Abstract
The DNA damage response (DDR) of host cells is utilized by a number of viruses to establish and propagate their genomes in the infected cells. We examined the expression of the DDR genes during Kaposi's sarcoma-associated herpesvirus (KSHV) infection of human peripheral blood mononuclear cells (PBMCs). The genes were mostly downregulated, except H2AX, which was upregulated during infection. H2AX is important for gammaherpesvirus infectivity, and its phosphorylation at serine 139 is crucial for maintenance of latency during mouse gamma-herpesvirus 68 (MHV-68) infection. We now also observed phosphorylation of H2AX at serine 139 during KSHV infection. H2AX is a histone H2A isoform shown to interact with the latency-associated nuclear antigen (LANA) encoded by KSHV. Here, we show that LANA directly interacted with H2AX through domains at both its N and C termini. The phosphorylated form of H2AX (γH2AX) was shown to colocalize with LANA. Chromatin immunoprecipitation (ChIP) assays showed that a reduction in H2AX levels resulted in reduced binding of LANA with KSHV terminal repeats (TRs). Binding preferences of H2AX and γH2AX along the KSHV episome were examined by whole-episome ChIP analysis. We showed that γH2AX had a higher relative binding activity along the TR regions than that of the long unique region (LUR), which highlighted the importance of H2AX phosphorylation during KSHV infection. Furthermore, knockdown of H2AX resulted in decreased KSHV episome copy number. Notably, the C terminus of LANA contributed to phosphorylation of H2AX. However, phosphorylation was not dependent on the ability of LANA to drive KSHV-infected cells into S-phase. Thus, H2AX contributes to association of LANA with the TRs, and phosphorylation of H2AX is likely important for its increased density at the TRs.
INTRODUCTION
Viruses are molecular parasites, well known for utilizing the cellular machinery of hosts for their own benefit. These viruses utilize different cellular pathways to various degrees to complete their infection cycle. However, almost all viruses are completely dependent on the host's translational machinery for synthesis of viral proteins. Additional pathways are utilized to ensure the survival and propagation of the virus in the infected host cell. Studies over the past decade have shown that a number of viruses can modulate the DNA damage repair pathway of their host (1). Transduction and replication of viruses in mammalian cells lead to introduction of large amounts of foreign genetic material that induces signaling of the DNA damage response (DDR) pathway in response to infection (2). Additionally, a number of viral proteins have also been implicated in triggering the DDR through different mechanisms (2–4).
Human herpesvirus 8 (HHV8) is the causative agent of Kaposi's sarcoma (KS) and primary effusion lymphoma (PEL) and is associated with multicentric Castlemen's disease (MCD) (5). It is a linear double-stranded DNA virus with a biphasic life cycle in the infected host (5, 6). After an initial round of lytic replication, latent infection is established where the viral genome persists in the host as a circular episome (5). During host genome replication and cell division, viral episomes are replicated once per cycle and redistributed to the progeny cells (5, 6). Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded latency-associated nuclear antigen (LANA) is a major contributor to the mechanism whereby the episome is tethered to the host genome, which is important for redistribution of the episome to daughter cells during cell division (5, 7, 8). This function of LANA is crucial to the KSHV life cycle and is shown to be important, as demonstrated by a number of protein-protein and protein-DNA interactions associated with episome tethering (5, 9–11). LANA also forms complexes with the human origin recognition complex (ORC) proteins (59). However, a previous study from our group has shown that multiple regions of the KSHV genome can be utilized for initiating latent replication (60). Host molecules known to be associated with LANA and that function in episome tethering are histones H1, H2A, H2B, Brd2 (Ring3), Brd4, DEK, MeCP2, NuMA, CENP-F, and Bub1 (9–17). These proteins have been shown to contribute to LANA's interaction with host chromatin. However, episome binding of LANA is believed to be mostly a result of direct involvement of LANA with a large DNA-protein complex with the TR elements (6, 8, 10).
The histones are likely to be critical for linking LANA directly to host nucleosomes (12, 13). One variant of histone H2A is H2AX, a key molecule in the DNA damage repair pathway (18–20). It has a very high sequence homology with H2A (95% identity, 98% positive by NCBI blastx) and comprises approximately 10% to 15% of total cellular H2A (19). H2AX has a unique C-terminal tail that, in response to DNA damage, is phosphorylated extensively on a conserved serine (serine 139) residue (21). This phosphorylation is a hallmark of the DDR and is thought to be mediated by cellular kinases of the phosphoinositide-3-kinase-related protein kinase (PI3K) family, namely, ataxia telangiectasia mutated (ATM), ataxia telangiectasia- and Rad3-related protein (ATR), and the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) (21). A number of viral kinases are also known to phosphorylate H2AX during the course of infection (22, 23).
Infection by gammaherpesviruses, including KSHV, was previously shown to induce phosphorylation of H2AX during the course of their infections (3, 22, 23). However, these studies lacked the details of the mechanism responsible for triggering H2AX phosphorylation in the presence of KSHV infection. In mouse gamma-herpesvirus 68 (MHV-68), H2AX plays a crucial role in viral replication, as is evident by the fact that the MHV-68 titer in H2AX−/− macrophages is approximately 100-fold lower than in H2AX+/+ hosts (22). Another study on MHV-68 also reveals the importance of H2AX for efficient establishment of early MHV-68 latency (23). Further, in response to intranasal administration of MHV-68 virions, H2AX-deficient mice have 5-fold less viral genome-positive splenocytes than wild-type and heterozygous littermates (23).
Previous observations from the above-mentioned studies suggests that H2AX may play a role in episome persistence during gamma-herpesvirus latency. To examine the modulation of the host cell DDR during KSHV infection, we carried out gene expression profiling with a focus on the DDR-related genes. Most of the DDR genes showed downregulation. However, H2AX was clearly upregulated in response to KSHV infection. This molecule was also highly phosphorylated in response to KSHV infection. In this study, we show that H2AX and γH2AX bind and colocalize with LANA and TR and therefore contribute to a successful KSHV infection in a permissive cell background.
Phosphorylated H2AX has a significant level of affinity toward TRs even in cells with latently infected genomes. Furthermore, we show that LANA also has a role in phosphorylation of H2AX and that its C terminus is directly involved in contributing to the enhanced phosphorylation.
MATERIALS AND METHODS
Plasmids, cells, and antibodies.
The pBS-puro-TR plasmid contains three copies of the TR elements cloned in a pBluescript backbone (24). H2AX was cloned from RNA isolated from human peripheral blood mononuclear cells (PBMCs). The cDNA was synthesized as described previously (25). Specific primers containing restriction enzyme sites for cloning in pA3M and pGEX-2TK vectors (BamHI/XhoI for cloning in pA3M and BamHI/EcoRI for cloning in pGEX2T) were used for amplification of a specific product for cloning. The pA3F-LANA expresses FLAG-tagged LANA (16). pA3M-LANA, pA3M-N-LANA, and pA3M-GFP-C-LANA (where GFP is green fluorescent protein) express Myc-tagged, full-length LANA, N-terminal of LANA (amino acids [aa] 1 to 340), and C-terminal of LANA (aa 930 to 1162) with GFP tag, respectively, and have been described previously (16). LANA promoter construct pGL2-LANA has been described previously (26). For its expression as a glutathione S-transferase (GST) fusion protein, cDNA from PBMCs was amplified using primers specific for H2AX amplicon and cloned in pGEX2T vector. FLAG-H2AX and FLAG-H2AX(S-A) expression constructs were provided by Alvaro N. A. Monteiro (H. Lee Moffitt Cancer Center, Tampa, FL). An shH2AX construct was also provided by Titia de Lange (The Rockefeller University, New York). Sequences of all the constructs were verified by DNA sequencing (DNA Sequencing Facility, University of Pennsylvania).
For cell culture, a humidified incubator supplemented with 5% CO2 was used at 37°C. Human PBMCs were obtained from the CFAR Immunology Core at the University of Pennsylvania. An IRB-approved protocol was used by the CFAR Immunology Core as per declarations of Helsinki protocols, and a written, informed consent was obtained from each donor. KSHV-negative cell line BJAB and the KSHV-positive cell lines BCBL1 and BC3 were cultured in RPMI 1640 medium with 10% bovine growth serum (BGS) with additional supplements as described previously (14). Human embryonic kidney 293 (HEK-293) and HEK-293–BAC36–KSHV cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% BGS, penicillin-streptomycin (5 U/ml and 5 μg/ml, respectively), and 2 mM l-glutamine (14). ECV304 is an endothelial cell line provided by Harry Ischiropoulos (University of Pennsylvania, Philadelphia, PA) and has been described previously (27). ECV304 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), penicillin-streptomycin (5 U/ml and 5 μg/ml, respectively), and 2 mM l-glutamine.
Fluorescein isothiocyanate (FITC)-conjugated mouse anti-γH2AX (Ser139) monoclonal antibody (clone JBW301) was purchased from Millipore Inc. (Billerica, MA). Rabbit polyclonal anti-H2AX antibody was purchased from Bethyl Laboratories Inc. (Montgomery, TX), and unconjugated mouse anti-γH2AX monoclonal (2F3) antibody was purchased from BioLegend Inc. (San Diego, CA). Hybridoma culture supernatants were used as sources of anti-Myc (9E10) and anti-LANA (LANA1) antibodies. Mouse anti-FLAG monoclonal antibody (M2) was purchased from Sigma-Aldrich Corp. (St. Louis, MO).
Virus production and transduction of PBMCs.
A previously described protocol was used with minor changes (28). Approximately 500 million exponentially growing HEK-293–BAC36–KSHV cells were induced with 20 ng/ml of tetradecanoyl phorbol acetate (TPA) and 1.5 mM sodium butyrate (Sigma-Aldrich Corp., St. Louis, MO) for 4 to 5 days at 37°C in 5% CO2. Supernatant was centrifuged and filtered through a 0.45-μm-pore-size filter to remove the cell debris. Virions were collected by centrifuging the filtered supernatant at 23,500 rpm for 2 h at 4°C in a Sorvall discovery 90SE ultracentrifuge. Viruses from HEK-293–BAC36–KSHV were used to infect approximately 10 million PBMCs. Infection was carried out by incubating PBMCs with virus using 4 μg/ml Polybrene in RPMI 1640 medium (10% BGS) and 5% CO2 at 37°C for 3 h. Cells were centrifuged at 1,500 rpm for 5 min and resuspended in fresh RPMI 1640 medium (10% BGS) in a 6-well plate. The infection was examined by visualization of GFP expression using fluorescence microscopy.
