Abstract
In the plasma samples of hepatitis C virus (HCV)-infected patients, lipoviroparticles (LVPs), defined as (very-) low-density viral particles immunoprecipitated with anti-β-lipoproteins antibodies are observed. This HCV-lipoprotein association has major implications with respect to our understanding of HCV assembly, secretion, and entry. However, cell culture-grown HCV (HCVcc) virions produced in Huh7 cells, which are deficient for very-low-density lipoprotein (VLDL) secretion, are only associated with and dependent on apolipoprotein E (apoE), not apolipoprotein B (apoB), for assembly and infectivity. In contrast to Huh7, HepG2 cells can be stimulated to produce VLDL by both oleic acid treatment and inhibition of the MEK/extracellular signal-regulated kinase (ERK) pathway but are not permissive for persistent HCV replication. Here, we developed a new HCV cell culture model to study the interaction between HCV and lipoproteins, based on engineered HepG2 cells stably replicating a blasticidin-tagged HCV JFH1 strain (JB). Control Huh7.5-JB as well as HepG2-JB cell lines persistently replicated viral RNA and expressed viral proteins with a subcellular colocalization of double-stranded RNA (dsRNA), core, gpE2, and NS5A compatible with virion assembly. The intracellular RNA replication level was increased in HepG2-JB cells upon dimethyl sulfoxide (DMSO) treatment, MEK/ERK inhibition, and NS5A overexpression to a level similar to that observed in Huh7.5-JB cells. Both cell culture systems produced infectious virions, which were surprisingly biophysically and biochemically similar. They floated at similar densities on gradients, contained mainly apoE but not apoB, and were not neutralized by anti-apoB antibodies. This suggests that there is no correlation between the ability of cells to simultaneously replicate HCV as well as secrete VLDL and their capacity to produce LVPs.
INTRODUCTION
A remarkable feature of chronic hepatitis C (CHC) virus infection resides in the interplay between viral replication and host gluco-lipidic metabolism. CHC infection is associated with a high prevalence of insulin resistance (1, 2) and increased prevalence of type 2 diabetes mellitus (3, 4). CHC infection is also associated with an increased incidence of fatty liver (steatosis), which varies between 40% and 80% of patients depending on other risk factors (i.e., alcohol consumption, type 2 diabetes, or obesity) (5, 6). In addition to metabolic risk factors, hepatitis C virus (HCV) replication has been reported to be associated with altered serum lipid and lipoprotein levels (6, 7). Indeed, hypobetalipoproteinemia is observed in 5 to 50% of patients, depending on viral genotype (8, 9). Furthermore, HCV-infected patients present lower cholesterol, triglyceride, and low-density lipoprotein (LDL) levels (10), which normalize following successful antiviral treatment (11). These metabolic defects are more prevalent in genotype 3a-infected subjects and have important consequences for patient management as patients with CHC present a higher risk of atherosclerosis (12), whereas treatment responders may also have an increased risk of coronary heart disease due to elevated LDL and cholesterol levels (11). Recently, a report studying transgenic mice expressing the HCV polyprotein showed altered hepatocellular lipid and lipoprotein metabolism in these animals, with increased lipogenesis and decreased lipoprotein secretion, suggesting a direct role for the virus in modulating host lipoprotein metabolism (13).
Besides the clinical observation of the impact of HCV on lipoprotein metabolism, a more direct interaction between HCV virions and lipoproteins was first suggested in vivo in 1992 when Thomssen and colleagues observed that a substantial fraction of circulating HCV RNA could be immunoprecipitated by anti-β-lipoprotein antibodies (14). β-Lipoprotein-associated hybrid low-density HCV particles were reported to contain apolipoprotein B (apoB), HCV RNA, and the viral core protein (15) and have been termed “lipoviroparticles” (LVP). Further characterization of these LVP by immunoprecipitation studies revealed the presence of apolipoprotein E (apoE) in addition to apoB and HCV RNA, suggesting a close association of HCV particles with very-low-density lipoproteins (VLDL) (16). Interestingly, HCV particles seemed to be predominantly present in light, lipoprotein-rich serum fractions from patients after a high-fat meal (17). The concept of LVP is now widely accepted although no association between HCV and apoB has been reported in vitro (18, 19).
In vitro studies on HCV were mainly performed with the Huh7 (and derived) cell line infected by a cell culture-adapted JFH1 viral strain (20–22) or derived chimeras such as Jc1 (23). Using these cell culture models, many studies have characterized apoE as associating with HCV particles, concluding that apoE plays a role in infectious particle formation and entry into host cells (24–29). Interestingly, HCV particles produced in cell culture (HCVcc virions) have a relatively high density compared to their in vivo counterparts, with densities ranging from 1.10 to 1.18 g/ml. These particles are infectious for chimpanzees; however, passage of the virus in these animals generates particles with lower density and increased specific infectivity (i.e., density [d] of < 1.10 g/ml) (30), suggesting a role for host lipoproteins in the HCV life cycle. These studies highlight the need to develop in vitro cell culture models that synthesize LVP to understand their nature, composition, and role(s) in HCV replication.
Huh7 cells were recently reported to be deficient at producing mature VLDL (31, 32), limiting their usefulness as a model system to study the role of VLDL in HCV replication. In contrast, HepG2 hepatoma cells have been shown to assemble and secrete lipoproteins (33), and recent reports showed that oleic acid (OA) stimulation and inhibition of the MEK/extracellular signal-regulated kinase (ERK) pathway promote cells to secrete VLDL (32, 34). HepG2 cells have been extensively used to study HCV entry (35–39), replication (39–41), and the effect of cell polarization on HCV infection (42, 43). Several attempts to persistently and efficiently replicate HCV in HepG2 cells using either inducible vector transfection (44), HCV RNA transfection (45), baculoviral transduction (46), or infection with patient serum (47) have not been successful yet. Recently, it has been shown that ectopic expression of miR-122, a microRNA which is not endogenously expressed by HepG2 cells, promoted HCV JFH1 replication (41). However, the level of replication and viral production was insufficient to allow biochemical analysis of HCV particles, and therefore such analysis was not reported.
Given the ability of HepG2 cells to express VLDL, we established a stable cell line expressing a blasticidin-tagged JFH1 virus (HepG2-JB). HepG2-JB cells support genome replication and protein expression and generate infectious extracellular particles that allow us to study the role of the VLDL assembly pathway in the assembly and infectivity of HCV particles.
MATERIALS AND METHODS
Reagents, antibodies, and plasmids.
Chemicals were purchased from Sigma (St. Louis, MO, USA), unless otherwise specified. Tris, sucrose, and bovine serum albumin fraction V (BSA) were purchased from Euromedex (Mundolsheim, France). Complete protease inhibitor cocktail was purchased from Roche (Indianapolis, IN, USA). To-Pro-3 nucleic acid stain and HCS LipidTOX Deep Red neutral lipid stain were purchased from Molecular Probes (Life Technologies, Paisley, United Kingdom). The selection antibiotics blasticidin-S, zeocin, and G418 were obtained from InvivoGen (San Diego, CA, USA). Protein G-coated magnetic beads (BioAdembeads) were obtained from Ademtech (Pessac, France). HCV antiprotease (VX-950) and antipolymerase (2-methyl-cytidine) were kindly provided by J. Neyts (Rega Institute, Leuven, Belgium).
IgG from goat and mouse sera was obtained from Sigma. Goat polyclonal anti-apoB (ab27626) and anti-apoE (ab7620) antibodies, rabbit polyclonal anti-V5 tag (ab9116) and anti-BSD (ab38307), and mouse monoclonal anti-apoE (E6D7; ab1907), anti-apoE (D6E10; ab1906), and anti-HCV core protein (C7-50; ab2740) antibodies were purchased from Abcam (Cambridge, United Kingdom). Goat polyclonal anti-human apoB (AF3556) was purchased from R&D Systems (Minneapolis, MN, USA). Rabbit polyclonal anti-beta actin antibody (A0760-40A) was purchased from US Biological (Swampscott, MA, USA). Mouse monoclonal antibody (MMAb) anti-NS3 (217-A) and anti-double-stranded RNA (dsRNA) (J2) were purchased from Virogen (Watertown, MA, USA) and Scicons (Hungary). MMAb anti-HCV E2 (AP33) and anti-CD81 (JS81), were kindly given by Arvind H. Patel (Institute of Virology, Glasgow, United Kingdom) (48)and E. Rubinstein (INSERM U1004, Villejuif, France), respectively; anti-apoB (4G3) was purchased from the Heart Institute, Ottawa, Canada. Human monoclonal anti-HCV E2 antibody (CBH-5) was provided by Steven Foung (Stanford Blood Center, Palo Alto, CA, USA) (49). Control VLDL was obtained from the floating phase of ultracentrifuged human plasma samples (EFS, Lyon, France).
The plasmid enabling T7-mediated runoff transcription of the HCV JFH1 strain, pJFH1, was kindly given by T. Wakita (National Institute of Infectious Diseases, Tokyo, Japan) (21). This plasmid was first modified, to derive pJFH1-CSK, by insertion of three mutations by site-directed mutagenesis (leading to the following amino acids changes in the JFH1 polyprotein: F172C, P173S, and N534K) to obtain a cell culture-adapted strain, which was previously described (50). For simplicity, this strain will be referred to as JFH1 here. Next, the sequence encoding blasticidin resistance (BSD) was added within the NS5A region between amino acids (aa) 2395 and 2396 of the polyprotein, i.e., a position that was described as tolerant for insertion of foreign sequences without affecting HCV replication (51). To this end, the sequence AGATCTTCGCGAGTTTAAACGCGT, containing the restriction sites for BglII, NruI, PmeI, and MluI, was first inserted at position 7523 of the JFH1 genome by a PCR-based strategy (details on request). Then, a PCR product of the BSD open reading frame (ORF) was cloned between BglII and MluI to derive pJFH1-CSK-BSD. The protein sequence at the insertion site was modified from MPP2395LEG to MPP2395RSTM-BSD AA seq-KTRLEG, where BSD AA seq is blasticidin amino acid sequence. Again, for simplicity, this strain will be named the JFH1-BSD, or JB, strain here.
Cell culture.
Huh7.5 cells were a kind gift from Charles M. Rice (The Rockefeller University, NY, USA) (52). HepG2 cells stably transduced with a lentiviral vector expressing CD81 (HepG2-CD81) were previously described (35, 53). The same lentiviral vector was used to derive other CD81-expressing HepG2-based lines. HepG2 cells stably replicating a JFH1 subgenomic replicon (HepG2-SGR) were kindly provided by Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan) (40). Cell culture medium and reagents were purchased from Life Technologies (Paisley, United Kingdom), unless otherwise specified. Fetal bovine serum Fetal Clone II (FCII; Thermo Scientific, Waltham, MA, USA) was used for all cultures. Huh7.5 and HepG2 (and derived cells) were maintained in Dulbecco's modified Eagle medium (high-glucose DMEM) containing 10% FCII, 1× GlutaMAX, 1 mM sodium pyruvate, 100 units/ml penicillin, and 100 μg/ml streptomycin in a 5% CO2-normoxia incubator at 37°C. HepG2 cells were cultured on collagen-coated surfaces (type I collagen from rat tail; Sigma, Saint-Louis, MO, USA). HepG2-CD81 or HepG2-SGR cells and derived cell lines were maintained in the presence of 500 μg/ml G418 sulfate.
Viral production and cell line generation.
The plasmid pJFH1-CSK-BSD was linearized by XbaI, treated by mung bean exonuclease, and purified with a Macherey-Nagel DNA purification minicolumn. The linearized DNA was in vitro transcribed using a MEGAscript T7 kit from Ambion (Life Technologies). Viral RNA was then delivered to Huh7.5 cells by electroporation as previously described (52). After three cell passages, infectious supernatants were harvested, clarified by centrifugation, and filtered (0.45-μm pore size). Titers were determined, and samples were and stored at −80°C until used.
