Abstract
Maribavir (MBV) inhibits Epstein-Barr virus (EBV) replication and the enzymatic activity of the viral protein kinase BGLF4. MBV also inhibits expression of multiple EBV transcripts during EBV lytic infection. Here we demonstrate, with the use of a BGLF4 knockout virus, that effects of MBV on transcription take place primarily through inhibition of BGLF4. MBV inhibits viral genome copy numbers and infectivity to levels similar to and exceeding levels produced by BGLF4 knockout virus.
TEXT
Although a number of antiviral drugs are effective inhibitors of Epstein-Barr virus (EBV) replication and are used empirically, none is of proven effectiveness for treatment of EBV infection (1, 2). Maribavir (MBV), which is in late-stage clinical trials for use against human cytomegalovirus (HCMV) infection in allogeneic stem cell and bone-marrow transplant recipients (3, 4), is of special interest because it is also a potent inhibitor of EBV replication (5–7). In stem-cell and organ transplant recipients, EBV infection poses the hazard of generating B-cell lymphomas that are ultimately fatal. While no drugs are currently approved for treatment of EBV disease, several that inhibit EBV are available, and these can be divided into two main classes: those that target the viral DNA polymerase and those that function independently of it (8–12). Acyclic nucleoside and phosphonated nucleotide analogs, as well as pyrophosphate analogs, all target the viral polymerase.
A new class of HCMV inhibitors, benzimidazole compounds, with more specific antiviral properties and fewer adverse side effects, blocked HCMV DNA maturation and encapsidation processes and led to the design of 1-H-β-l-ribofuranoside-2-isopropylamino-5,6-dichlorobenzimidazole (maribavir [MBV]) (13–20). Unlike its parent compound, which inhibits HCMV replication but not EBV replication, MBV inhibits both (7, 21). Inhibitory effects of MBV are produced mainly through inhibition of the HCMV and EBV protein kinases (PK) (21–24). Previous phase 3 studies with a dosage of 100 mg twice a day (BID) did not have sufficient activity to prevent HCMV disease, but the safety profile and data from case studies suggested that higher doses would be clinically active (3). MBV is now in new phase 2 trials at doses of 400, 800, and 1,200 mg BID (3).
Maribavir selectively inhibits the HCMV protein kinase, UL97, determined by direct inhibition of kinase activity in vitro and by genetic mapping of the MBV-resistant phenotype (21). MBV also inhibits the EBV protein kinase (BGLF4), resulting in inhibition of phosphorylation of the EBV DNA processivity factor BMRF1, but does not seem to act directly on the EBV kinase in vitro (7, 24).
We have recently found that MBV also inhibits expression of multiple EBV transcripts, in contrast to acyclovir (ACV), which has little effect on EBV RNAs. Thus, MBV has a unique dual effect on viral DNA transcription as well as replication (25). In this study, we find that the inhibitory profile of MBV transcripts is similar to that produced by mutant EBV in which PK expression and activity have been knocked out (26). Thus, the results suggest that MBV largely affects EBV transcript levels through inhibition of BGLF4.
