI fragment containing exon 4 of the APE1 gene was replaced with a floxed puromycin selection cassette to obtain the final targeting vector, pLH5. Gene-targeting procedures have been described previously (21).
Genetic complementation.
APE1 coding region sequences were amplified from CH12F3 cDNA and cloned into the retroviral transfer vector pMSCV-puro (Clontech Laboratories, Inc.). Recombinant retroviruses were prepared in the Phoenix packaging cell line and used to infect APE1Δ/Δ/Δ cells (see Results). Infected cells were selected by puromycin and subjected to Western blot and CSR assays.
MMS sensitivity assay.
Cells were seeded at 100,000/ml per well in a 24-well plate, and methyl methanesulfonate (MMS) was added to various concentrations. After 48 h of growth, cell viability was determined by a colorimetric assay that measures the metabolic conversion of thiazolyl blue tetrazolium bromide (MTT) in the mitochondria of living cells. The details of the MTT assay have been described previously (21).
Switch junction analysis.
Individual IgA-positive clones were isolated by limiting dilutions of cytokine-stimulated cultures in 96-well plates. Switch junctions were amplified with primers KY761 (5′-AACTCTCCAGCCACAGTAATGACC-3′) and KY743 (5′-GAGCTCGTGGGAGTGTCAGTG-3′) as previously described (22–24). PCR products were sequenced at the Genomic Core Facility at Michigan State University.
RESULTS
Gene targeting of the APE1 gene in CH12F3 cells.
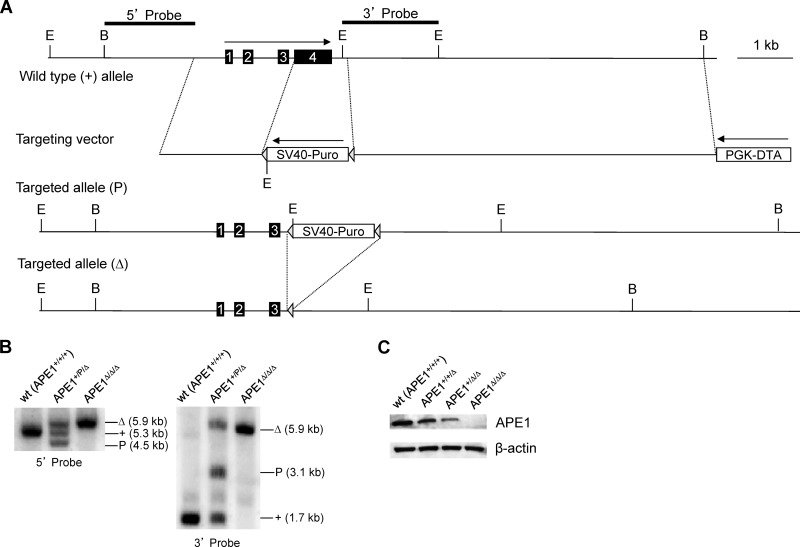
Gene targeting, facilitating specific and complete ablation of various cellular factors, has provided a powerful tool to test cellular function. Although it has been reported that depletion of APE1 results in cellular lethality in a number of human cell lines (15) and mouse fibroblasts (25), we considered that the lethality might be species or cell type specific or could be rescued by reducing oxidative stress (26) (e.g., growing cells in a low-oxygen environment and/or with antioxidants). We therefore designed a gene-targeting vector (Fig. 1A) to attempt disruption of the APE1 gene in a mouse B cell line (CH12F3) that is capable of robust cytokine-induced CSR. The gene-targeting strategy utilized has been described previously (21–24). Gene targeting would delete exon 4 of the APE1 gene, which contains ∼52% of the coding sequence (Fig. 1A). The targeted allele containing the puromycin selection cassette is designated “P.” Upon transient transfection of a Cre-expressing plasmid, the floxed puromycin marker can be excised to yield a delta (Δ) allele. The conversion from “P” to “Δ” allows another round of gene targeting using the same targeting vector. During the course of the experiment, it became apparent that CH12F3 cells have three copies of the APE1 gene (Fig. 1B). Therefore, three rounds of gene targeting were required to completely delete the APE1 gene from the CH12F3 genome. The sequential orders of genotypes during the gene-targeting experiment are as follows: +/+/+, +/+/P, +/+/Δ, +/P/Δ, +/Δ/Δ, P/Δ/Δ, and Δ/Δ/Δ. A progressive reduction in APE1 protein level was observed with the decreasing copy number of the APE1 gene as shown by Western blot analysis (Fig. 1C). APE1Δ/Δ/Δ cells have no detectable APE1 protein, indicating that the targeted allele is a null allele.