Real-time PCR.
Total RNA was extracted from cells using TRIzol reagent (Invitrogen Inc., Carlsbad, CA) as per the manufacturer's instruction. A high-capacity RNA-to-cDNA kit (Applied Biosystems Inc., Foster City, CA) was used to make cDNA. Real-time PCR was set up using SYBR green PCR master mix (Applied Biosystems Inc., Foster City, CA) as per the manufacturer's protocol. A StepOne real-time PCR cycler (Applied Biosystems) was used with the following cycling conditions: 10 min at 95°C for initial denaturation, 40 cycles each at 95°C for 20 s for denaturation, 54.5°C for 20 s for annealing, and 72°C for 20 s for polymerization. Data were collected at every cycle after the polymerization step. A melt curve analysis was also performed to ensure the specificity of amplified products; relative quantitation was carried out by the threshold cycle method. All reactions were set up in triplicate. Primers for real-time PCR are provided in the supplemental data (see Table S1).
GST pulldown assay.
For expression of GST and GST fusion proteins, bacterial culture was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at log phase (optical density at 600 nm [OD600] = 0.6), and cells were incubated with shaking at 37°C for 4 h. Bacterial lysis and purification of protein(s) with glutathione-Sepharose beads were carried out as described earlier (29). Quantification was done by SDS-PAGE of purified proteins with bovine serum albumin (BSA) standards followed by densitometry analysis of Coomassie brilliant blue-stained gel using an Odyssey imager (LiCor Inc., Lincoln, NE). GST fusion protein, bound to glutathione-Sepharose beads, was incubated with the cell lysates to allow binding of GST fusion protein with the target protein. After stringent washes, the beads were boiled in SDS-sample loading buffer to allow denaturation and dissociation of bound proteins. The sample was centrifuged, and the supernatant was subjected to SDS-PAGE fractionation followed by Western blotting.
Coimmunoprecipitation (Co-IP) and Western blotting.
From the lysates of approximately 20 million cells, immunoprecipitation (IP) of exogenously expressed protein was carried out using specific antibodies as has been described previously (14). Briefly, the lysates from cells expressing the probable interacting proteins was incubated with nonspecific IgG for 1 h and bead-loaded control, followed by specific antibodies against one of the interacting proteins, as indicated. Immune complexes were collected by binding with protein A/G-Sepharose beads. After stringent washes, the beads were boiled in SDS sample loading buffer and centrifuged, and supernatant was subjected to SDS-PAGE fractionation followed by Western blot (WB) analysis. The membrane was blocked with 5% skim milk in phosphate-buffered saline (PBS) for 1 h, followed by incubation with the appropriate dilution of primary antibodies (in PBS) for 2 h at room temperature. IR dye-tagged secondary antibodies were used to detect the binding of primary antibodies, and the membrane was scanned using an Odyssey imager (LiCor Inc., Lincoln, NE). Quantitation of the protein bands was carried out using Odyssey scanning software.
Immunofluorescence (IF) analysis.
Cells were washed in PBS, layered on poly-lysine-coated coverslips, and incubated for 1 h. To fix and permeabilize the cells, coverslips were submerged in 3% paraformaldehyde containing 0.1% Triton X-100 for 30 min at room temperature. Cells were washed twice with 1× PBS and blocked with 5% skim milk in PBS for 1 h. For LANA and γH2AX colocalization experiments, cells were incubated with mouse anti-LANA antibody and then probed with Alexa 594-linked goat anti-mouse antibody. Next, γH2AX was probed with FITC-labeled anti-γH2AX antibody. DAPI (4′,6-diamidino-2-phenylindole) was used for nuclear staining. After being washed with PBS, coverslips were mounted on glass slides using mounting medium and visualized with an Olympus FluoView 300 confocal microscope.
RNA interference.
For short hairpin RNA (shRNA) cloning, pGIPZ vector was purchased from Open Biosystems, Inc. (Huntsville, AL). Oligonucleotides were designed against a unique region of H2AX, 5′ CAACAAGAAGACGCGAATC 3′, and cloning was carried out according to the manufacturer's guidelines. A nonspecific shRNA sequence with no significant homology with other human mRNA was used as a control and has been described previously (11). To determine if there were any nonspecific effects of the shH2AX, we used the second shRNA set for H2AX, which has been reported earlier by Dimitrova and Lange (10).
Immunofluorescence in situ hybridization (immuno-FISH).
Probe preparation. The probe was prepared against the KSHV TR region by digesting the pBS-puro-TR vector with NotI enzyme. The TR region was gel extracted and labeled using the Bionick labeling system (Invitrogen, Carlsbad, CA), used to generate the biotinylated probes by nick translation as per the manufacturer's instruction. Briefly, the reaction for nick translation was set at 16°C for 2 h. After the reaction was stopped, unincorporated nucleotides were removed from the labeled probe by using the Zymoclean gel DNA recovery kit (Zymo Research Corp., Irvine, CA). The probe was eluted in TE (10 mM Tris-HCl, 1 mM EDTA, pH 7.5) buffer and stored at −20°C. The sequence of TR FISH probe is CTTGCTTTCGTTTTCTCCC.
Immuno-FISH was carried out as has been described previously (16), with a few modifications. In brief, cells grown on the coverslip were fixed and permeabilized with 4% paraformaldehyde containing 0.4% Triton X-100, at room temperature for 30 min. After two PBS washes, cells were treated with RNase A (100 μg/ml) in 2× SSC (0.3 M NaCl plus 0.03 M sodium citrate) at 37°C for 30 min. Coverslips containing cells were washed twice with 2× SSC, overlaid with warm (preheated to 72°C) prehybridization buffer (50% formamide, 10% dextran sulfate, and 2× SSC), and incubated at 72°C for 7 min. The probes (approximately 20 ng/ml) were denatured by heating in hybridization buffer (i.e., 1 mg/ml of sonicated salmon sperm DNA in prehybridization buffer) at 72°C for 7 min. Coverslips and probe were brought to 37°C, and coverslips were washed once with 2× SSC for 5 min. The probe was overlaid on coverslips and incubated in a humidity chamber at 37°C overnight. Coverslips were washed in 0.1× SSC at 42°C for 10 min and then with 2× SSC at 42°C for 10 min. Blocking was done with 4% skim milk for 30 min, and the following steps were carried out as described above. For detection, Alexa Fluor 594 goat anti-mouse IgG, Alexa Fluor 594 goat anti-rabbit IgG, and Alexa Fluor 680 streptavidin conjugate were used to detect γH2AX, H2AX, and TR, respectively. Cells were also counterstained with DAPI. An Olympus FluoView FV300 confocal microscope was used for visualization.
ChIP assay.
Chromatin immunoprecipitation (ChIP) was carried out as has been described earlier (24, 30). Briefly, after the cells were fixed, nuclei were isolated and sonicated in the nuclear lysis buffer to an average DNA length of 700 bp, as confirmed by agarose gel electrophoresis. Samples were precleared with salmon sperm DNA-protein A-Sepharose slurry for 30 min at 4°C with rotation. Ten percent of the total supernatant was saved for input control, and the remaining 90% was divided into two fractions and incubated with (i) control antibody (Sigma, Inc., St. Louis, MO) or (ii) mouse monoclonal anti-γH2AX, rabbit polyclonal anti-H2AX, or mouse monoclonal anti-FLAG tag antibody. The precipitated immune complex was washed for stringency, reverse cross-linked, and purified using a Zymo spin column from Zymo Research Corp. (Irvine, CA) after proteinase K treatment.
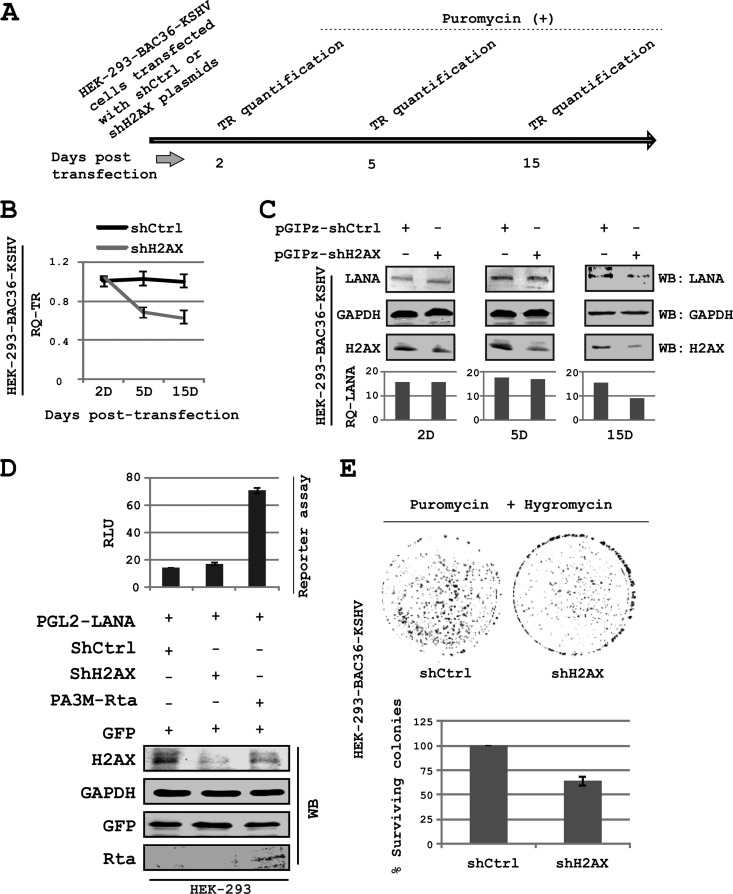
Episome retention assay.
