Abstract
HIV-1 proteins are synthesized from a single transcript in an unspliced form or following splicing, but the existence of an antisense protein (ASP) expressed from an antisense polyadenylated transcript has been suggested. Difficulties linked to the detection of this protein in mammalian cells led us to codon optimize its cDNA. Codon-optimized ASP was indeed efficiently detected in various transfected cell lines following flow cytometry and confocal microscopy analyses. Western blot analyses also led to the detection of optimized ASP in transfected cells but also provided evidence of its instability and high multimerization potential. ASP was mainly distributed in the cytoplasm in a punctate manner, which was reminiscent of autophagosomes. In agreement with this observation, a significant increase in ASP-positive cells and loss of its punctate distribution was observed in transfected cells when autophagy was inhibited at early steps. Induction of autophagy was confirmed by Western blot analyses that showed an ASP-mediated increase in levels of LC3b-II and Beclin 1, as well as colocalization and interaction between ASP and LC3. Interestingly, Myc-tagged ASP was detected in the context of proviral DNA following autophagy inhibition with a concomitant increase in the level and punctate distribution of LC3b-II. Finally, 3-methyladenine treatment of transfected or infected U937 cells decreased extracellular p24 levels in wild-type proviral DNA and to a much lesser extent in ASP-mutated proviral DNA. This study provides the first detection of ASP in mammalian cells by Western blotting. ASP-induced autophagy might explain the inherent difficulty in detecting this viral protein and might justify its presumed low abundance in infected cells.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) is a complex retrovirus which harbors all three common retroviral genes (gag, pol, and env), in addition to two regulatory genes (tat and rev) and four accessory genes (vif, vpr, vpu, and nef). All of these genes are expressed through a single transcript initiating from the promoter-harboring 5′ long terminal repeat (LTR) region. The various viral proteins are synthesized from unspliced, monospliced, or multispliced forms of this major transcript and then assume their functional roles in infected cells. Early studies had suggested, however, that another gene, encoding antisense protein (ASP), could be expressed through an antisense transcript (1–3).
Antisense transcription is a more common phenomenon than previously thought (4–7). The possible existence of antisense transcription in retroviruses had also been suggested (3, 8). More recent studies have further confirmed its existence in members of the human T cell leukemia virus retrovirus family and have further highlighted that these antisense transcripts are spliced and encode proteins with a capacity to modulate the transactivation potential of multiple cellular transcription activators (9–23). Based on the presence of a conserved open reading frame (ORF) on the antisense strand of HIV-1 proviral DNA, antisense transcription has also been suggested for HIV-1 (3). In fact, we and others have provided evidence for the existence of antisense transcripts overlapping the ASP ORF region with transcription initiation sites located near the 3′ LTR and the poly(A) signal in a complementary region of the pol gene (1, 2, 31).
Bioinformatic analyses have indicated that the presumed encoded ASP is highly hydrophobic, harboring a cysteine-rich amino region and potential transmembrane domains (3). However, early detection of ASP had been limited to electron microscopy (EM) studies of infected and transfected cells and in vitro translation studies (32). More recently, we have demonstrated that ASP could be analyzed by immunofluorescence microscopy and localized at the plasma membrane of T cells (33). Despite these studies, no functions have yet been suggested for this new viral protein due to the difficulties related to its detection. One explanation is linked to its cysteine-rich region, which might mediate strong agglomerated complexes. Protein aggregates have previously been linked to the degrading autophagic pathway (34).
Autophagy is a cellular homeostasis mechanism mediating degradation of long-lived proteins and cellular organelles. Typical autophagy (also known as macroautophagy) involves the formation of the autophagosome, a double membrane vesicle which, upon fusion with lysosomes, forms the autolysosome, thereby degrading delivered contents. Molecular studies of autophagy demonstrate a conserved mechanism during evolution, which involves several Atg proteins playing distinct roles during each step (35–37). One of the important autophagy-associated proteins is the microtubule-associated protein (MAP) light chain 3b (LC3b), an LC3 isoform known as a marker for autophagosomes (38). This LC3 isoform is cleaved as the cytoplasmic LC3-I form, which, upon induction of autophagy, is linked to phosphatidylethanolamine at its C terminus and is associated with the autophagosomal membrane as the LC3-II form (38, 39). In recent years, several studies have demonstrated the implications of autophagy in virus replication (40–42) and, more specifically, in HIV-1 infection and pathogenesis (43–49). In T cells, HIV-1-induced autophagy has been suggested to depend on the surface expression of Env proteins, leading to apoptosis of infected and uninfected CD4+ T cells (43, 45, 46, 50). In sharp contrast, in macrophages, HIV-1-induced autophagy increases viral replication and is inhibited at late stages, i.e., at the step of fusion of autophagosomes with lysosomes, a process which is driven by Nef through its interaction with Beclin 1, another induced autophagy marker (47). Several aspects of autophagy in macrophages have yet to be elucidated, and the actual inducer remains to be identified.
The study of ASP is highly important, as no information is currently available as to its function. Thus, it is important to seek a better understanding of how this protein could affect HIV-1 replication and/or cellular functions. In the present study, we used a codon-optimized version of ASP DNA to improve its detection. Using anti-ASP and anti-Myc antibodies, ASP was detected in different mammalian cell lines by flow cytometry and confocal microscopy, and was further shown to aggregate by Western blot analyses. In transfected cells, a punctate ASP signal was noted, which was reminiscent of autophagosomes. Interestingly, inhibition of autophagy at an early step increased the number of ASP-positive cells but led to a loss in the number of ASP puncta. In addition, the LC3b-II isoform was more abundant in ASP-expressing cells and colocalized and interacted with ASP. Our study further led to the detection of ASP in cells transfected with a Myc-tagged ASP-expressing proviral DNA construct upon autophagy inhibition. Finally, we show that HIV-1 replication in U937 cells was dependent on wild-type (WT) ASP expression and that autophagy played a distinct role.
MATERIALS AND METHODS
Cell lines, antibodies, and chemical products.
African green monkey kidney (COS-7), human embryonic kidney 293T fibroblast, and human cervical HeLaCD4-LTR/βgal cell lines were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (PAA Laboratories Inc., Toronto, Canada). The Jurkat E6.1 T and monocytic U937 cell lines were cultured in RPMI 1640 medium supplemented with 10% FBS. The anti-Myc antibody (hybridoma) was purchased from the ATCC, and the mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody was ordered from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). The anti-LC3b antibody (AP1802a) was purchased from Abgent (Brockville, Canada). The anti-Beclin 1 antibody (sc-48431) was purchased from Santa Cruz Biotechnology Inc. Enhanced chemiluminescence sheep anti-mouse IgG and anti-rabbit IgG antibodies coupled to horseradish peroxidase were obtained from GE Healthcare Inc. (Buckinghamshire, United Kingdom), and the goat anti-mouse IgG antibody coupled to the Alexa fluor 488 (A11001) was ordered from Invitrogen Canada Inc. (Burlington, Canada). Anti-HIV-1 ASP polyclonal antibodies were produced by immunizing rabbits with a synthetic peptide coupled to keyhole limpet hemocyanin. The selected peptide corresponds to an hydrophilic region of ASP, PAAPKNPRNKAPIPT (residues 47 to 61) (Fig. 1) (Eurogentec, Seraing, Belgium). Autophagy inhibitors 3-methyladenine (3-MA), chloroquine, and bafilomycin A1 (Baf A1) were all obtained from Sigma-Aldrich (Toronto, Canada).
Fig 1.
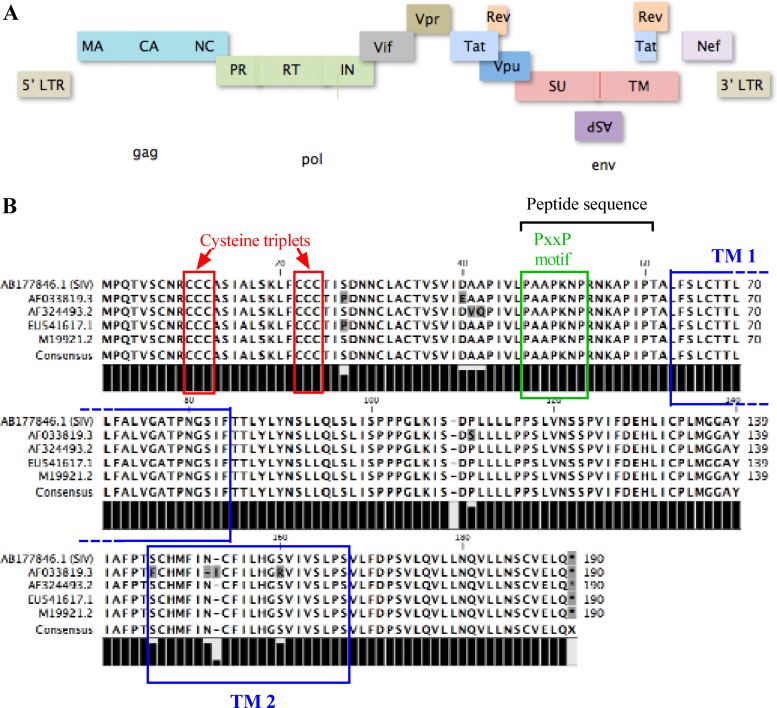
ASP ORF is localized on the minus strand of HIV-1 proviral DNA and is highly conserved between different HIV-1 strains. (A) Schematic localization of the ASP ORF in the proviral DNA of HIV-1 and position of other known HIV-1 genes. (B) Amino acid sequences of ASP from different strains (AP033819.3, AF324493.2, CUS41617.1, and M19921.2) were aligned using WorkBench 4.0. Potential transmembrane domains (TM) are indicated by blue boxes, while cysteine triplets and the PXXP motif are highlighted by red and green boxes, respectively. The position of the peptide sequence used for the production of the polyclonal anti-HIV-1 ASP antibody is indicated with a black bracket.
Purification of recombinant ASP.
The ASP cDNA was amplified by PCR with the addition of a Myc tag and a 6-His tag at the NH2 and COOH extremities, respectively, and then cloned into the pFastBac vector. DH10Bac transformation with this construct generated a recombinant bacmid, which was then transfected in Sf9 cells to generate high concentrations of recombinant baculoviruses expressing ASP, as previously described (51). Recombinant ASP was produced from baculovirus-infected Sf9 cells using the BAC-TO-BAC expression system (Invitrogen). Proteins were purified from 1-liter spinner flasks of Sf9 cells (1 × 106 per ml) infected with ASP-expressing baculoviruses (multiplicity of infection, ∼10) for 3 days at 27°C. Cells were harvested, frozen in dry ice, and stored at −20°C. The cell paste was resuspended in 80 ml of phosphate (P) buffer (50 mM NaPO4, pH 7.0, 0.5 M NaCl, 10% glycerol, 0.02% Triton X-100, 0.02% β-mercaptoethanol) containing 5 mM imidazole and protease inhibitors. The suspension was lysed using a Dounce homogenizer (10 strokes), sonicated, and then homogenized a second time. Insoluble material was removed by centrifugation at 30,000 rpm for 1 h in a T647.5 rotor. The supernatant was loaded on a 5-ml Talon column (BD Bioscience, Mississauga, Canada) and washed stepwise with P buffer containing 20 and 30 mM imidazole. ASP was then eluted with a 40-ml linear gradient of 0.05 to 0.5 M imidazole in P buffer. Carefully selected fractions were dialyzed for 90 min against storage buffer (20 mM Tris-Cl, pH 7.5, 200 mM NaCl, 10% glycerol, 1 mM dithiothreitol [DTT]) and stored in aliquots at −80°C (51).
Plasmids and transfection.
