Abstract
Membrane localization of Rho GTPases is essential for their biological functions and is dictated in part by a series of posttranslational modifications at a carboxyl-terminal CaaX motif: prenylation at cysteine, proteolysis of the aaX tripeptide, and carboxymethylation. The fidelity and variability of these CaaX processing steps are uncertain. The brain-specific splice variant of Cdc42 (bCdc42) terminates in a CCIF sequence. Here we show that brain Cdc42 undergoes two different types of posttranslational modification: classical CaaX processing or novel tandem prenylation and palmitoylation at the CCaX cysteines. In the dual lipidation pathway, bCdc42 was prenylated, but it bypassed proteolysis and carboxymethylation to undergo modification with palmitate at the second cysteine. The alternative postprenylation processing fates were conserved in the GTPases RalA and RalB and the phosphatase PRL-3, proteins terminating in a CCaX motif. The differentially modified forms of bCdc42 displayed functional differences. Prenylated and palmitoylated brain Cdc42 did not interact with RhoGDIα and was enriched in the plasma membrane relative to the classically processed form. The alternative processing of prenylated CCaX motif proteins by palmitoylation or by endoproteolysis and methylation expands the diversity of signaling GTPases and enables another level of regulation through reversible modification with palmitate.
INTRODUCTION
Cdc42 is a Rho GTPase that regulates diverse cellular functions, including cell polarity, migration, and progression through the cell cycle (1–3). Like all monomeric GTPases, Cdc42 functions as a molecular switch, cycling between the GDP-bound inactive state and the GTP-bound active state. This GTPase cycle is regulated by guanine nucleotide exchange factors that stimulate nucleotide exchange and GTPase-activating proteins that accelerate intrinsic GTPase activity. GTP-bound GTPases preferentially bind to effector proteins and activate downstream signaling events. An additional level of regulation is imposed on Rho GTPases by binding to RhoGDI, which sequesters inactive Rho proteins in the cytoplasm (4).
Localization at cell membranes is essential for the physiological functions of most members of the Ras superfamily of GTPases. For Rho GTPases, at least two signals within the C-terminal hypervariable region are involved in membrane targeting. The first is prenyl modification of the C-terminal CaaX motif, where C is cysteine, usually an aliphatic amino acid, and X dictates the identity of the prenyl group. The CaaX motif triggers three sequential posttranslational modifications: prenylation, proteolysis, and carboxymethylation. The CaaX cysteine of newly synthesized Rho GTPases is modified in the cytoplasm with a C-15 farnesyl or C-20 geranylgeranyl isoprenoid (5). Prenylated GTPases accumulate on endoplasmic reticulum (ER) membranes, where the C-terminal aaX tripeptide is cleaved by the endoprotease Rce1 (Ras-converting enzyme 1), and the carboxyl group of the newly exposed prenyl cysteine is carboxymethylated by Icmt (isoprenylcysteine carboxyl methyltransferase) (6). The variability and fidelity of this multistep modification are unclear. Proteomic analysis of bovine brain Gγ subunits that have a CaaX motif identified significant variation of their C-terminal processing (7). Functionally, all three CaaX processing steps are required for proper localization and biological activity of farnesylated Ras GTPases (8, 9), whereas there appear to be differential requirements for postprenylation processing of Rho GTPases for their localization (9, 10). Rho-mediated cell motility in cancer cells is sensitive to chemical inhibition by Icmt, pointing to the potential importance of methylation in metastasis (11). Postprenylation processing of Rheb is required for ER/Golgi localization but not for mTOR signaling (12). Thus, evidence suggests that variability of CaaX processing can affect protein function.
In addition to the CaaX motif, a second signal is also required for proper membrane targeting and biological activity (6). For Rho GTPases, a polybasic sequence in the hypervariable region increases affinity for negatively charged membranes and has consequences for specific biological outcomes. For example, the di-arginine motif of Cdc42 is important for its association with phosphatidylinositol 4,5-bisphosphate and its ability to oncogenically transform cells (13). Several Ras and Rho GTPases are modified with a second lipid modification, palmitoylation at cysteine residues in the hypervariable domain (14, 15). Reversible modification with palmitate dynamically regulates GTPase association with membranes, facilitates association with lipid rafts, and potentiates signaling (16, 17).
In vertebrates, two Cdc42 isoforms arise from alternative splicing: the ubiquitously expressed canonical Cdc42 and the brain-specifically expressed Cdc42 (bCdc42) (18). These isoforms share 95% identity and only have a different C-terminal exon encoding the hypervariable region (Fig. 1A). Canonical Cdc42 undergoes classical CaaX processing (geranylgeranylation, proteolysis, and carboxymethylation) at its C-terminal CVLL, whereas bCdc42 has the double-cysteine CCaX (CCIF) motif. Canonical Cdc42 has been extensively studied for its biological function in various cells and tissues. Although Cdc42 and bCdc42 were isolated more than 20 years ago (19, 20), the functional differences between canonical Cdc42 and bCdc42 are unclear. A recent report of neural palmitoyl-proteomics demonstrated that bCdc42 but not Cdc42 is palmitoylated in neurons and that bCdc42 plays a prominent role in dendritic spine formation (21). Mutation of the CCIF motif to SSIF blocks bCdc42 palmitoylation and abolishes its ability to induce spine formation. These results suggest that spine formation could be dynamically regulated by the palmitoylation cycle of bCdc42 and that both cysteines of the CCIF motif are sites of palmitoylation. Consistent with this prediction, Wrch-1, which has a C-terminal CCFV motif, is not a substrate for prenylation but is modified with palmitate in a manner that is dependent on the second cysteine (22). However, it has been reported that other CCaX motif-containing proteins undergo canonical posttranslational CaaX processing (Table 1).
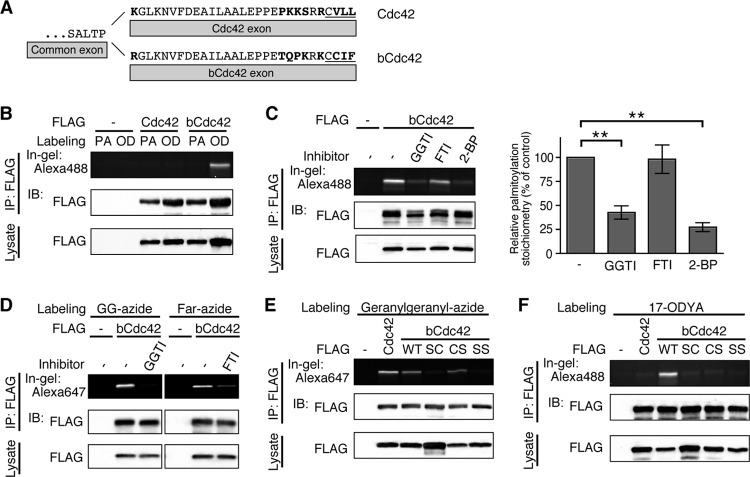
Fig 1.
The first and the second cysteine residues of the CCaX motif of bCdc42 are prenylated and palmitoylated, respectively. (A) The C-terminal amino acid sequences of human Cdc42 and bCdc42 are shown. Amino acids distinguishing the two isoforms are shown in boldface type. The CaaX motif is underlined. (B) HEK293 cells transfected with FLAG-Cdc42 or FLAG-bCdc42 were incubated in medium containing 100 μM palmitic acid (PA) or 100 μM 17-ODYA (OD) for 6 h. Lysates were immunoprecipitated (IP) with anti-FLAG antibody, and 17-ODYA incorporated into bCdc42 was detected with Alexa Fluor 488-azide using click chemistry. bCdc42 labeled with Alexa Fluor 488 was separated by SDS-PAGE and analyzed by in-gel fluorescence. Cdc42 and bCdc42 protein expression in the immunoprecipitates and cell lysates was analyzed by immunoblotting (IB). (C) HEK293 cells expressing FLAG-bCdc42 were labeled with 100 μM 17-ODYA for 6 h. Geranylgeranyltransferase inhibitor (5 μM; GGTI) or farnesyltransferase inhibitor (5 μM; FTI) was added to the medium 18 h prior to labeling with 17-ODYA. 2-Bromopalmitate (100 μM; 2-BP) or dimethyl sulfoxide (DMSO) was added to the medium 1 h prior to labeling with 17-ODYA. 17-ODYA incorporated into bCdc42 was detected with Alexa Fluor 488-azide. Palmitoylated bCdc42 was quantified (bar graph). Data represent the means ± SDs (n = 3). **, P < 0.01 (Student's t test). (D) HEK293 cells expressing FLAG-bCdc42 were labeled with 30 μM geranylgeranyl-azide (GG-azide) or farnesyl-azide (Far-azide) for 24 h in the presence or absence of 5 μM GGTI or FTI. Geranylgeranyl-azide or farnesyl-azide incorporated into bCdc42 was detected with Alexa Fluor 647-alkyne. (E and F) HEK293 cells transiently transfected with FLAG-Cdc42, wild-type FLAG-bCdc42 (WT), or CCaX motif mutants of FLAG-bCdc42 (SC, CS, and SS) were labeled with 30 μM geranylgeranyl-azide (D) or 100 μM 17-ODYA (E) for 24 h or 6 h, respectively. Geranylgeranylation and palmitoylation were detected with Alexa Fluor 647-alkyne and Alexa Fluor 488-azide, respectively.
Table 1.
| Protein | CCaX | Prenylationc | Palmitoylation | Methylation | Reference(s) and source or accession no. |
|---|---|---|---|---|---|
| bCdc42 | CCIF | GG/Far | + | + | 21, this study |
| Wrch-1 | CCFV | − | +d | 22 | |
| RalA | CCIL | GG | + | + | 26, 29, this study |
| RalB | CCLL | GG | + | 29, this study | |
| PRL-1 | CCIQ | Far | 30 | ||
| PRL-2 | CCVQ | Far | 30 | ||
| PRL-3 | CCVM | Far | + | + | 30, this study |
| PDE6α | CCIQ | Far | + | 41 | |
| PDE6β | CCIL | GG | +e | 41 | |
| PLA2γ | CCLA | Far | +f | 44 | |
| SLFNL1 | CCVL | ? | ? | ? | NP_001161719 |
| ASPA | CCLH | ? | ? | ? | NP_000040 |
| MPI | CCLL | ? | ? | ? | NP_002426 |
The list of proteins was derived from known and hypothetical CaaX prenyltransferase substrates (60) and PRENbase (http://mendel.imp.ac.at/PrePS/PRENbase/).
Modifications identified in this study are in boldface type.
Far, farnesylation; GG, geranylgeranylation.
The second cysteine of the CCFV motif of Wrch-1 is palmitoylated.
Carboxymethylation of PDE6β is approximately 20 times lower than that of PDE6α.
The palmitoylation site(s) of PLA2γ was not identified.