HEK-293–BAC36–KSHV cells were transfected with pGIPz-shCtrl or pGIPz-shH2AX plasmids. After 2 days posttransfection, the cells were grown in DMEM containing 1× puromycin (2 μg/ml), and the medium was changed on alternate days. Cells were passed at approximately 80% confluence. Episome copies were determined by real-time PCR on the 2nd, 5th, and 15th days posttransfection. To isolate the episomal DNA for real-time PCR, a modified Hirt's DNA isolation procedure was used (14). The KSHV TR region was amplified to measure the episome copies using specific primers. An amplicon along the genomic DNA coding for XRCC1 was used as the internal control for real-time PCR.
Colony formation assay.
Approximately 3,000 cells from each clone of HEK-293–BAC36–KSHV, stably harboring pGIPz-shCtrl or pGIPz-shH2AX plasmids were inoculated in a 100-mm2 petri plate and grown in DMEM containing puromycin (2 μg/ml) and hygromycin (150 μg/ml). Medium was changed on alternate days. After 2 weeks, the medium was aspirated and the cell layer was washed twice in PBS and fixed in 4% paraformaldehyde (PFA) for 20 min in the dark. Cells were stained with 0.1% crystal violet (Sigma-Aldrich Corp., St. Louis, MO). Plates were photographed using an Odyssey scanner (LiCor Inc., Lincoln, NE) using the 680 channel, and the number of colonies was counted using ImageJ software (National Institutes of Health, Bethesda, MD) (31).
Reporter assay.
Reporter assays were carried out as described previously with minor modifications (11). HEK-293 cells were transfected with plasmids as indicated. Transfection with the pEGFP-C1 construct was carried out for monitoring the transfection efficiency. Cells were lysed in reporter lysis buffer (Promega Inc., Madison, WI) 36 h after transfection, and the luciferase activity was measured using LMaxII384 luminometer (Molecular Devices, Sunnyvale, CA). In this assay, we used the basic pGL2 plasmid as a negative control, which reflects the differences between negative controls compared to the positive LANA promoter. The graphs were drawn in reference of LANA promoter to calculate the fold change.
H2AX phosphorylation and cell cycle analysis.
For the cytometric determination of H2AX phosphorylation, HEK-293 cells were transfected with 15 μg of the indicated plasmids. After 36 h posttransfection, H2AX phosphorylation (on serine 139) along various cell cycle phases was determined by dual labeling of cells with antibodies specific for γH2AX and propidium iodide (PI) staining. The process was carried out as previously described by Huang and Darzynkiewicz with slight modifications (32). Briefly, cells were trypsinized, resuspended in DMEM, and washed in PBS twice. Approximately 5 million cells were fixed with 2% PFA at room temperature for 10 min. PFA was removed by washing with PBS. Cells were resuspended in ice chilled with 70% ethanol and kept at −20°C for 2 h. Cells were collected by centrifugation and washed 3 times with 5 ml of BSA-T-PBS (1% bovine serum albumin [wt/vol] and 0.2% Triton X-100 [vol/vol] in phosphate-buffered saline). For primary antibody treatment, cells were resuspended in 100 μl of BSA-T-PBS containing 1.5 μg of anti-γH2AX antibody and incubated overnight at 4°C. Cells were washed twice with 1 ml of BSA-T-PBS and incubated with 100 μl of secondary antibody solution (goat anti-mouse Alexa-647 diluted 1:500 in BSA-T-PBS) in the dark at room temperature for 1 h. Unbound antibody was removed by 2 washes of BSA-T-PBS. For cell cycle analysis, cells were simultaneously stained with propidium iodide staining solution (50 mg/ml propidium iodide and 500 U/ml RNase A in 10 mM Tris [pH 7.5]) for 2 h in the dark.
RESULTS
Expression of DDR genes and phosphorylation status of H2AX during KSHV infection.
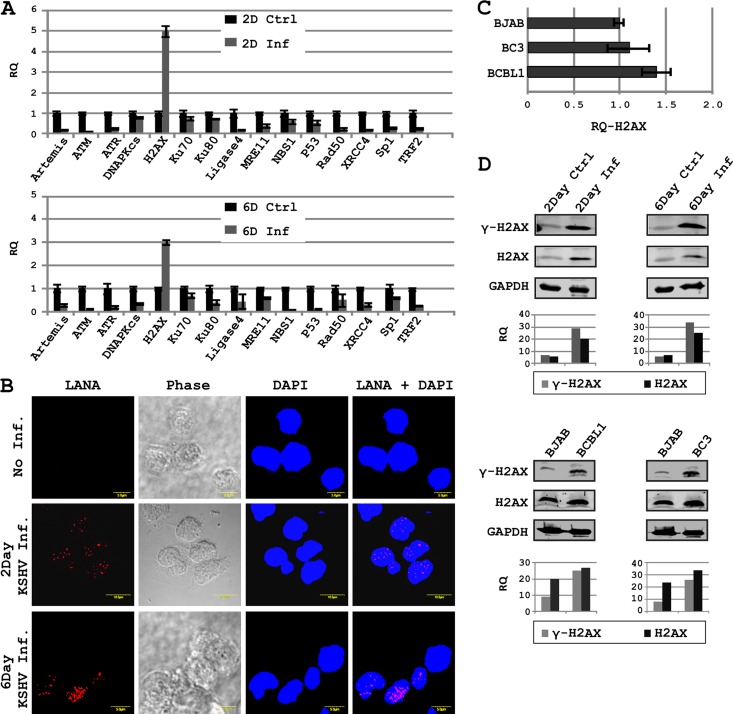
KSHV is a linear double-stranded DNA virus which infects primarily endothelial and lymphatic cell types (6). We examined the expression status of a total of 15 DDR-related genes in response to KSHV infection of human PBMCs by real-time PCR (Fig. 1A). Infection of PBMCs by KSHV was confirmed by examining the infected cells for LANA expression at day 2 and day 6 by immunofluorescence assays (Fig. 1B). The DDR molecules included in the present study are Artemis, ATM, ATR, DNA-PKcs, H2AX, Ku70, Ku80, Ligase4, MRE11, NBS1, p53, Rad50, XRCC4, Sp1, and TRF2. These molecules are important constituents of the mammalian DDR machinery (2, 33–35). Interestingly, most of the genes were downregulated, except H2AX, which was found to be significantly upregulated on day 2 as well as on day 6 of infection albeit at a lower level, which was about 50% of that seen on day 2 (Fig. 1A). Expression of LANA in primary cells (PBMCs) was also monitored up to 6 days postinfection with KSHV by immunofluorescence. An increase in LANA signal was seen which supported infection of the cells at 6 days postinfection (Fig. 1B). Furthermore, Artemis, ATM, ATR, NBS1, p53, XRCC4, and TRF2 were all downregulated by as much as 5-fold or greater (Fig. 1A). H2AX has previously been implicated in gamma-herpesvirus pathogenicity (36). However, we wanted to examine if H2AX expression is dependent on the initial phase of KSHV infection. Therefore, we examined H2AX levels in B-cell lines latently infected with KSHV. Both BC3 and BCBL1 cell lines showed a slight trend for upregulation of H2AX transcripts compared to the KSHV-negative Burkitt's lymphoma BJAB cell line (Fig. 1C). It should be noted that the fold change seen in primary cells infected compared to uninfected was much greater, as seen above, suggesting an increase in H2AX levels in the KSHV-negative Burkitt's cell line above that in human PBMCs, probably due to their transformed state.
Fig 1.
KSHV infection modulates expression and phosphorylation of H2AX. (A) Expression of H2AX and/or other DDR genes was examined by real-time PCR in KSHV-infected PBMCs; (B) KSHV infection of PBMCs was confirmed by probing LANA by immunofluorescence assays; (C) H2AX transcript levels were also examined in KSHV-positive cell lines by real-time PCR; (D) phosphorylation of H2AX in KSHV-positive cell lines and KSHV-infected PBMCs was examined by Western blotting with specific antibodies. Bar diagrams show densitometry-based relative quantification of H2AX or γH2AX normalized against GAPDH.
H2AX phosphorylated at serine 139 is considered the functionally active form which is known to bind to DNA breaks and is a hallmark of DDR induction (21). We examined the phosphorylation status of H2AX at serine 139 during PBMC infection and KSHV-infected B-cell lines BCBL1 and BC3. Interestingly levels of phosphorylated H2AX were increased in response to KSHV infection (Fig. 1D), suggesting that during KSHV infection, an increase in H2AX expression is also accompanied by its phosphorylation.
Phosphorylation of H2AX in KSHV lesions has previously been reported (3). An increase in the number of p53 binding protein 1 (53 BP1) foci in infected endothelial cells was also monitored as an indirect indication of H2AX phosphorylation in response to KSHV infection (3, 37). The KSHV latent protein vCyclin has been implicated in H2AX phosphorylation (3). However, the major latent protein LANA is known to interact with histones, including the H2AX isoform H2A (9, 13). H2AX and H2A have very high sequence homology (95% identity, 98% positive by NCBI blastx). Therefore, we examined the interaction between LANA and H2AX and the implication for contributing to episome persistence as previously described for LANA and H2A interaction (12).
LANA interacts with H2AX through its amino- and carboxy-terminal domains.