For detection in mammalian cells, the ASP ORF sequence was PCR amplified from pNL4.3 proviral DNA using the primers 5′-AT GAA TTC ATG GAA GAG CAG AAG CTG ATC AGC GAG GAG GAC CTG ATG CCC CAG ACT GTG AGT TGC AAC-3′ (forward) and 5′-AT GGT ACC CTA CTG TAA TTC AAC ACA ACT GTT TAA TAG TAC-3′ (reverse) with a Myc tag added at the N-terminal end (boldface). The amplified DNA was next cloned into the pcDNA3.1 expression vector under the cytomegalovirus (CMV) promoter. For codon optimization of the ASP ORF, an optimized cDNA sequence with a Myc tag at the N-terminal end was generated by Genscript Corp. (Piscataway, NJ) and equally cloned into pcDNA3.1 following digestion with EcoRI and KpnI. As an additional construct, the optimized ASP was also PCR amplified from the Myc-tagged optimized ASP construct using the primers 5′-ATG AAT TCA TGT GTT GTC CCG GCT GTT GTC CCC AGA CCG GTA-3′ (forward) and 5′-ATG GTA CCT CAC TGC AGC TCC AC-3′ (reverse) with a tetracysteine (TC) tag added the N-terminal end (boldface). The amplicon was cloned into pcDNA3.1 as described above. The pGFP-optimized ASP expression vector was generated by cloning the optimized ASP cDNA in the EcoRI/KpnI-digested peGFP-C1 vector downstream and in frame with the green fluorescent protein (GFP) ORF. The NL4.3Δ5′LTR ASP-Myc construct was derived from the pNL4.3Δ5′LTR ASP-Flag construct previously described (33). The inserted Flag tag at the COOH end of the ASP sequence was removed following NcoI/XbaI digestion of the ASP-containing NheI/BamHI fragment cloned into the pGL3basic vector and replaced by the Myc tag. The resulting NheI/BamHI fragment was cloned back into the pNL4.3Δ5′LTR vector. The ASP-mutated pNL4.3Δ5′LTR-ASP67stop-Myc proviral DNA was also generated from a Flag-tagged version of an ASP-mutated NL4.3 proviral DNA, which has also been previously described (33) and contains a stop codon at codon 67 (TGC to TGA). The GFP-LC3 expression vector (24) (plasmid 22405; Addgene) was generated by Jayanta Debnath and provided by Addgene (Cambridge, MA). The vector expresses a GFP-LC3 chimeric protein. The pRcActin-LacZ plasmid expressed the β-galactosidase gene under the control of the β-actin promoter. COS-7 and 293T cells were transfected using Lipofectamine 2000 (Invitrogen) according to recommendations of the manufacturers. Briefly, 1 day before transfection, 293T or COS-7 cells (5 × 105 cells per well) were plated. Cells were transfected with 5 μg DNA in the presence of 10 μl of Lipofectamine 2000. Jurkat cells (1.5 × 105) were microporated with the MP-100 device (a single pulse of 1,350 V for 30 ms; Digital Bio, Montreal, Canada) with 1 μg of expression vectors. U937 cells (1 × 106) were transfected with 2 μg plasmids with the Nucleofector II apparatus according to the manufacturer's recommendations (Amaxa Biosystems). For certain experiments, transfected COS-7 and U937 cells were treated with 3-methyladenine (10 and 2.5 mM, respectively) for 4 to 24 h or with bafilomycin A1 (100 nM) for 2 h.
Preparation of cellular extracts.
Transfected cells were washed in phosphate-buffered saline (PBS), and proteins were isolated in different lysis buffers: buffer A (50 mM Tris-HCl, 150 mM NaCl, 0.5% NP-40, and protease inhibitor cocktail in tablets), radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS). In addition, hydrophobic cell fractions were also prepared using the Mem-PER Eukaryotic Membrane Protein Extraction kit (Thermo Fisher Scientific, Rochester NY) according to the manufacturer's recommendations. Protein concentrations were determined with the bicinchoninic acid (BCA) protein assay (Thermo Fisher Scientific Inc.).
Immunoprecipitation experiments and Western blot analyses.
Dynabeads protein G (Invitrogen) were used according to the manufacturer's recommendations. Briefly, 50 μl of Dynabeads protein G was incubated with 5 μg of antibodies. Cellular extracts were then incubated for 2 h with antibody-coupled Dynabeads. Protein-antibody-bead complexes were washed and eluted in denaturing conditions using NuPAGE LDS sample buffer (Invitrogen) and NuPAGE sample reducing agent (Invitrogen) according to the manufacturer's instructions. Cellular extracts and immunoprecipitated samples were migrated on a 12% SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane. Membranes were next blocked in 5% milk and incubated with an anti-Myc antibody (1/250) in the presence of Signal Boost (Millipore, CA) and with an anti-LC3b (1/500), anti-Beclin 1 (1/500), or anti-GAPDH (1/1000) antibody. Membranes were further incubated with a goat anti-mouse IgG antibody conjugated with horseradish peroxidase (1/5,000) or with a similarly coupled goat anti-rabbit IgG antibody (1/10,000). Signals were detected with the BM Chemiluminescence Blotting Substrate kit (Roche Diagnostics), and membranes were subsequently analyzed with Fusion FX7 (Vilber Lourmat, France).
Confocal microscopy.
COS-7 and 293T cells were seeded in 24-well plates containing a 1.5-mm-thick coverslip for 24 h and then transfected with pcDNA3.1 (as a control) and expression vectors for nonoptimized or optimized Myc-tagged ASP cDNA. At 48 h posttransfection, cells were washed with PBS, fixed with 4% formaldehyde for 10 min, and permeabilized with 0.1% Triton X-100 for 5 min at room temperature. Cells were then washed three times with PBS and incubated with the anti-Myc antibody (dilution, 1:250) overnight at 4°C. Cells were washed three times with PBS and incubated with goat anti-mouse IgG coupled to Alexa fluor 488 for 1 h at room temperature. Cells were washed twice with PBS and incubated in a 2.5 μg/ml propidium iodide (PI) solution. Coverslips were then mounted in a drop of ProLong Antifade (Invitrogen). Jurkat and U937 cells were transfected with pcDNA3.1, peGFP, pGFP-optimized ASP, or the TC-tagged optimized ASP expression vector. U937 cells were then incubated with phorbol myristate acetate (PMA; 50 ng/ml) for 18 h prior to analyses. At 40 h posttransfection, cells were either directly observed for GFP fluorescence or labeled with the TC-FlAsH II In-Cell Tetracysteine Tag Detection kit (Invitrogen) for TC-tagged ASP expression vectors. Briefly, cells were washed twice with RPMI 1640 and incubated with 0.5 μM FIAsH solution for 5 min at 37°C. Cells were washed in BAL wash buffer and in RPMI 1640 as a final wash. For TC detection, Jurkat cells were initially plated on coverslips coated with poly-l-lysine before labeling, as previously described by Clerc et al. (33). For colocalization analysis with LC3, cells were first cotransfected with GFP-LC3 (Addgene, Cambridge, MA) and an ASP expression vector and then incubated with the anti-Myc antibody (dilution, 1:250), followed by the goat anti-mouse IgG antibody coupled to Alexa fluor 568, as indicated above. Cells were analyzed at 48 h posttransfection. All cell samples were visualized with an MRC1024 laser-scanning confocal microscope (Bio-Rad, Hercules, CA) at room temperature using a 60× objective under oil immersion and with a numerical aperture of 1.4 or with a Nikon A1 laser-scanning confocal microscope (Nikon Canada, Mississauga, Canada).
Flow cytometry analyses.
Transfected COS-7, 293T, and U937 cells were washed with PBS, fixed with 4% formaldehyde for 10 min, and permeabilized with 0.1% Triton X-100 for 5 min at room temperature. Cells were then washed three times with PBS and incubated with the anti-Myc antibody (dilution, 1:250) overnight at 4°C. After three additional washes with PBS, cells were incubated with goat anti-mouse IgG antibodies coupled to Alexa fluor 488 for 1 h at 4°C. Cells were fixed with 1% formaldehyde and incubated overnight at 4°C before analysis with the FACScan device (BD Biosciences).
Multimerization assay.
Purified ASP (5 μg) was incubated with increasing concentrations of glutaraldehyde (from 0.005 to 1%) during 30 min at room temperature. As a second series of conditions, purified ASP was incubated with 0.05% glutaraldehyde for 2, 5, 10, 20, and 30 min at room temperature. All reactions were stopped by adding 1 M Tris-HCl, pH 8.0. ASP complexes were analyzed by migration on 12% SDS-PAGE and Western blot detection with anti-Myc antibodies.
Production of pseudotyped virions for infection.
Pseudotyped HIV-1 virions were produced as previously described (27, 33). 293T cells were plated 16 h before transfection and then transfected with WT or ASP67stop-mutated pNL4.3-env− proviral DNA (30 μg) and the vesicular stomatitis virus G envelope protein (VSVg) expression vector (15 μg) using the Lipofectamine 2000 reagent according to the manufacturer's instructions. Twenty-four hours after transfection, culture media were renewed. After 48 h of transfection, supernatants were harvested and filtered through a 0.45-μm-pore size cellulose acetate membrane. Virus stocks then were aliquoted and frozen at −80°C. All virus stocks underwent a single freeze-thaw cycle before use in infection studies. Virus stock titers were determined by a p24 enzyme-linked immunosorbent assay (ELISA) as previously described (27).
Detection of viral p24 capsid protein in transfected and HIV-1-infected cells.
293T, HeLaCD4-LTR/βgal, U937, and Jurkat cells were cotransfected with pcDNA3.1, pNL4.3-ASP WT, or pNL4.3-ASP67stop and pRcActin-lacZ (for normalization of transfection experiments). Treatment by 3-MA was performed for 4 h prior to analyses. At 48 h posttransfection, supernatants were harvested and levels of the HIV-1 p24 protein determined by a p24 ELISA, as previously described (52). For infection experiments, U937 cells were induced to differentiate for 18 h in PMA. Fresh medium was added, and after 48 h, 100 ng of p24 of VSVg-pseudotyped NL4.3-ASP WT or NL4.3-ASP67stop was incubated with 1 × 106 cells for 5 h. Cells were washed twice with 1× PBS and cultured for 6 days. At days 3 and 6, 3-MA treatment (2.5 mM) was performed for 4 h prior to analyses. Supernatants were harvested and p24 levels were determined with a p24 ELISA.
Statistical analysis.
Experiments were performed in triplicate and independently conducted two to three times. For flow cytometry experiments and counts of ASP-positive cells or GFP-LC3+ puncta in confocal microscopy experiments, results are expressed as means ± standard deviations (SD) and statistically analyzed using GraphPad Prism software through a one-way analysis of variance (ANOVA) followed by a Tukey test (P < 0.05 was considered significant). Densitometry analyses were performed in certain Western blot experiments and statistically analyzed using GraphPad Prism software through a one-way ANOVA followed by a Tukey test (P < 0.05 was considered significant). Graphs represent the results for one experiment, which is representative of repeated experiments.
RESULTS
Detection of ASP in Sf9 cells.
Our previous study had demonstrated that antisense transcripts initiating at the border of the 3′ LTR and terminating in the pol region was produced as an unspliced form and therefore could mediate synthesis of ASP (27). The ASP ORF is located in the complementary segment of the env gene encompassing the cleavage site needed for the generation of the two Env subunits, gp120 and gp41, and the rev-responsive element (RRE), an RNA stem-loop structure responsible for shuttling of unspliced and singly spliced HIV-1 sense transcripts (Fig. 1A). The predicted amino acid sequence is highly conserved among HIV-1 isolates and is also conserved in certain simian immunodeficiency virus (SIV) genomes (Fig. 1B). Interesting features of this predicted protein include a cysteine-rich region, potential transmembrane domains (which could be membrane-associated), and a PXXP motif.
