Abstract
Inflammasomes are multiprotein complexes that recognize pathogens and pathogen- or danger-associated molecular patterns. They induce the maturation and secretion of powerful proinflammatory interleukin-1B (IL-1β), IL-18, and IL-33 cytokines, which in turn activate expression of other immune genes and lymphocyte recruitment to the site of primary infection, thereby controlling invading pathogens. Inflammasomes are comprised of cytoplasmic sensor molecules, such as NLRP3 and AIM2 or nuclear sensor IFI16, the adaptor protein ASC (apoptosis-associated speck-like protein containing CARD), and the effector protein procaspase-1. Herpes simplex virus 1 (HSV-1), a ubiquitous virus that infects humans and establishes life-long latency, has evolved numerous mechanisms to evade host detection and immune responses. Here, we show that early during in vitro infection of human foreskin fibroblasts (2 to 4 h), HSV-1 induced the activation of the IFI16 and NLRP3 inflammasomes and maturation of IL-1β. Independent of viral gene expression, IFI16 recognized the HSV-1 genome in infected cell nuclei, relocalized, and colocalized with ASC in the cytoplasm. However, HSV-1 specifically targeted IFI16 for rapid proteasomic degradation at later times postinfection, which was dependent on the expression of ICP0, an immediate early protein of HSV-1. In contrast, NLRP3, AIM2, and ASC levels were not decreased. Also, caspase-1 was “trapped” in actin clusters at later time points that likely blocked the NLRP3/IFI16 inflammasome activity. In addition, the secretion of mature IL-1β was inhibited. These results suggest that though the host cell responds to HSV-1 infection by IFI16 and NLRP3 inflammasomes early during infection, HSV-1 has evolved mechanisms to shut down these responses to evade the proinflammatory consequences.
INTRODUCTION
The inflammasome is a multiprotein proinflammatory complex that is an important bridge between the innate and adaptive immune responses. Inflammasome complexes assemble after recognition of pathogen- or danger-associated molecular patterns (PAMPs or DAMPs, respectively) and include the adaptor molecule apoptosis-associated speck-like protein containing a caspase activation and recruitment domain (CARD, or ASC, for apoptosis-associated speck-like protein containing CARD), the effector molecule procaspase-1, and a sensor protein, which varies to confer specificity. Inflammasome sensor proteins associate with ASC via interactions through their respective pyrin domains (PYDs), and ASC interacts with caspase-1 via CARD-CARD interactions. The sensor proteins that have been described to recognize viral stimuli thus far include nucleotide binding and oligomerization domain (NOD)-like receptor family pyrin domain-containing 3 (NLRP3, also called NALP3), absent in melanoma 2 (AIM2), and gamma interferon-inducible protein 16 (IFI16) (reviewed in reference 1). Activation of the inflammasome complex results in the autoproteolytic cleavage of procaspase-1, which in turn cleaves prointerleukin-1β (pro-IL-1β), pro-IL-18, and pro-IL-33 (2). Mature, secreted IL-1β, IL-18, and IL-33 mediate inflammatory responses by activating lymphocytes and facilitating their infiltration to the site of primary infection and by inducing expression of interferon (IFN) and other proinflammatory cytokines (3, 4).
The NLRP3 inflammasome has been the most widely studied, possibly due to the breadth of activating stimuli: infection with DNA and RNA viruses such as encephalomyocarditis virus (EMCV), vaccinia virus, influenza A virus, and adenovirus (5–8); the generation of reactive oxygen species (ROS); cation flux; fungal infection; and exposure to particulate matter, including uric acid, silica, aluminum salts, and asbestos (9–11). Activation of the NLRP3 inflammasome occurs through a two-step model: (i) transcriptional activation to produce autorepressed cytoplasmic NLRP3 protein and (ii) activation, which involves sensing of cytoplasmic cellular stress followed by multimerization and inflammasome assembly. The mechanisms of the second step of NLRP3 inflammasome activation are unknown as of yet (12). Because of the range of stimuli that activate the NLRP3 inflammasome, it is hypothesized that NLRP3 does not directly recognize all of its agonists but, instead, senses a change or changes in its direct environment that is a shared result of the stimuli (12). One such shared stimulus may be ROS, which are induced by fungal infection (13), by infection with influenza virus (14), adenovirus (15), or EMCV (16), and by exposure to silica (17).
Human hematopoietic interferon-inducible nuclear proteins with a 200-amino-acid repeat (HIN200) domain-containing proteins, AIM2, IFI16, Marek's disease virus nuclear antigen (MDNA), and IFIX have long been known to be transcriptional regulators involved in apoptosis, autoimmunity, and cell cycle regulation and differentiation (18–23). Recently, a role in microbial DNA sensing has been appreciated for AIM2 and IFI16. Both contain pyrin and HIN domains (PYHINs); they can associate with ASC through their pyrin domains and with DNA through their HIN200 domains. Like NLRP3, AIM2 and IFI16 exist in an autorepressed state (24) until stimulation by DNA binding. IFI16 is predominantly nuclear though it has been shown to translocate to the cytoplasm following recognition of some stimuli (19, 25, 26), while AIM2 is usually cytoplasmic (19). AIM2 was the first member of the family discovered to have role in the inflammasome response: the AIM2 inflammasome is activated in the presence of cytosolic, transfected, or microbial DNA (27–29).
It was widely believed that the intracellular innate response to viral DNA was largely restricted to the cytoplasm. However, our recent studies showed that IFI16 acts as a nuclear sensor molecule for inflammasome activation during Kaposi's sarcoma-associated herpesvirus (KSHV) infection (25). Unlike alpha- and betaherpesvirus infections, in vitro KSHV primary infection of adherent target cells and THP-1 cells does not result in a productive lytic cycle and progeny viral particle formation. Instead, the virus enters into latency with limited viral gene expression. After primary infection of human microvascular dermal endothelial cells (HMVEC-d) with KSHV, IFI16 recognized the KSHV genome and interacted with ASC in the nucleus and cytoplasm, resulting in caspase-1 and IL-1β activation (25).
Herpes simplex virus 1 (HSV-1) is a ubiquitous (30) and highly contagious virus that causes a productive infection in many cell types in vitro, in which over 80 gene products are expressed. HSV-1 also establishes a latent infection in the trigeminal ganglia of host organisms, from which it can be periodically reactivated for recurrent lesions at the site of primary infection (31). Like the KSHV genome, the HSV-1 genome is quickly delivered to the nuclei of infected cells, where gene expression and replication occur (31). HSV-1 has evolved several mechanisms to evade the host innate immune responses, including the prevention of interferon (IFN) response factor 3 (IRF3) activation by the HSV-1 immediate-early protein ICP0 (32–34), inhibition of type I IFN signaling by ICP27 (35–37), and the counteracting of PKR activity by γ134.5 (38). In two recent studies, HSV-1 induction of IFN-β expression in human foreskin fibroblast (HFF) cells (34) and CCL3 and CXCL10 expression in monocyte-derived macrophages (39) were shown to occur via IFI16 recognition. However, neither study addressed inflammasome induction.
Our studies with KSHV demonstrating the activation of the IFI16 inflammasome (25) suggested that other herpesviruses might similarly induce inflammasome activation. Previously, HSV-1 infection has been shown to stimulate IL-1β maturation (40). Earlier reports also suggested that IL-1β, IL-18, and IFI16 were effective restriction factors for HSV-1 infection (41–43). In this study, we set out to determine (i) whether the host cell responds to HSV-1 infection through inflammasome activation, (ii) the identity of the sensor(s), and (iii) the downstream effects. Our comprehensive studies, for the first time, report that though in vitro HSV-1 infection of HFF cells induces the activation of the IFI16 and NLRP3 inflammasomes early during infection, HSV-1 subsequently stimulates a selective degradation of IFI16 and suppression of the NLRP3 inflammasome.
MATERIALS AND METHODS
Cells and viruses.
Human foreskin fibroblasts (HFF cells; American Type Culture Collection, ATCC Manassas, VA) were grown in Dulbecco's modified Eagle Medium (DMEM) supplemented with Glutamax (Gibco, Grand Island, NY), 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA), and 1% penicillin/streptomycin (Gibco, Grand Island, NY). They were routinely tested for mycoplasma using a Mycoalert kit (Lonza, NJ), according to the manufacturer's instructions, and were found to be negative. KOS strain HSV-1 was propagated, and titers were determined by plaque assay on Vero cells, as described previously (44). DNA copy number was determined using quantitative real-time PCR with primers for gB (forward primer, 5′-TGTGTACATGTCCCCGTTTTACG-3′; reverse primer, 5′-ACACCAGCTACGCCGCC-3′). The mutant d106 and d109 HSV-1 viruses were a generous gift from Neal DeLuca (University of Pittsburgh) (45). Purification of KSHV from BCBL-1 cells and determination of copy number were carried out as described previously (46, 47).
Virus infection.
HFF cells were incubated with HSV-1 (multiplicity of infection [MOI] of 1 PFU/cell, approximately 25 HSV-1 genome copies/cell) or KSHV (30 KSHV genome copies/cell) for 2 h in serum-free DMEM, washed with phosphate-buffered saline (PBS), and incubated in DMEM supplemented with 2% FBS until the times indicated in the figures.
Antibodies.
The following antibodies were used in Western blotting and immunofluorescence analysis: anti-IFI16 and total caspase-1 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA); Rab27a, TATA binding protein (TBP), and AIM2 (Abcam, Cambridge, MA); IL-1β (Cell Signaling Technology, Beverly, MA; Millipore, Billerica, MA); NLRP3 (Enzo Life Sciences, Farmingdale, NY); ASC (MBL Laboratories, Woods Hole, MA); ICP0 (Virusys, Taneytown, MD); actin and tubulin (Sigma, St. Louis, MO); or HSV-1 (48). Rabbit anti-IFI16 was a generous gift from Santo Landolfo (University of Turin), and the J17 ICP0 antibody was a generous gift from David Davido (University of Kansas, Lawrence, KS).
Immunoprecipitations.
Cells were washed with PBS, lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with protease inhibitor cocktail (Sigma, St. Louis, MO), sonicated, clarified by centrifugation, and normalized to equal amounts of total protein. The lysate was precleared by incubation with protein G-Sepharose beads (GE Healthcare Life Sciences, Buckinghamshire, United Kingdom) for 20 min. Precleared lysates were incubated overnight with anti-ASC and protein G-Sepharose beads. Beads were washed three times with PBS and subjected to Western blot analysis, as described below.
Western blotting.
Cells were lysed in RIPA buffer supplemented with protease inhibitor cocktail (Sigma, St. Louis, MO), sonicated, and clarified by centrifugation at 16,000 × g for 10 min before separation by SDS-PAGE. Equal amounts of protein were electrophoretically transferred to nitrocellulose membranes and incubated with primary antibodies and secondary antibodies conjugated to horseradish peroxidase (KPL, Gaithersburg, MD). Immunoreactive bands were visualized using ECL Western blotting substrate (Pierce, Rockford, IL).
Nuclear and cytoplasmic extract preparation.
Extracts were prepared with a nuclear extract kit (Active Motif, Carlsbad, CA), as described previously (25). Antibodies to TBP and actin were used to determine extract purity.
Immunofluorescence.
Cells were fixed in 4% paraformaldehyde, permeabilized in 0.2% Triton X-100, and blocked with Image-IT (Life Technologies, Grand Island, NY) before being stained with primary antibodies and Alexa Fluor-conjugated secondary antibodies (Life Technologies, Grand Island, NY). Actin was visualized using Alexa Fluor-conjugated phalloidin. Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI). Cells were imaged using a Nikon Eclipse 80i fluorescence microscope and Metamorph software (Molecular Devices, Silicon Valley, CA).
Fluorescence in situ hybridization (FISH).