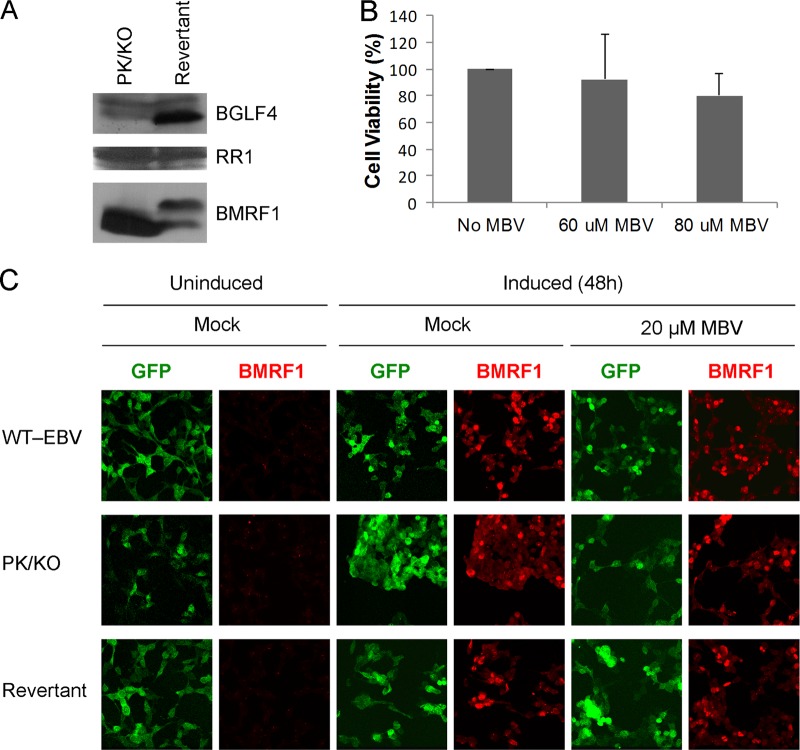
To determine if the profile of viral transcripts produced by MBV is mediated by the viral kinase, we utilized BGLF4 knockout (KO) (dBGLF4/NeoST) and revertant (dBGLF4/NeoSt/R) viruses constructed and characterized by Murata et al. (26). 293 cells maintaining wild-type (WT), BGLF4 knockout, and revertant EBV genomes (27) were induced into the lytic cycle by transfecting the EBV immediate early transactivator BZLF1, and lysates were probed by Western blotting after 48 h. Figure 1A demonstrates that expression of BGLF4 is abolished in the PK knockout but not the revertant cell line. Expression of the early EBV ribonucleotide reductase large subunit (RR1), used as a control, was unaffected in both cell lines. In contrast, phosphorylation of BMRF1, used as an indicator of BGLF4 activity, was detected only upon expression of BGLF4 (upper band, BMRF1 panel). Immunofluorescence staining of induced cell lines shows efficient viral induction as indicated by the detection of BMRF1 (Fig. 1C). These findings confirm nonexpression of BGLF4 in the knockout virus, inhibition of phosphorylation of its natural substrate BMRF1, and efficient induction of the lytic cycle.
Fig 1.
Protein expression in induced PK knockout and revertant cell lines. (A) Viral reactivation in PK/KO and revertant cell lines was induced with EBV BZLF1 for 48 h. Total lysates were immunoblotted with antibodies against BGLF4, RR1, and BMRF1. The faint bands in the BGLF4 panel are likely nonspecific. (B) Cell viability assays of 293 EBV WT cells. Cells were treated with indicated amounts of MBV for 48 h, and viable cells were counted. Results indicate no loss in viability with MBV concentrations less than 80 μM. (C) Immunofluorescence staining of induced PK knockout and revertant cell lines. Cell lines containing green fluorescent protein (GFP)-labeled EBV genome are shown in green. BMRF1 staining is visualized in red. BMRF1 is not detected in uninduced samples, but efficient induction, reflected by BMRF1 expression, is observed at 48 h postinduction and is unaffected by MBV.
To measure the effects of MBV and the PK knockout virus on EBV transcripts, we profiled EBV mRNA using real-time quantitative PCR (qPCR) as described before (25). Cell viability assays were performed with MBV concentrations up to 80 μM; no evidence of toxicity for the cell lines used was detected below 80 μM MBV (Fig. 1B). Twenty micromolar MBV is used in these studies as before and was not toxic at the concentration used.