With the goal of elucidating the functional significance of bCdc42 palmitoylation, we first sought to confirm the palmitoylation status of bCdc42. Contrary to expectations, we found that bCdc42 was both prenylated and palmitoylated in a sequence context that has not been previously reported. Additionally, we demonstrate that bCdc42 and other CCaX motif proteins undergo alternative posttranslational processing pathways, generating two populations of differentially modified proteins. The two mature species of bCdc42, a dual prenyl and palmitoyl form and a CaaX-processed form, display different affinities for RhoGDIα and capacities to activate downstream signaling.
MATERIALS AND METHODS
Reagents.
17-Octadecynoic acid (17-ODYA) was purchased from Cayman Chemical (Ann Arbor, MI). Geranylgeranyl-azide, farnesyl-azide, Alexa Fluor 488-azide, and Alexa Fluor 647-alkyne were purchased from Invitrogen (San Diego, CA). GGTI-298 and FTI-277 were purchased from Calbiochem (La Jolla, CA). bCdc42-specific antibody (21) was a kind gift from the laboratory of the late A. El-Husseini (University of British Columbia). The following antibodies were purchased: anti-FLAG M2, from Stratagene; anti-Cdc42, from Cell Signaling; anti-RalA, from BD Biosciences; anti-GODZ (DHHC3), from Millipore; anti-RhoGDI (G-3), from Santa Cruz Biotechnology; and anti-transferrin receptor, from Invitrogen.
Plasmid construction.
The cDNAs of mouse Cdc42 and bCdc42 were kind gifts from R. A. Cerione (Cornell University). RalA (Addgene plasmid 15251), RalB (Addgene plasmid 19720), and PRL-3 (Addgene plasmid 16618) were purchased from Addgene (Cambridge, MA). The cDNA of human Wrch-1 was a kind gift from A. D. Cox (University of North Carolina). These cDNAs were subcloned into the mammalian FLAG tag expression vector pCMV5-FLAG and the green fluorescent protein (GFP) tag expression vector pEGFP-C1. The CCaX motif mutants of bCdc42—SC (C188S), CS (C189S), and SS (C188,189S)—were generated by site-directed PCR mutagenesis. bCdc42 F28L and R66A mutants were also generated by site-directed PCR mutagenesis. The integrity of all plasmid constructs was confirmed by DNA sequence analysis. The retroviral vector pBabe-puro was a kind gift from S. Gonzalo (St. Louis University). FLAG-tagged bCdc42 was subcloned into pBabe-puro. The cDNA of human DHHC3 was subcloned into pmEGFP-N3 to generate the C-terminal monomeric GFP fusion protein. For bioluminescence resonance energy transfer (BRET) assays, Venus-tagged PTP1b, giantin, and K-Ras were kind gifts from N. A. Lambert. The N terminus of bCdc42 was fused to humanized Renilla luciferase with a glycine-serine linker (GGGGS). The pGEX-RhoGDIα prokaryotic expression construct was provided by R. A. Cerione.
Cell culture.
HEK293 and HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C with 5% CO2. Rce1−/−, Icmt−/−, and Icmt+/+ mouse embryonic fibroblasts (MEFs) were kindly provided by S. G. Young (UCLA). Plasmid DNAs were transiently transfected into HEK293 and HeLa cells using Effectene (Qiagen). For MEFs, Lipofectamine 2000 or retroviral infection was used.
Cu(I)-catalyzed azide-alkyne cycloaddition reaction (click chemistry).
Transfected HEK293 cells were cultured in DMEM with 10% FBS for 42 h. Cells were then incubated in DMEM containing 10% dialyzed FBS and 100 μM 17-ODYA or palmitic acid for 6 h. For inhibitor treatment, 5 μM geranylgeranyltransferase inhibitor (GGTI-298) or farnesyltransferase inhibitor (FTI-277) was added to cells 18 h prior to 17-ODYA labeling; 2-bromopalmitate was added to cells 1 h prior to 17-ODYA labeling. To detect prenylation, transfected cells were cultured for 8 h and then incubated in DMEM containing 10% dialyzed FBS and 30 μM geranylgeranyl-azide or farnesyl-azide for 24 h. Cells were washed with phosphate-buffered saline (PBS) and lysed with RIPA buffer (20 mM HEPES-NaOH [pH 7.4], 100 mM NaCl, 3 mM MgCl2, 1% NP-40, 0.5% deoxycholate, and 0.1% SDS) supplemented with protease inhibitors (3 μg/ml of leupeptin and 1 mM phenylmethylsulfonyl fluoride [PMSF]). Cleared lysates were immunoprecipitated with anti-FLAG and protein G-Sepharose (GE Healthcare) for 4 h. The immunoprecipitates were washed 3 times with RIPA buffer and suspended in 94 μl of PBS, and 6 μl of freshly premixed click chemistry reagent (final concentrations of 10 μM Alexa Fluor 488-azide or Alexa Fluor 647-alkyne, 1 mM tris(2-carboxyethyl)phosphine (TCEP), 100 μM tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA), and 1 mM CuSO4) was added (23, 24). After 1 h at room temperature, the immunoprecipitates were washed twice with PBS containing 1% NP-40 and treated with sample buffer for SDS-PAGE. Probe-labeled proteins were detected by in-gel fluorescence.
Detection of dually prenylated and palmitoylated proteins.
Infected Sf9 cells were cultured in TriEx medium (Novagen) for 24 h. Cells were then incubated in TriEx medium containing 5% dialyzed FBS and 30 μM geranylgeranyl-azide for 24 h. Cells were washed with PBS and disrupted with lysis buffer (20 mM HEPES-NaOH [pH 7.4], 100 mM NaCl, 3 mM MgCl2, 15 mM imidazole, 1% NP-40, and 0.5% deoxycholate) supplemented with protease inhibitors. Cleared lysates were incubated with nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) for 1 h. The resin was washed 3 times with lysis buffer and boiled in blocking buffer (100 mM HEPES-NaCl [pH 7.4], 1 mM EDTA, and 2.5% SDS) for 5 min. Samples were recovered from the resin, and palmitoylated proteins were further purified using acyl-resin-assisted capture (acyl-RAC) as described previously (25). Briefly, samples were incubated in blocking buffer containing 0.1% methyl methanethiosulfonate (MMTS) at 42°C for 10 min, with frequent vortexing. Proteins were precipitated by adding 3 volumes of cold acetone. After centrifugation, protein pellets were washed with 70% cold acetone and suspended in binding buffer (100 mM HEPES-NaCl [pH 7.4], 1 mM EDTA, and 1% SDS). Samples were mixed with thiopropyl-Sepharose in the presence of 250 mM hydroxylamine to cleave thioester linkages. As a negative control, the same volume of NaCl was added instead of hydroxylamine. After 5 h of rotation at room temperature, the resins were washed four times with binding buffer and then mixed with click chemistry reagent as described above. After click chemistry, samples were eluted in buffer containing 50 mM Tris-HCl (pH 6.8), 50 mM dithiothreitol (DTT), 10% glycerol, and 1% SDS.
Carboxymethylation assay in cultured cells.
The in vivo carboxymethylation assay was performed as described previously (26, 27). At 8 h after transfection, HEK293 cells were incubated in labeling medium (90% methionine-free and 10% complete DMEM) containing 10% dialyzed FBS and 0.1 mCi/ml of l-[methyl-3H]methionine for 20 h. Cells were washed with PBS and lysed with RIPA buffer supplemented with protease inhibitors. FLAG-tagged proteins were immunoprecipitated from the cleared lysates. To analyze the carboxymethylation of palmitoylated proteins, infected Sf9 cells were starved in labeling medium (95% methionine-free and 5% complete Grace's medium) containing 10% dialyzed FBS. At 3 h after starvation, 0.1 mCi/ml of l-[methyl-3H]methionine was added and cells were incubated for 30 h. Palmitoylated proteins of interest were purified by Ni-NTA agarose, followed by acyl-RAC as described above. Samples were separated by SDS-PAGE, and the gel was then stained with Coomassie brilliant blue (CBB). The bands of interest were excised and dried in a vacuum centrifuge. Methyl-esterified proteins were analyzed by an alkali hydrolysis/diffusion assay as described previously (26, 27).
Protein purification.
His-bCdc42 was purified from membrane fractions of baculovirus-infected Sf9 cells using Ni-NTA agarose. Infected Sf9 cells were incubated with TriEX medium containing 5% dialyzed FBS and 100 μM palmitic acid or 17-ODYA for 6 h. Glutathione S-transferase (GST) and GST-RhoGDIα were purified from bacterial strain BL21(DE3) cells using glutathione-Sepharose.
Binding assay.
For the pulldown assay, transfected cells were washed with PBS and lysed with RIPA buffer supplemented with protease inhibitors. Cleared lysates were incubated for 2 h at 4°C with 1 μM GST or GST-RhoGDIα precoupled to glutathione-Sepharose. After three washes with RIPA buffer, the bound proteins were analyzed by immunoblotting. For the in vitro binding assay, 1 μM palmitate- or 17-ODYA-labeled His-bCdc42 was incubated with 1 μM GST or GST-RhoGDIα in binding buffer B (20 mM HEPES-NaOH [pH 7.4], 100 mM NaCl, 3 mM MgCl2, and 1% NP-40) for 1.5 h at 30°C. Samples were further incubated with glutathione-Sepharose for 1 h at 4°C. The resins were collected by centrifugation, and the supernatants were recovered as the glutathione-Sepharose unbound fraction. After three washes with binding buffer B, the bound proteins (bound fraction) were eluted by boiling in binding buffer B containing 1% SDS. SDS was added to the unbound fraction at a final concentration of 1%, and the total volume of each fraction was equalized. The bound and unbound fractions were subjected to click chemistry and immunoblotting.
Liposome binding assay.
17-ODYA-labeled bCdc42 (1 μM) was incubated with 1 mg/ml of liposomes containing 35% phosphatidylethanolamine, 25% phosphatidylserine, 5% phosphatidylinositol, and 35% cholesterol (Avanti Polar Lipids) for 30 min and centrifuged at 16,000 × g for 20 min. The liposome pellets were suspended in binding buffer C (50 mM HEPES-NaOH [pH 7.4], 100 mM NaCl, 3 mM MgCl2) and incubated with GST-RhoGDIα for 30 min. After a final centrifugation, the pellets were suspended in binding buffer C containing 1% SDS. The supernatants were supplemented with SDS (final concentration, 1%), and each sample was used for click chemistry. After click chemistry, samples were precipitated with methanol-chloroform and dissolved in sample buffer. Probe-labeled bCdc42 and total bCdc42 were detected by in-gel fluorescence and CBB stain, respectively.
Fractionation assay.