Fig 1.
Gene targeting of APE1 in CH12F3 cells. (A) Genomic organization of the wild-type and targeted APE1 allele. The map is drawn to scale. Exons are indicated by numbered boxes. Arrows indicate transcription orientations of expression cassettes of the puromycin-resistant gene (Puro) and diphtheria toxin A chain (DTA), respectively. SV40, simian virus 40 early promoter; PGK, phosphoglycerate kinase promoter. Restriction sites: B, BglII; E, EcoRI. Probes used in Southern blot analysis are depicted at the top. A “+” sign indicates the wild-type allele. “P” and “Δ” indicate a targeted allele with or without the puromycin resistance gene cassette, respectively. Triangles indicate loxP sites. (B) Southern blot analysis of EcoRI-digested genomic DNA from wild-type (wt) and targeted cells. Genotypes, sizes of bands, and probes are indicated. (C) Western blot analysis of the APE1 protein level in cells containing three (+/+/+), two (+/+/Δ), one (+/Δ/Δ), and zero (Δ/Δ/Δ) copies of APE1, respectively.
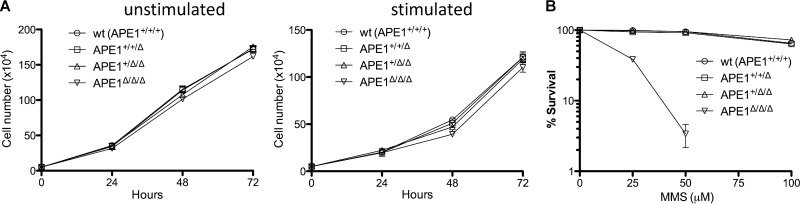
It is well appreciated that cells with proliferative defects switch poorly; thus, to determine whether APE1 deficiency affects proliferation of CH12F3 cells, live cells were counted over a span of 3 days in cultures with (stimulated) or without (unstimulated) cytokines. As can be seen, normal cell proliferation was observed in all cultures regardless of the APE1 copy number (Fig. 2A). APE1 is a core component of the BER pathway that is essential for repair of DNA base damage (e.g., base adducts). As expected, APE1-null CH12F3 cells are remarkably hypersensitive to the DNA-alkylating agent methyl methanesulfonate (Fig. 2B), consistent with a defect in BER.
Fig 2.
Proliferation and base excision repair in APE1-deficient cells. (A) Live cell (trypan exclusion) counts over a span of 3 days in unstimulated or stimulated cultures. Error bars indicate standard errors of three independent experiments. wt, wild type. (B) Cells were grown in the presence of various concentrations of methyl methanesulfonate for 2 days, and cell survival was determined by metabolic conversion of thiazolyl blue tetrazolium bromide (MTT) in the mitochondria of living cells.
Class switching in APE1-null cells.
