Abstract
For many insect-vectored plant viruses, the relationship between feeding behavior and vector competence may prove integral to an understanding of the epidemiology of the resulting plant disease. While plant-infecting viruses are well known to change host plant physiology in a way that makes them more attractive to vectors, viral manipulation of the vectors themselves has only recently been reported. Previous research suggested that the rapid spread of Tomato yellow leaf curl virus (TYLCV) throughout China has been facilitated by its primary vector, the whitefly Bemisia tabaci. We conducted two experiments testing the impact of TYLCV infection of the host plant (tomato) and vector (B. tabaci biotypes B and Q) on whitefly feeding behavior. Whiteflies of biotypes B and Q both appeared to find TYLCV-infected plants more attractive, probing them more quickly and having a greater number of feeding bouts; this did not, however, alter the total time spent feeding. Viruliferous whiteflies fed more readily than uninfected whiteflies and spent more time salivating into sieve tube elements. Because vector salivation is essential for viral transmission, this virally mediated alteration of behavior should provide TYLCV a direct fitness benefit. This is the first report of such manipulation by a nonpropagative virus that belongs to an exclusively plant-infecting family of viruses (Geminiviridae). In the context of previous research showing that feeding on TYLCV-infected plants harms biotype B but helps biotype Q, the fact that both biotypes were equally affected by TYLCV also suggests that the virus may alter the biotype B-biotype Q competitive interaction in favor of biotype Q.
INTRODUCTION
The ability of insects and other arthropods to act as vectors is tightly linked to a given organism's feeding behavior (1, 2). While an array of animal-infecting viruses have been shown to modify the behavior and feeding processes of their vectors (3), such alterations have been far less studied in plant-infecting viruses (4). This lacuna is notable because since these viruses are often dependent on their vectors for transmission, such “parasitic manipulation” (3) should strongly benefit vector-modifying viruses. Evidence for indirect manipulation (i.e., virally mediated changes in host plant physiology) comes from previous work showing that insect vectors of Barley yellow dwarf virus feed differently on infected versus healthy oats (5). Similarly, Potato leafroll virus infection of potato alters the feeding behavior of its aphid vector (6). Other work has shown preferential attraction of vectors to infected plants (7, 8), even when virus-free plants are actually better hosts for the vector (9).
A recent paper by Stafford et al. (10) provided the first evidence that plant-infecting viruses can also directly alter vector feeding behavior. Those authors found that infection of the thrips vector Frankliniella occidentalis (Pergande) by Tomato spotted wilt virus makes male thrips spend more time feeding, thus increasing their vector competence (10). Tomato spotted wilt virus is a circulative propagative virus that is a member of the Bunyaviridae, a group consisting primarily of animal-infecting viruses that increase biting rates and feeding behaviors in their vectors (11). Those researchers suggested that direct viral manipulation of vectors may be an evolutionarily conserved trait in the Bunyaviridae that increases viral transmission (2, 10). While this work demonstrates that such alterations are possible, it does not address whether it occurs in the many viral families that exclusively infect plants or in plant-infecting viruses that do not replicate within their vectors.
Tomato yellow leaf curl virus (TYLCV) (family Geminiviridae, genus Begomovirus) is a complex of circular, single-stranded DNA plant geminiviruses (12). When it infects tomatoes, Solanum lycopersicum (L.), TYLCV causes a devastating disease that has severely reduced cultivated tomato production worldwide (13). It is vectored by the sweetpotato whitefly, Bemisia tabaci (Gennadius), in a circulative and persistent manner (14–16). Whiteflies feeding on the phloem of infected plants ingest viral particles that first cross the midgut wall barrier into the hemolymph and then cross the salivary gland barrier into the saliva. Within 8 h of feeding on an infected plant, the now-viruliferous whiteflies are themselves capable of transmitting TYLCV to uninfected plants (16–18).
Bemisia tabaci is an agricultural pest and virus vector that, although formally a single species, contains multiple “biotypes” that differ in areas such as behavior, degree of vector competence, endosymbiont communities, and genetic makeup (19–21). Although there has been much debate (22–25) over whether B. tabaci is “a complex species or species complex” (23), the most recent molecular evidence suggests that B. tabaci actually includes at least 24 genetically distinct but morphologically indistinguishable cryptic species. Two of the most widely distributed members of the species complex are the East-Minor Asia 1 genetic group (biotype B) and the Mediterranean genetic group (biotype Q). Bemisia tabaci damages plants both directly, by stylet probing, and indirectly, by acting as a vector for TYLCV and more than 100 other begomoviruses (26).
After biotype B was first detected in China in the mid-1990s, it replaced the indigenous whitefly species and became the dominant whitefly in both greenhouse and field crops (27, 28). This situation persisted until 2003, when biotype Q was found in Yunnan Province (27); by 2007, biotype Q had replaced biotype B as the dominant whitefly in China (29, 30). TYLCV was first detected in tomato plants in Shanghai in 2006 (31) and has since quickly spread throughout China (32, 33). The fact that the introduction and spread of TYLCV followed the 2003 introduction of biotype Q, rather than the earlier introduction of biotype B, suggests that biotype Q may be a more competent vector of TYLCV.
We tested whether Tomato yellow leaf curl virus directly and/or indirectly manipulates the feeding behavior of Bemisia tabaci biotypes B and Q in a manner consistent with improved viral acquisition and transmission. Specifically, we describe the results of two experiments assessing whitefly feeding behavior in response to TYLCV infection of the viral vectors (B. tabaci biotypes B and Q) and/or the host plant (tomato). The first experiment assessed whether uninfected biotype Q and B whiteflies fed differently on virus-free and TYLCV-infected tomato plants: this tested for indirect (i.e., plant-mediated) viral modification of whitefly feeding behavior. The second experiment assessed whether viruliferous and uninfected biotype Q and B whiteflies fed differently on virus-free tomato plants: this tested for direct modification of whitefly feeding.
MATERIALS AND METHODS
Plants.
Tomato plants (cv. Zhongza 9) were grown in pots in a greenhouse under natural lighting and controlled temperature (26°C ± 2°C). TYLCV-infected plants were produced by agroinoculation at the 3- to 4-true-leaf stage with Agrobacterium tumefaciens-mediated TYLCV clones originally isolated from Shanghai, China (31). Infection was determined visually and confirmed by PCR validation (34, 35). All plants used in experiment 1 were at the 6- to 7-true-leaf stage; all plants used in experiment 2 were at the 2- to 3-true-leaf stage. The same cultivar was used in both experiments 1 and 2.
Laboratory whitefly populations.
The laboratory population of B. tabaci biotype B was collected in 2004 from cabbage, Brassica oleracea var. Jingfeng1, growing in the Haidian District of Beijing, China. Bemisia tabaci biotype Q whiteflies were collected in 2009 from poinsettia, Euphorbia pulcherrima Wild. ex Klotz., growing in the same area. Colonies of biotypes B and Q were maintained on cabbage and poinsettia, respectively, in a greenhouse in separate screen cages under natural lighting and ambient temperature (26°C ± 2°C). Because both biotypes appear identical (i.e., same appearance and stylet length, etc.), we ensured the purity of each colony according to standard protocols (28) and by sampling the mitochondrial cytochrome oxidase I (mtCOI) region of 15 adult whiteflies every generation.
