Abstract
Simian foamy viruses (SFVs) are thought to infect virtually any adult nonhuman primate (NHP). While many data have accumulated about patterns of codivergence with their hosts and cross-species transmission events, little is known about the modalities of SFV transmission within NHP species, especially in the wild. Here we provide a detailed investigation of the dynamics of SFV circulation in a wild community of Western chimpanzees (Pan troglodytes verus). We demonstrate that mother-offspring (vertical) SFV transmission is common and hypothesize that it accounts for a number of primary infections. We also show that multiple infections with several chimpanzee-specific SFV strains (i.e., superinfection) commonly happen in adult chimpanzees, which might point to adult-specific aggressive behaviors as a lifelong source of SFV infection. Our data give evidence for complex SFV dynamics in wild chimpanzees, even at a single community scale, and show that linking wild NHP social interactions and their microorganisms' dynamics is feasible.
INTRODUCTION
Retroviruses are arguably the best-studied microorganisms infecting wild nonhuman primates (NHP) (1). Simian immunodeficiency viruses (SIVs) and simian T-cell leukemia viruses (STLVs) have naturally received considerable attention due to their proximity to human pathogens, but presumably apathogenic simian foamy viruses (SFVs) have also been intensively studied. It was quickly realized that SFVs infect nearly all living wild NHPs, which applies at different scales. That is, not only do SFVs infect most NHP species in the wild, they also reach extremely high prevalence in wild NHP adults, e.g., close to 100% in Western red colobus (Piliocolobus badius badius) (2) or chimpanzees (Pan troglodytes) (3, 4).
Several important questions related to the biology of SFVs have already been tackled. For example, a striking pattern of cospeciation of SFVs and their primate hosts could be demonstrated (5), which is now known to be embedded into larger-scale codivergence of FVs with their vertebrate hosts (6, 7). Another focus of SFV research has been on the risk of cross-species transmission. This was documented in natural hunter-prey systems in sub-Saharan Africa such as those involving chimpanzees and their red colobus prey (8) or humans and their numerous primate prey (more particularly great apes) (e.g., see reference 9). Although a significant amount of information has accumulated, much of the biology of SFVs still remains unknown. In particular, while they are, as mentioned above, known to infect virtually any adult individual in any infected species, the modalities and routes of transmission are still relatively unclear.
In this respect, a key question is whether in their primate hosts, which are highly social, SFVs are transmitted horizontally, vertically, or both, vertical transmission being here understood as mother-offspring transmission (whenever it occurs). Increases in seroprevalence and/or SFV nucleic acid detection rate have been documented in captive or semicaptive colonies of macaques (Macaca tonkeana [10]), baboons (Papio sp. [11, 12]), and mandrills (Mandrillus sphinx [13]). In the wild, comparable data were produced only for chimpanzees (Pan troglodytes schweinfurthii), evidencing the same trend (3). This was generally taken as pointing at horizontal transmission, e.g., through aggressive contacts during adulthood, being a favored route of transmission (10). However, vertical transmission was only rarely directly investigated. Anecdotal reports were made concerning chimpanzees, with one captive (14) and one wild (3) mother-offspring pair being found to harbor indistinguishable SFV strains. Calattini et al. (10) addressed the question more specifically, focusing on a captive colony of macaques. Showing that 8 of 11 mother-offspring pairs harbored different SFV strains, these authors argued that vertical transmission was likely not a major route of infection (10). Given the high prevalence of SFVs, it is, however, likely that many individuals will get infected with different strains over their lifetime (i.e., superinfections), a fact that might pass unnoticed when using bulk PCR product sequencing (15). Further insights into in-host SFV variability might therefore lead to different conclusions. Clearly, the possibility of vertical transmission of SFV warrants further investigation, in particular in wild primate groups and using finer tools.
Here, we performed a study on a wild community of habituated Western chimpanzees (P. troglodytes verus from the Taï forest, Côte d'Ivoire) known to be infected with SFVs (4, 8). Taking advantage of the availability of a fecal sampling of individuals with known pedigree, we investigated in depth the influence of the mother-offspring bond on SFV transmission in comparison to relatively weaker social bonds linking offspring to their fathers or nonparental members of the same group. To increase the power of our analyses, we estimated SFV diversity within all samples. This also allowed us to tackle the questions of SFV persistence and accumulation dynamics. Our results provide a detailed view of transmission patterns of SFVs in a wild NHP community and highlight complex patterns of individual accumulation of SFVs, to which mother-offspring transmission events contribute significantly.
MATERIALS AND METHODS
Sample description.
The study was performed on noninvasive samples stemming from a habituated community of P. troglodytes verus living in the Taï forest, Côte d'Ivoire. This community has been continuously monitored for more than 30 years (38). This resulted in behaviorally determined identification of pedigrees, which were further refined genetically (16, 17). All fecal samples were collected immediately after defecation by trained personnel, thereby allowing for immediate individual identification of the defecating chimpanzees. In total, 208 fecal samples were collected from 37 chimpanzees of the community. An initial screening for SFV showed that 32 individuals were infected. We performed most following analyses on 37 fecal samples stemming from 23 individuals (Table 1). Those were selected to maximize the number of mother-offspring and father-offspring pairs included in the study, ending up with 12 mother-offspring pairs and 7 father-offspring pairs (Table 2) (17); altogether, individuals corresponding to the same sample formed a total of 257 non-mother-offspring non-father-offspring dyads. The 12 offspring had been sampled between 3 and 19 years of age, while the range within the complete data set was 3 to 44 years. For 11 subjects, longitudinal sampling was available (8 subjects with 2 samples, 3 subjects with 3 samples).
Table 1.
Individual sample characteristics
| Individual | Sex | Birth date | Sample | Sampling date | Age at sampling | No. of clone sequences |
|---|---|---|---|---|---|---|
| Caramel | Male | 2002 | A | 2005 | 3 | 25 |
| Celine | Female | 1995 | A | 2005 | 10 | 25 |
| Coco | Female | 1980 | A | 2004 | 24 | 25 |
| Coco* | Female | 1980 | B | 2005 | 25 | 25 |
| Gogol | Male | 1991 | A | 2001 | 10 | 25 |
| Gogol | Male | 1991 | B | 2008 | 17 | 25 |
| Jacobo | Male | 1998 | A | 2004 | 6 | 25 |
| Julia* | Female | 1970 | A | 2004 | 34 | 25 |
| Kabisha | Female | 1977 | A | 2001 | 24 | 25 |
| Kabisha | Female | 1977 | B | 2002 | 25 | 25 |
| Kaos | Male | 1977 | A | 2006 | 29 | 25 |
| Kinshasa | Female | 1990 | A | 2001 | 11 | 25 |
| Kinshasa | Female | 1990 | B | 2007 | 17 | 25 |
| Kiriku | Male | 2005 | A | 2008 | 3 | 25 |
| Louise | Female | 1980 | A | 2005 | 25 | 25 |
| Lula* | Male | 2003 | A | 2007 | 4 | 25 |
| Rebecca | Female | 1995 | A | 2002 | 7 | 25 |
| Romario* | Male | 1999 | A | 2004 | 5 | 25 |
| Romario | Male | 1999 | B | 2008 | 9 | 25 |
| Rubra | Female | 1970 | A | 2001 | 31 | 25 |
| Rubra | Female | 1970 | B | 2005 | 35 | 25 |
| Rubra | Female | 1970 | C | 2008 | 38 | 25 |
| Sagu | Male | 1989 | A | 2002 | 13 | 25 |
| Sagu | Male | 1989 | B | 2006 | 17 | 25 |
| Sagu | Male | 1989 | C | 2008 | 19 | 25 |
| Settut* | Female | 1996 | A | 2003 | 7 | 25 |
| Shogun | Male | 2001 | A | 2008 | 7 | 25 |
| Sumatra | Female | 1965 | A | 2003 | 38 | 15 |
| Sumatra | Female | 1965 | B | 2005 | 40 | 25 |
| Utan | Male | 1994 | A | 2001 | 7 | 25 |
| Utan | Male | 1994 | B | 2005 | 11 | 25 |
| Yao | Male | 1995 | A | 2003 | 8 | 25 |
| Yucca | Female | 1970 | A | 2002 | 32 | 25 |
| Yucca | Female | 1970 | B | 2004 | 34 | 25 |
| Zyon | Male | 1964 | A | 2001 | 37 | 25 |
| Zyon | Male | 1964 | B | 2006 | 42 | 25 |
| Zyon | Male | 1964 | C | 2008 | 44 | 25 |
An asterisk marks samples which have been excluded from statistical analysis of superinfection status in relation to age and sex because of their ambiguous infection status.