Given that detection of ASP in mammalian cells remained problematic, we first opted for the frequently used insect Sf9 cell line for overexpression studies. Sf9 cells were infected with Myc/His-tagged ASP-expressing baculoviruses and analyzed at 24 and 48 h postinfection. Confocal microscopy and flow cytometry analyses using an anti-Myc antibody led us to first confirm expression of the tagged ASP in infected cells (data not shown). Confocal microscopy presented a strong signal at the cellular periphery and in the cytoplasmic region, which was excluded from the nuclear region. Staining of infected cells with the CellMask membrane marker suggested that ASP could localize to the plasma membrane (data not shown). Importantly, no similar Myc-specific signals were obtained in uninfected Sf9 cells or cells infected with baculoviruses expressing His-tagged MRE11, an unrelated protein. Western blot analyses were next performed using total and soluble fractions. Importantly, two major signals were observed at 48 h postinfection, demonstrating the monomeric form in addition to a high-molecular-mass form (data not shown). At later time points (72 and 96 h), a variety of signals was observed and again suggested the presence of multimers.
These results indicated that ASP could indeed be detected in eukaryotic cells and presented the first successful Western blot detection of ASP in a eukaryotic cell. Furthermore, our results also argued for a heterogeneous distribution of ASP, from the plasma membrane to the cytoplasm.
Detection of ASP in mammalian cells following codon optimization of its cDNA.
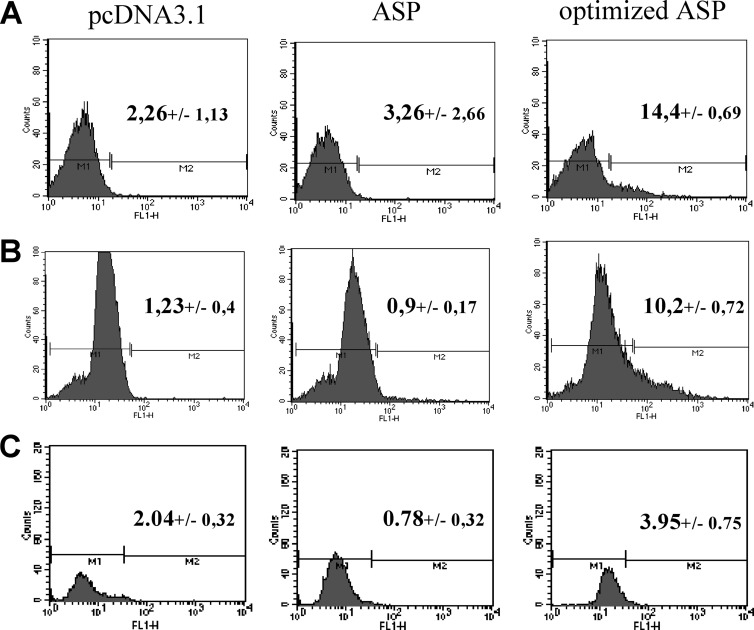
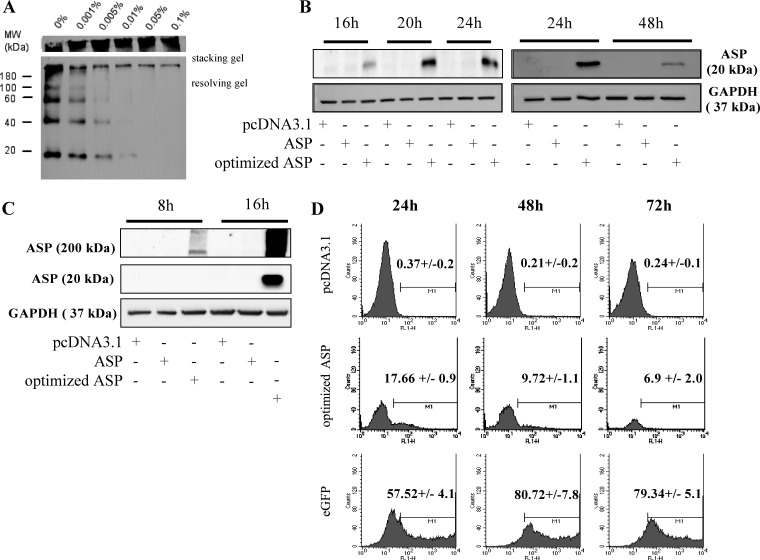
We next tested various mammalian cell lines for ASP detection upon expression of Myc-tagged ASP. Our initial results demonstrated that a very limited number of cells were positive in transfected COS-7 cells, and that ASP detection was not possible by either Western blot or flow cytometry analysis. We opted for an optimization of codons of the ASP cDNA, an approach which had also been previously undertaken for other HIV-1 viral genes (53, 54). To determine if codon optimization of the ASP cDNA could improve protein expression, nonoptimized ASP and optimized ASP expression vectors were transfected in COS-7, and cells were analyzed by flow cytometry (Fig. 2A). In cells transfected with the optimized ASP expression vector, a significant percentage of cells was positive (14.4 versus 2.26% for pcDNA3.1-transfected cells), while positive cells in nonoptimized transfected samples were modest. Similar results were obtained from 293T cells (Fig. 2B). In U937 cells, 3.95% of ASP-positive cells were detected (Fig. 2C). When Jurkat T cells were transfected with ASP-expressing vectors, the percentage of ASP-positive cells was low but reproducible (1.5% for the optimized ASP-expressing cells) (data not shown).
Fig 2.
Codon optimization of the ASP cDNA increases the number of positive transfected cells in different mammalian cell lines. COS-7 (A), 293T (B), and U937 (C) cells were transiently transfected with pcDNA3.1 or with nonoptimized (ASP) or optimized Myc-tagged ASP expression vectors. At 48 h posttransfection, cells were fixed and permeabilized with formaldehyde and Triton X-100, as described in Materials and Methods. Cells were analyzed by flow cytometry using a FACScan with an anti-Myc antibody, followed by the addition of a goat anti-mouse IgG antibody coupled to Alexa fluor 488. Data were acquired with CellQuest software. Percentages represent the averages ± SD from three independent experiments.
Taken together, these results demonstrated that ASP could be detected at low levels in different mammalian cell lines in overexpression conditions, and that codon optimization of its cDNA significantly improved its detection.
Localization of ASP in mammalian cells.
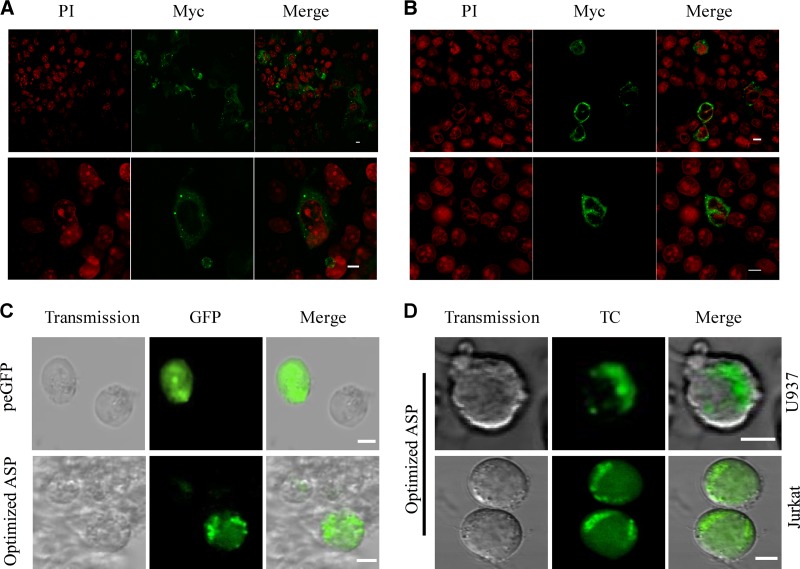
Since the optimized ASP-expressing vector provided us with an improved signal in tested cell lines, this cDNA version was used to study the localization of ASP in mammalian cell lines. COS-7 and 293T cells were transfected with the optimized Myc-tagged ASP expression vector. After 48 h, ASP was detected with an anti-Myc antibody, while nuclei were concomitantly stained with propidium iodide. Confocal microscopy analyses revealed a major cytoplasmic distribution of ASP in these two cells lines, with a partial localization at the plasma membrane (Fig. 3A and B). Localization of ASP was also determined in a monocytic and a T cell line. To improve detection of ASP in these cell lines showing lower transfection efficiency, optimized ASP cDNAs were fused with either a tetracysteine (TC) tag or GFP. Transfected U937 and Jurkat cells again presented similar cytoplasmic punctate patterns in the majority of positive cells in these two cell lines, regardless of the nature of the fusion (Fig. 3C and D). Interestingly, in both cell types a polarized signal was apparent, further confirming our previous results (31, 33), although we were not capable of discerning whether part of the signal was plasma membrane related.
Fig 3.
ASP is mainly distributed in the cytoplasm of positive cells in a punctate manner. COS-7 (A) and 293T (B) cells were transiently transfected with pcDNA3.1 or with the optimized Myc-tagged ASP expression vector. At 48 h posttransfection, cells were permeabilized and stained as described in Materials and Methods. Nuclei were stained with propidium iodide (PI), and ASP was detected by an anti-Myc antibody followed by goat anti-mouse IgG coupled to Alexa fluor 488. Merged signals from ASP (green) and nuclei (red) are also presented. Cells were observed with a Bio-Rad laser-scanning confocal microscope (MRC-1024ES) using a 60× objective under oil immersion and with a numerical aperture of 1.4. LaserSharp software was used for image acquisition. Scale bars, 10 μm. (C) U937 cells were transiently transfected with pcDNA3.1, peGFP-N1, or the GFP-optimized ASP expression vector. At 24 h posttransfection, cells were treated with 50 ng/ml PMA. After 18 h of treatment, cells were observed using the Nikon A1 laser-scanning confocal microscope using a 20× objective and with a numerical aperture of 1.4. The NIS software was used for image acquisition. (D) U937 cells (upper) and Jurkat cells (lower) were transiently transfected with the TC-optimized ASP expression vector. After 48 h of transfection, Jurkat cells were plated on a coverslip coated with poly-l-lysine. Jurkat cells were then labeled using the TC detection kit, while U937 cells were directly labeled in suspension before being centrifuged on a coverslip. Cells were visualized with the Bio-Rad laser scanning confocal microscope (MRC-1024ES) using a 60× objective under oil immersion and with a numerical aperture of 1.4. LaserSharp software was used for image acquisition. Scale bars, 5 μm.
These results demonstrated that cellular localization of ASP in various mammalian cell lines showed important similarities and highlighted a punctate cytoplasmic distribution.
Western blot detection of ASP in mammalian cells.
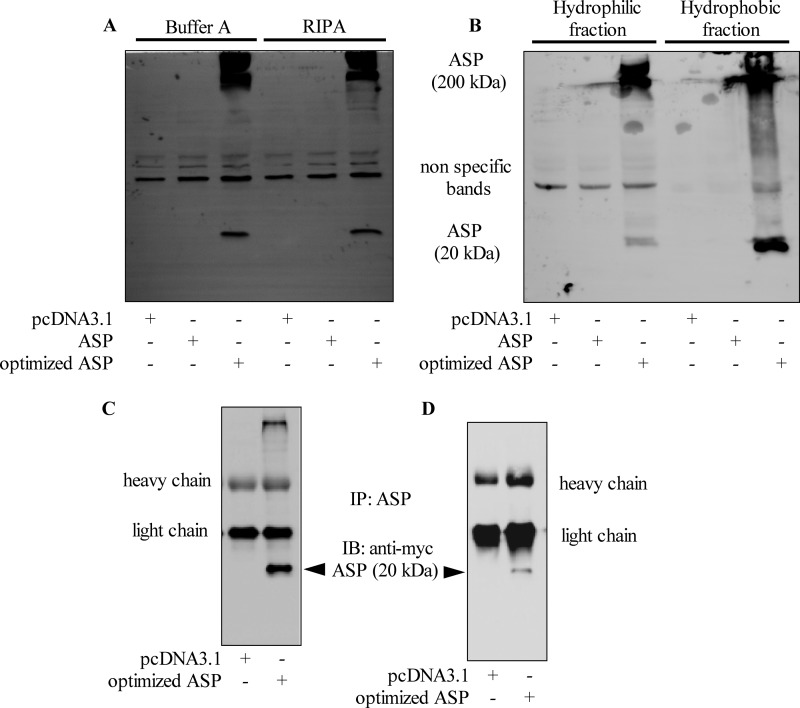
Western blot approaches were undertaken to confirm that, indeed, ASP could be detected in mammalian cell lines. Although Western blot analysis of ASP expression has been difficult in previous studies (32, 33), we reasoned that increased levels of expression obtained with our codon-optimized ASP expression vector, as observed by flow cytometry and confocal microscopy, could lead to its detection by Western blotting. Different buffers were tested in parallel for the preparation of total extracts from COS-7 cells transfected with nonoptimized and optimized ASP expression vectors. Knowing that ASP is predicted to be highly hydrophobic, extracts enriched for membrane proteins were also tested. For all analyzed extracts, a signal at the expected 20-kDa molecular mass was detected solely in lysates of cells transfected with the optimized Myc-tagged ASP expression vector (Fig. 4A). In addition, the signal was more pronounced in extracts enriched with membrane proteins than residual extracts, thereby suggesting a membrane-associated characteristic (Fig. 4B). These analyses further revealed the abundant presence of high-molecular-mass bands in transfected mammalian cells, possibly indicating a multimerized form of ASP, as indicated in similar ASP detection analyses in infected insect cells (data not shown).