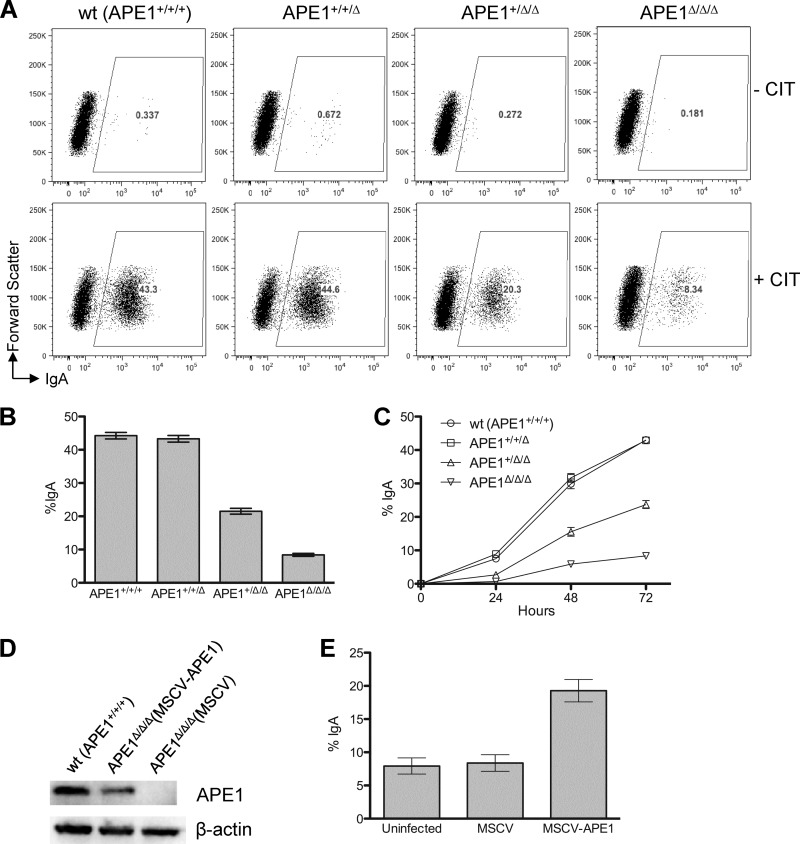
To determine whether APE1 is required for CSR, CSR efficiency was determined for CH12F3 cells containing three (+/+/+), two (+/+/Δ), one (+/Δ/Δ), and zero (Δ/Δ/Δ) copies of APE1. As can be seen, reduction of APE1 copy numbers from three (+/+/+) to two (+/+/Δ) reduces the APE1 protein level by ∼30% (Fig. 1C), but has little effect on cell proliferation (Fig. 2A) or CSR efficiency (Fig. 3A and B). Consistent with the study from Guikema et al. (18), further reduction to one copy (+/Δ/Δ) reduces the protein level by ∼60% (Fig. 1C) and reduces CSR efficiency by ∼50% (Fig. 3A and B). Importantly, complete deletion of APE1 (Δ/Δ/Δ) reduces CSR efficiency to ∼20% of the wild-type level (Fig. 3A and B), providing direct evidence that APE1 is required for efficient CSR. To analyze the kinetics of CSR in cells with different levels of APE1, CSR efficiency was measured at a 24-h interval over a span of 3 days. As can be seen, the impact of APE1 deficiency on CSR efficiency is even more apparent at earlier (∼10-fold at 24 h) time points (Fig. 3C).
Fig 3.
APE1 affects CSR efficiency. (A) Fluorescence-activated cell sorter (FACS) analysis of CSR by cell surface staining of IgA after 3 days of cell growth with (+CIT) or without (−CIT) cytokines. CIT, anti-CD40, interleukin-4, and TGF-β1; wt, wild type. (B) CSR efficiency (determined as the percentage of IgA+ cells) in CH12F3 cells containing 0 to 3 copies of APE1. Error bars indicate standard errors of 10 independent experiments. (C) APE1 affects CSR kinetics. CSR was monitored every 24 h over a span of 3 days. (D) Genetic complementation of APE1-null cells. Western blot analysis of transgenic APE1 expression in retrovirus-transduced APE1-null cells. (E) CSR in retrovirus-transduced APE1-null cells.
To confirm that the observed CSR defect is indeed due to the lack of APE1, a genetic complementation assay was performed. A mouse stem cell virus (MSCV)-based retroviral vector was used to transduce the mouse APE1 gene back into APE1Δ/Δ/Δ cells. In this experiment, expression of the APE1 transgene does not reach the level of the wild-type cells (APE1+/+/+), but is comparable to that of the APE1+/Δ/Δ cells (Fig. 3D). Correspondingly, CSR efficiency is restored to the level of the APE1+/Δ/Δ cells (Fig. 3E). In contrast, APE1Δ/Δ/Δ cells transduced with an empty vector show no increase in CSR efficiency (Fig. 3E). These data confirm that the CSR defect in APE1Δ/Δ/Δ cells is intrinsic to the level of APE1.
