Abstract
G1 cyclins, in association with a cyclin-dependent kinase (CDK), are universal activators of the transcriptional G1-S machinery during entry into the cell cycle. Regulation of cyclin degradation is crucial for coordinating progression through the cell cycle, but the mechanisms that modulate cyclin stability to control cell cycle entry are still unknown. Here, we show that a lack of phosphate downregulates Cln3 cyclin and leads to G1 arrest in Saccharomyces cerevisiae. The stability of Cln3 protein is diminished in strains with low activity of Pho85, a phosphate-sensing CDK. Cln3 is an in vitro substrate of Pho85, and both proteins interact in vivo. More interestingly, cells that carry a CLN3 allele encoding aspartic acid substitutions at the sites of Pho85 phosphorylation maintain high levels of Cln3 independently of Pho85 activity. Moreover, these cells do not properly arrest in G1 in the absence of phosphate and they die prematurely. Finally, the activity of Pho85 is essential for accumulating Cln3 and for reentering the cell cycle after phosphate refeeding. Taken together, our data indicate that Cln3 is a molecular target of the Pho85 kinase that is required to modulate cell cycle entry in response to environmental changes in nutrient availability.
INTRODUCTION
When environmental conditions change, Saccharomyces cerevisiae, like other organisms, takes several coordinated decisions about growth, cell division, and maintaining homeostasis. Nutrient status is among the most important environmental conditions that must be accurately sensed and responded to in order to ensure cell survival. In the absence of nutrients, cells arrest in G1 phase, and this regulation of cell cycle becomes essential in the adaptation process. However, nothing is known about how nutrients impinge on the cell cycle machinery (1).
In budding yeast, the commitment to a new round of cell division takes place in late G1, at a point called Start, where transcriptional activation of more than 200 G1-specific genes occurs (reviewed in reference 2). This includes the transcription of major cell cycle regulators, including the G1 cyclins Cln1, Cln2, Clb5, and Clb6, as well as numerous genes with functions related to DNA metabolism, budding, spindle pole body duplication, and cell wall synthesis (3, 4). Many of these transcribed genes are targets of the heterodimeric transcription factors SBF and MBF, each of which contains a Swi6 subunit and a distinct DNA-binding subunit: Swi4 and Mbp1, respectively (5). It is interesting to note that, when SBF and MBF are poised at their target promoters during much of G1 phase, they cannot activate transcription; rather, they repress it (6, 7). To become activators at Start, they need an upstream element: the Cdc28/Cln3 complex.
Cdc28 is a cyclin-dependent kinase (CDK) that governs all of the cell cycle progression, and Cln3 is a highly unstable cyclin that remains fairly constant throughout the cell cycle (8, 9). When Cdc28 is associated with Cln3 cyclin, it unfolds the activation of transcriptional G1 machinery, chiefly through two mechanisms: phosphorylation of the Whi5 repressor (and presumably of Swi6) at multiple residues (10–12) and control of the Stb1 dual protein and the histone deacetylases (13). In addition to Cln3, other elements involved in G1 transcription firing have recently been demonstrated: for example, a positive-feedback mechanism involving Cln1 and Cln2 could reinforce SBF/MBF activation (14). At the end of G1, the rise in Cln/Cdc28 activity results in the phosphorylation and targeting for degradation of Sic1, an inhibitor of the S-phase cyclins, thereby enabling robust entry into S-phase (15).
Nutrients are important trophic factors that control the passage through Start by activating several signaling pathways, including the protein kinase A and Snf1 pathways, which positively regulate cell proliferation in response to glucose availability (16), and the TORC1 pathway, which controls the cell cycle according to nitrogen levels (17). Consequently, inactivation of any of three major pathways results in a cell cycle blockade and, in other typical phenotypes, of the G0-like growth arrest program, even in the presence of other abundant nutrients. Due to its role as the most upstream activator, Cln3 has emerged as a good target for these pathways. Indeed, it has been demonstrated that this cyclin becomes less stable under nitrogen deprivation, although the mechanism(s) underlying this effect remains obscure (18).
Phosphate is sensed by another signaling pathway, the PHO pathway, which has progressively been uncovered by the O'Shea group (19). The central element of this pathway is another CDK: Pho85, which associates with a family of 10 cyclins, each of which can direct Pho85 to different substrates (for a review, see reference 20). Pho80 is the main cyclin of the PHO pathway, which controls phosphate homeostasis, and Pho81 is the corresponding CDK inhibitor (21). The phosphate sensor that controls this pathway is still unknown: however, a low-Pi signal is known to be transmitted via certain inositol polyphosphate (IP) species (e.g., heptakisphosphate [IP7]), which are synthesized by Vip1 IP6 kinase. IP7 apparently interacts noncovalently with the Pho80/Pho85/Pho81 complex and induces additional interactions between Pho81 and Pho80/Pho85, preventing substrates from accessing the active site of the kinase (19, 22). Thus, the ternary complex Pho80/Pho81/Pho85 is active in rich medium but becomes inactive upon phosphate starvation, leading to the migration of unphosphorylated Pho4 transcription factor into the nucleus and enabling the expression or repression of PHO genes (21, 23, 24). This transcriptional response ultimately results in maintenance of the internal phosphate levels.
Besides being constitutively associated with Pho80, Pho85 is also bound to other cyclins (e.g., Pcl1, Pcl2, and Pcl9) (20). In fact, the four Pho85 complexes should be considered different enzymes: they recognize different substrates, they are localized in different subcellular regions, and the respective activities of the Pcl1/Pho85, Pcl2/Pho85, and Pcl9/Pho85 complexes are not regulated by external phosphate. When Pho85 is associated with these Pcl cyclins, it cooperates with Cdc28 in specific morphogenetic events during the G1-S transition.
It is logical to wonder whether Pho80/Pho85 complexes could also help control the cell cycle. Indeed, there is evidence that Pho80/Pho85 phosphorylates and inhibits Rim15, a PAS kinase that promotes the entry into the G0 program in stationary cells (25), although whether this mechanism is involved in cell cycle-induced arrest is unknown. In addition, it has been demonstrated that Pho80/Pho85 is essential to restart the cell cycle after G1 arrest due to DNA damage (26), suggesting that the Pho85 activity is essential when Cdc28 activity is absent (20).
Here, we demonstrate that the lack of phosphate leads to downregulation of Cln3 protein levels and to G1 arrest. Interestingly, neither effect is observed in cells that overexpress Pho85 or in cells that cannot inhibit it (i.e., vip1Δ or pho81Δ cells). In accordance, we also demonstrate that Cln3 is less stable in strains with low Pho85/Pho80 activity and that it is phosphorylated in vitro by Pho85/Pho80 complexes at S449 and T520. More interestingly, we report that cells carrying a CLN3 allele encoding aspartic acid substitutions at these sites maintain high levels of Cln3 independently of Pho85 activity. Accordingly, the nonphosphorylatable alanine mutant displays the same low levels as the pho85Δ mutant. Finally, we show that when nutrient levels drop, downregulation of Cln3 is essential to establish proper G1 arrest and that once these levels recuperate, activation of Pho85 is essential to restart the cell cycle from the G0 state. Together, our findings indicate that phosphate levels regulate the amount of Cln3 by controlling Pho85/Pho80 kinase activity.
MATERIALS AND METHODS
Strains.
The strains used are indicated in Table 1. The pho85-as strain (YAM67, which carries an analog-sensitive allele of PHO85) was provided by Erin O'Shea and was used for inhibition of Pho85 in the presence of the inhibitor 1-Na PP1; (4-amino-1-tert-butyl-3-(1′-naphthylpyrazolo[3,4-d]pyrimidine) kindly provided by K. Shokat) as described previously (27). The Pho85 inhibition was confirmed by following the entry of Pho4-green fluorescent protein (GFP) into the nucleus.
Table 1.
Yeast strains used in this study
| Strain | Background | Genotype | Source |
|---|---|---|---|
| BY4741 | BY4741 | MATa his3Δ1 leu2Δ200 met15Δ0 ura3Δ0 | Euroscarf |
| YNR55 | BY4741 | CLN3-MYC-KanMX | This study |
| YAM78 | BY4741 | pho85Δ::LEU2 | This study |
| YAM78 | BY4741 | pho85Δ::LEU2 CLN3-MYC-KanMX | This study |
| YAM113 | BY4741 | pRS416-CLN3-MYC | This study |
| YAM114 | BY4741 | pho85Δ::LEU2 pRS416-CLN3-MYC | This study |
| YAM115 | BY4741 | pcl1Δ::LEU2 pRS416-CLN3-MYC | This study |
| YAM119 | BY4741 | pcl2Δ::KanMX pcl1Δ::LEU2 pRS416-CLN3-MYC | This study |
| YAM116 | BY4741 | pcl6Δ::KanMX pRS416-CLN3-MYC | This study |
| YAM117 | BY4741 | pcl7Δ::KanMX pRS416-CLN3-MYC | This study |
| YAM118 | BY4741 | pcl8Δ::KanMX pRS416-CLN3-MYC | This study |
| YAM99 | BY4741 | pcl9Δ::KanMX pRS416-CLN3-MYC | This study |
| YAM111 | BY4741 | pho80Δ::LEU2 pRS416-CLN3-MYC | This study |
| YPC702 | BY4741 | SWI6-TAP-KanMX | This study |
| YNR11 | BY4741 | WHI5-TAP-KanMX | This study |
| YAM150 | BY4741 | vip1Δ::KanMX pRS416-CLN3-MYC | This study |
| YNR82 | BY4741 | pho81Δ::LEU2 CLN3-MYC-KanMX | This study |
| YAM63 | BY4741 | CLN3-MYC-KanMX pEG(KT)-GAL-PHO85 | This study |
| YAM91 | BY4741 | pRS416-cln3-1-HA | This study |
| YAM92 | BY4741 | pho85Δ::LEU2 pRS416-CLN3-1-HA | This study |
| YAM151 | BY4741 | ubc4Δ::LEU2 CLN3-MYC-KanMX | This study |
| YAM142 | BY4741 | pho85Δ::URA ubc4Δ::LEU2 CLN3-MYC-KanMX | This study |
| YAM146 | BY4741 | PHO85-TAP-KanMX pRS416-CLN3-MYC | This study |
| YNR13 | BY4741 | PHO85-TAP-KanMX | This study |
| YAM113 | BY4741 | pRS416-CLN3-MYC | This study |
| YAM163 | BY4741 | pRS416-CLN3-(T520A S449A-MYC) | This study |
| YAM149 | BY4741 | pho85Δ::LEU2 pRS416-CLN3-(T519D T520D I448D S449D-MYC) | This study |
| YAM103 | BY4741 | pho80Δ::LEU2 | This study |
| YAM45 | BY4741 | pho81Δ::LEU2 | This study |
| YNR46 | BY4741 | vip1Δ::KanMX | This study |
| YPC631 | BY4741 | cln3Δ::KanMX | This study |
| YSH10 | BY4741 | atg1Δ::LEU2 CLN3-MYC-KanMX | This study |
| YSH11 | BY4741 | pho85Δ::URA atg1::LEU2 CLN3-MYC-KanMX | This study |
| YAM67 | K699 | pho85Δ::LEU2 ADE2 PHO4-GFP trp1::PHO85(F82G)-TRP1 | E. O'Shea |
| YAN32 | W303 | CLN2-HA-URA3 SIC1-MYC-KanMX CLB5-TAP-LEU2 | F. Posas |
Plasmids.