The subgenomic JFH1 replicon was “cured” from HepG2-SGR cells by treatment over five passages with both alpha interferon (IFN-α; 100 IU/ml) and an anti-HCV polymerase drug (i.e., 2-methyl-cytidine at 10 μM). The derived cell line, HepG2-cured (HepG2c), was then rendered positive for CD81 expression by stable transduction with a lentivirus carrying the CD81 gene together with a neomycin resistance gene as described previously (35). The resulting HepG2c-CD81 cell line was further modified by transduction with a lentivirus carrying a gene encoding an N-terminal truncated form of interferon regulatory factor 3 (IRF3) (54); the resulting HepG2c-CD81-IRF3(ΔN) cell line is deficient for the production of IFN-β upon stimulation with the dsRNA prototypic ligand poly(I · C) (data not shown). The NS5A gene from JFH1 was PCR amplified and cloned into the pLenti4-TO-zeocin vector from Invitrogen. Lentiviruses were generated with this plasmid according to the manufacturer's protocol and used to generate cell lines with an enforced expression of a V5-tagged (in the N terminus) version of NS5A. Two cell lines were generated: Huh7.5-NS5A and HepG2c-CD81-IRF3(ΔN)-NS5A.
Wild-type and various engineered Huh7.5 and HepG2 cells were infected with JFH1-BSD at a multiplicity of infection (MOI) of 1, and persistently replicating cells were selected with blasticidin at 20 μg/ml. To prevent the selection of a particular clone, which might have a genotype different from that of the parental cells, persistently replicating cell lines were kept polyclonal (i.e., multiple colonies were mixed when a line was generated).
Density gradient separation.
Cells at confluence were treated for 3 days with 2% dimethyl sulfoxide (DMSO) and then incubated for another 24 h in a 5% CO2-normoxia incubator at 37°C with fresh medium deprived of serum and containing either 10% (vol/vol) BSA-coupled oleic acid (10 mg/ml BSA and 300 μM OA final concentrations) and 1 μM U0126 (20 mM stock, diluted in DMSO) or 10% (vol/vol) BSA (10 mg/ml final concentration) as a control. Supernatants were harvested (10 to 12 ml for a culture surface of 75 cm2), spun at 11,000 × g for 5 min to remove cell debris, filtered (0.45-μm pore size), supplemented or not with protease inhibitors, and loaded on Amicon Ultracel 100K centrifugal filter units (Millipore, Billerica, MA, USA) to concentrate them, according to the manufacturer's protocol. Concentrated supernatant (1 ml) was loaded on precast iodixanol-sucrose density gradients (see below for details) and ultracentrifuged for 10 h at 55,000 rpm (368,000 maximum relative centrifugal force [rcfmax]) or at 40,000 rpm (274,000 rcfmax) (lipoprotein or viral supernatant separation, respectively) and 4°C using an SW 55 Ti rotor or SW 41 Ti rotor in an Optima L-90K centrifuge (Beckman, Indianapolis, IN). Fractions were harvested from the top of the tube, and their densities were determined with a refractometer. Fraction volumes for lipoprotein or virus separation were, respectively, 400 μl or 500 μl for the four upper fractions and 1 ml for the others. Discontinuous iodixanol-sucrose density gradients were prepared by successive pouring and freezing of 1-ml fractions ranging from 35% or 54% iodixanol to 6% iodixanol. Each fraction contained 10 mM Tris-HCl, pH 8, 2 mM EDTA, pH 8, iodixanol (OptiPrep density medium), and sucrose (each fraction was filled up to 1 ml with 0.25 M sucrose to keep osmolarity). These precast density gradients were thawed for 24 h at 4°C before use to generate continuous density gradients by diffusion.
apoB100 quantification.
apoB100 was quantified in each fraction or in filtered culture supernatants using a commercial enzyme-linked immunosorbent assay (ELISA) kit from AlerCHEK (Springvale, ME, USA) and following the manufacturer's protocol.
Apolipoprotein immunoprecipitations.
Harvested and filtered supernatants (0.5 to 1 ml) (see above) were incubated with 4 μg/ml of anti-apoB or anti-apoE antibody (ab27626 or ab7620, respectively) or control IgG for 1h 30 min at room temperature under agitation (1,000 rpm). Washed, protein G-coated magnetic beads (10 μl for 1 μg of antibody) were then added, and the supernatants were incubated for 1h 30 min at room temperature under agitation. Magnetic beads were washed five times and then resuspended with 50 to 100 μl of cold PBS supplemented with protease inhibitors for HCV RNA quantification.
Drug treatment and HCV RNA quantification.
JFH1-BSD-replicating cells were seeded on six-well culture plates and kept for 3 days after confluence by the addition of 2% DMSO to the culture medium. Then cells were incubated for another 24 h with medium containing 2% DMSO and different drug concentrations to assess the effect of drug treatment on HCV RNA levels. Briefly, cells were treated with either 0, 0.5, 1, and 2% ethanol or with 0, 25, 50, and 100 μM chenodeoxycholic acid (CDCA) or with 0, 2.5, 5, and 10 μM U0126, with or without 10% oleic acid.
Total intracellular RNA was extracted using Extract-All (Eurobio, Courtaboeuf, France) according to the manufacturer's protocol. Extracellular RNA was extracted from 150 μl of filtered (0.45-μm pore size) supernatants, from 150 μl of each fraction of density gradients, or from 10 to 20 μl of immunoprecipitated samples using a NucleoSpin RNA Virus kit (Macherey-Nagel, Düren, Germany) according to the manufacturer's protocol. HCV RNA absolute quantification was performed by quantitative reverse transcription-PCR (RT-qPCR) with a OneStep Express SYBR GreenER RT-qPCR SuperMix kit (Life Technologies, Paisley, United Kingdom) on a MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad, Hercules, CA). The amplification was performed with previously described RC1 and RC21 primers (55).
Viral infectivity determination.
Viral infectivity was determined in each fraction of the density gradients using the 50% tissue culture infective dose (TCID50), determined as previously described (20). Viral infectivity in culture supernatants was determined as the number of focus-forming units (FFU/ml). Briefly, 2 × 104 Huh7.5 cells per cm2 were seeded on 48-well culture plates and infected the next day with serial dilutions of infectious supernatants. Three days after infection, cells were fixed with an ice-cold methanol-acetone (1/1) solution for 30 min at −20°C and processed for immunofluorescent detection of HCV core antigen. Core-positive infectious foci were quantified for highest dilutions, and the infectious titer was thus determined.
Neutralization assay.
Two different readouts were used to analyze antibody-mediated neutralization of viral supernatant infection: supernatant infectivity (FFU/ml) and viral RNA entry (intracellular RT-qPCR after a 4-h infection). Briefly, naïve Huh7.5 cells were seeded on 48-well or 6-well plates (2 ×104 cells/cm2 or 8 × 104 cells/cm2, respectively). The next day, serum-free viral supernatants were incubated for 2 h at room temperature with neutralizing antibodies or control IgG, i.e., with MMAb anti-apoB (4G3) (10, 2, and 0.4 μg/ml), MMAb anti-apoE (D6E10) (10, 2, and 0.4 μg/ml), both anti-apoB and anti-apoE antibodies, MMAb anti-CD81 (JS-81) (10 μg/ml), or control IgG from mouse (10 μg/ml). For CD81 neutralization, naïve cells were also incubated with anti-CD81 antibodies (1 μg/ml) before and during infection. The virus-antibody mixture was then incubated for 4 h at 37°C with naïve Huh7.5 cells (100 μl of serial dilutions for infectivity determination and 500 μl for RNA entry determination). Cells were then washed with phosphate-buffered saline (PBS) and processed for FFU count determination or washed three times with PBS and processed for intracellular RNA quantification.
Northern blotting.
Purified RNA was glyoxylated, migrated into agarose gel in phosphate buffer, transferred onto positively charged nylon membrane in 20× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and subjected to Northern blotting with a radioactively labeled (32P-labeled CTP) negative probe (i.e., complementary to the HCV positive strand and corresponding to the 5′ untranslated region [UTR] sequence) following standard procedures (56). Briefly, a PCR amplicon was generated with primers T3-JFH1-fr (AAAATCTAGAAATTAACCCTCACTAAAGGGACCTGCCCCTAATAGGGGCGACACTCC) and T7-JFH1-rv (AAAATCTAGATAATACGACTCACTATAGGGAGAGCCTCGGGGGGACAGGAGCCATCC), which contain, respectively, T3 and T7 polymerase sequences (underlined). The PCR amplicon was used as the template for in vitro transcription using MAXIscript kits from Ambion (Life Technologies) in the presence of radioactive [P32]CTP (GE Healthcare) according to the manufacturer's instructions. A negative probe (i.e., hybridizing with positive-strand HCV RNA) was generated with the T7 polymerase. Prehybridization and hybridization were performed in hybridization buffer from Ambion at 65°C in a rotary oven, and the probe was used at least at 106 cpm/ml. Washes were performed with nonstringent buffer (2× SSC–1% SDS) at room temperature and stringent buffer (0.1× SSC–1% SDS) at 65°C. The membrane was then exposed to Kodak film and developed with conventional methods.
Western blotting.
Cell lysates, density gradient fractions, or immunoprecipitates were denatured for 5 min at 95°C in Laemmli buffer, and proteins were separated using SDS-PAGE. After samples were blotted on a nitrocellulose membrane, apoB100 and apoE proteins were revealed using, respectively, a goat polyclonal anti-apoB antibody (AF3556; 1:1,000 dilution) and a mouse monoclonal anti-apoE (E6D7) antibody (1:500 dilution). HCV proteins core, E2, and NS3 were revealed using mouse monoclonal antibodies (1:500 dilutions). V5-tagged or BSD-tagged NS5A proteins were revealed using rabbit polyclonal antibodies (1:500 dilutions). For cell lysates, equal loading was controlled by revealing β-actin with a rabbit polyclonal antibody (1:250 dilution). Anti-goat, anti-rabbit, and anti-mouse antibodies coupled to horseradish peroxidase were used to detect proteins by enhanced chemiluminescence.
Immunofluorescence (IF).
Cells were seeded on glass coverslips and kept for 3 days after confluence by adding 2% DMSO to the medium. Coverslips were then washed in PBS and fixed for 20 min at room temperature with 4% paraformaldehyde (PFA) in PBS and permeabilized with saponin. Fixed cells were washed in PBS, and a blocking step in PBS containing 4% bovine serum albumin (PBS-BSA) was performed, followed by incubation with primary antibodies in PBS-BSA for 1h 30 min. Mouse monoclonal anti-HCV core (C7-50; 1:400 dilution) and anti-dsRNA (J2; 0.5 μg/ml) antibodies, human monoclonal anti-HCV E2 (CBH-5; 1:500 dilution) antibody, and rabbit polyclonal anti-V5 tag (1:500 dilution) and anti-BSD (1:500 dilution) antibodies were used. Cells were washed three times in PBS and incubated with respective secondary antibodies coupled to either Alexa-488 or Alexa-555 fluorescent probe (Life Technologies) diluted 1:2,000 in PBS-BSA for 1 h. Cells were then washed twice with PBS and incubated for another 30 min in PBS containing either 1:400 LipidTOX neutral lipid stain or 1:2,000 To-Pro-3 to stain lipid droplets or nucleic acid, respectively. Coverslips were finally washed in PBS and mounted on glass slides using fluorescence mounting medium (Dako, Glostrup, Denmark) for analysis. Pictures were taken with a Zeiss LSM 510 Meta confocal microscope (Carl Zeiss MicroImaging GmbH, Jena, Germany). Cell counting, image processing, and montage were performed with ImageJ software (National Institutes of Health, USA).
RESULTS
HepG2-CD81 cells secrete apoB/apoE-containing very-low-density lipoproteins.