Fig 4.
Detection of ASP by Western blotting. (A and B) COS-7 cells were transfected with pcDNA3.1 or expression vectors for nonoptimized or optimized Myc-tagged ASP. (A) At 48 h posttransfection, cellular extracts were prepared using Buffer A or the RIPA buffer. (B) Cellular extracts were isolated as hydrophobic or hydrophilic fractions. Western blot analyses were performed using an anti-Myc antibody. (C and D) COS-7 (C) and 293T (D) cells were transfected with pcDNA3.1 or the Myc-tagged optimized ASP expression vector. After 48 h of transfection, cellular extracts were prepared and immunoprecipitation was performed with the anti-ASP antibody prior to Western blot analyses with anti-Myc antibodies. Signals corresponding to the heavy and light immunoglobulin chains are indicated next to each panel. IP, immunoprecipitation; IB, immunoblot.
In order to more convincingly detect ASP, an anti-ASP antibody was generated against a single peptide located in a rare hydrophilic region of the protein (Fig. 1). Initial Western blot analyses using this antibody revealed that numerous nonspecific signals were detected in addition to the expected 20-kDa band (data not shown). Thus, we opted to use it in immunoprecipitation experiments. Hence, cellular extracts from COS-7 and 293T cells transfected with expression vectors for nonoptimized or optimized ASP were immunoprecipitated with our anti-ASP antibody, and signals were subsequently revealed with anti-Myc antibodies. Again, a specific 20-kDa band was detected only when COS-7 and 293T cells were transfected with the optimized ASP cDNA vector (Fig. 4C and D). In addition, the high-molecular-mass signal was present in immunoprecipitated complexes, confirming that ASP seemed to multimerize in mammalian cells.
These results showed that ASP could be detected by Western blotting in mammalian cells transfected with an expression vector for a codon-optimized ASP cDNA, and that multimerization of ASP was again suggested under these conditions.
ASP can multimerize in vitro and is unstable in mammalian cells.
The Western blot analyses described above suggested that ASP could multimerize at a very high molecular mass. To confirm that ASP could form aggregates, a multimerization assay was conducted in the presence of glutaraldehyde. ASP purified from the baculovirus-infected Sf9 cells described above was used in this assay. In the absence of glutaraldehyde, based on the estimated resulting molecular mass, ASP formed monomers, dimers, trimers, tetramers, and complexes of higher molecular mass (Fig. 5A). In the presence of increased concentrations of glutaraldehyde, low-molecular-mass complexes were gradually less abundant, while high-molecular-mass multimers located in the stacking gel increased in intensity (Fig. 5A). In a time kinetic experiment, we observed that high-molecular-mass multimers augmented very rapidly and in abundance (within 10 min following the addition of glutaraldehyde), confirming the high potential for multimerization of this protein (data not shown).
Fig 5.
ASP multimerizes in vitro and is unstable in mammalian cells. (A) Purified ASP isolated from baculovirus-infected Sf9 cells was incubated with different concentrations of glutaraldehyde for 30 min and then analyzed by Western blotting with an anti-Myc antibody. The molecular mass marker is indicated on the left side of the panel. (B and C) COS-7 cells were transfected with pcDNA3.1 or with nonoptimized or optimized Myc-tagged ASP expression vectors. From 8 to 48 h posttransfection, cellular extracts were prepared using the RIPA buffer and analyzed by Western blotting using an anti-Myc antibody. In panel C, the high-molecular-mass signal was also compared between 8 and 16 h posttransfection. In these latter experiments, GAPDH was used as a loading control. (D) COS-7 cells were transiently transfected with pcDNA3.1, peGFP-N1, or the optimized Myc-tagged ASP expression vector. At 24, 48, and 72 h posttransfection, cells were fixed and permeabilized with formaldehyde and Triton X-100, as described in Materials and Methods. Cells were analyzed by flow cytometry using a FACScan with an anti-Myc antibody, followed by the addition of a goat anti-mouse IgG antibody coupled to Alexa fluor 488 (for cells transfected with pcDNA3.1 or with the optimized Myc-tagged ASP expression vector). Data were acquired with CellQuest software. Values presented in each flow cytometry analysis represent percentage of positive cells compared to similarly analyzed pcDNA3.1-transfected cells. MW, molecular mass (in kilodaltons).
Based on these results, we thus reasoned that these high-molecular-mass signals, which were equally detected in insect and mammalian cells, render ASP unstable in expressing cells. In order to understand why ASP is poorly detected in mammalian cells, expression of codon-optimized ASP was monitored in transfected COS-7 cells in a time kinetic experiment. Cells transfected with codon-optimized ASP expression vectors presented an increasing signal up to 24 h, which quickly diminished and became faint at 48 h posttransfection (Fig. 5B). A concomitant increase in high-molecular-mass complexes was also apparent in transfected COS-7 cells (Fig. 5C). Data from these time kinetic experiments were also confirmed in COS-7 cells transfected with the codon-optimized ASP expression vector and analyzed by flow cytometry (Fig. 5D). A significant decrease in the percentage of ASP-positive cells was observed (from 17.66% at 24 h posttransfection to 9.72% at 48 h). However, no comparable decrease in signals was noted in cells expressing only enhanced GFP.
These results showed that ASP had a high potential for multimerization. Furthermore, codon-optimized ASP was detected only at early time points following transfection of COS-7 cells and was likely unstable. This might involve the formation of the multimer complexes.
Autophagy is activated when ASP is expressed in mammalian cells.
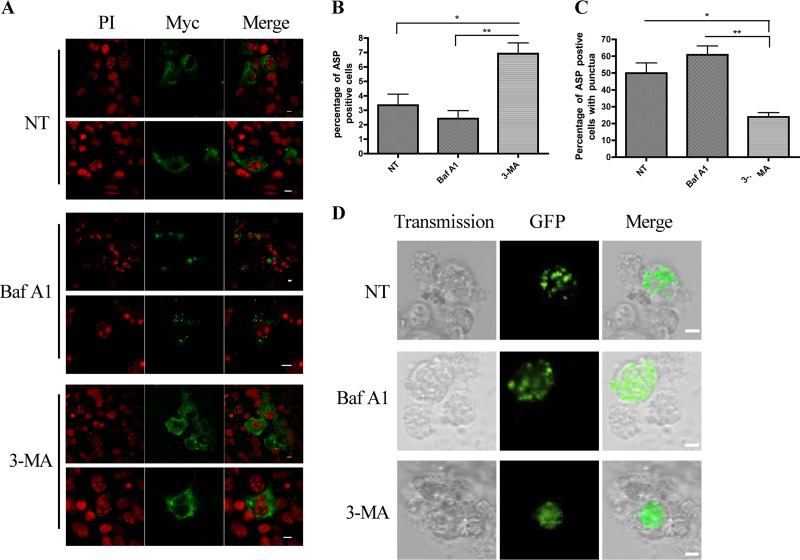
Since our data argued that ASP could multimerize in expressing cells, we reasoned that these aggregates could contribute to the instability of the protein. We first tested the proteasome inhibitor MG132 in transfected cells and found no improvement in ASP detection (data not shown). As autophagy is a cellular mechanism linked to the degradation of aggregated proteins, we next investigated its implication in ASP instability. Autophagy was first inhibited with 3-MA, an inhibitor blocking the early step of the formation of the phagophore (55). COS-7 cells were transfected with the expression vector of codon-optimized ASP and subsequently treated with 3-MA (Fig. 6). Confocal microscopy revealed that 3-MA treatment indeed led to an improved detection of ASP in terms of both intensity and number of positive cells (Fig. 6A). Importantly, the punctate nature of the ASP signal was greatly reduced in 3-MA-treated cells compared to control cells, while the percentage of ASP-positive cells significantly increased when cell counts were performed (Fig. 6B and C). The inhibitor of late-stage autophagy, bafilomycin A1 (56), was also tested, and upon analyses by confocal microscopy, cells treated with this inhibitor did show an increase in the number of punctate-positive cells, although the increase did not show statistical significance (Fig. 6B and C). Similar analyses were conducted in GFP-optASP-expressing U937 cells (Fig. 6D) and Myc-tagged optimized ASP-expressing 293T cells (data not shown), which revealed again a loss of punctate signals with a concomitant increase in the number of ASP-positive cells upon 3-MA treatment in addition to an increase in the number of punctate ASP-positive cells in bafilomycin A1-treated cells. An increase in the punctate nature of the ASP signal following late-step inhibition of autophagy was further demonstrated in COS-7 cells using another similar inhibitor, chloroquine (data not shown).
Fig 6.
Induction of autophagy following ASP expression in COS-7 cells. (A) COS-7 cells were transfected with pcDNA3.1 or the optimized Myc-tagged ASP expression vector. Cells were then left untreated (NT) or were treated with 3-MA (10 mM) for 24 h or Baf A1 (100 nM) for 2 h. At 36 h posttransfection, cells were stained with PI (red) and incubated in the presence of anti-Myc antibodies, followed by goat anti-mouse IgG coupled to Alexa fluor 488. Cells were then observed by laser-scanning confocal microscopy using a 60× objective under oil immersion and with a numerical aperture of 1.4. The LaserSharp software was used for image acquisition. Merged signals are also presented on the right. Scale bars, 10 μm. (B and C) Untreated or treated transfected cells were counted in terms of number of positive cells (B) or of positive cells with punctate signals (C). Three images were used per condition, and values are expressed as percentages ± SD from the total cell number evaluated as averages from triplicates and two independent experiments. *, P < 0.05; **, P < 0.01. (D) U937 cells were transfected with pcDNA3.1 or the GFP-optimized ASP expression vector. At 24 h posttransfection, cells were plated on coverslips in the presence of 50 ng/ml PMA for 18 h. Cells were then left untreated (NT) or were treated with 3-MA (2.5 mM) for 4 h or Baf A1 (100 nM) for 2 h and visualized with the Nikon A1 laser-scanning confocal microscope using a 20× objective and with a numerical aperture of 1.4. The NIS software was used for image acquisition. Scale bars, 5 μm.