The plasmids used in this work are listed in Table 2. CLN3-MYC was expressed from its own promoter in the centromeric plasmid pRS416; this plasmid expresses CLN3 at a level similar to the expression of the genomic tagged version. To express MYC-tagged mutated versions of CLN3, we replaced S449 and T520 with alanine or used the substitutions I448D S449D and T519 T520D to mimic the double-negative charge of the phosphate group. PHO85 and Pho80 were overexpressed under the control of the GAL1 promoter in the pEG(KG) plasmid. YAM91 carries a plasmid with CLN3 hemagglutinin (HA)-tagged protein lacking the PEST sequence of CLN3 (the cln3-1-HA allele). The CLN2 open reading frame (ORF) was placed under the control of the Schizosaccharomyces pombe adh1 promoter in YCplac22 (28), resulting in plasmid pCM64. Recombinant proteins were expressed from the pGEX6P1 plasmid. Due to its toxicity in Escherichia coli cells, full-length Cln3 could not be expressed, and the C-terminal half of Cln3 (from Met 347 to Arg 580) was cloned into the BamHI site of pGEX6P1 (pJC1154).
Table 2.
Plasmids used in this study
| Name | Relevant characteristics | Reference or source |
|---|---|---|
| pCM64 | YCplac22-ADHp-CLN2 | 14 |
| pJC1065 | pEG(KG)-PHO85 (cloned in BamHI) | This study |
| pJC1437 | pEG(KG)-PHO80 (cloned in BamHI) | This study |
| pJC1280 | pRS416-CLN3-MYC | This study |
| pJC1161 | pGEX6P1-PHO80 (cloned in BamHI) | This study |
| pJC1164 | pGEX6P1-PHO85 (cloned in BamHI) | This study |
| pJC1154 | pGEX6P1-CLN3 (from nucleotide 1038 to stop codon) | This study |
| pJC1448 | pGEX6P1-CLN3 (from nucleotide 1038 to stop codon) with the 2 putative Pho85 sites (S449 and T520) mutated to Asp | This study |
| pJC1456 | pGEX6P1-CLN3 (from nucleotide 1038 to stop codon) with the 2 putative Pho85 sites (S449 and T520) mutated to Ala | This study |
| pJC1449 | pGEX6P1-CLN3 (from nucleotide 1159 to stop codon) with the 8 putative Cdc28 sites (T420, S455, S462, S466, S466, T478, S514, and T517) mutated to Ala | This study |
Growth conditions.
Cells were grown in either YPD medium (1% yeast extract, 2% peptone, and 2% glucose) or complete synthetic dextrose (SD) medium (0.67% yeast nitrogen base and 2% glucose) containing amino acids for auxotrophic requirements (15 μg/ml leucine, 5 μg/ml histidine, and 10 μg/ml tryptophan).
Yeast nitrogen base without phosphate was used as recommended by the manufacturer (MP Biomedicals) to prepare SD medium without a phosphate source. Phosphate deprivation experiments were done with cells growing exponentially in SD for 14 to 16 h until they reached an optical density at 600 nm (OD600) of 0.3 to 0.4, at which point the cells were collected by filtration and, after a quick wash, resuspended at the same cellular concentration in prewarmed medium lacking the phosphate source, as previously described (18). The nitrogen deprivation experiment was performed under the same conditions using a yeast nitrogen base without ammonium sulfate (Difco). α-Factor cell synchrony experiments were done as previously described (29).
DNA content, cell volume, cell number, and budding index measurements.
Approximately 1 × 107 cells were collected and processed as described previously (30). DNA was stained with SYBR green and then analyzed in a FACSCalibur cytometer (Becton Dickinson). Cell number and volume were quantified using a Scepter cell counter (Millipore). Budding was analyzed microscopically by scoring a minimum of 200 cells.
Immunoblot analysis.
The primary anti-Myc or anti-HA monoclonal antibodies (kindly provided by F. Posas) were used at 1:100 and followed by anti-mouse-horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibody (1:20,000), using SuperSignal West Femto chemiluminescent substrate (Thermo Scientific). Anti-glucose-6-phosphate dehydrogenase (anti-G6PDH; Sigma) was used at 1:500.
Coimmunoprecipitation of PHO85-TAP and CLN3-MYC.
Exponential-phase yeast cells were harvested (500 ml at an OD600 of 0.7) and resuspended in 5 ml of cold extraction buffer A (50 mM Tris, pH 8, 15 mM EDTA, 15 mM EGTA, 0.1% Triton X-100) containing protease inhibitors (2 μg/ml each of pepstatin, leupeptin, phenylmethylsulfonyl fluoride [PMSF], and benzamidine) and phosphatase inhibitors (10 mM sodium orthovanadate and 250 mM β-glycerophosphate). Cells were ruptured by vortexing with glass beads, and the resulting extract was centrifuged at 4°C for 1 h at 12,000 rpm. Amounts of 3 mg of crude extracts were incubated for 2 h at 4°C with 150 μl of IgG-Sepharose beads (Amersham Biosciences). After washing with extraction buffer, the proteins bound to the beads were resuspended in 30 μl of SDS-PAGE sample buffer, heated at 95°C for 5 min, and loaded onto SDS-PAGE gels.
Recombinant protein purification.
For expression of glutathione S-transferase (GST) or GST fusion proteins, Escherichia coli strain BL21(DE3) (Stratagene) was transformed with the corresponding plasmids. Protein expression was induced with 1 mM isopropyl β-d-thiogalactopyranoside for 3 h at 25°C. Cells were collected by centrifugation, resuspended in 600 μl of phosphate-buffered saline with 0.1% Triton X-100 (PBS-T) supplemented with a protease inhibitor mixture (Roche Applied Science), and subjected to mechanical rupture. The cell debris was removed by centrifugation, and the supernatants were purified using glutathione-Sepharose column chromatography, as described in the manufacturer's protocol. After incubation for 2 h at 4°C with rotation, the beads were collected by centrifugation (1,000 rpm for 1 min at 4°C) and washed with PBS-T three times. The elution was performed by adding 10 mM glutathione.
Kinase assays.
Kinase assays were performed essentially as described previously (31). GST-Pho85 and GST-Pho80 were purified from bacteria as described above. A fragment of GST-Cln3 was expressed (plasmid pJC1154) and purified from bacteria. Cdc28-TAP was purified using IgG Sepharose beads (Sigma). Phosphorylated proteins were detected by using the Pro-Q diamond phosphoprotein gel stain kit (Invitrogen).
Viability assays.
The viability experiments were done as described in reference 32. Stationary-phase cells were diluted, plated in YPD, and incubated for 2 days.
RNA isolation and analysis.
Cells were harvested and stored at −80°C. Total RNA was isolated by hot phenol extraction and quantified spectrophotometrically. An amount of 2 μg of total RNA was incubated with DNase and reverse transcribed using Quanta qScript cDNA supermix following the manufacturer's instructions. The cDNA was subjected to reverse transcription (RT)-PCR on a C1000 thermal cycler-CFX96 real-time PCR system, and expression was normalized to that of CDC28.
RESULTS
The G1 arrest caused by phosphate depletion may involve active mechanisms to downregulate Cdc28-Cln activity.