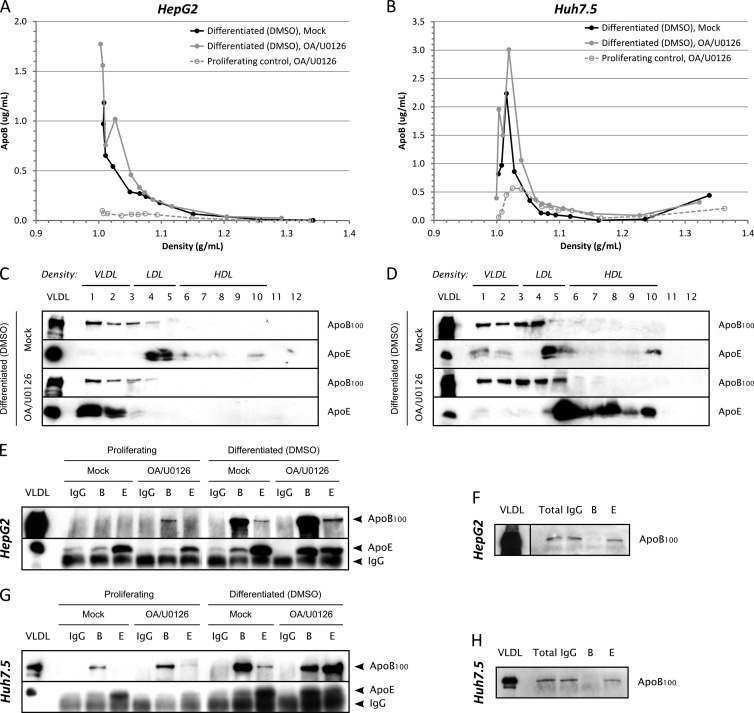
HepG2 cells have been shown to produce apoB100 in the very-low-density range, following stimulation with oleic acid (OA) and MEK/ERK inhibitor (U0126) (32, 34). In contrast, Huh7 cells produce mainly intermediate-density lipoprotein (IDL) and are insensitive to MEK/ERK inhibition (32). HepG2 cells may therefore represent a better model to study the assembly of LVP (i.e., apoB/apoE/HCV-positive hybrid particles) in vitro. To confirm these observations with our cell lines (i.e., HepG2-CD81 and Huh7.5 cells), we analyzed apoB100 and apoE secretion levels and density by immunoblotting gradient fractions. Indeed, proper VLDL production is characterized by cosedimentation of apoB100 and apoE in the lightest fractions of the gradient (d < 1.006 g/ml). Proliferative HepG2-CD81 cells produced VLDL only after OA/U0126 treatment, whereas Huh7.5 did not (data not shown), as previously described in the literature for HepG2 and Huh7 cells (32). However, in vivo, VLDL are secreted by differentiated hepatocytes. When cultured under basal conditions and kept in proliferation, Huh7 and HepG2 cells have rather dedifferentiated phenotypes, but it has been shown that DMSO could be used to promote hepatoma differentiation (57, 58). Thus, to characterize VLDL production under more physiological conditions, we analyzed apoB100 and apoE secretion and cosedimentation in more differentiated HepG2-CD81 and Huh7.5 cells after exposing them for 4 days to 2% DMSO. We first quantified apoB in each fraction of the density gradient using ELISAs. DMSO-induced differentiation greatly improved both the amount of secreted apoB and its repartition in the lowest density in HepG2-CD81 cells independently of OA/U0126 treatment (Fig. 1A). Differentiation also increased apoB secretion by Huh7.5 cells but resulted in no or little decrease in apoB density (Fig. 1B). The differences between HepG2-CD81 and Huh7.5 cells were further highlighted by assessing apoB100 and apoE density repartition by immunoblotting (Fig. 1C and D). Only differentiated and OA/U0126-treated HepG2-CD81 cells showed total cosedimentation between apoB100 and apoE in the very first fractions of the gradient, suggesting that only these cells were able to secrete proper, mature VLDL (Fig. 1C).
Fig 1.
apoB100 and apoE secretion in partially redifferentiated hepatoma cells. HepG2-CD81 (panels A, C, E, and F) or Huh7.5 (panels B, D, G, and H) cells were seeded on 100-mm petri dishes. At confluence, cells were exposed for 3 days to 2% DMSO to promote their differentiation and incubated another 24 h with medium without serum containing 2% DMSO, 10% (vol/vol) OA-BSA, and 1 μM U0126 (OA/U0126) or containing 2% DMSO and 10% (vol/vol) BSA (Mock). Supernatants (10 ml) were harvested, filtered (0.45-μm pore size), and subjected either to density separation on iodixanol-sucrose gradients (panels A to D) or to immunoprecipitation (panels E to H). (A) ApoB quantification using ELISAs in each fraction of the density gradient for DMSO-differentiated HepG2-CD81 cells. apoB secretion in subconfluent proliferative and OA/U0126-treated cells was also quantified for comparison (dashed gray curve). (B) apoB quantification using ELISAs in each fraction of the density gradient for DMSO-differentiated Huh7.5 cells. apoB secretion in subconfluent proliferative and OA/U0126-treated cells was also quantified for comparison (dashed gray curve). (C) apoB100 and apoE immunoblotting in each fraction for DMSO-differentiated HepG2-CD81 cells. (D) apoB100 and apoE immunoblotting in each fraction for DMSO-differentiated Huh7.5 cells. (E to H) Immunoprecipitations were performed with 4 μg/ml of anti-apoB100 or anti-apoE antibodies, using protein G-coated magnetic beads, in HepG2-CD81 (E) and Huh7.5 (G) cells. IgG from goat serum was used as a negative control. Immunoprecipitated apoB100 or apoE was detected by immunoblotting with specific antibodies. apoB100 was also detected in total supernatants (Total) and IP flowthrough by immunoblotting to check for immunoprecipitation efficiency (F and H). B, IP with anti-apoB antibodies; E, IP with anti-apoE antibodies; VLDL, control VLDL from human serum.
One feature of VLDL is the presence of both apoB100 and apoE apolipoproteins. Therefore, we investigated the presence of both markers on the same lipoprotein particle by performing coimmunoprecipitation (co-IP) assays with anti-apoB and anti-apoE antibodies. Anti-apoB antibodies were able to immunoprecipitate the vast majority of apoB100 molecules since none were detected by immunoblotting in the flowthrough fractions for both differentiated HepG2-CD81 (Fig. 1F) and Huh7.5 cells (Fig. 1H). No specific co-IP was observed between apoB100 and apoE in culture supernatants from proliferating HepG2-CD81 (Fig. 1E) or Huh7.5 cells (Fig. 1G). However, when HepG2-CD81 cell differentiation was induced by DMSO and when cells were treated with OA and U0126 compound, we could observe a potent co-IP between both apoB100 and apoE (Fig. 1E). This result confirms that HepG2-CD81 cells are able to produce proper VLDL under specific culture conditions. Surprisingly, DMSO-differentiated and OA/U0126-treated Huh7.5 cells also showed a strong co-IP between apoB100 and apoE (Fig. 1G), arguing for the production of apoB/apoE-positive particles. However, these particles have too high a density to be qualified as VLDL, thus suggesting lipidation defects in Huh7 cells.
In conclusion, DMSO- and OA/U0126-treated HepG2 cells represent an ideal model to study HCV assembly and morphogenesis and to further characterize VLDL biogenesis co-option by the virus. Therefore, we focused on establishing persistent HCV replication in these HepG2 cells to analyze viral particle production after partial cell redifferentiation with DMSO.
Generation of blasticidin-tagged JFH1 virus.
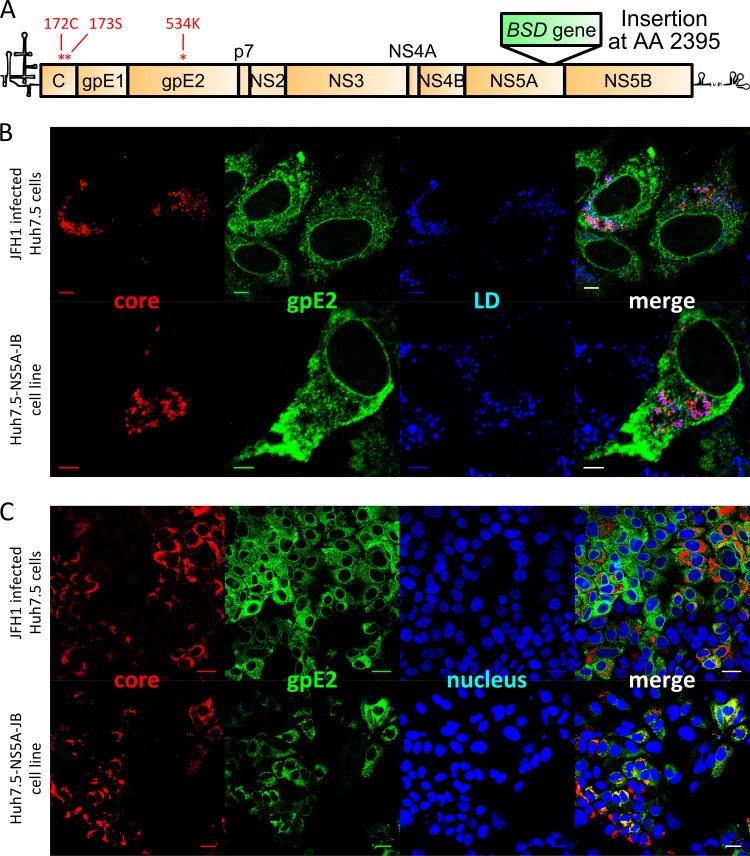
To establish chronically infected HepG2 cells, we engineered a cell culture-adapted blasticidin-tagged JFH1 virus as described in the Materials and Methods section and schematized in Fig. 2A. The blasticidin resistance gene was inserted at position 2395 of the polyprotein in domain III of NS5A since this insertion position was previously described by other investigators and did not impair HCV RNA replication (51). However, as Appel and colleagues reported that domain III was required for HCV assembly (59), we anticipated a hypothetical assembly defect in replicating cells due to BSD insertion and created various Huh7.5 and HepG2 (and derived) cell lines overexpressing a V5-tagged version of NS5A. The expression of NS5A-V5 was verified in these cells by immunofluorescence and Western blotting (data not shown).
Fig 2.
Viral protein expression in JB-replicating Huh7.5 cells. Naïve Huh7.5 cells were infected with JFH1 virus, and immunofluorescence (IF) staining was performed to detect viral structural proteins. Naïve Huh7.5-NS5A cells were infected with JB virus, stable viral replication was selected by blasticidin, and IF staining was performed to detect viral structural proteins. (A) Schematic representation of JB viral genome containing BSD resistance gene insertion in the C-terminal domain of NS5A at position 2395 (in polyprotein amino acid sequence). (B) Immunofluorescent detection of HCV core (red) and gpE2 (green) proteins with lipid droplet stain (blue). Scale bar, 5 μm. (C) Immunofluorescent detection of HCV core (red) and gpE2 (green) proteins with nucleus counterstain (blue). Scale bar, 25 μm.
To check the functionality of our recombinant JFH1-BSD (referred to here as JB) strain, in vitro transcribed RNA was electroporated into Huh7.5 cells, and supernatants were collected over a period of 1 week and pooled to generate an infectious viral stock. Naïve Huh7.5-NS5A cells were infected with various dilutions of JB virus, and cells supporting viral replication were selected with blasticidin in colony-forming assays. A population cell line, named Huh7.5-NS5A-JB, was established by pooling blasticidin-resistant clones. These cells were maintained under antibiotic selection and supported HCV RNA replication at a level similar to that of JFH1-infected Huh7.5 cells (106 to 107 viral RNA copies per μg of total cellular RNA). Comparison of stably replicating Huh7.5-NS5A-JB cells with freshly JFH1-infected Huh7.5 cells showed comparable levels of expression as well as viral protein E2 and core localization (Fig. 2B and C). Indeed, HCV core protein, when detected, was mainly localized around lipid droplets (LD), whereas gpE2 glycoprotein showed a reticular localization pattern (Fig. 2B). Huh7.5-NS5A-JB cells produced infectious viral particles although the viral titer was 10 times lower than that of JFH1-infected Huh7.5 cells (1.5 × 104 FFU/ml compared to 105 FFU/ml).
To summarize, we have shown that the JFH1-BSD (JB) strain is infectious in Huh7.5 cells and can establish a persistent infection following blasticidin selection, leading to continuous infectious viral particle secretion in Huh7.5 cells.
Cured HepG2-CD81 cells can stably replicate a blasticidin-tagged JFH1 strain.
To obtain stably replicating HepG2 cells, we infected various HepG2-CD81 lines with Huh7.5-produced JB virus and selected them with blasticidin. We observed transient replication in both HepG2-CD81 and HepG2-CD81-shIFN-β-shIFNAR1 (a cell line impaired in IFN responses as previously reported [60]) cells, suggesting that ATCC-derived HepG2 cells, with a knocked down IFN response, do not support sustained HCV replication (Table 1). Furthermore, differentiated HepaRG cells, a hepatic bipotent progenitor cell line (61, 62), and freshly isolated primary human hepatocytes (PHH) showed transient resistance to blasticidin, with no stable cell lines being generated under our experimental conditions (Table 1). Stably replicating HepG2 cells were obtained only with cured HepG2 cells, which had previously replicated subgenomic HCV replicons. These cured HepG2 cells were obtained by removing the JFH1 subgenomic replicon from HepG2-SGR cells (40) using IFN-α-2b and anti-polymerase treatments. As observed for Huh7.5 cells, which were obtained by IFN curing of an HCV subgenomic replicon, cured HepG2 cells showed cellular adaptations to viral replication (63). Additionally, HepG2 cells were engineered to express CD81, and/or NS5A and/or IRF3(ΔN), a dominant negative form of interferon regulatory factor 3 (IRF3) lacking its DNA binding domain and blocking synthesis of IFN-β mRNA (54). The most permissive HepG2 lines were HepG2c-CD81-IRF3(ΔN) cells and HepG2c-CD81-IRF3(ΔN)-NS5A cells expressing V5-tagged NS5A (named here HepG2c and HepG2c-NS5A, respectively) (Table 1). Three HepG2 cell lines were established, defined as HepG2c-JB, HepG2c-NS5A-JB1, and HepG2c-NS5A-JB2, and used to study HCV replication and viral particle production in VLDL-producing cells.