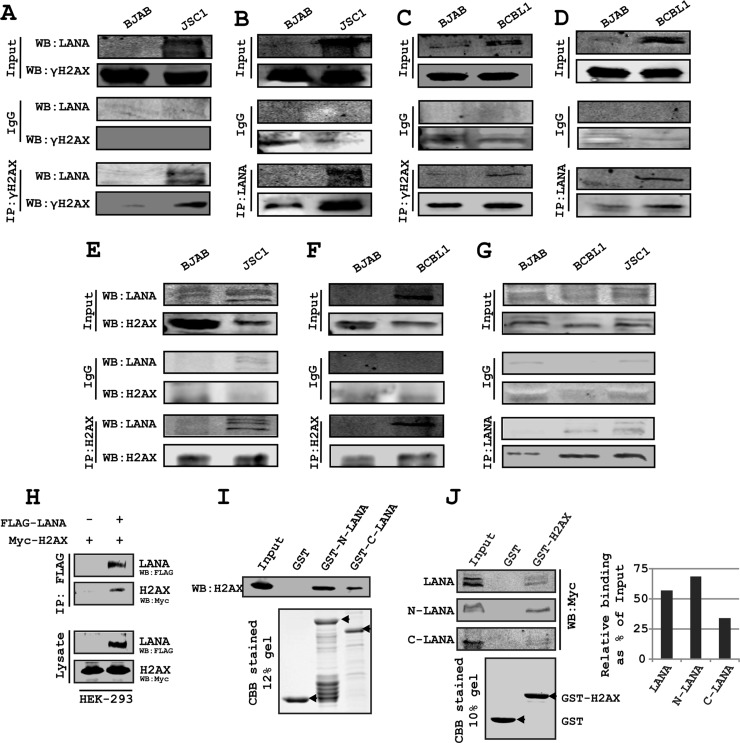
LANA binding with histones is linked to its episome retention function (9). We further examined the association of H2AX/γH2AX with LANA by coimmunoprecipitation and GST pulldown assays. We performed immunoprecipitation assays for γH2AX Co-IP with LANA and the reverse assay for LANA Co-IP with γH2AX in KSHV-positive cells. These results demonstrated that γH2AX strongly associated with LANA and that the phosphorylated form of H2AX can definitely associate with LANA (Fig. 2A to D). To support the results from our exogenously expressed proteins, we monitored formation of complexes between LANA and H2AX by immunoprecipitation of LANA using H2AX-specific antibodies in KSHV-positive cell lines BCBL1 and JSC-1 and the reverse (Fig. 2E to G). Western blotting for H2AX specifically showed the presence of H2AX in the LANA-associated protein complex from the KSHV-positive PEL cell lines (Fig. 2A to H). In HEK-293 cells, immunoprecipitation of LANA tagged with Flag epitope showed its association with H2AX tagged with a Myc epitope, as determined by Western blot analysis (Fig. 2H). Further, the GST pulldown assay revealed independent expression of either the amino (aa 1 to 340) or carboxy (aa 930 to 1162) terminus of LANA as GST fusion proteins formed a complex with H2AX expressed in HEK-293 cells in vitro (Fig. 2I). To examine the relative binding of full-length N and C termini of LANA with H2AX, we carried out a GST pulldown assay using GST-H2AX (Fig. 2J). The results showed similar binding of full-length LANA and the N terminus of LANA with H2AX; however, the C terminus bound to LANA with less intensity (Fig. 2J). Binding of the N terminus of LANA to histone H2AX may contribute to anchoring of LANA to the host genome, as has been shown for its isoform H2A (12). However, binding of the C terminus of LANA to H2AX in the context of the previously described role of the LANA C terminus in binding to the KSHV TR suggests a potential role for H2AX in a complex containing LANA and the TR. However, in a Co-IP experiment, the binding of LANA with wild-type and serine 139-alanine-mutated H2AX was similar (data not shown). This is justified and could be attributed mainly to the N terminus of LANA that showed similar binding with H2AX as full-length LANA (data not shown). Relatively higher levels of colocalization of LANA were seen with γH2AX, which may be the result of differential localization of γH2AX and H2AX in the cell. LANA is known to display a punctuate expression pattern in the presence of KSHV TR. In the above-described studies, we now show that LANA colocalizes with γH2AX. Therefore, we wanted to further evaluate the potential complex formed between the KSHV TRs and either H2AX or γH2AX in the background of KSHV infection.
Fig 2.
LANA interacts with H2AX and γH2AX. (A, C) Coimmunoprecipitation of LANA with γH2AX was examined with endogenous proteins in JSC-1 and BCBL1 cells; (B, D) coimmunoprecipitation of γH2AX with LANA was examined with endogenous proteins in JSC-1 and BCBL1 cells; (E, F) coimmunoprecipitation of LANA with H2AX was examined with endogenous proteins in JSC-1 and BCBL1 cells; (G) coimmunoprecipitation of H2AX with LANA was examined with endogenous proteins in JSC-1 and BCBL1 cells. In panels A to G, nonspecific IgG control antibody was used before specific antibody binding. (H) Coimmunoprecipitation of H2AX with LANA was examined with exogenously expressed proteins in HEK-293 cells. (I) GST pulldown assay shows binding of H2AX with the N (aa 1 to 340) and C (aa 930 to 1162) termini of LANA. GST-N-LANA and GST-C-LANA bound to glutathione-Sepharose beads were used to pull down H2AX from pA3M-H2AX-transfected 293 cell lysate. (J) GST pulldown assay to show relative binding of the full-length N terminus (aa 1 to 340) and C terminus (aa 930 to 1162) of LANA with H2AX. GST-H2AX bound to glutathione-Sepharose beads was used to pull down LANA and its truncations expressed in HEK-293 cells. Ten percent of input samples and 10% of elution fraction were loaded. Bar diagrams show densitometry-based relative quantification of LANA and its N and C terminus polypeptide.
H2AX can mediate the interaction between LANA and TR.
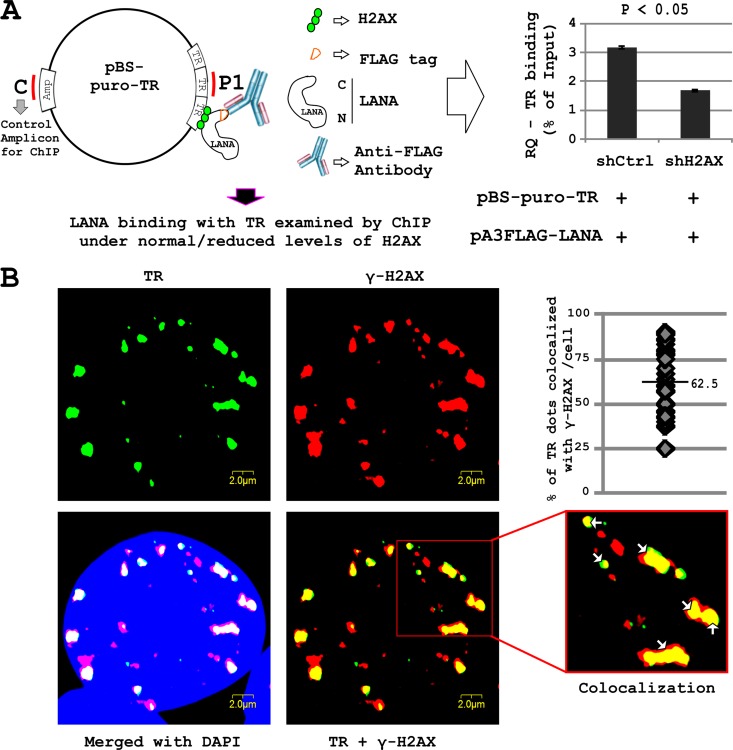
Previous studies showed an interaction of LANA with histones, and more specifically the C terminus of LANA with the TR (8, 38–44). This prompted us to explore the role of H2AX (or γH2AX) in mediating the interaction between LANA and the TR. To investigate further, we transfected HEK-293 cells with the pA3F-LANA plasmid expressing LANA, the pBS-puro-TR plasmid containing 3 copies of the KSHV TR, and either the shRNA expression plasmid pGIPz-shCtrl (control plasmid) or pGIPz-shH2AX (which knocks down H2AX). After 48 h of transfection, we performed a ChIP assay by immunoprecipitating LANA using anti-FLAG antibody under normal and reduced levels (by shRNA knockdown) of H2AX. The binding of LANA to TR was quantified by real-time PCR (Fig. 3A). In HEK-293 cells knocked down for H2AX, the binding of LANA to the TR was reduced by approximately 30 to 40%. A region from the ampicillin open reading frame (ORF) of pBS-puro-TR was used as a control region in the ChIP assay. Choosing the control region within the same plasmid nullifies the variations imposed due to various degrees of episome loss during cell division. The results suggested that H2AX is an important contributor to the interaction of LANA with the TR (Fig. 3A).
Fig 3.
(A) Role of H2AX in LANA binding with TR. HEK-293 cells were transfected with pA3FL-LANA, pBS-puro-TR, and pGIPz-shCtrl or pGIPz-shH2AX constructs. After 24 h posttransfection, cells were fixed and ChIP was carried out using M2 antibody. Levels of TRs were compared in the different samples. Data have been normalized using an amplicon in the Ampr region of pBS as a reference sequence. (B) Immunofluorescent in situ hybridization (immuno-FISH) was carried out to examine the localization of KSHV TR and γH2AX in HEK-293–BAC36–KSHV cells. Cells were hybridized with the biotinylated KSHV TR probe, followed by incubation with specific antibody against γ-H2AX. The staining was detected by incubation with Alexa Fluor 594 anti-mouse IgG and Alexa Fluor 680 streptavidin conjugate. Alexa Flour 680 staining is shown as a green pseudocolor. Cells were also counterstained with DAPI.
The presence of γH2AX on the TRs was also validated by immuno-FISH analysis carried out on KSHV-positive HEK-293 cells. The results showed significant colocalization (62.5%) between the TR and γH2AX (Fig. 3B) and again corroborated the above-described studies showing that phosphorylated H2AX increased in the presence of the KSHV genome. Further, our in vitro ChIP analysis showed that under reduced levels of H2AX, LANA binding to the TR was also reduced. This supports our hypothesis and suggests that phosphorylated H2AX may be localized to KSHV TR and can contribute to binding of LANA to the TRs.
The relative occupancy of H2AX and γH2AX on the KSHV episome suggests specificity of γH2AX to regions of the KSHV episome.