Establishment of uninfected and viruliferous B. tabaci colonies.
We created four whitefly colonies: uninfected biotype B, uninfected biotype Q, viruliferous biotype B, and viruliferous biotype Q. We created viruliferous colonies by placing four TYLCV-infected tomato plants into each of two cages. We then transferred 300 uninfected biotype B and Q adults into each cage, one biotype per cage. We simultaneously established uninfected biotype B and Q colonies by placing 300 uninfected biotype B and Q adults into cages with virus-free tomato plants, one biotype per cage. All colonies were maintained for 2 whitefly generations in a controlled-temperature (26°C ± 2°C) greenhouse with a 14-h-light–10-h-dark photoperiod. At the start of the third generation, we selected newly emerged female whiteflies (2 to 5 days old) from each colony for use in the experiments (36).
Design of experiment 1.
Experiment 1 tested whether the feeding behavior of whiteflies of biotypes B and Q differed on TYLCV-infected versus virus-free tomato plants. It was a 2-by-2 factorial experiment, with B. tabaci biotype (B or Q) crossed with plant infection status (TYLCV-infected tomato plant or virus-free tomato plant). There were thus four treatments: biotype B feeding on a TYLCV-infected tomato plant (n = 29 replicates), biotype B feeding on a virus-free tomato plant (n = 29), biotype Q feeding on a TYLCV-infected tomato plant (n = 29), and biotype Q feeding on a virus-free tomato plant (n = 30). Although we tested the same number of whiteflies for every treatment, experimental difficulties (e.g., whiteflies dying or escaping or probes becoming detached) resulted in slightly different levels of replication in the four treatments.
Design of experiment 2.
Experiment 2 tested whether viruliferous versus uninfected whiteflies of biotypes B and Q fed differently on a virus-free tomato plant. It was a 2-by-2 factorial experiment, with B. tabaci biotype (B or Q) crossed with whitefly infection status (viruliferous whitefly or uninfected whitefly) for a total of four treatments: viruliferous biotype B (n = 30 replicates), uninfected biotype B (n = 28), viruliferous biotype Q (n = 33), and uninfected biotype Q (n = 30). Again, various experimental difficulties resulted in slightly different replication levels.
Electrical penetration graph recording.
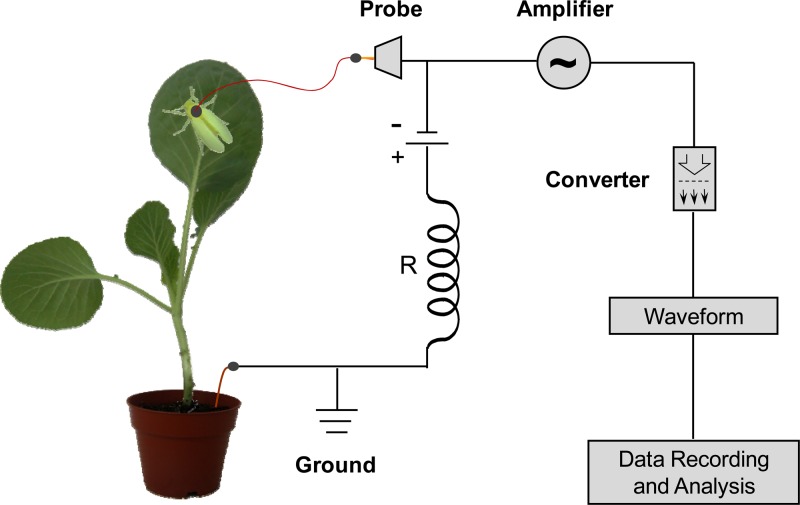
We recorded whitefly electrical penetration graphs (EPGs) using a Giga-8 direct-current EPG (DC-EPG) system (Wageningen University, Netherlands) with a 109-Ω input resistance (Fig. 1). Briefly, the EPG turns a phloem-sucking insect and its host plant into part of an electrical circuit that is completed when the insect's mouthparts penetrate the plant (37, 38). The resulting electrical signals are amplified and digitized. Fluctuations in voltage and electrical resistance are recorded on a computer hard disk and can be matched to specific feeding events (e.g., ingestion, salivation, and penetration of sieve elements, etc.) in order to monitor feeding behavior. Prior to recording, female whiteflies were immobilized in an ice-chilled glass dish. We then attached a gold wire (1.5 cm long and 12.5 μm in diameter) to the whitefly's dorsum using a drop of water-based silver glue. The wired whiteflies were then connected to the Giga-8 probe input and placed onto the lower surface of the bottom leaf, which was fixed on a stick vertically inserted into the pot. Each whitefly-plant-probe combination was placed into an electrically grounded Faraday cage to shield against external electrical noise. The EPG signals were digitized with a DI710-UL analogue-to-digital converter (Dataq Instruments, Akron, OH), and the output was acquired and stored with PROBE3.4 software. Six hours of EPGs were continuously recorded for each replicate (i.e., a fresh whitefly-plant-probe combination). All experiments were carried out at 26°C ± 2°C, at 70% relative humidity (RH), and under artificial light (1,500 lx) with a 14-h-light–10-h-dark regime.
Fig 1.
Schematic drawing of a direct-current electrical penetration graph (DC-EPG)-based behavioral analysis system. This EPG system turns a phloem-sucking insect and its host plant into part of an electrical circuit that is completed when the insect's mouthparts penetrate the plant. The electrical signal is amplified by an amplifier and digitized by a converter. Fluctuations in voltage and electrical resistance are recorded and can be matched to specific feeding events.
Data analysis.
Waveform patterns were categorized as described previously by Jiang et al. (36, 39). We identified five distinct waveforms: nonprobing (NP); pathway (C); potential drop (pd); and the phloem phases E(pd)1, salivation into a sieve element, and E(pd)2, ingestion of sieve element sap. Waveforms F (presumed penetration difficulties) and G (xylem sap ingestion) were rare and grouped into waveform C. The time from the start to the end of each waveform was recorded and exported by using PROBE3.4 software (Wageningen University, Netherlands). We used this information to calculate 10 non-phloem-phase parameters (parameters A to J) (see Tables 1 and 3) and 10 phloem-phase parameters (parameters K to T) (see Tables 2 and 4). Each parameter was calculated for each replicate, and the replicates were averaged to derive treatment-level means and standard errors. Prior to analysis, data were checked for normality and homogeneity of variance and transformed where necessary to improve model fit (log10 transformation for duration). All statistical analyses were done with SPSS 11.5 (SPSS Inc., Chicago, IL).
Table 1.