Table 2.
Parental relationships
| Individual | Mother | Father |
|---|---|---|
| Caramel | Coco | Sagu |
| Celine | Coco | Kaos |
| Coco | ||
| Gogol | ||
| Jacobo | Julia | |
| Julia | ||
| Kabisha | ||
| Kaos | ||
| Kinshasa | Kabisha | |
| Kiriku | Kinshasa | |
| Louise | ||
| Lula | Louise | Sagu |
| Rebecca | Rubra | |
| Romario | Rubra | Kaos |
| Rubra | ||
| Sagu | Sumatra | |
| Settut | Sumatra | Kaos |
| Shogun | Sumatra | Zyon |
| Sumatra | ||
| Utan | ||
| Yao | Yucca | Zyon |
| Yucca | ||
| Zyon |
All necessary permissions had been obtained beforehand from the Ivorian Ministry of the Environment and Forests, the Ivorian Ministry of Research, and the directorship of the Taï National Park. After collection, samples were immediately placed on ice before being stored in liquid nitrogen, as previously described (18, 19).
Molecular biology.
We followed the same methodology as in reference 15. Briefly, commercial kits were used to extract nucleic acids from all samples, which were then used to generate cDNA (from RNA). All extracts were screened for SFV using a generic nested PCR system (first round: PFVint1s, 5′-GCCACCCAAGGGAGTTATGTGG-3′, and PFVint2as, 5′-GCTGCACCCTGATCAGAGTG-3′; second round: PFVint3s, 5′-CCTGGATGCAGAGTTGGATC-3′, and PFVint4as, 5′-GAAGGAGCCTTAGTGGGGTA-3′) targeting 470 bp of the integrase region of the polymerase gene (3, 20). Previous analyses had demonstrated that SFV nucleic acids could be detected through this assay when initial template numbers were <5 (15), making it an ideal tool for samples with low viral loads, e.g., feces. To minimize sampling bias, we generated three to five independent PCR products for the 37 selected samples. All PCR products were then cloned using the TOPO-TA cloning kit (Invitrogen, Karlsruhe, Germany). Following colony PCR, five clones per product were sequenced on both strands according to Sanger's methodology, generating a total of 915 clone sequences.
Initial sequence analysis.
All clone sequences generated in this study were first confirmed by BLAST (22) to be chimpanzee specific (more precisely P. troglodytes verus specific). After having been aligned using the MUSCLE algorithm (23) as implemented in SeaView v4 (24), all sequences were checked for evidence of recombination using the RDP, GENECONV, MaxChi, Chimaera, SiScan, and 3Seq tools, as available in RDP3 v3.44 (25). No recombinant sequence could be identified.
Mismatch distribution-based identification of single/superinfections.
We applied the algorithm described in reference 15 to differentiate between infections with a single SFV strain (single infection) and infections with two or more SFV strains (superinfection). Briefly, the diagnosis for single/superinfection was based on the frequency distribution of the number of mismatches in individual clone sequence alignments (i.e., mismatch distributions). Practically, several models (one or two Poisson or normal distributions) were fitted to the observed mismatch distributions in a maximum likelihood framework. Models assuming a unimodal distribution (one Poisson or normal distribution; single infection) or a bimodal distribution (two Poisson or normal distributions; superinfection) were then compared, using Akaike's information criterion (26). The resulting diagnosis (single or superinfection) was confirmed by visual inspection of mismatch distributions and networks. It should be noted here that this method is by nature independent of the definition of a cutoff number of mismatches that would make two sequences unlikely to be derived from each other: it relies only on shape analysis. This shape analysis essentially allows for the identification of deviations from a single infection scenario and not for the determination of the number of SFV strains involved in multiple infections with >2 SFV strains (but note that multiple infections with >2 SFV strains are properly identified as cases of superinfection) (15). Examination of individual networks and phylogenetic analyses did not evidence unambiguous instances of infection with >2 SFV strains (data not shown).
Individual networks and network-based identification of the founder SFV strains.
Many clone sequences derived from a bulk PCR product will comprise Taq-induced errors (15, 27). Hence, much of the observed variation will be artifactual. To identify sequences more likely to be biological, we used a network-based statistic, the outgroup probability (OP) (15). Individual networks were generated using median joining (28) and statistical parsimony (29), as implemented in Network (Fluxus Technology, Clare, Suffolk, United Kingdom) and TCS (30), named according to the three inventors of the method (Templeton, Crandall, and Sing [29]). In all cases, there was good agreement between networks computed by the two softwares. For all individual sequences, OP values, which summarize both frequency and connectivity, were recovered from TCS analyses. For single infections, the sequence(s) with the highest OP was considered biological; for superinfections, the two sequences with the highest OP were considered biological. Later on, these sequences are referred to as founder sequences (15).
Other networks.
