Abstract
Picorna-like viruses in the Picornavirales order are a large group of positive-strand RNA viruses that include numerous important pathogens for plants, insects, and humans. In these viruses, nonstructural protein 2C is one of the most conserved proteins and contains ATPase activity and putative RNA helicase activity. Here we expressed 2C protein of Ectropis obliqua picorna-like virus (EoV; genus Iflavirus, family Iflaviridae, order Picornavirales) in a eukaryotic expression system and determined that EoV 2C displays ATP-independent nucleic acid helix destabilizing and strand annealing acceleration activity in a concentration-dependent manner, indicating that this picornaviral 2C is more like an RNA chaperone than like the previously predicted RNA helicase. Our further characterization of EoV 2C revealed that divalent metal ions, such as Mg2+ and Zn2+, inhibit 2C-mediated helix destabilization to different extents. Moreover, we determined that EoV 2C also contains ATPase activity like that of other picornaviral 2C proteins and further assessed the functional relevance between its RNA chaperone-like and ATPase activities using mutational analysis as well as their responses to Mg2+. Our data show that, when one of the two 2C activities was dramatically inhibited or almost abolished, the other activity could remain intact, showing that the RNA chaperone-like and ATPase activities of EoV 2C can be functionally separated. This report reveals that a picorna-like virus 2C protein displays RNA helix destabilizing and strand annealing acceleration activity, which may be critical for picornaviral replication and pathogenesis, and should foster our understanding of picorna-like viruses and viral RNA chaperones.
INTRODUCTION
The picorna-like virus superfamily is a loosely defined group of positive-strand RNA viruses that currently includes 14 families of viruses and several unclassified genera and species (1). Among them, five families of viruses (Picornaviridae, Iflaviridae, Dicistroviridae, Marnaviridae, and Secoviridae) are classified into the Picornavirales order. Picorna-like viruses in the Picornavirales order include numerous important pathogens that are responsible for diseases in humans, such as poliomyelitis (2), hand-foot-and-mouth disease (3), hepatitis A (4), upper respiratory illness (5), myocarditis (6), diseases in insects, such as sacbrood (7), and diseases in plants, such as sharka (8). All of these viruses share several common features. The viral genomes of the Picornavirales order are characterized by a set of highly conserved genes, including an RNA-dependent RNA polymerase (RdRP) for viral RNA replication, a chymotrypsin-like 3C protease for proteolytic processing of picornaviral polyproteins into separate proteins, and a putative helicase (nonstructural protein 2C) (9–11).
Nonstructural protein 2C is one of the most conserved proteins within the Picornavirales order and has long been predicted to be a superfamily 3 (SF3) helicase on the basis of its conserved SF3 signature A, B, and C motifs (12). In addition, like other helicases, ATPase activities have been reported for 2C proteins in Picornaviridae and are believed to provide the energy generated from ATP hydrolysis for RNA helix unwinding (13, 14). Thus, picornaviral 2C is often referred to as 2CATPase in the literature (15, 16). In addition to its putative helicase activity, picornaviral 2C proteins are also involved in RNA binding (17), membrane anchoring (18, 19), intracellular membrane rearrangements (20), encapsidation and viral morphogenesis (15, 16), and autophagy inhibition (21). In recent years, because of their high conservation within Picornavirales and unambiguous importance for picornaviral life cycles, 2C proteins have attracted much attention as ideal targets for developing antiviral drugs against picorna-like viruses (22, 23).
For both viruses and host cells, most RNA molecules require proper tertiary structures/folding for their functions. However, the self-folding of RNA molecules is a challenging process, since RNAs can easily become trapped in inactive intermediate structures that are thermodynamically stable (kinetic trap), resulting in only a fraction of RNAs reaching their native and functional conformation (24, 25). In response, cells or viruses encode a variety of RNA remodeling proteins that help RNA overcome the thermodynamic barriers of kinetically trapped RNAs for refolding (24–26). RNA remodeling proteins generally include RNA helicases, RNA chaperones, and RNA annealers (25, 27). RNA helicases are highly similar to DNA helicases, contain ATPase activity, and use the energy provided by ATP hydrolysis to melt base pairs. They are thought to be involved in most ATP-dependent rearrangements of structured RNA molecules and are classified into six superfamilies based on their conserved common motifs (28). RNA chaperones are a heterogeneous group of proteins that do not share consensus sequences or motifs but are able to destabilize RNA helixes/duplexes and promote the formation of more globally stable structures by facilitating the escape of misfolded RNAs from kinetic traps. Since many RNA chaperones also show helix destabilizing and annealing acceleration activity with respect to DNA molecules in vitro, these proteins are also often referred to as nucleic acid chaperones. RNA chaperones do not require ATP hydrolysis or ATP binding for their activities (25, 27). RNA annealers are the proteins that accelerate annealing of cRNA strands, and their activities are also ATP independent. Actually, the distinction between RNA chaperones and RNA annealers is blurred, since many RNA chaperones also exhibit annealing activity, and some previously defined RNA annealers are often reported in the literature as RNA chaperones (25, 27, 29–31). It has been reported that many RNA viruses encode their own RNA helicases and/or chaperones (12, 26), whereas some RNA viruses also take advantage of host RNA helicases or chaperones (26, 32, 33). For Picornavirales, it has been reported that poliovirus 3AB displays in vitro nucleic acid chaperone activity (34, 35) and that host RNA helicase A (RHA) is required for the replication of foot-and-mouth disease virus (FMDV) (31, 32). On the other hand, despite the medical and economic importance of picornaviruses and picorna-like viruses, as well as the potential importance of RNA remodeling for RNA viruses (26), the RNA remodeling activity of picornaviral 2C protein remains putative and has never been formally determined (22, 36, 37). This obvious gap has hindered our understanding of this large group of important viruses.
The Iflaviridae family is a newly classified member of the Picornavirales order (http://www.ictvonline.org/virusTaxonomy.asp?version=2009) and currently includes the sole genus Iflavirus (38, 39). Based on its viral structure and genome organization, Iflaviridae is the family closest to Picornaviridae within Picornavirales, as the two families share features of N-terminal structural proteins and the conserved C-terminal nonstructural 2CATPase-3CPro-3DPol module (9). Ectropis obliqua picorna-like virus (EoV) was initially identified by our group in 2000 and was classified as a member of Iflaviridae in 2010 (39, 40). It can cause lethal granulosis in the tea looper (Ectropis obliqua), which is an important agricultural pest for tea cultivation. Thus far, the RNA genome of EoV has been completely sequenced (40), and the internal ribosome entry site, RdRP, and 3C protease of EoV have been studied (41–44). In addition to EoV, Iflaviridae comprises other insect picorna-like viruses, including infectious flacherie virus (IFV) of the silkworm, sacbrood virus (SBV) of the honeybee, deformed wing virus (DWV), Perina nuda virus (PnV), and Varroa destructor virus-1 (VDV-1), as well as some tentative members (38, 39). So far, the iflavirus 2C proteins have not been described in terms of their putative helicase or RNA remodeling activities or their ATPase activities.
In this study, we expressed the 2C domain of EoV polyprotein in a eukaryotic expression system and determined that EoV 2C protein displays nonspecific nucleic acid helix destabilizing and annealing acceleration activities in an ATP-independent manner, indicating that this picornaviral 2C is more like an RNA chaperone than like the previously predicted RNA helicase. Our further characterization of EoV 2C revealed that divalent metal ions, such as Mg2+ and Zn2+, inhibit 2C-mediated helix destabilization to different extents. Moreover, we determined that EoV 2C also contains ATPase activity similar to that of other picornaviral 2C proteins, and we further examined the functional relevance between its RNA chaperone-like and ATPase activities. Using a mutational approach, we showed that when one of the two 2C activities was dramatically inhibited by point mutation, the other activity remained intact, showing that the RNA chaperone and ATPase activities of EoV 2C can be functionally separated. Our report is the first to show that a picorna-like virus 2C protein displays an RNA chaperone-like activity.
MATERIALS AND METHODS
Construction of recombinant baculoviruses.
Standard procedures were used for extraction of viral genome RNA and reverse transcription-PCR (40). A cDNA fragment of the putative EoV helicase domain (amino acids [aa] 1374 to 1789 of polyprotein open reading frame [ORF]) of hepatitis C virus (HCV) NS3 protein was inserted into the vector pFastBacHTA-MBP. Briefly, pFastBacHTA-MBP vector originated from the vector pFastBacHTA (Invitrogen, Carlsbad, CA), into which the maltose-binding protein (MBP) was N-terminally fused by our laboratory as previously described (45–48). Mutations were generated as described previously (44). Briefly, point mutations were introduced into the corresponding templates via PCR-mediated mutagenesis with appropriate primers containing the desired nucleotide changes. The primers used in this study are shown in Table 1. The resulting plasmids were subjected to the Bac-to-Bac baculovirus expression system to express the fusion proteins with an MBP tag at the N terminus of the EoV helicase domain (MBP-2C).
Table 1.
List of primersa
| Primer | Sequence (5′–3′) |
|---|---|
| Hel-F | GAATTCACTTTCGCCAAAGGTGAAGAC (EcoRI) |
| Hel-R | GTCGACTTAACCCGTGGGTGC (SalI) |
| NS3-F | GGATCCCCGGTGCTCTGCCCTAGGGGCCACG (BamHI) |
| NS3-R | AAGCTTTTAATTGGTAATAGGGCCCAAACGGT (HindIII) |
| G1557A-F* | GTATGGACACTCAgcgTGTGGAAAG |
| G1557A-R* | CACAcgcTGAGTGTCCATACACCC |
| GK1559AA-F* | CTCAGGGTGTgcagcgTCACATGTTTG |
| GK1559AA-R* | GTGAcgctgcACACCCTGAGTGTCC |
| H1562A-F* | GGAAAGTCAgcaGTTTGTGACAATG |
| H1562A-R* | CACAAACtgcTGACTTTCCACACCC |
| Q1599A-F* | CACAGGAgcgAAATTAATATCGTGGC |
| Q1599A-R* | GATATTAATTTcgcTCCTGTGTAACC |
| D1606A-F* | CGTGGCAAgccTTTGCGAAAATAAC |
| D1606A-R* | CGCAAAggcTTGCCACGATATTAAT |
| K1642A-F* | GAAGATgcaAGGAAAATAGCGGATG |
| K1642A-R* | CTATTTTCCTtgcATCTTCTAAAGC |
Underlined characters indicate restriction endonuclease sites, and the types are shown in parentheses. Primers designated with an asterisk were designed for overlapping PCR. Nucleotides substituted for mutagenesis are shown with lowercase characters.
Expression and purification of recombinant proteins.
The expression and purification of MBP alone, MBP-NS3, and MBP-2C and its derivates were performed as previously described (47). Briefly, the recombinant proteins were purified from Sf9 cells 3 days after infection. After being resuspended, cells were lysed via sonication and then debris was removed by centrifugation for 30 min at 11,000 × g. The protein in the supernatant was purified using amylose affinity chromatography (New England BioLabs, Ipswich, MA) according to the manufacturer's protocol and then concentrated using Amicon Ultra-15 filters (Millipore, Schwalbach, Germany). All proteins were quantified by the Bradford method and stored at −70°C in aliquots.
