Abstract
Galectin-9 is a pleiotropic immune modulator affecting numerous cell types of innate and adaptive immunity. Patients with chronic infection with either hepatitis C virus (HCV) or HIV have elevated circulating levels. Limited data exist on the regulation of natural killer (NK) cell function through interaction with galectin-9. We found that galectin-9 ligation downregulates multiple immune-activating genes, including eight involved in the NK cell-mediated cytotoxicity pathway, impairs lymphokine-activated killing, and decreases the proportion of gamma interferon (IFN-γ)-producing NK cells that had been stimulated with interleukin-12 (IL-12)/IL-15. We demonstrate that the transcriptional and functional changes induced by galectin-9 are independent of Tim-3. Consistent with these results for humans, we find that the genetic absence of galectin-9 in mice is associated with greater IFN-γ production by NK cells and enhanced degranulation. We also show that in the setting of a short-term (4-day) murine cytomegalovirus infection, terminally differentiated NKs accumulate in the livers of galectin-9 knockout mice, and that hepatic NKs spontaneously produce significantly more IFN-γ in this setting. Taken together, our results indicate that galectin-9 engagement impairs the function of NK cells, including cytotoxicity and cytokine production.
INTRODUCTION
Galectin-9 (Gal-9) is a member of the galectin family of carbohydrate-binding proteins, which are characterized by the presence of two or more conserved carbohydrate-recognition domains (CRD) that bind galactose (1). Gal-9 is widely distributed throughout various tissues, being particularly abundant in the liver (2). We have demonstrated that Gal-9 circulates at very high levels in the serum, and its hepatic expression is significantly increased in patients with chronic hepatitis C virus (HCV) compared to normal controls (3), and others have found high levels of Gal-9 in patients with HIV (4). Gal-9 interacts with various ligands, the best characterized of which is the T cell immunoglobulin mucin 3 (Tim-3) cell surface molecule, widely expressed on innate and adaptive immune cells (5). Various other molecules are known to interact with Gal-9 with functional consequences, including the Epstein-Barr virus latent membrane protein-1 (6) and several members of the protein disulfide isomerase (PDI) family (7). Originally described as an eosinophil chemoattractant, Gal-9 is now known to be an important pleiotropic immune modulator affecting numerous immune cell types (reviewed in references 8 and 9). Gal-9 is thought to be involved in the activation of innate immune responses (3, 10) and the downregulation of Th17 (11) and Th1 responses (12, 13). Gal-9 can promote inflammation through triggering proinflammatory cytokine production from monocytes (3) and inducing maturation of macrophages (14) and monocyte-derived dendritic cells (mDC) (10). Conversely, Gal-9 has a major anti-inflammatory role in the induction of apoptosis in activated T cell subsets (13), as well as in promoting the differentiation of regulatory T cells (Treg) expressing FoxP3 (3, 15).
Natural killer (NK) cells constitute the first line of host defense against viral pathogens (16, 17), eliminating virus-infected cells both directly via cytolytic mechanisms and indirectly by secreting cytokines such as gamma interferon (IFN-γ) (18, 19). NK cell activity is stringently controlled by membrane-expressed inhibitory NK receptors (NKRs) that, in steady-state conditions, override signals provided by engagement of activating receptors (20, 21). In a recent study, Gal-9 was shown to act on NK cells as an activating ligand (22), but only when NK cells had been preprimed with proinflammatory cytokines (interleukin-12 [IL-12]/IL-18). The stimulatory effect of Gal-9 on NK cells was found to be mediated through Tim-3 (22), which is expressed preferentially on activated NK cells (23).
Several studies have shown Tim-3-independent immune cell regulation by Gal-9 (7, 11, 13, 24, 25), indicating that other pathways are involved in Gal-9 modulation of the immune response. Gal-9 binding to PDI on Th2 cells results in increased cell migration (7). Through interaction with an as-yet unidentified glycoprotein, the development of Th17 cells is inhibited in Gal-9 knockout (KO) mice (11). Gal-9 can induce the production of proinflammatory cytokines from Th cells in a manner that does not require Tim-3 (24). These studies suggest that immune cell regulation by Gal-9 is complex and can be mediated by additional receptors as well as Tim-3. Currently, we do not know the effects of Tim-3-independent Gal-9 signaling in NK cells. As mentioned above, Gal-9 recognizes carbohydrates, and recent reports suggest that differential glycosylation of NK cell receptors represents an important receptor regulatory mechanism for control of NK cell function (26–28). In the current study, we investigated the effect of Gal-9 on human NK cell transcription and function and the NK cell phenotype and function of Gal-9 KO mice. Taken together, our results indicate that Gal-9 engagement can impair the function of NK cells, including cytotoxicity and cytokine production.
(Part of this work was presented as an oral presentation at the American Association for the Study of Liver Diseases, November 2010.)
MATERIALS AND METHODS
Sample collection and storage.
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood by cellular preparation tubes (anticoagulant sodium citrate; Becton, Dickinson [BD]). PBMCs were viably frozen in 80% fetal bovine serum (FBS) (BioWhittaker), 10% dimethyl sulfoxide (DMSO), and 10% RPMI 1640 medium (Life Technologies) and stored in liquid nitrogen for subsequent analyses.
Antibodies for detection and fluorescence-activated cell sorter (FACS) analysis of surface antigen expression.
Multiparameter flow cytometry was performed using a BD FACSCanto II instrument, compensated with single fluorochromes, and analyzed using Diva software (BD Biosciences). Lymphocyte populations were identified by their characteristic forward scatter/side scatter (FSC:SSC) properties. For the human studies, fluorochrome-labeled (peridinin chlorophyll protein [PerCP]/allophycocyanin [APC]) monoclonal antibodies (MAb) specific for CD3 and CD56 (BD Biosciences) were used to identify NK (CD3− CD56+), NT (CD3+ CD56+), and conventional T (CD3+ CD56−) cells within the overall lymphocyte population. Anti-NKG2C-APC was purchased from R&D Systems. Anti-NKR MAb-APC NKG2A was obtained from Immunotech (Beckman Coulter), and NKp30 and NKp46 were obtained from BD Biosciences. For the murine studies, anti-NK1.1-PerCP (BD Biosciences) and anti-CD3e-APC-EF780 (eBioscience) were used to identify NK cells. Anti-mouse CD27-phycoerythrin (PE), NKG2D-APC, and NKp46-EF450 were purchased from eBiosciences. CD11b-V450 was obtained from BD (Horizon). Thawed cells (1 × 106 to 2 × 106) were stained for cell surface antigen expression at 4°C in the dark for 30 min. They were then washed twice in 2 ml phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) and 0.01% sodium azide (FACS wash) and subsequently fixed in 200 μl of 2% paraformaldehyde (PFA; Sigma-Aldrich). Isotype-matched control antibodies were used to determine background levels of staining.
Microarray and gene set enrichment analyses of purified human NK cells.
Human NK cells were isolated (n = 4 normal control subjects) using an NK Isolation Kit II from Miltenyi Biotec according to the manufacturer's instructions. Median purity of NKs was >90% in all cases. Following isolation, the NKs were cultured at 2 million cells/ml with or without Gal-9 (5 μg/ml; GalPharma) for 24 h at 37°C and 5% CO2. After culture, NK cells were washed and RNA extracted from cell pellets using an PicoPure RNA isolation kit (Arcturus; Applied Biosystems). Differential expression was assayed (in triplicate for each subject) using Agilent 4×44K human microarrays according to the manufacturer's instructions (Agilent Technologies). Gene set enrichment analysis was performed using the DAVID Bioinformatics Resource (29). Agilent identifiers (IDs) were used as identifiers to investigate both the upregulated and downregulated gene sets in order to identify enriched biological annotations and pathways facilitating interpretation of the microarray data. Four hundred forty-four of the 480 downregulated genes and 222 of the 341 upregulated genes corresponded to DAVID IDs for enrichment analysis. Ingenuity pathway analysis software was also used to examine the differentially expressed genes within the context of networks and pathways.
Animals.
Ten- to 12-week-old mice were used for all experiments, with the exception of the murine cytomegalovirus (MCMV) infection experiments, which used 7-week-old mice. Control C57BL/6 mice were purchased from Jackson Laboratories. The Gal-9 KO mice were kindly provided by GalPharma and were described elsewhere (15). All animal experiments were performed humanely under a protocol approved by the Animal Care and Use Committee at the University of Colorado–Denver.
MCMV infection.