Transfected cells were cultured for 8 h and then incubated in the medium containing 30 μM geranylgeranyl-azide and 17-ODYA for 24 h and 6 h, respectively. Cells were harvested with buffer A (20 mM HEPES-NaCl [pH 7.4], 5 mM KCl, and 2 mM EDTA) containing protease inhibitors. Cells were homogenized on ice by 20 passes through a 27-gauge syringe needle. Nuclei and intact cells were removed by centrifugation at 800 × g for 5 min. Postnuclear supernatants were subjected to centrifugation at 100,000 × g for 30 min, and the pellets were suspended in buffer A containing 1% NP-40, 0.5% deoxycholate, and 0.1% SDS. The same detergents were directly added to supernatants. After 1 h of incubation, the samples were subjected to ultracentrifugation and cleared lysates were used for immunoprecipitation, followed by click chemistry.
Confocal microscopy.
HeLa cells were grown on glass bottom dishes and transiently transfected. At 6 h after transfection, cells were incubated in phenol red-free DMEM with 10% FBS and 25 mM HEPES overnight. Cells were imaged on Zeiss LSM510 META confocal microscope using a Plan-Apochromat 63×/1.4-numerical-aperture (NA) objective lens.
BRET.
HEK293 cells were seeded on a 12-well plate 24 h prior to transfection. Rluc-bCdc42 wild type or mutants (donor) were transfected alone or with Venus-PTP1b, Venus-giantin, or Venus-K-Ras (acceptor). At 24 h posttransfection, cells were washed with PBS, detached from the plate with PBS containing 2 mM EDTA, and collected by centrifugation at 400 × g for 5 min. Cells were suspended in PBS containing CaCl2 and MgCl2 and transferred to a 96-well plate. Coelenterazine-h (5 μM) was added 10 min before BRET measurement. Luminescence and fluorescence signals were detected using multimode microplate reader Synergy 2 (BioTek). The BRET ratio was calculated by dividing the fluorescence signal (528/20 emission filter) by the luminescence signal (460/40 emission filter). Net BRET was this ratio minus the same ratio measured from cells expressing the donor construct only.
Luciferase assay.
HEK293 cells were seeded on a 12-well plate and transfected with pEGFP-bCdc42 or mutant bCdc42 plasmids, pSRE-Luc, and pEF-Rluc. At 7 h posttransfection, the medium was replaced with DMEM containing 1% FBS. After 17 h, cells were lysed and subjected to the Dual-Glo luciferase assay system (Promega). Expression levels of bCdc42 and its mutants were confirmed and quantified by immunoblotting. Firefly luciferase activity derived from pSRE-Luc was normalized to the Renilla luciferase activity derived from pEF-Rluc. Data were normalized to the expression level of bCdc42 and its mutants.
Stoichiometry of endogenous palmitoylated bCdc42 in mouse neonatal brain.
For the acyl-RAC assay, mouse neonatal brain or adult kidney was homogenized in buffer A containing protease inhibitors with a Dounce homogenizer. Nuclei and intact cells were removed by centrifugation at 800 × g for 7 min. Postnuclear supernatants were subjected to centrifugation at 100,000 × g for 30 min, and the pellets were suspended and incubated in blocking buffer containing 0.1% MMTS at 42°C for 30 min with rotation. Proteins were precipitated by adding 3 volumes of cold acetone. After centrifugation, protein pellets were washed with 70% cold acetone and suspended in binding buffer. An aliquot was saved for the input lane. The remaining sample was used for an acyl-RAC assay as described above. The acyl-RAC samples and input (10% of total) were subjected to immunoblotting with bCdc42-specific and Cdc42 antibodies. The Cdc42 antibody detects both Cdc42 and bCdc42 isoforms. For the RhoGDIα pulldown assay, postnuclear lysates of neonatal brain and adult kidney were subjected to centrifugation at 100,000 × g for 30 min, and the pellets were suspended in binding buffer B. After 1 h of incubation, the samples were subjected to ultracentrifugation, and cleared lysates (2 mg/ml) were incubated for 2 h at 4°C with 0.4 μM GST-RhoGDIα precoupled to glutathione-Sepharose. GST-RhoGDIα unbound and bound fractions were collected. bCdc42 was detected by immunoblotting.
Statistics.
Data are presented as means ± standard deviations (SDs) of results of three or more independent experiments. Two-tailed Student's t tests were performed using GraphPad Prism 4 (GraphPad Software).
RESULTS
bCdc42 is geranylgeranylated and palmitoylated at its CCaX motif.
To examine the posttranslational modifications of the C-terminal CCaX motif of bCdc42, we first confirmed palmitoylation of bCdc42 in HEK293 cells. Cells were incubated with the palmitic acid analog 17-octadecynoic acid (17-ODYA) or palmitic acid as a negative control. Protein incorporating 17-ODYA was detected by selective labeling with Alexa Fluor 488-azide via Cu(I)-catalyzed azide-alkyne cycloaddition (click) chemistry (23, 24). As previously reported (21), bCdc42 but not Cdc42 was palmitoylated (Fig. 1B). Palmitoylation of bCdc42 was inhibited by 2-bromopalmitate, a nonmetabolizable fatty acid that inhibits palmitoylation (28) (Fig. 1C). Interestingly, a geranylgeranyl transferase inhibitor also blocked the incorporation of palmitate into bCdc42 (Fig. 1C), suggesting that geranylgeranyl modification might be a prerequisite for bCdc42 palmitoylation. To confirm prenylation of bCdc42, cells were incubated with geranylgeranyl-azide or farnesyl-azide. bCdc42 incorporated both prenyl lipid analogs, and incorporation was blocked by prenyltransferase-specific inhibitors (Fig. 1D).
To identify the prenylated and palmitoylated cysteine residues of bCdc42, we generated cysteine-to-serine mutants of the CCaX motif (SC, C188S; CS, C189S; and SS, C188,189S). Consistent with the canonical CaaX motif, mutation of the first but not the second cysteine completely inhibited the geranylgeranylation of bCdc42 (Fig. 1E). Mutation of either cysteine resulted in the loss of palmitoylation (Fig. 1F). The results of the mutagenesis provide further support for the hypothesis that prenylation of the first cysteine residue of bCdc42 CCaX motif is required for subsequent palmitoylation at the second cysteine.
In the case of the canonical CaaX motif, the C-terminal aaX tripeptide of prenylated CaaX is cleaved by the endoprotease Rce1, and the prenyl cysteine is then carboxymethylated by Icmt. Our data suggest that prenylated bCdc42 can bypass CCaX proteolysis and methylation and instead is modified with palmitate at the second cysteine. To more directly detect prenylation and palmitoylation on the same bCdc42 molecules, we measured prenylation of isolated palmitoyl bCdc42. H-Ras was used as a proof of principle for the experimental strategy. H-Ras is farnesylated at the CaaX motif and palmitoylated at two cysteine residues N terminal to the farnesylated cysteine. His-tagged H-Ras was purified from Sf9 cells labeled with farnesyl-azide using Ni-NTA agarose. Palmitoylated H-Ras was purified using acyl protein resin-assisted capture (acyl-RAC) (25). In this approach, free cysteines of H-Ras were first blocked by MMTS. The thioester-linked palmitate groups were then cleaved by hydroxylamine (NH2OH), and newly generated SH groups were conjugated with thiopropyl-Sepharose. Before elution from thipropyl-Sepharose, palmitoyl H-Ras was reacted with Alexa Fluor 647-alkyne to detect incorporation of farnesyl-azide into palmitoylated H-Ras. Dually farnesylated and palmitoylated H-Ras was detected in a hydroxylamine-dependent manner (Fig. 2A). As shown in Fig. 2B, bCdc42 was also dually geranylgeranylated and palmitoylated. Analysis of the bCdc42CS mutant (Fig. 2C) showed that detection of prenylated bCdc42 following acyl-RAC was dependent upon palmitoylation at the second cysteine. The absence of unmodified bCdc42 (bCdc42SC) in the acyl-RAC eluate ruled out nonspecific association of bCdc42 with the resin. These results provide additional evidence that bCdc42 is modified by tandem lipid groups, prenylation at the CaaX cysteine and palmitoylation of the second cysteine in the CCaX motif.
Fig 2.
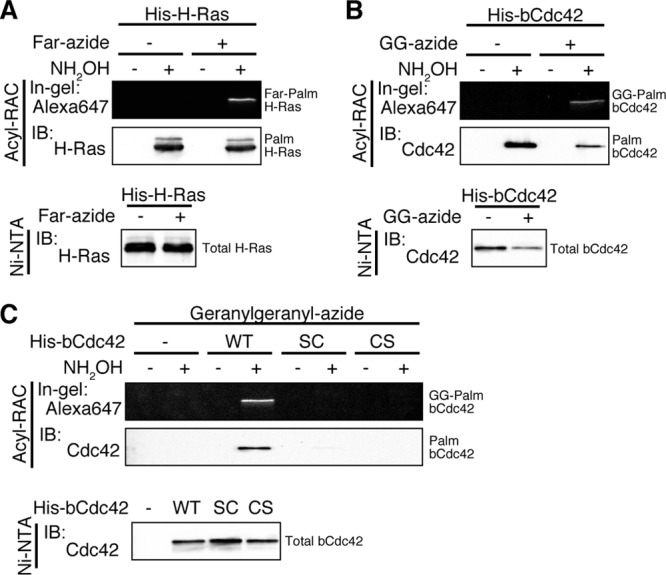
The CCaX motif is modified with a prenyl group and a palmitate on the same molecule. Sf9 cells expressing His-H-Ras (A) or His-bCdc42 (B) and its CCaX motif mutants (C) were incubated in medium containing 30 μM farnesyl-azide (Far-azide) or geranylgeranyl-azide (GG-azide) for 24 h. Lysates were incubated with Ni-NTA agarose, and His-tagged proteins were eluted from the resin. Then, hydroxylamine (NH2OH)-sensitive palmitoylated proteins were specifically immobilized on thiopropyl-Sepharose resin using acyl-RAC, and prenylation of captured proteins was detected with Alexa Fluor 647-alkyne using click chemistry. Dually prenylated and palmitoylated H-Ras (Far-Palm H-Ras) or bCdc42 (GG-Palm bCdc42) was analyzed by in-gel fluorescence.
Other CCaX motif-containing proteins have dual lipid modifications.
The results shown in Fig. 2 support the identification of a novel pattern of tandem lipid modifications at a CaaX motif with double cysteines. We next investigated whether the dual prenyl and palmitoyl modification occurs on other proteins that terminate in CCaX. The small GTPases RalA and RalB and protein tyrosine phosphatase PRL-3 have a C-terminal CCaX sequence (RalA, CCIL; RalB, CCLL; and PRL-3, CCVM), and prenylation of these proteins has been reported previously (29, 30). HEK293 cells transfected with FLAG-RalA, -RalB, or -PRL-3 were incubated with 17-ODYA or palmitic acid, and click chemistry was performed. All of these proteins were modified with palmitate (Fig. 3A). To confirm the lipid modification site, CCaX motif mutants of RalA and PRL-3 were generated. Prenylation and palmitoylation of CCaX motif mutants of RalA (Fig. 3B and C) and PRL-3 (Fig. 3D and E) showed results similar to those of bCdc42 (Fig. 1E and F), suggesting that the dual prenyl and palmitoyl modification is conserved in not only bCdc42 but also various CCaX motif-containing proteins. Not all CCaX motif proteins have the prenyl modification. We confirmed prior results (22) showing the Wrch-1 does not incorporate prenyl analogs and is palmitoylated only at the C-terminal CCFV sequence (data not shown).