To determine if APE1 deficiency affects either the targeting or joining of DSBs, switch junctions were isolated from postswitched IgA cell clones and sequenced. No significant difference was noted with regard to nucleotide overlaps (see Fig. S1A in the supplemental material; Mann-Whitney test, P = 0.88) or breakpoint locations within switch regions (see Fig. S1B and C).
APE2 does not contribute to class switch recombination.
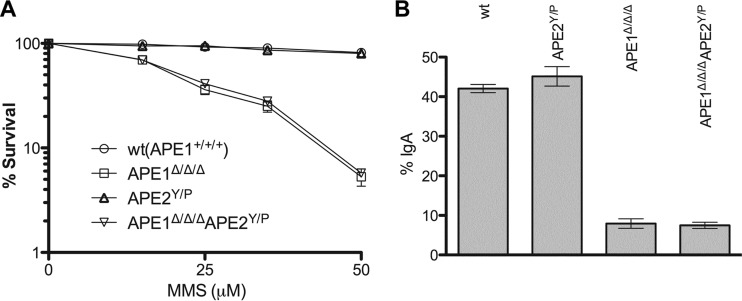
Mammals have two type II AP endonucleases (that are homologous to the prototype Escherichia coli Exo III). While APE1 is the dominant (>95%) activity in the cell (12), APE2 is a very weak endonuclease (17). Because deletion of APE1 in CH12F3 does not completely abolish CSR, we considered whether the residual CSR activity is mediated by APE2. To address this possibility, we ablated the APE2 gene in both wild-type and APE1-null CH12F3 cells. APE2 is located on the X chromosome (27). CH12F3 cells are male derived (XY); thus, one round of gene targeting was sufficient to delete APE2. The resulting line is designated “Y/P” as the puromycin-resistant gene cassette on the targeted X chromosome was not removed (Fig. 4A and B). APE2Y/P cells contain no detectable APE2 protein (Fig. 4C), indicating that gene targeting generates a null allele. It has been difficult to determine whether APE2 plays any role in BER because of the presence of the more dominant activity of APE1. To determine if APE2 participates in BER in CH12F3 cells, MMS sensitivity was measured for cells that are deficient for both APE1 and APE2. As can be seen, deletion of APE2 in APE1-deficient cells does not further increase cell sensitivity to MMS (Fig. 5A), suggesting that APE2 does not function in BER. We next assessed the impact of APE2 on CSR in CH12F3 cells. As can be seen, APE2Y/P cells switch as efficiently as wild-type cells (Fig. 5B). More importantly, APE1Δ/Δ/Δ APE2Y/P cells switch as efficiently as APE1Δ/Δ/Δ cells (Fig. 5B). We conclude that APE2 is not involved in CSR in the CH12F3 cell line.
Fig 4.
Gene targeting of APE2 in CH12F3 cells. (A) Genomic organization of the wild-type and targeted APE2 allele on the X chromosome. The map is drawn to scale. The symbols used here are the same as in Fig. 1. Restriction site: B, BamHI. (B) Southern blot analysis of BamHI-digested genomic DNA. Genotypes, sizes of bands, and probes are indicated. (C) Western blot analysis of the protein level in cells deficient for APE1, APE2, or both. WT, wild type.
Fig 5.
BER and CSR in APE2-null cells. (A) Cells were grown in the presence of various concentrations of methyl methanesulfonate for 2 days, and cell survival was determined by metabolic conversion of thiazolyl blue tetrazolium bromide (MTT) in the mitochondria of living cells. (B) CSR in cells deficient for APE1, APE2, or both.