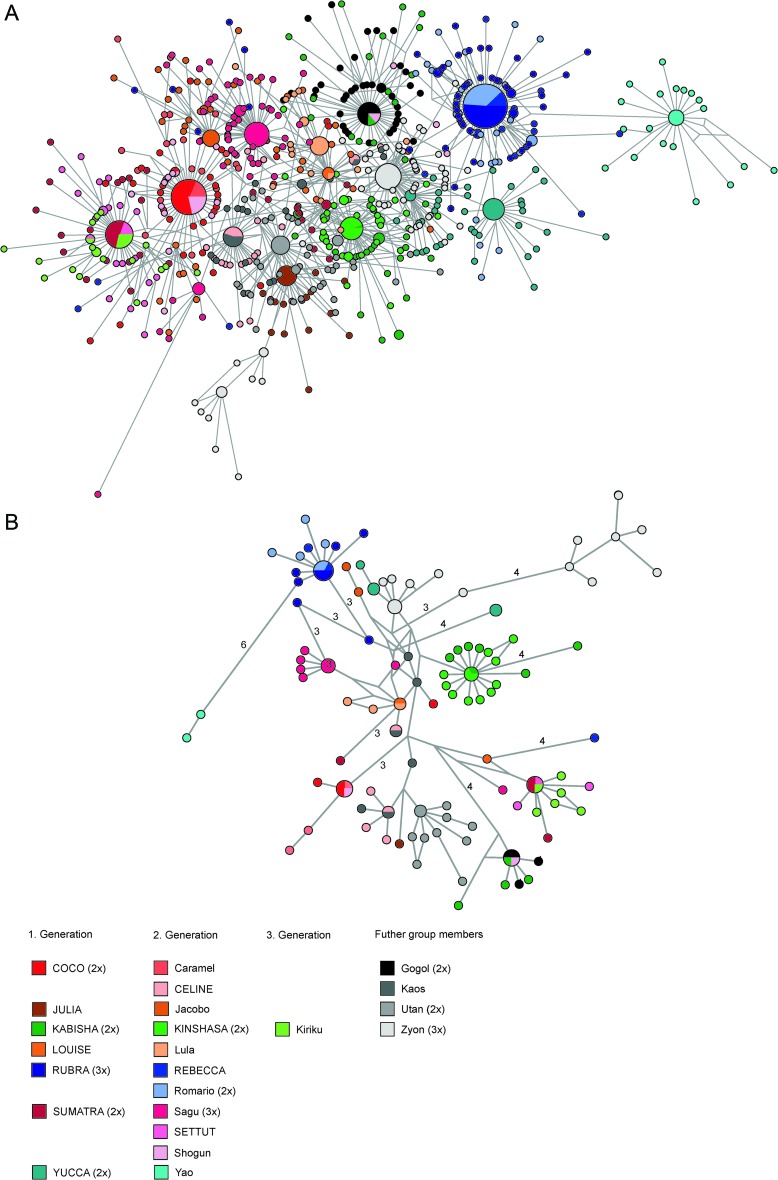
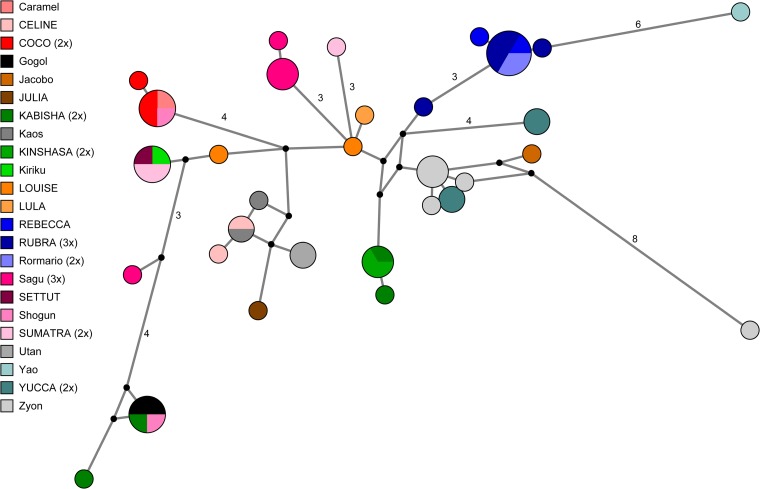
In addition to the 37 individual sample networks, networks were produced which included all samples taken from a same individual (9 networks), all samples from mothers and their offspring (7 networks), and all samples included in this study (1 global network out of 915 sequences) (Fig. 1A). Two alternative versions of the global network were also produced: (i) a streamlined network, which did not comprise clone sequences that exhibited minimal OP values in individual sample networks, that is, excluding these sequences more likely to be PCR-borne variants (128 sequences) (Fig. 1B), and (ii) a “founder” network including all founder sequences (n = 55) (Fig. 2). These networks were also computed with Network and TCS, which yielded very comparable network structures.
Fig 1.
Networks of all SFV sequences generated for this study. (A) Comprehensive version. (B) Streamlined version. Only those SFV sequences less likely to comprise “noise,” Taq-induced mutations, were included. The legend illustrates matrilineal lines included in this study. Females' names are in capital letters. When individuals were repeatedly sampled, number of samples is given between brackets in the legend. Within the networks, node size is proportional to the frequency of sequence occurrence. Branch lengths are directly related to the number of mutations between sequences (total length of aligned sequences: 432 bp [A], 426 bp [B]). Please note that the streamlined network still comprise all but two of the shared SFV sequences, the two exceptions consisting of sequences closely related to other shared sequences (1 bp difference). Data set reduction therefore did not result in any loss of information about SFV transmission.
Fig 2.
Network of founder sequences. Node size is proportional to the frequency of sequence occurrence in the data set. Branch lengths are directly related to the number of mutations between sequences, with values noted for differences greater than two base pairs. Legend conventions are the same as in Fig. 1.
Molecular evolution and substitution processes.
All sequences were examined for the presence of frameshift-inducing indels and in-frame stop codons. In addition, the proportions of variable sites at the first and second codon positions and at the third codon position were also determined as a proxy of selective forces operating on these sequences. Finally, mutations occurring as the first step away from founder sequences were recorded in all TCS-derived individual networks, allowing for the determination of a complete nonreversible nucleotide substitution matrix.
Estimation of virus persistence.
For 9 individuals, it was possible to unambiguously determine the infection status (single or superinfection) from two to three samples (n = 21) collected 1 to 7 years apart (mean, 3.3 years). Considering that some individuals were superinfected over multiple sampling points and that each of the underlying major strains constituted an independent observation, we determined the number of viral years. Here we define viral years as the minimum number of consecutive years over which one SFV strain can be assumed to have infected an individual. Practically, this means that if strain A and strain B are detected during year n and strain A and strain B in year n + 1 while only strain A is detected in year n + 2, strain A will have persisted over 2 years (2 viral years) while strain B will have persisted over 1 year (1 viral year). It should be noted here that the assumption is made that two consecutive observations of the same strain are explained by viral persistence rather than other processes, e.g., reinfection.
Phylogenetic analysis.
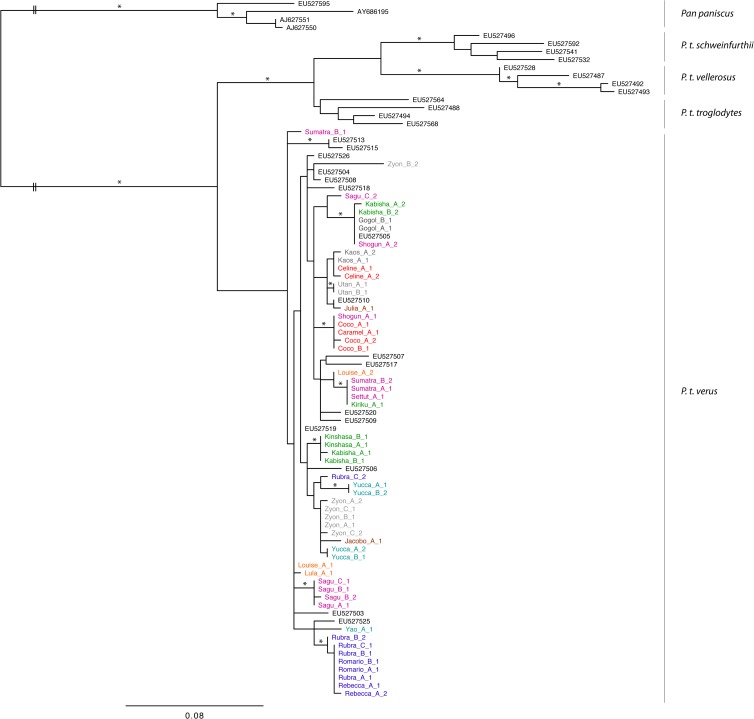
A maximum likelihood (ML) analysis of a data set comprising all founder sequences as well as sequences from all chimpanzee subspecies (P. troglodytes verus, P. troglodytes ellioti, P. troglodytes troglodytes, and P. troglodytes schweinfurthii) and from bonobos (Pan paniscus) was performed. Model selection was first conducted using jModelTest v0.1 (32), which resulted in selecting a global time-reversible (GTR) matrix of substitution together with across-site rate variation (+G) and a proportion of invariant sites (+I). Tree reconstruction was then performed under this model using PhyML v3.0 (33). Branch robustness was assessed through nonparametric bootstrapping (500 pseudoreplicates) (Fig. 3).
Fig 3.
Maximum likelihood tree of founder sequences. Asterisks mark branches with bootstrap support of >70. Please note that most inner branches are not statistically supported. Legend conventions are the same as in Fig. 1.
Statistical modeling.
