Abstract
Koala retrovirus (KoRV) is a gammaretrovirus that is currently endogenizing into koalas. Studies on KoRV infection have been hampered by the lack of a replication-competent molecular clone. In this study, we constructed an infectious molecular clone, termed plasmid pKoRV522, of a KoRV isolate (strain Aki) from a koala reared in a Japanese zoo. The virus KoRV522, derived from pKoRV522, grew efficiently in human embryonic kidney (HEK293T) cells, attaining 106 focus-forming units/ml. Several mutations in the Gag (L domain) and Env regions reported to be involved in reduction in viral infection/production in vitro are found in pKoRV522, yet KoRV522 replicated well, suggesting that any effects of these mutations are limited. Indeed, a reporter virus pseudotyped with pKoRV522 Env was found to infect human, feline, and mink cell lines efficiently. Analyses of KoRV L-domain mutants showed that an additional PPXY sequence, PPPY, in Gag plays a critical role in KoRV budding. Altogether, our results demonstrate the construction and characterization of the first infectious molecular clone of KoRV. The infectious clone reported here will be useful for elucidating the mechanism of endogenization of the virus in koalas and screening for antiretroviral drugs for KoRV-infected koalas.
INTRODUCTION
Retroviral elements, termed endogenous retroviruses (ERVs), occupy about 8 to 10% of mammalian genomes (1). Most ERVs are defective due to genomic mutations and deletions, or their expression is epigenetically suppressed. However, some ERVs retain functionality and contribute to host physiological processes, exemplified by the human syncytins in placentation (2). In this regard, ERVs are believed to play a role in the evolution of mammals, yet the process of endogenization of retroviruses, resulting in the establishment of ERVs, has not been elucidated. The koala retrovirus (KoRV), found in koalas (Phascolarctus cinereus), is a gammaretrovirus that is believed to be currently endogenizing into its host, thus providing us with a rare opportunity to investigate the mechanisms involved in retrovirus endogenization (3, 4). Genetically and phylogenetically, KoRV is closely related to gibbon ape leukemia virus (GALV), which is an exogenous gammaretrovirus that induces leukemia/lymphoma in gibbons (5). In addition, KoRV shares the viral receptor Pit-1 (a phosphate transporter) with GALV when it infects cells (6). GALV is considered to be derived from an endogenous retrovirus of the Asian mouse Mus caroli or a related species (7). However, the origin of KoRV is still unknown, because gibbons and Asian mice do not live in Australia.
In addition to benefits provided by ERVs, there are also negative consequences of harboring them in the host genome. Indeed, increased levels of KoRV infection in koalas have been associated with several diseases. For instance, koalas suffer from leukemia and lymphoma at a rate of 3 to 5% in the wild and an even higher rate of up to 60% in some captive colonies (8, 9). Tarlinton et al. reported that, using quantitative real-time reverse transcriptase (RT) PCR, KoRV RNA levels in plasma were significantly increased in koalas suffering from leukemia or lymphoma compared with healthy koalas (10). Furthermore, increased levels of KoRV were also seen in Queensland koalas with clinical chlamydiosis, a disease that is associated with immunosuppression (10–13), although a definite link between KoRV and chlamydiosis has been disputed (http://espace.library.uq.edu.au/view/UQ:244963). Altogether, these observations suggest that KoRV may be linked to oncogenesis and immunosuppression in koalas, making the study of the virus important for understanding its pathogenesis.
To date, studies on KoRV infection have been limited due to the lack of a replication-competent molecular clone and to the fact that it is not considered ethical to infect naïve koalas with the virus. In one study, individual genes derived from a molecular clone of KoRV, termed pcindy, were investigated for their impacts on KoRV-pseudotyped virus infection (6). The authors reported that several mutations in the Gag and Env regions were involved in reduction in viral infection/production in vitro during endogenization, yet the negative impact of these and other mutations on viral replication has not been evaluated. In this study, we constructed an infectious molecular clone of KoRV isolated from a koala reared in Hirakawa Zoological Park (Kagoshima, Japan) and characterized it in terms of virus infectivity and budding. We found that the infectious clone is highly infectious in a human cell line despite containing mutations reportedly involved in reduction in viral infection/production in vitro.
MATERIALS AND METHODS
Cell cultures.
Human embryonic kidney (HEK293T) cells (ATCC CRL-11268), TE671 (human rhabdomyosarcoma) cells (ATCC CRL-8805), G355-5 (feline glial) cells (ATCC CRL-2033), Chinese hamster ovary (CHO) cells (ATCC CCL-61), Mv1-Lu (mink lung epithelial) cells (ATCC CCL-64), NIH 3T3 cells (mouse fibroblasts) (ATCC CRL-1658), Mus dunni tail fibroblasts (MDTF) (ATCC, CRL-2017), TELCeB6 cells (14), TELCeB6/GALV cells (15), and TELCeB6/pFBFeLV-B cells (16) were cultured in Dulbecco's modified Eagle's medium (Sigma, Tokyo, Japan) supplemented with 10% heat-inactivated fetal serum, penicillin (100 units/ml), and streptomycin (100 mg/ml) (Invitrogen, Carlsbad, CA). The cells were cultured at 37°C in a humidified atmosphere of 5% CO2 in air.
Virus isolation.
A heparinized blood sample was taken by venipuncture from a Queensland koala (named Aki) reared in Hirakawa Zoological Park (Kagoshima, Japan). The procedure of virus isolation was described previously (17), and the virus isolate was named strain Aki.
Construction of an infectious molecular clone of strain Aki.