Statistical analysis of nonphloem EPG parameters A to J of uninfected B. tabaci biotypes B and Q probing TYLCV-infected and virus-free tomato plants (experiment 1)a
| Parameter, description |
P value |
||
|---|---|---|---|
| Biotype | Infection | Interaction | |
| A, time when first probe begins | 0.063 | 0.012 | 0.623 |
| B, no. of probes | 0.001 | 0.012 | 0.243 |
| C, total duration of probes (total time in waveform) | 0.001 | 0.014 | 0.677 |
| D, mean duration of probes (avg probing waveform duration) | <0.001 | 0.957 | 0.179 |
| E, total duration of NP | 0.001 | 0.014 | 0.677 |
| F, total duration of C | 0.687 | 0.004 | 0.989 |
| G, duration of first probe | 0.035 | 0.264 | 0.8 |
| H, time from first probe to first E(pd) | 0.015 | 0.295 | 0.209 |
| I, no. of probes before first E(pd) | <0.001 | 0.463 | 0.437 |
| J, time from beginning of probe to first E(pd) within that probeb | 0.003 | 0.452 | 0.64 |
Parameter letters correspond to those in Fig. 2. P values were calculated by using two-way ANOVA with the main effects “biotype” (B or Q), “infection” (TYLCV-infected plant or virus-free plant), and their interaction. P values in boldface type are significant at an α value of 0.05.
Calculation excludes whiteflies that did not enter phloem phase within the 6-h recording period.
Table 3.
Statistical analysis of nonphloem EPG parameters A to J of viruliferous and uninfected B. tabaci biotypes B and Q probing virus-free tomato plants (experiment 2)a
| Parameter, description |
P value |
||
|---|---|---|---|
| Biotype | Infection | Interaction | |
| A, time when first probe begins | 0.673 | <0.001 | 0.336 |
| B, no. of probes | 0.106 | 0.004 | 0.941 |
| C, total duration of probes (total time in waveform) | 0.022 | 0.105 | 0.817 |
| D, mean duration of probes (probing waveform duration) | <0.001 | 0.226 | 0.767 |
| E, total duration of NP | 0.01 | 0.027 | 0.946 |
| F, total duration of C | 0.163 | 0.054 | 0.432 |
| G, duration of first probe | 0.441 | 0.035 | 0.629 |
| H, time from first probe to first E(pd) | 0.386 | 0.275 | 0.178 |
| I, no. of probes before first E(pd) | 0.187 | 0.557 | 0.289 |
| J, time from beginning of probe to first E(pd) within probeb | 0.173 | 0.315 | 0.452 |
Parameter letters correspond to those in Fig. 4. P values were calculated by using two-way ANOVA with the main effects “biotype” (B or Q), “infection” (viruliferous whitefly or uninfected whitefly), and their interaction. P values in boldface type are significant at an α value of 0.05.
Calculation excludes whiteflies that did not enter phloem phase within the 6-h recording period.
Table 2.
Statistical analysis of phloem EPG parameters K to T of uninfected B. tabaci biotypes B and Q probing TYLCV-infected and virus-free tomato plants (experiment 1)a
| Parameter, description |
P value |
||
|---|---|---|---|
| Biotype | Infection | Interaction | |
| K, total duration of E(pd)1 | 0.034 | 0.001 | 0.159 |
| L, total no. of E(pd)1 | 0.151 | 0.008 | 0.899 |
| M, mean duration of E(pd)1 | <0.001 | 0.926 | 0.523 |
| N, total duration of E(pd)2 | <0.001 | 0.423 | 0.922 |
| O, total no. of E(pd)2 | 0.16 | 0.034 | 0.701 |
| P, mean duration of E(pd)2 | <0.001 | 0.582 | 0.859 |
| Q, potential E(pd)2 indexb | <0.001 | 0.994 | 0.806 |
| R, % of non-E(pd) time in NPc | 0.061 | 0.006 | 0.689 |
| S, % of probes reaching phloem phase | 0.019 | 0.78 | 0.203 |
| T, % of phloem phases reaching waveform E(pd)2d | 0.805 | 0.024 | 0.461 |
Parameter letters correspond to those in Fig. 3. P values were calculated by using two-way ANOVA with the main effects “biotype” (B or Q), “infection” (TYLCV-infected plant or virus-free plant), and their interaction. P values in boldface type are significant at an α value 0.05.
Calculated as [total time in E(pd)2]/[recording time − time to first E(pd)].
Calculated as (total time in NP)/[total recording time − time in E(pd)1 and E(pd)2].
Calculated as [number of times reaching E(pd)2]/[number of times reaching E(pd)].
Table 4.
Statistical analysis of phloem EPG parameters K to T of viruliferous and uninfected B. tabaci biotypes B and Q probing virus-free tomato plants (experiment 2)a
| Parameter, description |
P value |
||
|---|---|---|---|
| Biotype | Infection | Interaction | |
| K, total duration of E(pd)1 | 0.346 | 0.012 | 0.241 |
| L, total no. of E(pd)1 | 0.526 | 0.001 | 0.342 |
| M, mean duration of E(pd)1 | 0.105 | 0.546 | 0.784 |
| N, total duration of E(pd)2 | 0.001 | 0.213 | 0.926 |
| O, total no. of E(pd)2 | 0.277 | 0.004 | 0.625 |
| P, mean duration of E(pd)2 | <0.001 | 0.243 | 0.586 |
| Q, potential E(pd)2 indexb | 0.001 | 0.342 | 0.688 |
| R, % of non-E(pd) time in NPc | 0.086 | 0.034 | 0.851 |
| S, % of probes reaching phloem phase | 0.36 | 0.92 | 0.148 |
| T, % of phloem phases reaching waveform E(pd)2d | 0.022 | 0.049 | 0.012 |
Parameter letters correspond to those in Fig. 5. P values were calculated by using two-way ANOVA with the main effects “biotype” (B or Q), “infection” (viruliferous whitefly or uninfected whitefly), and their interaction. P values in boldface type are significant at an α value of 0.05.
Calculated as [total time in E(pd)2]/[recording time − time to first E(pd)].
Calculated as (total time in NP)/[total recording time − time in E(pd)1 and E(pd)2].
Calculated as [number of times reaching E(pd)2]/[number of times reaching E(pd)].
For experiment 1, we used two-way analysis of variance (ANOVA) to analyze the effect of biotype (B or Q), plant infection (TYLCV infected or virus free), and their interaction on non-phloem-phase parameters A to J (Table 1) and phloem-phase parameters K to T (Table 2). A main effect of biotype means that whiteflies of biotypes B and Q differ in their feeding behavior, a main effect of plant infection means that whiteflies feed differently on TYLCV-infected versus virus-free plants, and an interaction means that whiteflies of biotypes B and Q respond differently to TYLCV-infected versus virus-free plants.
For experiment 2, we again used two-way ANOVA to analyze the effect of biotype (B or Q), whitefly infection (viruliferous or uninfected), and their interaction on non-phloem-phase parameters A to J (Table 3) and phloem-phase parameters K to T (Table 4). A main effect of biotype means that whiteflies of biotypes B and Q differ in their feeding behavior, a main effect of whitefly infection means that viruliferous and uninfected whiteflies feed differently on virus-free plants, and an interaction means that TYLCV affects the behavior of whiteflies of biotypes B and Q differently.
RESULTS
Experiment 1: does biotype B and Q feeding behavior differ on TYLCV-infected versus virus-free tomato plants?