We used a generalized linear mixed model (GLMM) (34) with binominal error structure and logit link function to analyze the combined influence of sex and age (categorical and continuous predictors) and their interaction (fixed effects) on the superinfection status (yes or no). We included subject identity as a random effect because for several subjects we had samples from different ages. The total number of data points analyzed was reduced to 32 samples from 20 subjects, as the infection status of five samples was ambiguous (see Fig. S1 in the supplemental material). Into this model we entered all variables simultaneously. We checked for various diagnostics of model validity and stability. For example, colinearity could be excluded by inspection of variance inflation factors (VIF) (both = 1.18, determined using the function vif of the R package car) (35). Model stability was assessed by excluding data points one by one and comparing the derived coefficients. This revealed no obviously influential cases. To establish the significance of the full model we used a likelihood ratio test (36), comparing its deviance with that of the null model comprising only the intercept and the random effect. To test the significance of the interaction between age and sex, we used the standard z-test provided by the function lmer (37).
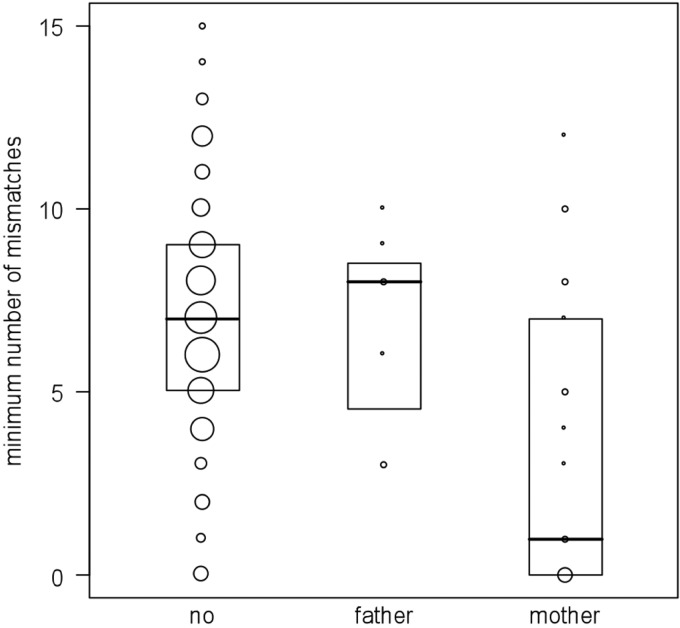
To test if close social relationships influenced the virus circulation, we compared the virus pool of available offspring to their mother, father, and nonparental group members. As a measure of dissimilarity between the SFV strains of samples from two individuals, we simply determined the minimum number of mismatches between any pair of their respective SFV sequences. We then used a GLMM to test whether this similarity measure corresponded to the familial relationship (mother-offspring, father-offspring, or non-mother-offspring non-father-offspring) between the dyad's members. In this model we considered the similarity measures between all samples of each of the 12 offspring, on the one hand, and samples from all group members, on the other hand, given they were collected prior to the respective sample of the offspring. In addition to the relation between the two individuals we included the following predictors, in order to control for their effects: the total time the two individuals have spent together in the same community until the first of the two samples was collected, assuming that longer times spent together might lead to increased similarity of their infecting strains; absolute difference between their birthdays (number of days), assuming that individuals born close to one another might be more similar; the sex of the offspring, assuming female and male offspring might differ in their social/play behavior; the sex of the other individual, assuming that interactions between offspring and other group members might be more likely when the other is a female; the difference between the two sampling dates, assuming that larger differences lead to decreased similarity; and, finally, offspring age, assuming that older offspring might have SFV strains more similar to those of others than younger ones. In addition, we included the two-way interactions between time spent together, on the one hand, and relation, difference between birthdays, and sex of the other individual, on the other hand, in the model. We included the interaction between time spent together and relation because we hypothesized that offspring SFV population would generally become more similar to that of any other individuals with increasing age but that this increase in similarity would be more pronounced in mother-offspring dyads. The other two interactions we included to control for their potential effects, assuming that a potentially increased similarity between offspring born close to one another in time and offspring with females would show up only after some time spent together. All these terms were included as fixed effects into the model. To control for the identity of the offspring and the other individual we included them as random effects into the model. Prior to running the model we inspected all numerical predictors for their distribution and, as a consequence, square root transformed the difference between birthdays to achieve an approximately symmetrical distribution. Subsequently, we z-transformed all numerical predictors to a mean of zero and a standard deviation of one. To test the overall effect of the factor relation and its interaction, we compared the full model (as described above) with a null model not comprising relation or its interaction with time spent together but including all other terms present in the full model. This comparison was based on a likelihood ratio test (36). We fit the model with Poisson error structure and log link function. Using a Poisson error, structure was justified, as the response (dissimilarity) comprised only integer numbers ≥0 and since overdispersion was not an issue (dispersion parameter: 0.97; χ2 = 300.3; df = 310; P = 0.64). Also, colinearity was no issue (largest VIF, 1.93, determined using the function vif of the R package car) (35).
All analyses were performed using R (version 2.10.1; R Development Core Team, 2010). The GLMM was run using the function lmer of the R package lme4 (37), VIF were calculated using the functions vif of the R package car (35), and likelihood ratio tests were conducted with the R function anova with the argument test set to chisq.
Nucleotide sequence accession numbers.
All founder sequences have been deposited in the EMBL depository under accession numbers HF568879 through HF568933. The complete sequence data set, comprising all clone sequences, has been deposited in DRYAD under doi:10.5061/dryad.bb8r6.
RESULTS
For this study, we generated a total of 183 PCR products from 37 fecal samples obtained from 23 wild chimpanzees belonging to the same community. Because our specific objective was to better investigate the possibility of vertical transmission of SFVs in the wild, all PCR products were cloned, resulting in a total of 915 clone sequences. Phylogenetic analyses of founder sequences evidenced that most inner branches were poorly supported (for a definition of founder sequences please refer to Materials and Methods) (Fig. 3). We interpreted this as reflecting the inappropriateness of phylogenetic methods in depicting reticulate evolution and most notably here the fact that some sequences are likely ancestral to others. Therefore, we implemented network analyses on three data sets: (i) a full data set comprising the 652 unique sequences (Fig. 1A), (ii) a streamlined data set, from which sequences likely to stand only for “methodological noise” had been removed (Fig. 1B), and (iii) a data set only comprising founder sequences (Fig. 2). All networks, including both streamlined versions, exhibited considerable complexity, despite a maximum observed pairwise distance of only 6% (Fig. 1A and B; Fig. 2).
SFV transmission pattern.