Western blot analysis.
Western blot assays were performed as described previously (44). The anti-MBP polyclonal antibody was purchased from New England BioLabs and used at a dilution of 1:10,000.
Preparation of oligonucleotide helices.
RNA helices/duplexes consisted of two complementary oligonucleotide strands that were annealed and gel purified. Of the two strands, the longer one was unlabeled and is referred to as the template strand. The shorter strand, referred to as the release strand, was labeled at the 5′ end with hexachlorofluorescein (HEX) (TaKaRa, Dalian) (49). All unlabeled DNA strands were purchased from Invitrogen, and HEX-labeled DNA and RNA strands were purchased from TaKaRa. Unlabeled RNA strands were synthesized from the in vitro transcription using T7 RNA polymerase (Promega, Madison, WI). The posttranscriptional RNAs were electrophoresed in a 5% polyacrylamide–8 M urea gel and further purified by the use of a Poly-Gel RNA extraction kit (Omega Bio-Tek) according to the manufacturer's instructions.
To generate oligonucleotide helices, the labeled release strand and unlabeled template strand were incubated at a proper ratio in a 10-μl reaction mixture containing 25 mM HEPES-KOH (pH 8.0)–25 mM NaCl. The mixture was heated to 95°C for 5 min and was then cooled gradually to 4°C. A standard RNA helix substrate with both 5′ and 3′ tails was annealed with RNA1 and RNA2, R*/D substrate was annealed with RNA1 and DNA1, D*/D substrate was annealed with DNA2 and DNA3, 3′-tailed RNA helix substrate was annealed with RNA1 and RNA3, 5′-tailed RNA helix substrate was annealed with RNA1 and RNA4, and blunt-ended substrate was annealed with RNA1 and RNA5. The 3′-tailed RNA helix substrates with different sizes of 3′ tails were annealed with RNA1 and RNA3-2, RNA3-3, RNA3-4, RNA3-5, and RNA3-6. The R*/R substrates with 42 matched base pairs were annealed with RNA6 and RNA7. All DNA and RNA oligonucleotides are listed in Table 2.
Table 2.
List of oligonucleotides
| Oligonucleotide | Sequence (5′–3′)a |
|---|---|
| RNA1 | GUGUAGUAAUCGUCCAU |
| RNA2 | AGUUUAAAACGCACGAGACACAAUGGACGAUUACUACACCAACAGAUCAAUUGAGCAAUCCG |
| RNA3 | AUGGACGAUUACUACACCAACAGAUCAAUUGAGCAAUCCG |
| RNA3-2 | AUGGACGAUUACUACACCAA |
| RNA3-3 | AUGGACGAUUACUACACCAACAGA |
| RNA3-4 | AUGGACGAUUACUACACCAACAGAUCAAU |
| RNA3-5 | AUGGACGAUUACUACACCAACAGAUCAAUUGAGC |
| RNA3-6 | AUGGACGAUUACUACACCAACAGAUCAAUUGAGCAAU |
| RNA4 | AGUUUAAAACGCACGAGACACAAUGGACGAUUACUACAC |
| RNA5 | AUGGACGAUUACUACAC |
| RNA6 | CAUUAUCGGAUAGUGGAACCUAGCUUCGACUAUCGGAUAAUC |
| RNA7 | UUUUUUUUUUUUUUUUUUUUGAUUAUCCGAUAGUCGAAGCUAGGUUCCACUAUCCGAUAAUGUUUUUUUUUUUUUUUUUUUU |
| DNA1 | AGTTTAAAACGCACGAGACACAATGGACGATTACTACACCAACAGATCAATTGAGCAATCCG |
| DNA2 | CACCACAACCACCACCACCACACCATGG |
| DNA3 | TTTTTTTTTTTTTTTCCATGGTGTGGTGGTGGTGGTTGTGGTGTTTTTTTTTTTTTTT |
The synthesized single strands with florescence labeling are indicated with boldface characters.
Nucleic acid helix destabilizing assays.
In a standard helix destabilizing assay, unless otherwise indicated, 10 pmol of recombinant protein was added to 0.1 pmol of HEX-labeled helix substrate in a buffer containing 50 mM HEPES-KOH (pH 8.0), 2.5 mM MgCl2, 2 mM dithiothreitol (DTT), 0.01% bovine serum albumin [BSA], 1.5 U/μl RNasin, and 2.5 mM ATP. After incubation at 37°C for 1 h or the indicated time period, the reaction mixture was treated with proteinase K (final concentration of 1 μg/μl) at 37°C for 15 min. The digestion reaction was terminated by the addition of 5× loading buffer (100 mM Tris-HCl, 20 mM EDTA, 1% SDS, 0.04% Triton X-100, 50% glycerol, and bromophenol blue [pH 7.5]). Mixtures were then resolved on 12% native-PAGE gels. Gels were scanned with a Typhoon 9200 imager (GE Healthcare, Piscataway, NJ).
RNA strand hybridization assays.
The indicated amount of recombinant protein was incubated with 0.1 pmol of HEX-labeled and unlabeled RNA strands at 37°C in a buffer containing 50 mM HEPES-KOH (pH 8.0), 2.5 mM MgCl2, 2 mM DTT, 0.01% BSA, and 1.5 U/μl RNasin. The reaction was terminated and analyzed as described above. For the hybridization assay of stem-loop-structured RNA strands, the sequences of the HEX-labeled and unlabeled RNA strands were processed as indicated (see Fig. 7A). The stem-loop structures were predicted by the use of mfold (http://mfold.rna.albany.edu/?q=mfold). For the hybridization of 46-nucleotide (nt) and 146-nt RNA strands, the shorter HEX-labeled strand was 5′-GCGGAUAACAAUUUCACACAGGAAACAGCUAUGACCAUGAUUACGA-3′, while the longer unlabeled strand was 5′-CGGCAAGUGGACGAUUAUCUCCAGAGGAUCGCCGGGAACCGAGGACGAGUUCGUAAUCAUGGUCAUAGCUGUUUCCUGUGUGAAAUUGUUAUCCGCUCACAAUUCCACACAACAUACGAGCCGGAAGCAUAAAGUGUAAAGCCUGG-3′. Mixtures were then also resolved on 12% native-PAGE gels. Gels were scanned with a Typhoon 9200 imager (GE Healthcare, Piscataway, NJ).
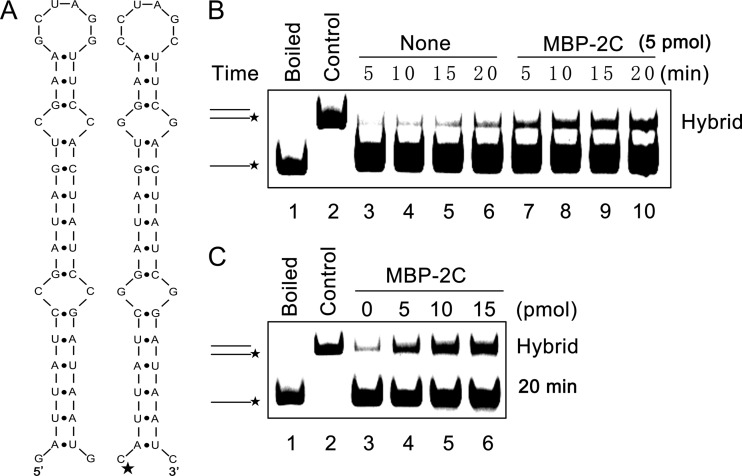
Fig 7.
EoV 2C destabilizes structured RNA strands. (A) Schematic illustrations of the stem-loop structures of the two 42-nt RNA substrates predicted by mfold. The two RNA strands are complementary. One strand has a HEX-labeled 5′ end as indicated by asterisk (right), while the other strand was not labeled (left). (B) The two complementary strands were mixed (0.1 pmol each) and reacted in the absence (lanes 3 to 6) or presence (lanes 7 to 10) of MBP-2C (5 pmol) for the indicated times (5 to 20 min). (C) The two complementary strands were mixed (0.1 pmol each) and reacted with increasing amounts of MBP-2C (0 to 15 pmol) for 20 min. For panels B and C, the mixture of the two strands was boiled (to prevent spontaneous annealing) (lane 1) or treated with a thermal cycler (lane 2) as a negative or positive control, respectively. Samples were subjected to gel electrophoresis (see Materials and Methods).
NTPase assay.
NTPase activities were determined via measuring the released inorganic phosphate during NTP or dNTP hydrolysis using a direct colorimetric assay (6, 7). Briefly, a typical NTPase assay was carried out in 25-μl reaction volumes containing 50 mM HEPES-KOH (pH 8.0), 25 mM NaCl, 2.5 mM MgCl2, 2.5 mM NTP, and 10 pmol protein. The reaction mixtures were incubated at 37°C for 30 min, and then 80 μl of malachite green-ammonium molybdate reagent was added to the reaction mixture, and reactions were further incubated at room temperature for 5 min. After that, 10 μl 34% sodium citrate was added to the reactions for 15 min of incubation, and the absorbance at 620 nm (A620) was then measured. The concentrations of inorganic phosphate were determined via matching the A620 in a standard curve of A620 versus known standard phosphate concentrations. All of the values given with this quantitative assay are averages of the results of three repeated experiments.
RESULTS
EoV 2C protein can destabilize both RNA and DNA helices.
As previously reported, both RNA helicase and RNA chaperone contain the activity to destabilize or unwind short RNA helices (27). Moreover, comparison of the predicted 2C domain (aa 1374 to 1789) of EoV polyprotein (Fig. 1A) with those of the 2C proteins from other picorna-like viruses in the Iflaviridae and Picornaviridae families indicated that, like other 2C proteins, EoV 2C contains all the three conserved SF3 signature motifs (Fig. 1B). To determine whether EoV 2C contains the RNA helix destabilizing activity, we expressed the EoV 2C domain (aa 1374 to 1789) as an MBP-fusion protein (MBP-2C) in a eukaryotic (baculovirus) expression system and then purified the protein. To assess the helix destabilizing activity of EoV 2C, a short HEX-labeled RNA (RNA1) and a long nonlabeled RNA (RNA2) were annealed to generate a standard RNA helix substrate with both 5′ (22 bases) and 3′ (23 bases) single-stranded tails (Fig. 2A and Table 2). The helix destabilizing assay was performed by incubating the RNA helix substrate with purified MBP-2C in a standard destabilizing reaction mixture and then carrying out gel electrophoresis. As shown in Fig. 2A, the HEX-labeled RNA strand was released from the RNA helix substrate in the presence of MBP-2C, MgCl2, and ATP (lane 3), whereas the same substrate was stable when the negative control (MBP) was added to the reaction mixture (lane 2). In this and subsequent experiments, boiled helix substrates were used as positive controls. The results indicated that EoV 2C has RNA helix destabilizing activity.