The genomic DNA of HEK293T cells infected with KoRV strain Aki was isolated by using a QIAamp DNA Blood Kit (Qiagen, Valencia, CA). We amplified two fragments covering the entire KoRV genome using primer sets 1 (Fw1, AATGAAGGAGGCAGAAATCATGAGGC, and Rv1, AAGTGATCTGATTATAAGCATGTTC) and 2 (Fw2, AGGGAAGGGAATTCATTCAACGGCTG, and Rv2, AATGAAAGACCCCAATGTTCGGGTAG) with PrimeStar GXL DNA polymerase (TaKaRa, Ohtsu, Shiga, Japan) according to the manufacturer's instructions (Fig. 1). We succeeded in cloning two KoRV fragments (fragments 1 and 2), which overlapped in a region corresponding to C-terminal pol and N-terminal env genes. The two fragments were ligated using the EcoRI site (present in the overlapping region) to obtain a full-length KoRV genome. Then, we cloned the full-length KoRV into a pSP73 vector (Promega, Fitchburg, WI) to obtain pKoRV522 (Fig. 1). The detailed procedure for the plasmid construction is available upon request.
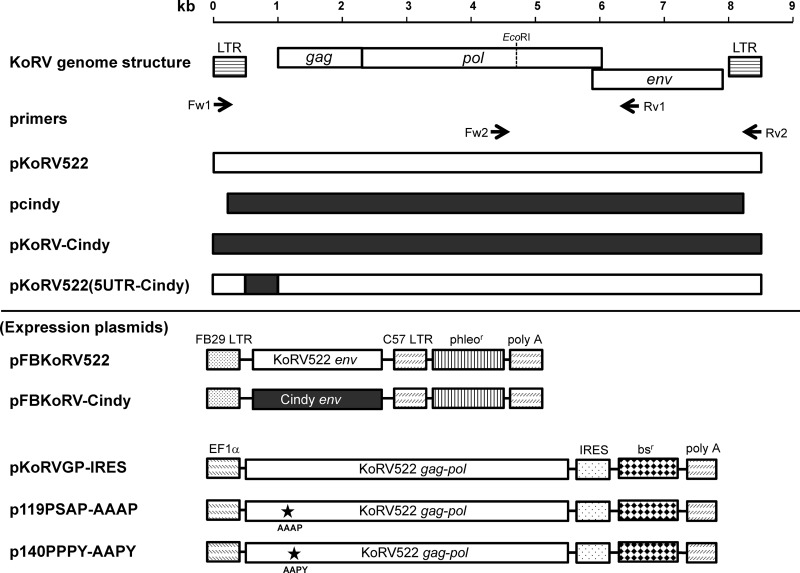
Fig 1.
Structures of infectious molecular clones of KoRV and expression plasmids used in this study. The arrows indicate primer sets (Fw1, Rv1, Fw2, and Rv2) used for PCR cloning. The white and black bars indicate the sequences derived from pKoRV522 and pcindy, respectively. The asterisks indicate amino acid mutations introduced. phleor, phleomycin resistance gene; bsr, blasticidin resistance gene.
Virus preparation.
The supernatant of HEK293T cells transfected with pKoRV522 or HEK293T cells cocultured with peripheral blood mononuclear cells (PBMCs) of the koala named Aki was collected and then filtered through a 0.45-μm filter unit (Acrodisc; PALL Corporation, Ann Arbor, MI). The stock viruses were designated strain KoRV522 and strain Aki, respectively, and kept at −80°C until they were used. Infection via viral stocks was performed in the presence of Polybrene (2 μg/ml; Sigma-Aldrich, St. Louis, MO) in the culture medium for viral adsorption as described previously (18).
Plasmids.
To construct expression plasmids of KoRV Envs, the env genes of pKoRV522 and pcindy were amplified by PCR using PrimeStar GXL DNA polymerase (TaKaRa) with primers KoRVenvFw (5′-AATCTAGACCACCATGCTTCTCATCTCAAACCC-3′) and KoRVenvRv (5′-ACGCCCATCGATTTAAAGGTTATCCTCGTTGT-3′). The PCR fragments were digested with XbaI and ClaI and then cloned into pFBPERV-A (15) to produce pFBKoRV522 and pFBKoRV-Cindy, respectively (Fig. 1).
A pGEM-T Easy vector containing the complete KoRV proviral genome (pcindy) was obtained from Jon Hanger (Royal Society for the Prevention of Cruelty to Animals [RSPCA], Queensland, Australia). Because pcindy is not a proviral form, we reconstituted the clone into a proviral form of KoRV having long terminal repeats (LTRs) at both ends in the backbone of the pSP73 vector and named it pKoRV-Cindy (Fig. 1). A chimeric clone, termed pKoRV522(5UTR-Cindy), was constructed by exchanging the entire 5′ untranslated region (UTR) of pKoRV522 with that of pcindy using an In-Fusion HD cloning kit (Clontech, Mountain View, CA) according to the manufacturer's instructions. The detailed procedures for the construction of pKoRV-Cindy and pKoRV(5UTR-Cindy) are available on request.
To construct the expression plasmid for the KoRV gag-pol genes, named pKoRVGP-IRES (Fig. 1), the entire KoRV gag-pol region was amplified by PCR using PrimeStar GXL polymerase (TaKaRa) with primers KoRVGPFw (5′-AAGTTTAAACCACCATGGGACAGGGTGAGTCGAC-3′) and KoRVGPRv (5′-TGTATGCGGCGCTTATGCTGTTGATTCATTTC-3′). The amplicon was used to replace the murine leukemia virus (MLV) gag-pol genes in the internal ribosome entry site (IRES) expression plasmid, termed pGP-IRES, in the Plat-E retroviral packaging system (19). To generate the expression plasmids for the KoRV L-domain mutants, p119PSAP-AAAP and p140PPPY-AAPY (Fig. 1), alanine substitutions were introduced into pKoRVGP-IRES by site-directed mutagenesis using a KOD-Plus-Mutagenesis Kit (Toyobo, Osaka, Japan).
To prepare HEK293T cells infected with Friend MLV and feline leukemia virus subgroup B (FeLV-B), pBluescriptIISK+/LTR-Friend-LTR (20) and pGAHF (21), respectively, were transfected into HEK293T cells.
LacZ pseudotype assay.