Cells were stained with antibodies against IFI16 as above, fixed again with 3% paraformaldehyde for 10 min, permeabilized in 0.2% Triton X-100 for 5 min, and treated with 0.1 M Tris-HCl (pH 7.0) for 2 min and with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) twice for 2 min. DNA was denatured by incubation in 70% formamide in 2× SSC at 70°C for 2 min; samples were then washed and dehydrated in increasing concentrations of ethanol. In situ hybridization was performed using a biotinylated HSV-1 bioprobe (Enzo Life Sciences, Farmingdale, NY) in cDenhyb hybridization solution (Insitus Biotechnologies, Albuquerque, NM) at 37°C overnight in a humid chamber. Cells were washed with 2× SSC and probed with Alexa Fluor-conjugated streptavidin (Life Technologies, Grand Island, NY). Nuclei were stained with DAPI. Cells were imaged as described above.
RESULTS
HSV-1 transiently induces inflammasome activation early during infection.
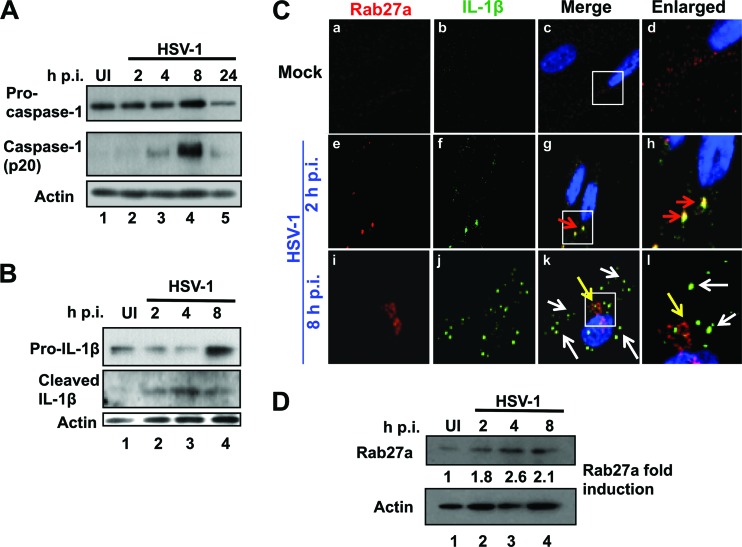
To determine whether the host cell responds to HSV-1 infection by inflammasome activation, HFF cells were mock infected or infected with HSV-1 at an MOI of 1 PFU/cell (approximately 25 DNA copies/cell) for different time periods, as indicated in Fig. 1. We observed the induction of procaspase-1 cleavage to active caspase-1 as early as 4 h postinfection (h p.i.) (Fig. 1A, lanes 2 and 3). Maximal cleavage was detected at 8 h p.i. (Fig. 1A, lane 4) and was reduced at 24 h p.i. (Fig. 1A, lane 5). Procaspase-1 levels increased moderately up to 8 h p.i., and the dramatic decrease observed at 24 h p.i. could be due to cell death. HSV-1 infection induced pro-IL-1β levels at 8 h p.i. (Fig. 1B, lane 4) and cleavage of pro-IL-1β to mature IL-1β at 2 h p.i., peaking at 4 h p.i. (Fig. 1B, lanes 2 to 4). These results are consistent with the earlier observation that MacIntire strain HSV-1 infection induced the maturation of IL-1β and caspase-1 in THP-1 cells (40).
Fig 1.
HSV-1 infection induces the inflammasome early during infection. (A) HSV-1 infection induces the cleavage of caspase 1. HFF cells were mock infected or infected with HSV-1 (1 PFU/cell; 25 HSV-1 DNA copies/cell) and harvested at the times indicated. Western blot analysis was done with antibodies against procaspase-1 and cleaved caspase-1. Actin was used as a loading control. (B) HSV-1 infection causes the cleavage of IL-1β. HFF cells were mock infected or infected with HSV-1 and harvested at the times indicated. Western blot analysis was done with antibodies to total IL-1β and cleaved IL-1β. (C) HSV-1 infection induces transient colocalization of IL-1β with Rab27a. HFF cells were mock infected or infected with HSV-1 as indicated. Immunofluorescence analysis was done with antibodies to Rab27a (red) and IL-1β (green). Red arrows indicate colocalization between Rab27a and IL-1β, white arrows indicate IL-1β puncta with no Rab27a, and yellow arrows show Rab27a in the absence of IL-1β. (D) HSV-1 infection does not cause degradation of Rab27a. HFF cells were mock infected or infected with HSV-1 for the indicated time points, and total cell lysates were analyzed by Western blotting with antibodies against Rab27a and actin. The bands were analyzed as fold induction of Rab27a calculated by considering the uninfected cell level as 1 with actin as a loading control. UI, uninfected.
Secretion of IL-1β is an important outcome of inflammasome activation that leads to NF-κB activation and the expression of proinflammatory cytokines. We next determined whether IL-1β is secreted following HSV-1 infection. Though intracellular IL-1β levels appeared to be somewhat elevated following infection (Fig. 1B), the secreted levels were too close to the detection threshold in our assays to be conclusive (data not shown). Because IL-1β is secreted from cells through exolysosomes (49), we examined the association of IL-1β with Rab27a, a marker for exosomes and secretory lysosomes (50), by immunofluorescence. In mock-infected cells there was minimal staining for both Rab27a and IL-1β and little, if any, colocalization (Fig. 1C, a-d). After 2 h of HSV-1 infection we detected significant colocalization of IL-1β with Rab27a (Fig. 1C, frames e to h, red arrows). However, though we detected elevated IL-1β as evidenced by the numerous punctate spots at 8 h p.i. in the cytoplasm of infected cells (Fig. 1C, frames i to l, white arrows), they were not associated with Rab27a, which was relocalized to the perinuclear region (Fig. 1C, frames i to l, yellow arrows). Because these analyses suggested that Rab27a levels increase following HSV-1 infection, we performed Western blot analysis. Indeed, Rab27a levels increased over the course of 8 h of HSV-1 infection (Fig. 1D). These data demonstrated that though HSV-1 primary infection of HFF cells induces the activation of an inflammasome complex early during infection, it prevents IL-1β from being secreted to detectable levels.
HSV-1 infection induces association of adaptor ASC molecules with sensor proteins IFI16 and NLRP3.
Inflammasome complexes assemble in response to pathogen infection, cation flux, or ROS generation. This complex is made of a sensor molecule (NLRP1, NLRP3, NLRC4, AIM2, or IFI16), an adaptor molecule (ASC), and an effector molecule (procaspase-1) (1). Existing evidence suggests that HSV-1 may activate a number of inflammasome sensor proteins. HSV-1 infection has been shown to generate elevated ROS levels (51), which in unrelated studies were shown to activate the NLRP3 inflammasome (9, 10). Overexpressed IFI16 has also been shown to associate with HSV-1 DNA and translocate into the cytoplasm (26). IFI16 has been shown to recognize HSV-1 infection and activate IRF3 to stimulate IFN-β expression (34, 52). Finally, AIM2 has been shown to be a DNA sensor for inflammasome activation in response to the presence of foreign cytoplasmic DNA (53) and therefore may play a role in the induction of inflammasome activation by HSV-1 infection.
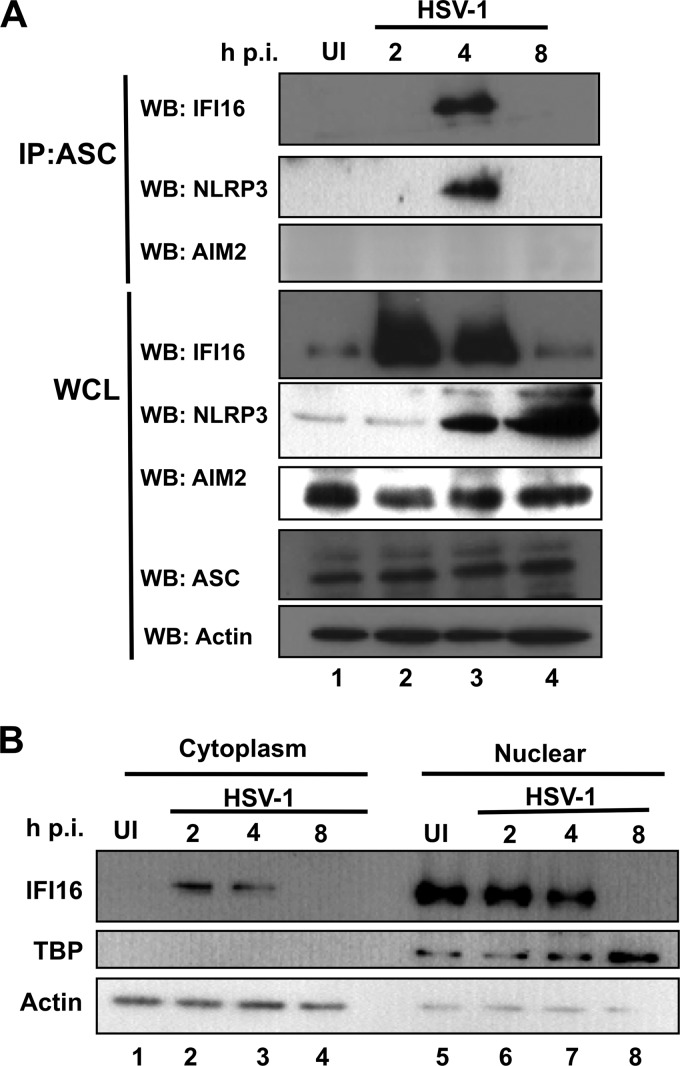
To determine the sensor protein(s) responsible for inflammasome activation during HSV-1 infection, mock-infected or HSV-1-infected HFF cells were immunoprecipitated with ASC antibodies at different times p.i. IFI16 and NLRP3 coprecipitated with ASC only at 4 h p.i. but not in uninfected cells or at later time points of infection (Fig. 2A, top two panels, lane 3). Interestingly, total IFI16 protein increased at early times p.i. (Fig. 2A, fourth panel, lanes 2 and 3) and then decreased dramatically at 8 h p.i. (Fig. 2A, fourth panel, lane 4). In contrast to IFI16, total NLRP3 levels continually increased beginning at 4 h p.i. (Fig. 2A, fifth panel, lanes 3 and 4). There was no coprecipitation of AIM2 with ASC at any of the times tested (Fig. 2A, third panel), and the total levels of AIM2 and ASC remained unchanged throughout the course of infection (Fig. 2A, sixth and seventh panels). These results suggested that IFI16 and NLRP3 act as sensor proteins early during HSV-1 infection of HFF cells. These results also demonstrated that HSV-1 infection induces the reduction in IFI16 levels and induction in NLRP3 levels at 8 h p.i.
Fig 2.
HSV-1 infection induces IFI16-dependent inflammasome activation early during infection. (A) HSV-1 infection induces the association of IFI16 with ASC. HFF cells were mock infected or infected with HSV-1, as indicated. ASC was immunoprecipitated IP) from the lysates. Western blot (WB) analysis of ASC immunoprecipitates was done for IFI16, NLRP3, and AIM2. Whole-cell lysates (WCL) were probed for IFI16, NLRP3, AIM2, ASC, and actin. (B) IFI16 accumulates in the cytoplasm of HSV-1-infected cells at early points postinfection. HFF cells were mock infected or infected with HSV-1 as indicated and separated into nuclear and cytoplasmic fractions before SDS-PAGE. Western blot analysis was done for IFI16, TBP, and actin.
HSV-1 infection induces subcellular redistribution of IFI16.
IFI16 is typically localized to the nucleus (28) but translocates to the cytoplasm in association with ASC following KSHV infection of endothelial cells (25). To determine the subcellular localization of IFI16 following infection with HSV-1, HFF cells were mock infected or infected with HSV-1 and fractionated to nuclear and cytoplasmic components. The purity of nuclear fractions was monitored by Western blotting for TATA binding protein (TBP) (Fig. 2B). In mock-infected cells, IFI16 was exclusively nuclear (Fig. 2B, lanes 1 and 5). However, at 2 to 4 h p.i., IFI16 was also detected in appreciable levels in the cytoplasm of HSV-1 infected cells (Fig. 2B, lanes 2 and 3). At 8 h p.i., IFI16 levels decreased dramatically in both the nuclei and cytoplasm of infected cells (Fig. 2B, lanes 4 and 8). These results demonstrated that HSV-1 induces the cytoplasmic translocation of IFI16 early during infection.