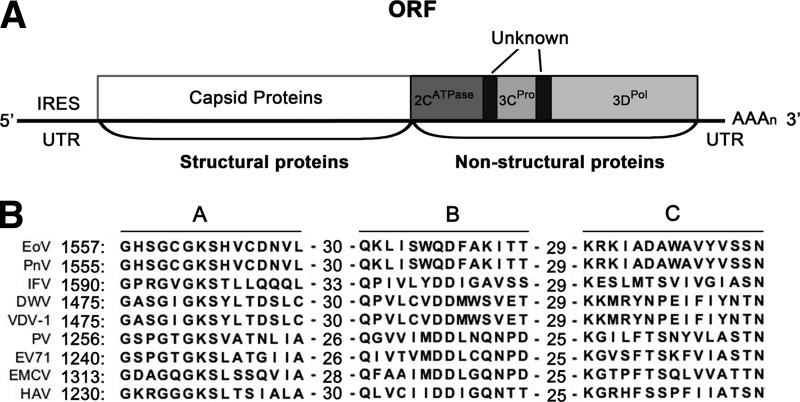
Fig 1.
Amino acid sequence alignment of 2C proteins of EoV and other picorna-like viruses. (A) Schematic representation of the EoV genome. (B) The three conserved motifs (A, B, and C) for SF3 helicases are indicated. The left numbers indicate the starting amino acid positions of the aligned sequences. The middle numbers indicate the numbers of amino acids between the conserved motif regions. PnV, Perina nuda virus; IFV, flacherie virus of silkworm; DWV, deformed wing virus; VDV-1, Varroa destructor virus-1; PV, poliovirus; EV71, enterovirus 71; EWCV, encephalomyocarditis virus; HAV, hepatitis A virus.
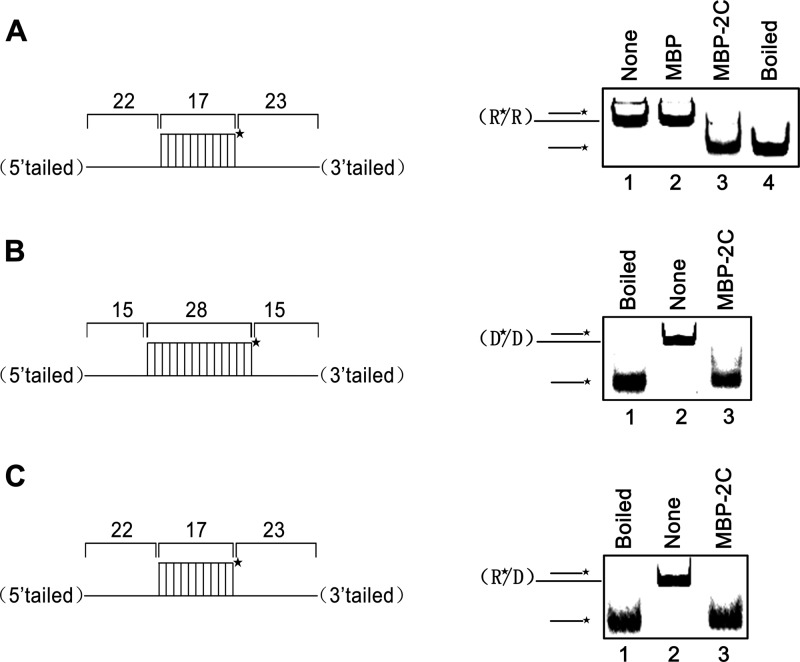
Fig 2.
EoV 2C destabilizes both RNA and DNA helices. Purified MBP-2C was incubated with standard RNA helix (R*/R substrate), DNA helix (D*/D substrate), or RNA/DNA hybrid helix (R*/D substrate) as illustrated in the left panels. Asterisks indicate the HEX-labeled strand. The preparations of destabilizing substrates are indicated in Materials and Methods. Substrate (0.1 pmol) was incubated in standard reaction mixtures in the presence or absence of 10 pmol MBP-2C as indicated, and the destabilizing activity was assessed via gel electrophoresis and scanning on a Typhoon 9200 imager. (A) R*/R substrate (left panel). Lane 1, reaction mixture without 2C addition; lane 2, complete reaction mixture with negative-control MBP alone; lane 3, complete reaction mixture with MBP-2C; lane 4, boiled reaction mixture without 2C addition. (B) D*/D substrate (left panel). Lanes 1 and 2, boiled (lane 1) or native (lane 2) reaction mixtures without 2C addition; lane 3, complete reaction mixture with MBP-2C. (C) R*/D substrate (left panel). Lanes 1 and 2, boiled (lane 1) or native (lane 2) reaction mixtures without 2C addition; lane 3, complete reaction mixture with MBP-2C.
We also sought to determine whether MBP-2C could destabilize helix substrates containing DNA. For this purpose, we constructed two different substrates, D*/D (Fig. 2B) by annealing a short HEX-labeled DNA (DNA1) and a long nonlabeled DNA (DNA3) and R*/D (Fig. 2C) by annealing a short HEX-labeled RNA (RNA1) and a long nonlabeled DNA (DNA2) (Table 2). Each helix substrate was incubated with MBP-2C under the same reaction conditions as were used for the experiment represented by Fig. 2A. MBP-2C was able to destabilize the two kinds of helix substrates (Fig. 2B and C, lane 3). Taken together, our results show that EoV 2C contains functional nucleic acid helix destabilizing activity that does not distinguish RNA and DNA.
2C destabilizes RNA helices in a bidirectional manner.
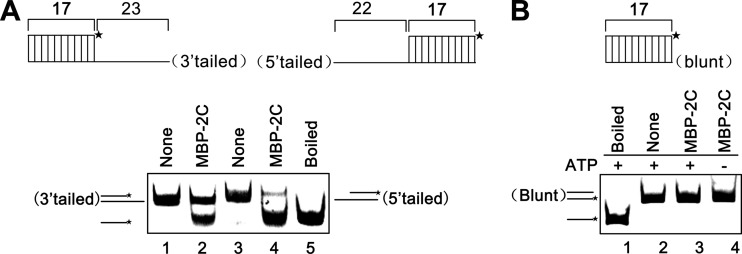
For helicases, the directionality of helix unwinding or destabilization is one of their fundamental characteristics (28). Since picornaviral 2C was long predicted to be a putative SF3 helicase, after determining that EoV 2C has nucleic acid helix destabilizing activity, we sought to assess its destabilizing directionality. Of note, although EoV 2C can destabilize both DNA and RNA helices, we used RNA helices as substrates in the following experiments, because the life cycle of picorna-like viruses does not involve a DNA stage. To determine the directionality, we generated three different RNA helix substrates, containing a 3′ single-stranded tail (23 bases), a 5′ single-stranded tail (22 bases), and blunt ends (as illustrated in the top panels of Fig. 3A and B). These substrates were then incubated with MBP-2C in our standard destabilizing reaction mixture. Our results showed that MBP-2C destabilized both 3′-tailed and 5′-tailed RNA helices (Fig. 3A, top), and its destabilizing activity corresponding to the 5′-tailed RNA helix was apparently higher than the activity corresponding to the 3′-tailed substrate (Fig. 3A, bottom [lane 4 versus lane 2]). The blunt-ended RNA helix could not be destabilized by MBP-2C (Fig. 3B, bottom). These experiments have been independently repeated several times. On the basis of these results, we concluded that EoV 2C protein destabilizes RNA helices in both the 3′-to-5′ and 5′-to-3′ directions and that a 5′ single-stranded tail is preferred.
Fig 3.
EoV 2C destabilizes RNA helices in a bidirectional manner. (A) MBP-2C (10 pmol) was reacted with 3′-tailed (lane 2) or 5′-tailed (lane 4) RNA helix substrate (0.1 pmol) as illustrated in the upper panel, respectively, under standard reaction conditions. Asterisks indicate the HEX-labeled strand. Each substrate alone (lanes 1 and 3) or boiled 5′-tailed substrate (lane 5) was used as a control. (B) MBP-2C (10 pmol) was reacted with blunt-ended RNA helix (0.1 pmol) as illustrated in the upper panel in the absence (lanes 3) or presence (lane 4) of ATP.
The RNA helix destabilizing activity of EoV 2C is NTP independent.
Our previously described results (Fig. 3) show that the RNA helix destabilizing activity of EoV 2C is bidirectional, which is apparently distinct from the activity of known viral RNA helicases that exhibit either 3′-to-5′ or 5′-to-3′ directionality (28). This observation led us to question whether EoV 2C is really an RNA helicase.
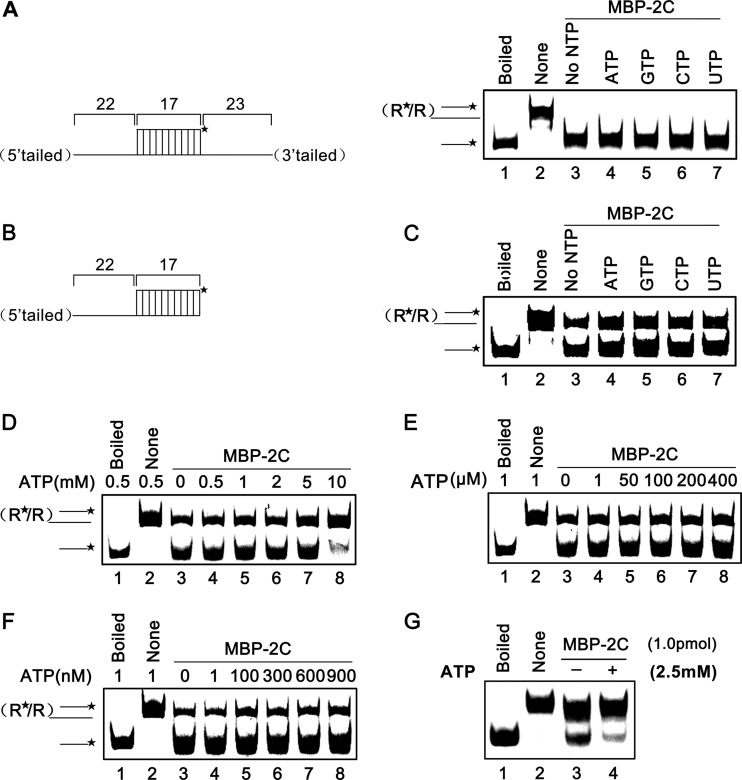
The RNA helicase is not the only type of protein that can destabilize RNA helix, and RNA chaperones can also exhibit this activity in an ATP-free manner. Thus, we assessed the RNA helix destabilizing activity of EoV 2C in the presence or absence of the four different NTPs at a concentration of 2.5 mM. Our results showed that the presence of different NTPs had no effect on the helix destabilizing activity of MBP-2C for either a 3′–plus-5′-tailed (Fig. 4A, right, lanes 4 to 7) or a 5′-tailed (Fig. 4B and lanes 4 to 7 of Fig. 4C) RNA helix substrate; more interestingly, MBP-2C could destabilize both kinds of RNA helix substrates in the absence of NTP (Fig. 4A, right, lane 3; Fig. 4C, lane 3), and the helix destabilizing activity was not promoted by the presence of 2.5 mM ATP or other NTPs (Fig. 4A and C).
Fig 4.