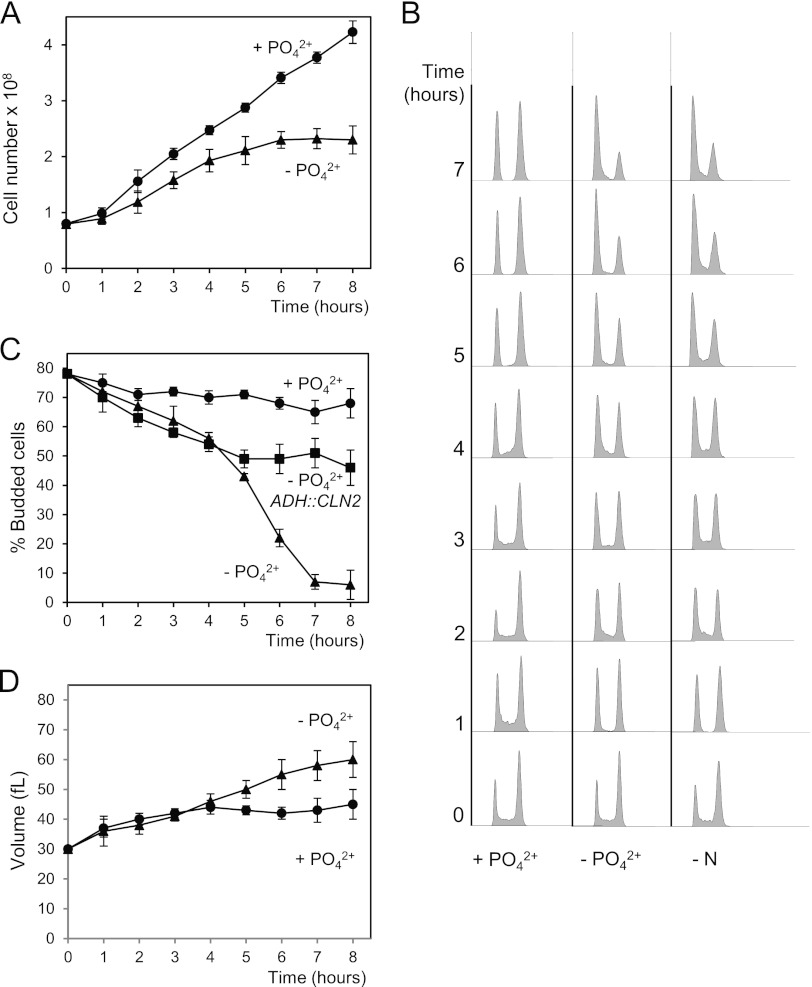
Nutrients are important trophic factors in all organisms. When deprived of a nitrogen or carbon source, yeast cells use accumulated reserves to complete the current cycle and arrest at the following G1 phase. We sought to determine whether phosphate starvation also provokes G1 arrest. To this end, we transferred exponentially growing yeast cells from complete medium to a medium lacking a phosphate source. The rate of division declined slowly over the next 3 h (Fig. 1A) and, finally, the cells arrested in G1, as indicated by their DNA content and budding index (Fig. 1B and C). Interestingly, the cell cycle arrest was comparable to that observed in the absence of nitrogen (Fig. 1B) and was not merely a consequence of the inability to meet the growth requirements of the G1/S transition, as suggested by the fact that the cell volume still increases under phosphate deprivation (Fig. 1D).
Fig 1.
Phosphate deprivation leads to G1 arrest. Wild-type cells were grown exponentially in synthetic complete medium. At time zero, cells were harvested and incubated either in the same medium (+PO42−) or in a medium without phosphate (−PO42−). At the indicated times, samples were collected and then subjected to several analyses: total cell number (A), DNA content (B), percentage of budding (C), and cell volume (D). (B) The DNA contents of wild-type cells incubated in a medium without a nitrogen source (−N) are shown. (C) The percentages of budding of wild-type cells carrying a centromeric plasmid with the CLN2 gene expressed from an ADH promoter are shown. Data ± standard deviations from three independent experiments are shown.
Considering that cyclins are fundamental to progression through G1, we asked if the arrest caused by phosphate deprivation requires downregulation of the activity of G1 cyclins. Cells expressing CLN2 from a constitutive promoter continue to grow in size (not shown) and, more importantly, cells cross Start despite the absence of phosphate, as deduced from the budding index (Fig. 1C). This indicates that the downregulation of Cln activity may be essential in order to achieve a proper G1 arrest under phosphate-fasting conditions. Taken together, these results strongly suggest that the G1 arrest caused by phosphate deprivation is mediated by specific mechanisms, some of which involve the downregulation of Cln proteins, and cannot be explained simply as a passive consequence of a growth arrest that would prevent cells from reaching the critical cell mass necessary to bud and to initiate S phase.
External phosphate controls the levels of Cln3 protein.
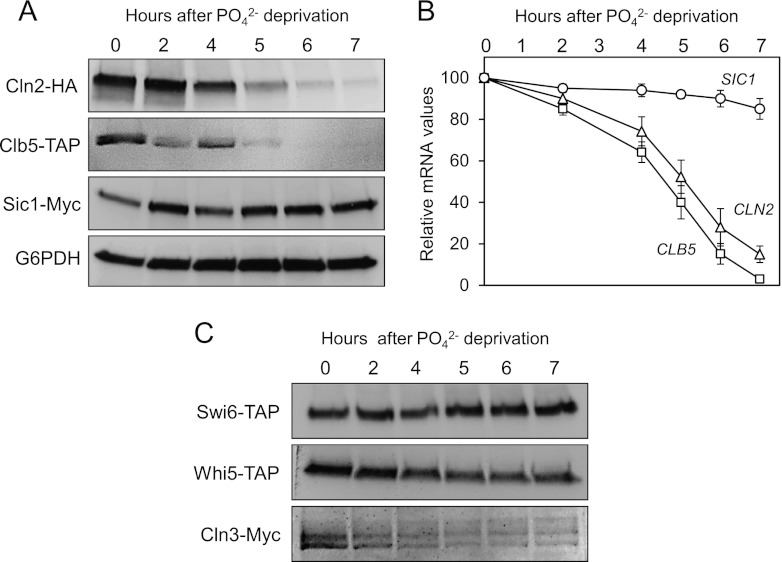
Since constitutive expression of Cln2 could partially reverse the blockade in G1, we examined whether the absence of phosphate could affect the levels of proteins controlling the G1/S transition. The results in Figure 2A clearly show that the protein levels of Cln2 and Clb5 (essential for budding and S-phase entry) decrease after 4 h of growth in phosphate-free medium, whereas the levels of the protein Sic1 (essential for arresting the cell cycle before S phase) remain constant. We then analyzed the mRNA levels of all these genes (Fig. 2B), finding that the mRNA of CLN2 and CLB5 were also progressively depleted, whereas the transcripts of SIC1 remained constant. These results suggest that the absence of phosphate may inhibit the transcription of genes required for the G1/S transition.
Fig 2.
Phosphate deprivation leads to the downregulation of Cln3p. (A) Strain YAN32 (triple tagged: Cln2-HA, Clb5-TAP, and Sic1-Myc) was grown exponentially in synthetic complete medium. At time zero, the cells were harvested and incubated in phosphate (PO42−)-free medium. At the indicated times, samples were recovered and then analyzed for different proteins by immunoblotting using specific antibodies. (B) The wild-type strain was treated and sampled as described in Materials and Methods. Transcripts were analyzed using RT-PCR with specific primers. Data ± standard deviations from three independent experiments are shown. (C) Strains YPC702 (SWI6-TAP), YNR11 (WHI5-TAP), and YNR55 (CLN3-MYC) were treated, sampled, and analyzed as described for panel A.
The G1/S transcriptional wave is controlled by the SBF and MBF factors, which may be regulated by phosphate levels. As observed from the results in Figure 2C, the protein levels of Swi6 are not affected during the first 6 h after phosphate deprivation (nor are those of Swi4 or Mbp1 [not shown]), thereby excluding the possibility that SBF and MBF are controlled at the expression level. Whi5 and Cln3 are known to play a key role in the activation of SBF and MBF complexes in late G1 (10). The results in Figure 2C show that, after 3 h of phosphate depletion, the amount of Cln3 protein decreases rapidly, while the levels of Whi5 repressor remain fairly constant. These results support the idea that phosphate deprivation downregulates SBF and MBF activity as a result of the loss of Cln3. Considering that a triple cln mutant cannot undergo the G1/S transition unless Sic1 is removed (33), we propose here that the absence of Cln proteins and the sustained presence of Sic1 may explain the G1 arrest produced by phosphate deprivation in terms of molecular requirements.
Pho85 activity regulates the levels of Cln3 protein.