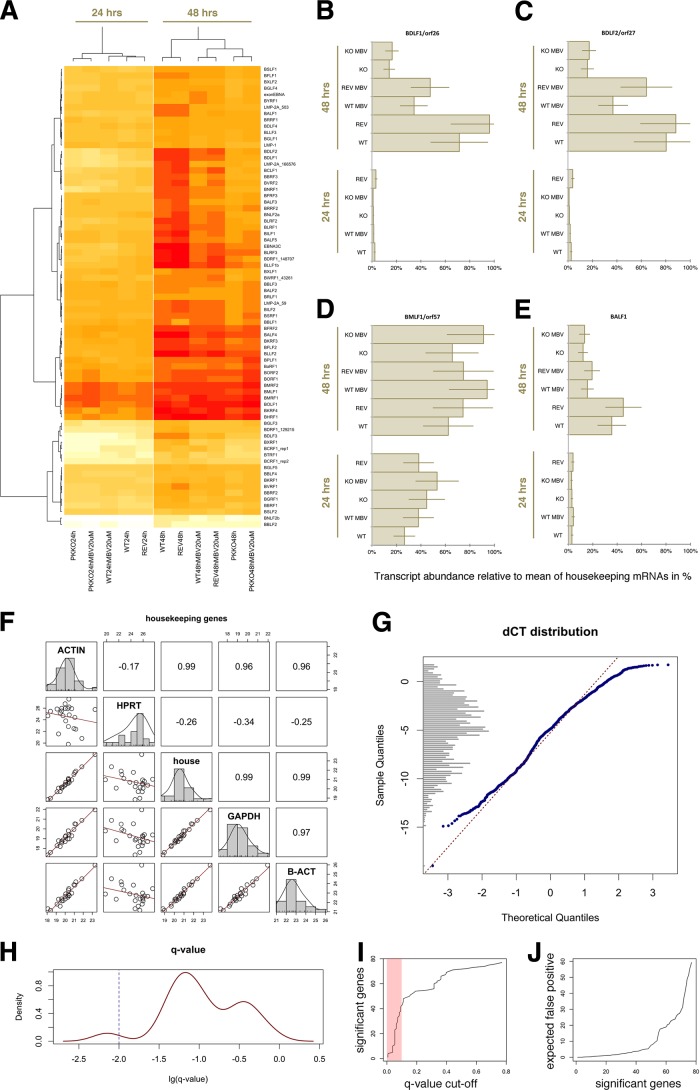
As expected, most EBV mRNAs were maximally induced 48 h after lytic cycle induction with BZLF1, compared with findings at 24 h (Fig. 2A). WT and revertant (REV) virus showed the same pattern of gene expression. MBV reduced overall EBV mRNA levels at 48 h, except for immediate early genes, such as BMLF1 (Fig. 2D). The PK KO virus exhibited significantly reduced gene expression at 48 h compared with findings for WT and revertant viruses. We observed two classes composed of late transcripts, i.e., those genes not detected at 24 h but strongly upregulated at 48 h. Class I RNAs, such as BALF1, were equally inhibited by either MBV or PK KO (Fig. 2E). Class II RNAs, although inhibited by MBV compared with results with WT or REV, were even more strongly inhibited with the PK KO virus alone (Fig. 2B and C). All data were normalized to the mean for three housekeeping genes, which correlated well across all experiments (Fig. 2F). The relative expression levels (delta cycle threshold [dCT]) were normally distributed (Fig. 2G), which allowed use of a t test for individual comparisons. We adjusted for multiple comparisons, since we used multiple genes, using q-value method (28) (Fig. 2H to J). We interpret these data to mean that MBV inhibits the kinase activity of PK and thus the kinase-dependent functions of PK, but there may also be kinase-independent functions of PK, which would not be affected. In terms of temporal viral mRNA transcription, the PK KO virus is more inhibited than wild-type virus by treatment with MBV.
Fig 2.
Transcription profiles of EBV mRNAs at 24 and 48 h after induction for different viruses, wild-type (WT), revertant (REV), and PK knockout (PKKO), in the presence of 20 μM maribavir or with mock treatment. (A) Heat map of relative mRNA levels (dCT), color coded: red corresponds to the highest, orange to the middle, and yellow to the lowest mRNA levels across the experiments. Raw CT values were normalized to geometric means for three cellular mRNAs to yield dCT values. Genes are listed on the vertical axis and conditions on the horizontal axis. (B to E) Individual transcription profiles representing the main patterns of changes in mRNA levels. Error bars represent the most conservative estimates of biological variation: twice % standard deviation of dCT for housekeeping genes (actin, beta actin, and gapdh) measured across n = 11 independent biological replicates. (F) Correlation of housekeeping mRNAs to each other and the geometric means, labeled “house,” of the “ACTIN,” “GAPDH,” and “B-ACT” qPCR assays. Shown are pairwise scatter plots with a fitted linear regression line (red) on the lower left, univariate CT distribution as a histogram with overlaid kernel density estimate on the diagonal, and pairwise Pearson correlation coefficient on the upper left. (G) Quantile-quantile plot comparing the distribution of dCT on the vertical to expected values under an assumption of normal distribution. A histogram is shown on the vertical axis. Also shown is a linear regression fit, which indicates that the data follow a normal distribution and thus the t test is appropriate for individual comparisons. (H) Distribution of log (lg) q value tests the ability of individual primers to distinguish between experimental conditions; the q value is a measure of statistical significance adjusted for multiple comparisons using a false discovery rate approach (28). The blue line indicates q = 0.01. (I) Number of significant genes for a given q-value cutoff: 40 genes had q ≤ 0.1, as indicated by red shading (corresponding to P ≤ 0.05 by single comparison). (J) Expected number of false-positive genes on the vertical axis given a set of significant genes. We expect ≤4 false-positive genes in the top 40 genes that individually discriminate among the experimental conditions: MBV exposure, viral PK status, and time postinduction.