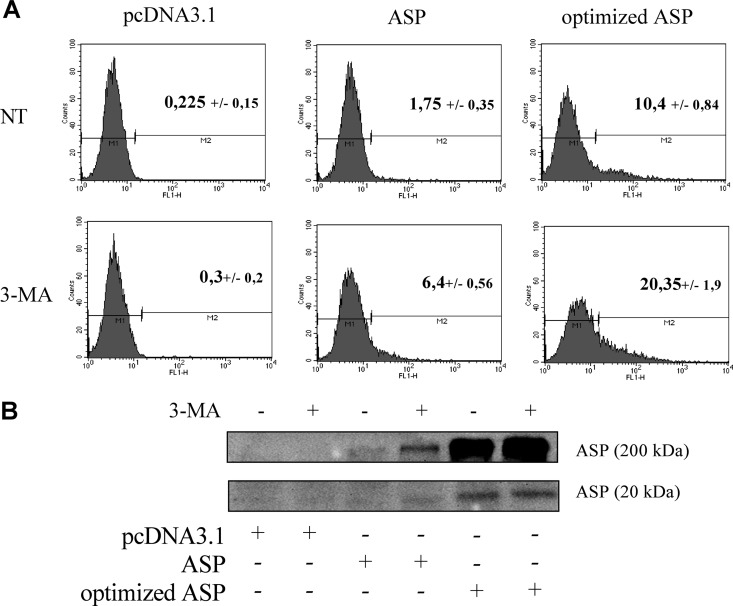
To further quantify changes in the level of ASP expression in 3-MA-treated COS-7 cells, flow cytometry analyses were performed and revealed a 2- to 4-fold increase in the percentage of non-codon-optimized or codon-optimized ASP-positive cells (Fig. 7A). Similar results were observed in transfected 293T cells following flow cytometry analyses (data not shown). Western blot analyses confirmed the detection of monomeric ASP in both untreated and 3-MA-treated COS-7 cells, although no clear increase was noted; on the other hand, 3-MA treatment led to the detection of the nonoptimized form of ASP, albeit at a low level (Fig. 7B). However, an increase in ASP levels following 3-MA treatment was more prominently observed with respect to high-molecular-mass signals.
Fig 7.
Inhibition of autophagy increases the number of ASP-positive cells. (A) COS-7 cells were transfected with pcDNA3.1 or with nonoptimized or optimized Myc-tagged ASP expression vectors. At 16 h posttransfection, cells were left untreated (NT) or were treated with 3-MA (10 mM) during 24 h. Cells were then analyzed by flow cytometry using an anti-Myc antibody followed by a goat anti-mouse IgG antibody coupled to Alexa fluor 488. Data were acquired with CellQuest software. Percentages represent the averages ± SD from three independent experiments. (B) COS-7 cells were transfected with pcDNA3.1 or with nonoptimized ASP or optimized Myc-tagged ASP expression vectors. At 16 h posttransfection, cells were treated with 3-MA (10 mM), and after 24 h of treatment, cells were analyzed by Western blotting using an anti-Myc antibody. High-molecular-mass signals are presented in the top panel while the monomeric ASP form is shown in the bottom panel.
Taken together, these results indicated that ASP was likely degraded by autophagy, which contributed to strongly reducing the stability of this viral protein in mammalian cells.
ASP expression leads to increased levels of the LC3-II isoform.
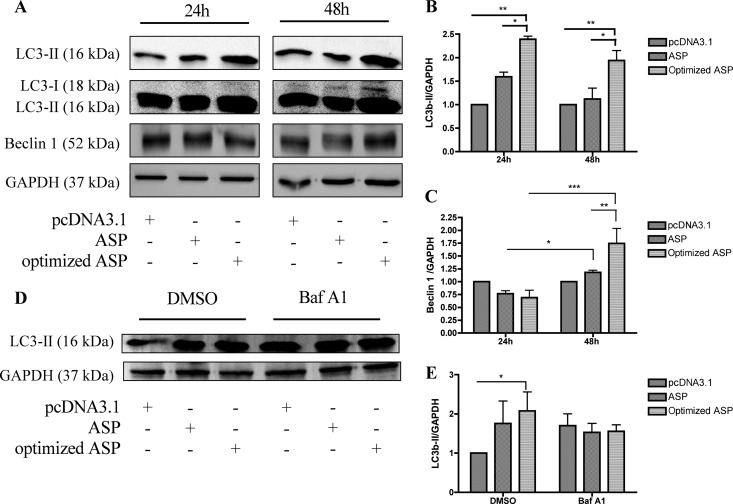
Autophagy has recently been associated with HIV-1 replication in macrophages but has also been suggested to contribute to CD4+ T cell death (43, 46, 47, 50, 57). We next wanted to determine if ASP could contribute to HIV-1-induced autophagy by further addressing its induction potential. Expression levels of two autophagy markers, LC3b-II and Beclin 1, was assessed in COS-7 cells transfected with either non-codon-optimized or codon-optimized ASP expression vectors. Western blot analyses indicated an increase in both LC3b-II and Beclin 1 upon transfection of either ASP expression vector (Fig. 8A), which is better illustrated following densitometry analyses (Fig. 8B and C). Upon high exposure of the Western blot, the LC3b-I isoform, commonly weak in abundance in COS-7 cells, was also increased in intensity upon ASP expression, an observation previously reported for other induced autophagic pathways (58–60). To distinguish between an increase in the basal autophagic flux versus induction of the autophagic pathway, COS-7 cells were transfected with non-codon-optimized or codon-optimized ASP expression vectors, followed by treatment with bafilomycin A1. As expected, an increase in the abundance of the LC3b-II form was noted when ASP was expressed, and this was confirmed by densitometry analyses (Fig. 8D and E). However, when autophagy was inhibited by Baf A1 in optimized ASP-expressing cells, LC3b-II levels were poorly induced, thereby suggesting that ASP itself induced autophagy and does not act solely by modulating basal autophagy.
Fig 8.
Increase in expression levels of autophagic markers LC3b-II and Beclin 1 upon ASP expression. (A) COS-7 cells were transfected with pcDNA3.1 or with nonoptimized or optimized Myc-tagged ASP expression vectors. At 24 and 48 h posttransfection, cellular extracts were analyzed by Western blotting using anti-LC3b, anti-Beclin 1, and anti-GAPDH antibodies. A high exposure of the membrane is also presented in the second upper panels for visualization of the LC3b-I signal. (B and C) Densitometry analyses of LC3b-II and Beclin 1 expression levels were calculated and normalized to the GAPDH signal using Quantity One and GraphPad Prism software. (D) COS-7 cells were transfected with pcDNA3.1 or with nonoptimized or optimized Myc-tagged ASP expression vectors. At 34 h posttransfection, cells were left untreated (NT) or were treated with Baf A1 (100 nM) for 2 h. Cellular extracts were then prepared and analyzed by Western blotting using anti-LC3b and anti-GAPDH antibodies. The LC3b-II form is presented in the top panel. GAPDH is shown as a control in the bottom panel. (E) Densitometry analyses of LC3b-II expression levels were calculated and normalized to GAPDH signals using Quantity One and GraphPad Prism softwares. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Taken together, these results confirmed that expression of ASP leads to the induction of autophagy, and that this induction is blocked by 3-MA.
ASP interacts with LC3 and colocalizes in a punctate manner.
Given that the LC3-II marker was indeed induced upon ASP expression, it was important to determine if ASP localized in autophagosomes. Confocal microscopy analyses were performed in COS-7 cells cotransfected with codon-optimized ASP and GFP-LC3 expression vectors (38, 39). As expected, greater than 90% of cells were positive for GFP-LC3 only (Fig. 9A). In these cells, the LC3 signal presented a punctate distribution, although a more heterogeneous cytoplasmic signal was also observed. In double-positive cells, ASP and LC3 showed a significant level of colocalization, and their distribution was often shared in punctate structures. Since the GFP signal is sensitive to low pH, these structures were identified as being autophagosomes as opposed to more acidic autolysosomes (59, 61, 62). When autophagy was inhibited at late stages by Baf A1 treatment, colocalization between ASP and GFP-LC3 was more clearly noted (Fig. 9B). We also conducted coimmunoprecipitation experiments in COS-7 cells transfected with the Myc-tagged optimized ASP expression vector. Upon immunoprecipitation of ASP, LC3b-II was detected, suggesting that it interacted with ASP (Fig. 9C).
Fig 9.
ASP-specific punctate signals colocalize and interact with LC3b. (A and B) COS-7 cells were cotransfected with optimized Myc-tagged ASP and GFP-LC3 expression vectors. Cells were either left untreated (A) or were treated with Baf A1 (100 nM) for 2 h prior to analyses (B). After 36 h of transfection, fixed cells were incubated with anti-Myc antibodies followed by a goat anti-mouse IgG antibody coupled to Alexa fluor 488. Cells were observed by laser-scanning confocal microscopy using a 60× objective under oil immersion and with a numerical aperture of 1.4. Colocalization analyses were performed using ImageJ software. Merging of ASP (red) and LC3 (green) signals is depicted on the right. Scale bars, 10 μm. (C). COS-7 cells were cotransfected with pcDNA3.1 or the optimized Myc-tagged ASP expression vector. After 36 h of transfection, cellular extracts were first immunoprecipitated with anti-ASP antibodies and then analyzed by Western blotting using anti-LC3b antibodies. Total cellular extracts were similarly analyzed (bottom panel).
These results indicated that ASP expression indeed induces autophagy, and that ASP at least partially localizes in autophagosomes along with the LC3 marker and, further, was part of a common complex.
ASP expressed from the proviral DNA induces autophagy and can be detected upon 3-MA treatment.
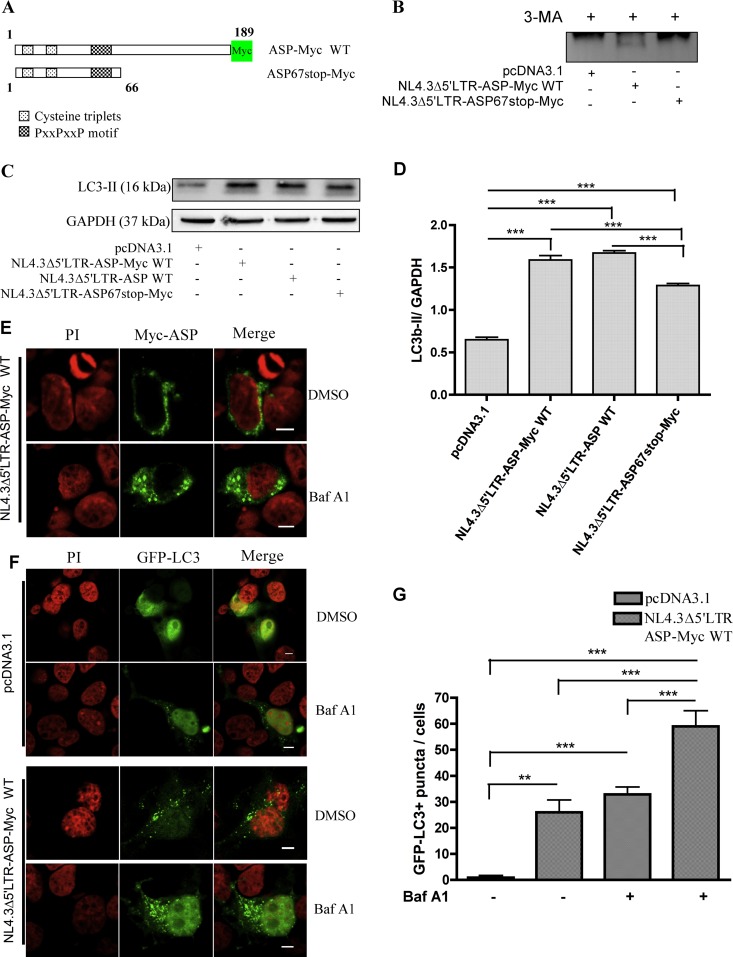
We have previously demonstrated that antisense transcription is greatly increased in HIV-1 proviral DNA deleted of its 5′ LTR (27). As autophagy inhibition by 3-MA treatment improved the detection of ASP, we reasoned that the use of such a proviral DNA clone upon autophagy inhibition would lead to ASP detection. COS-7 cells were transfected with pNL4.3Δ5′LTR-ASP-Myc WT, an NL4.3 version deleted of its 5′ LTR in which a Myc tag was inserted at the COOH end of the ASP ORF (Fig. 10A). For comparison, an equivalent 5′ LTR-deleted construct expressing a Myc-tagged truncated ASP, resulting from the insertion of a stop codon at amino acid position 67, was also transfected in COS-7 cells. Cells were treated with 3-MA, and cellular extracts were then subjected to immunoprecipitation with anti-ASP and analyzed with anti-Myc antibodies by Western blotting. Interestingly, a band at the expected molecular mass was observed in COS-7 cells transfected with pNL4.3Δ5′LTR-ASP-Myc WT but not in cells transfected with the mutated version of the proviral DNA clone (Fig. 10B).