Fig 3.
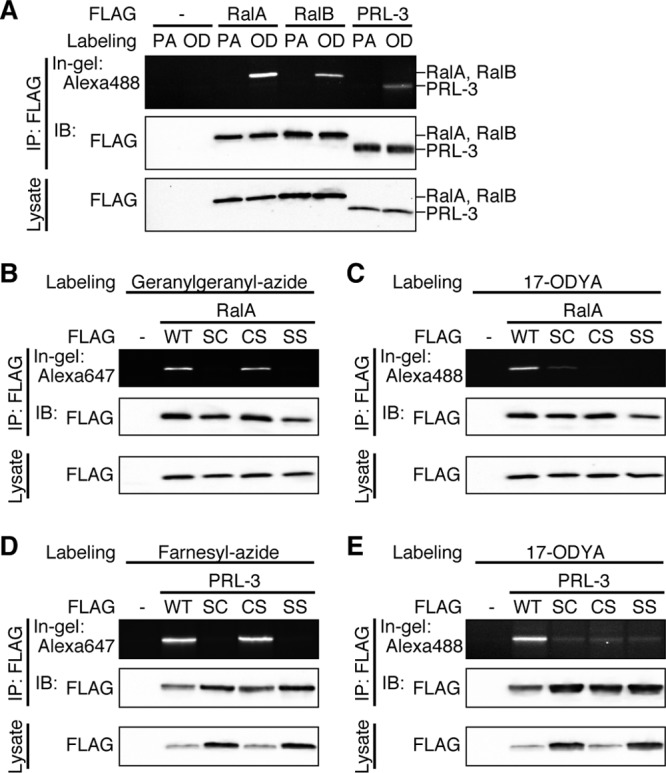
The dual prenyl-palmitoyl modification is conserved in other CCaX motif proteins. (A) HEK293 cells transfected with FLAG-RalA, FLAG-RalB, or FLAG-PRL-3 were incubated in medium containing 100 μM palmitic acid (PA) or 100 μM 17-ODYA (OD) for 6 h. Lysates were immunoprecipitated with anti-FLAG antibody, and 17-ODYA incorporation was detected with Alexa Fluor 488-azide. (B and C) HEK293 cells transiently expressing FLAG-RalA or its CCaX motif mutants were labeled with 30 μM geranylgeranyl-azide (B) or 100 μM 17-ODYA (C) for 24 h or 6 h, respectively. Geranylgeranylation and palmitoylation were detected with Alexa Fluor 647-alkyne or Alexa Fluor 488-azide, respectively. (D and E) HEK293 cells transiently expressing FLAG-PRL-3 or its CCaX motif mutants were labeled with 30 μM farnesyl-azide (D) or 100 μM 17-ODYA (E) for 24 h or 6 h, respectively. Farnesylation and palmitoylation were detected with Alexa Fluor 647-alkyne or Alexa Fluor 488-azide, respectively.
bCdc42 that is not palmitoylated is carboxymethylated in cells.
Detection of the dual prenyl and palmitoyl modification at the CCaX motif predicts that CCaX motif-containing proteins are not processed by proteolysis and carboxymethylation. However, it has previously been reported that RalA is carboxymethylated similarly to canonical CaaX proteins (26). To examine the carboxymethylation status of bCdc42 and PRL-3 in intact cells, HEK293 cells transfected with FLAG-bCdc42 or FLAG-PRL-3 were incubated with l-[methyl-3H]methionine, and FLAG-tagged proteins were immunoprecipitated from lysates. The amount of carboxymethylated protein was analyzed by measuring the level of [3H]methanol released after alkaline hydrolysis (26, 27). As shown in Fig. 4A, both bCdc42 and PRL-3 were carboxymethylated compared with nonprenylated SC mutants. We hypothesized two models for the posttranslational modification of the CCaX motif. In the first model, the CCaX motif proteins have two different mature forms: a CaaX-processed form and a dual prenyl and palmitoyl form. In the second model, the -aX dipeptide is cleaved following prenylation and palmitoylation by an endoprotease and the palmitoyl cysteine is methylated. To test these hypotheses, we measured the carboxymethylation of palmitoylated bCdc42. Sf9 cells infected with His-bCdc42 were labeled with l-[methyl-3H]methionine. H-Ras was used as a positive control. bCdc42 or H-Ras was purified from radiolabeled lysates by Ni-NTA agarose. An aliquot of this pool was reserved for analysis of carboxylmethylation (Fig. 4B, Total). Palmitoylated bCdc42 or H-Ras was separated from the nonpalmitoylated protein using acyl-RAC. Carboxymethylation of palmitoylated bCdc42 was dramatically reduced compared with total bCdc42 (Fig. 4B), whereas the stoichiometry of palmitoylated H-Ras methylation was the same as that of total H-Ras. We were not able to determine whether the residual methylation observed for bCdc42 represents a small pool of palmitoylated protein that is methylated or is an artifact.
Fig 4.
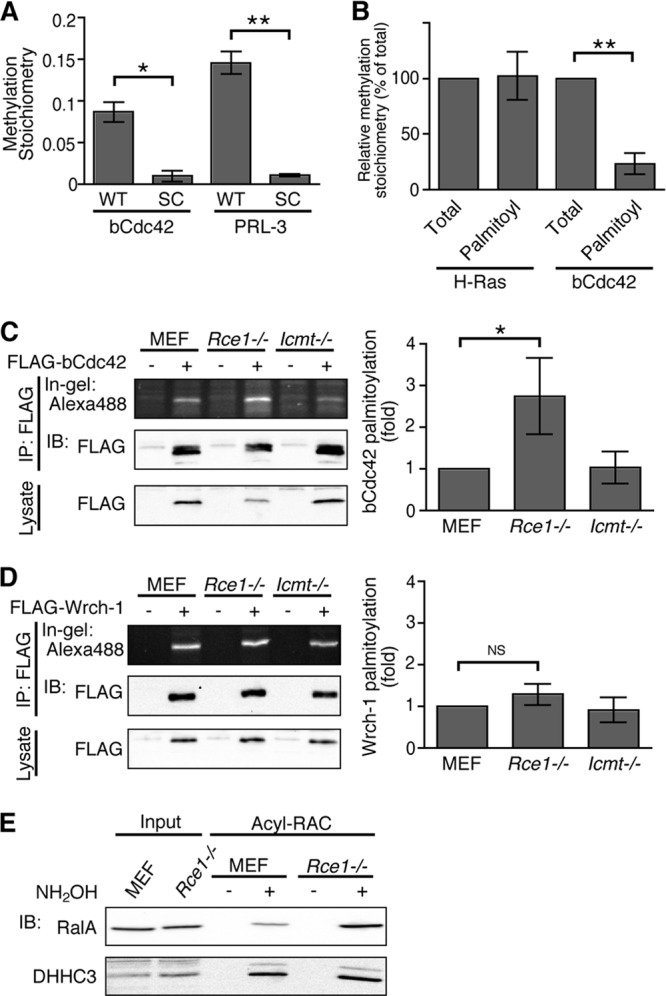
The CCaX motif is carboxymethylated in cultured cells. (A) HEK293 cells transiently transfected with FLAG-bCdc42, -bCdc42SC, -PRL-3, or -PRL-3SC were labeled with 0.1 mCi/ml of l-[methyl-3H]methionine for 20 h. Lysates were immunoprecipitated, and carboxymethylation was measured by an alkaline hydrolysis assay, as described in Materials and Methods, and quantitated by liquid scintillation spectrometry. The methylation stoichiometry was calculated by a ratio of the alkali-labile counts per minute to the alkali-stable counts per minute. Data represent the means ± SDs (n = 3). *, P < 0.05; **, P < 0.01. (B) Sf9 cells transiently expressing His-H-Ras or His-bCdc42 were labeled with 0.1 mCi/ml of l-[methyl-3H]methionine for 30 h. Total H-Ras and bCdc42 were pulled down using Ni-NTA agarose, and palmitoylated forms were further purified using acyl-RAC. Carboxymethylation was measured by an alkaline hydrolysis assay. Data represent the means ± SDs (n = 3). **, P < 0.01. (C and D) Wild-type (Icmt+/+), Rce1−/−, and Icmt−/− MEFs expressing FLAG-bCdc42 (C) or FLAG-Wrch-1 (D) were incubated in medium containing 100 μM 17-ODYA for 6 h. Lysates were immunoprecipitated with anti-FLAG antibody, and 17-ODYA incorporation was detected with Alexa Fluor 488-azide. Data represent the means ± SDs (n = 3 to 4). *, P < 0.05; NS, not significant. (E) Lysates from wild-type or Rce1−/− MEFs were treated or not with hydroxylamine (NH2OH), and NH2OH-sensitive palmitoylated proteins were immobilized on thiopropyl-Sepharose resin using acyl-RAC. Palmitoylation of endogenous RalA and DHHC3 was analyzed by immunoblotting.
To investigate whether the inhibition of CaaX processing pathway affects palmitoylation of bCdc42, we compared bCdc42 palmitoylation in wild-type mouse embryo fibroblasts (MEFs) and MEFs lacking Rce1 and Icmt. bCdc42 palmitoylation was increased in Rce1−/− MEFs (Fig. 4C), whereas palmitoylation of nonprenylated Wrch-1 (22) was unchanged (Fig. 4D), suggesting that in the absence of Rce1 or Icmt, more prenylated bCdc42 is available for palmitoylation. To exclude the possibility that the presence of two differentially modified populations of CCaX proteins is an artifact of high levels of expression, we characterized an endogenous CCaX protein. We could not directly measure the carboxymethylation of endogenous bCdc42, RalA, or PRL-3 because of low immunoprecipitation efficiency. Instead, the degrees of palmitoylation of endogenous RalA were compared in wild-type and Rce1−/− MEFs. Cellular palmitoylated proteins were isolated by acyl-RAC, and the palmitoylation of endogenous RalA was detected by immunoblotting. RalA palmitoylation was significantly enhanced in Rce1−/− MEFs, whereas palmitoylation of the integral membrane protein DHHC3 was unchanged (Fig. 4E). These results further support the existence of two modes of posttranslational processing of CCaX proteins: canonical CaaX processing and dual prenyl and palmitoyl lipid modification.
Palmitoylation of bCdc42 prevents its binding to RhoGDIα.