Wild-type (WT) MCMV was derived from pSM3fr, a bacterial artificial chromosome (BAC) clone of the Smith strain (30). Mouse embryonic fibroblasts (10.1) (31) were propagated in Dulbecco's modified Eagle medium (DMEM) with 10% fetal calf serum. The titer of MCMV was determined by plaque assay on 10.1 cells (32). Seven-week-old female Gal-9 KO or C57BL/6 mice were infected intraperitoneally (i.p.) with virus (2 × 106 PFU) in a volume of 200 μl. Mice were euthanized on day 4 after infection, and their spleens and livers were harvested. Tissue samples were weighed and homogenized in 2 ml of DMEM. Homogenates then were cleared by centrifuging the samples at 2,500 rpm for 10 min. Virus yields were determined by plaque assay on 10.1 fibroblast cells and reported as PFU per gram of tissue.
Isolation of murine splenic and intrahepatic leukocytes.
For isolation of intrahepatic leukocytes, explanted tissue was finely minced and homogenized with a 10-ml syringe, followed by 1 h of incubation with digestion medium (0.02% [wt/vol] collagenase IV, 0.002% DNase) at 37°C. Red blood cells were lysed in Red Blood Cell Lysis Buffer (eBioscience), and the cells were washed twice in RPMI 1640 (Gibco, Life Technologies) with 5% fetal bovine serum (HyClone, Thermo Scientific). Cells were then filtered through a 70-μm filter and, if not being used immediately, were frozen in FBS plus 10% DMSO. Mouse splenocytes were harvested in a similar manner but without collagenase digestion.
NK cytotoxicity assay.
Thawed mononuclear cell suspensions were enriched for NKs using NK Isolation Kit II from Miltenyi Biotec according to the manufacturer's instructions. Median purity of NKs was >90% in all cases. Following isolation, the NKs were cultured with or without IL-2 (25 ng/ml; R&D Systems) for 48 h at 37°C and 5% CO2. Following culture, carboxy fluorescein succinimidyl ester (CFSE)-labeled target cells (K562s) were added to the NKs at effector-to-target (E:T) concentrations of 0:1 (negative control) and 10:1 (test) and incubated at 37°C for 4 h. After incubation, cytotoxicity was measured using the flow cytometry-based Total Cytotoxicity & Apoptosis Detection kit from Immunochemistry as previously described (33). Natural cytotoxicity is measured in the absence of exogenous IL-2, and lymphokine-activated killing (LAK) is measured after culture with IL-2 (as described above). Immediately before acquisition, 7-aminoactinomycin D (7-AAD) was added to E:T populations and incubated for 15 min on ice. Cells treated with 1% Tween 20 served as positive controls.
Cell culture, degranulation, and intracellular IFN-γ production by NK cells.
Human NKs were enriched using magnetic beads and surface stained for CD3 and CD56 as described above. Isolated NKs were incubated for 24 h with IL-12 (1 ng/ml; R&D Systems) and IL-15 (10 ng/ml; R&D Systems) or medium alone. Four hours before the end of the culture, brefeldin A (BFA; 10 μg/ml; Sigma) was added to all wells. Cells were harvested, surface stained for CD3/CD56, fixed for 30 min at 4°C in 100 μl Fix and Perm Medium A (Caltag), permeabilized using 100 μl Fix and Perm Medium B (Caltag), and incubated with anti-IFN-γ (BD) MAb for 1 h. Cell suspensions were then washed in PBS-BSA-azide, fixed in 200 μl 1% PFA, and acquired after 1 h. Murine spleen cells were cultured for 6 h in the presence or absence of phorbol myristate acetate (PMA; 10 ng/ml; Sigma) and ionomycin (1 μg/ml; Sigma) (CD107a degranulation assay) or IL-12 (1 ng/ml; R&D Systems) and IL-18 (10 ng/ml; MBL International) for 24 h (IFN-γ assay).
Real-time PCR.
Gene expression was assessed using the Step One Plus real-time PCR system using the Fast SYBR green Master Mix protocol (Applied Biosystems, Life Technologies). QuantiTect primer assays for use with SYBR green detection were purchased from Qiagen/Superarray.
Tim-3 blocking assays.
Human NK cells were isolated (n = 4 normal control subjects) and cultured under the same conditions as those for the microarray assays, with the addition of 10 μg/ml anti-Tim-3 (MABB006; clone 344823; R&D Systems) or IgG control antibody (MABB006; clone 5448; R&D Systems) for 1 h prior to the addition of Gal-9 (5 μg/ml). Differential expression was assayed (in triplicate for each individual subject) using real-time PCR. Intracellular IFN-γ production was assessed as described above, with the addition of anti-Tim-3 or IgG (10 μg/ml) before the addition of Gal-9. For the cytokine assays on sorted NK cell subsets, bead-purified NK cells were incubated overnight with IL-12 (1 ng/ml) and IL-15 (10 ng/ml) and FACS sorted on the expression of Tim-3 (high- and low/negative-expression [Tim-3high and Tim-3low/neg, respectively] subsets). Anti-Tim-3/IgG (10 μg/ml) was added to NK cell subsets (2 million/ml) for 4 h, followed by the addition of Gal-9 (5 μg/ml) overnight. BFA was added, and 4 h later intracellular staining for IFN-γ was carried out as described above. For cytotoxicity assays, bead-purified NK cells were incubated and rested overnight, FACS sorted on the expression of Tim-3 (Tim-3high and Tim-3low/neg subsets), and then incubated for 48 h with IL-2 (25 ng/ml). Anti-Tim-3/IgG (10 μg/ml) was added to NK cell subsets for 1 h followed by the addition of Gal-9 (5 μg/ml) for 1 h, and the cytotoxicity assay was carried out as described above.
Statistics.
Results are expressed as medians (ranges). The nonparametric Mann-Whitney U or Student's t test (where appropriate) was used to compare differences between groups. Significance was defined as P < 0.05. The JMP 6.0 (SAS Institute) statistical software package was used.
RESULTS
Microarray analysis of human NK cells stimulated by Gal-9.
We investigated the cellular response of human NK cells from normal subjects to 24 h of Gal-9 stimulation by examining differential expression using Agilent 4×44K human microarrays. A complete list of the 820 differentially expressed genes with a P value of less than 0.05 and a 2-fold expression threshold are shown in Tables S1 (downregulated genes) and S2 (upregulated genes) in the supplemental material. Microarray analysis of NK cells stimulated with Gal-9, followed by gene set enrichment analysis (29), revealed that Gal-9 stimulation of NK cells results in a substantial downregulation of genes involved in DNA replication, DNA repair, and cell cycle, including 7 members of the minichromosome maintenance complex (mcm 3 to 8 and mcm 10), chromatin assembly factor subunit A and B, DNA-directed polymerase alpha 2 subunit and iota, and CDC45 cell division protein (Table 1). Of note, pathway analysis revealed that 8 genes involved in the NK cell-mediated cytotoxicity pathway are downregulated, including natural cytotoxicity triggering receptors 1 (NCR1) and 3 (NCR3), CD48 (the ligand for 2B4), the Fc fragment of IgG receptor FCGR3A (CD16), the killer cell immunoglobulin-like receptor KIR3DL2, the lymphocyte cytosolic protein LCP2, the perforin 1 pore-forming protein PRF1 (perforin), and the v-Ha-ras Harvey rat sarcoma viral oncogene homolog HRAS (Table 2 and Fig. 1). These data suggest that the cytolytic function of NK cells is inhibited by Gal-9.
Table 1.
Galectin-9 downregulated genes, annotation enrichment
| Category | Terma | Countb | %c | P value |
|---|---|---|---|---|
| GOTERM_BP_FAT | DNA metabolic process | 45 | 10.7 | 2.40E−15 |
| GOTERM_BP_FAT | DNA replication | 28 | 6.7 | 7.20E−15 |
| SP_PIR_KEYWORDS | DNA replication | 20 | 4.8 | 2.40E−14 |
| SP_PIR_KEYWORDS | Cell cycle | 34 | 8.1 | 1.10E−09 |
| GOTERM_BP_FAT | Cell cycle | 46 | 10.9 | 1.40E−09 |
| SP_PIR_KEYWORDS | Acetylation | 100 | 23.8 | 4.50E−09 |
| GOTERM_CC_FAT | Chromosome | 32 | 7.6 | 1.10E−08 |
| GOTERM_BP_FAT | Response to DNA damage stimulus | 28 | 6.7 | 4.40E−08 |
| GOTERM_BP_FAT | DNA repair | 24 | 5.7 | 6.10E−08 |
| GOTERM_BP_FAT | Cellular response to stress | 35 | 8.3 | 7.60E−08 |
| SP_PIR_KEYWORDS | DNA damage | 20 | 4.8 | 8.00E−08 |
| KEGG_PATHWAY | DNA replication | 10 | 2.4 | 1.40E−07 |
| UP_SEQ_FEATURE | Domain:MCM | 6 | 1.4 | 2.30E−07 |
| INTERPRO | DNA-dependent ATPase | 6 | 1.4 | 2.40E−07 |
| SMART | MCM | 6 | 1.4 | 3.90E−07 |
| INTERPRO | DNA-dependent ATPase MCM | 6 | 1.4 | 5.20E−07 |
| GOTERM_CC_FAT | Chromosomal part | 26 | 6.2 | 6.60E−07 |
| SP_PIR_KEYWORDS | DNA repair | 18 | 4.3 | 6.90E−07 |
| GOTERM_CC_FAT | Nucleoplasm | 41 | 9.7 | 3.50E−06 |
| GOTERM_BP_FAT | Cell cycle phase | 26 | 6.2 | 4.00E−06 |
| INTERPRO | AIG1 | 5 | 1.2 | 7.00E−06 |
Term from KEGG_PATHWAY category.