Table 1.
Generation of HCV-replicating Huh7.5 and HepG2 cell lines
| Naïve cell line | No. of BSD-resistant coloniesa | BSD-resistant cell lineb |
|---|---|---|
| Huh7.5 | >10,000 | Yes |
| Huh7.5-NS5A | >10,000 | Yes |
| HepG2-CD81 | Transient survivalc | No |
| HepG2-CD81-shINFβ-shINFR1 | 25 | No |
| HepG2c-CD81 | 500 | Yes |
| HepG2c-CD81-NS5A | 300 | Yes |
| HepG2c-CD81-IRF3(ΔN) | 800 | Yes |
| HepG2c-CD81-IRF3(ΔN)-NS5A | 800 | Yes |
| Differentiated HepaRG cells | Transient survivalc | No |
| PHH | Transient survivalc | No |
Approximate number of BSD-resistant colonies (extrapolated from the highest positive viral dilution).
Successful establishment of a BSD-resistant polyclonal cell line.
Transient survival to BSD for up to 1 week, followed by cell death without stable resistance.
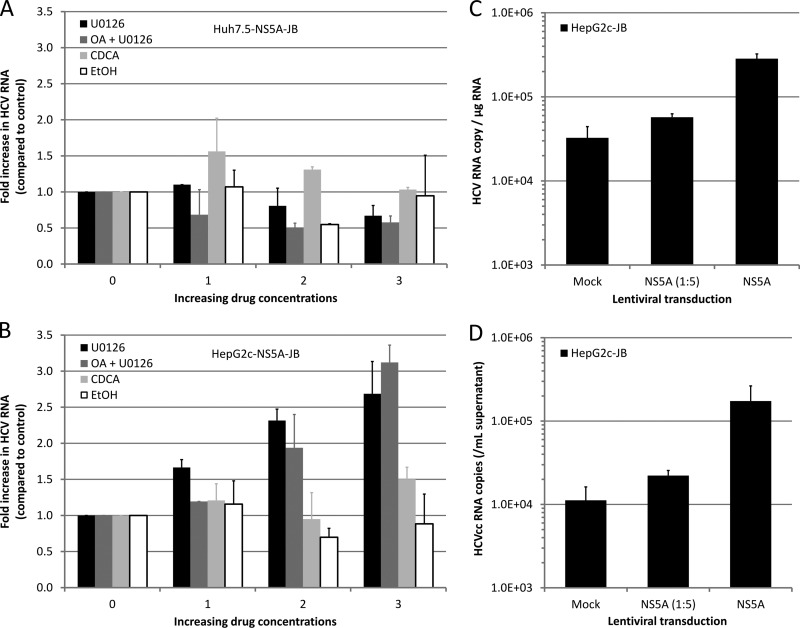
HepG2c-JB cells replicate HCV at lower levels than Huh7.5-NS5A-JB cells.
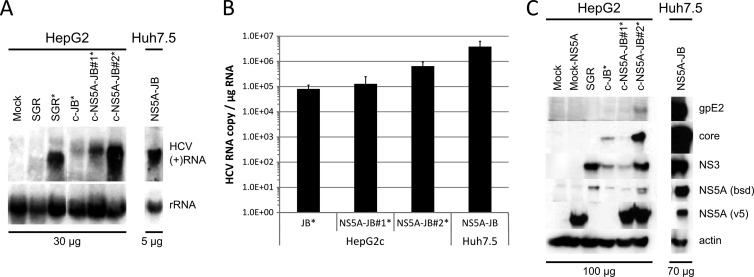
With these tools in hands, we characterized HCV replication in Huh7.5-NS5A-JB cells and in HepG2c-JB cells, expressing or not V5-tagged NS5A protein, in terms of viral RNA replication and viral protein expression. HCV positive-strand RNA [(+)RNA] was detected by Northern blotting in all cell lines at the expected size (Fig. 3A). Levels of accumulated intracellular viral (+)RNA were different for each cell line, with a higher level for NS5A-transcomplemented HepG2c-JB cells. HCV RNA quantification using one-step quantitative RT-PCR showed similar results with up to 5 × 105 viral RNA copies per μg of cellular RNA for HepG2c-NS5A-JB2 cells (Fig. 3B). However, HCV RNA levels were lower than in HepG2-SGR cells or Huh7.5-NS5A-JB cells.
Fig 3.
Replication of JB virus in HepG2c cell lines. Control Huh7.5 and HepG2 cells, HepG2-SGR cells, or JB-replicating Huh7.5 and HepG2c cell lines transcomplemented or not with V5-tagged NS5A were left untreated or treated (*) for 3 days with DMSO. RNA and proteins were extracted as described in Materials and Methods. (A) HCV (+)RNA was detected by Northern blotting with a full-length genomic minus-strand RNA [(−)RNA] probe. Total cellular RNAs were loaded for HepG2c (30 μg) and Huh7.5-NS5A-JB (5 μg) cell lines. (B) HCV RNA was quantified by one-step quantitative RT-PCR. Results were normalized with total RNA (μg). (C) Structural proteins (E2 and core) and nonstructural proteins (NS3 and BSD-tagged NS5A), as well as V5-tagged NS5A used for transcomplementation, were detected by immunoblotting with specific antibodies. Total cellular proteins were loaded for HepG2c-derived (100 μg) and Huh7.5-derived (70 μg) cell lines. Actin immunoblotting was performed to check for equal loadings.
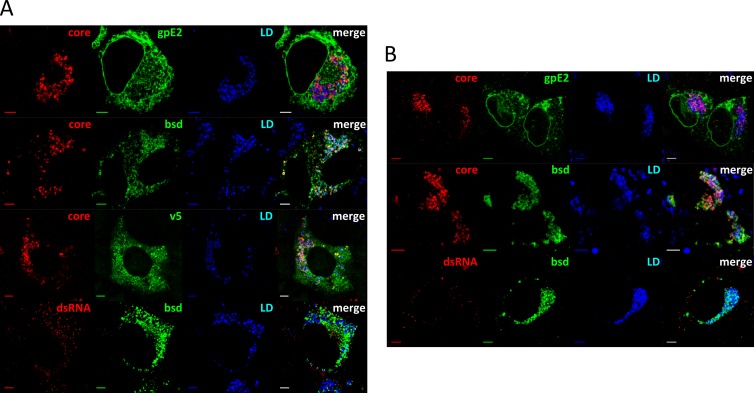
Next, we investigated viral protein expression and localization. Structural proteins gpE2 and core, as well as nonstructural proteins NS3 and BSD-tagged NS5A, were highly expressed in Huh7.5-NS5A-JB cell lines, as shown by protein immunoblotting (Fig. 3C). V5-tagged NS5A protein was also detected in transcomplemented Huh7.5-NS5A-JB and HepG2c-NS5A-JB cells. In contrast, we observed an overall weaker viral protein expression in HepG2c-JB cell types (Fig. 3C). Nevertheless, HCV gpE2, core, NS3, and BSD-tagged NS5A proteins could be detected in HepG2c-NS5A-JB2 cells, and this cell line was therefore used for future studies and is here referred to as HepG2c-NS5A-JB for simplicity. With respect to subcellular localization, HCV core protein accumulated at the periphery of lipid droplets (LD) and colocalized with BSD-tagged NS5A protein in both Huh7.5-NS5A-JB and HepG2c-NS5A-JB cell lines (Fig. 4A and B). Moreover, HCV E2 glycoprotein showed a reticular localization pattern, and HCV double-stranded RNA (J2 antibody stain) colocalized partially with BSD-tagged NS5A in close proximity to LD (Fig. 4A and B). It is worth noting that the majority of HepG2c-NS5A-JB cells replicated HCV RNA and expressed BSD-tagged NS5A protein, as shown by IF staining with anti-dsRNA and anti-BSD antibodies (data not shown). Together, these observations suggest efficient HCV replication in JB replicating cell lines with a functional interaction between dsRNA, core, and NS5A proteins that could lead to infectious viral production.
Fig 4.
Subcellular localization of viral components in JB-replicating hepatoma cell lines. Huh7.5-NS5A-JB (A) and HepG2c-NS5A-JB (B) cells were treated for 3 days with DMSO, fixed, and stained for HCV core protein (core, red), gpE2 protein (E2, green), BSD-tagged NS5A protein (BSD, green), V5-tagged NS5A protein used for transcomplementation (V5, green), or double-stranded RNA (dsRNA, red) using specific antibodies. Lipid droplet (LD, blue) staining was performed in parallel to determine viral protein localization. Scale bar, 5 μm. ND, not determined.
MEK/ERK inhibition and NS5A transcomplementation increase HCV replication in HepG2c-JB cells.
As the basal level of HCV replication was lower in HepG2 than in Huh7.5 cell lines, we sought to determine whether we could enhance it by treating cells with different drugs. Indeed, several drugs have been reported to enhance HCV replication in vitro. For instance, either ethanol, chenodeoxycholic acid (CDCA; a bile acid), or MEK/ERK inhibitor U0126 was shown to increase HCV replication in both HCV subgenomic replicon cell lines (64, 65) and JFH1-infected cells (66). Therefore, we evaluated the effects of these drugs on HCV replication within Huh7.5-NS5A-JB and HepG2c-NS5A-JB cells. Neither alcohol, CDCA, nor MEK/ERK inhibition had a significant effect on HCV RNA replication in Huh7.5-NS5A-JB cells (Fig. 5A). In contrast, viral RNA replication was increased following MEK/ERK inhibition in HepG2c-NS5A-JB cells, with or without oleic acid treatment (Fig. 5B). No effect of either ethanol or bile acids was detected in HepG2c-NS5A-JB cells. Interestingly, the best conditions to produce VLDL in HepG2 cells (Fig. 1) were also the best conditions to increase HCV replication in HepG2 cells, using OA stimulation and MEK/ERK inhibition. We also investigated the effect of NS5A transcomplementation in HepG2c-JB cells. Transient or stable NS5A expression using lentiviral transduction increased HCV RNA levels by 10-fold (Fig. 3B and 5C), thus demonstrating a partial deficiency of BSD-tagged NS5A protein with respect to viral RNA replication. Importantly, NS5A transcomplementation also increased viral RNA secretion (Fig. 5D).
Fig 5.
Modulation of HCV RNA replication in JB-replicating hepatoma cell lines. Huh7.5-NS5A-JB (A) or HepG2c-NS5A-JB (B) cells were treated with increasing concentrations of drugs for 24 h, and HCV RNA replication was assessed by one-step quantitative RT-PCR on cellular RNAs. Results were normalized with total RNA concentration and glucuronidase gene expression. Data are presented as fold increase compared to untreated cells. Error bars represent standard deviations of two independent experiments. Ethanol (EtOH), 0, 0.5, 1, and 2%; chenodeoxycholic acid (CDCA), 0, 25, 50 and 100 mM; U0126, 0, 2.5, 5 and 10 μM; oleic acid (OA), 10% (vol/vol). (C) HepG2c-JB cells (without V5-tagged NS5A) were transcomplemented with NS5A using a lentiviral vector. Six days after lentiviral transduction, intracellular HCV RNA was quantified by quantitative RT-PCR in transcomplemented and control cells. (D) At the same time, extracellular HCV RNA was also quantified by quantitative RT-PCR.
In summary, HCV replication in HepG2c-JB cells could be potentiated by OA/U0126 treatment, together with NS5A transcomplementation. Therefore, we used HepG2c-NS5A-JB cells, treated with DMSO, OA, and U0126, to analyze viral particle secretion and association with lipoproteins in an HCV-replicating and VLDL-competent cell culture system.
Characterization of infectious HCV particles secreted from HepG2c-NS5A-JB cells.