To investigate whether the two mature forms of bCdc42 have functional differences, we analyzed their interactions with binding partners, focusing in this study on RhoGDIα. CaaX processing of Rho GTPases is important for RhoGDIα binding. The geranylgeranyl moiety of a Rho GTPase is inserted into the hydrophobic pocket formed by the C-terminal immunoglobulin-like domain of RhoGDIα (31), facilitating the formation of a high-affinity complex. We first compared the interactions of the bCdc42 wild type and the nonpalmitoylated bCdc42CS mutant with recombinant GST-RhoGDIα using a GST pulldown assay. FLAG-tagged bCdc42 was pulled down from cell lysates using recombinant GST-RhoGDIα. The bCdc42CS mutant displayed more binding to RhoGDIα than did the bCdc42 wild type (Fig. 5A). As expected, nonprenylated bCdc42SC and bCdc42(R66A), a mutant defective in binding RhoGDI (32), bound weakly or not at all to GST-RhoGDIα. Next, we assessed whether increasing the palmitoylation level of bCdc42 altered its binding to RhoGDIα. DHHC3 is a protein acyltransferase (PAT) with broad substrate specificity (33). Coexpression of DHHC3 with bCdc42 in HEK293 cells significantly increased bCdc42 palmitoylation (data not shown). RhoGDIα binding to bCdc42 was substantially reduced by exogenous expression of DHHC3 (Fig. 5B). To investigate this in more detail, we purified bCdc42 from Sf9 cells incubated with 17-ODYA to enable detection of the palmitoylated form and performed an in vitro GST binding assay. Although most of the bCdc42 protein bound to GST-RhoGDIα, the 17-ODYA-labeled bCdc42 was found nearly exclusively in the unbound fraction. These results indicate that palmitoylation inhibits bCdc42 interaction with RhoGDIα.
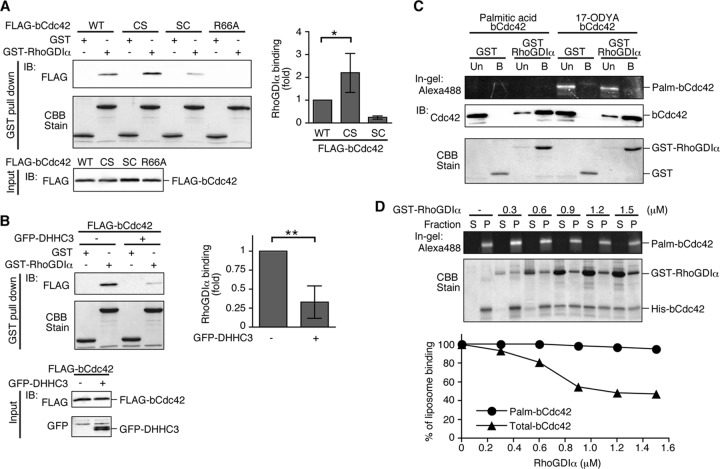
Fig 5.
Palmitoylation of bCdc42 inhibits its interaction with RhoGDIα. (A) HEK293 cells were transfected with FLAG-bCdc42 wild-type (WT), CCaX mutants (CS and SC), or the GDI-binding-deficient mutant R66A. bCdc42 pulled down by GST or GST-RhoGDIα bound to glutathione-Sepharose (top) or in the starting lysates (bottom) was detected by FLAG immunoblotting. GST or GST-RhoGDIα was visualized by CBB stain. bCdc42 bound to RhoGDIα was quantified and plotted as a bar graph. Data represent the means ± SDs (n = 5). *, P < 0.05. (B) HEK293 cells were transfected with FLAG-bCdc42 with or without GFP-DHHC3. bCdc42 was pulled down from lysates by GST or GST-RhoGDIα bound to glutathione-Sepharose (top two blots). Expression of FLAG-bCdc42 and GFP-DHHC3 in starting lysates was detected by immunoblotting (bottom two blots). Data represent the means ± SDs (n = 4). **, P < 0.01. (C) Recombinant bCdc42 was purified from Sf9 cells expressing His-bCdc42 in medium with 100 μM 17-ODYA or palmitic acid as a control. Control or 17-ODYA-labeled bCdc42 was pulled down with GST or GST-RhoGDIα and unbound (Un) and bound (B) fractions were collected. 17-ODYA-labeled bCdc42 was detected with Alexa Fluor 488-azide using click chemistry. (D) Liposomes containing recombinant 17-ODYA-labeled bCdc42 were incubated with the indicated concentrations of GST-RhoGDIα for 30 min and separated into soluble and particulate fractions. 17-ODYA-labeled bCdc42 from the liposome pellet (P) and supernatant (S) was detected with Alexa Fluor 488-azide. The recovery of 17-ODYA-labeled bCdc42 and unlabeled bCdc42 was analyzed by in-gel fluorescence and CBB stain, respectively.
Next, we analyzed the effect of RhoGDIα on the dissociation of palmitoylated bCdc42 from liposomes. bCdc42 labeled with 17-ODYA and reconstituted into liposomes was incubated with increasing concentrations of RhoGDIα. The liposomes were recovered by centrifugation, and the recovery of 17-ODYA-labeled bCdc42 and total bCdc42 in the pellet and supernatant fractions was analyzed. Consistent with the binding assays (Fig. 5C), association of palmitoylated bCdc42 with liposomes was not affected by RhoGDIα, whereas total bCdc42 was shifted to the supernatant fraction in a RhoGDIα concentration-dependent manner (Fig. 5D). We acknowledge that the absence of carboxylmethylation of the prenyl and palmitoyl form of bCdc42 may contribute to its reduced affinity for RhoGDIα. The affinity of RhoA and Rac1 for RhoGDI was modestly increased in cells treated with an Icmt inhibitor (11). A similar reduction in affinity was reported for Rac1 in Icmt null MEFs (9).
Two mature forms of bCdc42 show different subcellular distributions and signaling activities.
Our in vitro results that palmitoylation of bCdc42 inhibits its binding to RhoGDIα predicts a different subcellular distribution of the CaaX-processed and the dual prenyl and palmitoyl forms of bCdc42. To test this prediction, HEK293 cells were incubated with 17-ODYA and geranylgeranyl-azide, and the subcellular distribution of bCdc42 labeled with each lipid analog was analyzed. Prenylated bCdc42 was detected in both cytosolic and membrane fractions. In contrast, palmitoylated bCdc42 was detected only in the membrane fraction (Fig. 6A). These results suggest that the CaaX-processed bCdc42 form is present in the cytoplasm, presumably through its interaction with RhoGDIα in the cells, whereas the dual prenyl and palmitoyl form is stably associated with membranes. Consistent with this idea, the population of membrane-bound bCdc42 was higher than that of the nonpalmitoylated bCdc42CS mutant that is exclusively prenylated (Fig. 6B). We also checked the cellular localization of transfected bCdc42 and its mutants using confocal microscopy. As previously reported for MDCK and COS-1 cells, GFP-bCdc42 was observed in endomembranes and the plasma membrane in HeLa cells (Fig. 6C). GFP-bCdc42CS displayed a similar localization pattern, whereas GFP-bCdc42SC was distributed throughout the cytoplasm. To determine whether there was a quantitative difference between the localization of bCdc42 and the nonpalmitoylated bCdc42CS mutant, we used bioluminescence resonance energy transfer as a readout of colocalization in different membrane compartments (34). The BRET donor Renilla luciferase (Rluc) fused to bCdc42 was cotransfected with the BRET acceptor Venus fused to various organelle markers: PTP1b for the ER, giantin for the Golgi apparatus, or K-Ras for the plasma membrane. The colocalization of bCdc42 with each membrane compartment marker was evaluated by measuring BRET resulting from the random collision between BRET pairs (bystander BRET). Compared with nonlipidated Rluc-bCdc42SC, efficient BRET was detected between Rluc-bCdc42 with Venus-PTP1b, Venus-giantin, and Venus-K-Ras (Fig. 6D), indicating bCdc42 localization at the ER, Golgi apparatus, and plasma membrane. The BRET signal between nonpalmitoylated Rluc-bCdc42CS with the plasma membrane marker Venus-K-Ras was significantly lower than that of the wild-type protein, whereas localization on endomembrane compartments was unaffected. Thus, palmitoylation enriches bCdc42 at the plasma membrane.
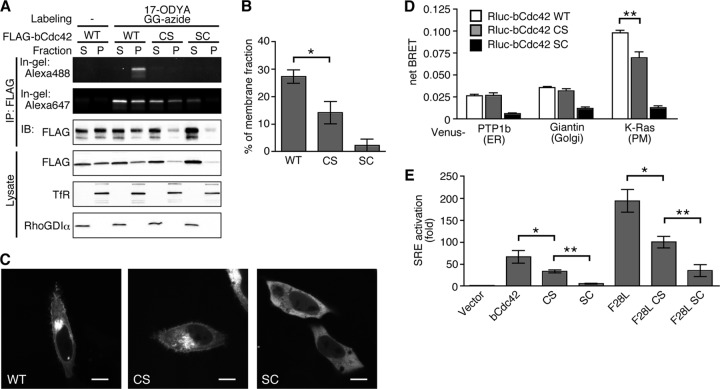
Fig 6.
Palmitoylation regulates the subcellular distribution and activity of bCdc42. (A) HEK293 cells transfected with FLAG-bCdc42, -bCdc42CS, or -bCdc42SC were incubated in medium containing 30 μM geranylgeranyl-azide for 24 h. 17-ODYA (100 μM) was added during the last 6 h of the incubation. Postnuclear lysates were fractionated by ultracentrifugation, and the supernatant (S) and pellet (P) fractions were immunoprecipitated with anti-FLAG antibody. Geranylgeranyl-azide and 17-ODYA incorporation were detected with Alexa Fluor 647-alkyne and Alexa Fluor 488-azide using sequential click chemistry. Transferrin receptor (TfR) and RhoGDIα were used as markers for membranes and cytoplasm, respectively. (B) Membrane-bound bCdc42 was quantified and plotted in the bar graph. Data represent the means ± SDs (n = 3). *, P < 0.05. (C) HeLa cells transiently transfected with GFP-bCdc42, -bCdc42CS, or -bCdc42SC were imaged by live-cell confocal microscopy. Scale bars, 10 μm. (D) Colocalization of bCdc42 with compartment-specific markers was analyzed by BRET. The net BRET ratio was calculated from cells expressing the BRET donor Renilla luciferase-bCdc42 (RLuc-bCdc42), RLuc-bCdc42CS, or RLuc-bCdc42SC, together with each BRET acceptor Venus-PTP1b (ER marker), Venus-giantin (Golgi marker), or Venus-K-Ras (PM marker). Data represent the means ± SDs (n = 3). **, P < 0.01. (E) An SRE transcriptional reporter construct was used to assess the activities of wild-type and mutant bCdc42. The lipidation mutants were assessed in the context of wild-type bCdc42 and the fast-cycling F28L mutant. The SRE activity is shown as the fold increase of normalized luciferase over that of the vector control. Data represent the means ± SDs of duplicate samples (n = 4). *, P < 0.05; **, P < 0.01.