Whiteflies fed differently on TYLCV-infected and virus-free plants, and whiteflies of biotypes B and Q differed in their feeding behavior. However, the two biotypes did not differ in their response to host plant infection (P > 0.05 for all biotype-infection interactions).
(i) Nonphloem EPG measurements (parameters A to J).
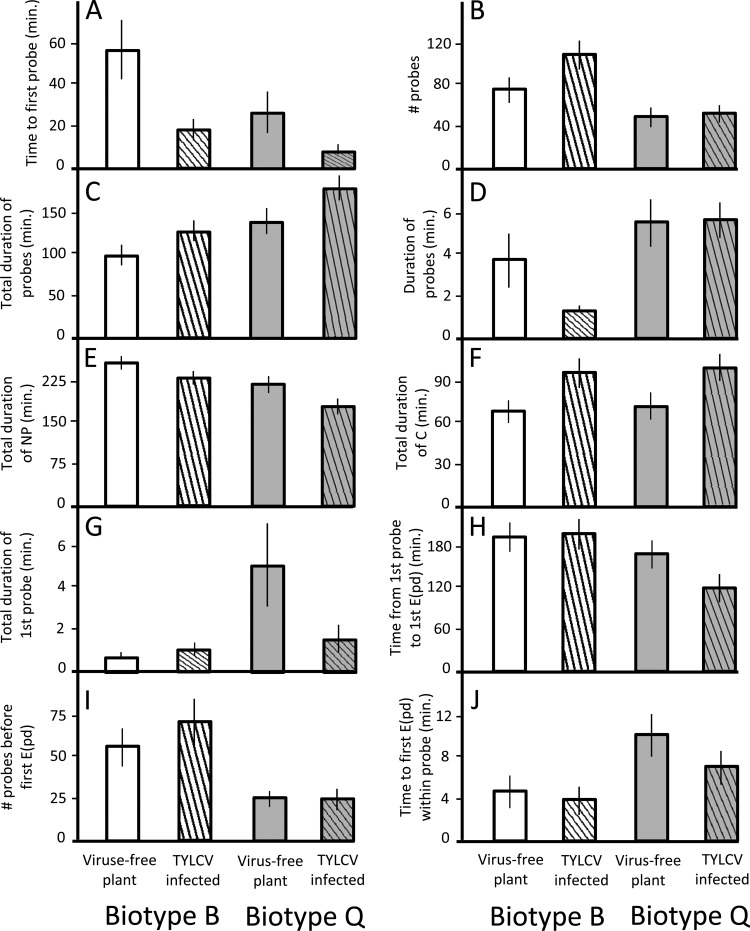
Both B. tabaci biotypes preferred TYLCV-infected over virus-free plants. Whiteflies took three times longer to start probing virus-free plants and attempted 29% more probes on infected plants (parameters A and B) (Table 1 and Fig. 2). As a result, they spent 22% less time probing virus-free plants overall (parameter C) and 38% more time in the pathway waveform on infected plants (parameter F).
Fig 2.
Mean values ± standard errors for nonphloem EPG parameters A to J of B. tabaci biotype B and Q whiteflies feeding on TYLCV-infected and virus-free tomato plants (experiment 1). Parameter letters correspond to parameter letters in Table 1. White bars, biotype B feeding on virus-free plant; white striped bars, biotype B feeding on TYLCV-infected plant; gray bars, biotype Q feeding on virus-free plant; gray striped bars, biotype Q feeding on TYLCV-infected plant.
Whiteflies of biotype B attempted 80% more probes than did whiteflies of biotype Q in experiment 1 and 2.5× more before the first E(pd)1 (parameters B and I) (Table 1 and Fig. 2). Despite this, whiteflies of biotype B also spent 22% more time “resting” (i.e., in NP) and took 35% longer to initiate feeding (parameters E and H). Biotype Q whiteflies had 41% longer total and 2.2×-longer mean probe durations than biotype B whiteflies and spent almost four times longer on the first probe (parameters C, D, and G). Biotype Q whiteflies did, however, take twice as long as biotype B whiteflies to start feeding within a given probe (parameter J).
(ii) Phloem EPG measurements (parameters K to T).
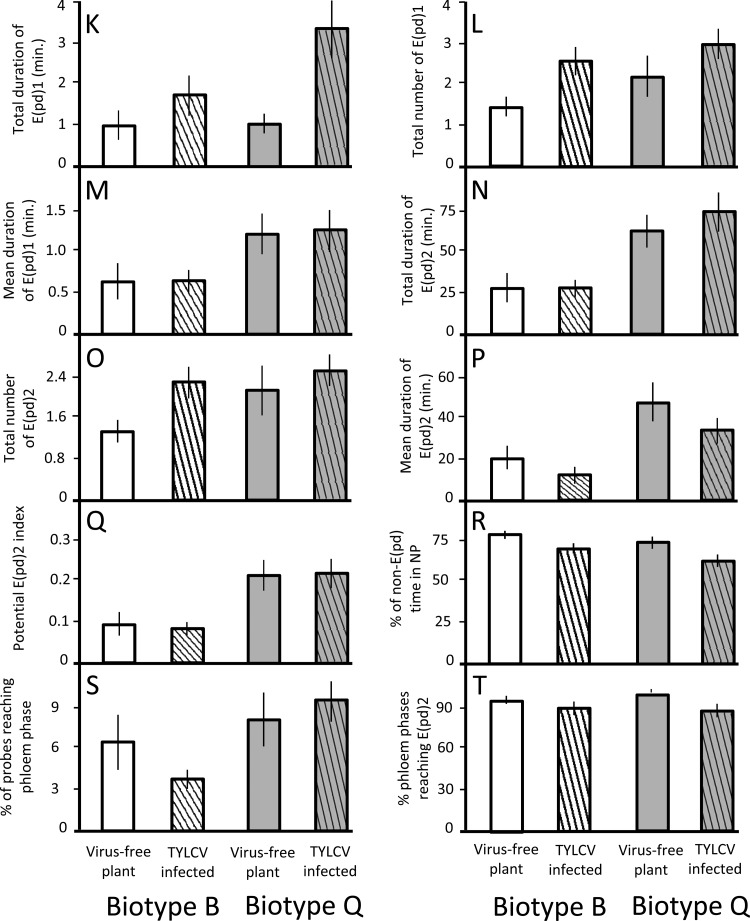
Whiteflies spent more time salivating into the sieve elements of TYLCV-infected versus virus-free plants. The total duration of E(pd)1 was 2.5× longer on infected plants, and the total number of E(pd)1 probes was 51% higher (parameters K and L) (Table 2 and Fig. 3). Whiteflies also fed more often on infected plants, with 38% more probes reaching E(pd)2 and 12% less non-E(pd) time spent resting (parameters O and R). Despite this, however, there was no difference in the total time spent feeding from TYLCV-infected versus virus-free plants (parameter N).
Fig 3.