To prepare the LacZ pseudotype viruses, termed lacZ(KoRV522) and lacZ(KoRV-Cindy), we transfected pFBKoRV522 and pFBKoRV-Cindy into TELCeB6 cells, which express high titers of MLV core particles incorporating an nlsLacZ gene (14), by using polyethylenimine as described previously (22). Two days after transfection, culture supernatant was harvested from the transfected cells, filtered through a 0.45-μm membrane filter, and used immediately in the LacZ assay. To prepare helper-free LacZ pseudotype viruses bearing GALV and FeLV-B, termed lacZ(GALV) and lacZ(FeLV-B), culture supernatants of TELCeB6/GALV (15) and TELCeB6/pFBFeLV-B (16) were harvested, filtered through a 0.45-μm filter unit, and used immediately in the LacZ assay. We inoculated lacZ pseudotype viruses into target cells and cultured them for 2 days. Then, the cells were fixed with 1% glutaraldehyde and stained with 1 mg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as described previously (15). We determined the virus titers in target cells by counting lacZ-positive foci and expressed the virus titers as focus-forming units (FFU)/ml.
LacZ marker rescue assay.
The LacZ marker rescue assay was performed as described previously (23). Briefly, prior to the LacZ marker rescue assay, HEK293T cells and TE671 cells were transduced with the nlsLacZ gene to become HEK293T(LacZ) and TE671(LacZ) cells, respectively. From the transgene, mRNA of the LacZ gene with a packaging signal from MLV is constitutively expressed. Because KoRV has viral core proteins belonging to gammaretroviruses, the mRNA with the packaging signal from MLV can be incorporated with the KoRV core and rescued. To monitor KoRV growth in HEK293T and TE671 cells, the virus was inoculated onto HEK293T(LacZ) or TE671(LacZ) cells, and the culture supernatants were collected at the times indicated in Fig. 2. Then, they were filtered through a 0.45-μm filter unit, and serially diluted samples were immediately inoculated into naïve 293T cells. Two days after inoculation, the cells were stained with β-Gal, and lacZ-positive foci were counted.
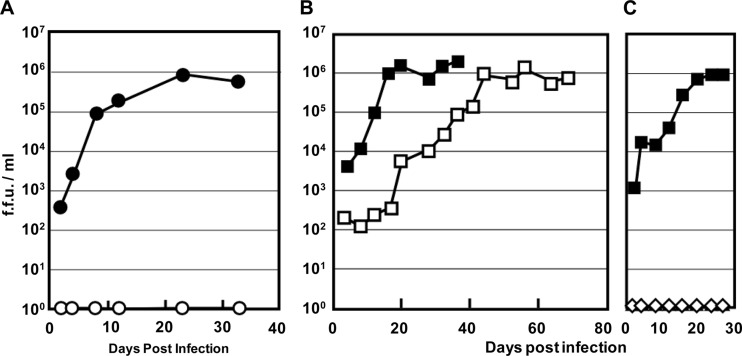
Fig 2.
Infectivity of KoRV522 derived from pKoRV522. (A) Growth of KoRV522 in HEK293T and TE671 cells. KoRV522 was inoculated onto HEK293T(LacZ) (●) and TE671(LacZ) (○) cells, and virus production was determined by the LacZ marker rescue assay. (B) Growth kinetics of KoRV522 and strain Aki in HEK293T cells. HEK293T(LacZ) cells were inoculated with strain Aki (□) and KoRV522 (■), which were adjusted by RT activity, and virus production was determined by the LacZ marker rescue assay. (C) Growth kinetics of pKoRV522 (■) and pKoRV-Cindy (♢). The plasmids were transfected into HEK293T(LacZ) cells, and virus production was determined by the LacZ marker rescue assay.
RT assay.
Single-tube fluorescent product-enhanced RT (STF-PERT) assays were performed as described previously (24). Briefly, lysates of viral supernatants were added to RT reaction mixtures containing MS2 RNA template, MS2-specific primer, and deoxynucleoside triphosphates (dNTPs) in 8-well tubes. Each RT reaction mixture was separated within the same well with AmpliWax (Applied Biosystems [ABI], Foster City, CA) from a real-time PCR mixture containing two MS2-specific primers, a fluorescent probe, dNTPs, and TaqMan Universal Master Mix (ABI). All samples were run in triplicate. A standard curve was created using serially diluted MLV RT enzyme (Roche, Basel, Switzerland) in the same buffer used for the samples and assayed in triplicate. The Prism 7700 (ABI) was used to perform the reactions and quantify the amounts of RT per sample. The reaction conditions were 48°C for 50 min (RT reaction), 95°C for 10 min (melting the wax to combine reactions), and 40 cycles of 95°C for 15 s and 60°C for 1 min (PCR).
Antibody preparation.
Polyclonal antibodies against capsid (CA) and surface unit (SU) of the Env protein of KoRV were prepared by a protocol reported previously (25, 26). Briefly, rabbits were immunized with the mixed synthetic peptides (WKSNHPSFSENPTGC and CRRDRRQEKNLTKIL for KoRV CA and CSQQARPPPDSNYEHAY and CSYPRARTRIARSQ for KoRV SU Env) conjugated with keyhole limpet hemocyanin (KLH).
Immunoblotting.
Cells were collected and lysed with RIPA buffer (50 mM Tris-HCl buffer, pH 7.6, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, protease inhibitor cocktail [Roche]), and the cell lysates were subjected to SDS-PAGE. Viruses in culture supernatants were collected by centrifugation (6,300 × g) for 12 h, and the concentrated supernatants were subjected to SDS-PAGE. To detect virus components (CA and SU Env), we used anti-KoRV CA or anti-KoRV SU Env antibody, respectively, as a primary antibody (1 μg/ml) and an anti-rabbit IgG antibody conjugated with horseradish peroxidase as a secondary antibody. Proteins recognized by the antibodies were visualized with Supersignal WestFemto (Thermo Scientific, Rockford, IL).
Virus budding assay.
Virus budding assays were performed as described previously (27). Briefly, HEK293T cells (2.5 × 105) were transfected with 1 μg of the expression plasmid for KoRV (pKoRVGP-IRES) or L-domain mutants, using Trans-IT LT-1 (Mirus Bio Corp., Madison, WI). At 48 h after transfection, the supernatants were separated from cell debris by centrifugation (10,000 × g) for 10 min, and then virions were pelleted by ultracentrifugation. Cell lysates and virions were analyzed by immunoblotting using anti-KoRV CA and SU Env antibodies and anti-actin antibody (Sigma).