Membrane localization of Rho GTPases is important for activation of downstream signaling pathways (35). Rho GTPases, including Cdc42, Rac1, and RhoA, regulate the transcriptional activity of the c-fos serum response element (SRE) (36). The different membrane binding affinities of the two mature forms of bCdc42 may impact signaling activity. To test this hypothesis, we measured SRE transcriptional activity of bCdc42 and its mutants. Expression of bCdc42 robustly stimulated the SRE reporter, producing 66-fold activation over vector (Fig. 6E). bCdc42CS also retained the ability to potentiate SRE activation (34-fold), but it did so less strongly than bCdc42. Nonprenylated bCdc42SC, which is predominately cytoplasmic, displayed little SRE activity. The absence of palmitoylation in the bCdc42CS mutation also reduced SRE activation induced by the fast-cycling, oncogenic bCdc42 F28L mutant (37). These results indicate that the prenyl, palmitoyl form of bCdc42 has a higher capacity to stimulate downstream signaling than the CaaX-processed form. The correlation of membrane binding affinity and plasma membrane association with signaling activity strongly suggests that the effect of palmitoylation on bCdc42 activity is due to enhanced access to membrane-bound effectors.
Palmitoylation stoichiometry of bCdc42.
Our finding that bCdc42 is differentially modified at the C terminus raises the question of the relative abundance of the fully CaaX-processed and the prenyl, palmitoyl form. Turnover of palmitate on bCdc42 (21) also yields a third population, the prenylated and depalmitoylated full-length protein. To estimate the abundance of palmitoylated bCdc42, we performed acyl-RAC on neonatal mouse brain, which expresses higher levels of bCdc42 than adult brain (data not shown). Approximately 5 to 10% of total bCdc42 from membrane fraction was recovered on the thiopropyl-Sepharose column (Fig. 7A). We have observed that recovery of palmitoylated proteins by acyl-RAC is incomplete and may give an underestimate of the abundance of palmitoylated bCdc42. Accordingly, we used RhoGDIα binding as a second method to estimate the stoichiometry of palmitoylation of endogenous bCdc42 (Fig. 7B). Immunoblotting of soluble and particulate fractions revealed that approximately 35% of bCdc42 and total Cdc42 is in the particulate fractions of neonatal brain. In contrast, adult kidney, where only canonical Cdc42 is expressed, most Cdc42 (84%) is in the soluble fraction. The membrane fractions were further processed by detergent solubilization and subjected to RhoGDIα pulldown assays. Approximately 25% of bCdc42 from the particulate fraction of neonatal brain did not bind to RhoGDIα, compared to 6% of Cdc42 from adult kidney. Using the RhoGDIα-binding assay, we estimate that 15 to 20% of the membrane-associated pool of bCdc42 is palmitoylated in neonatal brain. Thus, the palmitoylated population of bCdc42 likely represents 5 to 20% of membrane-associated bCdc42.
Fig 7.
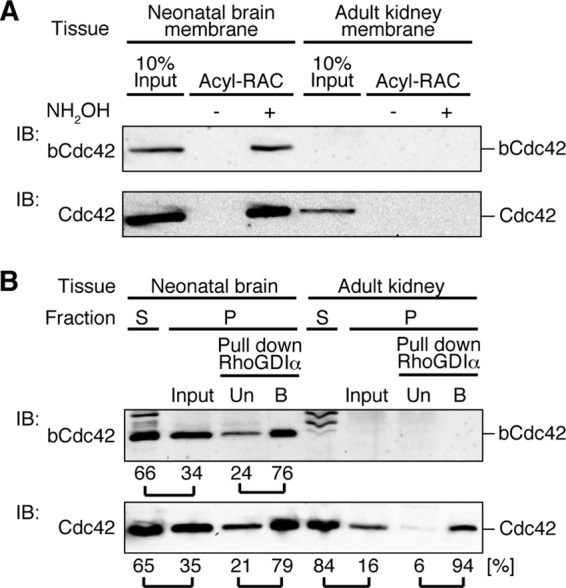
Estimate of the abundance of endogenous palmitoylated bCdc42 in neonatal brain. (A) Postnuclear lysates of mouse neonatal brain and adult kidney were fractionated by ultracentrifugation. The pellet was subjected to acyl-RAC as described in Materials and Methods. Acyl-RAC samples and input (10% of total) were subjected to immunoblotting using bCdc42-specific and Cdc42 antibodies. Cdc42 antibody detects both Cdc42 and bCdc42 isoforms. Band intensity was quantified. (B) Postnuclear lysates from neonatal brain and adult kidney were separated by ultracentrifugation into supernatant (S) and pellet (P) fractions. Proteins were extracted from the pellet fraction and mixed with GST-RhoGDIα bound to glutathione-Sepharose. GST-RhoGDIα unbound (Un) and bound (B) fractions were collected. bCdc42 and Cdc42 were detected by immunoblotting. Band intensity was quantified, and the relative proportions in each fraction are shown.
DISCUSSION
Of the hundreds of human proteins known or predicted to be substrates for CaaX prenyltransferases, a small subset have a cysteine at the second position of the CaaX motif (Table 1). In this study, we identified a novel posttranslational CaaX processing pathway for proteins terminating in a CCaX motif. We demonstrated that the brain-specific isoform of Cdc42, the GTPases RalA and RalB, and the phosphatase PRL-3 are prenylated and palmitoylated in a tandem fashion at the C terminus, bypassing the postprenylation steps of proteolysis and carboxymethylation. A prior study detected bCdc42 in the neural palmitoylome and confirmed that its palmitoylation is dependent on the two cysteines near the C terminus (21). Our experiments using pharmacological inhibitors and site-directed mutagenesis showed that palmitoylation occurs at the second cysteine residue and is dependent upon prenylation at the CaaX cysteine (Fig. 1 and 3). Independent evidence that our findings of RalA and RalB palmitoylation extend to their endogenous counterparts comes from their presence in the palmitoylomes of neural stem cells (38), B cells (39), and T cells (40).
Conservation of the CCaX sequence motif of bCdc42, Ral GTPases, and PRL phosphatases throughout vertebrates suggests a role for palmitoylation as a regulatory modification in the function of these proteins. Interestingly, the prenyl and palmitoyl modification we found on bCdc42 is not conserved in all proteins with a CCaX motif (Table 1). We confirmed an earlier study showing that Wrch-1 (RhoU), which terminates in CCFV, is palmitoylated at the second cysteine but not prenylated (22) (Table 1). Other proteins remain to be evaluated. PDE6α and PDE6β form an obligate heterodimer and are components of the photoreceptor cGMP phosphodiesterase. Canonical CaaX processing of PDE6α and -β is well documented (41), and it is known that RCE1 activity is required for trafficking of rod PDE6 to photoreceptor outer segments (42). Cytosolic phospholipase A2γ undergoes classical CaaX processing when expressed in insect cells (43). It is notable that incorporation of palmitate and oleate into this protein has been reported but the site (or sites) of fatty acylation is unknown (44).
Our study strongly supports the coexistence of two populations of CCaX proteins in cells, a dual prenyl, palmitoyl form and a classically CaaX-processed form. Palmitoylated bCdc42 isolated by acyl-RAC was labeled with a prenyl analog and depleted of carboxymethylation, consistent with a tandemly lipid-modified C terminus that does not undergo further postprenylation processing (Fig. 2 and 4). Prenylated but not proteolytically processed CaaX intermediates have been detected in proteomic analyses of native G protein γ subunits (7) and the α and β subunits of phosphorylase kinase (45), suggesting that the intermediates may be reasonably abundant in cells. Carboxymethylation was detected in total pools of bCdc42 and PRL-3, providing evidence for the canonically processed form (Fig. 4). The two different processing pathways are competitive because cells deficient in the protease Rce1 displayed increased palmitoylation levels of ectopically expressed bCdc42 and endogenous RalA (Fig. 4).
The identification of two posttranslational processing fates for bCdc42 and other CCaX proteins raises the question as to how the two pathways are regulated to maintain a steady-state population of both forms. Once proteolyzed, the canonically CaaX-processed form is no longer a substrate for palmitoylation. In contrast, the reversibility of palmitoylation generates a pool of mature protein that can be repalmitoylated or subjected to classical CaaX processing. The postprenylation processing enzymes, Rce1 and Icmt, are located in the ER. Following prenylation, nascent CaaX motif proteins are directed by an unknown mechanism to the ER, where they are proteolyzed and methylated. Palmitoylation is catalyzed by members of a family of protein acyltransferases with a DHHC-cysteine rich domain. Mammalian genomes encode at least 23 DHHC proteins, which show specific patterns of localization in the ER, Golgi apparatus, endosomes, and the plasma membrane (46). A current model (17) proposes that the Golgi apparatus is the major site of palmitoylation in mammalian cells for dually lipid-modified proteins that undergo reversible palmitoylation. The addition of palmitate to a prenylated protein at the Golgi apparatus stabilizes its association with membranes, enabling it to enter secretory vesicles for transport to the plasma membrane. Depalmitoylation returns the protein to a state where it interacts transiently and nonspecifically with endomembranes until it is trapped at the Golgi apparatus by palmitoylation. Given that the ER membrane is the most abundant membrane system in the cell, there must be mechanisms to avoid the terminal steps of CaaX processing if depalmitoylated CCaX proteins are sampling ER membranes during retrograde transport. The DHHC proteins that modify bCdc42 and the other proteins analyzed in this study are unknown, and their identification will be an important step in resolving this issue. A number of mammalian DHHC PATs are localized in the ER (46) and could compete with Rce1 for common substrates. ER-localized DHHC proteins in Saccharomyces cerevisiae are known to palmitoylate dually lipidated proteins (47, 48). However, as discussed below, it is also possible that cytoplasmic chaperones such as RhoGDI regulate the intracellular trafficking of depalmitoylated CCaX proteins and direct them to the Golgi apparatus or another membrane compartment for repalmitoylation (Fig. 8). The Golgi apparatus-localized DHHC3 is a candidate PAT for bCdc42; we observed increased palmitoylation of bCdc42 when the two proteins were coexpressed in tissue culture cells (data not shown).