Since knockout of BGLF4 had been shown to decrease viral infectivity 10-fold (26), we next determined whether MBV produced levels of viral genome copies and infectivity similar to those produced by knocking out BGLF4. 293EBV+ WT and PK knockout cell lines were induced with BZLF1, and supernatant fluids were collected at 72 h. Intracellular genome copies were determined as described before (29). Figure 3A demonstrates that MBV treatment of WT virus results in a 74% (±6.9%) reduction in genome copy numbers. Compared with results for the WT, induction of the PK knockout cell line yielded 64% (±4.7%) of viral genome copies. Treatment of PK KO cells with MBV reduced genome copies by an additional 25% (±4.4%). These results demonstrate that the PK KO virus is deficient in making viral genome copies and suggests that MBV also partially inhibits viral replication through a mechanism distinct from the viral PK activity.
Fig 3.
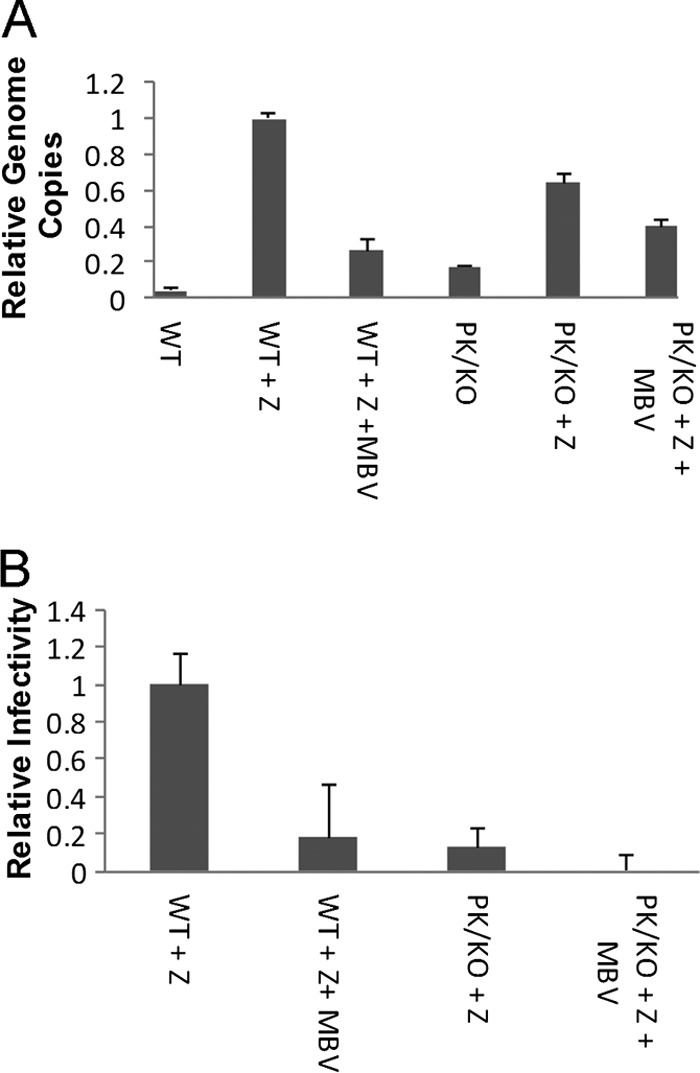
Viral genome copies and infectivity. (A) Intracellular genome copies relative to those for induced WT virus from triplicate samples. Genome copies are normalized to GAPDH. (B) Relative infectivity from supernatant fluids. Error bars represent standard errors of means.
Supernatant fluids were collected from 293EBV+ and PK KO cell lines, and titers of infectious virus were determined by infecting Raji cells (29). MBV resulted in an 82% decrease in viral infectivity—similar to the level observed in untreated PK knockout virus-infected cells (Fig. 3B). MBV treatment of the PK knockout line further decreased viral infectivity. This observation suggests that about half of the PK knockout genome copies are not released from the cell or are released but noninfectious. This is consistent with our earlier findings indicating that the viral PK is necessary for efficient viral egress (29).