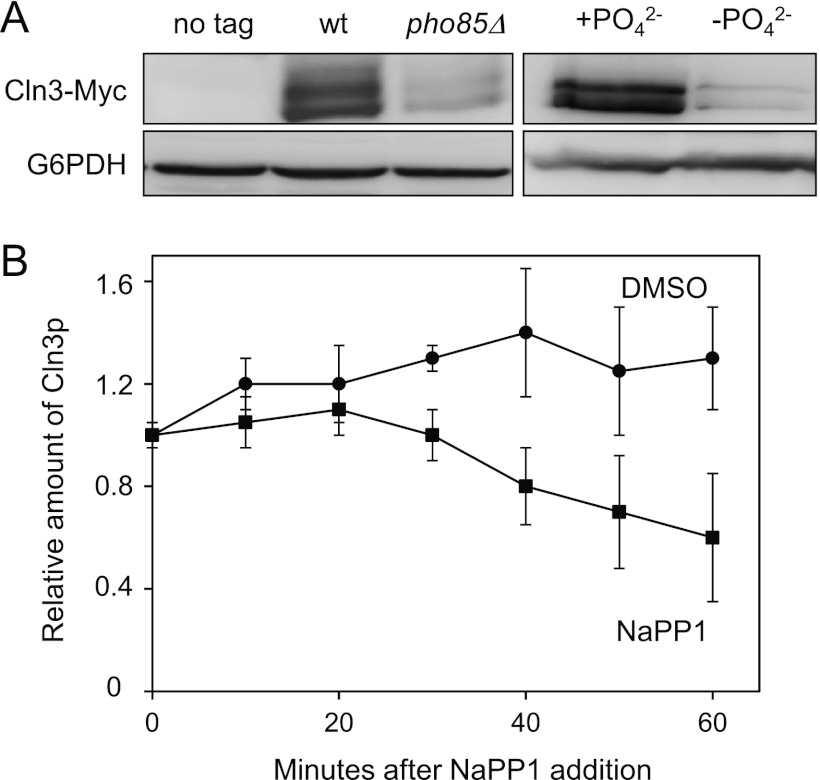
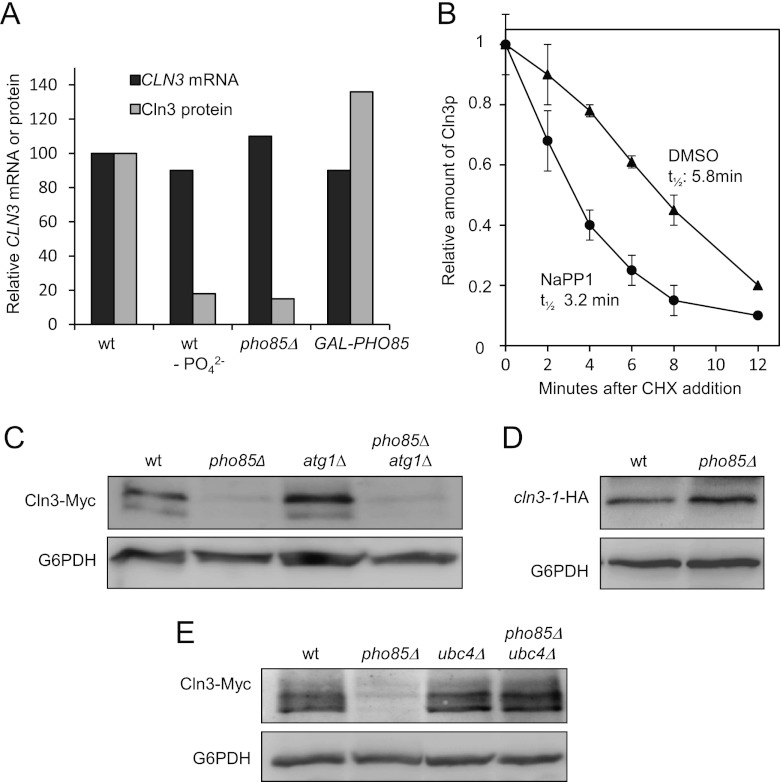
It is known that exogenous phosphate activates Pho85, a CDK that controls the homeostasis of phosphate, and we examined whether this kinase could also be involved in the control of Cln3 cyclin. As reflected in Figure 3A, PHO85-depleted cells growing exponentially in rich medium exhibit low levels of Cln3 protein. This decrease is very similar to that observed in wild-type cells grown in phosphate-free medium. PHO85 gene deletion results in a broad spectrum of defects, and it is possible that our pho85Δ laboratory strain bears suppressor mutations that alleviate such defects. To avoid this problem, we took advantage of the pho85-as strain, which behaved quite similarly to wild-type cells until the specific inhibitor 1-Na PP1 was added to the culture. As observed by the results in Figure 3B, the addition of 1-Na PP1 to the exponentially growing cells results in a progressive depletion of Cln3 relative to the amount in control cells. The localization of Pho4-GFP is a widely used readout to evaluate Pho85 activity (27), and Cln3 starts to decrease at the same time that Pho4-GFP protein enters the nucleus (not shown). The latter result shows a good correlation between Pho85 inactivation and the reduction of Cln3 protein and suggests that Pho85 activity (rather than the physical presence of the protein itself) is a key factor controlling cellular levels of Cln3.
Fig 3.
Pho85 inactivation leads to downregulation of Cln3. (A) Wild-type (wt) and pho85Δ cells were grown exponentially in YPD and then assessed for levels of Cln3-Myc (left), and wild-type cells were grown in synthetic complete medium with (+PO42−) or without (−PO42−) phosphate (right). Samples were taken after 6 h, and levels of Cln3-Myc were monitored. (B) Cells from the YAM67 strain were incubated with either 1-Na PP1 (a specific pho85-as inhibitor) or drug vehicle (dimethyl sulfoxide [DMSO]). Samples were taken at the indicated times, and Cln3-Myc was analyzed by immunoblotting. Data ± standard deviations from three independent experiments are shown. Cln3p, phosphorylated Cln3.
Upstream elements of the PHO pathway also affect Cln3 protein levels.
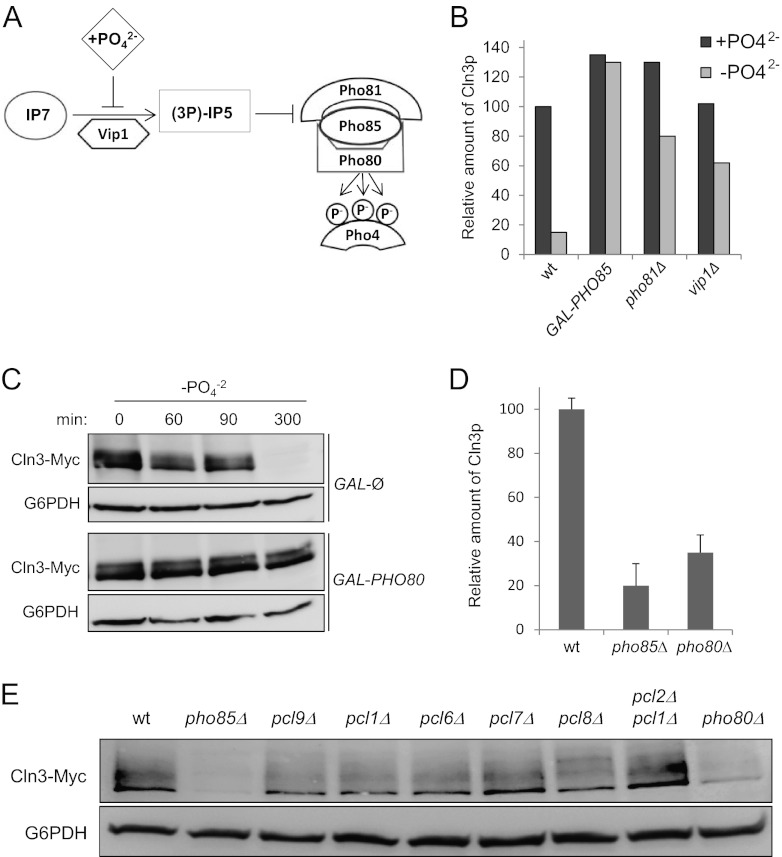
Pho85 activity is controlled by external phosphate through an upstream signaling transduction pathway that has been progressively elucidated by several research groups (Fig. 4A) (for a review, see reference 34). The vip1Δ and pho81Δ strains are unable to respond to changes in the external concentration of phosphate and therefore, cannot inhibit Pho85 in a deprivation situation. Thus, we hypothesized that these strains could not downregulate Cln3 protein levels in response to depletion of phosphate, which we confirmed experimentally (Fig. 4B). Consequently, the overexpression of PHO80 has the same effect (Fig. 4C) as the absence of the inhibitors. All these results strongly suggest that the phosphate source controls the quantity of Cln3 cyclin by modulating Pho85 activity.
Fig 4.
Phosphate controls cellular levels of Cln3 by modulating the PHO pathway. (A) Schematic of the PHO pathway. During phosphate starvation, Vip1 causes an increase in the levels of inositol heptakisphosphate (IP7), which binds to and changes the conformation state of Pho81, leading to the inactivation of Pho85/Pho80 complexes. (B) Relative amounts of Cln3-Myc in different strains. Wild-type (wt), pho81Δ, and vip1Δ strains were grown in synthetic complete medium with (+PO42−) or without (−PO42−) phosphate. After 5 h, the levels of Cln3-Myc were quantified by immunoblotting using monoclonal antibodies. Cells of the Gal1-PHO85 strain (a wild-type strain that carries a centromeric plasmid with PHO85 expressed under the Gal1 promoter) were grown for 5 h in synthetic complete medium with galactose as a carbon source, either with or without phosphate. (C) Pho80 is necessary to maintain high levels of Cln3. As described for panel B, wild-type (GAL-Ø) and GAL1-PHO80 strains were grown in synthetic complete medium in the presence of galactose without phosphate (−PO42−). Levels of Cln3-Myc were quantified by immunoblotting using monoclonal antibodies. (D) Pho80 is necessary to maintain high levels of Cln3. Quantification of data in panel E (data ± standard deviations from four independent experiments) is shown. (E) Pho80 is necessary to maintain high levels of Cln3. A plasmid with a Cln3-Myc epitope tag was introduced in strains with the indicated mutations for deficiency of the different Pho85 cyclins. After 3 h of exponential growth in YPD, the levels of Cln3-Myc were analyzed by immunoblotting.
Loss of a particular Pho85 cyclin can phenocopy some aspects of the phenotype of a pho85Δ mutant, and we decided to evaluate the levels of Cln3 protein of the different cyclin mutants. As shown by the results in Figure 4D and E, the pho80Δ strain is the only one that shows a reduction in the Cln3 content very similar to that observed in pho85Δ cells, suggesting that the effect of the CDK could be mediated by the Pho80 cyclin. This result is in agreement with the fact that the Pho80/Pho85 complexes (together with Pcl7/Pho85) are inhibited when deprived of phosphate (35), providing further evidence that such nutrients control the quantity of Cln3 by modulating the activity of Pho85 kinase.
Pho85 affects Cln3 stability.
Pho85/Pho80 complexes control the transcription of many genes, and we reasoned that these complexes could modulate the transcription of CLN3 in response to changes in phosphate levels. However, there are no differences in the levels of CLN3 mRNA in wild-type cells cultured in the presence or absence of phosphate (Fig. 5A).
Fig 5.