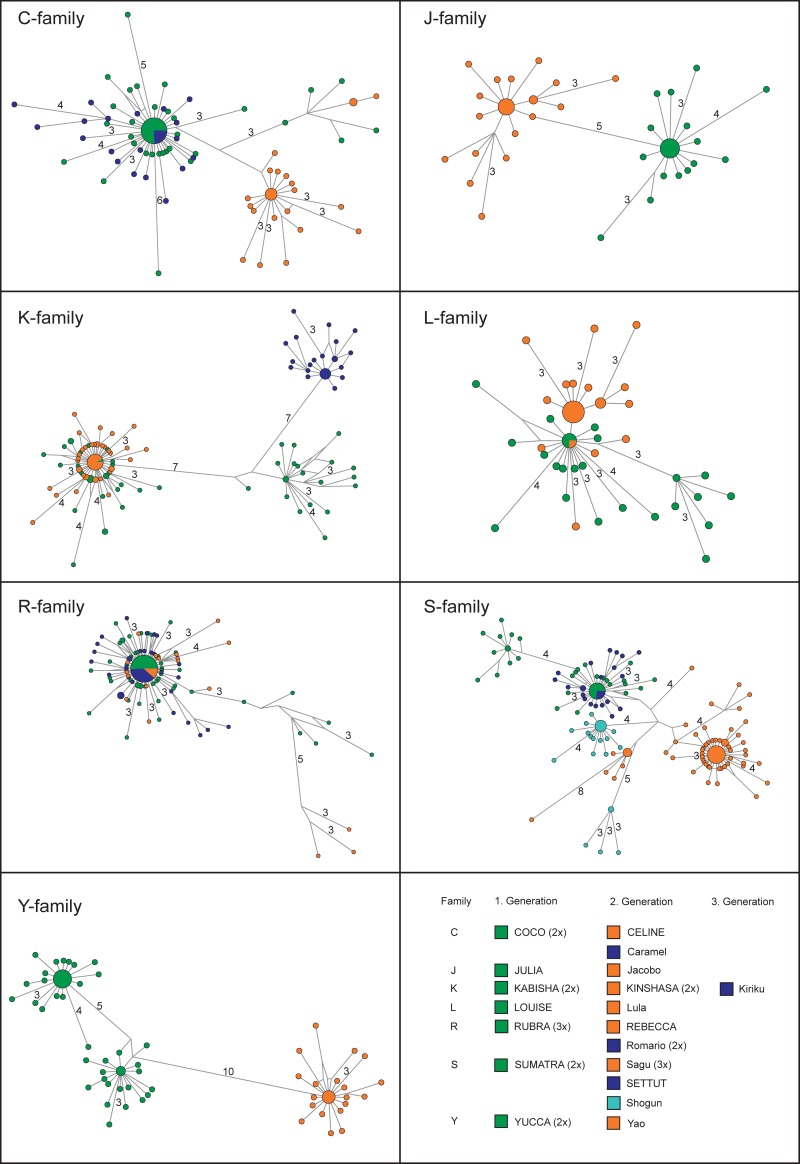
A number of individuals harbored identical SFV sequences/strains: 16/23 individuals did, 7 times with one, 3 times with two other group members (Fig. 3A and B). Among these, identical sequences were shared by six mother-offspring dyads (Fig. 4). A generalized linear mixed model (GLMM) revealed that similarity between the SFV strains found in samples was clearly influenced by the relatedness of the two respective individuals (likelihood ratio test comparing full model with null model not comprising relation or its interaction with time spent together but with all other terms present in the full model: χ2 = 37.9; df = 4; P < 0.001). No interaction was detected between relation and time spent together in the community (χ2 = 0.1193; df = 2; P = 0.94). After removing this interaction we found a clearly significant impact of the main effect relatedness (χ2 = 37.8; df = 2; P < 0.0001). Visual inspection of the data revealed that offspring SFV pools exhibited much more similarity to their mothers' than to their fathers' or unrelated individuals' (Fig. 5). None of the other predictors nor their interactions (time spent together and difference between birthdays, time spent together, and sex of the other) appeared to significantly influence similarity measures (smallest P = 0.14).
Fig 4.
Networks for all mother-offspring pairs. The bottom right illustrates matrilineal lines included in this study. The column family marks the individuals belonging to the family with the same initial (letter), followed by mothers to the left of their offspring (starting with the first generation). Females' names are in capital letters. When individuals were repeatedly sampled, the number of samples is given between brackets. Within each network, node size is proportional to the frequency of sequence occurrence. Branch lengths are directly related to the number of mutations between sequences, with values noted for differences greater than two base pairs (total length of aligned sequences: C, L, Y family, 425 bp; J, K family, 426 bp; R, S family, 428bp).
Fig 5.
SFV similarity as a function of host relatedness. Circle size represents the number of dyads exhibiting the corresponding relationship with the offspring.
SFV persistence.
Nine individuals included in this study were sampled at multiple time points. Altogether, this provided a record of 42 years of chimpanzee lifetime and 57 viral years (for a definition of viral years please see Materials and Methods) (Table 3). Over this period, nearly identical sequences (exhibiting <3 bp differences, i.e., 0.7% divergence) were found in consecutive samples spanning 55 viral years. Hence, 96% of viral years were characterized by a close-to-perfect SFV sequence stability.
Table 3.
SFV persistencea
| Individual | Age at sampling (yr) | Infection status | No. of virus strains | No. of virus strains transmitted | Time between samplings (yr) | Virus yr based on no. of strains and time between samplings | Virus stability (yr) |
|---|---|---|---|---|---|---|---|
| Gogol | 10 | Single infection | |||||
| Gogol | 17 | Single infection | 1 | 1 | 7 | 7 | 7 |
| Kabisha | 24 | Superinfection | |||||
| Kabisha | 25 | Superinfection | 2 | 2 | 1 | 2 | 2 |
| Kinshasa | 11 | Single infection | |||||
| Kinshasa | 17 | Single infection | 1 | 1 | 6 | 6 | 6 |
| Rubra | 31 | Single infection | |||||
| Rubra | 35 | Superinfection | 1 | 1 | 4 | 4 | 4 |
| Rubra | 38 | Superinfection | 2 | 2 | 3 | 6 | 6 |
| Sagu | 13 | Single infection | |||||
| Sagu | 17 | Superinfection | 1 | 1 | 4 | 4 | 4 |
| Sagu | 19 | Superinfection | 2 | 1 | 2 | 4 | 2 |
| Sumatra | 38 | Single infection | |||||
| Sumatra | 40 | Superinfection | 1 | 1 | 2 | 2 | 2 |
| Utan | 7 | Single infection | |||||
| Utan | 11 | Single infection | 1 | 1 | 4 | 4 | 4 |
| Yucca | 32 | Superinfection | |||||
| Yucca | 34 | Superinfection | 2 | 2 | 2 | 4 | 4 |
| Zyon | 37 | Superinfection | |||||
| Zyon | 42 | Superinfection | 2 | 2 | 5 | 10 | 10 |
| Zyon | 44 | Superinfection | 2 | 2 | 2 | 4 | 4 |
| Total | 42 | 57 | 55 | ||||
| Persistence (%) | 96 |
Individuals for which multiple samples were available were tested for virus stability. Strains identified in the earlier samples were compared with strains found in the later corresponding sample. A virus strain was defined using network-based analysis [identification of the founder sequence(s)/group of closely related sequence in the absence of the founder sequence]. Strains were defined as stable in the consecutive sample(s) if sequences revealed a maximal distance of 0.7% observed divergence (up to 3 bp difference). Of note, estimation of the number of independent infection events is a minimum estimate.
SFV accumulation dynamics.
The design of our study also allowed for investigating accumulation of SFV strains through time.