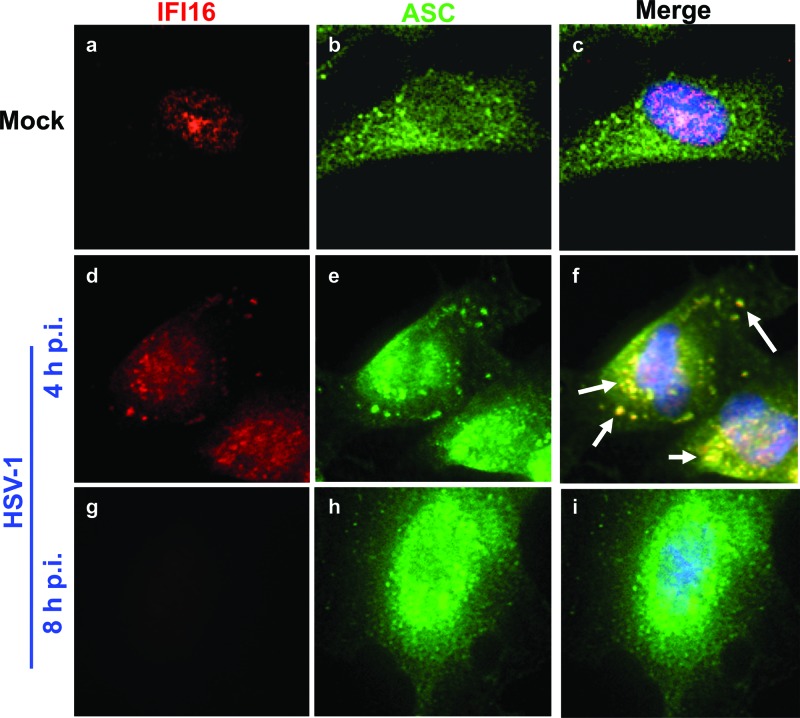
To confirm association of IFI16 and ASC and to determine the subcellular compartment in which this association takes place, we performed immunofluorescence assays on HFF cells that were mock infected or infected with HSV-1, as indicated in Fig. 3. In mock-infected cells ASC was predominantly cytoplasmic, and IFI16 was exclusively nuclear, as expected, with no colocalization (Fig. 3a to c). In contrast, at 4 h p.i., IFI16 and ASC colocalized in punctate spots, largely in the cytoplasm, such that nearly all cytoplasmic IFI16 colocalized with ASC (Fig. 3d to f, arrows). At 8 h p.i., consistent with the Western blot data in Fig. 2, IFI16 staining was lost but there was no appreciable change in ASC staining (Fig. 3g to i).
Fig 3.
IFI16 colocalizes with ASC at early points postinfection with HSV-1. HFF cells were mock infected or infected with HSV-1 (1 PFU/cell) as indicated. Immunofluorescence analysis was done with antibodies to IFI16 (red) and ASC (green). Cell nuclei were visualized by DAPI staining (blue). Arrows indicate colocalization between IFI16 and ASC. Magnification, ×40.
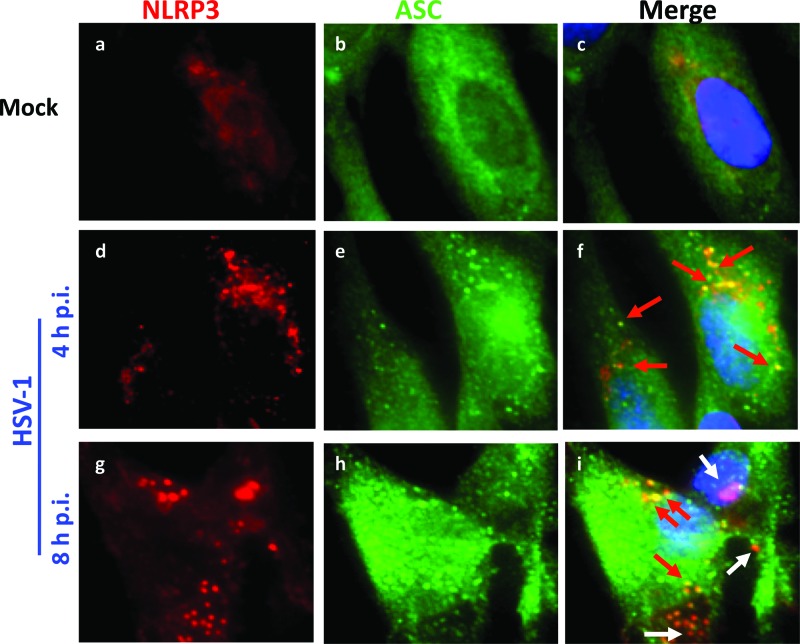
To confirm the HSV-induced association of ASC with NLRP3, immunofluorescence assays were performed. In mock-infected cells, NLRP3 levels were low, as expected from the Western blot analysis (Fig. 2), and there was little detectable colocalization with ASC (Fig. 4a to c). At 4 to 8 h p.i., NLRP3 levels increased (Fig. 4d and g), consistent with the Western blot analysis. In addition, at 4 h p.i., ASC and NLRP3 colocalized in punctate spots (Fig. 4f). However, there were fewer NLRP3/ASC colocalization spots than IFI16/ASC spots (Fig. 3). The loss of the ASC-NLRP3 interaction at late times postinfection (Fig. 2A) is accompanied by reduced colocalization of the two proteins (Fig. 4). Taken together, these data demonstrated that HSV-1 infection induces the transient activation of inflammasome complexes involving IFI16 and NLRP3 and a selective reduction in IFI16 levels.
Fig 4.
NLRP3 transiently colocalizes with ASC at early points postinfection with HSV-1. HFF cells were mock infected or infected with HSV-1 for the indicated time points. Immunofluorescence analysis was done with antibodies to NLRP3 and ASC. Red arrows indicate colocalization between NLRP3 and ASC, and white arrows indicate NLRP3 alone. Cell nuclei were visualized by DAPI staining (blue). Magnification, ×40.
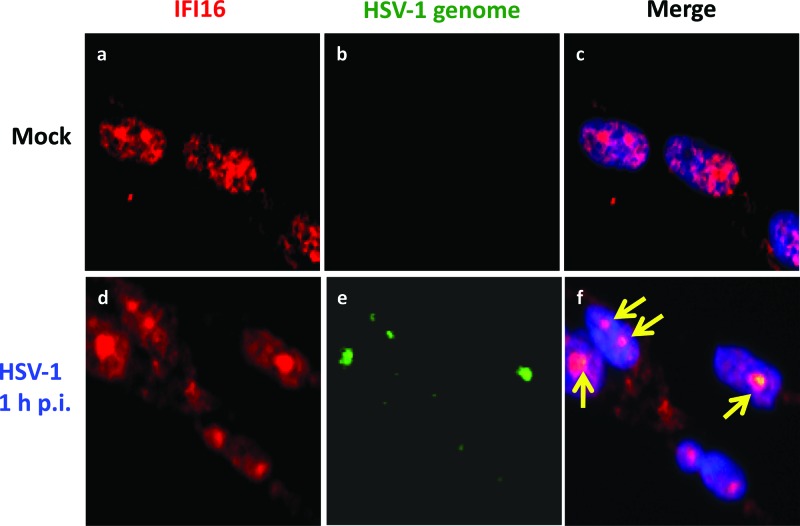
The HSV-1 genome colocalizes with IFI16.
Though IFI16 has been shown to interact with HSV-1 DNA in two recent studies, the authors of these studies used overexpressed IFI16 or a transfected 60-mer DNA sequence from the HSV-1 genome to detect the interaction (26, 52, 54). To determine if endogenous IFI16 interacts with HSV-1 DNA in the more physiological setting of infection, HFF cells were mock infected or infected with HSV-1 for 1 h. Immunofluorescence analysis was performed with IFI16 antibody, and FISH analysis was performed with a probe for the HSV-1 genome. In mock-infected cells, IFI16 was localized throughout the nucleus (Fig. 5a to c). After HSV-1 infection, nuclear IFI16 was enriched in distinct spots, and some IFI16 was also detected in the cytoplasm. The nuclear IFI16-enriched spots colocalized with the HSV-1 genome in the nuclei of infected cells at 1 h p.i. (Fig. 5d to f, arrows). This suggested that endogenous IFI16 associates with the HSV-1 genome early during primary infection of HFF cells.
Fig 5.
The HSV-1 genome is recognized by IFI16. IFI16 colocalizes with IFI16 after HSV-1 infection. HFF cells were mock infected or infected with HSV-1 (1 PFU/cell) for 1 h. Immunofluorescence analysis with an antibody to IFI16 (red) and FISH with a probe to the HSV-1 genome (green) were performed. Cell nuclei were visualized by DAPI staining (blue). Yellow arrows point to the colocalization of the HSV-1 genome and IFI16. Magnification, ×40.
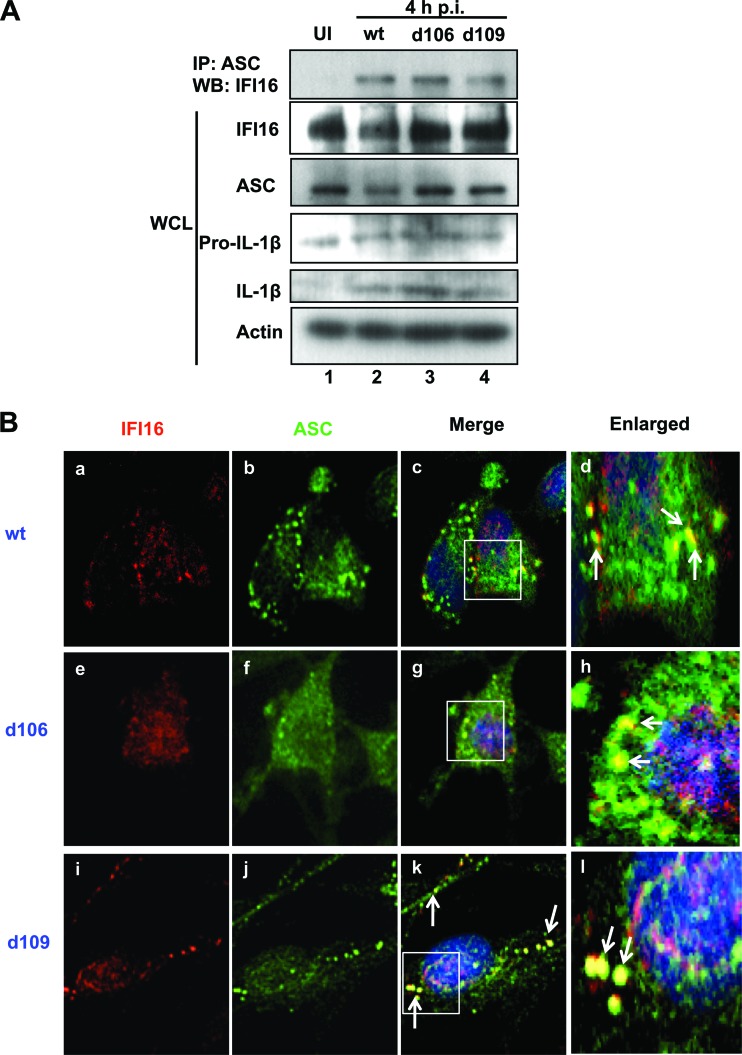
HSV-1 protein expression is not necessary for association of IFI16 and ASC.