The RNA helix destabilizing activity of EoV 2C is NTP independent. (A) Standard RNA helix substrate (3′ plus 5′ tailed) (0.1 pmol) as illustrated in the left panel was reacted with MBP-2C in the absence or presence of indicated NTP (2.5 mM). Native or boiled substrates without MBP-2C addition were used as controls. (B) Schematic illustration of the 5′-tailed RNA helix substrate that was used in the experiments represented by panels C to G. Asterisks indicate the HEX-labeled strand. (C) The 5′-tailed RNA helix (0.1 pmol) was reacted with MBP-2C (10 pmol) in the absence or presence of the indicated NTP (2.5 mM). Native or boiled substrates without 2C addition were used as controls. (D to F) The 5′-tailed RNA helix (0.1 pmol) was reacted with MBP-2C (10 pmol) in the presence of 0.5 to 10 mM ATP (D), 1 to 400 μM ATP (E), or 1 to 900 nM ATP (F). (G) The 5′-tailed RNA helix (0.1 pmol) was reacted with MBP-2C (1 pmol) in the absence or presence of 2.5 mM ATP.
To determine whether higher NTP concentrations could have a stimulating effect on the helix destabilizing activity of EoV 2C, we assessed the impact of ATP on MBP-2C helix destabilizing activity. To this end, the destabilizing activity of MBP-2C was assessed in the presence of 0.5 to 10 mM ATP, and our results showed that higher (>2 mM) ATP concentrations actually inhibited the RNA helix destabilization, and when ATP reached 10 mM, the helix destabilizing activity was dramatically inhibited (Fig. 4D). Because higher ATP concentrations had an inhibitory effect on the helix destabilizing activity, we further assessed the destabilizing activity of MBP-2C at various low ATP concentrations. The helix destabilizing activity of MBP-2C exhibited no obvious difference in the absence of ATP and in the presence of various low concentrations of ATP from 1 to 400 μM (Fig. 4E) or 1 to 900 nM (Fig. 4F).
For the RNA helix destabilizing assays, 10 pmol MBP-2C was used to destabilize 0.1 pmol RNA helix substrate, at a 100:1 molar ratio, as described for the previous experiments. Similar or higher molar ratios of 50:1 to 200:1 were often used by others in previous RNA helix destabilizing assays for helicases or chaperones (34, 35, 50–54). Here, we also examined the destabilizing activity of 1 pmol MBP-2C on 0.1 pmol RNA helix substrate, and the results showed that MBP-2C could efficiently destabilize the RNA helix substrate at the molar ratio of 10:1 (Fig. 4G, lane 3). Interestingly, the presence of 2.5 mM ATP dramatically inhibited the helix destabilizing activity of EoV 2C at the molar ratio of 10:1 (Fig. 4G, lane 4), suggesting that the inhibitory effect of ATP on the helix destabilizing activity of 2C is dependent on the ATP/2C ratio.
Taken together, our results show that the helix destabilizing activity of EoV 2C does not require the presence of ATP or any other NTPs and that the presence of ATP even exhibits an inhibitory effect on the helix destabilization at a relatively high ATP/2C ratio. These findings indicate that EoV 2C is not an RNA helicase but more likely is an RNA chaperone that exhibits RNA helix destabilizing activity.
Characterization of the RNA helix destabilizing activity of 2C.
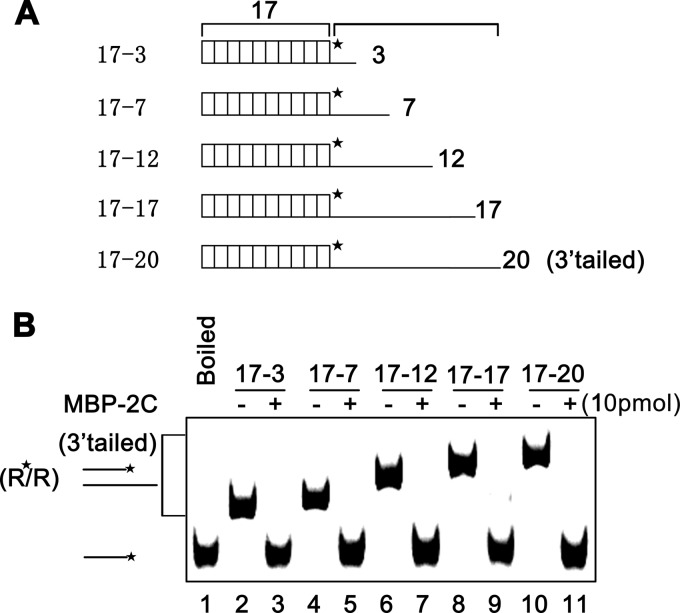
Previously, we have shown that the RNA helix destabilizing activity of 2C requires the presence of a 5′ or 3′ single-stranded tail (Fig. 3). Moreover, previous studies by us and others showed that some viral RNA helicases prefer longer single-stranded tails for their helix destabilizing activity (48, 49, 55). After determining that EoV 2C is not an RNA helicase, we further examined the impact of the length of the single-stranded tail on 2C helix destabilizing activity. As illustrated in Fig. 5A, a series of RNA helix substrates with progressively increasing lengths of 3′ single-stranded tails were generated and then reacted with MBP-2C in the absence of ATP or other NTPs. Interestingly, the length of the 3′ tail of the RNA helix substrates showed no obvious effect on the helix destabilizing activity of 2C (Fig. 5B).
Fig 5.
The length of 3′ tail of RNA helices shows no obvious effect on the helix destabilizing activity of EoV 2C. (A) Schematic illustration of RNA helix substrates with the indicated lengths of 3′ single strands. The shorter strand is HEX labeled. (B) A 0.1-pmol volume of the indicated 3′-tailed RNA helices was reacted with 10 pmol of MBP-2C. Neither ATP or another NTP was supplemented in the reaction mixture.
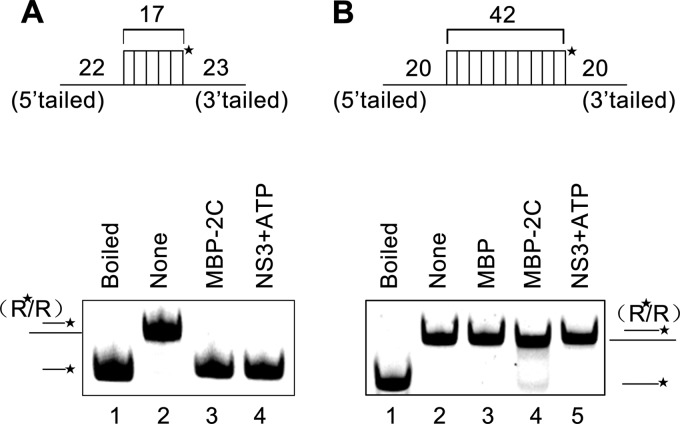
The main function of RNA chaperones is to overcome the kinetic trap of incorrectly folded RNAs to allow the formation of proper RNA folding, since the former structure has lower global energy but is locally stable, while the latter has higher global energy but forms more slowly (24). Thus, an RNA chaperone should have higher efficiency to destabilize a less stable RNA helix (with lower energy) but have lower efficiency to destabilize a more stable helix (with higher energy). To examine this, we designed two different RNA helix substrates as illustrated in Fig. 6A and B (top). The shorter RNA helix contains 17 matched base pairs together with a 5′ single-stranded tail (22 bases) and a 3′ single-stranded tail (23 bases) (Fig. 6A, top), while the longer one contains 42 matched base pairs together with a 5′ tail (20 bases) and a 3′ tail (20 bases) (Fig. 6B, top). Moreover, hepatitis C virus (HCV) NS3, a well-known RNA helicase (56, 57), was used as a control for the RNA helix destabilizing assay. Our results show that MBP-2C exhibited much higher efficiency to destabilize the 17-bp matched RNA helix (Fig. 6A, bottom, lane 3) than to destabilize the 42-bp matched one (Fig. 6B, bottom, lane 4). On the other hand, in the presence of ATP, HCV NS3 was able to destabilize the shorter RNA helix (Fig. 6A, bottom, lane 4) but showed no detectable activity to destabilize the longer RNA helix with 42 matched base pairs (Fig. 6B, bottom, lane 5). Moreover, in the absence of ATP, HCV NS3 showed no activity to destabilize either the shorter or longer RNA helix (data not shown).
Fig 6.
Characterization of the RNA helix destabilizing activity of 2C. Schematic illustrations of RNA helix substrates with indicated lengths of 3′ and 5′ tails and matched base pairs are shown in the upper panels. A 0.1-pmol volume of RNA helix substrate was reacted with 5 pmol MBP-2C in the absence of ATP or other NTP. A 5-pmol volume of MBP-fusion HCV NS3 (plus 2.5 mM ATP) and MBP alone were used as controls for the RNA helix destabilizing assay. Asterisks indicate the HEX-labeled strand.
2C demonstrates helix destabilizing activity to “unwind” structured RNA strands and stimulate annealing.
To further characterize the ability of EoV 2C to destabilize nucleic acid secondary structures, a classic assay, which was developed by DeStefano and colleagues for examining RNA chaperone activities of HIV NC and poliovirus 3AB (34, 35, 58), was adapted for EoV 2C. This assay measures helix destabilization/unwinding and annealing stimulation (35). As illustrated in Fig. 7A, two 42-nt cRNA strands that form defined stem-loop structures were used, and one strand was labeled with HEX at its 5′ end (Fig. 7A, right). The HEX (0.1 pmol)-labeled strand and nonlabeled strand were mixed in the presence or absence of 5 pmol MBP-2C, and then a gel shift assay was performed to measure the hybridization of the two complementary strands. As shown in Fig. 7B, in the absence of 2C, little hybridization was observed as the incubation time increased (lanes 3 to 6), while the presence of 2C dramatically promoted the hybridization of the two strands (lanes 7 to 10). Moreover, we found that the annealing stimulation by 2C was dose dependent, as the increasing dose of 2C resulted in an increase in hybrid formation (Fig. 7C). Overall, these results show that EoV 2C has RNA helix destabilizing activity that can destabilize RNA secondary structures and promote the formation of the more stable hybrids.
2C accelerates the annealing of RNA strands.
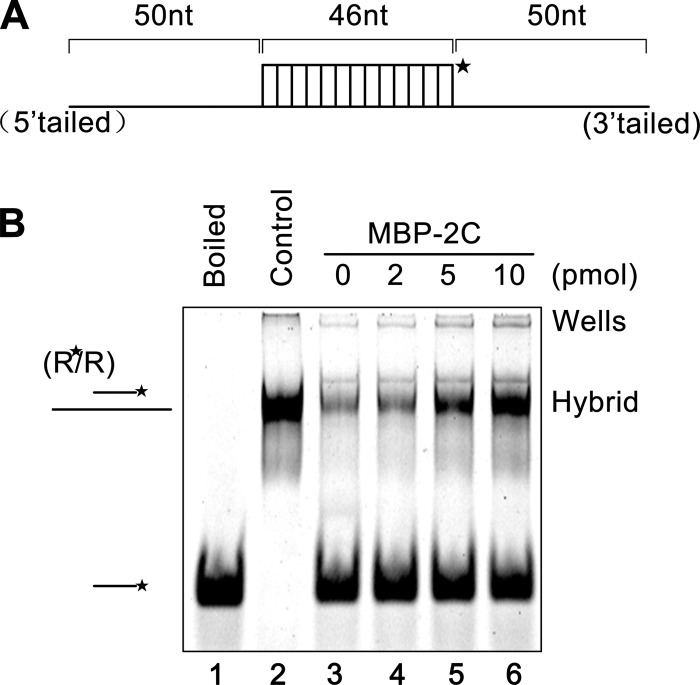
An important property of RNA chaperones is to stimulate annealing of cRNA strands. To determine if EoV 2C has this activity, a shorter (46-nt) labeled RNA strand was incubated with a longer (146-nt) nonlabeled RNA strand (as illustrated in Fig. 8A) for 30 min in the absence or presence of MBP-2C. Moreover, since the chaperone activity is not catalytic or enzymatic, a nucleic acid chaperone normally functions well when present in a large excess over the volume of its substrates (nucleic acid strands) (34, 59, 60). To examine if this is also the case for EoV 2C, increasing amounts of MBP-2C were reacted with 0.1 pmol of 46-nt and 146-nt RNA strands. A gel shift assay was performed to measure the annealing of the two complementary strands. As shown in Fig. 8B, in the absence of 2C, little annealing was observed (lane 3). On the other hand, although 2 pmol MBP-2C exhibited minimal but observable annealing stimulation (lane 4), significant stimulation was observed when the amount of MBP-2C reached 5 pmol (lane 5), and 10 pmol MBP-2C resulted in an even stronger annealing stimulation (lane 6). Overall, these results show that EoV 2C has annealing stimulation activity that is dependent on the dose of 2C.