Number of differentially regulated genes.
Percentage of total differentially regulated genes.
Table 2.
Galectin-9 downregulated genes, pathway enrichment
| Terma | Countb | %c | P value |
|---|---|---|---|
| DNA replication | 10 | 2.4 | 1.40E−07 |
| Cell cycle | 11 | 2.6 | 9.70E−04 |
| Mismatch repair | 5 | 1.2 | 2.20E−03 |
| One carbon pool by folate | 4 | 1 | 6.60E−03 |
| Nucleotide excision repair | 5 | 1.2 | 2.30E−02 |
| Pyrimidine metabolism | 7 | 1.7 | 3.00E−02 |
| Melanogenesis | 7 | 1.7 | 3.50E−02 |
| NK cell-mediated cytotoxicity | 8 | 1.9 | 4.60E−02 |
| Prostate cancer | 6 | 1.4 | 6.90E−02 |
| Glioma | 5 | 1.2 | 7.00E−02 |
| Leukocyte transendothelial migration | 7 | 1.7 | 7.20E−02 |
| p53 signaling pathway | 5 | 1.2 | 8.70E−02 |
Terms from the KEGG_PATHWAY category.
Number of differentially regulated genes.
Percentage of total differentially regulated genes.
Fig 1.
Downregulation of human NK cell-mediated cytotoxicity pathway genes in response to Gal-9. Among the genes downregulated in response to Gal-9 stimulation are genes of the NK cell-mediated cytotoxicity pathway, including the natural cytotoxicity triggering receptors 1 (NCR1/NKp46) and 3 (NCR3/NKp30) as well as CD48, CD16 (FCGR3A), the killer cell immunoglobulin-like receptor KIR3DL2, the lymphocyte cytosolic protein LCP2/SLP-76, the perforin 1 pore-forming gene PRF1, and the v-Ha-ras Harvey rat sarcoma viral oncogene homolog HRAS/Ras. This pathway was derived from the KEGG pathway database (50).
Real time RT-PCR analysis of human NK cells stimulated by Gal-9.
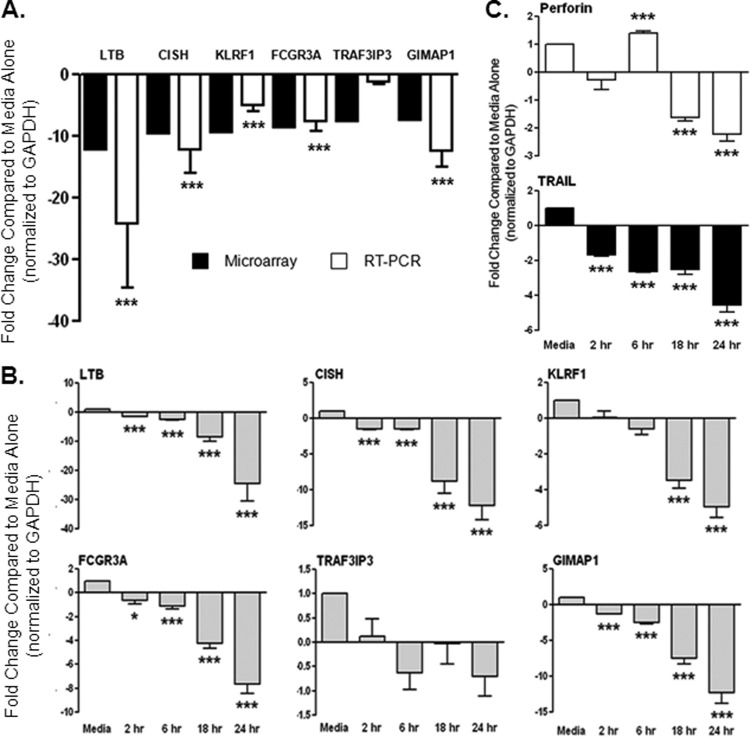
We used real-time reverse transcription-PCR (RT-PCR) to confirm the top 6 genes shown to be maximally differentially expressed (greater than 7-fold change) in the microarray analysis (Table 3). All of the six genes with greater than 7-fold change identified in the microarray analysis were downregulated. We confirmed downregulation of five of these six genes by PCR analysis of NK cells isolated from four normal control subjects (Fig. 2A). The five genes confirmed by PCR are involved in the immune response, and all, with the exception of one (CISH), play a stimulatory role. LTB (lymphotoxin beta) is a type II membrane protein of the tumor necrosis factor (TNF) family that is involved in the induction of the inflammatory response (34). KLRF1, also known as NKp80, encodes an activating homodimeric C-type lectin-like receptor expressed on nearly all NK cells and stimulates their cytotoxicity and cytokine release (35). The FCGR3A gene encodes a receptor for the Fc portion of immunoglobulin G (CD16), and it is involved in antibody-dependent cell cytotoxicity (ADCC). Mutations in this gene have been linked to susceptibility to recurrent viral infections (36). GIMAP1, GTPase IMAP family member 1, is thought to be involved in the differentiation of inflammatory T helper (Th) cells of the Th1 lineage (37, 38). CISH belongs to the cytokine-induced STAT inhibitor (CIS) family, also known as suppressor of cytokine signaling (SOCS) protein family, known to be cytokine-inducible negative regulators of cytokine signaling (39). We did not confirm the downregulation of TRAF3-interacting JNK-activating modulator protein, which mediates cell growth encoded by the TRAF3IP3 gene (40). In addition to verifying the microarray data (which was performed after 24 h of culture with Gal-9), we included earlier time points in our PCR analysis. Maximum gene downregulation was observed at 24 h; however, for the majority of these genes, significant downregulation was observed as early as 2 h after addition of Gal-9 to NK cells (Fig. 2B). We also used PCR to explore the effect of Gal-9 on NK cell genes involved in cytotoxicity, as pathway analysis of the microarray data suggested inhibition of this function of NK cells by Gal-9 (Fig. 1). We chose to look at expression of perforin (PRF1) and tumor necrosis factor ligand (TRAIL or TNFSF10), as these are the primary molecules involved in NK cell-mediated lysis of target cells. NK cells use perforin to mediate a secretory/necrotic killing, while TRAIL mediates a cell contact-dependent apoptotic cytotoxic mechanism (41). We had found perforin to be significantly downregulated (4.9-fold) in the microarray. TRAIL was downregulated at all time points tested, whereas perforin, after initial upregulation at 6 h, was downregulated at 18 and 24 h following addition of Gal-9 (Fig. 2C). Taken together, these data strongly suggest that exposure to Gal-9 inhibits the immune effector functions of NK cells.
Table 3.
Top six genes (by fold change) differentially expressed in microarray assay
| Probe name | Fold changea | P valueb | Gene designation | Entrez gene no. | Description |
|---|---|---|---|---|---|
| A_23_P93348 | 12.12 | 0.0073 | LTB | 4050 | Lymphotoxin beta (TNF superfamily, member 3) |
| A_23_P144096 | 9.53 | 0.0011 | CISH | 1154 | Cytokine inducible SH2-containing protein (CISH) |
| A_32_P158966 | 9.45 | 0.0413 | KLRF1 | 51348 | Killer cell lectin-like receptor subfamily F, member 1 |
| A_23_P200728 | 8.49 | 0.0176 | FCGR3A | 2214 | Fc fragment of IgG, low-affinity IIIa receptor (CD16a) |
| A_23_P334414 | 7.65 | 0.0092 | TRAF3IP3 | 80342 | TRAF3 interacting protein 3 (TRAF3IP3) |
| A_23_P427023 | 7.40 | 0.0097 | GIMAP1 | 170575 | GTPase, IMAP family member 1 (GIMAP1) |
All genes were downregulated.
P values were determined by t test.
Fig 2.