Nucleotide sequence accession number.
The sequence of pKoRV522 was deposited in GenBank and assigned accession number AB721500.
RESULTS
Construction of an infectious molecular clone of KoRV strain Aki.
To isolate an infectious clone of KoRV, PBMCs harboring a parental KoRV strain, Aki, were taken from an infected koala held in captivity, cocultured with HEK293T cells, and then harvested for integrated proviral DNA. Several proviral sequences were extracted and cloned into a pSP73 vector. Preliminary studies identified one highly infectious clone, which was designated pKoRV522 (data not shown). We then examined the replication kinetics of the virus derived from pKoRV522, designated KoRV522. This virus was inoculated onto HEK293T(LacZ) and TE671(LacZ) cells, and virus titers were measured in the culture supernatants by the LacZ marker rescue assay. We found that KoRV522 grew efficiently in HEK293T cells, but not in TE671 cells (Fig. 2A). To compare the growth kinetics of KoRV522 with those of the parental strain, Aki, the amounts of KoRV522 and strain Aki were adjusted by RT activity and inoculated onto HEK293T(LacZ) cells. The production of KoRV in the inoculated cells was monitored for 36 and 68 days for KoRV522 and strain Aki, respectively. KoRV522 and strain Aki grew efficiently, reaching plateaus of 106 FFU/ml at 20 and 44 days postinoculation, respectively (Fig. 2B). These data indicate that the virus derived from pKoRV522 is infectious in HEK293T cells. It should be noted that pKoRV522 might be derived from two different proviruses in the koala and recombined in the procedure for the construction of the molecular clone.
Infectivity of pKoRV-Cindy.
The infectivity of pKoRV-Cindy (a reconstituted clone of pcindy) (Fig. 1) was also evaluated by the LacZ marker rescue assay. pKoRV-Cindy and pKoRV522 were transfected into HEK293T(LacZ) cells, and the virus titers produced in culture supernatants were measured (Fig. 2C). The virus derived from pKoRV-Cindy did not grow in HEK293T(LacZ) cells, indicating that pKoRV-Cindy is defective.
Immunoblot analysis.
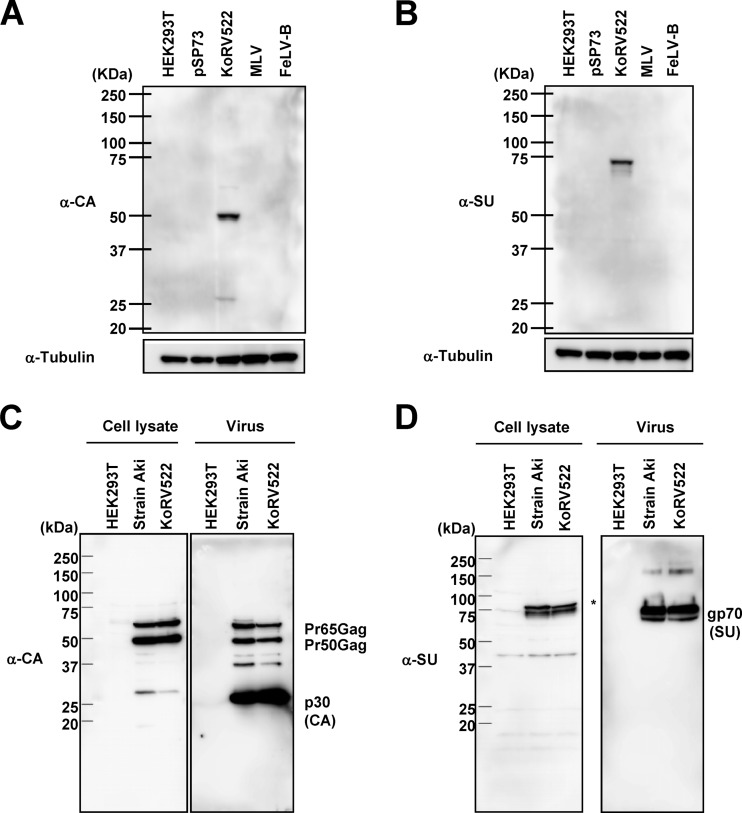
We next analyzed the protein expression profiles of KoRV522 by immunoblot analysis using anti-KoRV CA and SU Env antibodies, to confirm that viral proteins were being expressed. First, we confirmed that the anti-KoRV CA and SU Env antibodies detected KoRV CA and SU Env specifically and did not cross-react with MLV and FeLV-B (Fig. 3A and B). By using these antibodies, we detected precursor Gag proteins (Pr65 and Pr50) and mature CA protein (p30) in KoRV-infected cells (cell lysate) and culture supernatants in both strain Aki and KoRV522 (Fig. 3C). In culture supernatants, we detected mature CA (p30) as the predominant band. Furthermore, we detected SU Env protein in cell lysates and culture supernatants (Fig. 3D). No differences were observed in the protein sizes between KoRV522 and strain Aki. From these data, together with the replication kinetics, we conclude that pKoRV522 is a fully active and infectious molecular clone of strain Aki. We also examined the expression of viral proteins from pKoRV-Cindy. The pKoRV-Cindy was transfected into HEK293T cells, and 2 days after transfection, the cell lysates were examined for the presence of KoRV CA and SU Env proteins by immnoblot analysis. As a result, we could not detect any viral proteins in HEK293T cells transfected with pKoRV-Cindy (data not shown).
Fig 3.
(A and B) Specificity of anti-KoRV CA (α-CA) (A) and α-SU Env (B) antibodies. As controls, HEK293T cells transfected with pSP73 and cells infected with Friend MLV and FeLV-B were used. (C and D) Immunoblot analysis of KoRV522 and strain Aki. Cell lysates and concentrated culture supernatants (virus fraction) of HEK293T cells infected with strain Aki and KoRV522 were subjected to immunoblot analysis using anti-KoRV CA (C) and anti-SU Env (D) antibodies.