Fig 8.
EoV 2C accelerates the annealing of RNA strands. (A) Schematic illustration of the 46-nt RNA strand (upper) that is complementary to the 146-nt RNA strand. The 46-nt strand is HEX labeled as indicated by an asterisk. (B) The two strands were mixed (0.1 pmol for each strand) and reacted with increasing amounts of MBP-2C (2 to 10 pmol) for 20 min. The mix of the two strands was boiled (to prevent spontaneous annealing) (lane 1) or treated with a thermal cycler (lane 2) as a negative or positive control, respectively. Samples were subjected to gel electrophoresis (see Materials and Methods).
Characterization of biochemical reaction conditions for the RNA helix destabilizing activity of 2C.
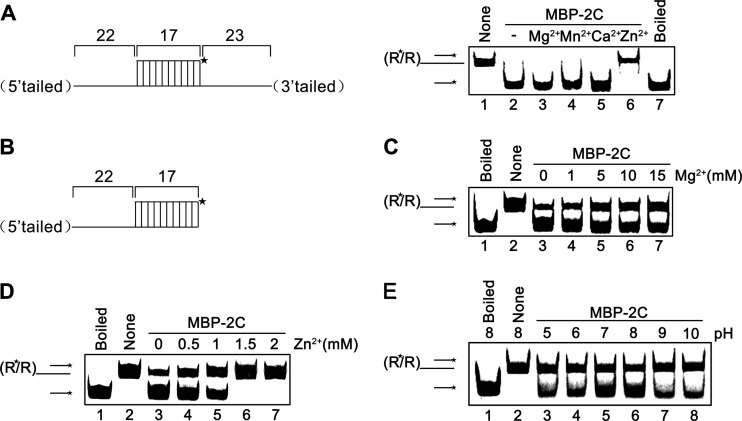
To further characterize the biochemical properties of MBP-2C, we assessed its RNA helix destabilizing activity under various conditions with different divalent metal ions, ion concentrations, and pH values. Here, we first examined the requirement for Mg2+ as well as three other divalent metal ions (Mn2+, Ca2+, and Zn2+) for the helix destabilizing activity of EoV 2C. In this experiment, the RNA helix substrates with both 5′ and 3′ tails were used (Fig. 9A, left). Interestingly, we found that MBP-2C did not require the presence of 2.5 mM Mg2+ or other divalent metal ions for its helix destabilizing activity (Fig. 9A, right, lanes 2 to 5), and the presence of 2.5 mM Zn2+ even abolished RNA helix destabilization (Fig. 9A, right, lane 6).
Fig 9.
Optimal conditions for EoV 2C RNA helix destabilization. (A) Standard RNA helix substrate (3′ plus 5′ tailed) (0.1 pmol) as illustrated in the left panel was reacted with MBP-2C in the presence of the indicated divalent metal ions at 2.5 mM. Native and boiled substrates without 2C addition were used as controls (lanes 1 and 7). (B) Schematic illustration of 5′-tailed RNA helix substrate that was used in the experiments represented by panels C to E. Asterisks indicate the HEX-labeled strand. (C and D) 5′-tailed RNA helix substrate (0.1 pmol) was reacted with MBP-2C in the presence of indicated concentrations of MgCl2 (C) or ZnCl2 (D). (E) Helix destabilizing activity was determined at the indicated pH in the absence of divalent metal ions.
After examining the requirement of different divalent ions, we further assessed the impact of Mg2+ and Zn2+ at various concentrations on the helix destabilizing activity of MBP-2C. For these experiments, a 5′-tailed RNA helix substrate was used (Fig. 9B). As shown in Fig. 9C, the highest unwinding activity was observed at 0 mM Mg2+ (lane 3), whereas the increase in Mg2+ concentration from 0 to 15 mM resulted in a gradual decrease in the helix destabilization. These results showed that Mg2+ was not required for the RNA helix destabilizing activity of EoV 2C and that high Mg2+ concentrations even had moderate inhibitory effects. Our results also showed that when the concentration of Zn2+ reached 1.5 mM, the helix destabilization by 2C was completely blocked (Fig. 9D, lanes 6 and 7); however, at lower (0.5 to 1 mM) Zn2+ concentrations, 2C was moderately inhibited but still retained its helix destabilizing activity (Fig. 9D, lanes 4 and 5). Moreover, the inhibitory effects of Mg2+ and Zn2+ were both rendered in a dose-response manner.
Furthermore, we examined the effects of various pH on the helix destabilizing activity of MBP-2C. Our results showed that EoV 2C prefers a mild basic pH as an optimal reaction condition, as MBP-2C exhibited the highest helix destabilizing activity with pH 8.0 (Fig. 9E).
Identification and characterization of the NTPase activity of EoV 2C.
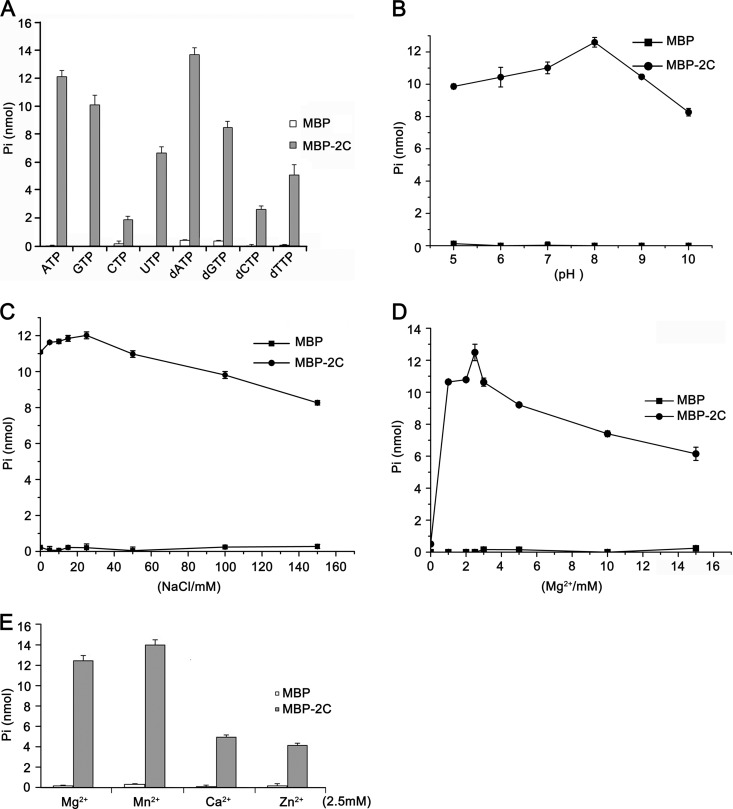
NTPase activity was previously determined in several picornaviral 2C proteins, such as poliovirus (61) and echovirus (62). To determine whether EoV 2C also has NTPase activity that preferentially hydrolyzes one or more of the NTPs or dNTPs, we incubated MBP-2C with ATP, CTP, GTP, UTP, dATP, dCTP, dGTP, and dTTP. The NTPase activity of MBP-2C was determined using a sensitive colorimetric assay that measures the total amount of orthophosphate released after each reaction. Our results showed that MBP-2C hydrolyzed all four types of NTPs and dNTPs but that its NTPase activity was highest for ATP and dATP, followed by GTP and dGTP (Fig. 10A). This ATP and GTP preference of EoV 2C is similar to that of poliovirus 2C (61, 63).
Fig 10.
EoV 2C has NTPase activity. (A) MBP-2C was reacted with the indicated NTP or dNTP. The NTPase activity of MBP-2C was measured as nanomoles of released inorganic phosphate. (B to E) The NTPase activity of EoV 2C was determined at the indicated pH (B), at the indicated concentrations of NaCl (C) orMgCl2 (D), or with indicated divalent metal ions at a concentration of 2.5 mM (E). For panels A to E, the complete reaction mixture with MBP alone was used as the negative control, and error bars represent standard deviation values from three separate experiments.
After determining that EoV 2C does have NTPase activity, we assessed its ATPase activity under various reaction conditions, such as different pH, salt content, and divalent metal ion levels, to further characterize its biochemical properties. Because EoV 2C NTPase prefers ATP and ATP is the major energy source in cells (64), we chose ATP as the hydrolysis substrate in the following experiments. As shown in Fig. 10B, C, and D, the optimal conditions for MBP-2C to hydrolyze ATP were pH 8.0 (Fig. 10B), 25 mM NaCl (Fig. 10C), and 2.5 mM Mg2+ (Fig. 10D). Moreover, we assessed the effects of different divalent metal ions (Mg2+, Mn2+, Ca2+, and Zn2+) on the ATPase activity of EoV 2C. Although Mg2+ was moderately inhibitory for the RNA helix destabilizing activity of EoV 2C (Fig. 9C), Mg2+ as well as Mn2+, Ca2+, and Zn2+ supported its ATPase activity, and their efficiencies were as follows: Mn2+ > Mg2+ > Ca2+ > Zn2+ (Fig. 10E). Moreover, our data show that this ATPase activity is Mg2+ dependent, as the absence of Mg2+ almost abolished the 2C ATPase activity (Fig. 10D). Compared with the optimal reaction conditions for EoV 2C RNA helix destabilizing activity (Fig. 4), the optimal pH for both helix destabilizing and ATP hydrolysis was the same (pH 8.0); interestingly, the helix destabilizing activity of 2C was Mg2+ dispensable (Fig. 9C) but the ATPase activity was Mg2+ indispensable (Fig. 10D). It is noteworthy that MBP protein was produced and purified in the exact same way as with MBP-2C and exhibited negligible NTPase activity in these experiments (Fig. 10A to E). Moreover, recombinant MBP-2C and MBP proteins were independently subjected to a further step of purification by gel filtration with Superdex G75 three times and then assayed for their ATPase activity. These further-purified MBP-2C proteins showed ATPase activity similar to that seen with the previously purified MBP-2C, and MBP alone still showed negligible ATPase activity (data not shown). These data exclude the possibility that a contaminant from the protein expression and purification was responsible for the NTPase activity of 2C.