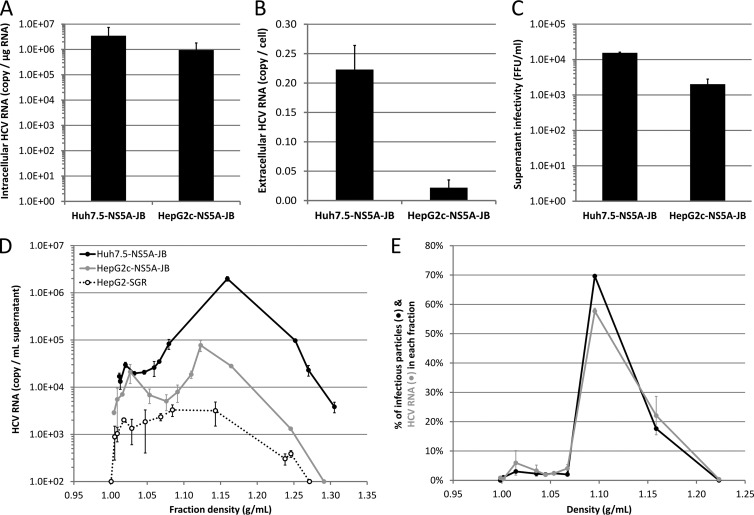
Under the optimized culture conditions, we observed a 3-fold difference in the steady-state levels of HCV RNA in Huh7.5-NS5A-JB and HepG2c-NS5A-JB cell lines (Fig. 6A). However, HepG2c-NS5A-JB cells secreted 10 times less HCV RNA than Huh7.5-NS5A-JB cells (Fig. 6B). In line with the latter result, Huh7.5-NS5A-JB and HepG2c-NS5A-JB cells were able to secrete infectious viral particles with titers of 104 and 103 focus forming units (FFU) per ml, respectively (Fig. 6C). The specific infectivity levels of virions produced by both cell types were not significantly different.
Fig 6.
JB-replicating hepatoma cell lines produce infectious viral particles. Huh7.5-NS5A-JB and HepG2c-NS5A-JB cells were treated for 3 days with DMSO and incubated another 24 h with DMSO, OA, and U0126 to harvest culture supernatants or quantify intracellular viral RNA. (A) Intracellular HCV RNA was quantified by quantitative RT-PCR, and results were normalized for total intracellular RNAs and glucuronidase gene expression. (B) Extracellular HCV RNA was quantified by quantitative RT-PCR, and results were normalized for cell number. (C) Supernatant infectivity was measured as focus-forming units (FFU/ml). Error bars represent standard deviations of three independent experiments. (D) Culture supernatants were loaded on iodixanol-sucrose density gradients, and HCV RNA concentration in each fraction was determined by quantitative RT-PCR (RT-qPCR). Results shown are representative of three independent experiments. (E) HCV RNA or infectious viral particles secreted by HepG2c-NS5A-JB cells are shown in each fraction as a percentage compared to the total amount of HCV RNA or infectious viral particles in the whole gradient, respectively. HCV RNA amount was determined by RT-qPCR. Infectious viral particles were quantified using the TCID50 method. Results are representative of two independent experiments.
To characterize the assembly and secretion of virus particles by JB-replicating hepatoma cells, we harvested the culture supernatant during optimal treatment to promote VLDL secretion, i.e., a 3-day differentiation with 2% DMSO, followed by 24 h with DMSO, OA, and U0126. Virus particle density was analyzed by iodixanol-sucrose gradients. Higher levels of HCV RNA were secreted from JB-replicating HepG2 and Huh7 cells than from control HepG2-SGR cells, suggesting bona fide viral particle release. The majority of HCV RNA secreted from both Huh7.5-NS5A-JB and HepG2c-NS5A-JB cells associated with high-density particles (between 1.1 and 1.2 g/ml) (Fig. 6D). A small amount of HCV RNA (around 5% in Huh7.5-NS5A-JB and 25% in HepG2c-NS5A-JB cells) was found in intermediate- to low-density fractions (1.006 to 1.019 g/ml and 1.019 to 1.063 g/ml, respectively), and less than 1 to 2% in very-low-density fractions (d < 1.006 g/ml). Importantly, in HepG2c-NS5A-JB supernatant, infectious particles and viral RNA cosegregated in the same density fractions (Fig. 6E), and no infectious particles were detected in the very-low-density fractions.
Huh7.5-NS5A-JB and HepG2c-NS5A-JB cells secrete mainly apoE-positive/apoB-negative viral particles.
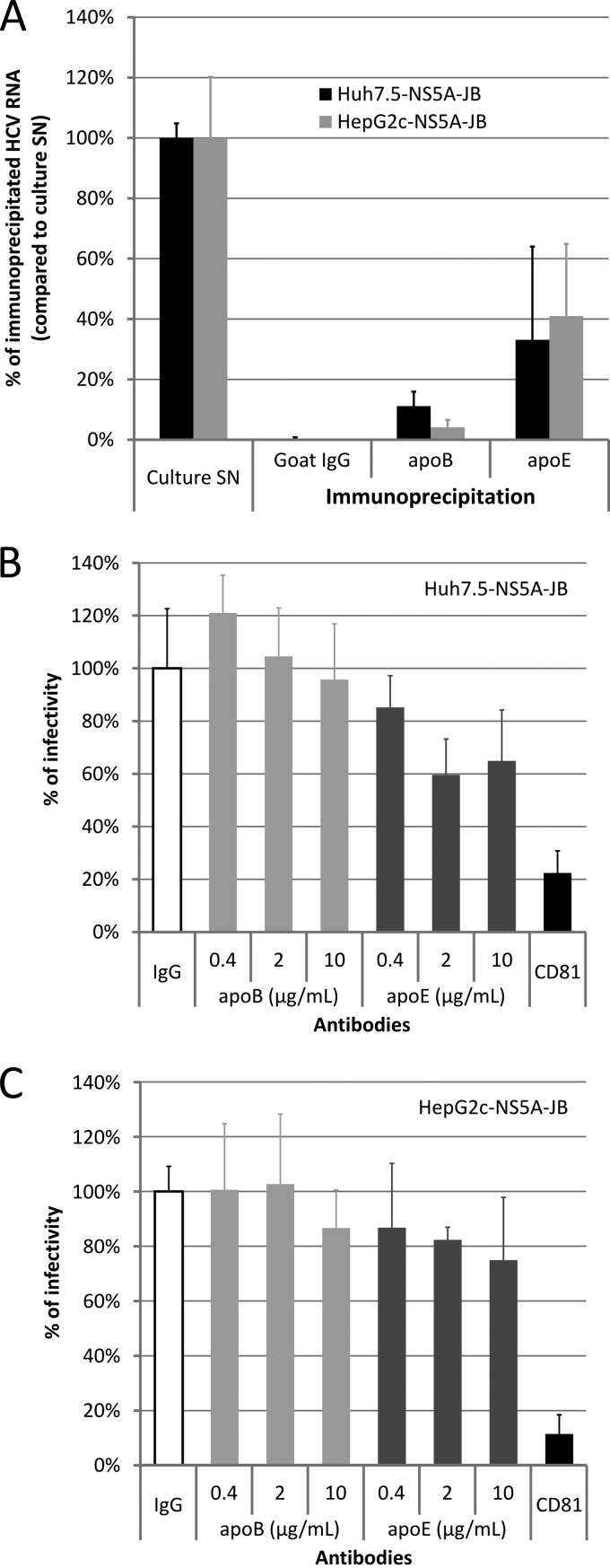
To investigate HCV RNA association with lipoproteins, we evaluated the ability of anti-apolipoprotein antibodies to immunoprecipitate HCV RNA by quantitative RT-PCR (IP/RT-qPCR). Viral supernatants were immunoprecipitated with either anti-apoB or anti-apoE antibodies, using purified goat IgG as a negative control, as previously performed on naïve cells (Fig. 1). Under our experimental conditions, all apoB100 was immunoprecipitated from both HepG2 and Huh7.5 supernatants as apoB100 was no longer detected by immunoblotting in the flowthrough fraction (Fig. 1F and H). In contrast, the anti-apoE antibody used for these experiments was unable to immunoprecipitate all apoE due to a high apoE secretion level (data not shown) (apoE secretion levels are shown in Fig. 1). Whereas a substantial amount of viral RNA (i.e., up to 40%) was associated with apoE, no or little viral RNA was associated with apoB (Fig. 7A). To ascertain whether, despite an undetectable (or weak) association, apoB could have any biological significance on HCVcc entry properties, we studied the effect of anti-apolipoprotein antibodies on virus infectivity. Culture supernatants from Huh7.5-NS5A-JB and HepG2c-NS5A-JB cells were harvested as previously described in serum-free medium and incubated with a mouse monoclonal anti-apoB antibody (4G3), previously reported to block apoB interaction with LDL receptor (67), or with a mouse monoclonal anti-apoE antibody (D6E10), previously reported to efficiently immunoprecipitate apoE (68). Neutralizing mouse monoclonal anti-CD81 antibody (JS81) (69) was used as a positive control. Neutralization was performed in serum-free medium, and two different readouts were used, i.e., cell-associated HCV RNA after 4 h of inoculation assessed by quantitative RT-PCR (data not shown) or quantification of infection and subsequent replication assessed by measuring focus-forming units (Fig. 7B and C). As expected, anti-CD81 antibodies had no effect on virus binding to cells after 4 h (data not shown) but efficiently neutralized infection, as shown by a 90% reduction in the infectivity of HepG2c-NS5A-JB-produced viral particles (Fig. 7C). Importantly, neither assay showed any effect of anti-apoB antibodies on virus binding or infection for virions produced by both Huh7.5-NS5A-JB and HepG2c-NS5A-JB (Fig. 7B and C). In contrast, although in a not statistically significant manner, anti-apoE antibodies were capable of slightly neutralizing viral infections, thus further supporting the presence of apoE on both Huh7- and HepG2-produced particles.
Fig 7.
Characterization of viral particles produced by JB-replicating cells. Huh7.5-NS5A-JB, HepG2-SGR, and HepG2c-NS5A-JB cells were treated for 3 days after confluence with 2% DMSO and incubated another 24 h with serum-free medium containing 2% DMSO, 10% (vol/vol) OA-BSA, and 1 μM U0126. Supernatants were harvested, filtered (0.45-μm pore size), and used for immunoprecipitation (IP) and neutralization experiments. (A) Harvested supernatants from Huh7.5-NS5A-JB and HepG2c-NS5A-JB cells were immunoprecipitated with goat polyclonal anti-apoB or anti-apoE antibodies, using protein G-coated magnetic beads. IgG from goat serum was used for a control. The percentage of HCV IP was assessed by quantifying HCV RNA using RT-qPCR in immunoprecipitated pellets, compared to HCV RNA in the culture supernatant (SN). Error bars represent standard deviations of two independent experiments. (B and C) Supernatants from Huh7.5-NS5A-JB and HepG2c-NS5A-JB cells were incubated for 2 h at room temperature with different concentrations of mouse monoclonal anti-apoB antibody (4G3) or with mouse monoclonal anti-apoE (D6E10). As a positive neutralization control, supernatants were incubated for 2 h at room temperature with 10 μg/ml of mouse monoclonal anti-CD81 antibody (JS81). As a negative control, supernatants were incubated for 2 h at room temperature with 10 μg/ml of IgG from mouse serum. Naïve Huh7.5 cells were then incubated for 4 h at 37°C with supernatant-antibody mixtures and further processed. Cells for CD81 neutralization experiments were incubated with 10 μg/ml of anti-CD81 antibody concomitantly. Cells were washed once with PBS and stained after 3 days for HCV core protein to determine viral titer (FFU). Error bars represent standard deviations of two independent experiments.
HCV replication does not alter VLDL secretion in HepG2 cells.