Mean values ± standard errors for phloem EPG parameters K to T of B. tabaci biotype B and Q whiteflies feeding on TYLCV-infected and virus-free tomato plants (experiment 1). Parameter letters correspond to parameter letters in Table 2. White bars, biotype B feeding on virus-free plant; white striped bars, biotype B feeding on TYLCV-infected plant; gray bars, biotype Q feeding on virus-free plant; gray striped bars, biotype Q feeding on TYLCV-infected plant.
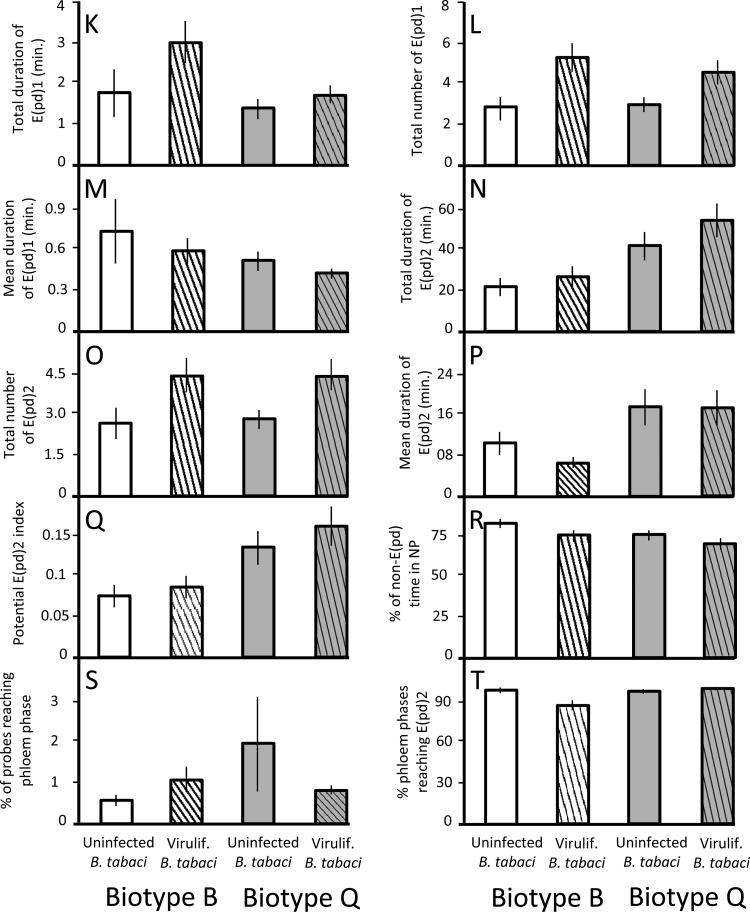
Biotype Q outperformed biotype B in 6 of 10 phloem-phase parameters in experiment 1. In terms of the E(pd)1 measurements (salivation into sieve elements) that Jiang et al. (36) classified as being essential for viral transmission, biotype Q had a 93% higher mean duration and a 60% higher total duration of time spent in E(pd)1 (parameters M and K) (Table 2 and Fig. 3). For the E(pd)2 measurements (ingestion of sieve element sap) that Jiang et al. (36) found to be essential for viral acquisition, the mean and total duration of time spent in E(pd)2 were 2.5× higher for biotype Q than for biotype B, and the potential E(pd)2 index was 2.4× higher (parameters P, N, and Q).
Experiment 2: do viruliferous and uninfected biotype B and Q whiteflies feed differently on virus-free tomato plants?
Viruliferous and uninfected whiteflies differed in their feeding behavior, and there were again differences between biotypes B and Q. For 9 of 10 feeding parameters, however, there was no interaction between biotype and whitefly infection status (all P > 0.05); the sole exception was parameter T.
(i) Nonphloem EPG measurements (parameters A to J).
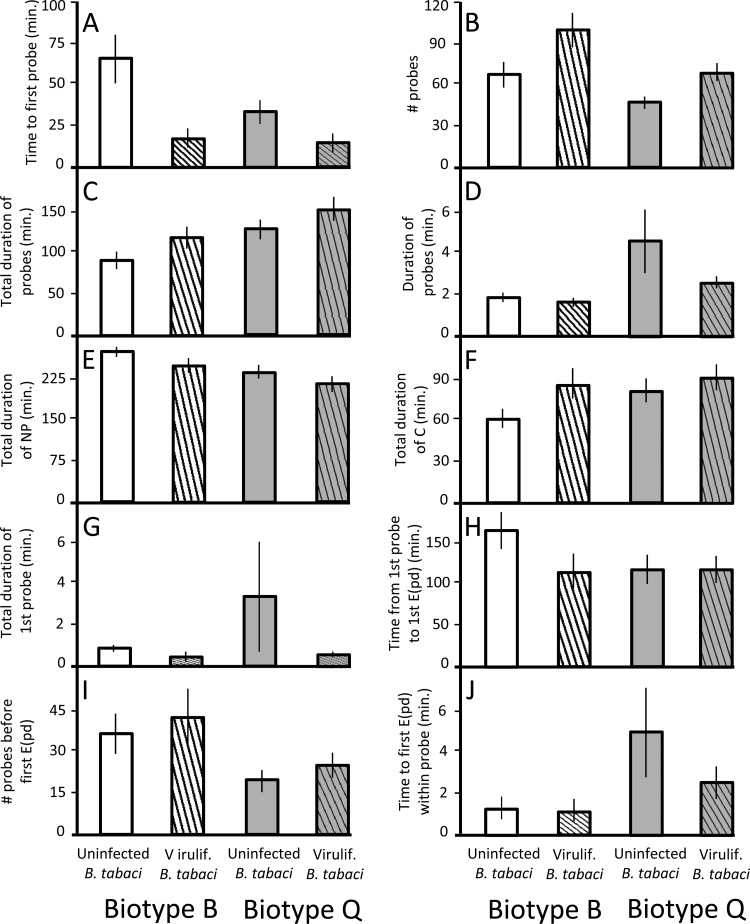
Viruliferous whiteflies fed more readily than did uninfected whiteflies. Uninfected whiteflies took three times longer to start probing virus-free plants, and viruliferous whiteflies attempted 47% more probes on virus-free plants (parameters A and B) (Table 3 and Fig. 4).
Fig 4.
Mean values ± standard errors for nonphloem EPG parameters A to J of viruliferous and uninfected B. tabaci biotype B and Q whiteflies feeding on virus-free tomato plants (experiment 2). Parameter letters correspond to parameter letters in Table 3. White bars, uninfected biotype B feeding on virus-free plant; white striped bars, viruliferous biotype B feeding on virus-free plant; gray bars, uninfected biotype Q feeding on virus-free plant; gray striped bars, viruliferous biotype Q feeding on virus-free plant.
As in experiment 1, whiteflies of biotypes B and Q differed in some aspects of their feeding behavior. Biotype Q whiteflies spent 32% more time probing than did biotype B whiteflies in experiment 2, with a mean probe duration that was more than double that of biotype B (parameters C and D). Biotype B whiteflies also spent 15% more time resting than did biotype Q whiteflies (parameter E).
(ii) Phloem EPG measurements (parameters K to T).