The RNA helix destabilizing and ATPase activities of EoV 2C can be separated.
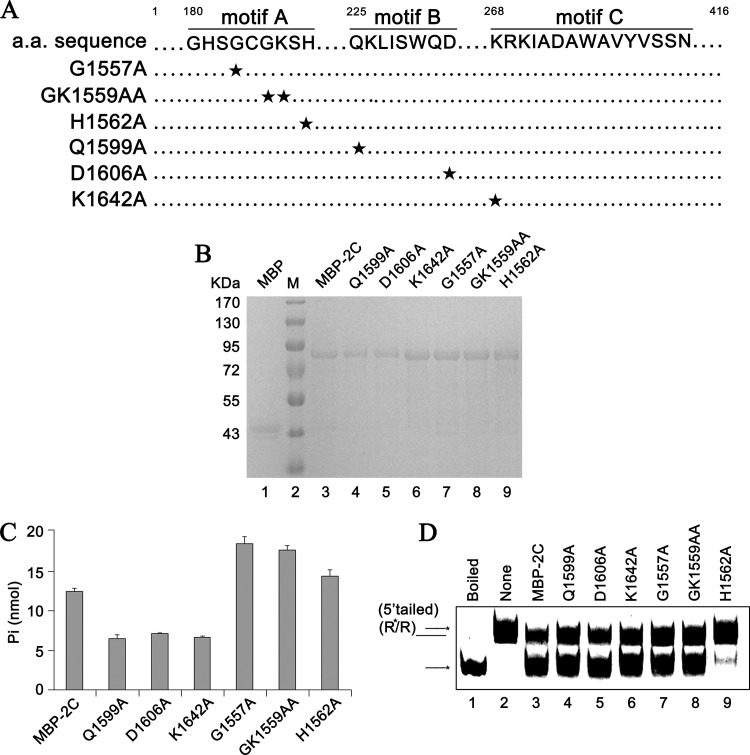
We found that EoV 2C has both RNA helix destabilizing and ATPase activities but that the helix destabilizing activity is independent of ATP. Thus, we sought to determine whether these two activities of EoV 2C could be functionally separated. To this end, a series of point mutations of the conserved residues within motifs A, B, and C (Q1599A, D1606A, K1642A, G1557A, GK1559AA, and H1562A) were constructed as indicated in Fig. 11A. It is noteworthy that the conserved motifs A and B correspond to canonical Walker A and B boxes, consisting of an NTP binding site and a divalent metal ion coordination site, respectively, while motif C is SF3 specific (28). Then, MBP-fusion wild-type and mutant 2C proteins were expressed in a eukaryotic expression system and subsequently purified (Fig. 11B). Afterward, the ATPase (ATP hydrolysis) and RNA helix destabilizing activities of MBP-2C wild-type and mutant strains were determined (Fig. 11C and D). The Q1599A, D1606A, and K1642A mutations significantly reduced the ATPase activity of MBP-2C to around half that of the MBP-2C wild-type strain (Fig. 11C), but these mutations had no effect on helix destabilization (Fig. 11D, lanes 4 to 6). The G1557A and GK1559AA mutations increased the ATPase activity but also had no effect on the RNA helix destabilizing activity (Fig. 11D, lanes 7 and 8). Moreover, we had previously found that in the absence of Mg2+, the ATPase activity of EoV 2C was almost abolished (Fig. 10D), but its helix destabilizing activity was even stronger than in the presence of Mg2+ (Fig. 9C). On the other hand, the H1562A mutation almost blocked the RNA helix destabilization by MBP-2C (Fig. 11D, lane 9) but moderately increased its ATPase activity (Fig. 11C).
Fig 11.
The RNA helix destabilizing and ATPase activities of EoV 2C can be separated. (A) Sequence analysis of the EoV 2C ORF and the mutagenesis strategy, with sites of replacement with alanine indicated by a star. (B) Expressed and purified eukaryotic proteins were subjected to 10% SDS-PAGE followed by Western blotting with anti-MBP polyclonal antibody. (C) Equal amounts of MBP-2C wild-type and mutant strains were reacted with ATP, and ATPase activity was measured as nanomoles of released inorganic phosphate. Error bars represent standard deviation values from three separate experiments. (D) 5′-tailed RNA helix substrate (as illustrated in Fig. 9B) was reacted with equal amounts of MBP-2C wild-type and mutant strains in the absence of NTP.
It is interesting that the important NTP binding residues (GK1559) in motif A of SF3 helicases are not important for the ATPase activity of EoV 2CATPase, which may lead to the speculation that other residues are involved in NTP binding of EoV 2CATPase. On the other hand, point mutation in H1562 in motif A may induce some conformational change that specifically inhibits this protein's helix destabilizing activity. Further crystallographic study of this protein should provide insight into how specific residues are involved in its helix destabilizing and ATPase activities.
Based on these data, our results show that the RNA helix destabilizing and ATPase activities of EoV 2C could be functionally separated.
DISCUSSION
In this report, we show that a picorna-like virus 2C protein has the ability to ATP-independently destabilize RNA helix and stimulate annealing of cRNA strands, which is consistent with the function of an RNA chaperone. The role of an RNA chaperone is to facilitate the escape of locally misfolded RNA molecules from “kinetic traps” and promote the formation of most thermodynamically stable and functional structures of RNAs. For RNA viruses, their viral RNA genomes can adopt various structures, some of which are kinetically trapped inactive intermediates, and it is believed that efficient viral RNA replication and translation often require RNA chaperone activities (25–27). The list of virus-encoded RNA chaperones is growing and includes nucleocapsid (NC) protein of retroviruses, HIV-1 Vif and Tat, poliovirus 3AB, flavivirus core protein, coronavirus nucleocapsid (N) protein, hantavirus nucleocapsid (N) protein, and hepatitis D virus small delta antigen (SdAg). Moreover, rotavirus nonstructural protein NSP2 has been reported to have an ATP-independent helix destabilizing activity but it is unknown if it has any strand annealing stimulation activity (50). In addition, poliovirus polymerase 3DPol has also been reported to display RNA helix unwinding activity that does not require ATP hydrolysis but does require an RNA chain elongation reaction, which is unlike classic RNA chaperone activity (65).
Our current report shows that EoV 2C displays RNA chaperone-like activity. In Picornavirales, poliovirus 3AB has also been reported to be an RNA chaperone (34, 35). Similarly with poliovirus 3AB and HIV-1 NC, EoV 2C destabilizes RNA helix and stimulates annealing in a dose-dependent manner, and large amounts of protein relative to nucleic acid strands are required to observe optimal activities. As previously reported, poliovirus 3AB exhibited optimal nucleic acid chaperone activity at a protein-to-nucleic-acid ratio of approximately 30:1 to 100:1 (34, 35), and HIV-1 NC had optimal chaperone activity at a ratio of approximately 20:1 to 50:1 for minus-strand transfer (51) or approximately 12.5:1 to 25:1 for helix destabilization (34). For EoV 2C, its optimal nucleic acid chaperone activity could be observed at a ratio of 10:1 to 50:1 (Fig. 4, 7, and 8). It is generally believed that nucleic acid chaperone proteins function by coating nucleic acid strands and thus need an excess of protein over nucleic acids (27). And it has also been suggested that poliovirus 3AB displays its chaperone activity in this manner, as large amounts of 3AB relative to nucleic acids are required (34). Our results suggest that EoV 2C exhibits RNA chaperone-like activity in a manner that is the same as or similar to that of poliovirus 3AB.
RNA remodeling proteins, including both RNA helicases and chaperones, function via binding to the single-stranded RNA (ssRNA) strands of RNA helices. For RNA chaperones, their ssRNA binding and chaperone activities are nonspecific. The main difference between an RNA chaperone and an ssRNA-binding protein, which stabilizes a determined RNA conformation, is that the ssRNA binding of RNA chaperones is weak, transient, and mainly of an electrostatic nature, while the latter remains bound to RNAs to ensure stabilization (25–27, 66). In the current study, EoV 2C was unable to shift ssRNA in a gel shift assay (data not shown). Since EoV 2C does display RNA chaperone-like activity, it is clear that this protein must interact with RNA strands. A possible explanation is that the binding of 2C to RNA is too weak and transient to be detected by the gel shift assay. In fact, FMDV 2C has been reported to have nonspecific ssRNA binding activity, but this binding was detected using a truncated form (amino acids 34 to 318) of FMDV 2C (13), and it is unclear if the N-terminal 33 amino acids of 2C have some inhibitory effect on its RNA-binding capacity. On the other hand, a previous study of poliovirus 3AB demonstrated that the combination of 3B plus the last 7 C-terminal amino acids of 3A (termed 3B + 7) is required and sufficient for RNA chaperone activity and that 3B + 7 was also unable to shift ssRNA in a gel shift assay (35). Moreover, studies of the bacterial chaperone protein StpA revealed that a StpA mutant with weakened RNA binding activity exhibits enhanced RNA chaperone activity, showing that weak and transient RNA binding could be beneficial to RNA chaperone activity (27, 66). Besides, for some other viral RNA chaperones, their ssRNA binding activities were not examined in their original reports, which prevented us from comparing the ssRNA binding capacities of different RNA chaperones.
Although RNA chaperones have been studied for many years, the mechanism(s) governing their ATP-independent helix destabilizing and annealing stimulation activity is still not well understood. To explain the activities of RNA chaperones, an “entropy transfer” model has been proposed in which the intrinsic disordered or unstructured regions of chaperone proteins can transfer their entropy or disorder to RNA molecules. Such transfer of disorder destabilizes misfolded RNAs, helps them escape kinetic traps, and facilitates the reformation of the correct RNA structure (26, 27). In reality, many viral RNA chaperones, including HIV-1 NC, Vif, and Tat, hantavirus N, flavivirus core, and coronavirus N, have been predicted to contain intrinsic disordered regions (26, 67, 68). The disordered regions of HIV-1 NC, Vif, and Tat, flavivirus core, and coronavirus N have been predicted using DisProt Predictor VL3 (26, 69, 70). The disorder of hantavirus N was predicted by another algorithm (VL-XT) (68), and we recalculated it using VL3 and predicted similar disordered regions (data not shown). However, the disorder prediction of EoV 2C as well as the 2C proteins of poliovirus, FMDV, and enterovirus 71 (EV71) determined using VL3 showed that these 2C proteins do not contain disordered regions (data not shown). Our further calculation of poliovirus 3AB using the same algorithm predicted that poliovirus 3AB also does not contain intrinsic disorder (data not shown). Moreover, intrinsic disorder has not been predicted for rotavirus NSP2 (data not shown), which has ATP-independent nucleic acid helix destabilizing activity (50). Considering that RNA chaperones lack sequence similarity, different specific RNA chaperones are likely to employ diverse mechanisms.
An alternative model is that some RNA chaperones achieve RNA remodeling through transient ionic or electrostatic interaction with RNA molecules. Due to the negative charge on the phosphodiester backbone of nucleic acids, ionic interactions may strongly affect the stability and dynamics of RNA molecules (24). In this case, chaperone proteins containing large positively charged regions could destabilize RNA structures and facilitate reformation of most stable structures, while multivalent metal cations, such as Mg2+, could inhibit RNA destabilization and strand annealing by stabilizing RNA conformation and competing with RNA chaperones, such as HIV-1 NC, for nonspecific RNA binding (24, 51, 71). In accordance with this, our results showed that Mg2+ exhibited moderate inhibition of EoV 2C RNA helix destabilizing activity (Fig. 9). Moreover, large positively charged regions have been predicted for EoV 2C using the Robetta server and the Swiss PDB Viewer (version 4.0.1) (data not shown), suggesting that transient ionic RNA-protein interaction is likely the mechanism employed by EoV 2C.