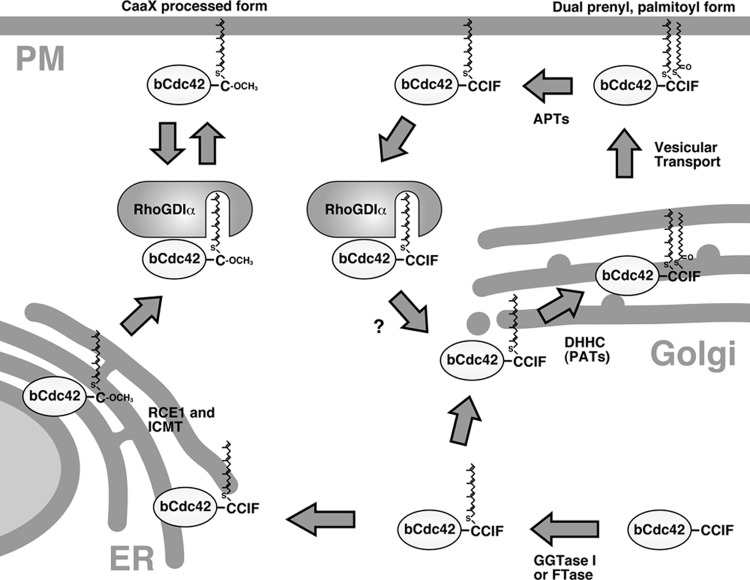
Fig 8.
Working model: differential posttranslational processing of bCdc42 yields two populations at steady state with distinct properties. Newly synthesized bCdc42 is prenylated in the cytoplasm. One population of bCdc42 undergoes endoproteolytic processing by RAS-converting enzyme 1 (Rce1) and carboxymethylation by isoprenylcysteine carboxyl methyltransferase (Icmt) in the endoplasmic reticulum. Trafficking of the CaaX-processed bCdc42 is regulated by the RhoGDIα binding cycle. A second population of bCdc42 bypasses the canonical CaaX processing pathway and instead is modified with palmitate at second cysteine residue of the CCaX motif by PATs in the Golgi apparatus or other membrane compartments. The dual prenyl-palmitoyl form is stably associated with membranes and would be translocated to the plasma membrane via vesicular transport. Palmitoylation of bCdc42 is reversed by the action of acylprotein thioesterases (APTs). The depalmitoylated form could be associated with RhoGDIα and may return to the Golgi apparatus for repalmitoylation.
Our observation that the prenyl, palmitoyl form of bCdc42 does not interact with RhoGDIα is consistent with the hypothesis of Michaelson and coworkers that palmitoylation negatively regulates the association of Rho GTPases with RhoGDI (49). They reported that mutation of the palmitoylation site in RhoB or TC10 increases binding to RhoGDIα, whereas introduction of a palmitoylation site into RhoA blocked binding. In the same study, they reported that bCdc42 binds to RhoGDIα. This conclusion is based on microscopy that showed a shift in the localization of bCdc42 from membranes to the cytoplasm when cooverexpressed with RhoGDIα. In light of our data, we suggest that both forms of bCdc42 were present in the cells and that it is the CaaX-processed form of bCdc42 that shifts from membranes to the cytoplasm when RhoGDIα is overexpressed. Interestingly, palmitoylation of Rac1 does not affect the interaction with RhoGDIα (15). This functional difference between Rac1 and other palmitoylated GTPases is likely due to the distance between the palmitoyl-cysteine and the prenyl-cysteine. In bCdc42, RhoB, and TC10, the palmitoylcysteine is either adjacent to or one residue away from the prenyl cysteine, whereas in Rac1 the lipidated residues are separated by 11 amino acids (10, 15). The structure of the RhoGDI-Cdc42 complex reveals that the geranylgeranyl group of Cdc42 is inserted into a hydrophobic pocket of RhoGDI (31). Palmitoylation adjacent to or nearby the prenylated cysteine may sterically impair binding of the dually lipidated form to RhoGDI (49).
Several lines of evidence support the conclusion that RhoGDI is required to target Rho GTPases to the appropriate membrane. RhoGDIα protects cytosolic Rho GTPases from proteolysis (4) and plays a role in facilitating their translocation to the plasma membrane upon cell stimulation. In the absence of RhoGDIα, Rho GTPases lose their plasma membrane localization and activation of downstream signaling (4, 32). These results suggest that membrane trafficking of CaaX-processed bCdc42 is controlled by RhoGDIα. Palmitoylation of bCdc42 stabilizes the association of the prenylated protein with membranes (Fig. 6), and trafficking of the prenyl, palmitoyl form between compartments will likely occur by vesicular transport. However, bCdc42 palmitoylation is dynamic (21). Accordingly, depalmitoylation will generate a form of bCdc42 that is available for regulation by RhoGDIα. There is increasing evidence that prenyl-binding proteins other than GDIs regulate the membrane interactions and trafficking of prenylated proteins. The δ subunit of PDE6 is a prenyl-binding protein that binds to and solubilizes the catalytic subunits of PDE6 from rod outer segment disc membranes in vitro (50). The importance of the δ subunit in trafficking of PDE6 is evident from the mislocalization of PDE6 in PDE6δ knockout mice (51). PDE6δ is broadly expressed and has been shown to bind to a number of proteins and regulate their trafficking (52–54). A recent report (55) shows that PDE6δ binding to depalmitoylated Ras facilitates its diffusion in the cytoplasm. When palmitoylation is inhibited, H-Ras accumulates nonspecifically on endomembranes. Expression of PDE6δ facilitates plasma membrane localization of H-Ras, presumably by extracting it from endomembranes until it is trapped at the Golgi apparatus by palmitoylation and subsequently moves to the plasma membrane by vesicular transport (55). We speculate that RhoGDI may function similarly to facilitate repalmitoylation of bCdc42 (Fig. 8).
Although the ubiquitously expressed and brain-specific isoforms of Cdc42 were purified and their cDNAs cloned more than 2 decades ago (19, 20), relatively little is known about how they differ functionally. The study that identified bCdc42 as a palmitoyl-protein showed that the two isoforms display distinct dendritic localizations, with bCdc42 more concentrated on dendritic spines (21). Constitutively active bCdc42 was more potent than canonical Cdc42 in an assay of spine induction in hippocampal neurons. The functional importance of palmitoylation in the localization and activity of bCdc42 in these assays is unclear because both prenylation and palmitoylation were blocked in the mutant that was studied. Other differences between bCdc42 and canonical Cdc42 may impact the distinct functions of the two isoforms. Spine induction was blocked by 2-bromopalmitate, an inhibitor of palmitoylation, but the pleiotropic effects of this compound require cautious interpretation of these results. Our findings of two populations of bCdc42 increase the complexity of parsing the neuronal functions of canonical Cdc42 and bCdc42. Localization and functional characterization of the nonpalmitoylated CS mutant in neurons will be informative. Identification of two populations of bCdc42 also raises the question about the proportion of palmitoylated bCdc42 compared to CaaX-processed bCdc42. We estimated that 5 to 20% of membrane-bound bCdc42 is palmitoylated in neonatal mouse brain (Fig. 7), pointing to a low basal level of palmitoylation, but also one that could be responsive to physiological changes. The palmitoylation status of bCdc42 is changed in response to glutamate stimulation of cortical neurons and in a mouse model of seizure, underscoring the potential importance of palmitoylation-dependent regulation of synapse architecture (21) and providing a strong impetus for further analysis.
Our studies show that palmitoylation enriches the amount of bCdc42 on the plasma membrane and potentiates its signaling activity (Fig. 6). The resistance of prenyl, palmitoyl bCdc42 to extraction by RhoGDIα implies that a larger pool of GTPase is available for activation by guanine nucletotide exchange factors at the membrane. An important goal for the future is to assess how the dynamics of the fatty acylation cycle coincide with nucleotide exchange and hydrolysis. Members of the RAL and PRL protein families are implicated in tumorigenesis and metastasis (56, 57). There is continued interest in the therapeutic potential of inhibition of CaaX processing enzymes, including Icmt (58, 59). Our study suggests that the prenyl, palmitoyl forms of CCaX proteins will be inert to inhibition by Icmt inhibitors but sensitive to inhibition of palmitoylation. Accordingly, it will be of interest to determine whether palmitoylation potentiates the activity of these proteins under normal and pathological conditions.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (GM051466).
We thank R. A. Cerione and members of his laboratory for reagents and advice, S. G. Young for Rce1 and Icmt null MEFs, A. D. Cox, N. A. Lambert, and S. Gonzalo for providing plasmids, Rujun Kang for the bCdc42 antibodies, and members of the Linder laboratory for helpful discussions.