Fig 10.
Detection of Myc-tagged ASP from NL4.3 proviral DNA and increase in LC3b-II levels. (A) Representation of Myc-tagged ASP and its mutated version containing an in-frame stop codon at codon 67, which was generated in the NL4.3 proviral DNA. (B) COS-7 cells were transfected with the pcDNA3.1, pNL4.3Δ5′LTR-ASP-Myc WT, or pNL4.3r5′LTR-ASP67stop-Myc proviral DNA construct. Cells were treated with 3-MA (10 mM) for 18 h prior to analyses. At 24 h posttransfection, immunoprecipitation was performed on cellular extracts with the anti-ASP antibody, and ASP was detected by Western blotting with anti-Myc antibodies. (C) COS-7 cells were transfected with pcDNA3.1, pNL4.3Δ5′LTR-ASP-Myc WT, pNL4.3Δ5′LTR-ASP WT, or pNL4.3Δ5′LTR stop-Myc proviral DNA construct. At 48 h posttransfection, Western blot analyses were performed using anti-LC3b and anti-GAPDH antibodies. (D) Densitometry analyses of LC3b-II expression levels were calculated and normalized with GAPDH signals using Quantity One and GraphPad Prism software. (E) COS-7 cells were transfected with pNL4.3Δ5′LTR-ASP-Myc and left untreated (DMSO) or were treated with Baf A1 (100 nM) for 2 h prior to analyses. At 36 h posttransfection, fixed cells were incubated with anti-Myc antibodies followed by a goat anti-mouse IgG antibody coupled to Alexa fluor 488. Nuclei were stained with PI. Cells were observed with a Nikon A1 laser-scanning confocal microscopy using a 60× objective under oil immersion and with a numerical aperture of 1.4. Scale bars, 5 μm. F. COS-7 cells were cotransfected with pGFP-LC3 along with pcDNA3.1 or pNL4.3Δ5′LTR-ASP-Myc WT. Cells were either left untreated (DMSO) or were treated with Baf A1 (100 nM) for 2 h prior to analyses. After 36 h of transfection, cells were fixed and nuclei were stained with PI. Cells were observed by Nikon A1 laser-scanning confocal microscopy using a 60× objective under oil immersion and with a numerical aperture of 1.4. G. GFP-LC3+ punctate signals were counted for each cell (for a total of approximately 30 GFP-positive cells per condition). **, P < 0.01; ***, P < 0.001.
Since these results also suggested that ASP was unstable even when expressed from proviral DNA, we sought to confirm that ASP in this context also mediated autophagy. Expression levels of LC3b-II were analyzed in COS-7 cells transfected with pNL4.3Δ5′LTR-ASP-Myc WT, pNL4.3Δ5′LTR-ASP67stop-Myc, and nontagged pNL4.3Δ5′LTR-ASP WT. Western blot analyses demonstrated a significant increase of LC3b-II after transfection for all tested proviral DNAs, which was confirmed by densitometry analyses (Fig. 10C and D). Interestingly, the mutated version of pNL4.3Δ5′LTR-ASP WT showed a significant reduction in the level of induced LC3b-II. These results represented the first evidence that proviral DNA-dependent ASP expression induces autophagy.
In light of these results, ASP localization was further analyzed in COS-7 cells transfected with pNL4.3Δ5′LTR-ASP-Myc WT. Confocal analyses revealed few ASP-positive cells, but importantly, a punctate expression pattern of ASP was observed under these conditions (Fig. 10E). After inhibition of the late steps of autophagy with Baf A1 treatment, the same pattern of ASP distribution was observed in COS-7 cells, although a more evident punctate pattern was noted (Fig. 10E). To further confirm autophagy induction in pNL4.3Δ5′LTR-ASP-Myc WT-transfected cells, LC3 distribution was assessed upon cotransfection of the GFP-LC3 expression vector in COS-7 cells. A significant increase in GFP-LC3+ punctate signals was observed in cells transfected with pNL4.3Δ5′LTR-ASP-Myc compared to cells similarly transfected with the control vector (Fig. 10F and G). When transfected cells were treated with Baf A1, a more substantial number of cells showing LC3 punctate signals was observed.
These results demonstrated that proviral DNA-expressing ASP can be detected and is sufficient to induce autophagy, which likely affects its stability in transfected cells.
ASP affects HIV-1 replication through induction of autophagy.
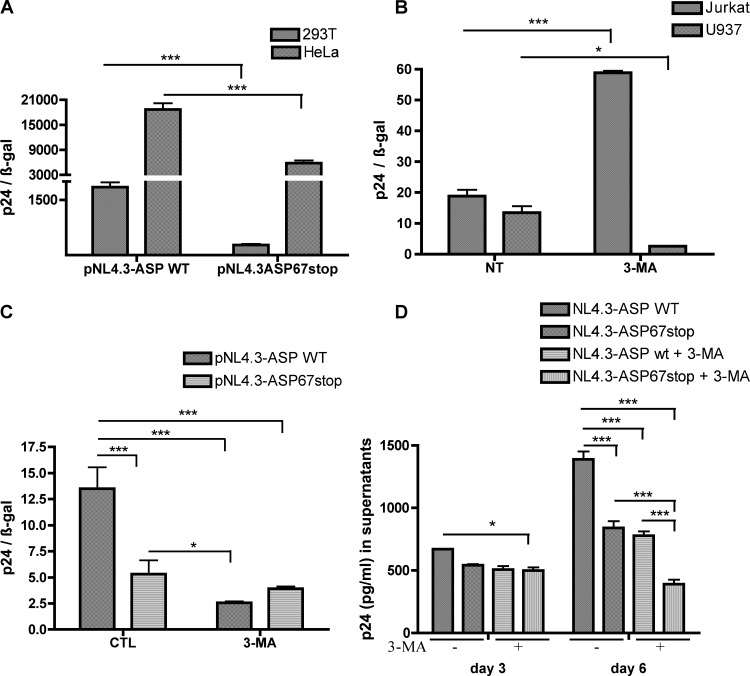
Since autophagy has been previously shown to affect HIV-1 replication in various cell lines (46, 63, 64), we tested whether the ASP mutant in the context of the full-length untagged proviral DNA affected HIV-1 replication. ASP-mutated pNL4.3ASP67stop was first compared to parental wild-type pNL4.3 in transfected 293T and HeLaCD4-LTR/βgal cells (Fig. 11A). These transfection experiments demonstrated that extracellular p24 levels were reduced in cells transfected with ASP-mutated proviral DNA, which was consistent with our previous studies using a differently ASP-mutated version of pNL4.3 (33). To study the impact of ASP on autophagy, both U937 and Jurkat cell lines were tested to confirm previous studies indicating that autophagy was affecting HIV-1 replication in opposing ways (46). As depicted in Fig. 11B, 3-MA-treated U937 cells showed reduced p24 production, while in Jurkat cells, 3-MA led to more pronounced extracellular p24 levels. As previous studies had indicated that monocytes were a more potent cell type for ASP expression (27, 31), U937 cells were selected to carry out transfection experiments with pNL4.3 or pNL4.3-ASP67stop proviral DNA (Fig. 11C). As expected, the ASP-mutated proviral DNA led to reduced HIV-1 replication, as measured by p24 ELISA. Upon 3-MA treatment, extracellular p24 levels were drastically reduced in pNL4.3-transfected cells, while p24 levels were less affected in ASP-mutated NL4.3-transfected cells. To further confirm these results, VSVg-pseudotyped NL4.3 virions and equivalent pseudotyped ASP-mutated NL4.3 virions were used to infect U937 cells (Fig. 11D). Again, NL4.3 WT-infected U937 cells were more efficient in replicating HIV-1 than ASP-mutated versions. Furthermore, 3-MA treatment led to a more pronounced reduction in WT NL4.3 virion production than for ASP-mutated NL4.3 virions, especially at day 3 postinfection.
Fig 11.
ASP modulates HIV-1 replication in U937 cells through autophagy. (A) HeLa and 293T cells were cotransfected with pcDNA3.1, pNL4.3-ASP WT, or pNL4.3-ASP67stop along with pRcActin-LacZ. Supernatants were harvested at 48 h posttransfection, quantified for p24 levels by ELISA, and normalized for β-galactosidase activity. (B) U937 and Jurkat cells were cotransfected with pNL4.3-ASP WT along with pRcActin-LacZ. Cells were left untreated (NT) or were treated with 3-MA for 4 h prior to harvest of supernatants and assessment of normalized extracellular p24 levels. (C) U937 cells were transfected with pcDNA3.1, pNL4.3ASP-WT, and pNL4.3-ASP67stop along with pRcActin-LacZ. Cells were left untreated (control [CTL]) or were treated with 3-MA for 4 h prior to harvest of supernatants and assessment of normalized extracellular p24 levels. (D) U937 cells were infected with VSV-g-pseudotyped NL4.3-env−-ASP WT or NL4.3-env−-ASP67stop virions and treated with 3-MA or left untreated. Supernatants were harvested at 3 or 6 days postinfection and assessed for p24 levels. *, P < 0.05; ***, P < 0.001.
These results strongly suggest that ASP has an important effect on HIV-1 replication through its capacity to induce autophagy.
DISCUSSION
Since the early identification of a conserved ORF in the antisense strand of the HIV-1 proviral DNA, numerous studies have provided evidence that antisense transcription in HIV-1 and the resulting encoded ASP are produced, although the detection of this viral protein has been a long-standing issue. Difficulty in the detection of this viral protein in mammalian cells is not an uncommon phenomenon, as, for example, certain HTLV-1 proteins are similarly difficult to detect in infected cells (65). It should also be stressed that its high level of conservation, observed in various sequenced isolates, makes this ORF likely to be translated into a viral protein. Based on this evidence and previous reports, in this study, our goal was to clearly detect the ASP in mammalian cells and to determine what mechanism causes its expression to be poorly detected. For the first time, we have indeed detected ASP in transfected mammalian cells by Western blotting. Although several reasons seem to be responsible for the low level of ASP detected in transfected cells, our results argue that the autophagy-inducing capacity of this protein, potentially linked to the formation of high-molecular-mass multimers, importantly and rapidly reduces intracellular levels, especially in overexpression conditions. However, it is important to note that ASP might also induce autophagy by other mechanisms. Refined cellular localization experiments indicated that ASP was indeed localized to autophagosomes.
The first objective of our study was to demonstrate that the identified antisense transcript, characterized by our team and others (1, 2, 25–31), could be translated in an antisense protein. The first tested eukaryotic system was the Sf9 insect cell line, which permitted us to detect ASP by confocal microscopy, flow cytometry, and Western blot analyses. Compared to other proteins produced in this model, ASP production was lower than the average level of production and in fact did not permit us to achieve levels high enough for antibody production. ASP was similarly weakly expressed in mammalian cell lines, as determined by confocal microscopy and flow cytometry analyses. We have thus opted for codon optimization of its cDNA, a method which had already been used for other HIV-1 proteins, such as Vpu and Vif (54). This approach led to improved detection of ASP by flow cytometry and confocal microscopy and allowed us to provide the first Western blot detection of the protein in mammalian cells. An immunoprecipitation experiment with an anti-ASP antibody was further optimized for its more pronounced detection. Although codon optimization is based on the fact that some codons are preferentially used for translation, earlier studies using codon optimization demonstrated that altered sequences in the mRNA led to greater RNA stability and more efficient nucleocytoplasmic RNA transport (53). Thus, our results suggest that inhibitory sequences, such as INS, IN, and CTS motifs, might be present in antisense transcripts and act upon nuclear export of mRNA. Alternatively, codon optimization of the ASP sequence might disrupt the formation of RNA stem-loop structures, such as the predicted partial antisense RRE structure, which would then lead to an improved translation capacity of these transcripts. Further experiments are needed to discern the exact mechanism by which codon optimization drastically improves ASP detection in mammalian cells.