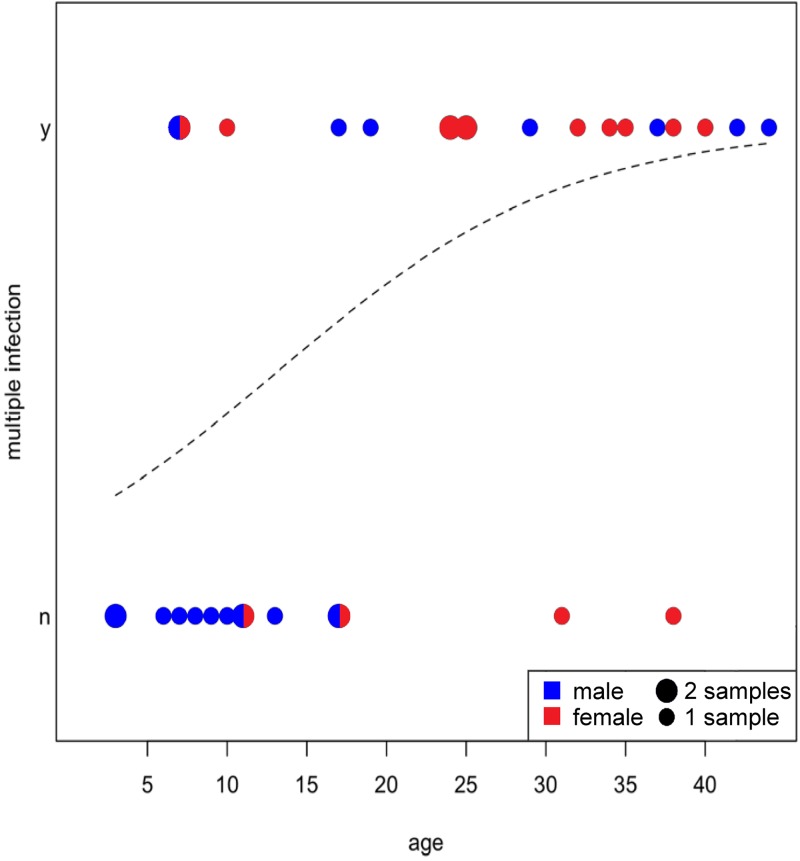
We first applied a statistical tool specifically meant to discriminate between single- and multiple-infection cases across all samples included in this study. For 32 samples from 20 individuals, clear assignment to either of the categories was possible (see Fig. S1 in the supplemental material). Only these samples were included in subsequent analyses on superinfection status. Most infected infants (0 to 4 year old), juveniles (5 to 9), and adolescents (10 to 14) were singly infected, while most adults showed evidence for superinfection (Fig. 6). Correcting for multiple sampling of individuals, that is, considering only one sample per individual when consecutive samples had the same status within the age class, revealed that 3/12 nonadults and 9/11 adults were superinfected. Correspondingly, a GLMM revealed that superinfection status was clearly influenced by age, sex, and their interaction (likelihood ratio test comparing the full with the null model comprising only the random effect subject identity: χ2 = 14.76; df = 3; P = 0.002). As there was no interaction between age and sex (estimate + standard error [SE] = 0.258 + 0.169; z = 1.522; P = 0.128), we removed it from the model. The final model confirmed that superinfection was significantly more likely with increasing age (estimate + SE = 0.110 + 0.047; z = 2.337; P = 0.019) but did not support any obvious effect of sex (estimate + SE = −0.815 + 1.050; z = −0.776; P = 0.438) (Fig. 6).
Fig 6.
SFV accumulation dynamics in wild chimpanzees from Taï (P. troglodytes verus). Superinfection status (no/yes) is shown as a function of age. Circle size represents the number of samples at the corresponding combination of age and infection status; colors indicate the sex of the individuals. The dashed line indicates the fitted model's prediction.
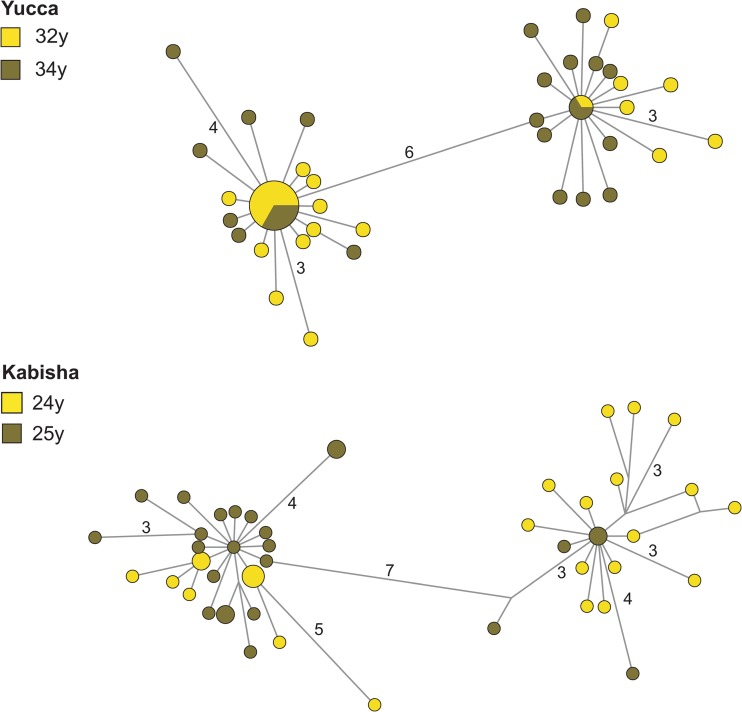
The cases of the 9 chimpanzees for which multiple samples were available also allowed for a longitudinal investigation of the dynamics of superinfection. Out of the six chimpanzees which were initially single infected, we identified a minimum of three superinfection events affecting Sagu, Rubra, and Sumatra, respectively (see Fig. S2 in the supplemental material). Confirming the trend derived from the cross-sectional analysis, these superinfection events were recorded close to or during adulthood, between 13 and 19, 31 and 35, and 38 and 40 years of age, respectively. Of special note, two of these superinfections actually occurred in relatively old individuals (>30 year old). Among individuals identified as superinfected over multiple sampling points, we observed marked frequency shifts in the number of clones belonging to one or the other clouds of sequences in two cases for which consecutive superinfected samples were available. For both Yucca and Kabisha, the smaller cloud of sequences indeed doubled its relative size between the two samplings, with frequencies rising from 28% to 56% and from 40% to 80%, respectively (Fig. 7).
Fig 7.
In-host SFV population dynamics. Chimpanzees Yucca and Kabisha were sampled at different time points (y, years of age). Within the networks, node size is proportional to the frequency of sequence occurrence. Branch lengths are directly related to the number of mutations between sequences, with values noted for differences greater than two base pairs (total length of aligned sequences: 425 bp).
SFV molecular evolution.
We examined all clone sequences and the 55 founder sequences for mutations indicating possibly defective viruses (i.e., frameshift-inducing indels and in-frame stop codons). While 14.1% of clone sequences were “defective,” the proportion dropped to 1.8% for founder sequences (Table 4), providing additional support in favor of these sequences' biological relevance. For founder sequences, variation occurred more often in the third codon position (mostly degenerate; 22.5%) than in the first and second codon positions (mostly nondegenerate; 6.7%), supporting an overall trend to purifying selection on this fragment (Table 4). Point mutations (other than nonsense) were also investigated to detect possible selective effects exerted by chimpanzee antiviral mechanisms, in particular through APOBEC proteins' editing activity (G-to-A mutations). Analysis of the first mutational step away from the founder sequence(s) identified in individual networks supported a strong dominance of transitions (82%) versus transversions (18%). Among the former, G-to-A mutations were not particularly frequent (11%) (Table 5).
Table 4.
Defective sequences and variable sites
| Group | No./total no. (%) |
|||
|---|---|---|---|---|
| Defective viruses |
Variable sitesa |
|||
| Indels | Stop codons | 1st and 2nd codon position | 3rd codon position | |
| All clones | 90/915 (9.8%) | 39/915 (4.3%) | 217/283 (76.7%) | 121/142 (85.2%) |
| Founder sequences | 1/55 (1.8%) | 0/55 (0%) | 19/283 (6.7%) | 32/142 (22.5%) |
Among apparently nondefective viral sequences.
Table 5.
Nucleotide substitution matrixa
| Initial/final state | Mean (SD) (%) |
|||
|---|---|---|---|---|
| A | C | T | G | |
| A | 2 (4) | 6 (7) | 28 (11) | |
| C | 1 (3) | 7 (8) | 0 (2) | |
| T | 5 (6) | 36 (13) | 2 (5) | |
| G | 11 (10) | 1 (2) | 1 (3) | |
Point mutations (n = 471) occurring as the first step away from founder sequences were recorded for all individual TCS networks. Mean and standard deviation (SD) of percentages are shown. Transitions are shown in gray boxes.