Interestingly, although at lower (≤1 mM) concentrations, the increase in the Zn2+ concentration resulted in a gradual and moderate inhibition of the helix destabilization by EoV 2C, similar to that seen with Mg2+, higher (≥1.5 mM) concentrations of Zn2+ completely abolished the helix destabilization (Fig. 9). This dramatic blocking effect of Zn2+ suggests that at higher concentrations, Zn2+ inhibits the RNA chaperone-like activity of EoV 2C in a mechanism different from that of Mg2+. It was reported by Wimmer and colleagues that poliovirus 2C contains a C-terminal cysteine-rich zinc finger-like motif that is able to bind Zn2+ in vitro. Point mutations at potential Zn2+ coordination sites of poliovirus 2C resulted in impaired viral RNA replication, but the ATPase activity of poliovirus 2C does not require Zn2+ or the Zn2+ binding motif (72). Considering that 2C is the most conserved picornaviral protein, it is plausible that Zn2+ is able to directly bind EoV 2C and regulate its RNA chaperone-like activity, possibly by inducing a conformational change of the protein.
Consistent with the results determined with other picornaviral 2C proteins, we determined that EoV 2C contains ATPase activity. Such an ATPase activity previously led to the speculation that picornaviral 2C proteins are RNA helicases that utilize the energy provided by ATP hydrolysis to unwind the RNA helix. Unexpectedly, our data in this work clearly showed that EoV 2C displays ATP-independent RNA chaperone-like activity which destabilizes RNA helix or stimulates strand annealing. Since this 2C protein contains both ATPase and RNA chaperone-like activities, we assessed the functional correlation between them. Our mutational analyses revealed that these two activities of EoV 2C can be functionally separated, as the Q1599A, D1606A, or K1642A mutation resulted in an approximately 50% reduction in ATPase activity but had no impact on RNA helix destabilization. In addition, the H1562A mutation, which dramatically inhibited the RNA helix destabilization, moderately increased ATPase activity (Fig. 11). Moreover, the other strong evidence is the opposing responses of the two 2C activities to Mg2+. In the absence of Mg2+, the ATPase activity of EoV 2C was almost abolished (Fig. 10D), but its RNA helix destabilizing activity was even stronger than in the presence of Mg2+ (Fig. 9C). Based on these findings, we can conclude that the ATPase and RNA chaperone-like activities of EoV 2C are independent of each other. EoV 2C is not the only viral protein that displays both ATPase and RNA chaperone-like activities, as rotavirus NSP2 also contains these two activities, as previously reported (50).
Within picornaviral RNA genomes, multiple cis-acting elements, including the 5′ cloverleaf, 3′ NTR-poly(A), internal origin of replication (oriI or cre), internal ribosomal entry site (IRES), and the cloverleaf at the 3′ end of the negative strand, which play essential or critical roles in the replication and translation of viral RNAs have been identified (73). These highly structured RNA elements may require RNA chaperone activities to aid their proper folding. Moreover, RNA chaperone activities may also be involved in switching the viral RNA conformation to different functions, such as RNA replication, translation, and encapsidation (26, 35).
In Picornavirales, besides EoV 2C, poliovirus 3AB has also been demonstrated to display RNA chaperone activity (34, 35). Although it is unclear if a picornavirus or picorna-like virus contains RNA chaperone activities in both 2C and 3AB, it is not uncommon for a virus to encode multiple RNA chaperones, as HIV-1 NC, Vif, and Tat have all been reported to display RNA chaperone activities (26). Comparison of the RNA chaperone activities of HIV-1 NC and Tat revealed that these proteins employ different mechanisms for their activities and may function at different steps of viral replication due to their differential levels of abundance during the HIV-1 life cycle (74). Likewise, 2C and 3AB proteins are similar in their activities in helix destabilization and strand annealing but different in their responses to Mg2+ (Fig. 9C) (34). Moreover, 3AB is proteolytically processed into 3A and 3B (VPg), neither of which contains RNA chaperone activity (35), but 2C is not further cleaved. Thus, if a picornavirus contains both 2C and 3AB chaperone activities, these two activities are likely to function differently during the picornaviral life cycle. It would be exciting to explore the potential spatial and temporal regulation of these RNA chaperone activities in viral replication.
This report reveals that a picorna-like virus 2C protein displays ATP-independent RNA helix destabilizing and strand annealing stimulation activity which is functionally separated from its ATPase activity. This should extend our understanding of Picornavirales and viral RNA chaperones. In the future, additional studies by our and other groups should reveal whether other picornaviral 2C proteins also have RNA chaperone-like activity. Moreover, specific point mutations, which abrogate RNA chaperone-like activity but have little effect on other 2C activities, should be used to study the role of 2C chaperone-like activity in the life cycle of picorna- or picorna-like viruses.
ACKNOWLEDGMENTS
We thank Markeda Wade for professionally editing the manuscript.
This work was supported by the National Natural Science Foundation of China (grants no. 31270190 [to X.Z.], 81201292 [to X.Z.], and 31270189 [to Y. Hu]) and by the Chinese 111 project (grant no. B06018).
Footnotes
Published ahead of print 28 February 2013
REFERENCES
- 1. Koonin EV, Wolf YI, Nagasaki K, Dolja VV. 2008. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 6:925–939 [DOI] [PubMed] [Google Scholar]
- 2. Kew O, Morris-Glasgow V, Landaverde M, Burns C, Shaw J, Garib Z, Andre J, Blackman E, Freeman CJ, Jorba J, Sutter R, Tambini G, Venczel L, Pedreira C, Laender F, Shimizu H, Yoneyama T, Miyamura T, van Der Avoort H, Oberste MS, Kilpatrick D, Cochi S, Pallansch M, de Quadros C. 2002. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science 296:356–359 [DOI] [PubMed] [Google Scholar]
- 3. Wong SS, Yip CC, Lau SK, Yuen KY. 2010. Human enterovirus 71 and hand, foot and mouth disease. Epidemiol. Infect. 138:1071–1089 [DOI] [PubMed] [Google Scholar]
- 4. Brundage SC, Fitzpatrick AN. 2006. Hepatitis A. Am. Fam. Physician 73:2162–2168 [PubMed] [Google Scholar]
- 5. de Almeida MB, Zerbinati RM, Tateno AF, Oliveira CM, Romao RM, Rodrigues JC, Pannuti CS, da Silva Filho LV. 2010. Rhinovirus C and respiratory exacerbations in children with cystic fibrosis. Emerg. Infect. Dis. 16:996–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wolthers KC, Benschop KS, Schinkel J, Molenkamp R, Bergevoet RM, Spijkerman IJ, Kraakman HC, Pajkrt D. 2008. Human parechoviruses as an important viral cause of sepsislike illness and meningitis in young children. Clin. Infect. Dis. 47:358–363 [DOI] [PubMed] [Google Scholar]
- 7. Bailey L, Gibbs AJ, Woods RD. 1964. Sacbrood virus of the larval honey bee (Apis mellifera Linnaeus). Virology 23:425–429 [DOI] [PubMed] [Google Scholar]
- 8. Laín S, Martin MT, Riechmann JL, Garcia JA. 1991. Novel catalytic activity associated with positive-strand RNA virus infection: nucleic acid-stimulated ATPase activity of the plum pox potyvirus helicaselike protein. J. Virol. 65:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Le Gall O, Christian P, Fauquet CM, King AM, Knowles NJ, Nakashima N, Stanway G, Gorbalenya AE. 2008. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture. Arch. Virol. 153:715–727 [DOI] [PubMed] [Google Scholar]
- 10. Banerjee R, Weidman MK, Echeverri A, Kundu P, Dasgupta A. 2004. Regulation of poliovirus 3C protease by the 2C polypeptide. J. Virol. 78:9243–9256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paul AV. 2002. Possible unifying mechanism of picornavirus genome replication, p 227–246 In Semler BL, Wimmer E. (ed), Molecular biology of picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 12. Kadaré G, Haenni AL. 1997. Virus-encoded RNA helicases. J. Virol. 71:2583–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sweeney TR, Cisnetto V, Bose D, Bailey M, Wilson JR, Zhang X, Belsham GJ, Curry S. 2010. Foot-and-mouth disease virus 2C is a hexameric AAA+ protein with a coordinated ATP hydrolysis mechanism. J. Biol. Chem. 285:24347–24359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Samuilova O, Krogerus C, Fabrichniy I, Hyypia T. 2006. ATP hydrolysis and AMP kinase activities of nonstructural protein 2C of human parechovirus 1. J. Virol. 80:1053–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu Y, Wang C, Mueller S, Paul AV, Wimmer E, Jiang P. 2010. Direct interaction between two viral proteins, the nonstructural protein 2C and the capsid protein VP3, is required for enterovirus morphogenesis. PLoS Pathog. 6:e1001066 doi:10.1371/journal.ppat.1001066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang C, Jiang P, Sand C, Paul AV, Wimmer E. 2012. Alanine scanning of poliovirus 2CATPase reveals new genetic evidence that capsid protein/2CATPase interactions are essential for morphogenesis. J. Virol. 86:9964–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rodríguez PL, Carrasco L. 1995. Poliovirus protein 2C contains two regions involved in RNA binding activity. J. Biol. Chem. 270:10105–10112 [DOI] [PubMed] [Google Scholar]
- 18. Echeverri A, Banerjee R, Dasgupta A. 1998. Amino-terminal region of poliovirus 2C protein is sufficient for membrane binding. Virus Res. 54:217–223 [DOI] [PubMed] [Google Scholar]
- 19. Krogerus C, Egger D, Samuilova O, Hyypia T, Bienz K. 2003. Replication complex of human parechovirus 1. J. Virol. 77:8512–8523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Teterina NL, Gorbalenya AE, Egger D, Bienz K, Ehrenfeld E. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71:8962–8972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gladue DP, O'Donnell V, Baker-Branstetter R, Holinka LG, Pacheco JM, Fernandez-Sainz I, Lu Z, Brocchi E, Baxt B, Piccone ME, Rodriguez L, Borca MV. 2012. Foot-and-mouth disease virus nonstructural protein 2C interacts with Beclin1, modulating virus replication. J. Virol. 86:12080–12090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Norder H, De Palma AM, Selisko B, Costenaro L, Papageorgiou N, Arnan C, Coutard B, Lantez V, De Lamballerie X, Baronti C, Sola M, Tan J, Neyts J, Canard B, Coll M, Gorbalenya AE, Hilgenfeld R. 2011. Picornavirus non-structural proteins as targets for new anti-virals with broad activity. Antiviral Res. 89:204–218 [DOI] [PubMed] [Google Scholar]