Real-time RT-PCR confirms downregulation of genes involved in the immune system in human NK cells treated with Gal-9. We used PCR to verify the 6 genes shown to be maximally differentially expressed (greater than 7-fold change) in the microarray analysis. (A) We confirmed downregulation of five of these six genes by PCR analysis of NK cells isolated from 4 normal control subjects after 24 h of culture with Gal-9, as performed for the microarray experiment. The five genes confirmed by PCR are involved in the immune response, and all, with the exception of one (CISH), play a stimulatory role. The microarray data are shown in the black bars for comparison to the PCR data shown in the white bars. (B) The time course for each of the six genes is shown. The majority of these genes were significantly downregulated after 2 h in culture with Gal-9. (C) In addition, we tested regulation of genes involved in the cytolytic activity of NK cells (perforin and TRAIL) and found them both to be significantly downregulated after 18 and 24 h of culture with Gal-9. The bars show means ± SEM, and Student's t test was used to test statistical significance.
Gal-9 inhibits the activity of human NK cells in vitro.
NK cells eliminate virus-infected cells directly via cytolytic mechanisms; therefore, we asked if Gal-9 altered the ability of bead-purified human NK cells to lyse NK cell-sensitive targets (K562) in the presence (LAK) or absence (natural cytotoxicity) of IL-2 using a standard flow-based assay (33). Addition of Gal-9 to cultures significantly inhibited LAK activity (Fig. 3A), in keeping with the human PCR and microarray data. No difference was seen in natural cytotoxicity (data not shown). NK cells also produce antiviral cytokines, predominantly IFN-γ. Therefore, we cultured human NK cells with cytokines IL-12 and IL-15, which are known to stimulate NK cytokine production (23). The addition of Gal-9 significantly decreased the proportion of IFN-γ-producing NK cells that had been stimulated with IL-12/IL-15 (Fig. 3B).
Fig 3.

Gal-9 functionally impairs human NK cells in vitro. (A) The IL-2-induced lymphokine-activated killing (LAK; 48 h of culture with 25 ng/ml IL-2) activity of isolated NK cells (n = 6 normal control subjects) against the NK cell-sensitive target cell line K562 is significantly reduced in the presence of Gal-9. (B) NK cells stimulated (IL-12, 1 ng/ml; IL-15, 10 ng/ml) in the presence of Gal-9 produce less IFN-γ, as evidenced by intracellular flow-cytometric staining. The bars show means ± SEM, and Student's t test was used to test statistical significance. *, P < 0.05.
Gal-9 regulation of human NK cell activity is independent of Tim-3.
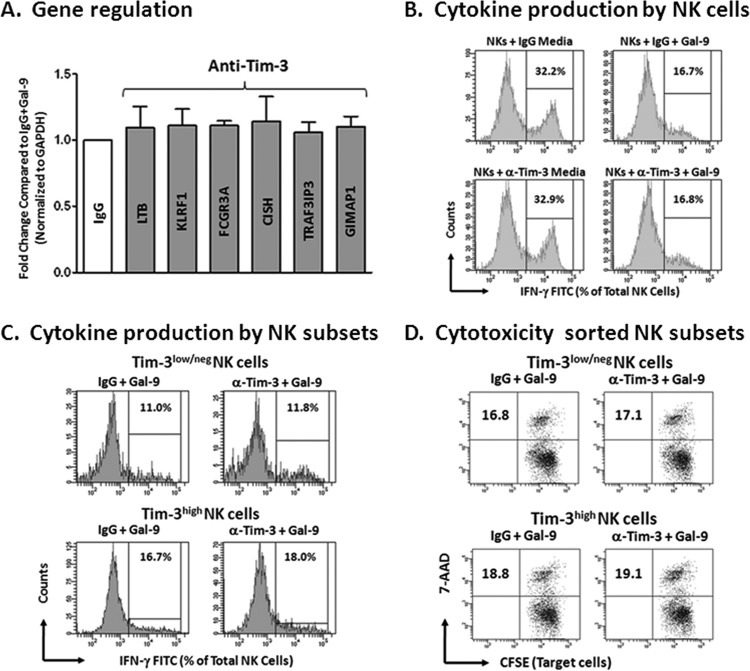
The best-described natural ligand for Gal-9 is T cell immunoglobulin and mucin domain-containing molecule 3 (Tim-3) (8), and evidence suggests a critical role for Gal-9–Tim-3 interactions in regulating the fate of immune cells during various phases of the immune response (12, 13, 15, 22, 23, 42). NK cells express higher levels of Tim-3 than T cells (23, 43); we therefore wanted to determine if the effects of Gal-9 on human NK cell function in our assays were mediated through interactions with Tim-3. We repeated the real-time RT-PCR analysis of human NK cells stimulated with Gal-9 (Fig. 2) after preincubation with anti-Tim-3 or control IgG antibodies (10 μg/ml; n = 4). Gene expression in the IgG condition was normalized to 1 for all genes, and expression in the anti-Tim-3 condition was compared to that of the IgG control condition. Blocking Tim-3 had no effect on gene transcription for the six genes tested (Fig. 4A). As shown in Fig. 3B, addition of 5 μg/ml Gal-9 resulted in inhibition of IL-12/IL-15-induced intracellular IFN-γ production. We performed the same intracellular cytokine assay after preincubation of isolated NK cells (n = 4) with anti-Tim-3 or control IgG antibodies (10 μg/ml). Representative flow-cytometric histograms of IFN-γ expression are shown in Fig. 4B for IL-12/IL-15-stimulated NK cells treated with IgG but without the addition of Gal-9 (upper left). IgG-treated NK cells with the addition of Gal-9 have reduced IFN-γ expression (upper right). Results for NKs treated with anti-Tim-3 in the absence (lower left) or presence (lower right) of Gal-9 were similar to those of their IgG-treated counterparts. As NK cells in the above-described assays were not primed with cytokines known to upregulate the expression of Tim-3 on NK cells before incubation with anti-Tim-3, we thought that the lack of efficacy of the blocking anti-Tim-3 antibody was related to the availability of Tim-3. Therefore, we FACS sorted NK cells (n = 3) into Tim-3high and Tim-3low/neg subsets and incubated these NK cell subsets with anti-Tim-3 or IgG before addition of Gal-9. We then performed intracellular cytokine assays and LAK assays (as previously shown in Fig. 3A and B). Blocking the interaction of Gal-9 and Tim-3 did not rescue NK cells from inhibition mediated by Gal-9 even after FACS sorting Tim-3high and Tim-3low/neg NK cell subsets (Fig. 4C and D). These data suggest that the effects of high-dose Gal-9 on NK cells in our assays are independent of Tim-3.
Fig 4.
Gal-9 inhibition of human NK cell activity is Tim-3 independent. Blocking experiments were carried out as described in Materials and Methods. (A) PCR detection of the 6 genes shown to be maximally differentially expressed (greater than 7-fold change) in the microarray analysis was performed on isolated NK cells (n = 4 normal subjects) after 24 h of incubation with Gal-9 in the presence of anti-Tim-3 or control IgG antibodies (10 μg/ml). Gene expression in the IgG condition was normalized to 1 for all genes (white bar), and expression in the anti-Tim condition was compared to that of IgG control samples. Blocking Tim-3 had no effect on gene transcription for all genes tested. (B) The top panel shows representative flow-cytometric histograms of IFN-γ production in IL-12/IL-15-stimulated NK cells in the presence of IgG with or without Gal-9, and the bottom panel shows the same samples in the presence of anti-Tim-3 antibody with or without Gal-9. Downregulation of IFN-γ was seen on the addition of Gal-9 (right 2 histograms); however, anti-Tim had no effect in this assay, as IFN-γ production was similar to that of IgG control samples. (C) Flow-cytometric histograms show IFN-γ production in IL-12/IL-15-stimulated, FACS-sorted Tim-3high and Tim-3low/neg NK cell subsets in the presence of IgG or anti-Tim-3, all treated with Gal-9. IFN-γ production in the anti-Tim-3 conditions were similar to those of IgG controls, suggesting that blocking Tim-3 does not block the effect of Gal-9 in this assay. (D) Flow-cytometric dot plots show the same sorted samples assayed for LAK activity (as described above). CFSE-stained K562 target cells are shown on the x axis, and 7-AAD is shown on the y axis. The percentages describe the double positive (dead) cells detected after 4 h of coincubation with the indicated NK cell subset at a ratio of 10 NK cells to 1 target cell. LAK activity in the anti-Tim-3 condition was similar to that of control IgG.
NK cells from Gal-9 KO mice demonstrate greater activity.