Pho85 activity increases the half-life of Cln3 protein. (A) Different strains with a genomic tagged version of CLN3 were sampled and then analyzed for CLN3 mRNA levels (by RT-PCR) or Cln3-Myc levels (by immunoblotting). Wild-type cells were grown exponentially in synthetic complete medium with (wt) or without (wt −PO42−) phosphate, pho85Δ strain cells were grown in the same complete medium with phosphate, and GAL-PHO85 cells were grown for 5 h in synthetic complete medium with galactose as a carbon source. (B) YAM67 strain cells were incubated with either 1-Na PP1 (a specific pho85-as inhibitor) or drug vehicle (DMSO). Forty minutes later (time zero), cycloheximide (CHX) was added to the cultures (final concentration, 10 μg/ml). At the indicated times, samples were taken and analyzed for Cln3-Myc levels by immunoblotting. Data ± standard deviations from four independent experiments are shown. t1/2, half-life. (C) Downregulation of autophagy does not restore the diminished Cln3 levels of pho85Δ mutants. Strains of the indicated genotypes were grown exponentially in phosphate-rich medium, and Cln3-Myc was analyzed by immunoblot assay using specific antibodies. (D) Absence of the PEST region restores Cln3 levels in pho85Δ cells. Wild-type or pho85Δ cells carrying a plasmid with a cln3-1 allele without the PEST region (41) were grown exponentially in YPD. Cln3-HA levels were measured by immunoblotting. (E) Absence of UBC4 restores Cln3 levels in a pho85Δ strain. Strains of the indicated genotypes carrying a genomic Myc-tagged version of Cln3 were grown exponentially in phosphate-rich medium. Cells were harvested, and Cln3 levels were evaluated by immunoblotting.
Likewise, we could not detect any changes in the mRNA levels of CLN3 when comparing wild-type cells, pho85Δ cells, and cells that overexpress PHO85. Despite the similar levels of mRNA, we again confirmed the differences in Cln3 protein levels under these new experimental conditions (Fig. 5A). These results suggest that Cln3 must be downregulated by posttranscriptional mechanisms.
Next, we tested whether Pho85 activity affects the stability of Cln3 by studying the levels of Cln3 in cycloheximide-treated cells. In wild-type cells, we estimated a Cln3 half-life of 8 min (data not shown), in accordance with values reported by others (18). Unfortunately, we were unable to measure the half-life of Cln3 in the pho85Δ strain because of the low levels in these cells. For this reason, we decided to assess the Cln3 stability in the pho85-as strain in the presence or absence of the specific inhibitor (Fig. 5B). Under such conditions, the pho85-as strain still retains low levels of Pho85 activity but clearly exhibits an apparent reduction in the half-life of Cln3 protein (5.8 min in control cells versus 3.2 min in the presence of the inhibitor). Although this result does not exclude the existence of other regulatory mechanisms (such as the control of the translation of Cln3 mRNA), it strongly suggests that Pho85 activity increases the stability of the cyclin.
Pho85 does not control Cln3 stability through regulation of autophagy.
It has been recently described that Pho85 negatively regulates autophagy through diverse mechanisms (36). Autophagy induces the destruction of many proteins under conditions of nutrient deprivation, and we hypothesized that Cln3 could be a likely candidate for destruction. To test this hypothesis, we quantified the Cln3 levels in a pho85Δ strain whose autophagy process has been abrogated the (pho85Δ atg1Δ strain) and found that the Cln3 protein levels still were clearly decreased (Fig. 5C). This result rules out the possibility that Pho85 controls Cln3 stability through the regulation of autophagy.
Pho85 affects Cln3 ubiquitin-dependent degradation.
Since Cln3 is constitutively degraded by a ubiquitination-dependent mechanism (9), we decided to test whether Pho85 controls this process. We found 2 pieces of experimental evidence to support this hypothesis: (i) deletion of the PEST region in CLN3 (the cln3-1 allele; see the legend to Fig. 6A), which has been shown to prevent ubiquitination, also stabilizes the protein, even in the pho85Δ strain (Fig. 5D), and (ii) deletion of the E2 ubiquitin ligase Ubc4, which is important for nutrient homeostasis and is involved in the ubiquitination of Cln3 (9), restores Cln3 levels in the pho85Δ strain (Fig. 5E). Therefore, Pho85 may somehow interfere with Cln3 ubiquitination and/or destruction processes. Bearing in mind that Pho85 is a protein kinase, one explanation could be that it phosphorylates Cln3 and thereby interferes with such processes.
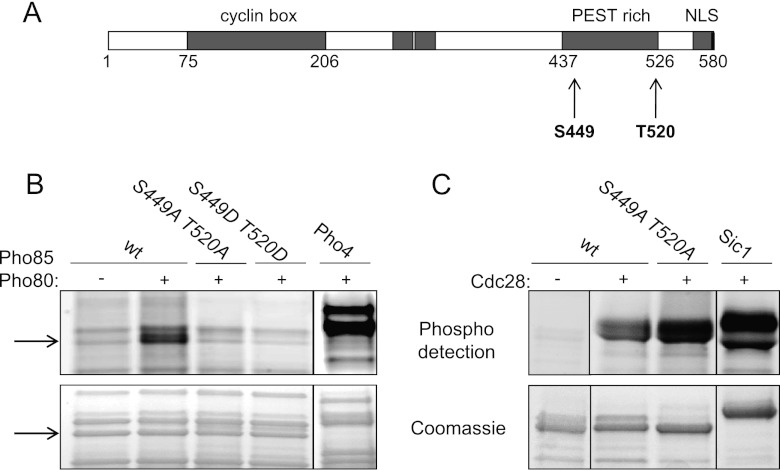
Fig 6.
Cln3 is phosphorylated in vitro by Pho85/Pho80. (A) Schematic representation of Cln3 with the different functional domains. The PEST region is indicated by shading, and the putative target residues for Pho85 by arrows. NLS, nuclear localization signal. (B) In vitro kinase assay of Pho85/Pho80 on Cln3. Recombinant Pho85 and GST-Pho80, purified from bacteria, were incubated with the C-terminal half of Cln3 (also from bacteria) containing the PEST region with the indicated mutations or the wild-type sequence (see Materials and Methods and Table 2). Pho4, a well-known substrate of Pho85/Pho80, was included as a control for the Pho85/Pho80 activity. The arrows indicate Cln3 protein. (C) In vitro kinase assay of Cdc28-TAP and the Cln3 mutants with the indicated mutations or the wild-type sequence. IgG-Sepharose beads were used to pull down Cdc28 either from a no-tag strain (−) or the Cdc28-TAP tag strain (+). Sic1 was included as a control for the Cdc28 kinase activity.
Cln3 is an in vitro substrate of Pho80/Pho85.
To test the latter hypothesis, we performed an in vitro phosphorylation experiment, for which we needed to obtain full-length Cln3 protein. We tried to purify Cln3 from wild-type yeast cells, but the cyclin is tightly associated with the endoplasmic reticulum (37) and remains with the particulate fraction of the cell extracts, making purification difficult. We also tried to purify it as a recombinant protein, but the expression of the full-length protein was highly toxic to E. coli cells, resulting in very poor expression levels. Finally, we expressed and purified a fragment of the recombinant GST-Cln3 protein (from Met347 to Arg580) containing the PEST region. This fragment of Cln3 was specifically phosphorylated by reconstituted Pho85/Pho80 complexes purified from E. coli cells (Fig. 6B). In contrast, we observed that the N-terminal half of the Cln3 protein (from Met1 to Met347) was not phosphorylated by Pho85 (not shown).
Pho85 is a proline-directed kinase that preferentially phosphorylates the consensus sequence S/T-P-X-I/L (17), and Cln3 contains two of these sites (Ser449 and Thr520) located precisely at the ends of the PEST region (Fig. 6A). We replaced both of these sites with either alanine or aspartic acid, rendering Cln3 that was no longer phosphorylated by Pho85 (Fig. 6B) and showing S449 and T520 to be the Pho85/Pho80 targets. A typical substrate of Pho85/Pho80, such as Pho4, was included as a control.
It has been suggested that Cdc28 is involved in the downregulation of Cln3 levels by acting through the CDK consensus phosphorylation sites in the PEST region of Cln3. Therefore, Cln3 phosphorylation by Cdc28 and Pho85 appears to have the opposite effect. We assayed the same Cln3 fragments against Cdc28-TAP immunopurified from yeast cells. The results in Figure 6C show that Cdc28 phosphorylates Cln3 (to a greater extent than Pho85, probably because it phosphorylates Cln3 at more sites in the PEST region). Interestingly, Cdc28 is still able to phosphorylate the S449A T520A cln3 mutant to the same extent as the wild type (Fig. 6C), indicating a differential site requirement between Cdc28 and Pho85 on Cln3, at least in vitro. A typical substrate of Cdc28, such as Sic1, is included as a control.
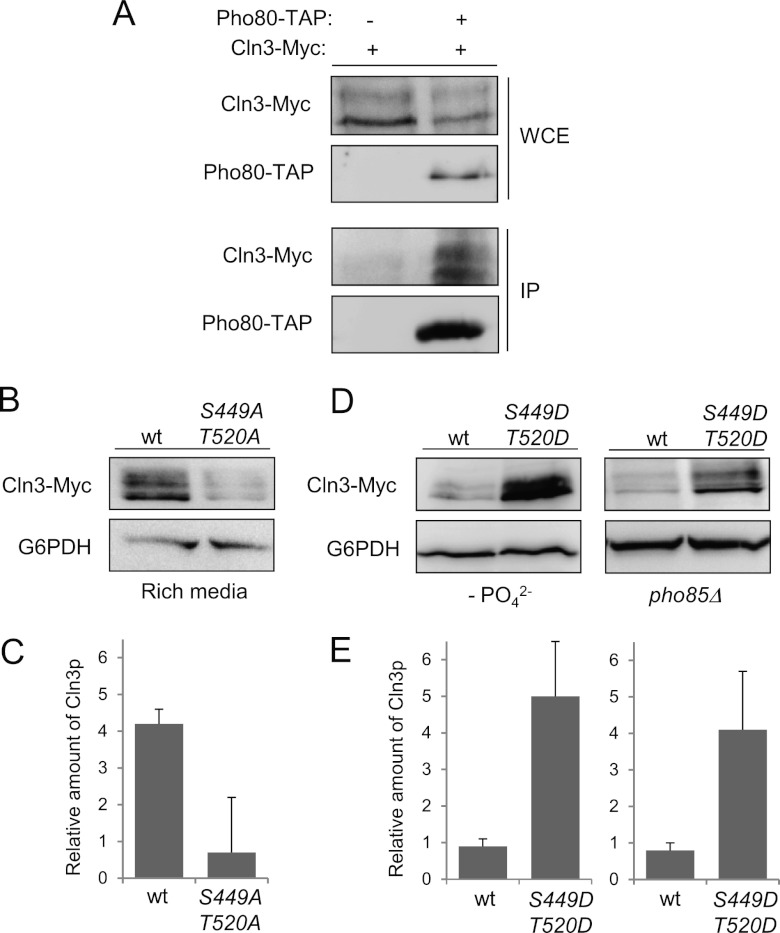
Because Cln3 and Pho80 interact in vivo, as indicated by the results of coimmunoprecipitation experiments (Fig. 7A), we examined whether Cln3 is also an in vivo substrate of Pho85.