- 23. De Palma AM, Heggermont W, Lanke K, Coutard B, Bergmann M, Monforte AM, Canard B, De Clercq E, Chimirri A, Purstinger G, Rohayem J, van Kuppeveld F, Neyts J. 2008. The thiazolobenzimidazole TBZE-029 inhibits enterovirus replication by targeting a short region immediately downstream from motif C in the nonstructural protein 2C. J. Virol. 82:4720–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Woodson SA. 2010. Taming free energy landscapes with RNA chaperones. RNA Biol. 7:677–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Musier-Forsyth K. 2010. RNA remodeling by chaperones and helicases. RNA Biol. 7:632–633 [DOI] [PubMed] [Google Scholar]
- 26. Zúñiga S, Sola I, Cruz JL, Enjuanes L. 2009. Role of RNA chaperones in virus replication. Virus Res. 139:253–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. 2007. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 4:118–130 [DOI] [PubMed] [Google Scholar]
- 28. Singleton MR, Dillingham MS, Wigley DB. 2007. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76:23–50 [DOI] [PubMed] [Google Scholar]
- 29. Hopkins JF, Panja S, McNeil SA, Woodson SA. 2009. Effect of salt and RNA structure on annealing and strand displacement by Hfq. Nucleic Acids Res. 37:6205–6213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ribeiro Ede A, Jr, Beich-Frandsen M, Konarev PV, Shang W, Vecerek B, Kontaxis G, Hammerle H, Peterlik H, Svergun DI, Blasi U, Djinovic-Carugo K. 2012. Structural flexibility of RNA as molecular basis for Hfq chaperone function. Nucleic Acids Res. 40:8072–8084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Doetsch M, Stampfl S, Furtig B, Beich-Frandsen M, Saxena K, Lybecker M, Schroeder R. 2013. Study of E. coli Hfq's RNA annealing acceleration and duplex destabilization activities using substrates with different GC-contents. Nucleic Acids Res. 41:487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lawrence P, Rieder E. 2009. Identification of RNA helicase A as a new host factor in the replication cycle of foot-and-mouth disease virus. J. Virol. 83:11356–11366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kovalev N, Pogany J, Nagy PD. 2012. A co-opted DEAD-box RNA helicase enhances tombusvirus plus-strand synthesis. PLoS Pathog. 8:e1002537 doi:10.1371/journal.ppat.1002537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DeStefano JJ, Titilope O. 2006. Poliovirus protein 3AB displays nucleic acid chaperone and helix-destabilizing activities. J. Virol. 80:1662–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gangaramani DR, Eden EL, Shah M, Destefano JJ. 2010. The twenty-nine amino acid C-terminal cytoplasmic domain of poliovirus 3AB is critical for nucleic acid chaperone activity. RNA Biol. 7:820–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Adams P, Kandiah E, Effantin G, Steven AC, Ehrenfeld E. 2009. Poliovirus 2C protein forms homo-oligomeric structures required for ATPase activity. J. Biol. Chem. 284:22012–22021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Papageorgiou N, Coutard B, Lantez V, Gautron E, Chauvet O, Baronti C, Norder H, de Lamballerie X, Heresanu V, Ferte N, Veesler S, Gorbalenya AE, Canard B. 2010. The 2C putative helicase of echovirus 30 adopts a hexameric ring-shaped structure. Acta Crystallogr. D Biol. Crystallogr. 66:1116–1120 [DOI] [PubMed] [Google Scholar]
- 38. Christian P, Carstens E, Domier L, Johnson K, Nakashima N, Scotti P, vander Wilk F. 2005. Iflavirus, p 779–782 In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball. LA. (ed), Virus taxonomy: eighth report of the International Committee on the Taxonomy of Viruses. Academic Press, San Diego, CA [Google Scholar]
- 39. van Oers MM. 2010. Genomics and biology of Iflaviruses, p 231–250 In Asgari S, Johnson K. (ed), Insect virology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 40. Wang X, Zhang J, Lu J, Yi F, Liu C, Hu Y. 2004. Sequence analysis and genomic organization of a new insect picorna-like virus, Ectropis obliqua picorna-like virus, isolated from Ectropis obliqua. J. Gen. Virol. 85:1145–1151 [DOI] [PubMed] [Google Scholar]
- 41. Lin M, Ye S, Xiong Y, Cai D, Zhang J, Hu Y. 2010. Expression and characterization of RNA-dependent RNA polymerase of Ectropis obliqua virus. BMB Rep. 43:284–290 [DOI] [PubMed] [Google Scholar]
- 42. Lu J, Zhang J, Wang X, Jiang H, Liu C, Hu Y. 2006. In vitro and in vivo identification of structural and sequence elements in the 5′ untranslated region of Ectropis obliqua picorna-like virus required for internal initiation. J. Gen. Virol. 87:3667–3677 [DOI] [PubMed] [Google Scholar]
- 43. Lu J, Hu Y, Hu L, Zong S, Cai D, Wang J, Yu H, Zhang J. 2007. Ectropis obliqua picorna-like virus IRES-driven internal initiation of translation in cell systems derived from different origins. J. Gen. Virol. 88:2834–2838 [DOI] [PubMed] [Google Scholar]
- 44. Ye S, Xia H, Dong C, Cheng Z, Xia X, Zhang J, Zhou X, Hu Y. 2012. Identification and characterization of Iflavirus 3C-like protease processing activities. Virology 428:136–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li SF, Wang HL, Hu ZH, Deng F. 2012. Genetic modification of baculovirus expression vectors. Virol. Sin. 27:71–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Han Y, Wang Q, Qiu Y, Wu W, He H, Zhang J, Hu Y, Zhou X. 2013. Periplaneta fuliginosa densovirus nonstructural protein NS1 contains an endonuclease activity that is regulated by its phosphorylation. Virology 437:1–11 [DOI] [PubMed] [Google Scholar]
- 47. Yang B, Zhang J, Cai D, Li D, Chen W, Jiang H, Hu Y. 2006. Biochemical characterization of Periplaneta fuliginosa densovirus non-structural protein NS1. Biochem. Biophys. Res. Commun. 342:1188–1196 [DOI] [PubMed] [Google Scholar]
- 48. Wang Q, Han Y, Qiu Y, Zhang S, Tang F, Wang Y, Zhang J, Hu Y, Zhou X. 2012. Identification and characterization of RNA duplex unwinding and ATPase activities of an alphatetravirus superfamily 1 helicase. Virology 433:440–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Warrener P, Collett MS. 1995. Pestivirus NS3 (p80) protein possesses RNA helicase activity. J. Virol. 69:1720–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Taraporewala ZF, Patton JT. 2001. Identification and characterization of the helix-destabilizing activity of rotavirus nonstructural protein NSP2. J. Virol. 75:4519–4527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu T, Heilman-Miller SL, Levin JG. 2007. Effects of nucleic acid local structure and magnesium ions on minus-strand transfer mediated by the nucleic acid chaperone activity of HIV-1 nucleocapsid protein. Nucleic Acids Res. 35:3974–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang Y, Liu ZR. 2002. The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J. Biol. Chem. 277:12810–12815 [DOI] [PubMed] [Google Scholar]
- 53. Gebhard LG, Kaufman SB, Gamarnik AV. 2012. Novel ATP-independent RNA annealing activity of the dengue virus NS3 helicase. PLoS One 7:e36244 doi:10.1371/journal.pone.0036244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jankowsky E. 2011. RNA helicases at work: binding and rearranging. Trends Biochem. Sci. 36:19–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Seybert A, Hegyi A, Siddell SG, Ziebuhr J. 2000. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA 6:1056–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gwack Y, Kim DW, Han JH, Choe J. 1996. Characterization of RNA binding activity and RNA helicase activity of the hepatitis C virus NS3 protein. Biochem. Biophys. Res. Commun. 225:654–659 [DOI] [PubMed] [Google Scholar]
- 57. Kim DW, Gwack Y, Han JH, Choe J. 1995. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem. Biophys. Res. Commun. 215:160–166 [DOI] [PubMed] [Google Scholar]
- 58. Heath MJ, Derebail SS, Gorelick RJ, DeStefano JJ. 2003. Differing roles of the N- and C-terminal zinc fingers in human immunodeficiency virus nucleocapsid protein-enhanced nucleic acid annealing. J. Biol. Chem. 278:30755–30763 [DOI] [PubMed] [Google Scholar]
- 59. Herschlag D. 1995. RNA chaperones and the RNA folding problem. J. Biol. Chem. 270:20871–20874 [DOI] [PubMed] [Google Scholar]
- 60. Levin JG, Guo J, Rouzina I, Musier-Forsyth K. 2005. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 80:217–286 [DOI] [PubMed] [Google Scholar]
- 61. Rodríguez PL, Carrasco L. 1993. Poliovirus protein 2C has ATPase and GTPase activities. J. Biol. Chem. 268:8105–8110 [PubMed] [Google Scholar]
- 62. Klein M, Eggers HJ, Nelsen-Salz B. 1999. Echovirus 9 strain barty non-structural protein 2C has NTPase activity. Virus Res. 65:155–160 [DOI] [PubMed] [Google Scholar]
- 63. Pfister T, Wimmer E. 1999. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 274:6992–7001 [DOI] [PubMed] [Google Scholar]
- 64. Hara KY. 2009. Permeable cell assay: a method for high-throughput measurement of cellular ATP synthetic activity. Methods Mol. Biol. 577:251–258 [DOI] [PubMed] [Google Scholar]
- 65. Cho MW, Richards OC, Dmitrieva TM, Agol V, Ehrenfeld E. 1993. RNA duplex unwinding activity of poliovirus RNA-dependent RNA polymerase 3Dpol. J. Virol. 67:3010–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mayer O, Rajkowitsch L, Lorenz C, Konrat R, Schroeder R. 2007. RNA chaperone activity and RNA-binding properties of the E. coli protein StpA. Nucleic Acids Res. 35:1257–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ivanyi-Nagy R, Lavergne JP, Gabus C, Ficheux D, Darlix JL. 2008. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res. 36:712–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Brown BA, Panganiban AT. 2010. Identification of a region of hantavirus nucleocapsid protein required for RNA chaperone activity. RNA Biol. 7:830–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Obradovic Z, Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK. 2003. Predicting intrinsic disorder from amino acid sequence. Proteins 53(Suppl 6):566–572 [DOI] [PubMed] [Google Scholar]
- 70. Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK, Obradovic Z. 2005. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 3:35–60 [DOI] [PubMed] [Google Scholar]
- 71. Vo MN, Barany G, Rouzina I, Musier-Forsyth K. 2006. Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. J. Mol. Biol. 363:244–261 [DOI] [PubMed] [Google Scholar]
- 72. Pfister T, Jones KW, Wimmer E. 2000. A cysteine-rich motif in poliovirus protein 2C(ATPase) is involved in RNA replication and binds zinc in vitro. J. Virol. 74:334–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Paul AV, Belov GA, Ehrenfeld E, Wimmer E. 2009. Model of picornavirus RNA replication, p 3–23 In Raney KD, Gotte M, Cameron CE. (ed), Viral genome replication. Springer, New York, NY [Google Scholar]
- 74. Godet J, Boudier C, Humbert N, Ivanyi-Nagy R, Darlix JL, Mely Y. 2012. Comparative nucleic acid chaperone properties of the nucleocapsid protein NCp7 and Tat protein of HIV-1. Virus Res. 169:349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]