To determine if viral gene expression is necessary for the observed association of IFI16 and ASC, HFF cells were mock infected or infected with the d106 or d109 mutant virus, which expresses only ICP0 or no immediate-early viral protein, respectively (45), or with wild-type (wt) HSV-1 at an MOI of 1 PFU/cell for 4 h, and lysates were immunoprecipitated for ASC. In mock-infected cells, ASC and IFI16 did not coprecipitate (Fig. 6A, top panel, lane 1). In contrast, IFI16 associated with ASC after infection not only with wt HSV-1 but also with the mutant d106 or d109 virus (Fig. 6A, top panel, lanes 2 to 4). ASC levels remained fairly constant, regardless of infection (Fig. 6A). These results demonstrated that HSV-1 induced IFI16 and ASC to interact, independent of viral gene expression (Fig. 6A).
Fig 6.
Viral gene expression is not required for HSV-1-induced association of IFI16 and ASC. HFF cells were mock infected or infected with wt HSV-1 or the mutants d106 and d109 (1 PFU/cell) for 4 h. (A) Immunoprecipitation was done using an ASC antibody; Western blot analysis was done for IFI16. Total cell lysate was subjected to Western blotting and probed for IFI16, ASC, pro-IL-1β, IL-1β, and actin. (B) Immunofluorescence was done with anti-IFI16 (red) and -ASC (green) antibodies. Cell nuclei were visualized by DAPI staining (blue). Arrows indicate colocalization between IFI16 and ASC. Magnification, ×40.
To determine whether a functional inflammasome is activated under these conditions, we probed the same lysates for IL-1β cleavage. Total pro-IL-1β levels were fairly constant, regardless of infection (Fig. 6A, pro-IL-1β), and cleaved IL-1β was present in cells infected with wt, d106, or d109 (Fig. 6A, IL-1β, lanes 2 to 4). Taken together, these results indicated that HSV-1 induction of inflammasome activation and the interaction of IFI16 and ASC are not dependent on viral gene expression.
To confirm the immunoprecipitation experiment of IFI16 association with ASC and to determine the subcellular localization of the interaction during the mutant HSV-1 virus infection, HFF cells were infected with wt HSV-1, d106, or d109 at an MOI of 1 PFU/cell for 4 h, and immunofluorescence assays were performed. As shown in Fig. 3, infection with wt HSV-1 induced translocation of IFI16 to the cytoplasm and association with ASC in punctate spots (Fig. 6B, frames a to d). Infection with either mutant virus also induced translocation of IFI16 to the cytoplasm and colocalization of cytoplasmic IFI16 with ASC (Fig. 6B, frames e to l, arrows), suggesting that the HSV-1 genome is sufficient to cause the association of IFI16 and ASC and activation of the inflammasome.
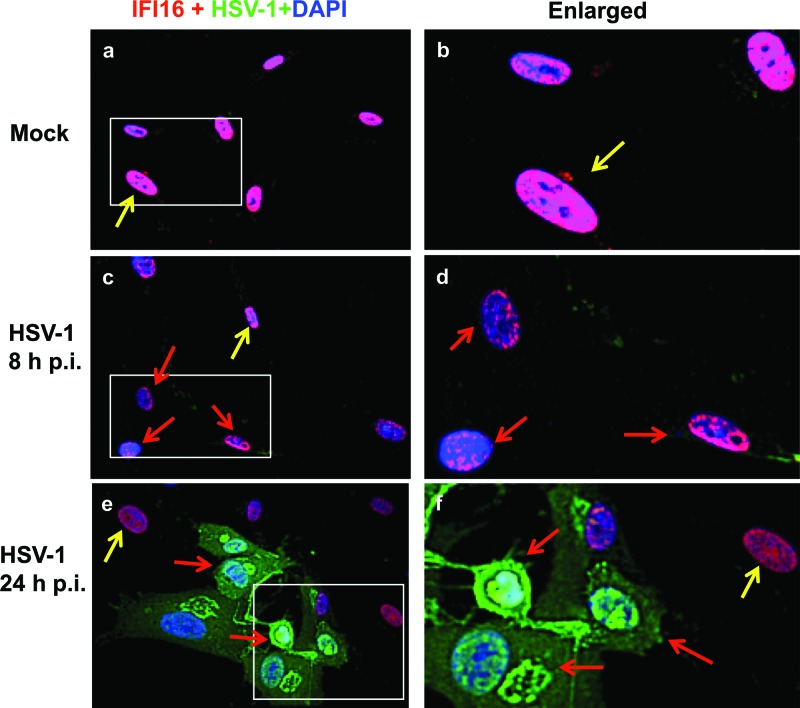
HSV-1 infection induces degradation of IFI16.
Western blot analysis of IFI16 after HSV-1 infection demonstrated decreased IFI16 levels at later times p.i. (Fig. 2). To confirm the HSV-1-induced decrease in IFI16, we mock infected or infected HFF cells with HSV-1 and performed immunofluorescence assays with antibodies against HSV-1-infected cell extracts and against IFI16. Mock-infected cells had high levels IFI16 staining throughout the nucleus (Fig. 7a and b, yellow arrows). In contrast, after 8 h of infection with HSV-1, the IFI16 levels had decreased dramatically, particularly in cells that were beginning to demonstrate HSV-1 staining (Fig. 7c and d, red arrows) but were still present at high levels in cells that were not HSV-1 positive (Fig. 7c, yellow arrow). However, by 24 h p.i., HSV-1-positive cells showed little to no IFI16 staining (Fig. 7e and f, red arrows), but uninfected cells in the same field still had elevated IFI16 levels (Fig. 7e and f, yellow arrows). These results along with the Western blot data showing loss of IFI16 at late times postinfection (Fig. 2) clearly demonstrated that HSV-1 infection causes decreased IFI16 levels. Though cell death may contribute to the decreased IFI16 levels at 24 h p.i., the loss of IFI16 apparent by Western blotting at 8 h p.i. (Fig. 2) and the presence of other cellular proteins such as NLRP3, ASC, caspase-1, and AIM2 suggest that IFI16 is specifically targeted.
Fig 7.
HSV-1 infection induces degradation of IFI16. HFF cells were mock infected or infected with HSV-1, as indicated. Immunofluorescence analysis was done with antibodies against IFI16 (red) and HSV-1 (green). Cell nuclei were visualized by DAPI staining (blue). Yellow arrows indicate nuclear IFI16 in uninfected cells. Red arrows indicate IFI16 degradation in HSV-1-infected cells. Magnification, ×40.
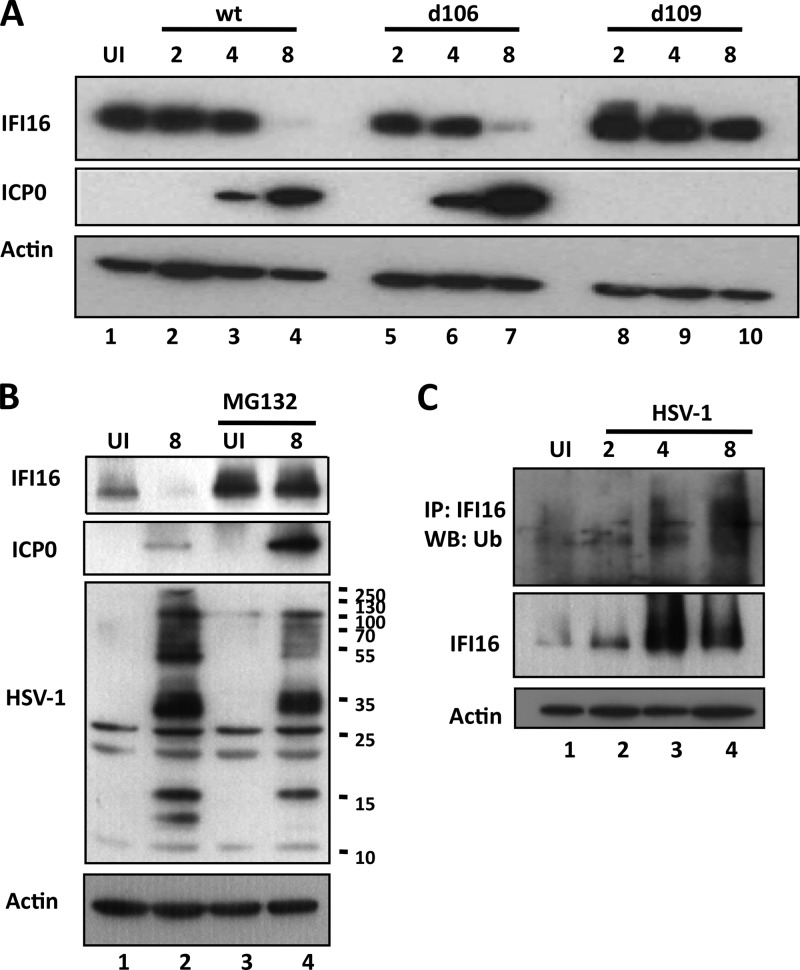
While the manuscript was being prepared, another study was published in which the authors examined the effects of the HSV-1 mutant viruses d106 and d109 on the IFI16-dependent induction of IFN-β. This study confirmed our results showing IFI16 degradation following infection with HSV-1 in an ICP0-dependent manner (34). To confirm their findings regarding the degradation of IFI16, to compare the mutant viruses they used with wt HSV-1 infection, and to determine the kinetics of degradation, we mock infected or infected HFF cells with wt HSV-1, d106, or d109. IFI16 levels decreased at 8 h p.i. in wt- and d106-infected cells (Fig. 8A, lanes 1 to 7). However, consistent with the previous report, IFI16 was stable over the course of infection with d109 (Fig. 8A, lanes 8 to 10). As a control, Western blot analysis of actin expression is shown (Fig, 8A). These results confirm a role for ICP0 in IFI16 degradation.
Fig 8.
HSV-1 infection induces proteasomal degradation of IFI16 in an ICP0-dependent manner. (A) ICP0 is necessary to induce IFI16 degradation. HFF cells were mock infected or infected with wt HSV-1, d106, or d109 (1 PFU/cell) as indicated. Total cell lysates were analyzed by Western blotting with antibodies to IFI16, ICP0, and actin. (B) IFI16 is proteasomally degraded during HSV-1 infection. HFF cells were mock infected or infected with wt HSV-1 in the presence or absence of 5 μM MG132, added at 1 h p.i., as indicated. Western blot analysis was done with antibodies to IFI16, ICP0, HSV-1-infected cell lysate, and actin. (C) IFI16 is ubiquitinated during HSV-1 infection. HFF cells were mock infected or infected with wt HSV-1 in the presence of 5 μM MG132 added at 1 h p.i. and immunoprecipitated with IFI16 antibody. Western blot analysis was done on immunoprecipitated samples for ubiquitin (Ub) and on total cell lysate for IFI16 and actin.
HSV-1 infection induces IFI16 degradation in an ICP0- and proteasome-dependent manner.
ICP0 is an E3 ubiquitin ligase that ubiquitinates several proteins via its RING finger domain, including PML, sp100, DNA-dependent protein kinase (DNA-PK), cellular ubiquitin ligases RNF8 and RNF168, and centromeric protein CENP-C (55–60). Orzalli et al. (34) showed that the degradation of IFI16 by HSV-1 is dependent on the RING-finger domain of ICP0, which contains its E3 ubiquitin ligase activity. To confirm that the HSV-1-induced degradation of IFI16 occurs via the proteasome, we mock infected or infected HFF cells in the presence or absence of the proteasome inhibitor MG132 for 8 h. In the absence of MG132, IFI16 was degraded at 8 h p.i. with HSV-1 (Fig. 8B, IFI16, lanes 1 and 2). In the presence of MG132, IFI16 levels remained constant through 8 h p.i., suggesting that the proteasome is important in the degradation of IFI16 (Fig. 8B, IFI16, lanes 3 and 4).