Fig 7.
In vivo phosphorylation of S449 and T520 is essential to maintain Cln3 levels. (A) Pho80 and Cln3 interact in vivo. Yeast extracts (WCE [whole-cell extracts]) containing untagged Pho80 or Pho80-TAP were pulled down with IgG-Sepharose. Tagged Cln3-Myc (from its chromosomal locus) was detected using specific antibodies (IP [immunoprecipitate]). (B) Alanine replacement of S449 and T520 destabilizes Cln3. Wild-type cells were transformed with a centromeric plasmid bearing a Cln3-Myc or a 2-Ala (S449A T520A) mutant version and grown for 4 h in rich medium. The amount of Cln3 was determined by immunoblotting. (C) Quantification of experiment whose results are shown in panel B. Data ± standard deviations of three independent experiments are shown. (D) Aspartic acid replacement of S449 and T520 stabilizes Cln3. Wild-type (in phosphate-deficient medium) or pho85Δ (in phosphate-rich medium) cells bearing a plasmid with different versions of CLN3 were grown exponentially for 4 h. Cln3 levels were analyzed by immunoblotting. To simplify the nomenclature, S449D means a double substitution of Asp into the Ser 449 and the previous residue to mimic the double-negative charge that represents the phosphate group (see Materials and Methods), and the same is true for T520D. (E) Quantification of experiments whose results are shown in panel D. Data ± standard deviations of three independent experiments are shown.
In vivo phosphorylation of S449 and T520 is essential to maintain Cln3 levels.
We verified the in vivo Cln3 phosphorylation status by using mobility shift analysis. However, unphosphorylated Cln3 is highly unstable and becomes nearly undetectable by Western blot analysis, making this experiment very challenging. We attempted to identify the shift using a pho85-as strain and adding 5 times more yeast extract to the electrophoresis gel, but the levels of Cln3 remained nearly undetectable (data not shown).
We decided to test the in vivo effects of phosphorylation by measuring the levels of different mutant versions of Cln3. We expressed Cln3 from its own promoter in a centromeric plasmid, and when we replaced S449 and T520 with alanines, the levels of Cln3 became nearly undetectable (Fig. 7B and C). We also replaced these 2 sites with aspartic acid, to mimic the effect of phosphorylation, predicting that they would restore Cln3 levels to some degree in a strain without Pho85 activity. The results in Figure 7D and E illustrate how in pho85Δ cells (or in wild-type cells growing in medium without phosphate), the levels of Cln3 carrying the S449 and T520 aspartic acid substitutions are higher than the levels of wild-type Cln3, suggesting that in vivo phosphorylation at these sites is essential for maintaining high levels of Cln3. Overall, these results suggest that the proposed phosphorylation sites are involved in the in vivo regulation of Cln3 levels.
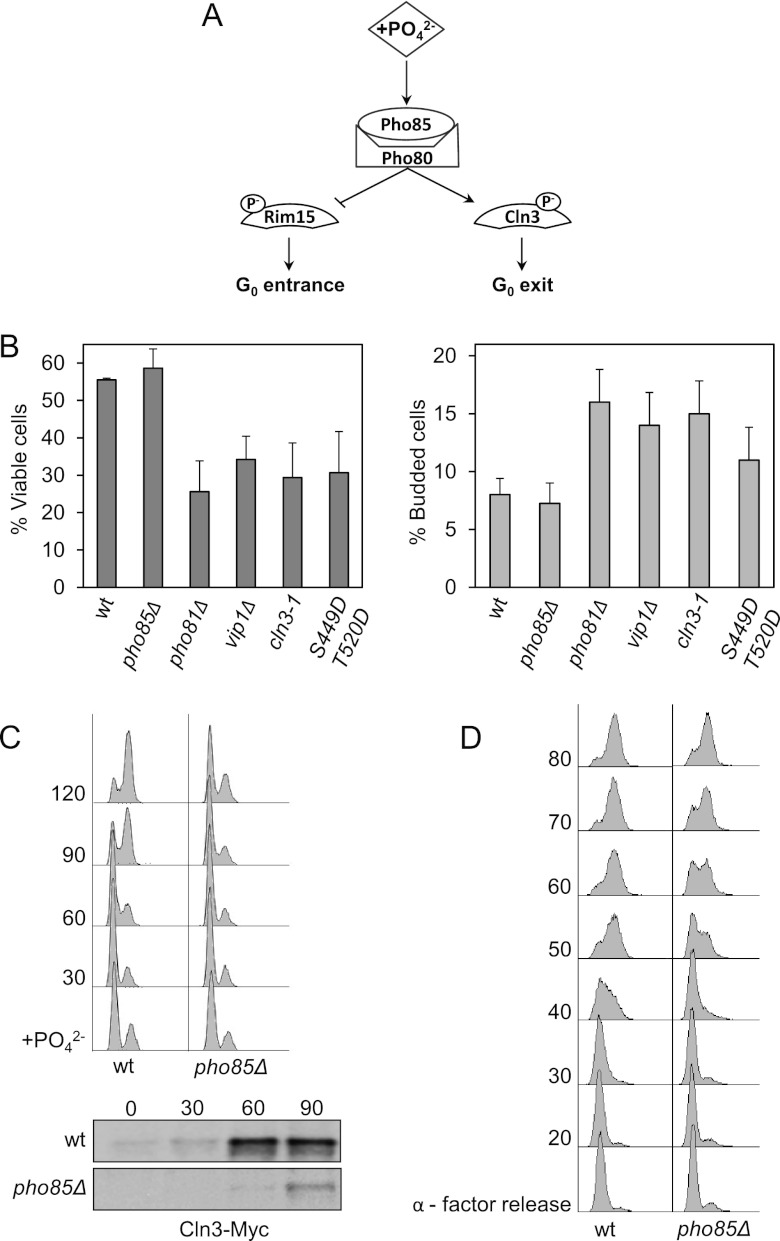
A possible scenario that emerges from all these results (Fig. 8A) is that in the absence of phosphate, Pho85 becomes inactive and Cln3 is no longer phosphorylated, which in turn would interfere with the ubiquitination and/or the destruction of the cyclin. At this point, we sought to test the physiological validity of our proposed model.
Fig 8.
Pho85 activity is necessary for proper G1 arrest and cell cycle reentry. (A) Proposed model of Pho85 activity. In phosphate-rich medium, Pho85/Pho80 complexes remain highly active. Under such conditions, Pho85 phosphorylates and inactivates Pho4 and Rim15 (25) and, conversely, activates cyclin Cln3. (B) Proper regulation of Pho85 activity is essential for survival under conditions of phosphate deprivation. Cells were incubated in synthetic complete medium for 7 days, at which point cells were collected and assessed for viability by colony counting (left). The percentage of budding was analyzed by counting no less than 200 cells under the microscope (right). Data ± standard deviations from three independent experiments are shown. (C) Pho85 activity is necessary for reentry into the cell cycle after refeeding. Cells transformed with a centromeric plasmid bearing Cln3-Myc were deprived of phosphate for 7 h (time zero) and then refed. At various times, samples were collected and analyzed for DNA content by flow cytometry (top). Times (min) are indicated at left; “+PO42−” indicates the initiation of refeeding. Cln3 levels were monitored by immunoblotting (bottom) at times (min) indicated above the gel. (D) pho85Δ progresses through G1, with a small delay, after α-factor exit. Wild-type and pho85Δ cells were synchronized with α-factor for 3 h and then released into fresh medium at 30°C. At various times, samples were collected. Times (min) are indicated at left; “α-factor release” indicates the time of release following synchronization. Total DNA content was measured as described in Materials and Methods, except that propidium iodide was used instead of SYBR green.
Pho85 inactivation is essential for proper G1 arrest.
Ectopic expression of Cln3 in G1-arrested cells (due to the presence of rapamycin or to nutrient depletion) leads to accidental entry into S phase and to diminished cell viability (38, 39). To ascertain the relevance of these observations to our work, we arrested cells in phosphate-depleted medium, and we found that, as predicted, cells with a hyperactive form of CLN3 (the cln3-1 allele) induced a diminished cell viability which correlated well with an increase in the number of cells that crossed Start, as indicated by the high percentage of budded cells (Fig. 8B). Moreover, as discussed above (Fig. 1), ectopic expression of Cln2 led to a clear increase in the number of budded cells and to a rise in the rate of cell death (not shown). Hence, it seems that downregulation of Cdc28 activity is essential for proper G1 arrest during phosphate deprivation.
According to our model, overactivation of Pho85 should also be detrimental to cell viability in G1-arrested cells, and our results demonstrated that this was indeed the case. As shown by the results in Figure 8B, wild-type cells incubated for 7 days in phosphate-free medium exhibited a viability rate of 55%. In contrast, vip1Δ or pho81Δ cells (which should retain Pho80/Pho85 activity under the same conditions of deprivation) showed an increase in improperly arrested cells (16 to 18% of budded cells) and a significant loss in viability (a total of 20%). Even more interesting is the fact that cells carrying the Cln3 with the Asp substitutions that mimic Pho85 phosphorylation also have low viability under phosphate-fasting conditions. Together, these results agree with the notion that, in the absence of phosphate, downregulation of Pho85 activity is essential for decreasing Cln3 levels and for proper G1 arrest.