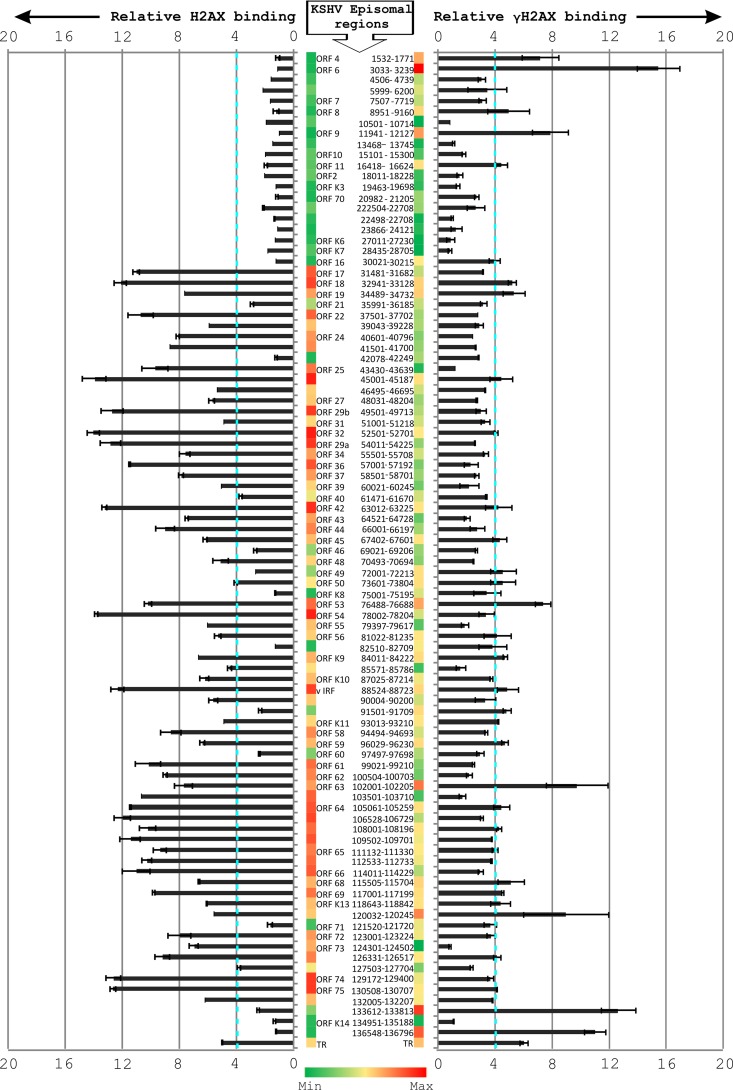
γH2AX can efficiently recognize sites of DNA damage (19). In view of its upregulation and phosphorylation in response to KSHV infection, we wanted to determine the abundance of this DDR protein associated with the KSHV episome. A whole-episome ChIP assay was carried out with KSHV-positive BCBL1 cells using KSHV primers across the KSHV genome and specific antibodies against H2AX and γH2AX. The results showed that H2AX and γH2AX displayed distinct patterns of occupancy along the KSHV episome (Fig. 4). An episome site-specific binding of phosphorylated H2AX along the KSHV genome was observed. Interestingly, both H2AX and γH2AX showed significant binding along the TR region. However, the binding of γH2AX to the TR region was definitely more prominent than in the rest of the KSHV episome. A number of other regions, including those corresponding to ORF9, ORF53, ORF63, and one region between ORF71 and ORFK13, also show a relatively high affinity for γH2AX, although the significance of these interactions is not yet understood. Interestingly, the region between 1 to 30 kb of the genome shows relatively less occupancy of H2AX than the rest of the genome.
Fig 4.
Relative abundance of H2AX and γH2AX along the KSHV episome. In BCBL1 cells, ChIP assay was carried out using specific antibodies against γH2AX and H2AX. The KSHV episome was examined at a resolution of approximately 1.6 kb to evaluate the binding of H2AX and γH2AX. Relative amplification was calculated by subtracting the amplification with control antibodies.
We compared our results with data from two independent ChIP-Seq analysis experiments where binding of LANA with KSHV episome was examined (45). Seven of the nine regions having a LANA binding peak score of >2 in ChIP-Seq analysis displayed a ≥2-fold binding with γH2AX in our experiments (Table 1). This further supports our data and suggests the cooccupancy of LANA and γH2AX at distinct regions of the KSHV episome. Nevertheless, H2AX is also likely to play a role in regulating KSHV genome maintenance, as there is enhanced binding activity of H2AX to regions between ORF75, ORF65, and ORF32. The significance of these differences is being explored.
Table 1.
Comparison of KSHV genes upregulating in LANA-ChIP and γH2AX-ChIP
| Gene name | Fold change |
|
|---|---|---|
| LANA-ChIP | γH2AX-ChIP | |
| vIRF | 2.08 | 5 |
| ORF71 | 2.43 | 3.8 |
| ORF72 | 3.06 | 3.85 |
| K15 3′UTR | 2.27 | 13 |
| K15 | 2.18 | 13 |
| TR | 7.72 | 10.5 |
| LBS1/2 | 13.16 | 6.2 |
H2AX contributes to LANA-mediated episome persistence.
In view of the role of H2AX in mediating LANA and TR interaction, we wanted to investigate whether or not H2AX was involved in KSHV persistence. We performed real-time PCR assays to evaluate the contribution of H2AX in LANA-mediated episome persistence. The results showed a significant reduction of approximately 40% in episome copy number as observed in HEK-293–BAC36–KSHV cells knocked down for H2AX expression compared to cells expressing control shRNA (Fig. 5A and B). Results from the real-time PCR assays showed that the reduction in episome copies was approximately 25 to 30% (average episome number, ∼1.15 × 104/10,000 cells) by 5 days and as much as 40% (average episome number, ∼0.96 × 104/10,000 cells) by 15 days postselection with puromycin, compared to the control. This suggests a contributory role for H2AX in mediating/strengthening LANA-TR interaction (Fig. 5A and B).
Fig 5.
H2AX is crucial for KSHV episome persistence. (A) Experimental scheme for episome retention experiment; (B) HEK-293–BAC36–KSHV cells were transfected with pGIPz-shCtrl or pGIPz-shH2AX plasmids. Episome copies have been quantified by real-time PCR on the 2nd, 5th, and 15th days posttransfection while keeping cells under puromycin selection. (C) Levels of LANA examined in the HEK-293–BAC36–KSHV cell line, stably knocked down for H2AX. Decrease in LANA level by day 15 may be indirect evidence of a decrease in the episome copies as a result of H2AX depletion. (D) Reporter assays showing that LANA promoter activity is unchanged by H2AX knockdown. Rta, a positive activator of LANA promoter, is transfected to serve as a positive control for promoter activity. Values shown in bar graphs are relative/normalized to a no-LANA negative control. (E) Colony formation assay carried out under dual antibiotic selection (puromycin and hygromycin) using the HEK-293–BAC36–KSHV cell line, stably expressing shRNA constructs for H2AX and control sequences. A significant decrease in the colony number shows that loss of H2AX also promotes loss of the KSHV episome.
A small reduction in LANA expression levels in H2AX knockdown cells was observed at day 15. However, there was negligible change in LANA expression on day 2 post-H2AX knockdown (Fig. 5C). This likely reflects an indirect effect of reduction on episome copies. However, to rule out the possibility that H2AX knockdown mediated LANA reduction, which led to a reduction in episome copies, we carried out reporter assays to examine the effect of H2AX knockdown on LANA promoter activity (Fig. 5D). Results of reporter assays reveal similar levels of LANA promoter activity when comparing the control to the H2AX knockdown. This suggests that there was no direct effect of H2AX knockdown on LANA expression. Cells expressing KSHV-Rta served as a positive control in the reporter assay, in view of the previously identified role of Rta in inducing the activity of the LANA promoter (26). These experiments were also repeated using a previously reported H2AX knockdown lentivirus (10), independent from our own shH2AX lentivirus and not as dramatic in terms of H2AX knockdown (60%) as seen by Western blotting (data not shown). Similar results were obtained supporting further a role for H2AX during KSHV infection (data not shown). Furthermore, in a colony formation assay using HEK-293–BAC36–KSHV cells, under dual antibiotic pressure (puromycin used for the shRNA-expressing constructs, and hygromycin used selecting for the HEK-293–BAC36–KSHV cells), a smaller number of colonies (40% reduction) was observed in H2AX knockdown cells than in the control cells (Fig. 5E). Thus, the results from the colony formation assay and LANA expression data nicely corroborated the results of the episome persistence experiment described above, and we previously showed that KSHV infection enhanced colony formation in HEK-293 cells (46). These results therefore strongly suggest an important role for H2AX in contributing to LANA-TR interaction and therefore maintenance of the KSHV episome.
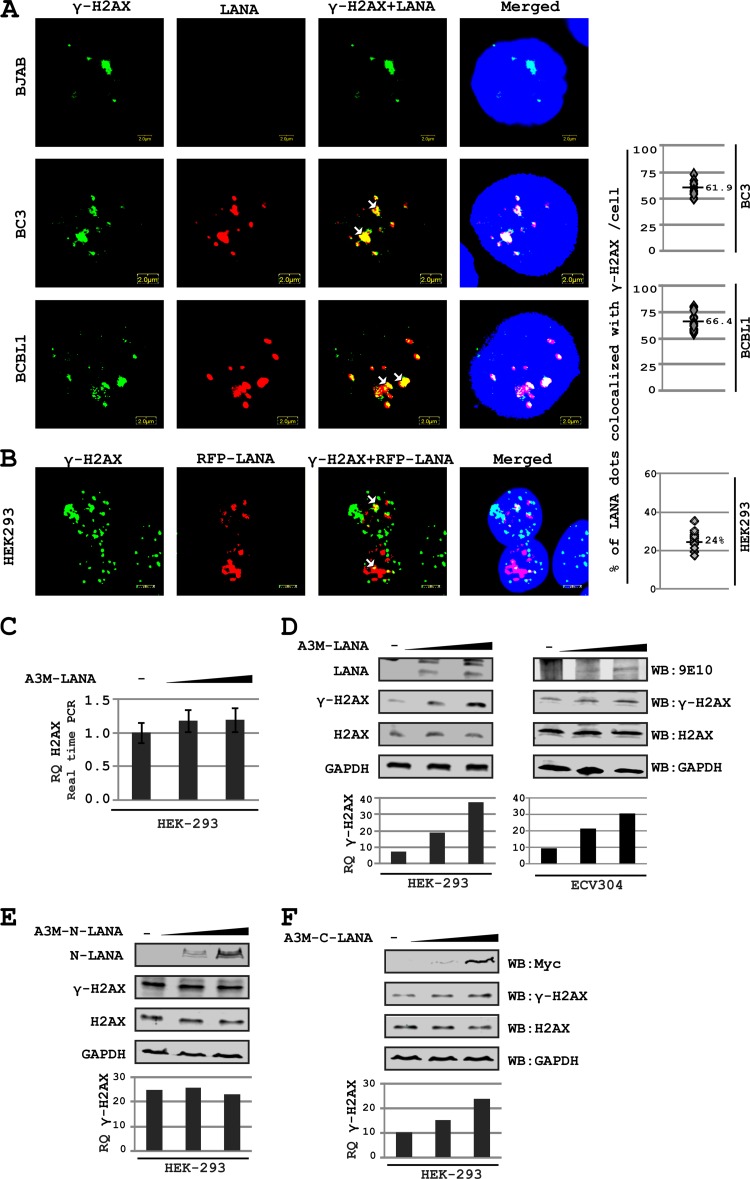
LANA colocalizes with γH2AX in KSHV-positive PEL cells.