The proteasome has been shown to be involved in HSV-1 entry (61) and ICP0 expression (62). To ensure that viral protein expression, including that of ICP0, was present in the MG132-treated samples, Western blot analysis was done with antibodies against ICP0 and against HSV-1-infected cell lysates. ICP0 was present at higher levels in MG132-treated cells (Fig. 8B, ICP0, lane 4) than in untreated cells (Fig. 8B, ICP0, lane 2). Though some viral proteins were at lower levels in the presence of MG132, several appeared to be at similar levels (Fig. 8B; compare HSV-1 lanes 2 and 4).
To determine if IFI16 is ubiquitinated, HFF cells were mock infected or infected with HSV-1 in the presence of MG132 and immunoprecipitated with IFI16 antibody. Western blot analysis with an antibody against ubiquitin shows that IFI16 is increasingly ubiquitinated, beginning at 2 to 4 h p.i. (Fig. 8C, top and middle panels, lanes 2 to 4). Taken together, these data confirm that IFI16 is degraded during HSV-1 infection in an ICP0- and proteasome-dependent manner.
HSV-1 ICP0 colocalizes with IFI16 in the infected-cell nuclei and cytoplasm.
To further confirm the role of ICP0 in IFI16 degradation, mock-infected cells or HFF cells infected with HSV-1 for 2 and 4 h p.i. were examined for the cellular localization of IFI16 and ICP0 by immunofluorescence assay (Fig. 9). In mock-infected cells IFI16 was detected throughout the nucleus (Fig. 9a, c, and d). At 2 h p.i., IFI16 and ICP0 colocalized in small puncta in the cytoplasm and nuclei of infected cells (Fig. 9e to h, white arrows), which became larger and less numerous at 4 h p.i. (Fig. 9i to l, white arrows). To determine if ICP0 and IFI16 physically interact with each other, we performed immunoprecipitation assays with antibodies to IFI16 and ICP0 and Western blot analysis for each. No interaction was detected under the conditions used (data not shown). These results suggested that IFI16 and ICP0 interact transiently directly or indirectly, resulting in the degradation of IFI16.
Fig 9.
HSV-1 immediate-early protein ICP0 colocalizes with IFI16. HFF cells were mock infected or infected with HSV-1 (1 PFU/cell) for the indicated time periods. Immunofluorescence analysis was done with antibodies to ICP0 and IFI16, and arrows indicate colocalization between IFI16 (red) and ICP0 (green). Cell nuclei were visualized by DAPI staining (blue). Magnification, ×40.
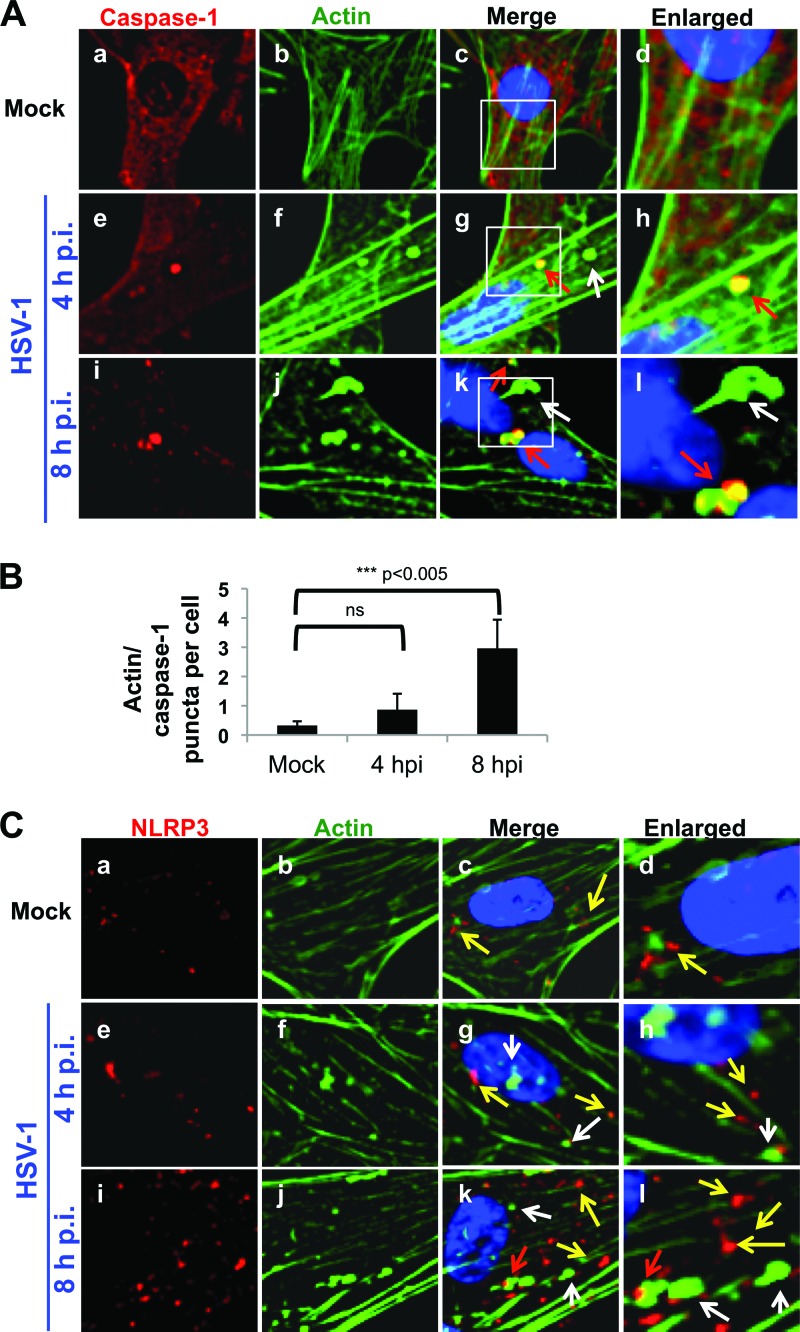
Caspase-1 associates with actin clusters in HSV-1-infected cells.
NLRP3 expression increases over the course of infection, but its association with ASC is transient (Fig. 2). It is interesting that the NLRP3 inflammasome has been shown to be inactivated by clusters of actin that “trap” caspase-1 (63) and that HSV-1 infection causes cytoskeletal reorganization, including relocalization of actin into similar clusters (64, 65). To determine if caspase-1 is trapped by actin clusters during HSV-1 infection, mock-infected or HSV-1-infected HFF cells were examined by immunofluorescence for caspase-1 and actin, using Alexa Fluor-conjugated phalloidin. Cells were scored for the number of colocalized caspase-1/actin puncta per cell. In mock-infected cells, there were no actin clusters and little or no colocalization of actin and caspase-1 (Fig. 10A, frames a to d). In contrast, at 4 h p.i., actin started to form clusters (Fig. 10A, frames f to h), some of which colocalized with caspase-1 puncta (Fig. 10A, frames e to h, red arrows, and B). By 8 h p.i., association of caspase-1 puncta with actin clusters was increased to approximately three colocalized puncta per cell (Fig. 10A, frames i to l, red arrows, and B) though there were some actin clusters that did not appear to contain caspase-1 (Fig. 10A, frames i to l, white arrows).
Fig 10.
HSV-1 infection induces caspase-1 association with actin clusters. (A and C) HFF cells were mock infected or infected with HSV-1 (1 PFU/cell) for the indicated times. Immunofluorescence analysis was done with antibody to caspase-1 (A) or NLRP3 (C). Actin was visualized using Alexa Fluor-conjugated phalloidin. Red arrows indicate colocalization between caspase-1 or NLRP3 and actin, white arrows indicate actin clusters devoid of caspase-1 or NLRP3, and yellow arrows indicate caspase-1 or NLRP3, as indicated. Cell nuclei were visualized by DAPI staining (blue). Magnification, ×40. (B). Caspase-1 and actin puncta were counted in 50 to 60 cells of four fields/condition and analyzed using a Student t test. ns, nonsignificant.
To determine if NLRP3 is also localized to actin clusters, we mock infected or infected HFF cells with HSV-1 and stained for NLRP3 and actin. In mock-infected cells, NLRP3 levels were very low, and there was little to no colocalization of NLRP3 and actin (Fig. 10C, frames a to d). At 4 h p.i., actin clusters had started to form (Fig. 10B, frames f to h, white arrows), but there was no colocalization of these clusters with NLRP3 puncta (Fig. 10C, g and h, yellow arrows). By 8 h p.i., though the NLRP3 puncta increased, there was minimal colocalization with actin clusters, as shown in Fig. 10B, frames i to l, where white arrows indicate actin alone, yellow arrows indicate NLRP3 alone, and red arrows indicate colocalized actin and NLRP3. These data suggested that caspase-1, but not NLRP3, is trapped by actin clusters in HSV-1-infected cells.
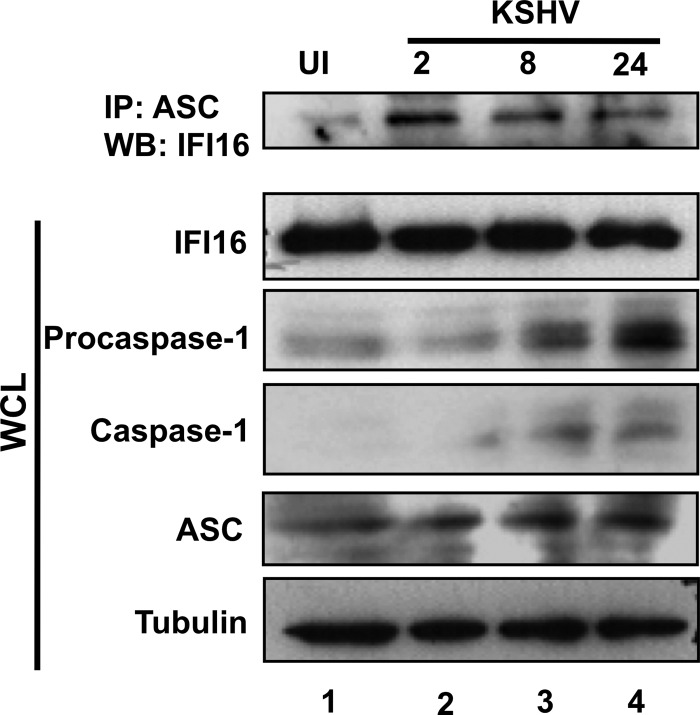
IFI16 degradation is not induced by infection with KSHV.
IFI16 was not degraded in endothelial cells infected with KSHV (25). However, there may be cell type differences that lead to the specific degradation of IFI16 by herpesviruses in fibroblasts and not endothelial cells. To determine if KSHV induces IFI16 degradation in HFF cells, we tested the stability of IFI16 in HFF cells infected with KSHV. HFF cells were mock infected or infected with KSHV for 2, 8, or 24 h and subjected to Western blotting for IFI16. As in endothelial cells, KSHV also induced the interaction of ASC and IFI16 as early as 2 h p.i. and for the duration of the 24-h infection tested (Fig. 11, top panel, lanes 2 to 4). In contrast to HSV-1 infection, KSHV infection did not cause a decrease in the IFI16 level through the course of a 24-h infection (Fig. 11, WCL IFI16, lanes 1 to 4). However, KSHV infection did result in increased ASC and procaspase-1 expression and increased caspase-1 maturation (Fig. 11, respective WCL panels, lanes 1 to 4). These results are consistent with the previous studies showing IFI16 stability and activation of the IFI16 inflammasome in endothelial cells during primary KSHV infection resulting in latency (25) and with the lack of a KSHV homolog for ICP0. Together, these results indicate that IFI16 degradation is specific to HSV-1 infection and not the cell type infected.
Fig 11.
KSHV infection results in activation of the IFI16 inflammasome but not in IFI16 degradation. HFF cells were mock infected or infected with KSHV (30 copies/cell), as indicated. Proteins from lysates were immunoprecipitated with antibodies against ASC and subjected to Western blot analysis with IFI16 antibody (top panel). Total cell lysates were analyzed by Western blotting for IFI16, procaspase-1, caspase-1, ASC, and tubulin, as indicated.