Pho85 activation is essential for proper cell cycle reentry.
It has been proposed that Pho85 activity is essential in situations with no Cdc28 activity, such as those involving cell cycle restart (20). This hypothesis is consistent with our model, which, if true, predicts that cells with low Pho85 activity should have difficulties reentering the cell cycle from G0. To test this prediction, we incubated yeast strains in phosphate-free medium and, after 7 h, transferred them to rich medium in order to monitor their growth. The results in Figure 8C show how, after 2 h of incubation in phosphate-rich medium, pho85Δ cells present low levels of Cln3 and consequently remain arrested, while wild-type cells already undergo mitosis. It is interesting to note that the differences between strains are only minor when we compare the ability of cell cycle reentry from α-factor arrest (Fig. 8D). This finding suggests that the observed delay in cell cycle reentry is specific rather than a general defect of cell cycle regulation.
In summary, these results reinforce the idea that Pho85 activity plays an indispensable role in exiting from G0 arrest.
DISCUSSION
Phosphate levels regulate cell cycle progression.
So far, there are several lines of evidence implicating glucose and nitrogen as critical elements in cell cycle control and that their absence leads to G1 arrest. Also, it is known that the absence of phosphate also stops proliferation. At first glance, one might assume that this defect is merely a nonspecific consequence arising from the reduction in the level of a particular metabolite (e.g., ATP). However, there is considerable evidence to confirm that the observed arrest is a controlled process, as follows. (i) Under phosphate-fasting conditions, the cells accumulate as mostly unbudded with 1N DNA content; if they had arrested accidentally, we should have found cells in various stages of the cycle. (ii) Metabolomic studies have shown that cells growing under phosphate-limiting conditions maintain a relatively constant free energy of ATP hydrolysis (40), suggesting that in the first moments of phosphate deprivation, cells might have enough energy to initiate an adaptive response, such as finishing one round of division until the next G1 phase. (iii) The volume of arrested cells continues to increase, indicating that they are metabolically active and not in a collapsed state. (iv) Finally, the arrest caused by lack of phosphate can be reversed with an increase in CDK/Cln2 activity. In conclusion, one of the contributions of the present work is to confirm that phosphate regulates Start through the control of G1/S transcriptional machinery in S. cerevisiae, as suggested some time ago by Johnston et al. (35).
It should be noted that, in our experiments, the cells did not arrest immediately: they gradually accumulated in the G1 phase, and finally, within approximately 6 h of deprivation, 90% of them had stopped dividing. This gradual response of cultures is probably due to polyphosphate reserves in the cells (41) which serve to buffer the sudden external changes.
Pho85 affects the stability of Cln3.
The central idea of this work is that phosphate levels dictate Cln3 stability by regulating the activity of the Pho85/Pho80 complex. It is interesting to note that the amounts of other cyclins (e.g., Cln2) are not affected by Pho85 activity (not shown), underscoring the specificity of the described effect.
Several studies indicate that Cln3 is downregulated by translational repression under conditions of nutrient deprivation (18, 42, 43). Although we cannot rule out the possibility that phosphate limitation has the same effect, our results show that the critical step targeted by phosphate availability on Cln3 is its stability. An analogous destabilization phenomenon has been described in nitrogen-deprived cells, although its mechanism remains unexplained (18). This same study showed that the half-life of Cln3 was again reduced by half. Thus, the destabilization of Cln3, along with its possible translational repression, could be a general response of the cell against the limitation of different nutrients. This would limit the activity of the Cln3/Cdc28 complex needed to pass Start in order to slow down the cell cycle and to adapt to the new conditions.
The finding that Cln3 is affected by Pho85/Pho80 activity is relevant for two reasons: on one hand, the Pho85/Pho80 route is shown for the first time to be involved in directly controlling the cell cycle machinery, suggesting coordination between phosphate homeostasis and the cell cycle, and on the other hand, to our knowledge, this is the first reported case of one CDK controlling the activity of another. Interestingly, during the revision process of the manuscript, the group of Kron and collaborators described that Pho85/Pcl2 controlled the amount of Cln3 through the activation of Hsp70 in response to nitrogen, pointing to Pho85 as a general controller of Cln3 in response to variations in nutrient levels (44).
How does Pho85 affect the stability of the cyclin Cln3? Having excluded the possibility of autophagy, we propose here that Cln3 phosphorylation by Pho85 may somehow hinder the normal degradation of Cln3. This raises the possibility that phosphorylation of Cln3 complicates its recruitment by the ubiquitination system, consequently expanding its lifetime. The idea that phosphorylation may prevent interaction with E3 ligases has already been suggested for other cell cycle-regulated proteins (45).
If Cdc28 and Pho85 have opposite effects on Cln3 stability, then it would be important to determine whether these CDKs phosphorylate different residues. The residues of Cln3 phosphorylated by Cdc28 are currently unknown, although some results point to S468 as one of the most likely candidates (9). However, we have demonstrated that a recombinant fragment of Cln3 that presents S449 and T520 mutated to alanine is still phosphorylated by Cdc28 and is no longer phosphorylated by Pho85 (Fig. 6B and C), indicating that the CDKs phosphorylate different residues of Cln3, at least in vitro. It is important to note that, although the consensus sequence for the CDK family is SP or TP, Cdc28 and Pho85 have distinct preferences for the sequence adjacent to these sites. Cdc28 prefers a positively charged lysine or arginine at the third position from the phosphoacceptor site, with the consensus site being S/TPXK/R (46, 47), whereas Pho80-Pho85 has the consensus phosphorylation site sequence SPXI/L (24, 48). Therefore, it is formally plausible that both kinases phosphorylate distinct sites, generating distinct effects. In this regard, it is interesting to note that only 2 of 10 SP/TP sites of the PEST region have the preferred sequence for Pho85 (the hydrophobic residue at +3), and these sites are the same two that appear to be phosphorylated by Pho85. There are other examples of proteins that are phosphorylated at SP/TP with opposite effects, such as Sic1, which must exceed a threshold of CDK phosphorylation in several SP/TP sites to be destroyed, but a single mitogen-activated protein kinase (MAPK) phosphorylation in one specific TP makes the protein stable (45).
Physiological significance of Cln3 regulation.
The underlying mechanism(s) that enables nutrient sensors to control cell cycle machinery remains unknown. Organisms may need to employ several convergent mechanisms to provoke complete G1 arrest, making the identification of a single molecular target rather difficult. We propose that modulation of Cln3 stability may be one of many principal targets in cell cycle regulation, at least in terms of the adaptive response to phosphate scarcity (the proposed model is shown in Fig. 8A). Cln3 cyclin is a good candidate because it is the most upstream control point in the cell cycle and because raising its cellular content by either increased transcription or altered protein turnover profoundly affects its capacity to pass through G1 (9, 49). Thus, the G1 arrest caused by phosphate deprivation could be explained by a simple model: due to the reduction in the Cln3 half-life, starved cells cannot reach the Cln3 threshold level required to execute the Start program. However, this mechanism might not be important in rich medium, given that cln3Δ cells can still progress through the entire cell cycle, albeit with a longer G1 phase.
According to our model, Cln3 destabilization is crucial during G1 arrest due to lack of phosphate. This downregulation could be important, for instance, in maintaining silencing of Swi6-dependent transcription in order to avoid the process of wall remodeling during nutrient deprivation (50). Moreover, Burhan's group has demonstrated that establishing and maintaining proper arrest in G1 is an important cellular response to nutrient deprivation (38); they report that cells that improperly halt at the S phase suffer replication stress and rapidly lose viability. Furthermore, they clearly demonstrate that ectopic expression of CLN3 increases the frequency with which nutrient-depleted cells arrest at the beginning of the S phase instead of G1. Our results confirm that the presence of a hyperstable Cln3 allele (cln3-1) also increases the number of S-phase-arrested cells and decreases cell viability. In this context, we propose that in response to the lack of phosphate, Pho85 inactivation is critical for stopping the cycle at G1, avoiding entry into the S phase with low levels of nucleotides. This assumption is clearly supported by evidence showing that mutants with high Pho85 activity (pho81Δ, vip1Δ, and Gal-PHO85) incubated in medium without phosphate (i) maintain high levels of Cln3, (ii) show large numbers of cells that have entered the cell cycle, and (iii) show a decrease in cell viability. Considering that under the same conditions of deprivation, cells with a CLN3 allele that encodes aspartic acid substitutions also die prematurely, we propose that such cellular defects chiefly derive from sustained and unscheduled phosphorylation of Cln3.
According to our model, Pho85 could also be important for restarting the cell cycle after refeeding. Our findings support this notion; either the absence of Cln3 or low Pho85 activity greatly hinders reentry into the cell cycle. Interestingly, this role for Pho85 in reentry seems to be specific, as reflected in the almost total lack of differences between wild-type cells and pho85Δ cells released from alpha factor arrest. These results are consistent with the hypothesis that Pho85 is essential when Cdc28 activity is lacking (20) and reinforce the idea that Pho85 activity is almost dispensable when yeast grows in nutrient-rich medium (although the cells still undergo a longer G1 phase), but is essential in other situations (e.g., exiting from nutrient-induced G0 arrest). Since wild yeast should thrive under diverse nutrient conditions (the availability of phosphate and other nutrients often varies widely), we postulate that Pho85 must be fundamental in controlling the constant cell cycle stalls and reentries that a yeast cell is subjected to under natural conditions.
ACKNOWLEDGMENTS
We gratefully acknowledge F. Posas, W. C. Burhans, J. Jiménez, and N. Casals for stimulating discussions, E. O'Shea for providing strains, K. Shokat for the 1-Na PP1 inhibitor, M. Pérez for technical assistance, and O. Mirallas for his collaboration in some of the experiments.