To validate the interaction between LANA and H2AX in KSHV-positive lymphoma cells, we examined the colocalization of LANA and γH2AX by immunofluorescence analysis. Approximately 61.9% and 66.4% of the LANA dots colocalized with γH2AX signals in BC3 and BCBL1 cells, respectively (Fig. 6A). The Pearson's correlation coefficient (0.54 and 0.56 for BC3 and BCBL1, respectively) and Mander's overlap coefficient (0.72 and 0.78 for BC3 and BCBL1, respectively) also demonstrated a significant level of colocalization of these two molecules (Fig. 6A). We examined the association of LANA with γH2AX in the absence of the KSHV episome by transfecting HEK-293 cells with red fluorescent protein (RFP)-LANA-expressing plasmid (Fig. 6B). A greater than 50% reduction was observed in LANA colocalization with γH2AX in the absence of episome, compared to that seen with BCBL1 and BC3 cells. However, in view of the variation in LANA staining pattern in the presence and absence of the KSHV episome, this comparison may not be ideal and requires further investigations. Interestingly the colocalization between LANA and H2AX was also dramatically less when total H2AX was probed instead of γH2AX (data not shown).
Fig 6.
H2AX phosphorylation is increased in response to LANA expression. (A) Immunofluorescence for examining colocalization of LANA and γH2AX in KSHV-positive and -negative cell lines. LANA was probed using anti-LANA mouse monoclonal antibody and Alexa Fluor 594-conjugated secondary antibody, whereas FITC-labeled monoclonal antibody against γH2AX was used to probe γH2AX (used after completion of secondary antibody treatment in LANA staining). (B) Binding of LANA with γH2AX in the absence of the KSHV episome was examined by transfecting HEK-293 cells with RFP-LANA-expressing plasmid. RFP-LANA shows a diffused staining pattern. The image contrast was set to high to visualize LANA-rich regions. (C) H2AX levels as affected by LANA expression. HEK-293 cells and ECV304 cells were transfected with increasing amounts of pA3M-LANA plasmid. H2AX levels were examined by real-time PCR 24 h posttransfection. (D) H2AX phosphorylation was examined in response to LANA expression. HEK-293 cells were transfected with increasing amounts of A3M-LANA constructs. After 24 h posttransfection, cells were harvested and H2AX phosphorylation (serine 139) was examined by Western blotting. (E) H2AX phosphorylation is examined in response to expression of the LANA N terminus in a dose-dependent manner. (F) H2AX phosphorylation is examined in response to expression of the LANA C terminus in a dose-dependent manner. HEK-293 cells were transfected with increasing amounts of the indicated plasmids. H2AX and γH2AX levels were examined by Western blotting 24 h posttransfection.
LANA promotes phosphorylation of H2AX.
Relatively high levels of H2AX phosphorylation were seen in the latently growing KSHV-positive cells (harboring the circular form of the viral genome). This suggests the involvement of factors besides that induced with the DNA-free ends introduced during infection, important for enhancing phosphorylation of H2AX. Previously, a latent protein, vCyclin, has been implicated in phosphorylation of H2AX by inducing replicative stress in endothelial cells, in response to KSHV infection (3). KSHV-encoded LANA is another major latent protein expressed in all stages of the KSHV infection cycle. LANA is also known to promote S-phase entry (39) and therefore capable of indirectly inducing H2AX phosphorylation due to the production of DNA replication intermediates in S-phase.
We next examined the phosphorylation of H2AX at serine 139 in response to LANA expression. A dose-dependent increase in the phosphorylation of H2AX was observed due to LANA expression, and its C-terminal residues (ss 993 to 1162) were the predominant residues implicated in this process (Fig. 6C to F). Importantly, no significant increase in the expression of H2AX at the transcript level was observed in response to LANA expression (Fig. 6C). Western blot analysis of H2AX and γH2AX showed a definite increase in γH2AX levels, compared to no detectable change in H2AX as observed with a dose-responsive increase as a result of an increase in LANA expression in HEK-293 cells as well as the endothelial cell line ECV304 (Fig. 6D). Expression of the N terminus of LANA showed no detectable change in γH2AX levels (Fig. 6E). However, when the C terminus of LANA was expressed, a similar increase in the level of γH2AX protein was seen compared to expression of full-length LANA (Fig. 6, compare panels D with E and F).
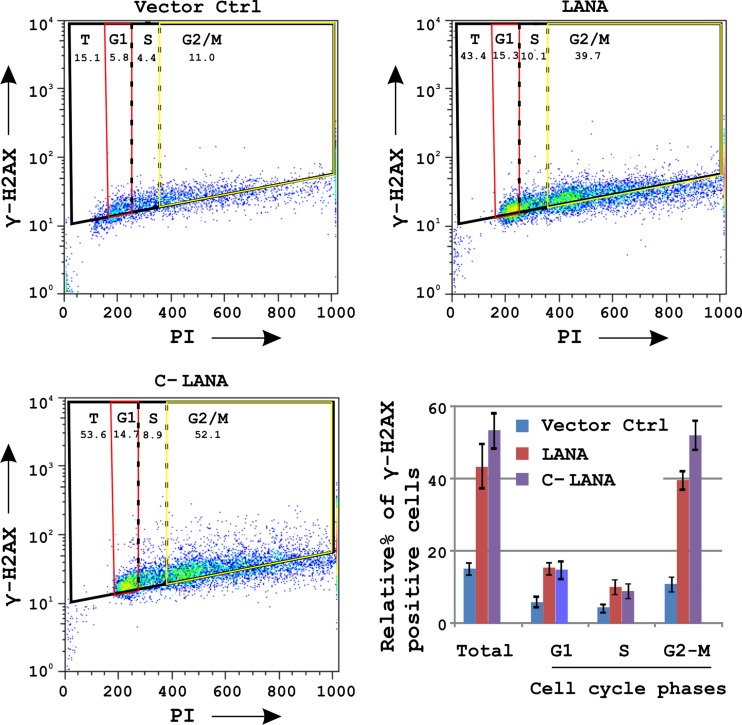
LANA-mediated phosphorylation of H2AX is not S-phase dependent.
A number of DNA replication intermediates are formed during the S-phase of the cell cycle, capable of inducing phosphorylation of H2AX (47). One of the KSHV proteins, vCyclin, has also been shown to induce replicative stress and phosphorylation of H2AX during KSHV infection (3). The major latent protein LANA is well known for its role in stimulating S-phase entry (39). We therefore investigated this phenomenon as a probable mechanism for H2AX-induced phosphorylation by LANA. Fluorescence-activated cell sorting (FACS) analysis of cells labeled with a specific antibody to detect γH2AX and propidium iodide to determine the DNA content during cell cycle phases were performed to examine the induction of H2AX phosphorylation by the full-length and C terminus of LANA. The results from these assays showed that both the full-length and C terminus of LANA showed significant phosphorylation of H2AX (Fig. 7). However, unexpectedly, the phosphorylation was not S-phase dependent, in that phosphorylation of H2AX was observed through the cell cycle in response to LANA or C-LANA expression, with the greatest induction occurring during the G2-M phase (Fig. 7).
Fig 7.
H2AX phosphorylation by LANA was examined during various stages of the cell cycle. HEK-293 cells were transfected with 15 μg of the indicated plasmids. H2AX phosphorylation in various phases of cell cycle was determined by labeling of cells with antibodies specific for γH2AX and propidium iodide (PI) staining.
DISCUSSION
Studies have shown that the mammalian DNA repair machinery do recognize incoming viral genetic material (2). Examples are available from different viruses, where the DDR has been shown to favor or regulate viral replication. It should be noted that viruses have also evolved to modulate components of the DDR signaling pathway by suppressing the activities of some but promoting others to the eventual goal of self-propagation (2).
Our results show that in response to KSHV infection, the majority of the molecules shown to be involved in DDR signaling are downregulated at the transcriptional level. These include ATM, ATR, DNA-PKcs, Ku70, and Ku80, which are major components in the pathway (34). Suppression of the DDR response to facilitate viral replication was previously observed in studies investigating adenovirus infection (2). In reference to adenovirus infection, DNA-PKcs, Ligase4, ATM, ATR, and the MRN complex (includes MRE11, Rad50, and NBS1) are known to favor concatemerization over viral replication (2). Furthermore, the MRN complex is a key component involved in ATM and ATR activation, and in the course of infection, adenovirus takes control by targeted degradation of the MRN complex via the activities of the viral E1b55K/E4orf6 proteins (48). This therefore favors viral replication over concatemerization (48). Degradation of the MRN complex is also known to abrogate the ATM-dependent G2/M checkpoint (2, 49). In another study on adenovirus, the E4 protein was identified as responsible for inactivating DNA-PKcs to block p53 activation and thereby prevent apoptosis of infected cells (50). Similarly, herpes simplex virus type 1 infection is also known to attenuate the DNA-PK activity by depleting the p350/DNA-PKcs catalytic subunit (51). In addition, Epstein-Barr virus (EBV) and cytomegalovirus (CMV) viral infection activates ATM, and abrogation of downstream ATM-mediated checkpoint signaling is crucial for cell cycle arrest and apoptosis (1, 52, 53). Downregulation of the DDR genes may be important for suppressing KSHV-induced apoptosis signal or clearing the cell cycle checkpoints to promote viral replication.