These findings demonstrate that MBV can efficiently inhibit viral transcription (25), genome replication, and infectivity, producing pleiotropic effects which are similar to those observed with viral PK knockout virus. These data are congruent with the function of MBV working largely but not entirely through inhibition of BGLF4. MBV likely also inhibits residual infectivity that is still observed in the absence of PK-mediated effects (Fig. 3B). Conversely, transcript levels of the PK KO virus are severely attenuated compared with those of the wild-type virus. Since BGLF4 has as many as 20 viral targets (30), MBV may also affect downstream targets indirectly.
ACKNOWLEDGMENTS
We thank T. Tsurumi and T. Murata for generously providing the PK knockout and revertant cell lines.
This work was supported by NIH grants CA019014 to J.S.P. and D.P.D., CA1095180 to J.S.P., and supplemental funding for AIDS malignancies to the Lineberger Comprehensive Cancer Center (CA016086).
Footnotes
Published ahead of print 28 February 2013
REFERENCES
- 1. Biron KK. 2006. Antiviral drugs for cytomegalovirus diseases. Antiviral Res. 71:154–163 [DOI] [PubMed] [Google Scholar]
- 2. Lowance D, Neumayer HH, Legendre CM, Squifflet JP, Kovarik J, Brennan PJ, Norman D, Mendez R, Keating MR, Coggon GL, Crisp A, Lee IC. 1999. Valacyclovir for the prevention of cytomegalovirus disease after renal transplantation. International Valacyclovir Cytomegalovirus Prophylaxis Transplantation Study Group. N. Engl. J. Med. 340:1462–1470 [DOI] [PubMed] [Google Scholar]
- 3. ViroPharma Incorporated 2012. ViroPharma announces initiation of clinical studies to evaluate maribavir for treatment of cytomegalovirus (CMV) infection. ViroPharma Incorporated, Exton, PA [Google Scholar]
- 4. Winston DJ, Young JA, Pullarkat V, Papanicolaou GA, Vij R, Vance E, Alangaden GJ, Chemaly RF, Petersen F, Chao N, Klein J, Sprague K, Villano SA, Boeckh M. 2008. Maribavir prophylaxis for prevention of cytomegalovirus infection in allogeneic stem cell transplant recipients: a multicenter, randomized, double-blind, placebo-controlled, dose-ranging study. Blood 111:5403–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Price NB, Prichard MN. 2011. Progress in the development of new therapies for herpesvirus infections. Curr. Opin. Virol. 1:548–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Williams SL, Hartline CB, Kushner NL, Harden EA, Bidanset DJ, Drach JC, Townsend LB, Underwood MR, Biron KK, Kern ER. 2003. In vitro activities of benzimidazole D- and L-ribonucleosides against herpesviruses. Antimicrob. Agents Chemother. 47:2186–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zacny VL, Gershburg E, Davis MG, Biron KK, Pagano JS. 1999. Inhibition of Epstein-Barr virus replication by a benzimidazole L-riboside: novel antiviral mechanism of 5,6-dichloro-2-(isopropylamino)-1-beta-L-ribofuranosyl-1H-benzimidazole. J. Virol. 73:7271–7277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gershburg E, Hong K, Pagano JS. 2004. Effects of maribavir and selected indolocarbazoles on Epstein-Barr virus protein kinase BGLF4 and on viral lytic replication. Antimicrob. Agents Chemother. 48:1900–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ljungman P. 2002. Prevention and treatment of viral infections in stem cell transplant recipients. Br. J. Haematol. 118:44–57 [DOI] [PubMed] [Google Scholar]
- 10. Pagano JS. 1995. Epstein-Barr virus: therapy of active and latent infection, p 155–195 In Jeffries DJ, De Clercq E. (ed), Antiviral chemotherapy. John Wiley & Sons Ltd., Hoboken, NJ [Google Scholar]
- 11. Pagano JS, Sixbey JW, Lin JC. 1983. Acyclovir and Epstein-Barr virus infection. J. Antimicrob. Chemother. 12(Suppl. B):113–121 [DOI] [PubMed] [Google Scholar]
- 12. Slifkin M, Doron S, Snydman DR. 2004. Viral prophylaxis in organ transplant patients. Drugs 64:2763–2792 [DOI] [PubMed] [Google Scholar]
- 13. Bogner E. 2002. Human cytomegalovirus terminase as a target for antiviral chemotherapy. Rev. Med. Virol. 12:115–127 [DOI] [PubMed] [Google Scholar]