DISCUSSION
In many microorganisms, deletion of the major AP endonuclease activity causes no apparent growth defect (15). In contrast, a complete deletion of APE1 appears to be incompatible with life in mammals (15, 25). Prior to this study, no APE1-null cell line had been established. An earlier study demonstrated cellular lethality in four different human cell lines upon prolonged inhibition of APE1 expression by retrovirus-transduced short hairpin RNA (shRNA) expression (15). Another study showed that excision of a floxed human APE1 transgene led to rapid cell death in mouse fibroblasts in which the endogenous APE1 gene had been deleted (25). Thus, the established consensus is that APE1 is indispensable for mammalian cell growth, even in culture. Because of this potential cellular lethality, the gene-targeting experiments described here were originally initiated with the integration of a tetracycline-regulated human APE1 transgene (“Tet-Off” system, prior to the deletion of the endogenous APE1 in CH12F3). However, it became apparent that the transgene could be completely turned off without affecting cell growth. Thus, for simplicity, the gene-targeting experiments were repeated without incorporating the human transgene first. We confirmed that APE1 is indeed dispensable for the viability of CH12F3 cells. The establishment of an APE1-null cell line should facilitate studies of APE1 function in other biological processes. Although the clear dispensability of APE1 in this B cell line (and not a variety of other cell lines) is intriguing, there are no obvious explanations for this cell type difference.
The cellular function of APE2 is unclear. Whether APE2 participates in BER has been difficult to assess because of its very weak AP endonuclease activity and the presence of the more dominant activity of APE1. The establishment of an APE1-null cell line allows, for the first time, a direct test of APE2 in BER in living cells. We show that APE2 deletion does not enhance cellular sensitivity to MMS in either APE1-proficient or -deficient cells, suggesting that APE2 does not have a significant role in BER.
Since the establishment of the DNA deaminase model for AID function in SHM and CSR, APE1 has been inferred as the enzyme that acts downstream of UNG2 to cleave DNA strands during CSR (28). However, definitive evidence supporting this hypothesis has been lacking. Indeed, two earlier studies on this topic reached contradictory conclusions (18–20). One study reported a small reduction of CSR efficiency in APE1-haplodeficient as well as APE2-deficient mice, suggesting participation of both APE1 and APE2 in CSR (18). However, another study (using siRNA) showed no effect on CSR in CH12F3 cells with reduced APE1 expression or in APE2-deficient mice. The latter study argued that neither APE1 nor APE2 is involved in CSR (19). In the present study, cell lines were generated with complete depletion of APE1, APE2, or both. We conclude that APE1 has an essential role in CSR and that APE2 does not contribute to CSR.
The severe CSR defect in APE1-null CH12F3 cells provides compelling evidence that APE1 confers the primary DNA incision activity during CSR; we conclude that the “AID-UNG2-APE1” axis is the major pathway to generate DSBs in switch regions. We recently showed that overlapping AID hot spots (WGCW) in switch regions are critical for efficient CSR (23). We hypothesized that this motif might facilitate DSB formation by colocalizing nicks generated via the AID-UNG2-APE1 pathway. The demonstration of the essential role of APE1 in CSR in this study is consistent with that hypothesis. Our data also provide insight into a previous debate as to whether UNG2 functions in CSR as a scaffold protein (29, 30) or as an active enzyme (31). Since APE1 is required for CSR, it is difficult to conceive how UNG2 could function in CSR without catalyzing uracil excision.
The DNA incision activity in APE1-null cells during CSR is currently unidentified. It is possible that some unidentified enzymatic activity or activities cleave at the abasic sites in a way analogous to APE1 or that abasic sites are just intrinsically unstable; however, previous reports have shown that mismatch recognition and base excision cooperate in processing uracils generated by AID during CSR. We think it is more likely that MSH2-dependent MMR (or a variation of MMR) accounts for the remaining CSR activity in UNG2- or APE1-deficient cells. In fact, the level of residual switching observed in APE1-deficient cells in this study is remarkably similar to that observed in cells deficient in UNG2. MMR has been shown to be particularly important when core switch repeats were deleted and overlapping AID hot spots were scarce (10, 32, 33). Among the MMR factors, the MutLα (MLH1/PMS2) complex has been reported to possess a latent endonuclease activity that is activated upon mismatch recognition (34). Exonuclease I, another MMR factor known to be involved in CSR (35), could be loaded at MutLα-inflicted nicks and initiate strand excision in the 5′-to-3′ direction. In this situation, converging Exo I excision on the two DNA strands would then yield a DSB to initiate CSR.