H2AX is the DDR molecule found to be highly upregulated in response to KSHV infection. It is one of the most well-studied DDR molecules, and its phosphorylation is considered one of the signatures of DNA damage (19). A number of viruses are known to induce phosphorylation of H2AX during their course of infection, including adenovirus (54), Epstein-Barr virus (22, 55), mouse HV68 (22, 23), human cytomegalovirus (HCMV) (46), and KSHV (3, 4). We have also observed high levels of phosphorylated H2AX during KSHV infection and in latently infected PEL cells. Previous studies suggested that H2AX phosphorylation is a marker of virus-induced DDR. Moreover, two studies with mouse HV68 explored the functional relevance of H2AX phosphorylation during the virus infection cycle in more detail (22, 23). Tarakanova et al. identified the ORF36 protein encoded by mouse HV68 as a kinase that can phosphorylate histone H2AX during infection (23). Seven days after infection, γHV68 titer in H2AX−/− macrophages was almost 100-fold lower than titers from wild-type H2AX+/+ cells (22, 56). This suggests that the virus can actively initiate the host DDR by targeted H2AX phosphorylation by a kinase that is encoded by the virus or an induced kinase in response to infection (56).
We now show that LANA interacts with H2AX at its N (aa 1 to 340)- and C (aa 930 to 1162)-terminal residues. This interaction was somewhat expected, as LANA was previously shown to interact with H2A at its N terminus, which is likely important for tethering to the host nucleosomes (12). The accepted model of LANA-mediated episome tethering shows that the C terminus of LANA forms a complex with the KSHV TR DNA, whereas the N terminus interacts with the host chromatin (9). However, the binding of the C terminus residues with the host chromatin-associated proteins have also been shown to contribute to episome tethering. None of the studies showed the binding of the corresponding N terminus to the KSHV TRs (11, 16). The interaction between the C terminus of LANA and H2AX presents a possible solution to this question and tempted us to analyze the distribution of H2AX along the KSHV episome, including the TR. LANA showed the typical punctuate pattern with significant colocalization with γH2AX as determined by immunofluorescence assays. However, relatively less colocalization was observed using an antibody against total H2AX in KSHV-positive BCBL1 and BC3 cells. The punctuate distribution of LANA was observed only in the presence of the KSHV episome, and without the KSHV episome it showed a diffused staining, as expected (7). Therefore, we explored the possibility of a model where phosphorylation of H2AX can increase its affinity for TR and thereby lead to direct binding and colocalization of γH2AX and LANA. Reduction of LANA binding to TR under reduced H2AX levels suggests a role for H2AX in contributing to the interaction of LANA with the TR elements. This, along with immunofluorescence and immuno-FISH data, strongly suggested that γH2AX is also a component of the LANA-TR complexes, which was supported by the data where enhanced association of γH2AX was seen at the TRs. To examine the relative density of H2AX and γH2AX on the KSHV genome, we did whole-genome ChIP assays using specific antibodies against total H2AX and γH2AX. Total H2AX shows a relatively homogenous distribution along the KSHV episome except for the region between ORF4 to ORF16, where the binding of total H2AX was surprisingly low. This may be due to the occupancy of other molecules on these regions. On the other hand, γH2AX binding was significantly biased toward regions close to TRs. These observations suggest the existence of a DNA-protein complex in KSHV-infected cells, with γH2AX partially mediating the LANA-TR interaction. H2AX−/− cells are hypersensitive toward double-stranded breaks (DSBs), and complementation of H2AX−/− mouse cells with phospho-acceptor serine-mutated H2AX did not rescue the DSB hypersensitivity (57). Phosphorylation of the H2AX C terminus tail is thought to create an epigenetic signal that is recognized by sensor protein(s) (36). Importantly, phosphorylation of H2AX is crucial for guiding protein complexes at the site of DNA damage (19). It is possible that similarly, phosphorylation of H2AX is crucial for guiding the episome-tethering molecules to the cis-acting element.
To determine a role for γH2AX, we measured the KSHV episome levels in cells with reduced levels of H2AX in comparison to control cells. H2AX knockdown cells showed a reduction in episome copy number in comparison to control cells. This reduction in episome copy number was consistent over a number of assays. The C terminus of LANA can directly bind to TRs at the LANA binding sequences (LBSs) (39), but the fact that the reduction was only partial suggests that γH2AX may contribute only partially to KSHV episome tethering, which is expected based on previous studies.
The results from colony formation assays also supported the results from the episome persistence studies. Moreover, the expression of LANA in the H2AX knockdown HEK-293–BAC36–KSHV cells was reduced and is most likely linked to the reduction in episome copies ultimately leading to a reduction in overall LANA protein levels.
In response to viral infection, phosphorylation of H2AX is thought to be induced by an unusual influx of nucleic acid (2). In addition, viral proteins have also been known to facilitate H2AX phosphorylation (2). These include the ORF36 protein of mHV68, BGLF4 of Epstein-Barr virus, and vCyclin of KSHV (2, 3, 22, 23). Phosphorylation of H2AX by vCyclin, a latent viral protein during KSHV infection, has been previously observed in response to an induction of replicative stress (3). LANA is also a major latent protein which is expressed in almost all phases of the KSHV life cycle (5). LANA is also known to induce replication stress by driving S-phase entry (39). We now show that phosphorylation of H2AX also increases in the presence of LANA in a dose-dependent manner and that this function is contributed mainly by the C terminus of LANA. LANA has no identified sequence homology with known kinases. Therefore, we evaluated the possibility of increased H2AX phosphorylation by LANA through induction of S-phase entry. LANA is known to stimulate S-phase entry by stabilizing β-catenin, a nuclear effector of the Wnt-signaling pathway (39). However, the data from cell-cycle-phase-dependent H2AX phosphorylation assays revealed that the H2AX phosphorylation was not restricted to the S-phase. Therefore, an active kinase (most likely cellular) induced by LANA is contributing to H2AX phosphorylation of H2AX throughout the cell cycle. It may be interesting to think of γH2AX as having a dual role in DDR through vCyclin-dependent phosphorylation and in episome tethering through induced phosphorylation by an unidentified cellular kinase.
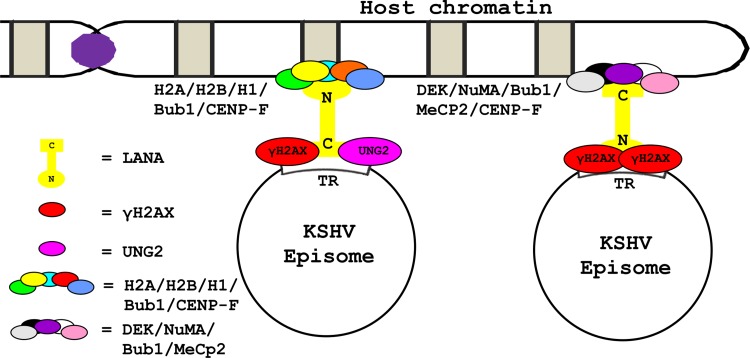
LANA not only binds to H2AX but also contributes to its phosphorylation. This probably serves as a molecular signal for TR identification. The observation that both N and C terminus residues of LANA are able to bind γH2AX allows a model to be proposed (see Fig. 8) where γH2AX contributes to the binding of LANA to the TRs. The opposite end of LANA may anchor to the host chromatin in association with N terminus binding proteins (e.g., H2A, H2B, H1, Bub1, CENP-F) or C terminus binding proteins (e.g., DEK, NuMA, Bub1, MeCP2, CENP-F) (11–14, 16, 30, 39). It must be noted that this model as proposed adds to the previous model where LANA directly binds to TRs at LBSs (12). Like H2AX, another human protein uracil DNA glycosylase 2 (UNG2) has previously been shown to contribute to the LANA-TR binding at the C terminus of LANA (30).
Fig 8.
Schematic presents a model by which γH2AX can contribute to the persistence of the viral genome after KSHV infection. As γH2AX is known to bind/localize with C-LANA and TR, along with partially assisting with the episome retention function, the model shows that γH2AX may strengthen the interaction between C-LANA and TR. This model is complementary to the classical model, where LANA by its C terminus binds with LANA binding sequences (LBSs) in the TRs. This model also presents a possible explanation for how the mammalian chromatin-associated proteins, known to bind to the C terminus of LANA, are able to contribute to LANA-mediated KSHV episome persistence after infection.
In the present study, we did not examine the role of H2AX in anchoring LANA to human chromatin. However, keeping in view the high degree of homology (95% identity, 98% positive by NCBI blastx) between histone H2A and H2AX, this possible interaction may present yet another important facet of H2AX in contributing to the KSHV episome tethering. Significant binding of the LANA N terminus to H2AX further strengthens this possibility. This study presents an excellent example of the evolution of KSHV to utilize the DDR, one of the most important cellular responses to infection by viruses. The levels of γH2AX in the infected cells are increased by induction of the transcription levels in addition to induction of its phosphorylation. Involvement of multiple viral proteins, including LANA, vCyclin, and possibly ORF36, as described for MHV68 may further ensure optimum phosphorylation levels under different cell environments (22, 58) and therefore enhance persistence of the virus in the infected cell.
Supplementary Material
ACKNOWLEDGMENTS
The wild-type KSHV BACmid, BAC36, was provided by S. J. Gao (University of Texas, San Antonio, TX). The ECV304 cell line was a kind gift from Harry Ischiropoulos (University of Pennsylvania).
This work was supported by public health service grants NCI CA072510, CA091792, and NIDCR DE017338 (to Erle S. Robertson). Erle S. Robertson is a scholar of the Leukemia and Lymphoma Society of America.
Footnotes
Published ahead of print 28 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03575-12.