DISCUSSION
The inflammasome sensor molecules NLRP3, AIM2, and IFI16 have been implicated in inflammasome activation following virus infection (5–7, 9, 10, 25). Once stimulated, these sensor molecules associate with the adaptor protein, ASC, and the effector protein, procaspase-1, to activate IL-1β, IL-33, and IL-18 by proteolytic cleavage. These cytokines cause signaling cascades that result in the expression of tumor necrosis factor alpha (TNF-α), IFN-γ, and other cytokines, which recruit lymphocytes and act as a vital bridge between the innate and adaptive immune responses to control the invading pathogens (3, 4).
Our studies here show that HSV-1 infection induces the inflammasome early during infection of HFF cells, characterized by the transient association of IFI16 and NLRP3, but not AIM2, with ASC in the cytoplasm of HSV-1-infected cells. IFI16 colocalization with the HSV-1 genome at early times postinfection suggests that recognition of the viral DNA drives IFI16 inflammasome assembly. Consistent with a previous report (34), HSV-1 infection of HFF cells induces the proteasomal degradation of IFI16 in an ICP0-dependent manner. Our studies also for the first time demonstrate the trapping of caspase-1 by actin clusters in HSV-1-infected cells, probably to block the NLRP3/IFI16 inflammasomes and the inhibition of IL-1β secretion. All of these collectively suggest that HSV-1 has evolved ways to effectively limit the antiviral and proinflammatory consequences of inflammasome activation though in vivo studies would be necessary to confirm this in host organisms. Additional studies would also be necessary to address the HSV-1-induced secretion of proinflammatory cytokines from sources other than the inflammasome. Because immunopathology of HSV-1 in host organisms is well documented (31), there must be other proinflammatory molecules. One possible stimulus for proinflammatory cytokines and recruitment of lymphocytes may be HSV-1-induced cell death.
We propose a model (Fig. 12) whereby HSV-1 enters the cell and the nucleocapsid travels along microtubules to the nucleus (66), during which time the viral genome is protected from recognition by AIM2 by the capsid proteins. Once in the nucleus, the HSV-1 genome circularizes and is recognized by IFI16, which exits the nucleus and assembles with ASC and procaspase-1 to form an inflammasome complex, causing the cleavage and activation of caspase-1 and IL-1β, concurrent with the cascade expression of HSV-1 immediate-early, early, and late genes. The IFI16-ASC inflammasome and NLRP3-ASC inflammasome (possibly generated by HSV-1-induced ROS or the degradation of IFI16) induce IL-1β cleavage, leading to association of mature IL-1β with Rab27a secretory vesicles. HSV-1-encoded ICP0 causes the degradation of IFI16. Rab27a is relocalized to the trans-Golgi network by an unknown HSV-1-encoded mechanism, leaving IL-1β in vesicles in the cytoplasm. Finally, actin clusters, formed by an unknown HSV-1-encoded mechanism, trap caspase-1 and suppress NLRP3 inflammasome activity.
Fig 12.
Schematic steps of HSV-1 infection-induced activation and subsequent deactivation of the IFI16 and NLRP3 inflammasomes during primary in vitro infection of human fibroblast cells. (Step 1) HSV-1 enters the cell, generating ROS, and the nucleocapsid travels along microtubules to the nucleus. Its genome is protected from recognition by AIM2 by the nucleocapsid. (Step 2) Once in the nucleus, the HSV-1 genome circularizes and is recognized by IFI16. (Step 3) IFI16 exits the nucleus and assembles with ASC and procaspase-1 to form an inflammasome complex, which causes the cleavage and activation of caspase-1 and IL-1β. (Step 4) HSV-1-encoded ICP0 causes the degradation of IFI16. (Step 5) ROS and/or the degradation of IFI16 or other host/HSV-1 factors activates the NLRP3 inflammasome, which activates IL-1β activation. (Step 6) IL-1β associates with Rab27a secretory vesicles. (Step 7) Rab27a is relocalized to the trans-Golgi network by an unknown HSV-1-encoded mechanism, leaving IL-1β in vesicles in the cytoplasm. (Step 8) Actin clusters formed by an unknown HSV-1-encoded mechanism trap caspase-1 and suppress NLRP3 inflammasome activity. Green arrows indicate HSV-1-encoded activities. Green T-bar indicates suppression.
Characteristics of HSV-1-induced inflammasomes during primary infection.
Our studies show that IFI16, but not AIM2, is a sensor of viral DNA that induces the assembly of inflammasome complexes in response to HSV-1 infection. One explanation for this is that HSV-1 DNA is released in the nucleus during a productive lytic infection, where it is accessible to IFI16 but not AIM2. In the cytoplasm, HSV-1 DNA is protected from AIM2 recognition by the viral capsid. Another possible explanation is the inhibition of AIM2 inflammasome activation by increased IFI16 levels (67). Though the expression of IFI16 and NLRP3 is stimulated by HSV-1 infection at early time points, HSV-1 selectively degrades IFI16 at later time points (Fig. 2).
IFI16 has long been known to bind DNA. Recently, reports have shown that IFI16 preferentially binds to cruciform or superhelical DNA (54), and a crystal structure of IFI16 bound to DNA showed that IFI16 recognizes the sugar-phosphate backbone in a non-sequence-specific manner (24). Because the IFI16 inflammasome is not constitutively active in uninfected cells but is activated after infection with HSV-1 (Fig. 1), this indicates that IFI16 recognizes the viral genome as distinct from cellular DNA. IFI16 is a known component of the DNA damage response (68) and may recognize the viral genome as “damaged” DNA. Additionally, the HSV-1 genome is known to circularize once it enters the host cell nuclei (69). It is possible that, during the process of circularization, there is a structure that resembles cruciform or superhelical DNA, which is recognized by IFI16. HSV-1 chromatin is less densely packed than host cell DNA (70), which could make the viral DNA more accessible to IFI16 recognition than host cell DNA. Other IFI16 binding partners involved in transcriptional repression, such as p53 (71, 72), may also cause changes in the conformation of IFI16 (23), rendering it unavailable to assemble in inflammasome complexes. The different topology of HSV-1 DNA may release structural constraints on IFI16, making it accessible to bind ASC. Additional extensive studies beyond the scope of the present study are required to further examine the interactions of IFI16 with host and viral DNA and to determine if there is a direct interaction of IFI16 with the HSV-1 genome or if the interaction occurs through other proteins known to interact with IFI16 and HSV-1 DNA, such as p53 (21), BRCA1 (73), or proteins in ND10 domains (74).
Recently, a study reported that recombinant varicella-zoster virus (VZV) infection induced inflammasome activation through NLRP3, resulting in the secretion of biologically active IL-1β (75). In this study, however, infected fibroblasts were cocultured with naïve fibroblasts to determine inflammasome activation. This infection system results in VZV infection likely mediated by fusion between infected and uninfected cells (76) rather than a primary infection. Therefore, NLRP3 inflammasome activation in this system could be due to a number of off-target effects caused by transfer of cell contents, induction of cation influx, or ROS generation rather than by infection itself.
Consistent with the accepted two-step model for NLRP3 inflammasome activation (1), total NLRP3 levels increased at 4 and 8 h p.i. (Fig. 2), possibly due to signaling through Toll-like receptor (TLR) and NF-κB activation (77). Though it is likely that the activation of the IFI16 inflammasome is due to recognition of the HSV-1 genome by IFI16 in the nucleus, the mechanism of activation of the NLRP3 inflammasome is more elusive. The HSV-1-induced generation of ROS (51) and the degradation of IFI16 (67) during infection may both induce NLRP3 inflammasome activation. Further investigation would need to be done to address the mechanism of activation of the NLRP3 inflammasome by HSV-1.
After HSV-1 infection the association of NLRP3 with ASC was limited to 4 h p.i. (Fig. 2). Our immunofluorescence assays suggest that some NLRP3 and ASC associate with each other at later times p.i. (Fig. 4); however, the amount of colocalized NLRP3 and ASC was significantly decreased compared with the total levels of either protein and with the degree of colocalization at 4 h p.i. This suggests that although there may be some interaction between NLRP3 and ASC at later times p.i., the proportion of the proteins in inflammasome complexes decreases as infection progresses and may be beneath the threshold of detection for our coimmunoprecipitation assays at later times p.i. Alternatively, this may indicate an indirect interaction of ASC and NLRP3, which may be interrupted by the stringency of our immunoprecipitation at later times.
Further studies need to be done to address the relative contributions of IFI16 and NLRP3 to HSV-1-induced caspase-1 and IL-1β maturation and to address whether or not the inflammasome is activated during latent HSV-1 infection.
HSV-1-mediated inflammasome suppression: IFI16 degradation.
Orzalli et al. (34) showed the degradation of IFI16 following HSV-1 infection in an ICP0-dependent manner and did not observe any cytoplasmic IFI16 after 4 h of infection. However, experiments with fractionated cells infected with wt HSV-1 were not carried out. Though we showed by immunofluorescence that IFI16 exited the nucleus of cells infected with d106 and d109 as well as wt HSV-1, translocation appeared to a lesser extent after infection with the mutant viruses than after infection with wt HSV-1, which may account for the discrepancy. In addition, Orzalli et al. (34) used a significantly higher MOI of 10 to 50 PFU/cell than the 1 PFU/cell in our studies, which could likely lead to some differences in the kinetics of IFI16 translocation and not detecting cytoplasmic IFI16 at their chosen time points. The capacity of IFI16 to localize to the cytoplasm has been demonstrated previously (25, 26, 52). The multiple antiviral roles of IFI16, such as the activation of IFN-β production (34, 39), inflammasome activation, and viral gene suppression (41), could be the potential reasons that HSV-1 has evolved to target IFI16 rather than other proteins such as ASC, AIM2, and NLRP3.
Addition of MG132 during infection with HSV-1 caused stabilization of IFI16. Though optimal transcription of ICP0 has been reported to depend on NF-κB activation, which is also dependent on proteasome activity (62), we detected high levels of ICP0 at late times p.i. in the presence of proteasome inhibitor (Fig. 8). This may be due to the predominantly nuclear localization of ICP0 during MG132 treatment (78), driving its transcription factor activity, and to a positive feedback loop of ICP0 expression (79). Because the proteasome has also been implicated in HSV-1 entry (61), we began the treatment with MG132 after exposure of the cells to HSV-1 and used a relatively low concentration of the drug to limit the effects of proteasome inhibition. We show that a large number of viral proteins, though not all, are expressed in the presence of proteasome inhibition. Decreased viral protein production in the presence of MG132 could be due to an important proteasome-dependent step later in viral replication. The proteasomal degradation of stalled RNA polymerase II has been shown to be important for efficient viral gene expression and replication compartment formation (80). HIN200 proteins are known to be modulators of transcription (18, 19), and hence the stabilization of IFI16 may affect the transcription of both cellular and viral genes. Though it is interesting to speculate about the effects of MG132-induced IFI16 stabilization on virus replication and inflammasome activation, the many potential effects of proteasome inhibition on the HSV-1 life cycle and on the cellular response to infection make it difficult to assess the effects of IFI16 stabilization in this system. Further studies are needed to determine the class(es) of viral protein affected by proteasome inhibition and to address the effects of IFI16 stabilization on inflammasome activation in the absence of chemical inhibitors.
We noted that ICP0 and IFI16 colocalized in punctate spots in the nuclei and cytoplasm of HSV-1-infected cells (Fig. 9). Colocalization of IFI16 and ICP0 was more distinct in the cytoplasm than in the nucleus, which may be due to the faster kinetics of IFI16 in the nucleus or to larger IFI16 aggregates in the cytoplasm. Though IFI16 and ICP0 colocalized in infected cells, they did not coimmunoprecipitate, which indicated that the interaction is transient or indirect. RING-finger E3 ubiquitin ligases, such as ICP0, have been known to bind transiently to their substrates; the E2 ubiquitin-conjugating enzymes are reported to act as scaffolds that mediate ubiquitination of the substrate (reviewed in reference 81), and some RING fingers can allosterically activate E2s with no substrate interaction (82). One of these ubiquitination mechanisms may be at play here.