Footnotes
Published ahead of print 28 January 2013
REFERENCES
- 1.Jaffe AB, Hall A. 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21:247–269 [DOI] [PubMed] [Google Scholar]
- 2.Erickson JW, Cerione RA. 2001. Multiple roles for Cdc42 in cell regulation. Curr. Opin. Cell Biol. 13:153–157 [DOI] [PubMed] [Google Scholar]
- 3.Heasman SJ, Ridley AJ. 2008. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9:690–701 [DOI] [PubMed] [Google Scholar]
- 4.Boulter E, Garcia-Mata R, Guilluy C, Dubash A, Rossi G, Brennwald PJ, Burridge K. 2010. Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat. Cell Biol. 12:477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adamson P, Marshall CJ, Hall A, Tilbrook PA. 1992. Post-translational modifications of p21rho proteins. J. Biol. Chem. 267:20033–20038 [PubMed] [Google Scholar]
- 6.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. 2012. Regulating the regulator: post-translational modification of RAS. Nat. Rev. Mol. Cell Biol. 13:39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook LA, Schey KL, Wilcox MD, Dingus J, Ettling R, Nelson T, Knapp DR, Hildebrandt JD. 2006. Proteomic analysis of bovine brain G protein gamma subunit processing heterogeneity. Mol. Cell. Proteomics 5:671–685 [DOI] [PubMed] [Google Scholar]
- 8.Bergo MO, Gavino BJ, Hong C, Beigneux AP, McMahon M, Casey PJ, Young SG. 2004. Inactivation of Icmt inhibits transformation by oncogenic K-Ras and B-Raf. J. Clin. Invest. 113:539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michaelson D, Ali W, Chiu VK, Bergo M, Silletti J, Wright L, Young SG, Philips M. 2005. Postprenylation CAAX processing is required for proper localization of Ras but not Rho GTPases. Mol. Biol. Cell 16:1606–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts PJ, Mitin N, Keller PJ, Chenette EJ, Madigan JP, Currin RO, Cox AD, Wilson O, Kirschmeier P, Der CJ. 2008. Rho Family GTPase modification and dependence on CAAX motif-signaled posttranslational modification. J. Biol. Chem. 283:25150–25163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cushman I, Casey PJ. 2009. Role of isoprenylcysteine carboxylmethyltransferase-catalyzed methylation in Rho function and migration. J. Biol. Chem. 284:27964–27973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanker AB, Mitin N, Wilder RS, Henske EP, Tamanoi F, Cox AD, Der CJ. 2010. Differential requirement of CAAX-mediated posttranslational processing for Rheb localization and signaling. Oncogene 29:380–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson JL, Erickson JW, Cerione RA. 2012. C-terminal di-arginine motif of Cdc42 protein is essential for binding to phosphatidylinositol 4,5-bisphosphate-containing membranes and inducing cellular transformation. J. Biol. Chem. 287:5764–5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang DA, Sebti SM. 2005. Palmitoylated cysteine 192 is required for RhoB tumor-suppressive and apoptotic activities. J. Biol. Chem. 280:19243–19249 [DOI] [PubMed] [Google Scholar]
- 15.Navarro-Lérida I, Sanchez-Perales S, Calvo M, Rentero C, Zheng Y, Enrich C, Del Pozo MA. 2012. A palmitoylation switch mechanism regulates Rac1 function and membrane organization. EMBO J. 31:534–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linder ME, Deschenes RJ. 2007. Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8:74–84 [DOI] [PubMed] [Google Scholar]
- 17.Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, Kuhlmann J, Brunsveld L, Chandra A, Ellinger B, Waldmann H, Bastiaens PI. 2010. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141:458–471 [DOI] [PubMed] [Google Scholar]
- 18.Marks PW, Kwiatkowski DJ. 1996. Genomic organization and chromosomal location of murine Cdc42. Genomics 38:13–18 [DOI] [PubMed] [Google Scholar]
- 19.Shinjo K, Koland JG, Hart MJ, Narasimhan V, Johnson DI, Evans T, Cerione RA. 1990. Molecular cloning of the gene for the human placental GTP-binding protein Gp (G25K): identification of this GTP-binding protein as the human homolog of the yeast cell-division-cycle protein CDC42. Proc. Natl. Acad. Sci. U. S. A. 87:9853–9857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munemitsu S, Innis MA, Clark R, McCormick F, Ullrich A, Polakis P. 1990. Molecular cloning and expression of a G25K cDNA, the human homolog of the yeast cell cycle gene CDC42. Mol. Cell. Biol. 10:5977–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R, Green WN, Yates JR, III, Davis NG, El-Husseini A. 2008. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456:904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berzat AC, Buss JE, Chenette EJ, Weinbaum CA, Shutes A, Der CJ, Minden A, Cox AD. 2005. Transforming activity of the Rho family GTPase, Wrch-1, a Wnt-regulated Cdc42 homolog, is dependent on a novel carboxyl-terminal palmitoylation motif. J. Biol. Chem. 280:33055–33065 [DOI] [PubMed] [Google Scholar]
- 23.Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, Hang HC. 2009. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J. Am. Chem. Soc. 131:4967–4975 [DOI] [PubMed] [Google Scholar]
- 24.Martin BR, Cravatt BF. 2009. Large-scale profiling of protein palmitoylation in mammalian cells. Nat. Methods 6:135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forrester MT, Hess DT, Thompson JW, Hultman R, Moseley MA, Stamler JS, Casey PJ. 2011. Site-specific analysis of protein S-acylation by resin-assisted capture. J. Lipid Res. 52:393–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leung KF, Baron R, Ali BR, Magee AI, Seabra MC. 2007. Rab GTPases containing a CAAX motif are processed post-geranylgeranylation by proteolysis and methylation. J. Biol. Chem. 282:1487–1497 [DOI] [PubMed] [Google Scholar]
- 27.Chelsky D, Gutterson NI, Koshland DE., Jr 1984. A diffusion assay for detection and quantitation of methyl-esterified proteins on polyacrylamide gels. Anal. Biochem. 141:143–148 [DOI] [PubMed] [Google Scholar]
- 28.Webb Y, Hermida-Matsumoto L, Resh MD. 2000. Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J. Biol. Chem. 275:261–270 [DOI] [PubMed] [Google Scholar]
- 29.Falsetti SC, Wang DA, Peng H, Carrico D, Cox AD, Der CJ, Hamilton AD, Sebti SM. 2007. Geranylgeranyltransferase I inhibitors target RalB to inhibit anchorage-dependent growth and induce apoptosis and RalA to inhibit anchorage-independent growth. Mol. Cell. Biol. 27:8003–8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng Q, Si X, Horstmann H, Xu Y, Hong W, Pallen CJ. 2000. Prenylation-dependent association of protein-tyrosine phosphatases PRL-1, -2, and -3 with the plasma membrane and the early endosome. J. Biol. Chem. 275:21444–21452 [DOI] [PubMed] [Google Scholar]
- 31.Hoffman GR, Nassar N, Cerione RA. 2000. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell 100:345–356 [DOI] [PubMed] [Google Scholar]
- 32.Lin Q, Fuji RN, Yang W, Cerione RA. 2003. RhoGDI is required for Cdc42-mediated cellular transformation. Curr. Biol. 13:1469–1479 [DOI] [PubMed] [Google Scholar]
- 33.Greaves J, Chamberlain LH. 2011. DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem. Sci. 36:245–253 [DOI] [PubMed] [Google Scholar]
- 34.Lan TH, Liu Q, Li C, Wu G, Lambert NA. 2012. Sensitive and high resolution localization and tracking of membrane proteins in live cells with BRET. Traffic 13:1450–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. 2004. Activation of endogenous Cdc42 visualized in living cells. Science 305:1615–1619 [DOI] [PubMed] [Google Scholar]
- 36.Hill CS, Wynne J, Treisman R. 1995. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell 81:1159–1170 [DOI] [PubMed] [Google Scholar]
- 37.Lin R, Bagrodia S, Cerione R, Manor D. 1997. A novel Cdc42Hs mutant induces cellular transformation. Curr. Biol. 7:794–797 [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Martin BR, Cravatt BF, Hofmann SL. 2012. DHHC5 protein palmitoylates flotillin-2 and is rapidly degraded on induction of neuronal differentiation in cultured cells. J. Biol. Chem. 287:523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ivaldi C, Martin BR, Kieffer-Jaquinod S, Chapel A, Levade T, Garin J, Journet A. 2012. Proteomic analysis of S-acylated proteins in human B cells reveals palmitoylation of the immune regulators CD20 and CD23. PLoS One 7:e37187 doi:10.1371/journal.pone.0037187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. 2012. Global profiling of dynamic protein palmitoylation. Nat. Methods 9:84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anant JS, Ong OC, Xie HY, Clarke S, O'Brien PJ, Fung BK. 1992. In vivo differential prenylation of retinal cyclic GMP phosphodiesterase catalytic subunits. J. Biol. Chem. 267:687–690 [PubMed] [Google Scholar]
- 42.Christiansen JR, Kolandaivelu S, Bergo MO, Ramamurthy V. 2011. RAS-converting enzyme 1-mediated endoproteolysis is required for trafficking of rod phosphodiesterase 6 to photoreceptor outer segments. Proc. Natl. Acad. Sci. U. S. A. 108:8862–8866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jenkins CM, Han X, Yang J, Mancuso DJ, Sims HF, Muslin AJ, Gross RW. 2003. Purification of recombinant human cPLA2 gamma and identification of C-terminal farnesylation, proteolytic processing, and carboxymethylation by MALDI-TOF-TOF analysis. Biochemistry 42:11798–11807 [DOI] [PubMed] [Google Scholar]
- 44.Tucker DE, Stewart A, Nallan L, Bendale P, Ghomashchi F, Gelb MH, Leslie CC. 2005. Group IVC cytosolic phospholipase A2gamma is farnesylated and palmitoylated in mammalian cells. J. Lipid Res. 46:2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heilmeyer LM, Jr, Serwe M, Weber C, Metzger J, Hoffmann-Posorske E, Meyer HE. 1992. Farnesylcysteine, a constituent of the alpha and beta subunits of rabbit skeletal muscle phosphorylase kinase: localization by conversion to S-ethylcysteine and by tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 89:9554–9558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohno Y, Kihara A, Sano T, Igarashi Y. 2006. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim. Biophys. Acta 1761:474–483 [DOI] [PubMed] [Google Scholar]
- 47.Lobo S, Greentree WK, Linder ME, Deschenes RJ. 2002. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 277:41268–41273 [DOI] [PubMed] [Google Scholar]
- 48.Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR, III, Davis NG. 2006. Global analysis of protein palmitoylation in yeast. Cell 125:1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michaelson D, Silletti J, Murphy G, D'Eustachio P, Rush M, Philips MR. 2001. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 152:111–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Norton AW, Hosier S, Terew JM, Li N, Dhingra A, Vardi N, Baehr W, Cote RH. 2005. Evaluation of the 17-kDa prenyl-binding protein as a regulatory protein for phototransduction in retinal photoreceptors. J. Biol. Chem. 280:1248–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang H, Li S, Doan T, Rieke F, Detwiler PB, Frederick JM, Baehr W. 2007. Deletion of PrBP/delta impedes transport of GRK1 and PDE6 catalytic subunits to photoreceptor outer segments. Proc. Natl. Acad. Sci. U. S. A. 104:8857–8862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nancy V, Callebaut I, El Marjou A, de Gunzburg J. 2002. The delta subunit of retinal rod cGMP phosphodiesterase regulates the membrane association of Ras and Rap GTPases. J. Biol. Chem. 277:15076–15084 [DOI] [PubMed] [Google Scholar]
- 53.Wilson SJ, Smyth EM. 2006. Internalization and recycling of the human prostacyclin receptor is modulated through its isoprenylation-dependent interaction with the delta subunit of cGMP phosphodiesterase 6. J. Biol. Chem. 281:11780–11786 [DOI] [PubMed] [Google Scholar]
- 54.Bhagatji P, Leventis R, Rich R, Lin CJ, Silvius JR. 2010. Multiple cellular proteins modulate the dynamics of K-ras association with the plasma membrane. Biophys. J. 99:3327–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chandra A, Grecco HE, Pisupati V, Perera D, Cassidy L, Skoulidis F, Ismail SA, Hedberg C, Hanzal-Bayer M, Venkitaraman AR, Wittinghofer A, Bastiaens PI. 2012. The GDI-like solubilizing factor PDEdelta sustains the spatial organization and signalling of Ras family proteins. Nat. Cell Biol. 14:148–158 [DOI] [PubMed] [Google Scholar]
- 56.Bodemann BO, White MA. 2008. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat. Rev. Cancer 8:133–140 [DOI] [PubMed] [Google Scholar]
- 57.Bessette DC, Qiu D, Pallen CJ. 2008. PRL PTPs: mediators and markers of cancer progression. Cancer Metastasis Rev. 27:231–252 [DOI] [PubMed] [Google Scholar]
- 58.Winter-Vann AM, Casey PJ. 2005. Post-prenylation-processing enzymes as new targets in oncogenesis. Nat. Rev. Cancer 5:405–412 [DOI] [PubMed] [Google Scholar]
- 59.Holstein SA, Hohl RJ. 2012. Is there a future for prenyltransferase inhibitors in cancer therapy? Curr. Opin. Pharmacol. 12:704–709 [DOI] [PubMed] [Google Scholar]
- 60.Reid TS, Terry KL, Casey PJ, Beese LS. 2004. Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity. J. Mol. Biol. 343:417–433 [DOI] [PubMed] [Google Scholar]