Viruliferous whiteflies spent more time than uninfected whiteflies salivating into sieve elements, a behavior identified previously by Jiang et al. (36) as being tightly correlated with vector-to-plant viral transmission. Viruliferous whiteflies spent 52% more time in E(pd)1 and had 72% more E(pd)1 episodes than did uninfected whiteflies (parameters K and L) (Table 4 and Fig. 5), and uninfected whiteflies spent more non-E(pd) time resting (parameter R). Although viruliferous whiteflies had 60% more feeding bouts [i.e., total number of E(pd)2] than did uninfected whiteflies, uninfected whiteflies had a higher percentage of probes reaching E(pd)2 (parameters O and T). The latter difference was driven entirely by the response of biotype B (i.e., a significant biotype-infection interaction) (Table 4 and Fig. 5T); viruliferous and uninfected B. tabaci biotype Q whiteflies did not differ in this parameter. Despite the differences in individual feeding behaviors, there was no effect of TYLCV infection on total time spent feeding (parameter N).
Fig 5.
Mean values ± standard errors for phloem EPG parameters K to T of viruliferous and uninfected B. tabaci biotype B and Q whiteflies feeding on virus-free tomato plants (experiment 2). Parameter letters correspond to parameter letters in Table 4. White bars, uninfected biotype B feeding on virus-free plant; white striped bars, viruliferous biotype B; gray bars, uninfected biotype Q; gray striped bars, viruliferous biotype Q.
Consistent with experiment 1, biotype Q outperformed biotype B on 4 of 10 phloem parameters. Biotype Q whiteflies fed for twice as long as biotype B whiteflies in experiment 2, had a greater mean duration of E(pd)2 probes and a higher E(pd)2 index, and had 6% more phloem probes reaching E(pd)2 (parameters N, P, Q, and T).
DISCUSSION
We found that Tomato yellow leaf curl virus directly and indirectly manipulates Bemisia tabaci feeding behavior. While Tomato spotted wilt virus was previously reported to manipulate the behavior of its thrips vector (10), this is the first report of manipulation by a virus that does not propagate in its vector and belongs to an exclusively plant-infecting group of viruses. Importantly, the altered behavior of viruliferous whiteflies seems likely to increase TYLCV transmission rates. Viruliferous B. tabaci spent more time salivating into sieve elements than did uninfected whiteflies, a behavior essential for viral inoculation of uninfected hosts (36). For the virus, the fitness benefits inherent in improved transmission thus provide an adaptive rationale for the observed behavioral modification (3).
The feeding behaviors altered by direct viral manipulation of B. tabaci may provide clues as to how TYLCV affects whitefly physiology. The fact that viruliferous whiteflies (i) attempted more probes, (ii) spent more time salivating, and (iii) engaged in more feeding bouts but (iv) spent the same total time in E(pd)2 suggests that TYLCV infection interferes with whitefly feeding. Specifically, the virus may reduce the ability of whitefly saliva to prevent sieve tube occlusion. This damage-induced plant response minimizes sap loss by sealing injured sieve tubes; in aphids, the watery saliva exuded during E1 counters this response and prevents occlusion (40). Although a similar sabotage of plant defense has not been reported for whiteflies, it occurs in a wide range of aphid species (41) and may be a “universal phenomenon” (41) in phloem-feeding hemipterans. Because B. tabaci salivation is essential for TYLCV transmission (36), greater time spent in E(pd)1, and a shift toward a larger number of short feeding bouts, should maximize viral inoculation. Interestingly, the deleterious effect of TYLCV infection on the number of probes reaching E(pd)2 was confined to biotype B whiteflies; it did not affect B. tabaci biotype Q. It is unclear why the two biotypes differed in their response to TYLCV vis-à-vis this parameter but not any others.
In addition to the direct viral manipulation, TYLCV-infected plants also appeared to be more attractive to B. tabaci: whiteflies probed infected plants more quickly and often, leading to a 22% increase in total probing time (Fig. 2C). Whiteflies also spent more time salivating into the sieve elements of infected plants and fed more often on them [i.e., probes reaching E(pd)2] (Fig. 3K and O). Similar virally mediated changes in host attractiveness to vectors have been documented for oats, potatoes, and tomatoes (5, 6, 8) and appear to be linked to the production of plant volatile blends characteristic of healthy plants (9). While preference for infected plants is often linked to higher vector fitness (5, 6, 8), we found that the apparent preference of B. tabaci for infected plants did not affect the total time spent feeding (Fig. 3N). While a longer recording period might have revealed a difference, our results thus provide no evidence that TYLCV-infected and virus-free plants differ in resource quality for whiteflies.
Bemisia tabaci biotypes B and Q differed in several aspects of their feeding behavior. In both experiments 1 and 2, biotype Q had significantly higher total and mean probe durations than did biotype B (Fig. 2C and D and 4C and D), while biotype B whiteflies spent more time resting than did biotype Q whiteflies (Fig. 2E and 4E). Biotype Q whiteflies also spent more time salivating into sieve elements (Fig. 3K) and fed for more than twice as long on phloem sap (Fig. 3N and 5N). This agrees with the results of two previous EPG-based studies (20, 39) that also found that, when feeding on tomato, biotype Q whiteflies spent more time on salivation and phloem ingestion than did biotype B whiteflies.
Although whiteflies of biotypes B and Q were affected similarly by the presence of TYLCV within themselves and the host plant, this does not mean that they are equally effective viral vectors. Although vector competence is defined by a wide array of factors (2), two of the most important are the vector's ability to acquire the virus from an infected host and, once viruliferous, its likelihood of transmitting the virus to a new host. Detailed EPG studies of TYLCV transmission in tomatoes by B. tabaci biotype B (36) found that viral acquisition was most strongly correlated with the total time spent feeding on sap [i.e., E(pd)2] (Fig. 3N and 5N). That same study found that viral transmission was most strongly correlated with the total time spent salivating into sieve elements [i.e., E(pd)1] (Fig. 3K and 5K). Our work confirms the results of two previous studies (20, 39), finding that biotype Q whiteflies spent more total time feeding than did biotype B whiteflies. In addition, biotype Q whiteflies spent more total time salivating into the sieve element sap in experiment 1; those previous studies also found the same result (20, 39). Our work, in combination with previous research, thus provides a mechanistic basis for the finding that biotype Q outperforms biotype B in both the acquisition and transmission of TYLCV on tomato (42).
The fact that B. tabaci biotypes B and Q responded similarly to infected plants is striking in light of the biotype-specific impact of TYLCV on whitefly fitness. Because feeding on infected tomatoes perturbs the cell cycle and metabolism of biotype B whiteflies (43), thus decreasing their survival and fecundity (44, 45), this biotype's attraction to an inferior host should affect population growth (9). Although we found no difference in whitefly feeding, recent work comparing the performance of B. tabaci biotypes B and Q on TYLCV-infected tomatoes confirmed the negative effect of infection on biotype B but found that feeding on infected tomatoes actually increased the survival and fecundity of biotype Q whiteflies (H. P. Pan, D. Chu, B. M. Liu, X. B. Shi, W. Xie, Y. Carriere, X. C. Li, and Y. J. Zhang, submitted for publication). The “mutualistic” relationship between TYLCV and B. tabaci biotype Q is notable in light of our finding that B. tabaci biotype B nonetheless appears attracted (Fig. 2C) to TYLCV-infected hosts of inferior quality. This suggests the possibility that indirect viral manipulation benefits TYLCV's “preferred” vector both directly (via increased fecundity and survival of B. tabaci biotype Q) and indirectly (via altered biotype B-biotype Q competitive interactions [Pan et al., submitted]). In China and elsewhere, the asymmetric impacts of TYLCV on B. tabaci biotypes B and Q may thus underlie both the TYLCV-biotype Q association (30) and the rapid displacement of one biotype by another (29, 30, 46).