Recently, IFI16 was shown to be a restriction factor for human cytomegalovirus (CMV), HSV-1, and HSV-2 infection (41). This study showed that IFI16 directly inhibited the expression of early and late CMV genes by repressing transcription of viral promoters containing Sp1 sites. The restriction of IFI16 on HSV-1 could be due to a similar mechanism, to the induction of interferon transcription through the IFI16-STING signaling pathway (34, 52), to activation of the inflammasome, or, very likely, to a combination of the three. Here, we show that the induction of inflammasome activation is transient and that the degradation of IFI16 provides a potential strategy by which the virus evades inflammasome-induced restriction. IFI16 is not degraded during infection with CMV or KSHV (25, 41, 83), suggesting that HSV-1, specifically, has evolved this particular detection evasion strategy during lytic infection.
HSV-1-mediated inflammasome suppression: NLRP3 inflammasome suppression.
In addition to IFI16 degradation, HSV-1 has probably evolved to prevent IL-1β cleavage by sequestration of inflammasome components from each other, as suggested by the dissociation of NLRP3 and ASC at later times p.i. (Fig. 2) and the punctate, ASC-free NLRP3 spots observed at later times p.i. (Fig. 4). Activation of the NLRP3 inflammasome was recently shown to be suppressed by association with clustered actin filaments in macrophages (63). HSV-1 infection induces dramatic actin rearrangement (64, 65). Colocalization of caspase-1 with actin clusters and the absence of NLRP3 in these clusters (Fig. 10) suggest an overall disassembly of the NLRP3 inflammasome at later times p.i. and indicate that a similar actin cluster trapping mechanism may be employed by HSV-1 to shutoff the NLRP3 inflammasome. Recently, the alphaherpesvirus US3 kinase has been implicated in actin relocalization after infection (84). Though this was shown for pseudorabiesvirus and not HSV-1, the proteins from the two viruses share 77% sequence homology, and this may be the mechanism by which HSV-1 induces actin rearrangement and NLRP3 inflammasome inactivation.
Inhibition of IL-1β secretion.
HSV-1 has evolved numerous layers of immune evasion strategies; it has been shown to inhibit interferon expression, signaling, and effects (32, 35, 37, 85). Our studies suggest that the secretion of mature IL-1β is also inhibited by HSV-1 infection (Fig. 1). Though IL-1β levels increased in response to infection, both in immunofluorescence assays and Western blot analysis (Fig. 1), IL-1β appeared as punctate cytoplasmic spots and was not secreted to detectable levels, consistent with previous reports in macrophages showing no IL-1β secretion during HSV-1 infection (86, 87). However, these studies did not address the cytoplasmic accumulation of IL-1β after HSV-1 infection.
Previous studies suggest that the Rab GTPases are necessary for HSV-1 assembly and egress (88–90). Our data show that IL-1β transiently associates with Rab27a-containing vesicles and then remains in the cytoplasm in punctate spots while Rab27a is relocalized to a perinuclear region (Fig. 1). Therefore, HSV-1 may be recruiting Rab GTPases from cellular functions to play roles in viral assembly, consistent with the model set forth by Bello-Morales et al. (90) of the recruitment of Rab GTPases to the endoplasmic reticulum (ER) and Golgi compartment, where they colocalize with HSV-1 viral glycoproteins, gD and gH, though the mechanism of recruitment is unclear.
The HSV-1-induced generation and cytoplasmic accumulation of mature IL-1β and lack of its secretion suggest that HSV-1 has evolved a mechanism to inhibit IL-1β secretion. Inhibition of the secretion of mature, biologically available IL-1β may be a viral strategy to subvert the proinflammatory and proapoptotic effects of IL-1β signaling. Taken together, these data suggest that HSV-1 infection causes the maturation of IL-1β but prevents its biological activity. Though the secretion of other inflammasome-activated proinflammatory cytokines cannot be ruled out by the IL-1β experiments, it is unlikely that their activation would be prolonged due to the dissociation of the inflammasome complexes.
Our studies clearly demonstrate that though the host cell responds to HSV-1 infection by activation of the IFI16 and NLRP3 inflammasomes early during infection, HSV-1 has evolved multiple strategies to block these responses to evade the proinflammatory consequences and facilitate replication. These studies form a framework to further explore the ways to overcome the capacity of HSV-1 to replicate and spread to neighboring cells.
ACKNOWLEDGMENTS
This study was supported, in part, by Public Health Service Grants CA 075911 and CA 168472 to B.C. and the RFUMS-H.M. Bligh Cancer Research Fund to B.C.
We thank Neal DeLuca (University of Pittsburgh) for his gift of the d106 and d109 mutant HSV-1 viruses, David Davido (University of Kansas, Lawrence, KS) for his gift of the J17 ICP0 antibody, and Santo Landolfo (University of Turin) for his gift of the rabbit antibody against IFI16.
Footnotes
Published ahead of print 20 February 2013
REFERENCES
- 1. Bauernfeind F, Ablasser A, Bartok E, Kim S, Schmid-Burgk J, Cavlar T, Hornung V. 2011. Inflammasomes: current understanding and open questions. Cell. Mol. Life Sci. 68:765–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wilson KP, Black JA, Thomson JA, Kim EE, Griffith JP, Navia MA, Murcko MA, Chambers SP, Aldape RA, Raybuck SA, Livingston DJ. 1994. Structure and mechanism of interleukin-1 beta converting enzyme. Nature 370:270–275 [DOI] [PubMed] [Google Scholar]
- 3. Okamura H, Nagata K, Komatsu T, Tanimoto T, Nukata Y, Tanabe F, Akita K, Torigoe K, Okura T, Fukuda S. 1995. A novel costimulatory factor for gamma interferon induction found in the livers of mice causes endotoxic shock. Infect. Immun. 63:3966–3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, Gorman DM, Bazan JF, Kastelein RA. 2005. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity 23:479–490 [DOI] [PubMed] [Google Scholar]
- 5. Delaloye J, Roger T, Steiner-Tardivel QG, Le Roy D, Knaup Reymond M, Akira S, Petrilli V, Gomez CE, Perdiguero B, Tschopp J, Pantaleo G, Esteban M, Calandra T. 2009. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathog. 5:e1000480 doi:10.1371/journal.ppat.1000480 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6. Ito M, Yanagi Y, Ichinohe T. 2012. Encephalomyocarditis virus viroporin 2B activates NLRP3 inflammasome. PLoS Pathog. 8:e1002857 doi:10.1371/journal.ppat.1002857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barlan AU, Griffin TM, McGuire KA, Wiethoff CM. 2011. Adenovirus membrane penetration activates the NLRP3 inflammasome. J. Virol. 85:146–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, Taraporewala ZF, Miller D, Patton JT, Inohara N, Nunez G. 2006. Critical role for cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J. Biol. Chem. 281:36560–36568 [DOI] [PubMed] [Google Scholar]
- 9. Benko S, Philpott DJ, Girardin SE. 2008. The microbial and danger signals that activate Nod-like receptors. Cytokine 43:368–373 [DOI] [PubMed] [Google Scholar]
- 10. Schroder K, Zhou R, Tschopp J. 2010. The NLRP3 inflammasome: a sensor for metabolic danger? Science 327:296–300 [DOI] [PubMed] [Google Scholar]
- 11. Joly S, Sutterwala FS. 2010. Fungal pathogen recognition by the NLRP3 inflammasome. Virulence 1:276–280 [DOI] [PubMed] [Google Scholar]
- 12. Tschopp J, Schroder K. 2010. NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nat. Rev. Immunol. 10:210–215 [DOI] [PubMed] [Google Scholar]
- 13. Said-Sadier N, Padilla E, Langsley G, Ojcius DM. 2010. Aspergillus fumigatus stimulates the NLRP3 inflammasome through a pathway requiring ROS production and the Syk tyrosine kinase. PLoS One 5:e10008 doi:10.1371/journal.pone.0010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peterhans E. 1997. Oxidants and antioxidants in viral diseases: disease mechanisms and metabolic regulation. J. Nutr. 127:962S–965S [DOI] [PubMed] [Google Scholar]
- 15. Livne A, Shtrichman R, Kleinberger T. 2001. Caspase activation by adenovirus E4orf4 protein is cell line specific and is mediated by the death receptor pathway. J. Virol. 75:789–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ano Y, Sakudo A, Kimata T, Uraki R, Sugiura K, Onodera T. 2010. Oxidative damage to neurons caused by the induction of microglial NADPH oxidase in encephalomyocarditis virus infection. Neurosci. Lett. 469:39–43 [DOI] [PubMed] [Google Scholar]
- 17. Hamilton RF, Jr, Thakur SA, Holian A. 2008. Silica binding and toxicity in alveolar macrophages. Free Radic. Biol. Med. 44:1246–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnstone RW, Trapani JA. 1999. Transcription and growth regulatory functions of the HIN-200 family of proteins. Mol. Cell. Biol. 19:5833–5838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ludlow LE, Johnstone RW, Clarke CJ. 2005. The HIN-200 family: more than interferon-inducible genes? Exp. Cell Res. 308:1–17 [DOI] [PubMed] [Google Scholar]
- 20. Choubey D, Deka R, Ho SM. 2008. Interferon-inducible IFI16 protein in human cancers and autoimmune diseases. Front. Biosci. 13:598–608 [DOI] [PubMed] [Google Scholar]
- 21. Ouchi M, Ouchi T. 2008. Role of IFI16 in DNA damage and checkpoint. Front. Biosci. 13:236–239 [DOI] [PubMed] [Google Scholar]
- 22. Mondini M, Costa S, Sponza S, Gugliesi F, Gariglio M, Landolfo S. 2010. The interferon-inducible HIN-200 gene family in apoptosis and inflammation: implication for autoimmunity. Autoimmunity 43:226–231 [DOI] [PubMed] [Google Scholar]
- 23. Johnstone RW, Kerry JA, Trapani JA. 1998. The human interferon-inducible protein, IFI 16, is a repressor of transcription. J. Biol. Chem. 273:17172–17177 [DOI] [PubMed] [Google Scholar]
- 24. Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, Hornung V, Latz E, Bowie AG, Fitzgerald KA, Xiao TS. 2012. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 36:561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. 2011. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe 9:363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li T, Diner BA, Chen J, Cristea IM. 2012. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc. Natl. Acad. Sci. U. S. A. 109:10558–10563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. 2009. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458:509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458:514–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, Fitzgerald KA. 2010. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11:395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu F, Sternberg MR, Kottiri BJ, McQuillan GM, Lee FK, Nahmias AJ, Berman SM, Markowitz LE. 2006. Trends in herpes simplex virus type 1 and type 2 seroprevalence in the United States. JAMA 296:964–973 [DOI] [PubMed] [Google Scholar]
- 31. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed). 2007. Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 32. Melroe GT, DeLuca NA, Knipe DM. 2004. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J. Virol. 78:8411–8420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Melroe GT, Silva L, Schaffer PA, Knipe DM. 2007. Recruitment of activated IRF-3 and CBP/p300 to herpes simplex virus ICP0 nuclear foci: Potential role in blocking IFN-beta induction. Virology 360:305–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Orzalli MH, Deluca NA, Knipe DM. 2012. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc. Natl. Acad. Sci. U. S. A. 109:E3008–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yokota S, Yokosawa N, Kubota T, Suzutani T, Yoshida I, Miura S, Jimbow K, Fujii N. 2001. Herpes simplex virus type 1 suppresses the interferon signaling pathway by inhibiting phosphorylation of STATs and Janus kinases during an early infection stage. Virology 286:119–124 [DOI] [PubMed] [Google Scholar]