Supplementary Material
ACKNOWLEDGMENTS
We thank Janet Stavnezer for the APE2 antibody.
This work is supported by a National Institutes of Health grant (R01 AI081817) to K. Yu.
We declare we have no conflicts of interest.
Footnotes
Published ahead of print 4 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00026-13.
REFERENCES
- 1. Chaudhuri J, Alt FW. 2004. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 4: 541– 552 [DOI] [PubMed] [Google Scholar]
- 2. Yu K, Lieber MR. 2003. Nucleic acid structures and enzymes in the immunoglobulin class switch recombination mechanism. DNA Repair (Amst.) 2: 1163– 1174 [DOI] [PubMed] [Google Scholar]
- 3. Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102: 553– 563 [DOI] [PubMed] [Google Scholar]
- 4. Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell 102: 565– 575 [DOI] [PubMed] [Google Scholar]
- 5. Maul RW, Saribasak H, Martomo SA, McClure RL, Yang W, Vaisman A, Gramlich HS, Schatz DG, Woodgate R, Wilson DM, III, Gearhart PJ. 2011. Uracil residues dependent on the deaminase AID in immunoglobulin gene variable and switch regions. Nat. Immunol. 12: 70– 76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ehrenstein MR, Neuberger MS. 1999. Deficiency in Msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombination: parallels with somatic hypermutation. EMBO J. 18: 3484– 3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12: 1748– 1755 [DOI] [PubMed] [Google Scholar]
- 8. Rada C, Di Noia JM, Neuberger MS. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell 16: 163– 171 [DOI] [PubMed] [Google Scholar]
- 9. Imai K, Catalan N, Plebani A, Marodi L, Sanal O, Kumaki S, Nagendran V, Wood P, Glastre C, Sarrot-Reynauld F, Hermine O, Forveille M, Revy P, Fischer A, Durandy A. 2003. Hyper-IgM syndrome type 4 with a B lymphocyte-intrinsic selective deficiency in Ig class-switch recombination. J. Clin. Invest. 112: 136– 142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Min IM, Rothlein LR, Schrader CE, Stavnezer J, Selsing E. 2005. Shifts in targeting of class switch recombination sites in mice that lack mu switch region tandem repeats or Msh2. J. Exp. Med. 201: 1885– 1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krokan HE, Drablos F, Slupphaug G. 2002. Uracil in DNA—occurrence, consequences and repair. Oncogene 21: 8935– 8948 [DOI] [PubMed] [Google Scholar]
- 12. Demple B, Harrison L. 1994. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 63: 915– 948 [DOI] [PubMed] [Google Scholar]
- 13. Ludwig DL, MacInnes MA, Takiguchi Y, Purtymun PE, Henrie M, Flannery M, Meneses J, Pedersen RA, Chen DJ. 1998. A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity. Mutat. Res. 409: 17– 29 [DOI] [PubMed] [Google Scholar]
- 14. Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. 1996. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl. Acad. Sci. U. S. A. 93: 8919– 8923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fung H, Demple B. 2005. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol. Cell 17: 463– 470 [DOI] [PubMed] [Google Scholar]
- 16. Hadi MZ, Wilson DM., III 2000. Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III. Environ. Mol. Mutagen. 36: 312– 324 [PubMed] [Google Scholar]
- 17. Burkovics P, Szukacsov V, Unk I, Haracska L. 2006. Human Ape2 protein has a 3′-5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res. 34: 2508– 2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guikema JE, Linehan EK, Tsuchimoto D, Nakabeppu Y, Strauss PR, Stavnezer J, Schrader CE. 2007. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J. Exp. Med. 204: 3017– 3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sabouri Z, Okazaki IM, Shinkura R, Begum N, Nagaoka H, Tsuchimoto D, Nakabeppu Y, Honjo T. 2009. Apex2 is required for efficient somatic hypermutation but not for class switch recombination of immunoglobulin genes. Int. Immunol. 21: 947– 955 [DOI] [PubMed] [Google Scholar]