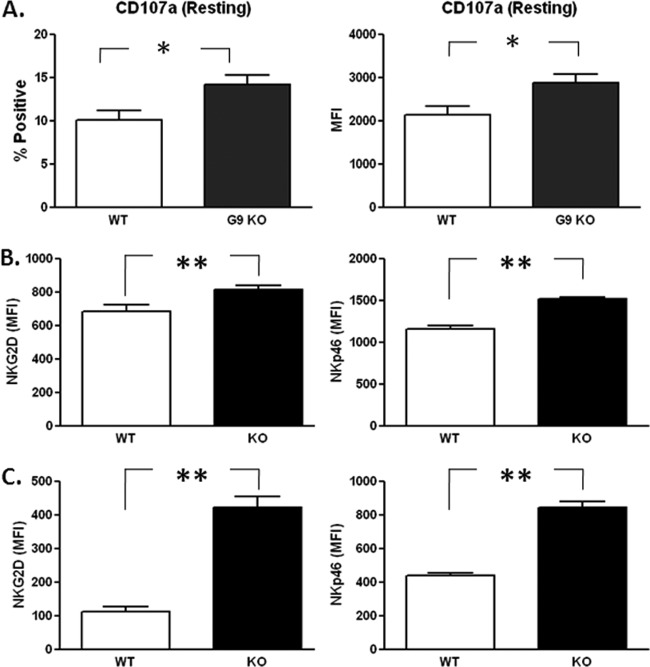
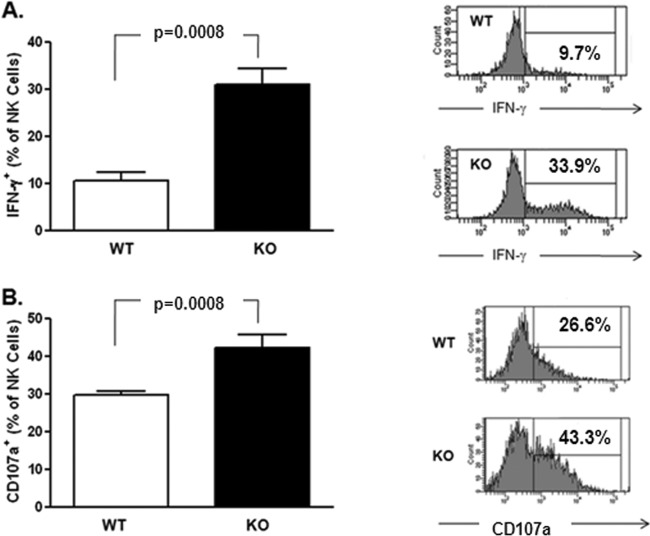
In order to test the hypothesis that the absence of Gal-9 is associated with more activated NK cells, we examined the phenotype and maturation stage of spleen- and liver-derived NK cells in Gal-9 KO mice. NK cell levels were similar in the spleens and livers of Gal-9 KO mice and control mice (data not shown). However, the mean fluorescence intensity (MFI) of CD107a, a lysosome-associated membrane glycoprotein expressed on the cell surface following release of cytotoxic granule contents (44), and the percentage of NK cells that spontaneously degranulated were greater in Gal-9 KO mice than WT mice (Fig. 5A). Accordingly, there were significant phenotypic differences; specifically, higher expression levels of activating receptors NKG2D and NKp46 were evident on both spleen- and liver-derived NK cells in the Gal-9 KO mice. The pattern was similar for both tissues, but the differences were more pronounced for liver-derived NK cells (Fig. 5B and C). No difference was observed for the expression of NKG2A (inhibitory receptor), NKG2C, or CD69 (activating receptors) (data not shown).
Fig 5.
Direct ex vivo characterization of NK cells from Gal-9 KO mice. (A) Flow-cytometric analysis of splenic NK cells (NK1.1+ CD3−) demonstrated that in the absence of exogenous stimulation, the percentage of NKs that degranulated (CD107a+) was higher in Gal-9 KO mice (KO). MFI analysis suggested that, in addition to a higher percentage of degranulating cells, each cell expresses more CD107a in the KO mice. (B) Splenic NK cells also expressed significantly higher levels of two major activating NK receptors (NKG2D and NKp46) than wild-type (WT) controls. The observed increase in NK receptor expression activity was also seen for liver-derived NK cells (C). The bars represent medians ± interquartile ranges (n = 5 mice/group). A nonparametric Mann-Whitney test was used to calculate P values between control (WT) and KO groups. *, P < 0.05; **, P < 0.005.
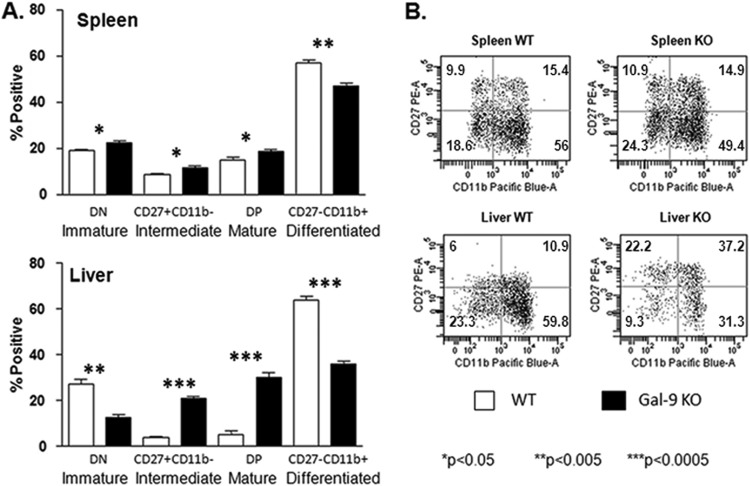
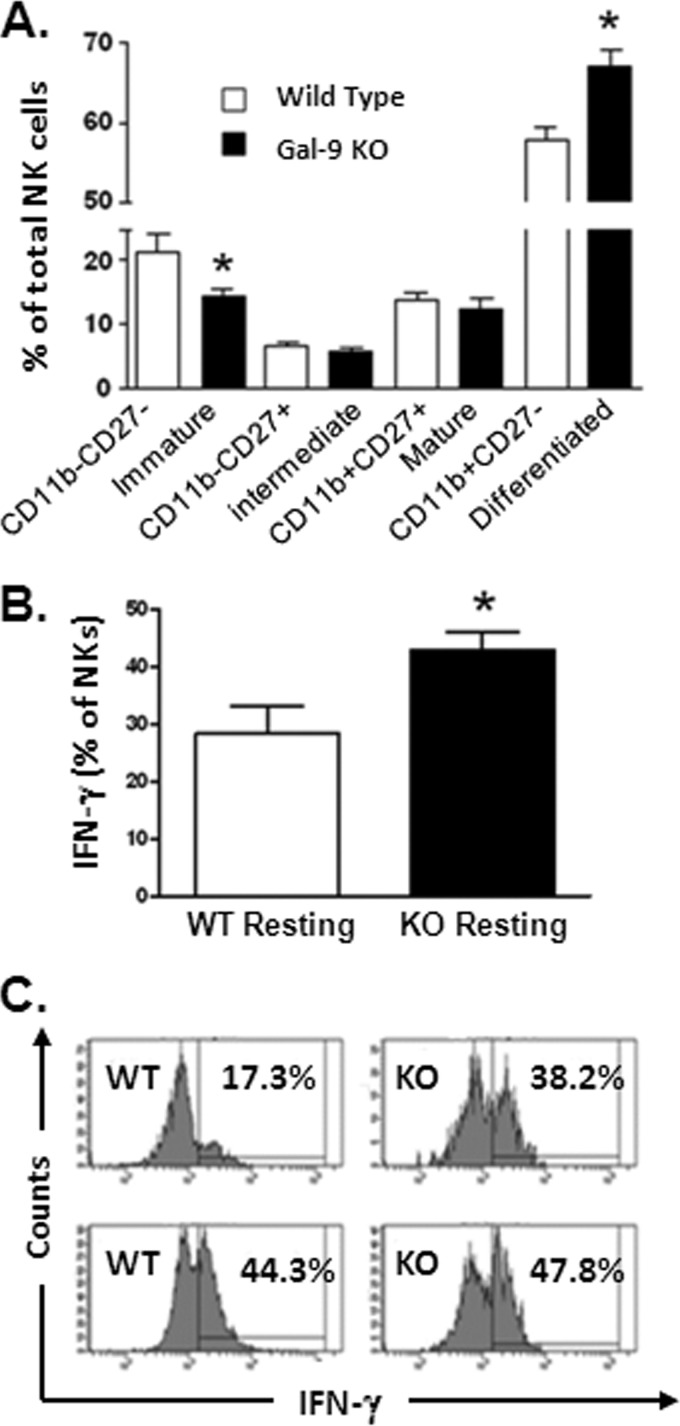
By convention, the cell surface expression of CD27 and CD11b is used to designate the maturation stage of murine NK cells. Immature CD27− CD11b− (double-negative [DN]) NK cells first acquire CD27 expression, become CD27+ CD11b−, and then acquire CD11b, yielding double-positive (DP) CD27+ CD11b+ NK cells, which eventually lose their CD27 expression to become fully differentiated CD27− CD11b+ NK cells (45, 46). Flow-cytometric analysis of these markers in Gal-9 KO mice showed that NK cell maturation was particularly affected in the livers of these mice, with accumulation at the CD27+ stages (Fig. 6). With regard to function, CD11b− CD27+ NK cells produce larger amounts of cytokines and show decreased cytotoxicity compared to CD11b+ CD27+ NK cells. In contrast, CD11b+ CD27− NK cells express higher levels of killer cell lectin-like receptor subfamily G, member 1 (KLRG1), which is induced following NK cell activation and proliferation. In addition, CD11b+ CD27− NK cells are predominant in the blood, while CD11b− CD27+ NK cells are found in a greater proportion in the liver (47). As these data provided equivocal information as to the functionality of NK cells derived from Gal-9 KO mice, we further addressed this question by assaying the activity of splenic NK cells in KO and WT mice (hepatic NK cells were limiting for functional studies). As shown in Fig. 7, production of IFN-γ in response to cytokine (IL-12/IL-18) stimulation, as well as degranulation in response to PMA and ionomycin, is enhanced in Gal-9 KO mice. Taken together, these data suggest that Gal-9 is involved in the restriction of NK cell activity in mice.