- 36. Chee AV, Roizman B. 2004. Herpes simplex virus 1 gene products occlude the interferon signaling pathway at multiple sites. J. Virol. 78:4185–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnson KE, Song B, Knipe DM. 2008. Role for herpes simplex virus 1 ICP27 in the inhibition of type I interferon signaling. Virology 374:487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He B, Chou J, Brandimarti R, Mohr I, Gluzman Y, Roizman B. 1997. Suppression of the phenotype of γ134.5− herpes simplex virus 1: failure of activated RNA-dependent protein kinase to shut off protein synthesis is associated with a deletion in the domain of the α47 gene. J. Virol. 71:6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Soby S, Laursen RR, Ostergaard L, Melchjorsen J. 2012. HSV-1-induced chemokine expression via IFI16-dependent and IFI16-independent pathways in human monocyte-derived macrophages. Herpesviridae 3:6 doi:10.1186/2042-4280-3-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, Parks RJ, Tschopp J. 2008. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature 452:103–107 [DOI] [PubMed] [Google Scholar]
- 41. Gariano GR, Dell'Oste V, Bronzini M, Gatti D, Luganini A, De Andrea M, Gribaudo G, Gariglio M, Landolfo S. 2012. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog. 8:e1002498 doi:10.1371/journal.ppat.1002498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sergerie Y, Rivest S, Boivin G. 2007. Tumor necrosis factor-alpha and interleukin-1 beta play a critical role in the resistance against lethal herpes simplex virus encephalitis. J. Infect. Dis. 196:853–860 [DOI] [PubMed] [Google Scholar]
- 43. Fujioka N, Akazawa R, Ohashi K, Fujii M, Ikeda M, Kurimoto M. 1999. Interleukin-18 protects mice against acute herpes simplex virus type 1 infection. J. Virol. 73:2401–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Knipe DM, Spang AE. 1982. Definition of a series of stages in the association of two herpesviral proteins with the cell nucleus. J. Virol. 43:314–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Samaniego LA, Neiderhiser L, DeLuca NA. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 72:3307–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krishnan HH, Naranatt PP, Smith MS, Zeng L, Bloomer C, Chandran B. 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J. Virol. 78:3601–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sharma-Walia N, Krishnan HH, Naranatt PP, Zeng L, Smith MS, Chandran B. 2005. ERK1/2 and MEK1/2 induced by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) early during infection of target cells are essential for expression of viral genes and for establishment of infection. J. Virol. 79:10308–10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Balachandran N, Oba DE, Hutt-Fletcher LM. 1987. Antigenic cross-reactions among herpes simplex virus types 1 and 2, Epstein-Barr virus, and cytomegalovirus. J. Virol. 61:1125–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Andrei C, Margiocco P, Poggi A, Lotti LV, Torrisi MR, Rubartelli A. 2004. Phospholipases C and A2 control lysosome-mediated IL-1 beta secretion: Implications for inflammatory processes. Proc. Natl. Acad. Sci. U. S. A. 101:9745–9750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Izumi T, Gomi H, Kasai K, Mizutani S, Torii S. 2003. The roles of Rab27 and its effectors in the regulated secretory pathways. Cell Struct. Funct. 28:465–474 [DOI] [PubMed] [Google Scholar]
- 51. Kavouras JH, Prandovszky E, Valyi-Nagy K, Kovacs SK, Tiwari V, Kovacs M, Shukla D, Valyi-Nagy T. 2007. Herpes simplex virus type 1 infection induces oxidative stress and the release of bioactive lipid peroxidation by-products in mouse P19N neural cell cultures. J. Neurovirol. 13:416–425 [DOI] [PubMed] [Google Scholar]
- 52. Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11:997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. 2009. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 10:266–272 [DOI] [PubMed] [Google Scholar]
- 54. Brazda V, Coufal J, Liao JC, Arrowsmith CH. 2012. Preferential binding of IFI16 protein to cruciform structure and superhelical DNA. Biochem. Biophys. Res. Commun. 422:716–720 [DOI] [PubMed] [Google Scholar]
- 55. Everett RD, Orr A, Preston CM. 1998. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 17:7161–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Everett RD, Earnshaw WC, Findlay J, Lomonte P. 1999. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 18:1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lomonte P, Sullivan KF, Everett RD. 2001. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J. Biol. Chem. 276:5829–5835 [DOI] [PubMed] [Google Scholar]
- 58. Parkinson J, Everett RD. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006–10017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Parkinson J, Lees-Miller SP, Everett RD. 1999. Herpes simplex virus type 1 immediate-early protein Vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lilley CE, Chaurushiya MS, Boutell C, Everett RD, Weitzman MD. 2011. The intrinsic antiviral defense to incoming HSV-1 genomes includes specific DNA repair proteins and is counteracted by the viral protein ICP0. PLoS Pathog. 7:e1002084 doi:10.1371/journal.ppat.1002084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Delboy MG, Roller DG, Nicola AV. 2008. Cellular proteasome activity facilitates herpes simplex virus entry at a postpenetration step. J. Virol. 82:3381–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. La Frazia S, Amici C, Santoro MG. 2006. Antiviral activity of proteasome inhibitors in herpes simplex virus-1 infection: role of nuclear factor-κB. Antivir. Ther. 11:995–1004 [PubMed] [Google Scholar]
- 63. Pelegrin P, Surprenant A. 2009. Dynamics of macrophage polarization reveal new mechanism to inhibit IL-1β release through pyrophosphates. EMBO J. 28:2114–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Norrild B, Lehto VP, Virtanen I. 1986. Organization of cytoskeleton elements during herpes simplex virus type 1 infection of human fibroblasts: an immunofluorescence study. J. Gen. Virol. 67:97–105 [DOI] [PubMed] [Google Scholar]
- 65. Heeg U, Dienes HP, Muller S, Falke D. 1986. Involvement of actin-containing microfilaments in HSV-induced cytopathology and the influence of inhibitors of glycosylation. Arch. Virol. 91:257–270 [DOI] [PubMed] [Google Scholar]
- 66. Akhtar J, Shukla D. 2009. Viral entry mechanisms: cellular and viral mediators of herpes simplex virus entry. FEBS J. 276:7228–7236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Veeranki S, Duan X, Panchanathan R, Liu H, Choubey D. 2011. IFI16 protein mediates the anti-inflammatory actions of the type-I interferons through suppression of activation of caspase-1 by inflammasomes. PLoS One 6:e27040 doi:10.1371/journal.pone.0027040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Aglipay JA, Lee SW, Okada S, Fujiuchi N, Ohtsuka T, Kwak JC, Wang Y, Johnstone RW, Deng C, Qin J, Ouchi T. 2003. A member of the Pyrin family, IFI16, is a novel BRCA1-associated protein involved in the p53-mediated apoptosis pathway. Oncogene 22:8931–8938 [DOI] [PubMed] [Google Scholar]
- 69. Garber DA, Beverley SM, Coen DM. 1993. Demonstration of circularization of herpes simplex virus DNA following infection using pulsed field gel electrophoresis. Virology 197:459–462 [DOI] [PubMed] [Google Scholar]
- 70. Cliffe AR, Knipe DM. 2008. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J. Virol. 82:12030–12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dawson MJ, Trapani JA. 1995. The interferon-inducible autoantigen, IFI 16: localization to the nucleolus and identification of a DNA-binding domain. Biochem. Biophys. Res. Commun. 214:152–162 [DOI] [PubMed] [Google Scholar]
- 72. Johnstone RW, Wei W, Greenway A, Trapani JA. 2000. Functional interaction between p53 and the interferon-inducible nucleoprotein IFI 16. Oncogene 19:6033–6042 [DOI] [PubMed] [Google Scholar]
- 73. Maul GG, Jensen DE, Ishov AM, Herlyn M, Rauscher FJ., III 1998. Nuclear redistribution of BRCA1 during viral infection. Cell Growth Differ. 9:743–755 [PubMed] [Google Scholar]
- 74. Pham TH, Kwon KM, Kim YE, Kim KK, Ahn JH. 2013. DNA sensing-independent inhibition of herpes simplex virus type-1 replication by DAI/ZBP1. J. Virol. 87:3076–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nour AM, Reichelt M, Ku CC, Ho MY, Heineman TC, Arvin AM. 2011. Varicella-zoster virus infection triggers formation of an interleukin-1β (IL-1β)-processing inflammasome complex. J. Biol. Chem. 286:17921–17933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Weller TH, Witton HM, Bell EJ. 1958. The etiologic agents of varicella and herpes zoster; isolation, propagation, and cultural characteristics in vitro. J. Exp. Med. 108:843–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. van Lint AL, Murawski MR, Goodbody RE, Severa M, Fitzgerald KA, Finberg RW, Knipe DM, Kurt-Jones EA. 2010. Herpes simplex virus immediate-early ICP0 protein inhibits Toll-like receptor 2-dependent inflammatory responses and NF-κB signaling. J. Virol. 84:10802–10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lopez P, Van Sant C, Roizman B. 2001. Requirements for the nuclear-cytoplasmic translocation of infected-cell protein 0 of herpes simplex virus 1. J. Virol. 75:3832–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Liu M, Rakowski B, Gershburg E, Weisend CM, Lucas O, Schmidt EE, Halford WP. 2010. ICP0 antagonizes ICP4-dependent silencing of the herpes simplex virus ICP0 gene. PLoS One 5:e8837 doi:10.1371/journal.pone.0008837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li L, Johnson LA, Dai-Ju JQ, Sandri-Goldin RM. 2008. Hsc70 focus formation at the periphery of HSV-1 transcription sites requires ICP27. PLoS One 3:e1491 doi:10.1371/journal.pone.0001491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Metzger MB, Hristova VA, Weissman AM. 2012. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 125:531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ozkan E, Yu H, Deisenhofer J. 2005. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc. Natl. Acad. Sci. U. S. A. 102:18890–18895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cristea IM, Moorman NJ, Terhune SS, Cuevas CD, O'Keefe ES, Rout MP, Chait BT, Shenk T. 2010. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J. Virol. 84:7803–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Jacob T, Van den Broeke C, van Troys M, Waterschoot D, Ampe C, Favoreel HW. 30 January 2013. Alphaherpesviral US3 kinase induces cofilin dephosphorylation to reorganize the actin cytoskeleton. J. Virol. doi:10.1128/JVI.03107-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Pasieka TJ, Baas T, Carter VS, Proll SC, Katze MG, Leib DA. 2006. Functional genomic analysis of herpes simplex virus type 1 counteraction of the host innate response. J. Virol. 80:7600–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Suzutani T, Nagamine M, Shibaki T, Ogasawara M, Yoshida I, Daikoku T, Nishiyama Y, Azuma M. 2000. The role of the UL41 gene of herpes simplex virus type 1 in evasion of non-specific host defence mechanisms during primary infection. J. Gen. Virol. 81:1763–1771 [DOI] [PubMed] [Google Scholar]
- 87. Miettinen JJ, Matikainen S, Nyman TA. 2012. Global secretome characterization of herpes simplex virus 1-infected human primary macrophages. J. Virol. 86:12770–12778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Roberts KL, Baines JD. 2010. Myosin Va enhances secretion of herpes simplex virus 1 virions and cell surface expression of viral glycoproteins. J. Virol. 84:9889–9896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hollinshead M, Johns HL, Sayers CL, Gonzalez-Lopez C, Smith GL, Elliott G. 2012. Endocytic tubules regulated by Rab GTPases 5 and 11 are used for envelopment of herpes simplex virus. EMBO J. 31:4204–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bello-Morales R, Crespillo AJ, Fraile-Ramos A, Tabares E, Alcina A, Lopez-Guerrero JA. 2012. Role of the small GTPase Rab27a during Herpes simplex virus infection of oligodendrocytic cells. BMC Microbiol. 12:265 doi:10.1186/1471-2180-12-265 [DOI] [PMC free article] [PubMed] [Google Scholar]