Nucleotide sequence comparison of pKoRV522 with pcindy.
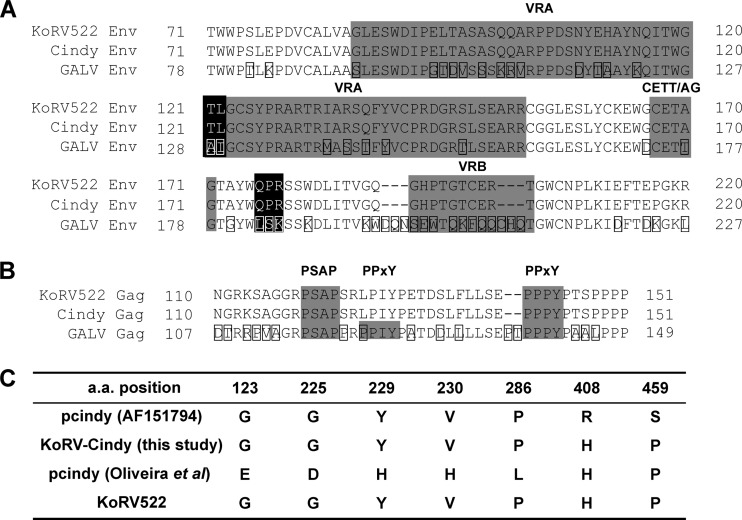
When we compared the nucleotide sequence of pKoRV522 with that of a previously characterized KoRV clone, pcindy (GenBank accession number AF151794) (9), we found that the sequence of pKoRV522 was nearly identical to that of pcindy in the coding and LTR regions (Table 1). However, the homology in the 5′ UTR of pKoRV522 was relatively low (97.4%), and pKoRV522 contained 6-bp and 3-bp insertions at positions 149 and 387 of pcindy, respectively. In a previous study (6), although both KoRV and GALV utilize the human Pit-1 molecule (a phosphate transporter) to infect human cells, pseudotyped KoRV Env infection was shown to be 1,000 times lower than that of GALV Env pseudotype on MDTF expressing human Pit-1. Further investigation led to the identification of several amino acids involved in the attenuation of Env functions, such as infectivity and fusion activity, of KoRV by comparing KoRV and GALV Env sequences (6). Consequently, replacing GALV Env residues A128, I129, L182, S183, and K184 with the corresponding KoRV Env residues T121, L122, Q176, P177, and R178 (Fig. 4A) resulted in markedly reduced virus titers (6). Furthermore, mutating the GALV Env residue T177 to the KoRV Env type (A170) in the CETTG motif resulted in decreased cell fusion activity (6). Contrary to our expectation, pKoRV522 also has these mutations, which were reported to be involved in a decrease in viral infection (6, 28) (Fig. 4A). Further, in a partial Env region containing a receptor binding domain encompassing variable regions A and B (VRA and VRB), we could not find any differences between pKoRV522 and pcindy (Fig. 3A). Gag proteins of many retroviruses include short peptide motifs required for virus budding, called L domains. To date, three types of L-domain motif have been identified in retroviruses: PT/SAP, PPXY, and YXXL (29). The disruption of the PPXY motif that is present 3 bases downstream of the PPXY motif in the L domain of KoRV was reported to be involved in the reduction of KoRV budding (6). The PPXY motif of pKoRV522 was also disrupted, like that of pcindy (Fig. 4B), but we found that an intact PPXY motif is also present 18 bases downstream of the PSAP motif (Fig. 4B). This additional PPXY motif is also present in a similar position in the GALV Gag sequence (Fig. 4B).
Table 1.
Comparison of nucleotide and amino acid sequences of pKoRV522 and pcindy
| Region | Identity (%) |
|
|---|---|---|
| Nucleotide | Amino acid | |
| LTR | 99.2 | |
| 5′ UTR | 97.4 | |
| gag (Gag) | 99.6 | 99.6 |
| pol (Pol) | 99.8 | 99.6 |
| env (Env) | 99.9 | 99.7 |
| 3′ UTR | 100.0 | |
Fig 4.
Alignment of amino acid sequences of KoRV (pKoRV522 and pKoRV-Cindy) and GALV. (A) Partial alignment of Env proteins of KoRV and GALV. The first methionine residue of the Env protein was defined as position 1. VRA and VRB and the CETTG motif are shaded in gray. Amino acid residues that are reported to be involved in reduction in viral infection/production in vitro (6) are shaded in black. Amino acid differences found in GALV are boxed. (B) Alignment of L domains of KoRV and GALV. The first methionine residue of Gag protein is defined as position 1. PSAP and PPXY motifs are shaded in gray. Amino acid differences found in GALV are boxed. (C) Amino acid differences in KoRVs. Sequences of pcindy, resequenced from the construct in our laboratory and from a published paper by Oliveira et al. (28), are derived from GenBank (AF151794).
The 5′ UTR is not a major determinant of the defectiveness of pKoRV-Cindy.
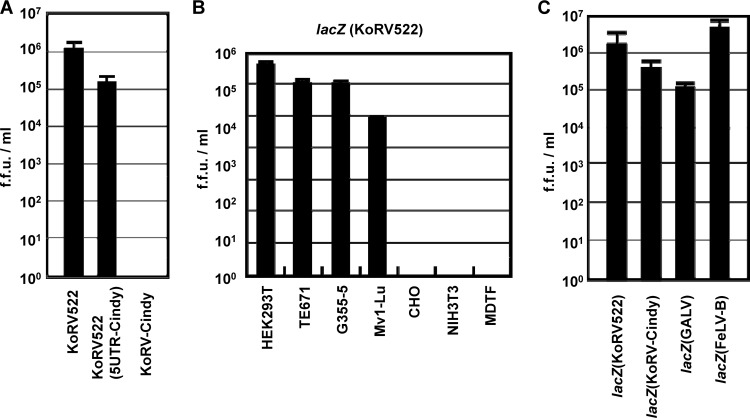
To explore the reason for the defectiveness of pKoRV-Cindy, we made a chimeric clone, termed pKoRV522(5UTR-Cindy) (Fig. 1), by exchanging the 5′ UTR of pKoRV522 with that of pKoRV-Cindy. pKoRV522, pKoRV522(5UTR-Cindy), and pKoRV-Cindy were transfected into HEK293T(LacZ) cells. Two days after transfection, KoRV in the culture supernatants was titrated by the LacZ marker rescue assay (Fig. 5A). Although the viral titer produced from pKoRV522(5UTR-Cindy) was slightly lower than that of the wild-type (WT) pKoRV522, the 5′ UTR mutant was still infectious. These data indicate that the 5′ UTR alone does not explain the defectiveness of pKoRV-Cindy.