Fig 6.
Gal-9 KO mice show proportionally greater frequency of NK cells demonstrating intermediate and mature phenotypes, particularly within the liver. Splenic and hepatic NK cells from wild-type (n = 5) and Gal-9 KO (n = 5) mice were assayed for maturation status using the expression patterns of CD27 and CD11b. (A) A subtle but significant difference in the subset distribution is evident in the spleen. Liver-derived NK cells from Gal-9 KO mice are highly enriched for CD27+ intermediate and mature NK subsets. Conversely, the proportion of CD27− immature and differentiated NK cells is decreased in Gal-9 KO mice. The bars represent medians ± interquartile ranges. The nonparametric Mann-Whitney test was used to calculate P values between control (WT) and KO groups. (B) Representative flow-cytometric dot plots of CD11b and CD27 expression on gated NK cells (CD3e− NK1.1+) from spleen and liver. The numbers in the quadrants represent the percentages of total NK cells positive in each subset.
Fig 7.
NK cells from Gal-9 KO mice respond to stimulation with enhanced cytokine production and degranulation. For the induction of IFN-γ, splenic cells were stimulated with cytokines IL-12 (1 ng/ml) and IL-18 (10 ng/ml) for 6 h in the presence of brefeldin A (BFA). After culture, cells were washed and stained for cell surface expression of CD3 and NK1.1 to identify NK cells. After fixation and permeabilization, intracellular staining for IFN-γ was performed. (A) Cytokine stimulation results in greater production of IFN-γ by NK cells from Gal-9 KO mice (n = 5 in each group). (B) Degranulation (CD107a expression) was measured after 6 h of stimulation with PMA and ionomycin in the presence of BFA. NK cells from KO mice demonstrated enhance degranulation compared to wild-type (WT) mice. The bars show means ± SEM, and Student's t test was used to test statistical significance. Representative flow-cytometric histograms are shown for cytokine-stimulated intracellular IFN-γ production and PMA-ionomycin-stimulated CD107a expression in WT and KO mice. The percentages represent the proportion of total NK cells positive in each subset.
Mouse model of viral infection.
We utilized the MCMV infection model, previously shown to engage inhibitory NK cell receptors through encoded viral proteins and activate NK cells through production of IFNs (48), in order to determine the in vivo effect of Gal-9 absence on NK cells following viral infection. We found that following 4 days of infection with MCMV, there was no difference in the PFU in either the livers or spleens between control and Gal-9 KO mice; however, the percentage of immature (CD11b/CD27 DN) NK cells within the liver was lower in the Gal-9 KO mice, in keeping with the uninfected mice depicted in Fig. 5. However, MCMV infection induced a greater percentage of intrahepatic NK cells that were terminally differentiated (CD11b+ CD27−). The proportion of NK cells that spontaneously produced IFN-γ following infection was greater in the KO mice than in the wild-type mice (Fig. 8). These data suggest that viral infection has a more profound functional effect on intrahepatic NK cells in the absence of Gal-9.
Fig 8.
MCMV infection in Gal-9 KO mice. (A) Hepatic mononuclear cells were isolated from the livers of control (n = 5) and Gal-9 KO (n = 5) mice after 4 days of infection with MCMV (as described in Materials and Methods). Flow-cytometric analysis was used to identify NK cells (CD3e− NK1.1+), and the expression of CD11b and CD27 on NK cells to assess the maturation stage was analyzed as described previously (45) and in Results. Gal-9 KO mice have a lower proportion of immature and a higher proportion of fully differentiated NK cells (denoted by an asterisk). (B) Hepatic mononuclear cells were rested overnight in the presence of BFA and stained for intracellular IFN-γ (as described in Materials and Methods). Spontaneous IFN-γ production by hepatic NK cells was significantly higher in Gal-9 KO mice than in wild-type controls. The bars represent medians ± interquartile ranges. Nonparametric Mann-Whitney test was used to calculate P values between control (WT) and KO groups. (C) Representative flow-cytometric histograms of spontaneous (upper) and IL-12/IL-18-stimulated (bottom) IFN-γ production by hepatic NK cells is shown. The numbers represent the percentages of total NK cells positive in control (WT) and Gal-9 KO mice. The level of IFN-γ production by WT and KO hepatic NK cells is similar with cytokine stimulation.
DISCUSSION
Gal-9 is highly expressed in tissues of the immune system, including bone marrow, lymph nodes, liver, thymus, and spleen, and patients with chronic viral infection have elevated circulating levels (3, 4). Gal-9 has complex immunomodulatory roles involving effector cells of both innate and adaptive immunity. This important pleiotropic immune modulator affects numerous immune cell types (reviewed in references 8 and 9). In general, Gal-9 is thought to be involved in the activation of innate immune responses (3, 10, 22) and the downregulation of Th17 (11) and Th1 responses (12, 13). Although a number of recent investigations have identified roles for Gal-9 in the inhibition of CD4+ Th1 and CD8+ T cells, expansion of regulatory T cells, secretion of proinflammatory cytokines from dendritic cells and macrophages (49), and promotion of Th2 cell migration (7), its role in mediating NK cell function is only now emerging (22).
The principal findings of the current study can be summarized as follows. (i) Gal-9 downregulates multiple genes involved in activation of the immune response, including those involved in the NK cytotoxicity pathway. (ii) Functional impairment of human NK cells by high-dose Gal-9 in vitro occurs independently of Tim-3. (iii) We provide novel phenotypic and functional data on NK cells in the Gal-9 KO mouse which overall suggest that in the genetic absence of Gal-9, NK cells are more active, although the maturation stage, particularly in the liver, is perturbed. (iv) We also show preliminary data that in the setting of a short-term (4-day) MCMV infection, terminally differentiated NKs accumulate in the livers of Gal-9 KO mice, and that hepatic NKs spontaneously produce significantly more IFN-γ in this setting than wild-type mice.
The microarray and PCR assays strongly suggested that, under the conditions used in our assays, Gal-9 inhibits the immune functions of human NK cells. This is in contrast to the current view of Gal-9 as an activator of innate immune cells (3, 10, 22). The best-described natural ligand for Gal-9 is Tim-3 (8), and evidence suggests a critical role for Gal-9–Tim-3 interactions in regulating the fate of immune cells during various phases of the immune response (12, 13, 15, 22, 23, 42). NK cells express higher levels of Tim-3 than T cells (23, 43); we therefore wanted to determine if the effects of Gal-9 on human NK cell function in our assays were mediated through interactions with Tim-3. Surprisingly, we found that inhibition of human NK cells was independent of Tim-3; this may be related to the relatively high concentration (5 μg/ml) of Gal-9 in our assays. Using an NK cell line that overexpressed Tim-3, as well as primary NKs, Gleason and colleagues found that engagement of Tim-3 by Gal-9 induced NK cells to produce IFN-γ in a Tim-3-dependent manner (22). However, in their study the concentration of Gal-9 was lower than that used in the present study; they also found that stimulation without initial priming with the addition of cytokines (IL-12/IL-18) was associated with only modest increases in IFN-γ production (22). We chose to use 5 μg/ml Gal-9 in our assays, as we have used this concentration previously to expand regulatory T cells and induce cytokine production from monocytes (3). Of note, Gal-9 at 5 μg/ml has also been shown to inhibit CD40 signaling in T cells in a Tim-3-independent manner (25).