Through flow cytometry, confocal microscopy, and Western blot analyses, our results suggest that localization of ASP is partially concordant with previously published electron microscopy results showing membrane association and cytoplasmic and plasma membrane distribution (31–33). However, in our hands, ASP did not localize to the nucleus and confirms our previous confocal microscopy results in transfected T cells (33). In addition, our results agreed with in silico analyses (3), which also suggested that ASP is associated with membrane (plasma membrane or intracellular membranes). Indeed, enrichment of our extracts with membrane proteins and the use of the RIPA lysis buffer all led to more pronounced ASP signals in our Western blot analyses.
Our Western blot analyses have also indicated that ASP formed very stable high-molecular-mass aggregates, even in loading buffer conditions containing β-mercaptoethanol, DTT, and SDS. These aggregates were also detectable subsequent to immunoprecipitation and were confirmed by our glutaraldehyde assay. Thus, ASP likely forms aggregates upon expression in cells. Recently, study on the β-amyloid protein and p53 demonstrated that formation of aggregates led to sequestering of essential cellular proteins (66, 67). It will be important in future studies to determine if ASP similarly has the capacity to interact with cellular proteins and thereby affect diverse cellular mechanisms.
Using inhibitors and the LC3-II marker, we provide evidence that ASP overexpression induces autophagy in transfected cells, which could represent a cellular mechanism linked to its instability. In fact, our confocal microscopy experiments revealed a punctate distribution in the cytoplasm of ASP-positive cells in all tested cell types, which was reminiscent of autophagosomes. The estimated size of puncta was similar to the known size of autophagosomes (ranging between 0.5 and 1.5 μm) (68). Inhibition of autophagy at its early step by treatment with 3-MA led to an important loss of ASP puncta, whereas treatment with Baf A1 and chloroquine, late-step inhibitors, tended to stabilize these ASP-positive punctate structures, thereby strongly suggesting the autophagosome nature of these signals. Additional results demonstrated that ASP induced the LC3b-II isoform and the Beclin 1 autophagy markers, and that ASP localized to autophagosomes, as illustrated by colocalization with LC3. Interestingly, our coimmunoprecipitation experiments also suggest that ASP and LC3b-II were interacting either directly or indirectly. This interaction might represent an alternative explanation for the autophagy-inducing capacity of ASP. This colocalization further argues that the increase in ASP levels in 3-MA-treated cells could be mediated by the inhibition of the final step of autophagy (formation of autolysosomes), which would lead to ASP degradation (38).
Autophagy has been implicated recently in HIV-1 replication (63, 69, 70). In viral infection, autophagy is a cellular mechanism mainly involved in the elimination of incoming viruses (71–74). However, DNA and RNA viruses can modulate autophagy, and this biological process can actually help in accentuating the progression of the infection (72). In fact, HIV-1 does interact with the autophagic pathway. A functional genomic screen has indeed identified Atg proteins (Atg7, Atg12, Atg16, etc.) as host proteins essential for HIV-1 infection (75). Interestingly, in CD4+ T cells, the Env protein, upon interaction with CD4 on the surface of uninfected T cells, mediates autophagy and subsequent apoptosis (45). In contrast, in macrophages, the induction of the initial steps of autophagy is required for the processing of gag and greatly contributes to increasing viral yield (47). However, HIV-1 inhibits autophagy at its late stage through the interaction of the viral protein Nef with Beclin 1 (47). Other studies have confirmed this divergent response toward HIV-1 according to cell type (46, 49).
More importantly, our results have also provided specific detection of ASP from an HIV-1 proviral DNA with a 5′ LTR-deleted version in order to lessen potential interference from sense transcription on antisense transcription, as we had previously suggested (27). In the context of the proviral DNA, inhibition of autophagy was required in order to detect ASP, which might reflect the fact that the proviral DNA was deleted of its 5′ LTR and thereby did not express Nef, a presumed inhibitor of the late autophagy step leading to the degradation pathway (47). We have further demonstrated that proviral DNA-expressed ASP can be a potential inducer of autophagy, since transfection of the 5′ LTR-deleted proviral DNA significantly increased levels of the LC3b-II form. This autophagy marker again demonstrated an ASP-dependent punctate distribution, which was further stabilized by Baf A1. Further evidence for autophagy was also indicated by the similar cellular distribution of ASP itself and the Baf A1-sensitive nature of the signal in these transfection experiments. Interestingly, the 5′ LTR-deleted version of pNL4.3 expressing the truncated form of ASP also led to the induction of autophagy, but to a more limited extent, based on LC3-II levels. These results suggest that the first 66 amino acids of ASP, although capable of inducing autophagy, did not lead to optimal induction. Possible partial autophagy induction by this mutant suggests a retained capacity to multimerize or other features of ASP, such as its capacity to interact with LC3b. Further study will be needed to identify this important ASP region.
In order to more clearly confirm that ASP was inducing autophagy in the context of full HIV-1 proviral DNA, we have further tested wild-type and ASP-truncated proviral DNAs in the U937 cell line. As opposed to transfected Jurkat T cells, WT HIV-1 proviral DNA-transfected U937 cells were strongly affected in their capacity to replicate HIV, in agreement with former studies on the effect of autophagy on HIV-1 replication in T versus monocytic cells (46, 47). Our result have also demonstrated that, in 293T, HeLa, and U937 cells, the mutated ASP version did not replicate HIV-1 as efficiently as the WT proviral DNA, thereby corroborating our previous report using a different ASP-mutated proviral DNA (33). Furthermore, in transfection experiments in U937 cells, p24 levels observed in 3-MA-treated cells were less affected in cells transfected with the mutated ASP proviral DNA than in WT NL4.3-transfected cells. A similar trend was also noted in infection experiments, albeit to a lesser extent (especially at later time points). Therefore, our results suggest that ASP is required for optimal HIV-1 replication in monocytic cells through induction of autophagy. Based on previous data from Kyei et al. (47), however, we predict that induced autophagy is blocked at the autophagosome-lysosome fusion step via Nef. Interestingly, previous data from our team and collaborators have suggested that monocytic cell lines seem to express higher levels of HIV-1 antisense transcripts than T cells (27, 31).
In conclusion, we demonstrate the detection of ASP by Western blot analyses using a codon-optimized expression vector of ASP and through inhibition of autophagy. We further speculate that this induced autophagy is important for HIV-1 replication in monocytic cells. Future studies will be needed to dissect the role of ASP in viral replication in different cell types and to further understand its link to autophagy.
ACKNOWLEDGMENTS
This work was supported by a grant from The Canadian Foundation for AIDS Research (CanFAR) (B.B.) and a grant from the Agence Nationale de Recherches sur le Sida et les Hépatites Virales (ANRS) (J.M.M.). C.T. was supported by an institutional FARE scholarship. S.L. was supported by a CIHR Ph.D. scholarship. B.B. holds a Canada Research Chair in Human Retrovirology (Tier 2).
We are thankful for the excellent technical support from Denis Flipo in confocal microscopy experiments.
Footnotes
Published ahead of print 20 February 2013
REFERENCES
- 1. Bukrinsky MI, Etkin AF. 1990. Plus strand of the HIV provirus DNA is expressed at early stages of infection. AIDS Res. Hum. Retrovir. 6:425–426 [DOI] [PubMed] [Google Scholar]
- 2. Michael NL, Vahey MT, d'Arcy L, Ehrenberg PK, Mosca JD, Rappaport J, Redfield RR. 1994. Negative-strand RNA transcripts are produced in human immunodeficiency virus type 1-infected cells and patients by a novel promoter downregulated by Tat. J. Virol. 68:979–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miller RH. 1988. Human immunodeficiency virus may encode a novel protein on the genomic DNA plus strand. Science 239:1420–1422 [DOI] [PubMed] [Google Scholar]
- 4. Knowling S, Morris KV. 2011. Non-coding RNA and antisense RNA. Nature's trash or treasure? Biochimie 93:1922–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lavorgna G, Dahary D, Lehner B, Sorek R, Sanderson CM, Casari G. 2004. In search of antisense. Trends Biochem. Sci. 29:88–94 [DOI] [PubMed] [Google Scholar]
- 6. Vanhee-Brossollet C, Vaquero C. 1998. Do natural antisense transcripts make sense in eukaryotes? Gene 211:1–9 [DOI] [PubMed] [Google Scholar]
- 7. Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, Nemzer S, Pinner E, Walach S, Bernstein J, Savitsky K, Rotman G. 2003. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 21:379–386 [DOI] [PubMed] [Google Scholar]
- 8. Larocca D, Chao LA, Seto MH, Brunck TK. 1989. Human T-cell leukemia virus minus strand transcription in infected T-cells. Biochem. Biophys. Res. Commun. 163:1006–1013 [DOI] [PubMed] [Google Scholar]
- 9. Basbous J, Arpin C, Gaudray G, Piechaczyk M, Devaux C, Mesnard JM. 2003. The HBZ factor of human T-cell leukemia virus type I dimerizes with transcription factors JunB and c-Jun and modulates their transcriptional activity. J. Biol. Chem. 278:43620–43627 [DOI] [PubMed] [Google Scholar]
- 10. Cavanagh MH, Landry S, Audet B, Arpin-Andre C, Hivin P, Pare ME, Thete J, Wattel E, Marriott SJ, Mesnard JM, Barbeau B. 2006. HTLV-I antisense transcripts initiating in the 3′LTR are alternatively spliced and polyadenylated. Retrovirology 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gaudray G, Gachon F, Basbous J, Biard-Piechaczyk M, Devaux C, Mesnard JM. 2002. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J. Virol. 76:12813–12822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hagiya K, Yasunaga J, Satou Y, Ohshima K, Matsuoka M. 2011. ATF3, an HTLV-1 bZip factor binding protein, promotes proliferation of adult T-cell leukemia cells. Retrovirology 8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Halin M, Douceron E, Clerc I, Journo C, Ko NL, Landry S, Murphy EL, Gessain A, Lemasson I, Mesnard JM, Barbeau B, Mahieux R. 2009. Human T-cell leukemia virus type 2 produces a spliced antisense transcript encoding a protein that lacks a classic bZIP domain but still inhibits Tax2-mediated transcription. Blood 114:2427–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hivin P, Arpin-Andre C, Clerc I, Barbeau B, Mesnard JM. 2006. A modified version of a Fos-associated cluster in HBZ affects Jun transcriptional potency. Nucleic Acids Res. 34:2761–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuhlmann AS, Villaudy J, Gazzolo L, Castellazzi M, Mesnard JM, Duc Dodon M. 2007. HTLV-1 HBZ cooperates with JunD to enhance transcription of the human telomerase reverse transcriptase gene (hTERT). Retrovirology 4:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Larocque E, Halin M, Landry S, Marriott SJ, Switzer WM, Barbeau B. 2011. Human T-cell lymphotropic virus type 3 (HTLV-3)- and HTLV-4-derived antisense transcripts encode proteins with similar Tax-inhibiting functions but distinct subcellular localization. J. Virol. 85:12673–12685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matsumoto J, Ohshima T, Isono O, Shimotohno K. 2005. HTLV-1 HBZ suppresses AP-1 activity by impairing both the DNA-binding ability and the stability of c-Jun protein. Oncogene 24:1001–1010 [DOI] [PubMed] [Google Scholar]
- 18. Mukai R, Ohshima T. 2011. Dual effects of HTLV-1 bZIP factor in suppression of interferon regulatory factor 1. Biochem. Biophys. Res. Commun. 409:328–332 [DOI] [PubMed] [Google Scholar]
- 19. Satou Y, Yasunaga J, Yoshida M, Matsuoka M. 2006. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc. Natl. Acad. Sci. U. S. A. 103:720–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Satou Y, Yasunaga J, Zhao T, Yoshida M, Miyazato P, Takai K, Shimizu K, Ohshima K, Green PL, Ohkura N, Yamaguchi T, Ono M, Sakaguchi S, Matsuoka M. 2011. HTLV-1 bZIP factor induces T-cell lymphoma and systemic inflammation in vivo. PLoS Pathog. 7:e1001274 doi:10.1371/journal.ppat.1001274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thebault S, Basbous J, Hivin P, Devaux C, Mesnard JM. 2004. HBZ interacts with JunD and stimulates its transcriptional activity. FEBS Lett. 562:165–170 [DOI] [PubMed] [Google Scholar]
- 22. Yoshida M, Satou Y, Yasunaga J, Fujisawa J, Matsuoka M. 2008. Transcriptional control of spliced and unspliced human T-cell leukemia virus type 1 bZIP factor (HBZ) gene. J. Virol. 82:9359–9368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao T, Yasunaga J, Satou Y, Nakao M, Takahashi M, Fujii M, Matsuoka M. 2009. Human T-cell leukemia virus type 1 bZIP factor selectively suppresses the classical pathway of NF-kappaB. Blood 113:2755–2764 [DOI] [PubMed] [Google Scholar]
- 24. Reference deleted.