- 14. Dittmer A, Drach JC, Townsend LB, Fischer A, Bogner E. 2005. Interaction of the putative human cytomegalovirus portal protein pUL104 with the large terminase subunit pUL56 and its inhibition by benzimidazole-D-ribonucleosides. J. Virol. 79:14660–14667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Evers DL, Komazin G, Ptak RG, Shin D, Emmer BT, Townsend LB, Drach JC. 2004. Inhibition of human cytomegalovirus replication by benzimidazole nucleosides involves three distinct mechanisms. Antimicrob. Agents Chemother. 48:3918–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kern ER, Hartline CB, Rybak RJ, Drach JC, Townsend LB, Biron KK, Bidanset DJ. 2004. Activities of benzimidazole D- and L-ribonucleosides in animal models of cytomegalovirus infections. Antimicrob. Agents Chemother. 48:1749–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krosky PM, Underwood MR, Turk SR, Feng KW, Jain RK, Ptak RG, Westerman AC, Biron KK, Townsend LB, Drach JC. 1998. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J. Virol. 72:4721–4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Prichard MN. 2009. Function of human cytomegalovirus UL97 kinase in viral infection and its inhibition by maribavir. Rev. Med. Virol. 19:215–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prichard MN, Frederick SL, Daily S, Borysko KZ, Townsend LB, Drach JC, Kern ER. 2011. Benzimidazole analogs inhibit human herpesvirus 6. Antimicrob. Agents Chemother. 55:2442–2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Underwood MR, Harvey RJ, Stanat SC, Hemphill ML, Miller T, Drach JC, Townsend LB, Biron KK. 1998. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Biron KK, Harvey RJ, Chamberlain SC, Good SS, Smith IA, Davis MG, Talarico CL, Miller WH, Ferris R, Dornsife RE, Stanat SC, Drach JC, Townsend LB, Koszalka GW. 2002. Potent and selective inhibition of human cytomegalovirus Replication by 1263W94, a benzimidazole L-riboside with a unique mode of action. Antimicrob. Agents Chemother. 46:2365–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Biron KK. 2011. Maribavir: a novel bensimidole ribonucleoside for the prevention and treatment of cytomegalovirus disease antiviral drug strategies. In De Clercq Erik. (ed), Antiviral drug strategies, 1st ed Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany: doi:10.1002/9783527635955.ch9 [Google Scholar]
- 23. Gershburg E, Pagano JS. 2005. Epstein-Barr virus infections: prospects for treatment. J. Antimicrob. Chemother. 56:277–281 [DOI] [PubMed] [Google Scholar]
- 24. Gershburg E, Pagano JS. 2002. Phosphorylation of the Epstein-Barr virus (EBV) DNA polymerase processivity factor EA-D by the EBV-encoded protein kinase and effects of the L-riboside benzimidazole 1263W94. J. Virol. 76:998–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang FZ, Roy D, Gershburg E, Whitehurst CB, Dittmer DP, Pagano JS. 2009. Maribavir inhibits Epstein-Barr virus transcription in addition to viral DNA replication. J. Virol. 83:12108–12117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murata T, Isomura H, Yamashita Y, Toyama S, Sato Y, Nakayama S, Kudoh A, Iwahori S, Kanda T, Tsurumi T. 2009. Efficient production of infectious viruses requires enzymatic activity of Epstein-Barr virus protein kinase. Virology 389:75–81 [DOI] [PubMed] [Google Scholar]
- 27. Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. U. S. A. 95:8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 100:9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gershburg E, Raffa S, Torrisi MR, Pagano JS. 2007. Epstein-Barr virus-encoded protein kinase (BGLF4) is involved in production of infectious virus. J. Virol. 81:5407–5412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu J, Liao G, Shan L, Zhang J, Chen MR, Hayward GS, Hayward SD, Desai P, Zhu H. 2009. Protein array identification of substrates of the Epstein-Barr virus protein kinase BGLF4. J. Virol. 83:5219–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]