Several studies have shown Tim-3-independent immune cell regulation by Gal-9 (7, 11, 13, 24, 25), indicating that other pathways are involved in Gal-9 modulation of the immune response. Gal-9 binding to protein disulfide isomerase (PDI) on Th2 cells results in increased cell migration (7). Through interaction with an as-yet unidentified glycoprotein, the development of Th17 cells is inhibited in Gal-9 KO mice (11). Gal-9 can induce the production of proinflammatory cytokines from Th cells in a manner that does not require Tim-3 (24). These studies suggest that immune cell regulation by Gal-9 is complex and can be mediated by additional receptors, as well as Tim-3. Gal-9 interaction with NK cells can be mediated by Tim-3 (22); however, we found that this interaction is not limited to Tim-3 and that Gal-9 can act on NK cells independently of Tim-3, extending prior work in T cells (13, 24, 25), to downregulate several genes involved in activation of the immune response, including NK cytotoxicity. Differences in results may be related to differences in experimental conditions, such as duration of culture and concentrations of Gal-9. Functionally, the addition of Gal-9 significantly inhibited lymphokine-activated killing and cytokine-induced production of IFN-γ, further supporting an inhibitory role for this galectin under certain conditions.
Moreover, in the genetic absence of Gal-9, murine NK cells are more active, demonstrating greater relative expression of the activating receptor NKG2D and higher spontaneous degranulation and IFN-γ production. Although these knockout mice demonstrated lower frequencies of terminally differentiated NK cells, MCMV induced a higher proportion of NK cells that spontaneously produced IFN-γ than the wild-type mice and expanded the terminally differentiated (CD11b+ CD27−) NK cells, suggesting that Gal-9 is dispensable for this phenotypic switch. That is, IFNs or other factors induced by infection might override the genetic absence of Gal-9. While we acknowledge that these murine studies are preliminary and do not examine the role of Tim-3 on NK cells in the Gal-9 KO mouse, they do support a role for Gal-9 in the inhibition of NK cells. Although the present study does not address the identification of the receptor on NK cells mediating inhibition by Gal-9, it is interesting to speculate that as Gal-9 recognizes carbohydrates, differential glycosylation of NK cell receptors is involved (26–28).
Gal-9 may regulate the antiviral response of NK cells, including IFN production, both negatively and positively (22, 23) depending on activation thresholds, stage of infection, inflammatory stimuli, and relative expression of cellular receptor(s). In situations where the collective effect of Gal-9 signaling in NK cells favors persistence of viral infections, the reversal of this pathway may represent a novel therapeutic target.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by U19 AI 1066328 (H.R.R.), K24AI083742 (H.R.R.), a VA Merit Review grant (H.R.R.), R21AIO76161 (B.E.P.), and R21AIO76161-01A2S1 (B.E.P.).
L.G.M. planned and performed experiments and wrote the paper; R.R., S.M., B.E.P., L.C., R.M., and C.K. planned and performed experiments; M.S. and S.M. analyzed data; M.H. and T.N. provided the galectin-9 KO mice; and H.R.R. designed the study and wrote the paper.
We declare no conflicts of interest.
Footnotes
Published ahead of print 13 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01085-12.
REFERENCES
- 1. Barondes SH, Cooper DN, Gitt MA, Leffler H. 1994. Galectins. Structure and function of a large family of animal lectins. J. Biol. Chem. 269:20807–20810 [PubMed] [Google Scholar]
- 2. Wada J, Kanwar YS. 1997. Identification and characterization of galectin-9, a novel beta-galactoside-binding mammalian lectin. J. Biol. Chem. 272:6078–6086 [DOI] [PubMed] [Google Scholar]
- 3. Mengshol JA, Golden-Mason L, Arikawa T, Smith M, Niki T, McWilliams R, Randall JA, McMahan R, Zimmerman MA, Rangachari M, Dobrinskikh E, Busson P, Polyak SJ, Hirashima M, Rosen HR. 2010. A crucial role for Kupffer cell-derived galectin-9 in regulation of T cell immunity in hepatitis C infection. PLoS One 5:e9504 doi:10.1371/journal.pone.0009504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Elahi S, Niki T, Hirashima M, Horton H. 2012. Galectin-9 binding to Tim-3 renders activated human CD4+ T cells less susceptible to HIV-1 infection. Blood 119:4192–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Freeman GJ, Casasnovas JM, Umetsu DT, DeKruyff RH. 2010. TIM genes: a family of cell surface phosphatidylserine receptors that regulate innate and adaptive immunity. Immunol. Rev. 235:172–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pioche-Durieu C, Keryer C, Souquere S, Bosq J, Faigle W, Loew D, Hirashima M, Nishi N, Middeldorp J, Busson P. 2005. In nasopharyngeal carcinoma cells, Epstein-Barr virus LMP1 interacts with galectin 9 in membrane raft elements resistant to simvastatin. J. Virol. 79:13326–13337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bi S, Hong PW, Lee B, Baum LG. 2011. Galectin-9 binding to cell surface protein disulfide isomerase regulates the redox environment to enhance T-cell migration and HIV entry. Proc. Natl. Acad. Sci. U. S. A. 108:10650–10655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rabinovich GA, Toscano MA. 2009. Turning “sweet” on immunity: galectin-glycan interactions in immune tolerance and inflammation. Nat. Rev. Immunol. 9:338–352 [DOI] [PubMed] [Google Scholar]
- 9. Wiersma VR, de Bruyn M, Helfrich W, Bremer E. 2011. Therapeutic potential of galectin-9 in human disease. Med. Res. Rev. doi:10.1002/med.20249. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 10. Dai SY, Nakagawa R, Itoh A, Murakami H, Kashio Y, Abe H, Katoh S, Kontani K, Kihara M, Zhang SL, Hata T, Nakamura T, Yamauchi A, Hirashima M. 2005. Galectin-9 induces maturation of human monocyte-derived dendritic cells. J. Immunol. 175:2974–2981 [DOI] [PubMed] [Google Scholar]
- 11. Oomizu S, Arikawa T, Niki T, Kadowaki T, Ueno M, Nishi N, Yamauchi A, Hirashima M. 2012. Galectin-9 suppresses Th17 cell development in an IL-2-dependent but Tim-3-independent manner. Clin. Immunol. 143:51–58 [DOI] [PubMed] [Google Scholar]
- 12. Klibi J, Niki T, Riedel A, Pioche-Durieu C, Souquere S, Rubinstein E, Le Moulec S, Guigay J, Hirashima M, Guemira F, Adhikary D, Mautner J, Busson P. 2009. Blood diffusion and Th1-suppressive effects of galectin-9-containing exosomes released by Epstein-Barr virus-infected nasopharyngeal carcinoma cells. Blood 113:1957–1966 [DOI] [PubMed] [Google Scholar]
- 13. Zhu C, Anderson AC, Schubart A, Xiong H, Imitola J, Khoury SJ, Zheng XX, Strom TB, Kuchroo VK. 2005. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat. Immunol. 6:1245–1252 [DOI] [PubMed] [Google Scholar]
- 14. Kadowaki T, Arikawa T, Shinonaga R, Oomizu S, Inagawa H, Soma G, Niki T, Hirashima M. 2012. Galectin-9 signaling prolongs survival in murine lung-cancer by inducing macrophages to differentiate into plasmacytoid dendritic cell-like macrophages. Clin. Immunol. 142:296–307 [DOI] [PubMed] [Google Scholar]
- 15. Seki M, Oomizu S, Sakata KM, Sakata A, Arikawa T, Watanabe K, Ito K, Takeshita K, Niki T, Saita N, Nishi N, Yamauchi A, Katoh S, Matsukawa A, Kuchroo V, Hirashima M. 2008. Galectin-9 suppresses the generation of Th17, promotes the induction of regulatory T cells, and regulates experimental autoimmune arthritis. Clin. Immunol. 127:78–88 [DOI] [PubMed] [Google Scholar]
- 16. Biron CA. 1999. Initial and innate responses to viral infections–pattern setting in immunity or disease. Curr. Opin. Microbiol. 2:374–381 [DOI] [PubMed] [Google Scholar]
- 17. Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S. 2011. Innate or adaptive immunity? The example of natural killer cells. Science 331:44–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Biron CA, Brossay L. 2001. NK cells and NKT cells in innate defense against viral infections. Curr. Opin. Immunol. 13:458–464 [DOI] [PubMed] [Google Scholar]
- 19. Cooper MA, Fehniger TA, Caligiuri MA. 2001. The biology of human natural killer-cell subsets. Trends Immunol. 22:633–640 [DOI] [PubMed] [Google Scholar]