Fig 5.
(A) The 5′ UTR is not a major determinant of the defectiveness of pKoRV-Cindy. Infectious molecular clones were transfected into HEK293T(LacZ) cells, and virus production was monitored by the LacZ marker rescue assay. (B) Host ranges of KoRV pseudotype virus. lacZ(KoRV522) was inoculated onto the indicated cell lines, and the virus titers were determined by the LacZ assay. (C) Comparison of viral titers of pseudotype viruses bearing KoRV, GALV, and FeLV-B Envs. Each pseudotype virus was inoculated onto naïve HEK293T cells, and the virus titer was determined by the LacZ assay. The data were obtained by three independent experiments, and the values are expressed as the mean ± 1 standard deviation (SD).
Host range of Env-pseudotyped virus derived from pKoRV522.
Since KoRV522 grew well in HEK293T cells but not in TE671 cells (Fig. 2A), we tried to determine the host range of KoRV522 using LacZ pseudotype virus bearing the envelope glycoprotein of pKoRV522, lacZ(KoRV522). HEK293T (human), TE671 (human), G355-5 (cat), Mv-1Lu (mink), NIH 3T3 (mouse), MDTF (mouse), and CHO (hamster) cells were inoculated with lacZ(KoRV522), and the virus titers were determined by the LacZ assay. We found that HEK293T, TE671, G355-5, and Mv-1Lu cells were susceptible to lacZ(KoRV522), while mouse cell lines, NIH 3T3 and MDTF, and a hamster cell line, CHO, were not (Fig. 5B). The titer of lacZ(KoRV522) was relatively high (more than 105 FFU/ml) in HEK293T, TE671 and G355-5 cells. These data indicate that human, cat, and mink cells express functional receptors, Pit-1, for KoRV. The pattern of the host range of KoRV522 was the same as those of GALV and FeLV-B, which utilize Pit-1 as a receptor (15, 16).
Comparison of viral titers of pseudotype viruses bearing KoRV, GALV, and FeLV-B Envs.
We compared the virus titers of pseudotype viruses bearing Envs from pKoRV522, pKoRV-Cindy, GALV, and FeLV-B by the LacZ assay (Fig. 5C). Titers of KoRV Env pseudotypes derived from both pKoRV522 and pKoRV-Cindy were comparable to those of GALV and FeLV-B Env pseudotype viruses. These data indicated that the function of KoRV Env is similar to that of exogenous gammaretroviruses in terms of infectivity.
The PPPY sequence functions as the major L domain in KoRV budding.
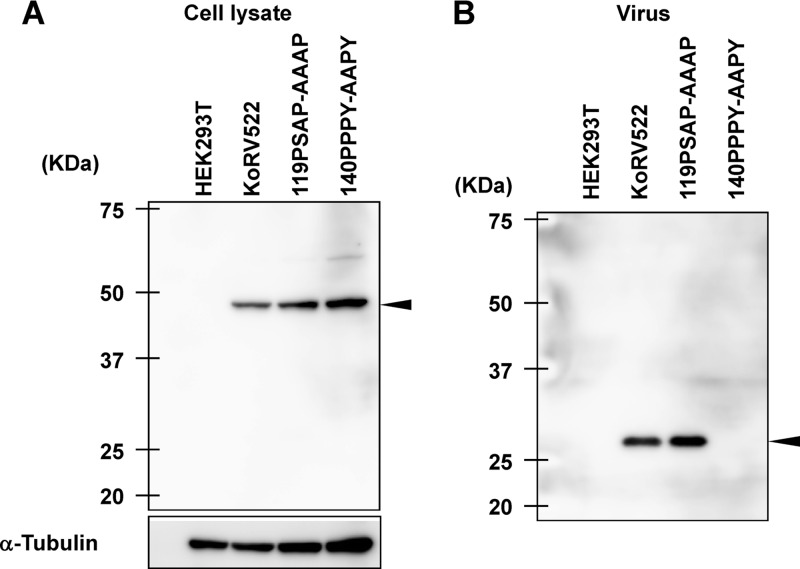
The clone pKoRV522 is highly infectious, although the PPXY motif that was reported to be involved in viral budding is disrupted (6). However, we found that an additional PPXY motif is present in the KoRV Gag sequence, as described above (Fig. 4B). To examine the functions of two putative L-domain motifs (PSAP and PPXY motifs) in KoRV Gag, we constructed expression plasmids for the KoRV mutants, which were mutated from PSAP and PPPY to AAAP and AAPY, respectively (Fig. 1). The expression plasmids for the WT and the mutants were transfected into HEK293T cells. At 72 h posttransfection, the amounts of intracellular Gag precursor (Pr65) and CA (p30) in virus particles were analyzed by immunoblotting using an anti-KoRV CA antibody. As shown in Fig. 6A, similar levels of Gag precursor were synthesized in cells expressing either WT or Gag mutants. The release of virus particles into the medium was greatly reduced by alanine substitutions of the PPPY motif, while the PSAP mutations had no apparent effect on virus production (Fig. 6B). These data suggest that the PPPY motif, but not the PSAP motif, plays a major role as the L domain in KoRV budding.
Fig 6.
Effects of mutations in the PSAP and PPXY motifs on KoRV budding. HEK293T cells were transfected with pKoRVGP-IRES or the L-domain mutants. Cells (A) and viruses (B) were collected at 48 h after transfection and analyzed by immunoblotting. Arrowheads indicate Pr50 Gag (A) and p30 CA (B) proteins, respectively.