- 25. Barbagallo MS, Birch KE, Deacon NJ, Mosse JA. 2012. Potential control of human immunodeficiency virus type 1 asp expression by alternative splicing in the upstream untranslated region. DNA Cell Biol. 31:1303–1313 [DOI] [PubMed] [Google Scholar]
- 26. Kobayashi-Ishihara M, Yamagishi M, Hara T, Matsuda Y, Takahashi R, Miyake A, Nakano K, Yamochi T, Ishida T, Watanabe T. 2012. HIV-1-encoded antisense RNA suppresses viral replication for a prolonged period. Retrovirology 9:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Landry S, Halin M, Lefort S, Audet B, Vaquero C, Mesnard JM, Barbeau B. 2007. Detection, characterization and regulation of antisense transcripts in HIV-1. Retrovirology 4:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lefebvre G, Desfarges S, Uyttebroeck F, Munoz M, Beerenwinkel N, Rougemont J, Telenti A, Ciuffi A. 2011. Analysis of HIV-1 expression level and sense of transcription by high-throughput sequencing of the infected cell. J. Virol. 85:6205–6211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ludwig LB, Ambrus JL, Jr, Krawczyk KA, Sharma S, Brooks S, Hsiao CB, Schwartz SA. 2006. Human immunodeficiency virus-type 1 LTR DNA contains an intrinsic gene producing antisense RNA and protein products. Retrovirology 3:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vanhee-Brossollet C, Thoreau H, Serpente N, D'Auriol L, Levy JP, Vaquero C. 1995. A natural antisense RNA derived from the HIV-1 env gene encodes a protein which is recognized by circulating antibodies of HIV+ individuals. Virology 206:196–202 [DOI] [PubMed] [Google Scholar]
- 31. Laverdure S, Gross A, Arpin-Andre C, Clerc I, Beaumelle B, Barbeau B, Mesnard JM. 2012. HIV-1 antisense transcription is preferentially activated in primary monocyte-derived cells. J. Virol. 86:13785–13789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Briquet S, Vaquero C. 2002. Immunolocalization studies of an antisense protein in HIV-1-infected cells and viral particles. Virology 292:177–184 [DOI] [PubMed] [Google Scholar]
- 33. Clerc I, Laverdure S, Torresilla C, Landry S, Borel S, Vargas A, Arpin-Andre C, Gay B, Briant L, Gross A, Barbeau B, Mesnard JM. 2011. Polarized expression of the membrane ASP protein derived from HIV-1 antisense transcription in T cells. Retrovirology 8:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wong E, Bejarano E, Rakshit M, Lee K, Hanson HH, Zaarur N, Phillips GR, Sherman MY, Cuervo AM. 2012. Molecular determinants of selective clearance of protein inclusions by autophagy. Nat. Commun. 3:1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Glick D, Barth S, Macleod KF. 2010. Autophagy: cellular and molecular mechanisms. J. Pathol. 221:3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang Z, Klionsky DJ. 2009. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 335:1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang Z, Klionsky DJ. 2010. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22:124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. 2005. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 1:84–91 [DOI] [PubMed] [Google Scholar]
- 40. Ren T, Dong W, Takahashi Y, Xiang D, Yuan Y, Liu X, Loughran TP, Jr, Sun SC, Wang HG, Cheng H. 2012. HTLV-2 Tax immortalizes human CD4+ memory T lymphocytes by oncogenic activation and dysregulation of autophagy. J. Biol. Chem. 287:34683–34693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tang SW, Chen CY, Klase Z, Zane L, Jeang KT. 2012. The cellular autophagy pathway modulates HTLV-1 replication. J. Virol. 87:1699–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tang SW, Ducroux A, Jeang KT, Neuveut C. 2012. Impact of cellular autophagy on viruses: insights from hepatitis B virus and human retroviruses. J. Biomed. Sci. 19:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Denizot M, Varbanov M, Espert L, Robert-Hebmann V, Sagnier S, Garcia E, Curriu M, Mamoun R, Blanco J, Biard-Piechaczyk M. 2008. HIV-1 gp41 fusogenic function triggers autophagy in uninfected cells. Autophagy 4:998–1008 [DOI] [PubMed] [Google Scholar]
- 44. Eekels JJ, Sagnier S, Geerts D, Jeeninga RE, Biard-Piechaczyk M, Berkhout B. 2012. Inhibition of HIV-1 replication with stable RNAi-mediated knockdown of autophagy factors. Virol. J. 9:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Espert L, Denizot M, Grimaldi M, Robert-Hebmann V, Gay B, Varbanov M, Codogno P, Biard-Piechaczyk M. 2006. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J. Clin. Investig. 116:2161–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Espert L, Varbanov M, Robert-Hebmann V, Sagnier S, Robbins I, Sanchez F, Lafont V, Biard-Piechaczyk M. 2009. Differential role of autophagy in CD4 T cells and macrophages during X4 and R5 HIV-1 infection. PLoS One 4:e5787 doi:10.1371/journal.pone.0005787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, Wu L, Kominami E, Ueno T, Yamamoto A, Federico M, Panganiban A, Vergne I, Deretic V. 2009. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 186:255–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou D, Kang KH, Spector SA. 2012. Production of interferon alpha by human immunodeficiency virus type 1 in human plasmacytoid dendritic cells is dependent on induction of autophagy. J. Infect. Dis. 205:1258–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou D, Spector SA. 2008. Human immunodeficiency virus type-1 infection inhibits autophagy. AIDS 22:695–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Molina L, Grimaldi M, Robert-Hebmann V, Espert L, Varbanov M, Devaux C, Granier C, Biard-Piechaczyk M. 2007. Proteomic analysis of the cellular responses induced in uninfected immune cells by cell-expressed X4 HIV-1 envelope. Proteomics 7:3116–3130 [DOI] [PubMed] [Google Scholar]
- 51. Boisvert FM, Dery U, Masson JY, Richard S. 2005. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 19:671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fortin JF, Cantin R, Lamontagne G, Tremblay M. 1997. Host-derived ICAM-1 glycoproteins incorporated on human immunodeficiency virus type 1 are biologically active and enhance viral infectivity. J. Virol. 71:3588–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ngumbela KC, Ryan KP, Sivamurthy R, Brockman MA, Gandhi RT, Bhardwaj N, Kavanagh DG. 2008. Quantitative effect of suboptimal codon usage on translational efficiency of mRNA encoding HIV-1 gag in intact T cells. PLoS One 3:e2356 doi:10.1371/journal.pone.0002356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nguyen KL, llano M, Akari H, Miyagi E, Poeschla EM, Strebel K, Bour S. 2004. Codon optimization of the HIV-1 vpu and vif genes stabilizes their mRNA and allows for highly efficient Rev-independent expression. Virology 319:163–175 [DOI] [PubMed] [Google Scholar]
- 55. Seglen PO, Gordon PB. 1982. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl. Acad. Sci. U. S. A. 79:1889–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. 1998. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 23:33–42 [DOI] [PubMed] [Google Scholar]
- 57. Wang X, Gao Y, Tan J, Devadas K, Ragupathy V, Takeda K, Zhao J, Hewlett I. 2012. HIV-1 and HIV-2 infections induce autophagy in Jurkat and CD4(+) T cells. Cell Signal. 24:1414–1419 [DOI] [PubMed] [Google Scholar]
- 58. Mizushima N. 2007. Autophagy: process and function. Genes Dev. 21:2861–2873 [DOI] [PubMed] [Google Scholar]
- 59. Mizushima N, Yoshimori T, Levine B. 2010. Methods in mammalian autophagy research. Cell 140:313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nara A, Mizushima N, Yamamoto A, Kabeya Y, Ohsumi Y, Yoshimori T. 2002. SKD1 AAA ATPase-dependent endosomal transport is involved in autolysosome formation. Cell Struct. Funct. 27:29–37 [DOI] [PubMed] [Google Scholar]
- 61. Hansen TE, Johansen T. 2011. Following autophagy step by step. BMC Biol. 9:39 doi:10.1186/1741-7007-9-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hundeshagen P, Hamacher-Brady A, Eils R, Brady NR. 2011. Concurrent detection of autolysosome formation and lysosomal degradation by flow cytometry in a high-content screen for inducers of autophagy. BMC Biol. 9:38 doi:10.1186/1741-7007-9-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Borel S, Espert L, Biard-Piechaczyk M. 2012. Macroautophagy regulation during HIV-1 infection of CD4+ T cells and macrophages. Front. Immunol. 3:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Killian MS. 2012. Dual role of autophagy in HIV-1 replication and pathogenesis. AIDS Res. Ther. 9:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Albrecht B, Lairmore MD. 2002. Critical role of human T-lymphotropic virus type 1 accessory proteins in viral replication and pathogenesis. Microbiol. Mol. Biol. Rev. 66:396–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. 2011. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144:67–78 [DOI] [PubMed] [Google Scholar]
- 67. Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S, Cornelis A, Rozenski J, Zwolinska A, Marine JC, Lambrechts D, Suh YA, Rousseau F, Schymkowitz J. 2011. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat. Chem. Biol. 7:285–295 [DOI] [PubMed] [Google Scholar]
- 68. Mizushima N, Ohsumi Y, Yoshimori T. 2002. Autophagosome formation in mammalian cells. Cell Struct. Funct. 27:421–429 [DOI] [PubMed] [Google Scholar]
- 69. Dinkins C, Arko-Mensah J, Deretic V. 2010. Autophagy and HIV. Semin. Cell Dev. Biol. 21:712–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Espert L, Codogno P, Biard-Piechaczyk M. 2008. What is the role of autophagy in HIV-1 infection? Autophagy 4:273–275 [DOI] [PubMed] [Google Scholar]
- 71. Deretic V. 2012. Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pattern recognition receptors. Curr. Opin. Immunol. 24:21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dreux M, Chisari FV. 2010. Viruses and the autophagy machinery. Cell Cycle 9:1295–1307 [DOI] [PubMed] [Google Scholar]
- 73. Kudchodkar SB, Levine B. 2009. Viruses and autophagy. Rev. Med. Virol. 19:359–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lin LT, Dawson PW, Richardson CD. 2010. Viral interactions with macroautophagy: a double-edged sword. Virology 402:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921–926 [DOI] [PubMed] [Google Scholar]