The fact that TYLCV both directly and indirectly manipulates its whitefly vector has far-reaching implications. While the observation that Tomato spotted wilt virus directly manipulates its vector (10) could be ascribed to the fact that it is a member of the primarily animal-infecting Bunyaviridae, TYLCV is a member of the exclusively plant-infecting Geminiviridae. The latter virus is also nonpropagative inside B. tabaci, eliminating the potential for intravector selection for viral strains capable of direct manipulation (but see reference 47). The fact that viral manipulation occurred despite these two points suggests that a similar manipulation of phloem-sucking insect vectors by viruses may be much more common than expected, perhaps even constituting the rule rather than the exception. The fact that both B. tabaci biotypes responded similarly to manipulation by a virus that harms biotype B (43, 45) but helps biotype Q (Pan et al., submitted) may also help explain the extraordinarily rapid replacement of biotype B by biotype Q throughout China and the concomitant increase in TYLCV distribution and damage. If so, the results of our research highlight the importance of vector-virus interactions in shaping population- and community-level processes (3).
ACKNOWLEDGMENTS
This work was funded by the National Science Fund for Distinguished Young Scholars (grant 31025020), the Key Project of Chinese National Programs for Fundamental Research and Development (grant 2013CB127602), the National Natural Science Foundation of China (grant 31171857), and the Beijing Key Laboratory for Pest Control and Sustainable Cultivation of Vegetables.
We thank G. Walker, J. Obrycki, D. Potter, A. Simon, F. M. Yan, X. G. Jiao, W. F. Tjallingii, and four anonymous reviewers for their helpful comments. We thank X. P. Zhou for providing TYLCV clones.
Footnotes
Published ahead of print 13 February 2013
REFERENCES
- 1. Fereres A, Moreno A. 2009. Behavioural aspects influencing plant virus transmission by homopteran insects. Virus Res. 141:158–168 [DOI] [PubMed] [Google Scholar]
- 2. Stafford CA, Walker GP, Ullman DE. 2012. Hitching a ride: vector feeding and virus transmission. Commun. Integr. Biol. 5:43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lefevre T, Thomas F. 2008. Behind the scene, something else is pulling the strings: emphasizing parasitic manipulation in vector-borne diseases. Infect. Genet. Evol. 8:504–519 [DOI] [PubMed] [Google Scholar]
- 4. Blanc S, Uzest M, Drucker M. 2011. New research horizons in vector-transmission of plant viruses. Curr. Opin. Microbiol. 14:483–491 [DOI] [PubMed] [Google Scholar]
- 5. Montllor C, Gildow F. 1986. Feeding responses of two grain aphids to Barley yellow dwarf virus-infected oats. Entomol. Exp. Appl. 42:63–69 [Google Scholar]
- 6. Alvarez A, Garzo E, Verbeek M, Vosman B, Dicke M, Tjallingii W. 2007. Infection of potato plants with potato leafroll virus changes attraction and feeding behaviour of Myzus persicae. Entomol. Exp. Appl. 125:135–144 [Google Scholar]
- 7. Eigenbrode S, Ding H, Shiel P, Berger P. 2002. Volatiles from potato plants infected with potato leafroll virus attract and arrest the virus vector, Myzus persicae (Homoptera: Aphididae). Proc. R. Soc. Lond. B 269:455–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maris P, Joosten N, Goldbach R, Peters D. 2004. Tomato spotted wilt virus infection improves host suitability for its vector Frankliniella occidentalis. Phytopathology 94:706–711 [DOI] [PubMed] [Google Scholar]
- 9. Mauck K, De Moraes C, Mescher M. 2010. Deceptive chemical signals induced by a plant virus attract insect vectors to inferior hosts. Proc. Natl. Acad. Sci. U. S. A. 107:3600–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stafford CA, Walker GP, Ullman DE. 2011. Infection with a plant virus modifies vector feeding behavior. Proc. Natl. Acad. Sci. U. S. A. 108:9350–9355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grimstad P, Ross Q, Craig G. 1980. Aedes triseriatus (Diptera: Culicidae) and La Crosse virus II. Modification of mosquito feeding behavior by virus infection. J. Med. Entomol. 17:1–7 [DOI] [PubMed] [Google Scholar]
- 12. Navot N, Pichersky E, Zeidan M, Zamir D, Czosnek H. 1991. Tomato yellow leaf curl virus: a whitefly-transmitted geminivirus with a single genomic component. Virology 185:151–161 [DOI] [PubMed] [Google Scholar]
- 13. Czosnek H, Laterrot H. 1997. A worldwide survey of tomato yellow leaf curl viruses. Arch. Virol. 142:1391–1406 [DOI] [PubMed] [Google Scholar]
- 14. Cohen S, Harpaz I. 1964. Periodic, rather than continual acquisition of a new tomato virus by its vector, the tobacco whitefly (Bemisia tabaci Gennadius). Entomol. Exp. Appl. 7:155–166 [Google Scholar]
- 15. Cohen S, Nitzany F. 1966. Transmission and host range of the Tomato yellow leaf curl virus. Phytopathology 56:1127–1131 [Google Scholar]
- 16. Ghanim M, Rosell R, Campbell L, Czosnek H, Brown J, Ullman D. 2001. Digestive, salivary, and reproductive organs of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) B type. J. Morphol. 248:22–40 [DOI] [PubMed] [Google Scholar]
- 17. Czosnek H, Ghanim M, Ghanim M. 2002. The circulative pathway of begomoviruses in the whitefly vector Bemisia tabaci—insights from studies with Tomato yellow leaf curl virus. Ann. Appl. Biol. 140:215–231 [Google Scholar]
- 18. Ghanim M, Morin S, Czosnek H. 2001. Rate of Tomato yellow leaf curl virus translocation in the circulative transmission pathway of its vector, the whitefly Bemisia tabaci. Phytopathology 91:188–196 [DOI] [PubMed] [Google Scholar]
- 19. Brown J, Frohlich D, Rosell R. 1995. The sweetpotato or silverleaf whiteflies: biotypes of Bemisia tabaci or a species complex? Annu. Rev. Entomol. 40:511–534 [Google Scholar]