REFERENCES
- 1. Chaurushiya MS, Weitzman MD. 2009. Viral manipulation of DNA repair and cell cycle checkpoints. DNA Repair (Amst.) 8:1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weitzman MD, Carson CT, Schwartz RA, Lilley CE. 2004. Interactions of viruses with the cellular DNA repair machinery. DNA Repair (Amst.) 3:1165–1173 [DOI] [PubMed] [Google Scholar]
- 3. Koopal S, Furuhjelm JH, Jarviluoma A, Jaamaa S, Pyakurel P, Pussinen C, Wirzenius M, Biberfeld P, Alitalo K, Laiho M, Ojala PM. 2007. Viral oncogene-induced DNA damage response is activated in Kaposi sarcoma tumorigenesis. PLoS Pathog. 3:1348–1360 doi:10.1371/journal.ppat.0030140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Park J, Lee D, Seo T, Chung J, Choe J. 2000. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) open reading frame 36 protein is a serine protein kinase. J. Gen. Virol. 81:1067–1071 [DOI] [PubMed] [Google Scholar]
- 5. Verma SC, Robertson ES. 2003. Molecular biology and pathogenesis of Kaposi sarcoma-associated herpesvirus. FEMS Microbiol. Lett. 222:155–163 [DOI] [PubMed] [Google Scholar]
- 6. Cai Q, Verma SC, Lu J, Robertson ES. 2010. Molecular biology of Kaposi's sarcoma-associated herpesvirus and related oncogenesis. Adv. Virus Res. 78:87–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ballestas ME, Chatis PA, Kaye KM. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641–644 [DOI] [PubMed] [Google Scholar]
- 8. Cotter MA, II, Subramanian C, Robertson ES. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291:241–259 [DOI] [PubMed] [Google Scholar]
- 9. Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Luger K, Kaye KM. 2006. Kaposi's sarcoma-associated herpesvirus LANA hitches a ride on the chromosome. Cell Cycle 5:1048–1052 [DOI] [PubMed] [Google Scholar]
- 10. Dimitrova N, Lange TD. 2006. MDC1 accelerates nonhomologous end-joining of dysfunctional telomeres. Genes Dev. 20:3238–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krithivas A, Fujimuro M, Weidner M, Young DB, Hayward SD. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596–11604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311:856–861 [DOI] [PubMed] [Google Scholar]
- 13. Cotter MA, II, Robertson ES. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254–264 [DOI] [PubMed] [Google Scholar]
- 14. Si H, Verma SC, Lampson MA, Cai Q, Robertson ES. 2008. Kaposi's sarcoma-associated herpesvirus-encoded LANA can interact with the nuclear mitotic apparatus protein to regulate genome maintenance and segregation. J. Virol. 82:6734–6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Viejo-Borbolla A, Ottinger M, Bruning E, Burger A, Konig R, Kati E, Sheldon JA, Schulz TF. 2005. Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1. J. Virol. 79:13618–13629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiao B, Verma SC, Cai Q, Kaul R, Lu J, Saha A, Robertson ES. 2010. Bub1 and CENP-F can contribute to Kaposi's sarcoma-associated herpesvirus genome persistence by targeting LANA to kinetochores. J. Virol. 84:9718–9732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. You J, Srinivasan V, Denis GV, Harrington WJ, Jr, Ballestas ME, Kaye KM, Howley PM. 2006. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J. Virol. 80:8909–8919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. 2008. GammaH2AX and cancer. Nat. Rev. Cancer 8:957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feeney KM, Parish JL. 2009. Targeting mitotic chromosomes: a conserved mechanism to ensure viral genome persistence. Proc. Biol. Sci. 276:1535–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. 2004. H2AX: the histone guardian of the genome. DNA Repair (Amst.) 3:959–967 [DOI] [PubMed] [Google Scholar]
- 21. Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. 2002. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12:162–169 [DOI] [PubMed] [Google Scholar]
- 22. Tarakanova VL, Leung-Pineda V, Hwang S, Yang CW, Matatall K, Basson M, Sun R, Piwnica-Worms H, Sleckman BP, Virgin HW., IV 2007. Gamma-herpesvirus kinase actively initiates a DNA damage response by inducing phosphorylation of H2AX to foster viral replication. Cell Host Microbe 1:275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tarakanova VL, Stanitsa E, Leonardo SM, Bigley TM, Gauld SB. 2010. Conserved gammaherpesvirus kinase and histone variant H2AX facilitate gammaherpesvirus latency in vivo. Virology 405:50–61 [DOI] [PubMed] [Google Scholar]
- 24. Verma SC, Lan K, Choudhuri T, Cotter MA, Robertson ES. 2007. An autonomous replicating element within the KSHV genome. Cell Host Microbe 2:106–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu J, Verma SC, Murakami M, Cai Q, Kumar P, Xiao B, Robertson ES. 2009. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus (KSHV) upregulates survivin expression in KSHV-associated B-lymphoma cells and contributes to their proliferation. J. Virol. 83:7129–7141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lan K, Kuppers DA, Verma SC, Sharma N, Murakami M, Robertson ES. 2005. Induction of Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen by the lytic transactivator RTA: a novel mechanism for establishment of latency. J. Virol. 79:7453–7465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paxinou E, Weisse M, Chen Q, Souza JM, Hertkorn C, Selak M, Daikhin E, Yudkoff M, Sowa G, Sessa WC, Ischiropoulos H. 2001. Dynamic regulation of metabolism and respiration by endogenously produced nitric oxide protects against oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 98:11575–11580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu J, Verma SC, Cai Q, Robertson ES. 2011. The single RBP-Jkappa site within the LANA promoter is crucial for establishing Kaposi's sarcoma-associated herpesvirus latency during primary infection. J. Virol. 85:6148–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kaul R, Verma SC, Robertson ES. 2007. Protein complexes associated with the Kaposi's sarcoma-associated herpesvirus-encoded LANA. Virology 364:317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Verma SC, Bajaj BG, Cai Q, Si H, Seelhammer T, Robertson ES. 2006. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus recruits uracil DNA glycosylase 2 at the terminal repeats and is important for latent persistence of the virus. J. Virol. 80:11178–11190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abramoff MD, Magalhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophoton. Int. 11:36–42 [Google Scholar]
- 32. Huang X, Darzynkiewicz Z. 2006. Cytometric assessment of histone H2AX phosphorylation: a reporter of DNA damage. Methods Mol. Biol. 314:73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li L, Olvera JM, Yoder KE, Mitchell RS, Butler SL, Lieber M, Martin SL, Bushman FD. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3272–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Riha K, Heacock ML, Shippen DE. 2006. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu. Rev. Genet. 40:237–277 [DOI] [PubMed] [Google Scholar]
- 35. Wyman C, Kanaar R. 2006. DNA double-strand break repair: all's well that ends well. Annu. Rev. Genet. 40:363–383 [DOI] [PubMed] [Google Scholar]
- 36. Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123:1213–1226 [DOI] [PubMed] [Google Scholar]
- 37. Ward IM, Chen J. 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276:47759–47762 [DOI] [PubMed] [Google Scholar]
- 38. Ballestas ME, Kaye KM. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fujimuro M, Wu FY, ApRhys C, Kajumbula H, Young DB, Hayward GS, Hayward SD. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9:300–306 [DOI] [PubMed] [Google Scholar]
- 40. Garber AC, Hu J, Renne R. 2002. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 277:27401–27411 [DOI] [PubMed] [Google Scholar]
- 41. Garber AC, Shu MA, Hu J, Renne R. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882–7892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grundhoff A, Ganem D. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 77:2779–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kelley-Clarke B, De Leon-Vazquez E, Slain K, Barbera AJ, Kaye KM. 2009. Role of Kaposi's sarcoma-associated herpesvirus C-terminal LANA chromosome binding in episome persistence. J. Virol. 83:4326–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ye FC, Zhou FC, Yoo SM, Xie JP, Browning PJ, Gao SJ. 2004. Disruption of Kaposi's sarcoma-associated herpesvirus latent nuclear antigen leads to abortive episome persistence. J. Virol. 78:11121–11129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lu F, Tsai K, Chen HS, Wikramasinghe P, Davuluri RV, Showe L, Domsic J, Marmorstein R, Lieberman PM. 2012. Identification of host-chromosome binding sites and candidate gene targets for Kaposi's sarcoma-associated herpesvirus LANA. J. Virol. 86:5752–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu J, Verma SC, Cai Q, Saha A, Dzeng RK, Robertson ES. 2012. The RBP-Jk binding sites within the RTA promoter regulate KSHV latent infection and cell proliferation. PLoS Pathog. 8:e1002479 doi:10.1371/journal.ppat.1002479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mirzoeva OK, Petrini JH. 2003. DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol. Cancer Res. 1:207–218 [PubMed] [Google Scholar]
- 48. Tauber B, Dobner T. 2001. Molecular regulation and biological function of adenovirus early genes: the E4 ORFs. Gene 278:1–23 [DOI] [PubMed] [Google Scholar]
- 49. Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22:6F600–6F620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Boyer J, Rohleder K, Ketner G. 1999. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263:307–312 [DOI] [PubMed] [Google Scholar]
- 51. Lees-Miller SP, Long MC, Kilvert MA, Lam V, Rice SA, Spencer CA. 1996. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J. Virol. 70:7471–7477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kudoh A, Fujita M, Zhang L, Shirata N, Daikoku T, Sugaya Y, Isomura H, Nishiyama Y, Tsurumi T. 2005. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J. Biol. Chem. 280:8156–8163 [DOI] [PubMed] [Google Scholar]
- 53. Luo MH, Rosenke K, Czornak K, Fortunato EA. 2007. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J. Virol. 81:1934–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nichols GJ, Schaack J, Ornelles DA. 2009. Widespread phosphorylation of histone H2AX by species C adenovirus infection requires viral DNA replication. J. Virol. 83:5987–5998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kamranvar SA, Gruhne B, Szeles A, Masucci MG. 2007. Epstein-Barr virus promotes genomic instability in Burkitt's lymphoma. Oncogene 26:5115–5123 [DOI] [PubMed] [Google Scholar]
- 56. Xie A, Scully R. 2007. Hijacking the DNA damage response to enhance viral replication: gamma-herpesvirus 68 orf36 phosphorylates histone H2AX. Mol. Cell 27:178–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A. 2003. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114:371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hoge AT, Hendrickson SB, Burns WH. 2000. Murine gammaherpesvirus 68 cyclin D homologue is required for efficient reactivation from latency. J. Virol. 74:7016–7023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Verma SC, Choudhuri T, Kaul R, Robertson ES. 2006. Latency-associated nuclear antigen (LANA) of Kaposi's sarcoma-associated herpesvirus interacts with origin recognition complexes at the LANA binding sequence within the terminal repeats. J. Virol. 80:2243–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Verma SC, Lu J, Cai Q, Kosiyatrakul S, McDowell ME, Schildkraut CL, Robertson ES. 2011. Single molecule analysis of replicated DNA reveals the usage of multiple KSHV genome regions for latent replication. PLoS Pathog. 7:e1002365 doi:10.1371/journal.ppat.1002365 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.