- 20. Lanier LL. 2005. NK cell recognition. Annu. Rev. Immunol. 23:225–274 [DOI] [PubMed] [Google Scholar]
- 21. Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, Biassoni R, Moretta L. 2001. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu. Rev. Immunol. 19:197–223 [DOI] [PubMed] [Google Scholar]
- 22. Gleason MK, Lenvik TR, McCullar V, Felices M, O'Brien MS, Cooley SA, Verneris MR, Cichocki F, Holman CJ, Panoskaltsis-Mortari A, Niki T, Hirashima M, Blazar BR, Miller JS. 2012. Tim-3 is an inducible human natural killer cell receptor that enhances interferon gamma production in response to galectin-9. Blood 119:3064–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ndhlovu LC, Lopez-Verges S, Barbour JD, Jones RB, Jha AR, Long BR, Schoeffler EC, Fujita T, Nixon DF, Lanier LL. 2012. Tim-3 marks human natural killer cell maturation and suppresses cell-mediated cytotoxicity. Blood 119:3734–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Su EW, Bi S, Kane LP. 2011. Galectin-9 regulates T helper cell function independently of Tim-3. Glycobiology 21:1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vaitaitis GM, Wagner DH., Jr 2012. Galectin-9 controls CD40 signaling through a Tim-3 independent mechanism and redirects the cytokine profile of pathogenic T cells in autoimmunity. PLoS One 7:e38708 doi:10.1371/journal.pone.0038708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hartmann J, Tran TV, Kaudeer J, Oberle K, Herrmann J, Quagliano I, Abel T, Cohnen A, Gatterdam V, Jacobs A, Wollscheid B, Tampe R, Watzl C, Diefenbach A, Koch J. 2012. The stalk domain and the glycosylation status of the activating natural killer cell receptor NKp30 are important for ligand binding. J. Biol. Chem. 287:31527–31539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Margraf-Schonfeld S, Bohm C, Watzl C. 2011. Glycosylation affects ligand binding and function of the activating natural killer cell receptor 2B4 (CD244) protein. J. Biol. Chem. 286:24142–24149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mason LH, Willette-Brown J, Anderson SK, Alvord WG, Klabansky RL, Young HA, Ortaldo JR. 2003. Receptor glycosylation regulates Ly-49 binding to MHC class I. J. Immunol. 171:4235–4242 [DOI] [PubMed] [Google Scholar]
- 29. Huang da W, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. 2007. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 35:W169–W175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith MG. 1954. Propagation of salivary gland virus of the mouse in tissue cultures. Proc. Soc. Exp. Biol. Med. 86:435–440 [DOI] [PubMed] [Google Scholar]
- 31. Harvey DM, Levine AJ. 1991. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev. 5:2375–2385 [DOI] [PubMed] [Google Scholar]
- 32. Wagner M, Jonjic S, Koszinowski UH, Messerle M. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056–7060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Golden-Mason L, Castelblanco N, O'Farrelly C, Rosen HR. 2007. Phenotypic and functional changes of cytotoxic CD56pos natural T cells determine outcome of acute hepatitis C virus infection. J. Virol. 81:9292–9298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haybaeck J, Zeller N, Wolf MJ, Weber A, Wagner U, Kurrer MO, Bremer J, Iezzi G, Graf R, Clavien PA, Thimme R, Blum H, Nedospasov SA, Zatloukal K, Ramzan M, Ciesek S, Pietschmann T, Marche PN, Karin M, Kopf M, Browning JL, Aguzzi A, Heikenwalder M. 2009. A lymphotoxin-driven pathway to hepatocellular carcinoma. Cancer Cell 16:295–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuttruff S, Koch S, Kelp A, Pawelec G, Rammensee HG, Steinle A. 2009. NKp80 defines and stimulates a reactive subset of CD8 T cells. Blood 113:358–369 [DOI] [PubMed] [Google Scholar]
- 36. Lichtfuss GF, Meehan AC, Cheng WJ, Cameron PU, Lewin SR, Crowe SM, Jaworowski A. 2011. HIV inhibits early signal transduction events triggered by CD16 cross-linking on NK cells, which are important for antibody-dependent cellular cytotoxicity. J. Leukoc. Biol. 89:149–158 [DOI] [PubMed] [Google Scholar]
- 37. Filen JJ, Filen S, Moulder R, Tuomela S, Ahlfors H, West A, Kouvonen P, Kantola S, Bjorkman M, Katajamaa M, Rasool O, Nyman TA, Lahesmaa R. 2009. Quantitative proteomics reveals GIMAP family proteins 1 and 4 to be differentially regulated during human T helper cell differentiation. Mol. Cell. Proteomics 8:32–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saunders A, Webb LM, Janas ML, Hutchings A, Pascall J, Carter C, Pugh N, Morgan G, Turner M, Butcher GW. 2010. Putative GTPase GIMAP1 is critical for the development of mature B and T lymphocytes. Blood 115:3249–3257 [DOI] [PubMed] [Google Scholar]
- 39. Khor CC, Vannberg FO, Chapman SJ, Guo H, Wong SH, Walley AJ, Vukcevic D, Rautanen A, Mills TC, Chang KC, Kam KM, Crampin AC, Ngwira B, Leung CC, Tam CM, Chan CY, Sung JJ, Yew WW, Toh KY, Tay SK, Kwiatkowski D, Lienhardt C, Hien TT, Day NP, Peshu N, Marsh K, Maitland K, Scott JA, Williams TN, Berkley JA, Floyd S, Tang NL, Fine PE, Goh DL, Hill AV. 2010. CISH and susceptibility to infectious diseases. N. Engl. J. Med. 362:2092–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ma X, Wang X, Gao X, Wang L, Lu Y, Gao P, Deng W, Yu P, Ma J, Guo J, Cheng H, Zhang C, Shi T, Ma D. 2007. Identification of five human novel genes associated with cell proliferation by cell-based screening from an expressed cDNA ORF library. Life Sci. 81:1141–1151 [DOI] [PubMed] [Google Scholar]
- 41. Vujanovic NL. 2011. Role of TNF superfamily ligands in innate immunity. Immunol. Res. 50:159–174 [DOI] [PubMed] [Google Scholar]
- 42. Sehrawat S, Reddy PB, Rajasagi N, Suryawanshi A, Hirashima M, Rouse BT. 2010. Galectin-9/TIM-3 interaction regulates virus-specific primary and memory CD8 T cell response. PLoS Pathog. 6:e1000882 doi:10.1371/journal.ppat.1000882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Khademi M, Illes Z, Gielen AW, Marta M, Takazawa N, Baecher-Allan C, Brundin L, Hannerz J, Martin C, Harris RA, Hafler DA, Kuchroo VK, Olsson T, Piehl F, Wallstrom E. 2004. T cell Ig-and mucin-domain-containing molecule-3 (TIM-3) and TIM-1 molecules are differentially expressed on human Th1 and Th2 cells and in cerebrospinal fluid-derived mononuclear cells in multiple sclerosis. J. Immunol. 172:7169–7176 [DOI] [PubMed] [Google Scholar]
- 44. Smyk-Pearson S, Tester IA, Klarquist J, Palmer BE, Pawlotsky JM, Golden-Mason L, Rosen HR. 2008. Spontaneous recovery in acute human hepatitis C virus infection: functional T-cell thresholds and relative importance of CD4 help. J. Virol. 82:1827–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, Walzer T. 2009. Maturation of mouse NK cells is a 4-stage developmental program. Blood 113:5488–5496 [DOI] [PubMed] [Google Scholar]
- 46. Jaeger BN, Donadieu J, Cognet C, Bernat C, Ordonez-Rueda D, Barlogis V, Mahlaoui N, Fenis A, Narni-Mancinelli E, Beaupain B, Bellanne-Chantelot C, Bajenoff M, Malissen B, Malissen M, Vivier E, Ugolini S. 2012. Neutrophil depletion impairs natural killer cell maturation, function, and homeostasis. J. Exp. Med. 209:565–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lassen MG, Lukens JR, Dolina JS, Brown MG, Hahn YS. 2010. Intrahepatic IL-10 maintains NKG2A+Ly49-liver NK cells in a functionally hyporesponsive state. J. Immunol. 184:2693–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mitrovic M, Arapovic J, Traven L, Krmpotic A, Jonjic S. 2012. Innate immunity regulates adaptive immune response: lessons learned from studying the interplay between NK and CD8(+) T cells during MCMV infection. Med. Microbiol. Immunol. 201:487–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Anderson AC, Anderson DE, Bregoli L, Hastings WD, Kassam N, Lei C, Chandwaskar R, Karman J, Su EW, Hirashima M, Bruce JN, Kane LP, Kuchroo VK, Hafler DA. 2007. Promotion of tissue inflammation by the immune receptor Tim-3 expressed on innate immune cells. Science 318:1141–1143 [DOI] [PubMed] [Google Scholar]
- 50. Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. 2006. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34:D354–D357 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.