- 20. Liu BM, Yan FM, Chu D, Pan HP, Jiao XG, Xie W, Wu QJ, Wang SL, Xu BY, Zhou XG, Zhang YJ. 2012. Difference in feeding behaviors of two invasive whiteflies on host plants with different suitability: implication for competitive displacement. Int. J. Biol. Sci. 8:697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pan HP, Li XC, Ge DQ, Wang SL, Wu QJ, Xie W, Jiao XG, Chu D, Liu BM, Xu BY, Zhang YJ. 2012. Factors affecting population dynamics of maternally transmitted endosymbionts in Bemisia tabaci. PLoS One 7:e30760 doi:10.1371/journal.pone.0030760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brown J. 2010. Phylogenetic biology of the Bemisia tabaci sibling species group, p 31–67 In Stansly PA, Naranjo SE. (ed), Bemisia: bionomics and management of a global pest. Springer, New York, NY [Google Scholar]
- 23. De Barro P, Liu S, Boykin L, Dinsdale A. 2011. Bemisia tabaci: a statement of species status. Annu. Rev. Entomol. 56:1–19 [DOI] [PubMed] [Google Scholar]
- 24. Dinsdale A, Cook L, Riginos C, Buckley Y, De Barro P. 2010. Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species-level genetic boundaries. Ann. Entomol. Soc. Am. 103:196–208 [Google Scholar]
- 25. Perring TM. 2001. The Bemisia tabaci species complex. Crop Prot. 20:725–737 [Google Scholar]
- 26. Jones D. 2003. Plant viruses transmitted by whiteflies. Eur. J. Plant Pathol. 109:195–219 [Google Scholar]
- 27. Chu D, Zhang YJ, Brown JK, Cong B, Xu BY, Wu QJ, Zhu GR. 2006. The introduction of the exotic Q biotype of Bemisia tabaci (Gennadius) from the Mediterranean region into China on ornamental crops. Fla. Entomol. 89:168–174 [Google Scholar]
- 28. Luo C, Yao Y, Wang RJ, Yan FM, Hu DX, Zhang ZL. 2002. The use of mitochondrial cytochrome oxidase I (mtCOI) gene sequences for the identification of biotype of Bemisia tabaci (Gennadius) in China. Acta Entomol. Sin. 45:759–763 [Google Scholar]
- 29. Chu D, Wan F, Zhang Y, Brown J. 2010. Change in the biotype composition of Bemisia tabaci in Shandong Province of China from 2005 to 2008. Environ. Entomol. 39:1028–1036 [DOI] [PubMed] [Google Scholar]
- 30. Pan HP, Chu D, Ge DQ, Wang SL, Wu QJ, Xie W, Jiao XG, Liu BM, Yang X, Yang NN, Su Q, Xu BY, Zhang YJ. 2011. Further spread of and domination by Bemisia tabaci (Hemiptera: Aleyrodidae) biotype Q on field crops in China. J. Econ. Entomol. 104:978–985 [DOI] [PubMed] [Google Scholar]
- 31. Wu JB, Dai FM, Zhou XP. 2006. First report of Tomato yellow leaf curl virus in China. Plant Dis. 90:1359 doi:10.1094/PD-90-1359C [DOI] [PubMed] [Google Scholar]
- 32. Ji Y, Xiong R, Cheng Z, Zhou T, Zhao T, Yu W, Fan Y, Zhou Y. 2008. Molecular diagnosis of tomato yellow leaf curl disease in Jiangsu Province. Acta Hortic. Sin. 35:1815–1818 [Google Scholar]
- 33. Mugiira RB, Liu SS, Zhou X. 2008. Tomato yellow leaf curl virus and Tomato leaf curl Taiwan virus invade south-east coast of China. J. Phytopathol. 156:217–221 [Google Scholar]
- 34. Ghanim M, Sobol I, Ghanim M, Czosnek H. 2007. Horizontal transmission of begomoviruses between Bemisia tabaci biotypes. Arthropod-Plant Interact. 1:195–204 [Google Scholar]
- 35. Xie Y, Zhou XP, Zhang ZK, Qi YJ. 2002. Tobacco curly shoot virus isolated in Yunnan is a distinct species of begomovirus. Chin. Sci. Bull. 47:197–200 [Google Scholar]
- 36. Jiang Y, de Blas C, Barrios L, Fereres A. 2000. Correlation between whitefly (Homoptera: Aleyrodidae) feeding behavior and transmission of Tomato yellow leaf curl virus. Ann. Entomol. Soc. Am. 93:573–579 [Google Scholar]
- 37. Tjallingii WF. 1978. Electronic recording of penetration behaviour by aphids. Entomol. Exp. Appl. 24:721–730 [Google Scholar]
- 38. Tjallingii WF. 1985. Electrical nature of recorded signals during stylet penetration by aphids. Entomol. Exp. Appl. 38:177–186 [Google Scholar]
- 39. Jiang Y, Lei H, Collar J, Martin B, Muniz M, Fereres A. 1999. Probing and feeding behavior of two distinct biotypes of Bemisia tabaci (Homoptera: Aleyrodidae) on tomato plants. J. Econ. Entomol. 92:357–366 [Google Scholar]
- 40. Will T, Tjallingii WF, Thönnessen A, van Bel AJ. 2007. Molecular sabotage of plant defense by aphid saliva. Proc. Natl. Acad. Sci. U. S. A. 104:10536–10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Will T, Kornemann SR, Furch AC, Tjallingii WF, van Bel AJ. 2009. Aphid watery saliva counteracts sieve-tube occlusion: a universal phenomenon? J. Exp. Biol. 212:3305–3312 [DOI] [PubMed] [Google Scholar]
- 42. Pan H, Chu D, Yan W, Su Q, Liu B, Wang S, Wu Q, Xie W, Jiao X, Li R, Yang N, Yang X, Xu B, Brown JK, Zhou X, Zhang Y. 2012. Rapid spread of Tomato yellow leaf curl virus in China is aided differentially by two invasive whiteflies. PLoS One 7:e34817 doi:10.1371/journal.pone.0034817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Luan JB, Li JM, Varela N, Wang YL, Li FF, Bao YY, Zhang CX, Liu SS, Wang XW. 2011. Global analysis of the transcriptional response of whitefly to Tomato yellow leaf curl China virus reveals the relationship of coevolved adaptations. J. Virol. 85:3330–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Czosnek H, Ghanim M, Morin S, Rubinstein G, Fridman V, Zeidan M. 2001. Whiteflies: vectors, and victims (?), of geminiviruses. Adv. Virus Res. 57:291–322 [DOI] [PubMed] [Google Scholar]
- 45. Rubinstein G, Czosnek H. 1997. Long-term association of Tomato yellow leaf curl virus with its whitefly vector Bemisia tabaci: effect on the insect transmission capacity, longevity and fecundity. J. Gen. Virol. 78:2683–2689 [DOI] [PubMed] [Google Scholar]
- 46. Teng X, Wan FH, Chu D. 2010. Bemisia tabaci biotype Q dominates other biotypes across China. Fla. Entomol. 93:363–368 [Google Scholar]
- 47. Seal SE, vanden Bosch F, Jeger MJ. 2006. Factors influencing begomovirus evolution and their increasing global significance: implications for sustainable control. Crit. Rev. Plant Sci. 25:23–46 [Google Scholar]