S. Hernández received a postgraduate Junior Faculty fellowship from the UIC and l'Obra Social la Caixa. This work was supported by grants from Ministerio de Ciencia e Innovación of the Spanish government (BFU 2009-09278).
Footnotes
Published ahead of print 22 January 2013
REFERENCES
- 1. De Virgilio C. 2012. The essence of yeast quiescence. FEMS Microbiol. Rev. 36:306–339 [DOI] [PubMed] [Google Scholar]
- 2. Wittenberg C, Reed SI. 2005. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene 24:2746–2755 [DOI] [PubMed] [Google Scholar]
- 3. Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ, Davis RW. 1998. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2:65–73 [DOI] [PubMed] [Google Scholar]
- 4. Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Koch C, Nasmyth K. 1994. Cell cycle regulated transcription in yeast. Curr. Opin. Cell Biol. 6:451–459 [DOI] [PubMed] [Google Scholar]
- 6. Cosma MP, Panizza S, Nasmyth K. 2001. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell 7:1213–1220 [DOI] [PubMed] [Google Scholar]
- 7. Koch C, Schleiffer A, Ammerer G, Nasmyth K. 1996. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 10:129–141 [DOI] [PubMed] [Google Scholar]
- 8. Cross FR, Blake CM. 1993. The yeast Cln3 protein is an unstable activator of Cdc28. Mol. Cell. Biol. 13:3266–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yaglom J, Linskens MH, Sadis S, Rubin DM, Futcher B, Finley D. 1995. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol. Cell. Biol. 15:731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M. 2004. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117:899–913 [DOI] [PubMed] [Google Scholar]
- 11. de Bruin RA, McDonald WH, Kalashnikova TI, Yates J, III, Wittenberg C. 2004. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117:887–898 [DOI] [PubMed] [Google Scholar]
- 12. Wagner MV, Smolka MB, de Bruin RA, Zhou H, Wittenberg C, Dowdy SF. 2009. Whi5 regulation by site specific CDK-phosphorylation in Saccharomyces cerevisiae. PLoS One 4:e4300 doi:10.1371/journal.pone.0004300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang H, Carey LB, Cai Y, Wijnen H, Futcher B. 2009. Recruitment of Cln3 cyclin to promoters controls cell cycle entry via histone deacetylase and other targets. PLoS Biol. 7:e1000189 doi:10.1371/journal.pbio.1000189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Skotheim JM, Di Talia S, Siggia ED, Cross FR. 2008. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature 454:291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cross FR, Schroeder L, Bean JM. 2007. Phosphorylation of the Sic1 inhibitor of B-type cyclins in Saccharomyces cerevisiae is not essential but contributes to cell cycle robustness. Genetics 176:1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. 2004. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68:187–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martin DE, Hall MN. 2005. The expanding TOR signaling network. Curr. Opin. Cell Biol. 17:158–166 [DOI] [PubMed] [Google Scholar]
- 18. Gallego C, Gari E, Colomina N, Herrero E, Aldea M. 1997. The Cln3 cyclin is down-regulated by translational repression and degradation during the G1 arrest caused by nitrogen deprivation in budding yeast. EMBO J. 16:7196–7206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee YS, Huang K, Quiocho FA, O'Shea EK. 2008. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat. Chem. Biol. 4:25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang D, Friesen H, Andrews B. 2007. Pho85, a multifunctional cyclin-dependent protein kinase in budding yeast. Mol. Microbiol. 66:303–314 [DOI] [PubMed] [Google Scholar]
- 21. Schneider KR, Smith RL, O'Shea EK. 1994. Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science 266:122–126 [DOI] [PubMed] [Google Scholar]
- 22. Lee YS, Mulugu S, York JD, O'Shea EK. 2007. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316:109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nishizawa M, Komai T, Katou Y, Shirahige K, Ito T, Toh-E A. 2008. Nutrient-regulated antisense and intragenic RNAs modulate a signal transduction pathway in yeast. PLoS Biol. 6:2817–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Neill EM, Kaffman A, Jolly ER, O'Shea EK. 1996. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 271:209–212 [DOI] [PubMed] [Google Scholar]
- 25. Wanke V, Pedruzzi I, Cameroni E, Dubouloz F, De Virgilio C. 2005. Regulation of G0 entry by the Pho80-Pho85 cyclin-CDK complex. EMBO J. 24:4271–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wysocki R, Javaheri A, Kristjansdottir K, Sha F, Kron SJ. 2006. CDK Pho85 targets CDK inhibitor Sic1 to relieve yeast G1 checkpoint arrest after DNA damage. Nat. Struct. Mol. Biol. 13:908–914 [DOI] [PubMed] [Google Scholar]
- 27. Carroll AS, Bishop AC, DeRisi JL, Shokat KM, O'Shea EK. 2001. Chemical inhibition of the Pho85 cyclin-dependent kinase reveals a role in the environmental stress response. Proc. Natl. Acad. Sci. U. S. A. 98:12578–12583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gietz RD, Sugino A. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527–534 [DOI] [PubMed] [Google Scholar]
- 29. Clotet J, Escote X, Adrover MA, Yaakov G, Gari E, Aldea M, de Nadal E, Posas F. 2006. Phosphorylation of Hsl1 by Hog1 leads to a G2 arrest essential for cell survival at high osmolarity. EMBO J. 25:2338–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haase SB, Reed SI. 2002. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle 1:132–136 [PubMed] [Google Scholar]
- 31. Hernandez-Ortega S, Bru S, Ricco N, Ramirez S, Casals N, Jimenez J, Isasa M, Crosas B, Clotet J. 21 December 2012. Defective in mitotic arrest 1 (Dma1) ubiquitin ligase controls G1 cyclin degradation. J. Biol. Chem. [Epub ahead of print.] doi:10.1074/jbc.M112.426593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fabrizio P, Longo VD. 2007. The chronological life span of Saccharomyces cerevisiae. Methods Mol. Biol. 371:89–95 [DOI] [PubMed] [Google Scholar]
- 33. Schneider BL, Yang QH, Futcher AB. 1996. Linkage of replication to start by the Cdk inhibitor Sic1. Science 272:560–562 [DOI] [PubMed] [Google Scholar]
- 34. Smets B, Ghillebert R, De Snijder P, Binda M, Swinnen E, De Virgilio C, Winderickx J. 2010. Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae. Curr. Genet. 56:1–32 [DOI] [PubMed] [Google Scholar]
- 35. Johnston GC, Pringle JR, Hartwell LH. 1977. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 105:79–98 [DOI] [PubMed] [Google Scholar]
- 36. Yang Z, Geng J, Yen WL, Wang K, Klionsky DJ. 2010. Positive or negative roles of different cyclin-dependent kinase Pho85-cyclin complexes orchestrate induction of autophagy in Saccharomyces cerevisiae. Mol. Cell 38:250–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Verges E, Colomina N, Gari E, Gallego C, Aldea M. 2007. Cyclin Cln3 is retained at the ER and released by the J chaperone Ydj1 in late G1 to trigger cell cycle entry. Mol. Cell 26:649–662 [DOI] [PubMed] [Google Scholar]
- 38. Weinberger M, Feng L, Paul A, Smith DL, Jr, Hontz RD, Smith JS, Vujcic M, Singh KK, Huberman JA, Burhans WC. 2007. DNA replication stress is a determinant of chronological lifespan in budding yeast. PLoS One 2:e748 doi:10.1371/journal.pone.0000748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zinzalla V, Graziola M, Mastriani A, Vanoni M, Alberghina L. 2007. Rapamycin-mediated G1 arrest involves regulation of the Cdk inhibitor Sic1 in Saccharomyces cerevisiae. Mol. Microbiol. 63:1482–1494 [DOI] [PubMed] [Google Scholar]
- 40. Boer VM, Crutchfield CA, Bradley PH, Botstein D, Rabinowitz JD. 2010. Growth-limiting intracellular metabolites in yeast growing under diverse nutrient limitations. Mol. Biol. Cell 21:198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thomas MR, O'Shea EK. 2005. An intracellular phosphate buffer filters transient fluctuations in extracellular phosphate levels. Proc. Natl. Acad. Sci. U. S. A. 102:9565–9570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hall DD, Markwardt DD, Parviz F, Heideman W. 1998. Regulation of the Cln3-Cdc28 kinase by cAMP in Saccharomyces cerevisiae. EMBO J. 17:4370–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Polymenis M, Schmidt EV. 1997. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 11:2522–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Truman AW, Kristjansdottir K, Wolfgeher D, Hasin N, Polier S, Zhang H, Perrett S, Prodromou C, Jones GW, Kron SJ. 2012. CDK-dependent Hsp70 phosphorylation controls G1 cyclin abundance and cell-cycle progression. Cell 151:1308–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Escote X, Zapater M, Clotet J, Posas F. 2004. Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat. Cell Biol. 6:997–1002 [DOI] [PubMed] [Google Scholar]
- 46. Holmes JK, Solomon MJ. 1996. A predictive scale for evaluating cyclin-dependent kinase substrates. A comparison of p34cdc2 and p33cdk2. J. Biol. Chem. 271:25240–25246 [DOI] [PubMed] [Google Scholar]
- 47. Songyang Z, Lu KP, Kwon YT, Tsai LH, Filhol O, Cochet C, Brickey DA, Soderling TR, Bartleson C, Graves DJ, DeMaggio AJ, Hoekstra MF, Blenis J, Hunter T, Cantley LC. 1996. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol. 16:6486–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang K, Ferrin-O'Connell I, Zhang W, Leonard GA, O'Shea EK, Quiocho FA. 2007. Structure of the Pho85-Pho80 CDK-cyclin complex of the phosphate-responsive signal transduction pathway. Mol. Cell 28:614–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nash R, Tokiwa G, Anand S, Erickson K, Futcher AB. 1988. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 7:4335–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Igual JC, Johnson AL, Johnston LH. 1996. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 15:5001–5013 [PMC free article] [PubMed] [Google Scholar]