DISCUSSION
Although the entire nucleotide sequence of KoRV has been determined and a KoRV clone has been isolated by long-range PCR using koala genomic DNA derived from PBMCs (9), studies on the virus have been hampered by the absence of an infectious molecular clone. To fill this void, we isolated a KoRV strain, termed Aki, from a koala reared in the Hirakawa Zoological Park and constructed an infectious molecular clone, designated pKoRV522. To the best of our knowledge, this is the first report on the construction and characterization of an infectious molecular clone of KoRV.
KoRV522, derived from the clone, grew efficiently in HEK293T cells but not in TE671 cells (Fig. 2A), although its pseudotype virus, lacZ(KoRV-522), was able to infect both HEK293T and TE671 cells (Fig. 5B), indicating that a block in viral growth in TE671 cells may have occurred at the postentry level. We also found that KoRV522 grew more efficiently than strain Aki in HEK293T cells (Fig. 2B). The slower growth of strain Aki may be explained by the presence of defective viruses. Although the coding sequences of pcindy are quite similar to those of pKoRV522, its reconstituted clone, pKoRV-Cindy, was not capable of producing any infectious viruses (Fig. 2C). Although the major difference between the nucleotide sequence of pKoRV522 and pKoRV-Cindy was found in the 5′ UTR, chimeric analysis by exchanging the 5′ UTR revealed that the 5′ UTR alone was not responsible for the inability of pKoRV-Cindy to replicate in cells.
In investigating the ability of pKoRV522 to infect various hosts, we found that lacZ(KoRV522)-infected cells derived from humans (HEK293T and TE671), cats (G355-5), and mink (Mv-1Lu), but not those of hamsters (CHO) and murine cell lines (NIH 3T3 and MDTF) (Fig. 5B). Consistent with our results, Fiebig et al. reported that KoRV strain Duisburg-Berlin, isolated from a koala reared in the Duisburg Zoo (Duisburg, Germany), infected HEK293T cells, human T cell lines (CEM and C8166), and a rat cell line (rat1), but not murine NIH 3T3 cells (30). In contrast to our results, Oliveira et al. demonstrated that an Env-pseudotyped virus derived from the Australian isolate, pcindy, was able to infect murine MDTF (28). The difference in murine cell susceptibility to these viruses may be attributed to five amino acid substitutions in the pcindy Env used to pseudotype virus (28) compared with pKoRV522 (Fig. 4C).
Oliveira et al. revealed several amino acid mutations involved in reduction in viral infection in vitro (28). Interestingly, even though pKoRV522 contains all of the mutations involved in the reduction of viral infectivity (Fig. 4A), the titers of the KoRV Env pseudotype viruses were comparable to those of exogenous retroviral Env pseudotype viruses in HEK293T cells (Fig. 5C). Therefore, we considered that the reduction of viral infectivity caused by these reported mutations is limited.
The disruption of the PPXY motif in the L domain of KoRV was reported to be involved in the reduction of KoRV budding. However, pKoRV-522 has the same mutations in this motif (Fig. 4B), yet it grew efficiently in HEK293T cells, reaching a maximum titer of 106 FFU/ml (Fig. 2). To explain this discrepancy, we compared the Gag sequences of KoRV and GALV and found an additional PPXY motif not previously reported. Mutations in the PSAP motif did not affect KoRV budding, whereas mutations in the novel PPXY motif had a significant impact (Fig. 6). Therefore, we conclude that the second PPXY motif is the major L-domain sequence for KoRV budding while the first PPXY is dispensable.
In conclusion, KoRV522 is highly active and replicates efficiently in HEK293T cells despite containing mutations believed to result in reduction in viral infection/production in vitro. Given that there is a relationship between the viral load in KoRV-infected koalas and leukemia, it is likely that KoRV induces diseases such as cancer and immunodeficiency in these animals, making antiretroviral drugs a viable therapeutic option for koalas in captivity. The infectious clone reported here may be a useful tool for the screening of antiretroviral drugs that can be administered to koalas.
ACKNOWLEDGMENTS
We are grateful to John Hunger (RSPCA, Brisbane, Queensland, Australia) and Atsushi Tanaka (Thailand-Japan Research Collaboration Center on Emerging and Reemerging Infections, Nonthaburi, Thailand) for providing pcindy and pBluescriptIISK+/LTR-Friend-LTR, respectively. We thank Hisashi Hashikawa (a koala specialist for the Japanese Association of Zoos and Aquariums) for helpful advice and staff at the Hirakawa Zoological Park (Kagoshima, Japan) for the collection of koala samples. We are grateful to Paul Young and Joanne Meers (University of Queensland, Brisbane, Queensland, Australia) for helpful discussions. We are grateful to M. Hattori (Kyoto University, Kyoto, Japan) for providing recombinant human interleukin 2 (IL-2)-producing Ltk−IL-2.23 cells.
This study was supported by grants from the Ministry of Education, Culture, Science and Sports of Japan and from the Bio-Oriented Technology Research Advancement Institution.