DISCUSSION
The data presented here provide unambiguous evidence that vertical transmission is an important modality of transmission in wild chimpanzees. Interestingly, this fits well with the assumed nonpathogenicity of SFVs. Because of their reliance on their host reproductive success, vertically transmitted microorganisms are indeed expected to evolve reduced virulence (39). A consequence of reduced virulence is that microorganisms' selective pressure on their host decreases; that is, a very efficient immune response to the infection is not a selective advantage anymore. In this study, two additional points are consistent with such a coevolved “taming” process: (i) SFVs infecting Taï chimpanzees do not seem to undergo diversifying selection, as expected under strong immune pressure (but note that the fragment examined here encodes the integrase), and (ii) APOBEC editing does not seem to influence strongly these SFVs' mutational patterns (in contrast to previously published results obtained in vitro) (40). It should, however, be noted that while mother-offspring SFV transmission is strongly supported here, other studies led on captive primates did not identify the same trend (10, 11), which might suggest that SFVs infecting distinct primate species are characterized by distinct transmission traits. The very long cospeciation history of SFV and their hosts would certainly have allowed for such divergence in SFV transmission patterns (5).
It is for the moment only possible to speculate about the possible medium or media supporting mother-offspring transmission. Mother-offspring dyads face numerous opportunities for body fluid exchanges. This includes intrauterine, perinatal, and/or breast-feeding-mediated transmission as observed for other exogenous retroviruses (e.g., deltaretroviruses, lentiviruses), saliva-saliva contacts through fruit sharing, and saliva-blood contact through grooming on wounds or bites and nips received by infants during weaning. Although saliva has not yet been proven to be the main shedding site of SF viral particles in chimpanzees (in contrast to findings for other primates [31, 41]), transmission of SFV from chimpanzees to humans following severe bites (9, 14, 42) and SF viral particle detection from chimpanzee fecal samples (3, 15) qualify it as the most likely medium for effective transmission of SFV among chimpanzees.
While we show here that mother-offspring transmission of SFV occurs frequently and therefore likely stands as a privileged route for primary infection, it is worth to note that not all siblings were infected with one of their mother's SFVs. This clearly points at the complexity of mother-offspring transmission patterns, which most likely depend on a variety of parameters, e.g., variation in maternal viral load and shedding as well as immune status of the offspring. Adding to the complexity of mother-offspring SFV transmission is the complexity of contemporary or subsequent horizontal transmissions. Assuming that saliva is the driving medium for infection, numerous opportunities of transmission will indeed also exist outside mother-offspring relationships. In this respect it is interesting to note that in this study father-offspring relationships did not measurably influence the circulation of SFVs. This does not come as a surprise, as in chimpanzees father-offspring relationships are notoriously weaker than mother-offspring relationships (21, 38, 43, 44). Other relationships, e.g., friendships, might be influential but could not be investigated here.
Whatever the quality of the relationships supporting their transmission, our data are also strongly supportive of a lifelong accumulation of SFV strains in chimpanzees. Although some of our results support the possibility of significant SFV population shifts after superinfection and a natural extension of this is the possibility that some SFVs get extinct, e.g., through the effects of genetic drift, we record a very high persistence rate. Lifelong persistence of SFV is therefore likely to be the rule in wild chimpanzees, which mirrors observations made on a similar time series of samples taken from captive Macaca tonkeana (10). In addition, we provide very strong evidence that SFV superinfection becomes more common with increasing age, which suggests continuous acquiring of new strains through horizontal transmission events. We note here that the well-known pattern of increasing sero- and/or genoprevalence with age found in other chimpanzee subspecies and primates (3, 10) may therefore hide very complex SFV dynamics as well.
An important question is that of the behaviors supporting horizontal transmission and therefore lifelong accumulation of SFVs, beyond a likely primary infection with mothers' SFVs. It has been hypothesized that truly violent aggressive behaviors, that is, those behaviors resulting in severe wounds providing opportunity for body fluid exchange, could fulfill the requirements for SFV transmission (10). In primates in general and in chimpanzees in particular, such aggressive behaviors usually involve subadults and adults, for either the defense of social rank, territory, or access to food or reproductive partners. Interestingly, the onset of these behaviors fits well with our observation that superinfection events mainly occur during adulthood. The finding that SFV accumulation is not influenced by sex might at first seem somewhat contradictory as males more frequently commit aggressive behaviors. However, it should not be considered that only males get involved into aggressive contacts or that aggressive contacts are systematically male-male or female-female events. For example, in Taï National Park, rates of male aggression toward females approach 0.08 per hour of female observation; that is, any female will be attacked by a male every 2 days (45). During these frequent aggressive attacks, opportunities exist for both male-to-female and female-to-male SFV transmission since females also reply aggressively. Taken together with effective vertical transmission, this contributes to making marked sex differences in SFV transmission unlikely, although slight differences cannot be ruled out.
Conclusions.
In addition to highlighting the interest and broad applicability of approaches focused on the depiction of within-host SFV diversity, our results are supportive of a new model of SFV transmission in wild chimpanzee communities.
In this model, primary infection with SFV would likely occur through the mother-offspring relationship, a strong social bond which we demonstrate here is a major route of SFV circulation in the community. Subsequent SFV infections (that is, superinfections, since SFVs persist in wild chimpanzees) would occur during adulthood, possibly as a consequence of the onset of aggressive interactions with other members of the group. Within this framework, different strains of SFV might experience different population dynamics within individuals.
Further investigation of SFV transmission patterns in the wild are clearly needed. For chimpanzees, studies investigating the influence of other social bonds, such as other forms of kinship or friendship, would be highly desirable. Determining likely modes of SFV transmission in other wild NHP species would also be a welcome addition and would help determine whether the model of transmission proposed here for chimpanzees is applicable across parts or the entire phylogenetic tree of NHP. Such studies will require gathering considerable amounts of behavioral and epizootic data. It might also provide useful information about possible patterns of SFV transmission within new hosts, including humans.
Supplementary Material
ACKNOWLEDGMENTS
We thank the authorities in Côte d'Ivoire for long-term support, especially the Ministry of Environment and Forests and the Ministry of Research, the directorship of the Taï National Park, the Office Ivoirien des Parcs et Réserves, and the Swiss Research Center in Abidjan. We also thank the Taï Chimpanzee Project for logistic support and S. Metzger, field assistants, and students for assistance in sample collection. We warmly thank Verena Keil, Corinna Blasse, and Franka Schaebs for their efficient assistance, the entire group Emerging Zoonoses for frequent and stimulating discussions, and Daniel Driscoll for proofreading.
A.B. was supported by a stipendium from the FAZIT foundation. This work was supported by the Deutsche Forschungsgemeinschaft (grant LE1813/4-1) and the Robert Koch Institute.
Footnotes
Published ahead of print 28 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02743-12.