- 20. Guikema JE, Stavnezer J, Schrader CE. 2010. The role of Apex2 in class-switch recombination of immunoglobulin genes. Int. Immunol. 22: 213– 214 (Letter.) [DOI] [PubMed] [Google Scholar]
- 21. Han L, Yu K. 2008. Altered kinetics of NHEJ and CSR in DNA ligase IV-deficient B cells. J. Exp. Med. 205: 2745– 2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Han L, Masani S, Yu K. 2010. Cutting edge: CTNNBL1 is dispensable for Ig class switch recombination. J. Immunol. 185: 1379– 1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han L, Masani S, Yu K. 2011. Overlapping activation-induced cytidine deaminase hotspot motifs in Ig class-switch recombination. Proc. Natl. Acad. Sci. U. S. A. 108: 11584– 11589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Han L, Mao W, Yu K. 2012. X-ray repair cross-complementing protein 1 (XRCC1) deficiency enhances class switch recombination and is permissive for alternative end joining. Proc. Natl. Acad. Sci. U. S. A. 109: 4604– 4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Izumi T, Brown DB, Naidu CV, Bhakat KK, Macinnes MA, Saito H, Chen DJ, Mitra S. 2005. Two essential but distinct functions of the mammalian abasic endonuclease. Proc. Natl. Acad. Sci. U. S. A. 102: 5739– 5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meira LB, Devaraj S, Kisby GE, Burns DK, Daniel RL, Hammer RE, Grundy S, Jialal I, Friedberg EC. 2001. Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Res. 61: 5552– 5557 [PubMed] [Google Scholar]
- 27. Ide Y, Tsuchimoto D, Tominaga Y, Iwamoto Y, Nakabeppu Y. 2003. Characterization of the genomic structure and expression of the mouse Apex2 gene. Genomics 81: 47– 57 [DOI] [PubMed] [Google Scholar]
- 28. Petersen-Mahrt SK, Harris RS, Neuberger MS. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418: 99– 103 [DOI] [PubMed] [Google Scholar]
- 29. Begum NA, Kinoshita K, Kakazu N, Muramatsu M, Nagaoka H, Shinkura R, Biniszkiewicz D, Boyer LA, Jaenisch R, Honjo T. 2004. Uracil DNA glycosylase activity is dispensable for immunoglobulin class switch. Science 305: 1160– 1163 [DOI] [PubMed] [Google Scholar]
- 30. Begum NA, Stanlie A, Doi T, Sasaki Y, Jin HW, Kim YS, Nagaoka H, Honjo T. 2009. Further evidence for involvement of a noncanonical function of uracil DNA glycosylase in class switch recombination. Proc. Natl. Acad. Sci. U. S. A. 106: 2752– 2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Di Noia JM, Williams GT, Chan DT, Buerstedde JM, Baldwin GS, Neuberger MS. 2007. Dependence of antibody gene diversification on uracil excision. J. Exp. Med. 204: 3209– 3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Min IM, Schrader CE, Vardo J, Luby TM, D'Avirro N, Stavnezer J, Selsing E. 2003. The Smu tandem repeat region is critical for Ig isotype switching in the absence of Msh2. Immunity 19: 515– 524 [DOI] [PubMed] [Google Scholar]
- 33. Min IM, Selsing E. 2005. Antibody class switch recombination: roles for switch sequences and mismatch repair proteins. Adv. Immunol. 87: 297– 328 [DOI] [PubMed] [Google Scholar]
- 34. Kadyrov FA, Dzantiev L, Constantin N, Modrich P. 2006. Endonucleolytic function of MutLalpha in human mismatch repair. Cell 126: 297– 308 [DOI] [PubMed] [Google Scholar]
- 35. Bardwell PD, Woo CJ, Wei K, Li Z, Martin A, Sack SZ, Parris T, Edelmann W, Scharff MD. 2004. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat. Immunol. 5: 224– 229 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.