Footnotes
Published ahead of print 20 February 2013
REFERENCES
- 1. Gifford R, Tristem M. 2003. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 26:291–315 [DOI] [PubMed] [Google Scholar]
- 2. Feschotte C, Gilbert C. 2012. Endogenous viruses: insights into viral evolution and impact on host biology. Nat. Rev. Genet. 13:283–296 [DOI] [PubMed] [Google Scholar]
- 3. Stoye JP. 2006. Koala retrovirus: a genome invasion in real time. Genome Biol. 7:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tarlinton RE, Meers J, Young PR. 2006. Retroviral invasion of the koala genome. Nature 442:79–81 [DOI] [PubMed] [Google Scholar]
- 5. Delassus S, Sonigo P, Wain-Hobson S. 1989. Genetic organization of gibbon ape leukemia virus. Virology 173:205–213 [DOI] [PubMed] [Google Scholar]
- 6. Oliveira NM, Satija H, Kouwenhoven IA, Eiden MV. 2007. Changes in viral protein function that accompany retroviral endogenization. Proc. Natl. Acad. Sci. U. S. A. 104:17506–17511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lieber MM, Sherr CJ, Todaro GJ, Benveniste RE, Callahan R, Coon HG. 1975. Isolation from the Asian mouse Mus caroli of an endogenous type C virus related to infectious primate type C viruses. Proc. Natl. Acad. Sci. U. S. A. 72:2315–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Canfield PJ, Sabine JM, Love DN. 1988. Virus particles associated with leukaemia in a koala. Aust. Vet. J. 65:327–328 [DOI] [PubMed] [Google Scholar]
- 9. Hanger JJ, Bromham LD, McKee JJ, O'Brien TM, Robinson WF. 2000. The nucleotide sequence of koala (Phascolarctos cinereus) retrovirus: a novel type C endogenous virus related to Gibbon ape leukemia virus. J. Virol. 74:4264–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tarlinton R, Meers J, Hanger J, Young P. 2005. Real-time reverse transcriptase PCR for the endogenous koala retrovirus reveals an association between plasma viral load and neoplastic disease in koalas. J. Gen. Virol. 86:783–787 [DOI] [PubMed] [Google Scholar]
- 11. Brown AS, Girjes AA, Lavin MF, Timms P, Woolcock JB. 1987. Chlamydial disease in koalas. Aust. Vet. J. 64:346–350 [DOI] [PubMed] [Google Scholar]
- 12. Cockram FA, Jackson AR. 1981. Keratoconjunctivitis of the koala, Phascolarctos cinereus, caused by Chlamydia psittaci. J. Wildl. Dis. 17:497–504 [DOI] [PubMed] [Google Scholar]
- 13. O'Dair HA, Hopper CD, Gruffydd-Jones TJ, Harbour DA, Waters L. 1994. Clinical aspects of Chlamydia psittaci infection in cats infected with feline immunodeficiency virus. Vet. Rec. 134:365–368 [DOI] [PubMed] [Google Scholar]
- 14. Cosset FL, Takeuchi Y, Battini JL, Weiss RA, Collins MK. 1995. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J. Virol. 69:7430–7436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takeuchi Y, Patience C, Magre S, Weiss RA, Banerjee PT, Le Tissier P, Stoye JP. 1998. Host range and interference studies of three classes of pig endogenous retrovirus. J. Virol. 72:9986–9991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nakata R, Miyazawa T, Shin YS, Watanabe R, Mikami T, Matsuura Y. 2003. Reevaluation of host ranges of feline leukemia virus subgroups. Microbes Infect. 5:947–950 [DOI] [PubMed] [Google Scholar]
- 17. Miyazawa T, Shojima T, Yoshikawa R, Ohata T. 2011. Isolation of koala retroviruses from koalas in Japan. J. Vet. Med. Sci. 73:65–70 [DOI] [PubMed] [Google Scholar]
- 18. Ishikawa M, Okada M, Baba K, Shojima T, Shiomojima M, Miura T, Miyazawa T. 2008. Establishment of a feline astrocyte-derived cell line (G355-5 cells) expressing feline CD134 and a rapid quantitative assay for T-lymphotropic feline immunodeficiency viruses. J. Virol. Methods 151:242–248 [DOI] [PubMed] [Google Scholar]
- 19. Morita S, Kojima T, Kitamura T. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063–1066 [DOI] [PubMed] [Google Scholar]
- 20. Tanaka A, Oka K, Tanaka K, Jinno A, Ruscetti SK, Kai K. 1998. The entire nucleotide sequence of friend-related and paralysis-inducing PVC-441 murine leukemia virus (MuLV) and its comparison with those of PVC-211 MuLV and Friend MuLV. J. Virol. 72:3423–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mullins JI, Casey JW, Nicolson MO, Burck KB, Davidson N. 1981. Sequence arrangement and biological activity of cloned feline leukemia virus proviruses from a virus-productive human cell line. J. Virol. 38:688–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wiseman JW, Goddard CA, McLelland D, Colledge WH. 2003. A comparison of linear and branched polyethylenimine (PEI) with DCChol/DOPE liposomes for gene delivery to epithelial cells in vitro and in vivo. Gene Ther. 10:1654–1662 [DOI] [PubMed] [Google Scholar]
- 23. Sakaguchi S, Okada M, Shojima T, Baba K, Miyazawa T. 2008. Establishment of a LacZ marker rescue assay to detect infectious RD114 virus. J. Vet. Med. Sci. 70:785–790 [DOI] [PubMed] [Google Scholar]
- 24. Ma YK, Khan AS. 2009. Evaluation of different RT enzyme standards for quantitation of retroviruses using the single-tube fluorescent product-enhanced reverse transcriptase assay. J. Virol. Methods 157:133–140 [DOI] [PubMed] [Google Scholar]
- 25. Fischer N, Krach U, Niebert M, Tönjes RR. 2003. Detection of porcine endogenous retrovirus (PERV) using highly specific antisera against Gag and Env. Virology 311:222–228 [DOI] [PubMed] [Google Scholar]
- 26. Krach U, Fischer N, Czauderna F, Kurth R, Tönjes RR. 2000. Generation and testing of a highly specific anti-serum directed against porcine endogenous retrovirus nucleocapsid. Xenotransplantation 7:221–229 [DOI] [PubMed] [Google Scholar]
- 27. Fukuma A, Abe M, Urata S, Yoshikawa R, Morikawa Y, Miyazawa T, Yasuda J. 2011. Viral and cellular requirements for the budding of feline endogenous retrovirus RD-114. Virol. J. 8:540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oliveira NM, Farrell KB, Eiden MV. 2006. In vitro characterization of a koala retrovirus. J. Virol. 80:3104–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bieniasz PD. 2006. Late budding domains and host proteins in enveloped virus release. Virology 344:55–63 [DOI] [PubMed] [Google Scholar]
- 30. Fiebig U, Hartmann MG, Bannert N, Kurth R, Denner J. 2006. Transspecies transmission of the endogenous koala retrovirus. J. Virol. 80:5651–5654 [DOI] [PMC free article] [PubMed] [Google Scholar]