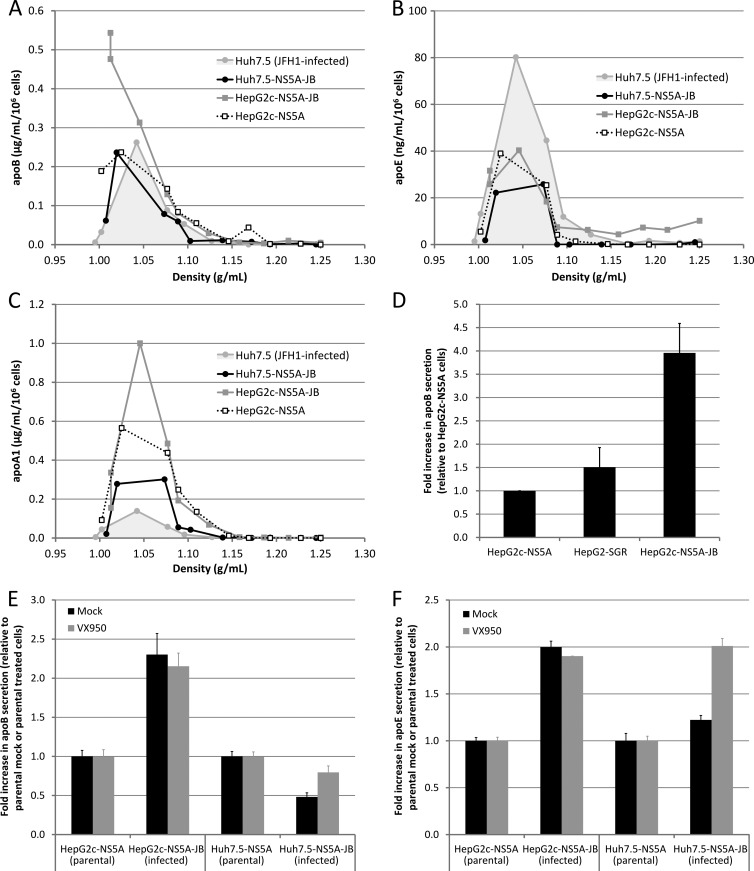
HCV is often associated in vivo with hypobetalipoproteinemia (8, 9), and HCV core protein has been shown in cell culture models to inhibit microsomal triglyceride transfer protein (MTP) activity, thus leading to decreased apoB secretion. Therefore, we sought to determine whether genomic HCV replication in VLDL-producing cells would quantitatively and qualitatively impact apoB and VLDL secretion. To this end, we analyzed apoB and apoE secretion profiles in our JB-replicating cell lines. We also analyzed apolipoprotein-A1 (apoA1) secretion as a high-density lipoprotein (HDL) secretion control. As previously, we harvested culture supernatants after a 3-day treatment with DMSO and another 24 h with DMSO, OA, and U0126; we then loaded harvested supernatants on iodixanol-sucrose density gradients and quantified apoB, apoE, and apoA1 in each gradient fraction by ELISA. Both HepG2c-NS5A and HepG2c-NS5A-JB cells secreted apoB in the VLDL density range (Fig. 8A), as previously shown for parental noninfected HepG2-CD81 cells (Fig. 1A). In contrast, JFH1-infected Huh7.5 and JB-replicating Huh7.5-NS5A-JB cells both secreted most of apoB in the IDL-LDL density range (Fig. 8A), as previously shown for parental noninfected Huh7.5 cells (Fig. 1B). Moreover, only HepG2c-NS5A-JB cells secreted significant amounts of apoE in the VLDL density range (Fig. 8B), suggesting that these cells were still able to secrete VLDL, even after the selection process, in contrast to Huh7.5 cells. Then, we wanted to discriminate between VLDL and HDL secretion by looking at an HDL-specific apolipoprotein, apoA1. For every cell type, apoA1 was not secreted in the VLDL density range; rather, it was secreted in the LDL-HDL density range, with a secretion profile similar to that of apoE (Fig. 8C). Interestingly, HCV replication seemed to increase apoB in HepG2c-NS5A cells (Fig. 8A and B). Indeed, apoB quantification in culture supernatants revealed that the levels of apoB secreted from HepG2c-NS5A-JB cells were significantly higher than from the HepG2-SGR and HepG2c counterparts (Fig. 8D), suggesting that full-length genomic HCV replication and/or structural protein expression may potentiate apoB secretion in vitro and not restrict it, as suggested in vivo by the hypobetalipoproteinemia status of some patients (8, 9). To confirm that this observation was directly attributable to viral replication and not to cell line selection issues, we treated Huh7.5-NS5A and HepG2c-NS5A cells (parental and JB-infected cells) with a viral protease inhibitor, VX-950. VX-950-treated cells showed a strong decrease in viral replication after 4 days and a concomitant decrease in apoB secretion (data not shown). However, the effect on apoB secretion was similar between parental and infected cells, suggesting a VX-950-mediated inhibition of apoB secretion. To clarify the reading of the results, we corrected for this off-target effect of the VX-950 compound by normalizing the data set against mock-treated as well as VX-950-treated conditions. As previously described in the literature, we could observe that HCV replication in Huh7.5-NS5A cells seemed to decrease apoB secretion since VX-950 was able to restore apoB secretion in Huh7.5-NS5A-JB cells (Fig. 8E). Similarly, apoE secretion was inhibited by HCV replication in Huh7.5-NS5A-JB cells (Fig. 8F). However, we could not demonstrate any specific effect of VX-950 on either apoB or apoE secretion levels in HepG2c-NS5A-JB cells (Fig. 8E and F), suggesting that the viral replication does not alter apolipoprotein secretion in HepG2 cells. These results highlight the functional differences between Huh7.5 and HepG2 cells with respect to lipoprotein metabolism and highlight HepG2 cells as a useful alternative model system to study virus-host interactions in general.
Fig 8.
VLDL/apoB secretion in JB-replicating cell lines. (A to C) Naive Huh7.5 cells were infected with JFH1 virus at an MOI of 0.01 and grown for 3 more days before being further processed. JFH1-infected Huh7.5, Huh7.5-NS5A-JB, HepG2c-NS5A, and HepG2c-NS5A-JB cells were treated for 3 days after confluence with 2% DMSO and incubated another 24 h with serum-free medium containing 2% DMSO, 10% (vol/vol) OA-BSA, and 1 μM U0126. Supernatants were harvested, filtered (0.45-μm pore size), and used for apoB quantification or concentrated and subjected to density separation on iodixanol-sucrose gradients. (A) apoB concentration was determined in each fraction with a commercial ELISA kit and plotted against density. Results are representative of three independent experiments. (B) apoE concentration was determined in each fraction with a commercial ELISA kit and plotted against density. (C) apoA1 concentration was determined in each fraction with a commercial ELISA kit and plotted against density. (D) apoB concentration was also determined in culture supernatants of HepG2-derived cells: HepG2c-NS5A, HepG2-SGR, and HepG2c-NS5A-JB. Error bars represent standard deviations of four independent experiments. (E and F) Huh7.5-NS5A-JB and HepG2c-NS5A-JB cells were treated for 3 days after confluence with 2% DMSO and 5 μM VX-950 and incubated another 24 h with serum-free medium containing 2% DMSO, 10% (vol/vol) OA-BSA, 1 μM U0126, and 5 μM VX-950. (E) apoB concentration was determined in supernatants by ELISA. For each condition (mock and VX-950-treated), fold change in apoB secretion compared to parental cells (HepG2c-NS5A and Huh7.5-NS5A) is shown. (F) apoE concentration was determined in supernatants by ELISA. For each condition (mock and VX-950-treated), fold change in apoE secretion compared to parental cells (HepG2c-NS5A and Huh7.5-NS5A) is shown.
DISCUSSION
One objective of this work was to develop a new cell culture model for HCV replication and production based on cells that are also competent for VLDL biogenesis. HepG2 cells were chosen for this purpose as we (Fig. 1) along with others (32, 34) have shown that HepG2 cells, in contrast to Huh7 (and derived) cells, are capable of secreting more physiologic VLDL, defined as apoB- and apoE-positive lipoprotein with a very low density (d < 1.006 g/ml).
If Huh7 and derived cells are fully permissive to HCV infection, leading to high viral replication and production of progeny virions, the permissivity of other hepatoma cell lines to HCV infection is lower and does not enable persistent replication of the virus. The high permissivity of Huh7.5 cells has been partially attributed to impaired innate immune responses in these cells, especially via RIG-I (retinoic acid inducible gene I) mutation and inactivation (63, 70). In contrast to Huh7.5 cells, HepG2 cells can mount an antiviral innate immune response against HCV infection via the production of type I interferon (data not shown). In this line, Sainz and collaborators recently suggested that the poor permissivity of several hepatoma cell lines could be partially attributed to HCV-induced innate signaling (39). Moreover, the high permissivity of Huh7 (and derived) cells has also been partially attributed to high expression levels of a liver-specific microRNA necessary for HCV RNA translation, i.e., miR-122 (71). Hence, the overexpression of miR-122 in otherwise negative HepG2 cells (41) or Hep3B cells (72) increased their permissivity to HCV infection. Moreover, ectopic expression of human miR-122 in mouse embryo fibroblasts rendered them permissive to HCV RNA replication when miR-122 expression is combined with inhibition of the IFN response (73), arguing for a critical role of miR-122 regarding viral tropism. Importantly, it has been shown that cells harboring HCV subgenomic replicons (and cells subsequently cured from those replicons) are positive for miR-122, arguing for a selection of an miR-122 phenotype during the establishment of the line (72). In this study, we have shown that cured HepG2 cells (HepG2c), deriving from HepG2-SGR cells (40) treated with IFN and anti-HCV-polymerase, together with an impaired IFN response, obtained by overexpression of IRF3(ΔN), were required to efficiently generate a stably HCV-replicating HepG2 cell line (Table 1). To select for cells persistently replicating HCV over multiple passages, we have opted for the use of a blasticidin (BSD)-tagged JFH1 strain (i.e., JFH1-BSD, or JB), and as the BSD tag was inserted into the NS5A area, transcomplementation with a V5-tagged version of NS5A was also performed. In fine, a cell line called HepG2c-NS5A-JB was generated and further used to analyze the interaction of HCV with lipoproteins. We demonstrated efficient viral RNA replication and viral protein production in HepG2c-NS5A-JB cells (Fig. 3). HCV proteins involved in viral assembly/morphogenesis were correctly sublocalized around LD in HepG2c-NS5A-JB cells (Fig. 4), and the secretion of infectious virions was also demonstrated (Fig. 6). Importantly, these cells could be maintained for several weeks in culture under blasticidin treatment, persistently replicating HCV and secreting infectious viral particles.
HepG2 cells are thought to be more mature hepatocytes than Huh7 cells. For instance, they can secrete VLDL (32) and are functionally polarized (74, 75). The polarization status of these cells has been shown to modify and to be modified by HCV infection (42, 43). Therefore, HepG2 might be a more relevant model than Huh7 to study HCV infection and its impact on cell physiology. The MEK/ERK signaling pathway, downstream of growth factor receptors, is implicated in crucial cellular processes, including proliferation, differentiation, angiogenesis, and survival. It is overactivated in many tumors and is thought to contribute to tumorigenesis (76). Interestingly, we showed an improvement in viral RNA replication after MEK/ERK inhibition in HepG2 cells, whereas no effect was observed in Huh7.5 cells (Fig. 5). MEK/ERK inhibition was also responsible for an improvement in VLDL secretion by HepG2 cells but not by Huh7.5 cells (Fig. 1). However, both effects might be nonrelated since VLDL secretion may be stimulated upon MEK/ERK inhibition by an increase in microsomal triglyceride transfer protein (MTP) and diacylglycerol acyltransferases (DGAT) expression (34), whereas HCV RNA translation could be improved by increased internal ribosome entry site (IRES)-mediated translation (66). Nevertheless, hepatocyte differentiation status, stimulated by exposure to DMSO, and mitogen activity, as shown by MEK/ERK pathway activity, may be of importance to study HCV replication and production in vitro. Without treatment, the overall viral replication level in HepG2 was lower than in Huh7.5 cells. This might be due to intrinsic characteristics of each cell line, as suggested before. However, we managed to increase the intracellular RNA replication level and viral particle secretion in HepG2 cells by combining DMSO-stimulated cell differentiation, MEK/ERK inhibition, and NS5A overexpression, thereby allowing analyzing secreted viral particles in this new model.