REFERENCES
- 1. Locatelli S, Peeters M. 2012. Cross-species transmission of simian retroviruses: how and why they could lead to the emergence of new diseases in the human population. AIDS 26:659–673 [DOI] [PubMed] [Google Scholar]
- 2. Leendertz SA, Junglen S, Hedemann C, Goffe A, Calvignac S, Boesch C, Leendertz FH. 2010. High prevalence, coinfection rate, and genetic diversity of retroviruses in wild red colobus monkeys (Piliocolobus badius badius) in Tai National Park, Cote d'Ivoire. J. Virol. 84:7427–7436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu W, Worobey M, Li Y, Keele BF, Bibollet-Ruche F, Guo Y, Goepfert PA, Santiago ML, Ndjango JB, Neel C, Clifford SL, Sanz C, Kamenya S, Wilson ML, Pusey AE, Gross-Camp N, Boesch C, Smith V, Zamma K, Huffman MA, Mitani JC, Watts DP, Peeters M, Shaw GM, Switzer WM, Sharp PM, Hahn BH. 2008. Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees. PLoS Pathog. 4:e1000097 doi:10.1371/journal.ppat.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morozov VA, Leendertz FH, Junglen S, Boesch C, Pauli G, Ellerbrok H. 2009. Frequent foamy virus infection in free-living chimpanzees of the Tai National Park (Cote d'Ivoire). J. Gen. Virol. 90:500–506 [DOI] [PubMed] [Google Scholar]
- 5. Switzer WM, Salemi M, Shanmugam V, Gao F, Cong ME, Kuiken C, Bhullar V, Beer BE, Vallet D, Gautier-Hion A, Tooze Z, Villinger F, Holmes EC, Heneine W. 2005. Ancient co-speciation of simian foamy viruses and primates. Nature 434:376–380 [DOI] [PubMed] [Google Scholar]
- 6. Han GZ, Worobey M. 2012. An endogenous foamy-like viral element in the coelacanth genome. PLoS Pathog. 8:e1002790 doi:10.1371/journal.ppat.1002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Han GZ, Worobey M. 2012. An endogenous foamy virus in the aye-aye (Daubentonia madagascariensis). J. Virol. 86:7696–7698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leendertz FH, Zirkel F, Couacy-Hymann E, Ellerbrok H, Morozov VA, Pauli G, Hedemann C, Formenty P, Jensen SA, Boesch C, Junglen S. 2008. Interspecies transmission of simian foamy virus in a natural predator-prey system. J. Virol. 82:7741–7744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Betsem E, Rua R, Tortevoye P, Froment A, Gessain A. 2011. Frequent and recent human acquisition of simian foamy viruses through apes' bites in central Africa. PLoS Pathog. 7:e1002306 doi:10.1371/journal.ppat.1002306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Calattini S, Wanert F, Thierry B, Schmitt C, Bassot S, Saib A, Herrenschmidt N, Gessain A. 2006. Modes of transmission and genetic diversity of foamy viruses in a Macaca tonkeana colony. Retrovirology 3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Blewett EL, Black DH, Lerche NW, White G, Eberle R. 2000. Simian foamy virus infections in a baboon breeding colony. Virology 278:183–193 [DOI] [PubMed] [Google Scholar]
- 12. Broussard SR, Comuzzie AG, Leighton KL, Leland MM, Whitehead EM, Allan JS. 1997. Characterization of new simian foamy viruses from African nonhuman primates. Virology 237:349–359 [DOI] [PubMed] [Google Scholar]
- 13. Mouinga-Ondeme A, Betsem E, Caron M, Makuwa M, Salle B, Renault N, Saib A, Telfer P, Marx P, Gessain A, Kazanji M. 2010. Two distinct variants of simian foamy virus in naturally infected mandrills (Mandrillus sphinx) and cross-species transmission to humans. Retrovirology 7:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Switzer WM, Bhullar V, Shanmugam V, Cong ME, Parekh B, Lerche NW, Yee JL, Ely JJ, Boneva R, Chapman LE, Folks TM, Heneine W. 2004. Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates. J. Virol. 78:2780–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goffe AS, Blasse A, Mundry R, Leendertz FH, Calvignac-Spencer S. 2012. Detection of retroviral super-infection from non-invasive samples. PLoS One 7:e36570 doi:10.1371/journal.pone.0036570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boesch C, Kohou G, Nene H, Vigilant L. 2006. Male competition and paternity in wild chimpanzees of the Tai forest. Am. J. Phys. Anthropol. 130:103–115 [DOI] [PubMed] [Google Scholar]
- 17. Vigilant L, Hofreiter M, Siedel H, Boesch C. 2001. Paternity and relatedness in wild chimpanzee communities. Proc. Natl. Acad. Sci. U. S. A. 98:12890–12895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kondgen S, Schenk S, Pauli G, Boesch C, Leendertz FH. 2010. Noninvasive monitoring of respiratory viruses in wild chimpanzees. EcoHealth 7:332–341 [DOI] [PubMed] [Google Scholar]
- 19. Leendertz FH, Pauli G, Maetz-Rensing K, Boardman W, Nunn C, Ellerbrok H, Jensen SA, Junglen S, Christophe B. 2006. Pathogens as drivers of population declines: the importance of systematic monitoring in great apes and other threatened mammals. Biol. Conserv. 131:325–337 [Google Scholar]
- 20. Schweizer M, Neumann-Haefelin D. 1995. Phylogenetic analysis of primate foamy viruses by comparison of pol sequences. Virology 207:577–582 [DOI] [PubMed] [Google Scholar]
- 21. Goodall J. 1986. The chimpanzees of Gombe: patterns of behaviour. Belknap Press, Cambridge, MA [Google Scholar]
- 22. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 23. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221–224 [DOI] [PubMed] [Google Scholar]
- 25. Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Burnham KP, Anderson DR. 2002. Model selection and multi-model inference: a practical information-theoretic approach. Springer-Verlag, New York, NY [Google Scholar]
- 27. Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, Manigart O, Mulenga J, Anderson JA, Swanstrom R, Haynes BF, Athreya GS, Korber BT, Sharp PM, Shaw GM, Hahn BH. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82:3952–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bandelt HJ, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16:37–48 [DOI] [PubMed] [Google Scholar]
- 29. Templeton AR, Crandall KA, Sing CF. 1992. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132:619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clement M, Posada D, Crandall KA. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9:1657–1659 [DOI] [PubMed] [Google Scholar]
- 31. Falcone V, Leupold J, Clotten J, Urbanyi E, Herchenroder O, Spatz W, Volk B, Bohm N, Toniolo A, Neumann-Haefelin D, Schweizer M. 1999. Sites of simian foamy virus persistence in naturally infected African green monkeys: latent provirus is ubiquitous, whereas viral replication is restricted to the oral mucosa. Virology 257:7–14 [DOI] [PubMed] [Google Scholar]
- 32. Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256 [DOI] [PubMed] [Google Scholar]
- 33. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 34. Baayen R. 2008. Analyzing linguistic data. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 35. Field A. 2005. Discovering statistics with SPSS. Sage Publications, London, United Kingdom [Google Scholar]
- 36. Dobson AJ. 2002. An introduction to generalized linear models. Chapman & Hall/CRC, Boca Raton, FL [Google Scholar]
- 37.Bates D, Maechler M, Bolker B.lme4: linear mixed-effects models using S4 classes, R package, 0.999375–41 ed. 2011. http://lme4.r-forge.r-project.org/
- 38. Boesch C, Boesch-Achermann H. 2001. The chimpanzees of the Taï Forest. Oxford University Press, New York, NY [Google Scholar]
- 39. Stewart AD, Logsdon JM, Jr, Kelley SE. 2005. An empirical study of the evolution of virulence under both horizontal and vertical transmission. Evolution 59:730–739 [PubMed] [Google Scholar]
- 40. Delebecque F, Suspene R, Calattini S, Casartelli N, Saib A, Froment A, Wain-Hobson S, Gessain A, Vartanian JP, Schwartz O. 2006. Restriction of foamy viruses by APOBEC cytidine deaminases. J. Virol. 80:605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Murray SM, Picker LJ, Axthelm MK, Hudkins K, Alpers CE, Linial ML. 2008. Replication in a superficial epithelial cell niche explains the lack of pathogenicity of primate foamy virus infections. J. Virol. 82:5981–5985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mouinga-Ondeme A, Caron M, Nkoghe D, Telfer P, Marx P, Saib A, Leroy E, Gonzalez JP, Gessain A, Kazanji M. 2012. Cross-species transmission of simian foamy virus to humans in rural Gabon, Central Africa. J. Virol. 86:1255–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nishida T. 1983. Alloparental behavior in wild chimpanzees from the Mahale mountains, Tanzania. Folia Primatol. 41:1–33 [Google Scholar]
- 44. Wroblewski EE, Johnson L, Murray CM, Henderson J, Stanley A, Pusey AE. 2010. Father-offspring relationships among the Gombe chimpanzess (Pan troglodytes schweinfurthii), abstr 125. Abstr Int. Primatol. Soc. 23rd Congr., Kyoto, Japan, 12 to 18 September 2010 [Google Scholar]
- 45. Stumpf RM, Boesch C. 2010. Male aggression and sexual coercion in wild West African chimpanzees, Pan troglodytes verus. Anim. Behav. 79:333–342 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.