In this study, we characterized for the first time HCV association with lipoproteins in HepG2 cells, which are competent for VLDL secretion upon MEK/ERK inhibition and oleate treatment (Fig. 1). Indeed, an interaction between HCV RNA, apoB, and apoE has been suggested in vivo by different groups (14–16). In vitro, however, most reports suggest a strong association of HCV particles with apoE but not, or only weakly, with apoB (27–29), which seems conflicting when the hypothesis of association between HCV and VLDL is concerned as VLDL are clearly both apoB and apoE positive. apoE seems necessary to infectious viral particle assembly and production (24, 27) via an interaction with NS5A protein (77). apoE is indeed a host factor present on viral particles (28, 29) and crucial for particle infectivity via its interaction with LDL receptor (LDLR) at the surface of hepatocytes (26, 29, 78). On the other hand, some contradictory studies suggested that apoB and MTP could be required for HCV secretion in vitro (79–81) even if no clear demonstration of the presence of apoB on viral particles was provided. Therefore, lipoprotein biogenesis and, more precisely, VLDL synthesis seem to have important roles regarding HCV particle assembly, secretion, and infectivity. If apoE is strongly associated with secreted viral particles in cell culture, data reflecting a strong and specific association of HCV with apoB remain absent since studies reported in the literature were performed only in VLDL-deficient Huh7.5 cells. As our HepG2c-NS5A-JB cells were capable of producing a significant amount of infectious particles (in fact only 10 times less than their Huh7.5 counterparts) (Fig. 6), they could be used to investigate association with lipoproteins. In this report, we show that HepG2-produced HCV particles, as previously described for Huh7.5-produced particles (28, 29, 82), are mostly apoE positive but apoB negative (Fig. 7A), and we show/confirm that HCVcc virions produced by both cell types are not neutralized by neutralizing anti-apoB antibodies (clone 4G3), whereas HCVcc particles can be neutralized to some extent by anti-apoE antibodies (Fig. 7B and C). Moreover, there were no significant differences between HepG2- and Huh7.5-produced particles in terms of density repartition of secreted RNA, with notably no RNA and infectious particles in the very-low-density range (Fig. 6D and E). Altogether, and additionally to what was previously described in the literature, these results suggest that the ability of cells to produce VLDL does not change the ability of the virus to associate with apoB in vitro. This result warrants reconsideration of the somewhat fragile results obtained in vivo, which suggest, more than demonstrate, that HCV (i.e., not only viral protein but also its genome) can associate to apoB-positive particles. There are two possible scenarios regarding the molecular mechanism underlying LVP formation in vivo. They could derive either from an intracellular fusion between nascent VLDL and viral precursors or from a postsecretion fusion between VLDL/LDL and HCV virions. In this respect, apoE could play an important role in the fusion process due to its exchangeable properties (83). A study by Gastaminza and colleagues demonstrated that, in Huh7 cells, intracellular HCV virions are high-density particles, whereas secreted ones are lighter, thus suggesting the acquisition of low-density features (e.g., apoE and triglycerides) during trafficking through the Golgi apparatus (84). The presence of apoE in Huh7-produced HCVcc particles has since been confirmed by many studies (28, 29, 82). Our results obtained in HepG2 cells, together with previously published studies in Huh7 cells, seem to indicate that in vitro produced HCV particles are apoE positive but likely apoB negative, thus suggesting that, either way, the intracellular or postsecretion fusion of lipoproteins and viral precursors does not happen, at least in hepatoma cells, which are transformed and nonphysiologic cells. In this respect the analysis of the association of HCV with lipoproteins remains to be analyzed in more physiologic cells in vitro, i.e., primary human hepatocytes. Moreover, these results warrant further study to properly demonstrate whether HCV does associate with apoB-positive lipoproteins under physiological conditions in vivo and how this association takes place. In this respect, in vivo data suggesting a strong association between apoB and HCV have been recently revisited. Scholtes and collaborators demonstrated that LVP could mainly be apoB/gpE1-gpE2 positive, but nucleocapsid-free, particles (85). This new subclass of particles could then be defined as defective subviral particles (named eLVP). Such eLVP containing only HCV glycoproteins and apoB had already been described in vitro in HepG2 and Caco-2 cell lines overexpressing gpE1 and gpE2 viral proteins (31).
Another interesting finding of our study in HCV-producing HepG2 cells is that viral replication in these cells does not seem to alter apolipoprotein secretion (Fig. 8), contrary to what is observed in HCV-producing Huh7.5 cells, with a virus-mediated decrease in apoB and apoE secretion, pointing out once again differences between both cell types. These results contrast with the hypobetalipoproteinemia observed in some patients (8, 9) and which is thought to be a consequence of virus replication. However, its underlying mechanisms remain mostly unknown.
To conclude, we engineered a unique HCV-replicating cell culture model in HepG2 cells using a blasticidin-tagged viral strain. A full replication cycle with infectious viral particle secretion could be obtained with these cells, which could be used as an alternative to commonly used Huh7 and derived cell lines, which present polarity and VLDL secretion defects. This cell line represents a novel cell culture model to study many aspects of HCV host-cell interactions and is very complementary of the currently used model based on Huh7 and derived cells. Using this model, we showed that apoE but not apoB is associated to infectious cell culture-grown HCV using, for the first time, cells which are competent to produce VLDL. This result challenges the relevance of in vivo observations, which suggested, more than demonstrated, an association between HCV and apoB-containing lipoproteins in HCV patients. In fact, it is not excluded that infectious HCV particles could harbor only apoE and not apoB. It remains to be determined whether other types of viral or subviral particles, such as eLVP (85), could be produced by HCV-replicating cells. In this respect the HCV-replicating HepG2 line developed here will be instrumental.
ACKNOWLEDGMENTS
We thank Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan) for the gift of the JFH1 strain and HepG2-SGR cells, Charles M. Rice (The Rockefeller University, NY, USA) for the gift of Huh7.5 cells, and Johan Neyts (Rega Institute, Leuven, Belgium) for the gift of anti-HCV-polymerase compounds. We are very grateful to Jane McKeating for her critical reading of the manuscript. We also thank Clarisse Benoit for her helpful and efficient technical assistance.
This work was supported by a grant from the French National Agency for Research against AIDS and Viral Hepatitis (AO1-2008-CSS4) and by an INSERM core grant. B.J. was the recipient of a bursary from the French Ministry of Research and Higher Education and also from the Association pour la Recherche sur le Cancer.
Footnotes
Published ahead of print 20 February 2013
REFERENCES
- 1. Knobler H, Zhornicky T, Sandler A, Haran N, Ashur Y, Schattner A. 2003. Tumor necrosis factor-alpha-induced insulin resistance may mediate the hepatitis C virus-diabetes association. Am. J. Gastroenterol. 98:2751–2756 [DOI] [PubMed] [Google Scholar]
- 2. Moucari R, Asselah T, Cazals-Hatem D, Voitot H, Boyer N, Ripault M-P, Sobesky R, Martinot-Peignoux M, Maylin S, Nicolas-Chanoine M-H, Paradis V, Vidaud M, Valla D, Bedossa P, Marcellin P. 2008. Insulin resistance in chronic hepatitis C: association with genotypes 1 and 4, serum HCV RNA level, and liver fibrosis. Gastroenterology 134:416–423 [DOI] [PubMed] [Google Scholar]
- 3. Mason AL, Lau JY, Hoang N, Qian K, Alexander GJ, Xu L, Guo L, Jacob S, Regenstein FG, Zimmerman R, Everhart JE, Wasserfall C, Maclaren NK, Perrillo RP. 1999. Association of diabetes mellitus and chronic hepatitis C virus infection. Hepatology 29:328–333 [DOI] [PubMed] [Google Scholar]
- 4. Serfaty L, Capeau J. 2009. Hepatitis C, insulin resistance and diabetes: clinical and pathogenic data. Liver Int. 29(Suppl 2):13–25 [DOI] [PubMed] [Google Scholar]
- 5. Asselah T, Rubbia-Brandt L, Marcellin P, Negro F. 2006. Steatosis in chronic hepatitis C: why does it really matter? Gut 55:123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Negro F. 2010. Abnormalities of lipid metabolism in hepatitis C virus infection. Gut 59:1279–1287 [DOI] [PubMed] [Google Scholar]
- 7. Negro F, Sanyal AJ. 2009. Hepatitis C virus, steatosis and lipid abnormalities: clinical and pathogenic data. Liver Int. 29(Suppl 2):26–37 [DOI] [PubMed] [Google Scholar]
- 8. Petit JM, Benichou M, Duvillard L, Jooste V, Bour JB, Minello A, Verges B, Brun JM, Gambert P, Hillon P. 2003. Hepatitis C virus-associated hypobetalipoproteinemia is correlated with plasma viral load, steatosis, and liver fibrosis. Am. J. Gastroenterol. 98:1150–1154 [DOI] [PubMed] [Google Scholar]
- 9. Serfaty L, Andreani T, Giral P, Carbonell N, Chazouillères O, Poupon R. 2001. Hepatitis C virus induced hypobetalipoproteinemia: a possible mechanism for steatosis in chronic hepatitis C. J. Hepatol. 34:428–434 [DOI] [PubMed] [Google Scholar]
- 10. Dai C-Y, Chuang W-L, Ho C-K, Hsieh M-Y, Huang J-F, Lee L-P, Hou N-J, Lin Z-Y, Chen S-C, Hsieh M-Y, Wang L-Y, Tsai J-F, Chang W-Y, Yu M-L. 2008. Associations between hepatitis C viremia and low serum triglyceride and cholesterol levels: a community-based study. J. Hepatol. 49:9–16 [DOI] [PubMed] [Google Scholar]
- 11. Corey KE, Kane E, Munroe C, Barlow LL, Zheng H, Chung RT. 2009. Hepatitis C virus infection and its clearance alter circulating lipids: implications for long-term follow-up. Hepatology 50:1030–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Adinolfi LE, Restivo L, Zampino R, Guerrera B, Lonardo A, Ruggiero L, Riello F, Loria P, Florio A. 2012. Chronic HCV infection is a risk of atherosclerosis. Role of HCV and HCV-related steatosis. Atherosclerosis 221:496–502 [DOI] [PubMed] [Google Scholar]
- 13. Lerat H, Kammoun HL, Hainault I, Mérour E, Higgs MR, Callens C, Lemon SM, Foufelle F, Pawlotsky J-M. 2009. Hepatitis C virus proteins induce lipogenesis and defective triglyceride secretion in transgenic mice. J. Biol. Chem. 284:33466–33474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thomssen R, Bonk S, Propfe C, Heermann KH, Köchel HG, Uy A. 1992. Association of hepatitis C virus in human sera with beta-lipoprotein. Med. Microbiol. Immunol. 181:293–300 [DOI] [PubMed] [Google Scholar]
- 15. André P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Bréchot C, Paranhos-Baccalà G, Lotteau V. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 76:6919–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nielsen SU, Bassendine MF, Burt AD, Martin C, Pumeechockchai W, Toms GL. 2006. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J. Virol. 80:2418–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Felmlee DJ, Sheridan DA, Bridge SH, Nielsen SU, Milne RW, Packard CJ, Caslake MJ, McLauchlan J, Toms GL, Neely RDG, Bassendine MF. 2010. Intravascular transfer contributes to postprandial increase in numbers of very-low-density hepatitis C virus particles. Gastroenterology 139:1774–1783 [DOI] [PubMed] [Google Scholar]
- 18. Bartenschlager R, Penin F, Lohmann V, André P. 2011. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 19:95–103 [DOI] [PubMed] [Google Scholar]
- 19. Herker E, Ott M. 2011. Unique ties between hepatitis C virus replication and intracellular lipids. Trends Endocrinol. Metab. 22:241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 21. Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich H-G, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, Abid K, Negro F, Dreux M, Cosset F-L, Bartenschlager R. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. U. S. A. 103:7408–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chang K-S, Jiang J, Cai Z, Luo G. 2007. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J. Virol. 81:13783–13793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cun W, Jiang J, Luo G. 2010. The C-terminal alpha-helix domain of apolipoprotein E is required for interaction with nonstructural protein 5A and assembly of hepatitis C virus. J. Virol. 84:11532–11541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hishiki T, Shimizu Y, Tobita R, Sugiyama K, Ogawa K, Funami K, Ohsaki Y, Fujimoto T, Takaku H, Wakita T, Baumert TF, Miyanari Y, Shimotohno K. 2010. Infectivity of hepatitis C virus is influenced by association with apolipoprotein E isoforms. J. Virol. 84:12048–12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang J, Luo G. 2009. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J. Virol. 83:12680–12691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Merz A, Long G, Hiet M-S, Brügger B, Chlanda P, Andre P, Wieland F, Krijnse-Locker J, Bartenschlager R. 2011. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J. Biol. Chem. 286:3018–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Owen DM, Huang H, Ye J, Gale M. 2009. Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology 394:99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lindenbach BD, Meuleman P, Ploss A, Vanwolleghem T, Syder AJ, McKeating JA, Lanford RE, Feinstone SM, Major ME, Leroux-Roels G, Rice CM. 2006. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl. Acad. Sci. U. S. A. 103:3805–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Icard V, Diaz O, Scholtes C, Perrin-Cocon L, Ramière C, Bartenschlager R, Penin F, Lotteau V, André P. 2009. Secretion of hepatitis C virus envelope glycoproteins depends on assembly of apolipoprotein B positive lipoproteins. PLoS One 4:e4233 doi:10.1371/journal.pone.0004233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meex SJR, Andreo U, Sparks JD, Fisher EA. 2011. Huh-7 or HepG2 cells: which is the better model for studying human apolipoprotein-B100 assembly and secretion? J. Lipid Res. 52:152–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dixon JL, Ginsberg HN. 1993. Regulation of hepatic secretion of apolipoprotein B-containing lipoproteins: information obtained from cultured liver cells. J. Lipid Res. 34:167–179 [PubMed] [Google Scholar]
- 34. Tsai J, Qiu W, Kohen-Avramoglu R, Adeli K. 2007. MEK-ERK inhibition corrects the defect in VLDL assembly in HepG2 cells: potential role of ERK in VLDL-ApoB100 particle assembly. Arterioscler. Thromb. Vasc. Biol. 27:211–218 [DOI] [PubMed] [Google Scholar]
- 35. Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, Scarselli E, Cortese R, Nicosia A, Cosset F-L. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624–41630 [DOI] [PubMed] [Google Scholar]
- 36. Fukuhara T, Tani H, Shiokawa M, Goto Y, Abe T, Taketomi A, Shirabe K, Maehara Y, Matsuura Y. 2011. Intracellular delivery of serum-derived hepatitis C virus. Microbes Infect. 13:405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Harris HJ, Davis C, Mullins JGL, Hu K, Goodall M, Farquhar MJ, Mee CJ, McCaffrey K, Young S, Drummer H, Balfe P, McKeating JA. 2010. Claudin association with CD81 defines hepatitis C virus entry. J. Biol. Chem. 285:21092–21102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, Royer C, Fischer B, Zahid MN, Lavillette D, Fresquet J, Cosset F-L, Rothenberg SM, Pietschmann T, Patel AH, Pessaux P, Doffoël M, Raffelsberger W, Poch O, McKeating JA, Brino L, Baumert TF. 2011. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat. Med. 17:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sainz B, Jr, Barretto N, Yu X, Corcoran P, Uprichard SL. 2012. Permissiveness of human hepatoma cell lines for HCV infection. Virol. J. 9:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Date T, Kato T, Miyamoto M, Zhao Z, Yasui K, Mizokami M, Wakita T. 2004. Genotype 2a hepatitis C virus subgenomic replicon can replicate in HepG2 and IMY-N9 cells. J. Biol. Chem. 279:22371–22376 [DOI] [PubMed] [Google Scholar]
- 41. Narbus CM, Israelow B, Sourisseau M, Michta ML, Hopcraft SE, Zeiner GM, Evans MJ. 2011. HepG2 cells expressing microRNA miR-122 support the entire hepatitis C virus life cycle. J. Virol. 85:12087–12092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mee CJ, Farquhar MJ, Harris HJ, Hu K, Ramma W, Ahmed A, Maurel P, Bicknell R, Balfe P, McKeating JA. 2010. Hepatitis C virus infection reduces hepatocellular polarity in a vascular endothelial growth factor-dependent manner. Gastroenterology 138:1134–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mee CJ, Harris HJ, Farquhar MJ, Wilson G, Reynolds G, Davis C, van IJzendoorn SCD, Balfe P, McKeating JA. 2009. Polarization restricts hepatitis C virus entry into HepG2 hepatoma cells. J. Virol. 83:6211–6221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lim SP, Soo HM, Tan YH, Brenner S, Horstmann H, MacKenzie JM, Ng ML, Lim SG, Wjin Hong. 2002. Inducible system in human hepatoma cell lines for hepatitis C virus production. Virology 303:79–99 [DOI] [PubMed] [Google Scholar]
- 45. Dash S, Halim AB, Tsuji H, Hiramatsu N, Gerber MA. 1997. Transfection of HepG2 cells with infectious hepatitis C virus genome. Am. J. Pathol. 151:363–373 [PMC free article] [PubMed] [Google Scholar]
- 46. McCormick CJ, Challinor L, Macdonald A, Rowlands DJ, Harris M. 2004. Introduction of replication-competent hepatitis C virus transcripts using a tetracycline-regulable baculovirus delivery system. J. Gen. Virol. 85:429–439 [DOI] [PubMed] [Google Scholar]
- 47. el-Awady MK, Tabll AA, el-Abd YS, Bahgat MM, Shoeb HA, Youssef SS, Bader el-Din NG, Redwan el-RM, el-Demellawy M, Omran MH, el-Garf WT, Goueli SA. 2006. HepG2 cells support viral replication and gene expression of hepatitis C virus genotype 4 in vitro. World J. Gastroenterol. 12:4836–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Owsianka A, Clayton RF, Loomis-Price LD, McKeating JA, Patel AH. 2001. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J. Gen. Virol. 82:1877–1883 [DOI] [PubMed] [Google Scholar]
- 49. Hadlock KG, Lanford RE, Perkins S, Rowe J, Yang Q, Levy S, Pileri P, Abrignani S, Foung SK. 2000. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J. Virol. 74:10407–10416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Delgrange D, Pillez A, Castelain S, Cocquerel L, Rouillé Y, Dubuisson J, Wakita T, Duverlie G, Wychowski C. 2007. Robust production of infectious viral particles in Huh-7 cells by introducing mutations in hepatitis C virus structural proteins. J. Gen. Virol. 88:2495–2503 [DOI] [PubMed] [Google Scholar]
- 51. Moradpour D, Evans MJ, Gosert R, Yuan Z, Blum HE, Goff SP, Lindenbach BD, Rice CM. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bartosch B, Cosset F-L. 2009. Studying HCV cell entry with HCV pseudoparticles (HCVpp). Methods Mol. Biol. 510:279–293 [DOI] [PubMed] [Google Scholar]
- 54. Lin R, Heylbroeck C, Pitha PM, Hiscott J. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Komurian-Pradel F, Paranhos-Baccalà G, Sodoyer M, Chevallier P, Mandrand B, Lotteau V, André P. 2001. Quantitation of HCV RNA using real-time PCR and fluorimetry. J. Virol. Methods 95:111–119 [DOI] [PubMed] [Google Scholar]
- 56. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 57. Galbraith RA, Sassa S, Fujita H. 1988. Regulation of heme synthesis in HepG2 human hepatoma cells by dimethyl sulfoxide. Biochem. Biophys. Res. Commun. 153:869–874 [DOI] [PubMed] [Google Scholar]
- 58. Sainz B, Chisari FV. 2006. Production of infectious hepatitis C virus by well-differentiated, growth-arrested human hepatoma-derived cells. J. Virol. 80:10253–10257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035 doi:10.1371/journal.ppat.1000035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lucifora J, Durantel D, Testoni B, Hantz O, Levrero M, Zoulim F. 2010. Control of hepatitis B virus replication by innate response of HepaRG cells. Hepatology 51:63–72 [DOI] [PubMed] [Google Scholar]
- 61. Marion M-J, Hantz O, Durantel D. 2010. The HepaRG cell line: biological properties and relevance as a tool for cell biology, drug metabolism, and virology studies. Methods Mol. Biol. 640:261–272 [DOI] [PubMed] [Google Scholar]
- 62. Parent R, Marion M-J, Furio L, Trépo C, Petit M-A. 2004. Origin and characterization of a human bipotent liver progenitor cell line. Gastroenterology 126:1147–1156 [DOI] [PubMed] [Google Scholar]
- 63. Sumpter R, Jr, Loo Y-M, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale, M 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chang K-O, George DW. 2007. Bile acids promote the expression of hepatitis C virus in replicon-harboring cells. J. Virol. 81:9633–9640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang T, Li Y, Lai J-P, Douglas SD, Metzger DS, O'Brien CP, Ho W-Z. 2003. Alcohol potentiates hepatitis C virus replicon expression. Hepatology 38:57–65 [DOI] [PubMed] [Google Scholar]
- 66. Ndjomou J, Park I-W, Liu Y, Mayo LD, He JJ. 2009. Up-Regulation of Hepatitis C Virus Replication and Production by Inhibition of MEK/ERK Signaling. PLoS One 4:e7498 doi:10.1371/journal.pone.0007498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Milne RW, Theolis R, Jr, Verdery RB, Marcel YL. 1983. Characterization of monoclonal antibodies against human low density lipoprotein. Arteriosclerosis. 3:23–30 [DOI] [PubMed] [Google Scholar]
- 68. Soler-López M, Zanzoni A, Lluís R, Stelzl U, Aloy P. 2011. Interactome mapping suggests new mechanistic details underlying Alzheimer's disease. Genome Res. 21:364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lavillette D, Tarr AW, Voisset C, Donot P, Bartosch B, Bain C, Patel AH, Dubuisson J, Ball JK, Cosset F-L. 2005. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41:265–274 [DOI] [PubMed] [Google Scholar]
- 70. Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172 [DOI] [PubMed] [Google Scholar]
- 71. Henke JI, Goergen D, Zheng J, Song Y, Schüttler CG, Fehr C, Jünemann C, Niepmann M. 2008. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 27:3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kambara H, Fukuhara T, Shiokawa M, Ono C, Ohara Y, Kamitani W, Matsuura Y. 2012. Establishment of a novel permissive cell line for the propagation of hepatitis C Virus by expression of microRNA miR122. J. Virol. 86:1382–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lin L-T, Noyce RS, Pham TNQ, Wilson JA, Sisson GR, Michalak TI, Mossman KL, Richardson CD. 2010. Replication of Subgenomic Hepatitis C virus replicons in mouse fibroblasts is facilitated by deletion of interferon regulatory factor 3 and expression of liver-specific microRNA 122. J. Virol. 84:9170–9180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. van IJzendoorn SC, Hoekstra D. 2000. Polarized sphingolipid transport from the subapical compartment changes during cell polarity development. Mol. Biol. Cell 11:1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zegers MM, Hoekstra D. 1998. Mechanisms and functional features of polarized membrane traffic in epithelial and hepatic cells. Biochem. J. 336:257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Whittaker S, Marais R, Zhu AX. 2010. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene 29:4989–5005 [DOI] [PubMed] [Google Scholar]
- 77. Benga WJA, Krieger SE, Dimitrova M, Zeisel MB, Parnot M, Lupberger J, Hildt E, Luo G, McLauchlan J, Baumert TF, Schuster C. 2010. Apolipoprotein E interacts with hepatitis C virus nonstructural protein 5A and determines assembly of infectious particles. Hepatology 51:43–53 [DOI] [PubMed] [Google Scholar]
- 78. Liu S, McCormick KD, Zhao W, Zhao T, Fan D, Wang T. 2012. Human apolipoprotein E peptides inhibit hepatitis C virus entry by blocking virus binding. Hepatology 56:484–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gastaminza P, Cheng G, Wieland S, Zhong J, Liao W, Chisari FV. 2008. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J. Virol. 82:2120–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M, Ye J. 2007. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 104:5848–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nahmias Y, Goldwasser J, Casali M, van Poll D, Wakita T, Chung RT, Yarmush ML. 2008. Apolipoprotein B-dependent hepatitis C virus secretion is inhibited by the grapefruit flavonoid naringenin. Hepatology 47:1437–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Akazawa D, Morikawa K, Omi N, Takahashi H, Nakamura N, Mochizuki H, Date T, Ishii K, Suzuki T, Wakita T. 2011. Production and characterization of HCV particles from serum-free culture. Vaccine 29:4821–4828 [DOI] [PubMed] [Google Scholar]
- 83. Saito H, Dhanasekaran P, Baldwin F, Weisgraber KH, Lund-Katz S, Phillips MC. 2001. Lipid binding-induced conformational change in human apolipoprotein E. Evidence for two lipid-bound states on spherical particles. J. Biol. Chem. 276:40949–40954 [DOI] [PubMed] [Google Scholar]
- 84. Gastaminza P, Kapadia SB, Chisari FV. 2006. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J. Virol. 80:11074–11081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Scholtes C, Ramière C, Rainteau D, Perrin-Cocon L, Wolf C, Humbert L, Carreras M, Guironnet-Paquet A, Zoulim F, Bartenschlager R, Lotteau V, André P, Diaz O. 2012. High plasma level of nucleocapsid-free envelope glycoprotein-positive lipoproteins in hepatitis C patients. Hepatology 56:39–48 [DOI] [PubMed] [Google Scholar]